Abstract
Replication protein A (RPA), a major eukaryotic ssDNA-binding protein, is essential for all metabolic processes that involve ssDNA, including DNA replication, repair, and damage signaling. To perform its functions, RPA binds ssDNA tightly. In contrast, it was presumed that RPA binds RNA weakly. However, recent data suggest that RPA may play a role in RNA metabolism. RPA stimulates RNA-templated DNA repair in vitro and associates in vivo with R-loops, the three-stranded structures consisting of an RNA-DNA hybrid and the displaced ssDNA strand. R-loops are common in the genomes of pro- and eukaryotes, including humans, and may play an important role in transcription-coupled homologous recombination and DNA replication restart. However, the mechanism of R-loop formation remains unknown. Here, we investigated the RNA-binding properties of human RPA and its possible role in R-loop formation. Using gel-retardation and RNA/DNA competition assays, we found that RPA binds RNA with an unexpectedly high affinity (KD ≈ 100 pm). Furthermore, RPA, by forming a complex with RNA, can promote R-loop formation with homologous dsDNA. In reconstitution experiments, we showed that human DNA polymerases can utilize RPA-generated R-loops for initiation of DNA synthesis, mimicking the process of replication restart in vivo. These results demonstrate that RPA binds RNA with high affinity, supporting the role of this protein in RNA metabolism and suggesting a mechanism of genome maintenance that depends on RPA-mediated DNA replication restart.
Keywords: RNA-binding protein, RNA, DNA repair, homologous recombination, R-loops, DNA polymerase, R-loop extension, DNA replication, DNA replication restart
Replication protein A (RPA) is a major ssDNA-binding protein in eukaryotes (1). It is a highly conserved trimeric protein composed of three subunits, RPA70, RPA32, and RPA14, which all are essential for cell viability (2). RPA plays a critical role in most, if not all, metabolic processes that involve ssDNA, including DNA replication, repair, transcription, and DNA damage signaling (2–5). RPA binding protects ssDNA from degradation and unfolds DNA secondary structures. RPA interacts with various proteins helping to coordinate different cellular processes.
Recently, it was found that RPA is closely associated with R-loops in vivo (6–8). R-loops are currently known to exist in the genomes of bacteria, yeast, and higher eukaryotes (9–11). In humans, R-loops occur over tens of thousands of genomic loci covering up to 5% of the genome (12, 13).
It was suggested that R-loops may play an important role during DNA repair by initiating transcription-coupled homologous recombination in actively transcribed genome regions (6, 14, 15). It was also proposed that R-loops may promote restart of replication forks stalled at damaged DNA (16, 17). The role of R-loops in priming replication was actually the first biological function proposed for this structure in bacteria (18). More recently, it was found that in eukaryotes, persistent RNA-DNA hybrids initiate DNA synthesis in ribosomal DNA in a replication origin–independent manner (19). Being an important regulator of cellular processes such as transcription, gene expression, DNA replication, and DNA repair, R-loops also represent a source of genome instability, if not timely processed or repaired (20, 21). The mechanism of R-loop formation in vivo remains to be understood.
RPA has a strong binding affinity to ssDNA (2, 22); therefore, it was thought that RPA association with R-loops is due to its binding to the displaced ssDNA strand generated during R-loop formation. Surprisingly, until recently, RPA binding to RNA had not been explored. It was presumed that RPA binds to RNA weakly, because in early studies the affinity of RPA for both RNA and dsDNA was estimated to be at least 3 orders of magnitude lower than for ssDNA (23).
However, our current data indicate that RPA binds to RNA much more strongly than was previously anticipated. We found that RPA binds RNA with high affinity (KD ≈ 100 pm). Furthermore, we demonstrate that RPA has a unique ability to form an active complex with RNA, which promotes formation of bona fide R-loops through invasion of RNA into homologous covalently closed duplex DNA. Using RPA-generated R-loops, we reconstituted DNA synthesis in vitro using human DNA polymerases, supporting the role of R-loops in the mechanism of DNA replication restart.
Results
RPA binds to RNA with high affinity
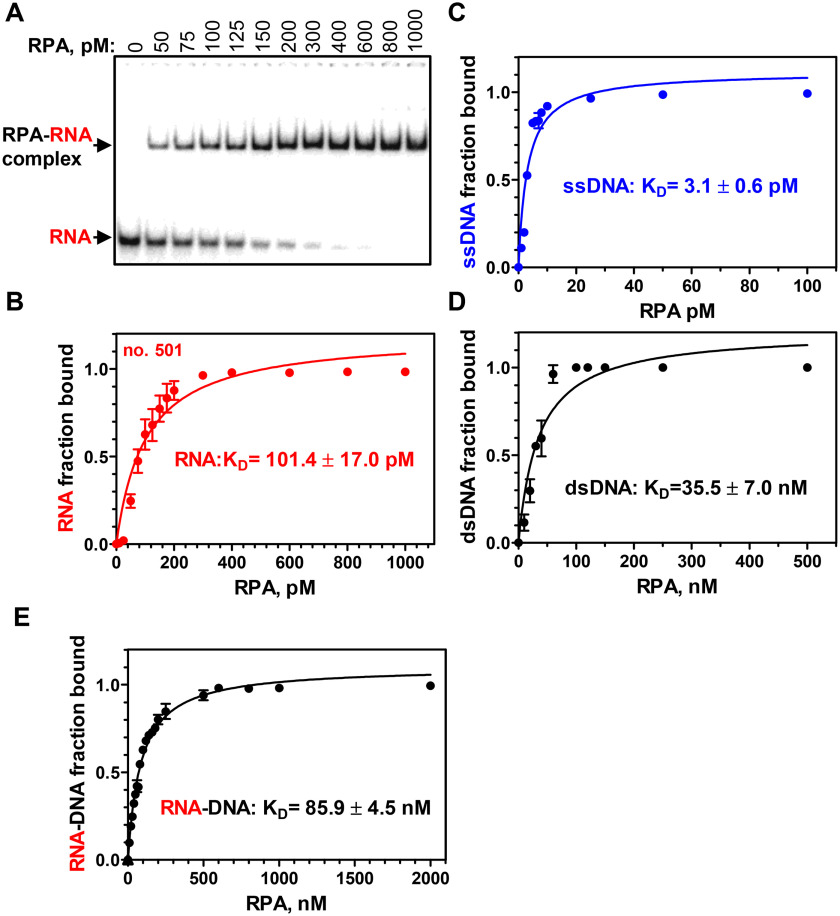
First, using electrophoretic mobility shift assay (EMSA), we examined the RPA affinity for RNA. Previously, it was reported that the RPA-binding affinity for RNA is approximately the same as for dsDNA and ∼1000-fold weaker than for ssDNA (23). Surprisingly, we found that RPA binding to a 48-nt RNA (no. 501; Table S1) (KD = 101.4 ± 17.0 pm) is 300–400-fold stronger than for homologous 48-bp dsDNA (nos. 211/212) (KD = 35.5 ± 7.0 nm) and only 30–60-fold weaker than for a 48-mer ssDNA of the identical sequence (no. 211) (KD = 3.1 ± 0.6 pm; Fig. 1 and Fig. S1). The presence of 100 mm NaCl, the condition that was used in previous studies, had no significant effect on the RPA affinity (KD = 72.0 ± 10.2 pm) for RNA (no. 501) (Fig. S2). We also tested the RPA-binding affinity for RNA-DNA hybrid (nos. 501/212), which appeared to be 2 times weaker (KD=85.9 ± 4.5 nm) than for dsDNA of identical sequence (nos. 211/212) (Fig. 1E and Fig. S1C).
Figure 1.
RPA binds to RNA with a high affinity. A, analysis of RPA binding to a 48-mer RNA (no. 501; 5 pm) using EMSA in a 6% polyacrylamide gel. B, data from A plotted as a graph. C–E, graphical representation of RPA binding to 48-mer substrates: ssDNA (no. 211; 0.5 pm), dsDNA (no. 211/212; 3 nm), and RNA-DNA hybrid (no. 501/212; 3 nm). Error bars, S.E.
Then we examined RPA binding to RNA using competitors. When RNA was used as a competitor against ssDNA, we found that the RPA affinity for RNA (no. 501) was ∼60-fold weaker than for ssDNA of identical sequence (no. 211) (Fig. S3, A and B). When nonhomologous supercoiled pHSG299 plasmid dsDNA was used as a competitor, the affinities of RPA for RNA and ssDNA were ∼500- and ∼33,000-fold, respectively, stronger than for plasmid dsDNA (Fig. S3, C and D). Thus, these results were consistent with the RPA KD values for RNA and DNA indicated above.
Then we tested the RPA binding to four other 48-nt RNAs of different sequences (Fig. S4). For three of them (nos. 3R, 7R, and 8R), the RPA-binding affinity was strong (KD in the range of 62.9–248.1 pm), and for one of them (no. 540), it was significantly weaker (KD > 4 nm). Inspection of the RNA structures showed that no. 540 has a much stronger propensity to form secondary structures than other tested RNAs (Table S2). We also tested the RPA binding to homopolymers: poly(rA) (no. 10R, 48 nt) and poly(rU) (no. 11R, 48 nt) (Fig. S5, A and B). For poly(rU), the RPA-binding affinity (KD = 184.4 ± 26.4 pm) was in the range with other tested RNA molecules of the same size, except for no. 540, whereas for poly(rA), it was significantly weaker (KD = 1.4 ± 0.2 nm). Moreover, poly(rA) appeared to be an extremely weak competitor against ssDNA (no. 211, 48 nt), even weaker than could be expected based on the KD measurements. Consistent with the reports from Wold's group (23), a 1000-fold excess of poly(rA) was not sufficient for a 2-fold decrease in RPA binding to ssDNA (Fig. S5C). An increase in incubation time up to 3 h had no apparent effect on the outcome of poly(rA) competition with ssDNA for RPA binding. We suggest that this weak competitiveness of poly(rA) may be related to the kinetics of RPA binding to this substrate. For instance, RPA-poly(rA) complexes may have a significantly shorter lifetime than the RPA-ssDNA complexes.
Overall, these data show that RPA binds RNA with high affinity. They also indicate that RPA binding to RNA is lowered by RNA secondary structures and by poly(rA) sequences.
RPA promotes R-loop formation in vitro
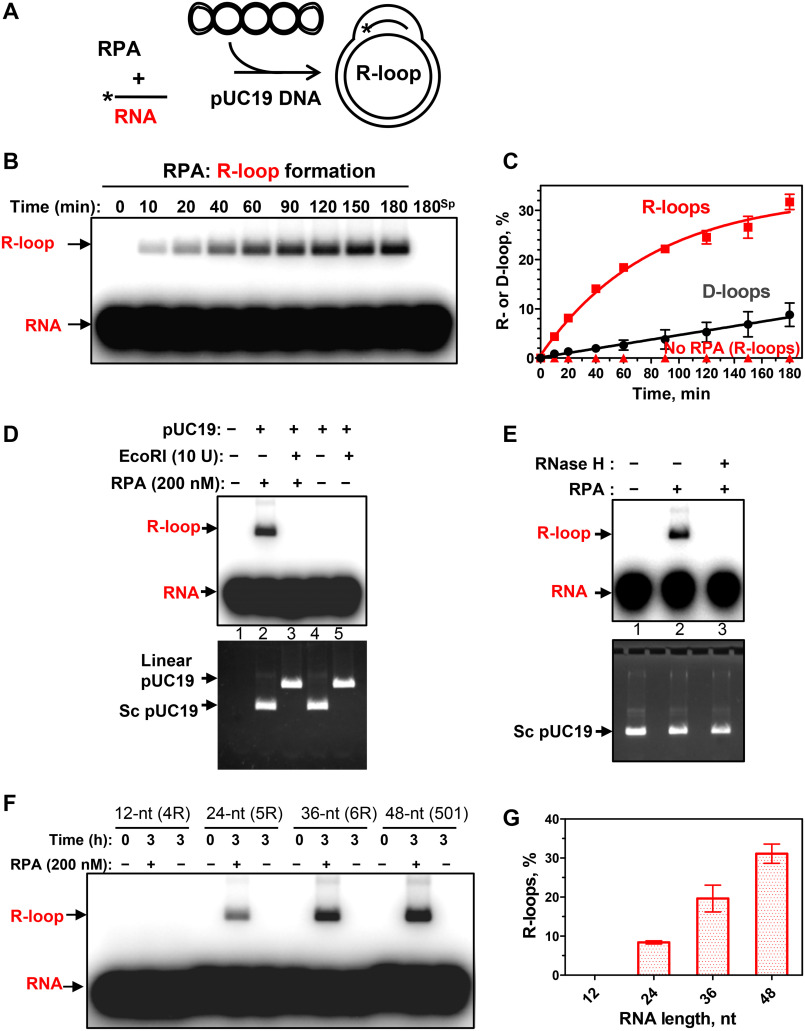
The finding that RPA binds RNA strongly, taken together with the known association of RPA with R-loops in vivo, prompted us to test whether RPA has the R-loop formation activity (Fig. 2A). Indeed, we found that RPA can promote R-loop formation between a 32P-labeled 48-mer RNA (no. 501) and homologous supercoiled pUC19 plasmid DNA (Fig. 2, B and C). In these experiments, the plasmid DNA was prepared by a nondenaturing method to avoid formation of irreversibly denatured plasmid DNA, a source of a potential artifact due to RNA/DNA annealing. We then tested the authenticity of the RPA-promoted R-loops. In contrast to RNA-DNA hybrids produced by annealing or RNA-protein complexes that can resist deproteinization, R-loops, similar to D-loops, are sensitive to plasmid dsDNA cleavage (outside of the R-loop region) with a restriction endonuclease (24). The cleavage causes loss of plasmid dsDNA superhelicity and R-loop dissociation due to DNA branch migration. We found that dsDNA linearization with EcoRI indeed causes R-loop dissociation, confirming their bona fide nature (Fig. 2D). As expected, the R-loops were also sensitive to RNase H, which digests the RNA moiety in the RNA-DNA hybrid (Fig. 2E).
Figure 2.
RPA promotes R-loop formation. A, reaction scheme. *, 32P label. B, kinetics of the R-loop formation by RPA (200 nm) analyzed by electrophoresis in a 1% agarose gel. 180sp, reaction (180 min) in the absence of RPA. The stoichiometric ratio of RNA/dsDNA was 5:1 (in molecules). C, graphical representation of R- and D-loop formation by RPA. D, sensitivity of R-loops to EcoRI cleavage. 32P-labeled RNA (no. 501; 3 μm, nt) was incubated with supercoiled pUC19 dsDNA (67.2 μm, nt) for 3 h in the presence of RPA (200 nm, lanes 2 and 3) or in its absence (lanes 4 and 5). The R-loops were then incubated with EcoRI (lane 3). The products were analyzed by electrophoresis in 1% agarose gel. Controls include [32P]RNA (no. 501; 3 μm, nt) (lane 1) and a mixture of 32P-labeled RNA (no. 501; 3 μm, nt) and pUC19 incubated with EcoRI storage buffer (lane 4) or with EcoRI (lane 5). The gel was autoradiographed (top) to visualize 32P-labeled RNA and R-loops and then stained with ethidium bromide (bottom) to monitor intactness of supercoiled pUC19 dsDNA and its cleavage by EcoRI. Note that R-loops co-migrate in the gel with supercoiled pUC19 DNA. E, sensitivity of R-loops to RNase H. The R-loops produced as in D (lane 2) were incubated with RNase H (5 units) (lane 3) or with the storage buffer (lane 2). The products were analyzed as in D. F, RNA length dependence of R-loop formation by RPA. R-loops were formed by RPA (200 nm) in pUC19 DNA (67.2 μm, nt) using 32P-labeled RNAs: 12-mer (no. 4R), 24-mer (no. 5R), 36-mer (no. 6R), and 48-mer (no. 501) (each 3 μm, nt). R-loops were analyzed by electrophoresis in a 1% agarose gel. G, data from F presented as a graph. Error bars, S.E.
When RNA was replaced with a 48-mer ssDNA of the identical sequence (no. 211), the efficiency of the reaction (D-loop formation) was reduced significantly, indicating that the RPA activity was specific for R-loop formation (Fig. 2C and Fig. S6A). R-loop formation strictly requires homology between RNA and dsDNA; no R-loops were formed with a nonhomologous RNA (no. 534, 48 nt) (Fig. S6B). Not all tested RNAs were equally proficient in RPA-promoted R-loop formation (Fig. S7). This proficiency inversely correlates with the RNA propensity to form secondary structures (Table S2). It does not generally correlate with the RPA-binding affinity for the tested RNAs, as RPA has similar KD for nos. 501 and 3R, which differ dramatically in their ability to support R-loop formation (Table S2). However, by titrating the RPA-RNA complexes with NaCl, we found that the RPA complex with RNA no. 501 is more stable than with RNA no. 3R (Fig. S8). Thus, the stability of RPA-RNA complexes may contribute to R-loop formation efficiency. The yield of R-loop formation rises with the increase of RNA length from 24 to 48 nt. No R-loops formed with a 12-nt RNA (Fig. 2, F and G), consistent with poor RPA binding to short RNAs (Fig. S9).
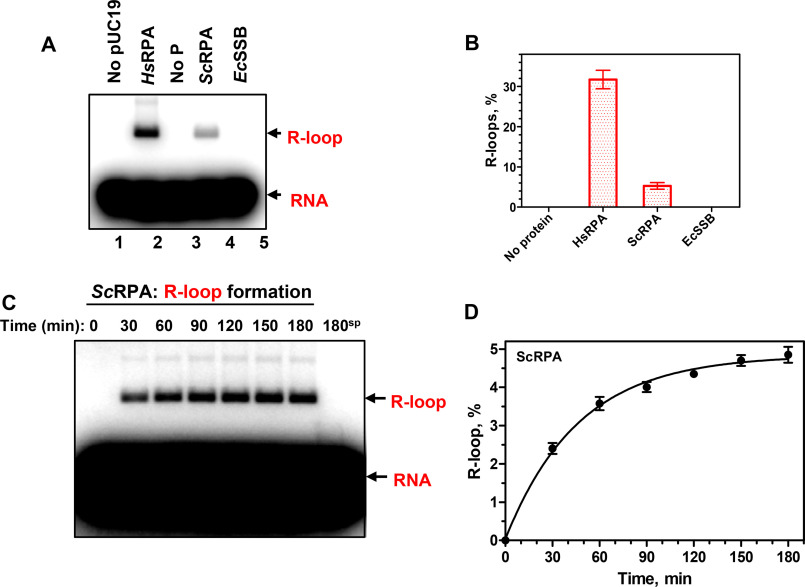
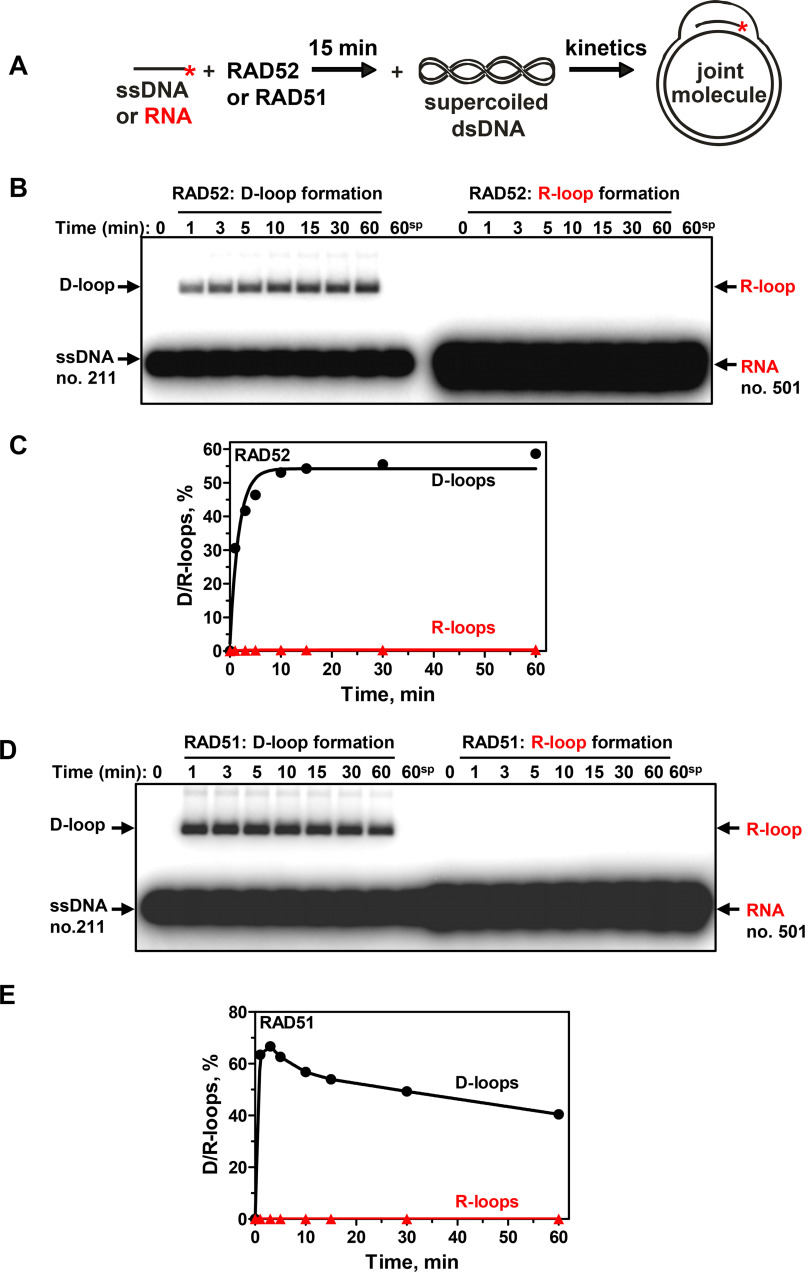
The R-loop–forming activity shows evolutionary conservation among RPA orthologs. Saccharomyces cerevisiae RPA (ScRPA) promotes R-loop formation, albeit with an ∼6-fold reduced efficiency. We also tested an RPA functional homolog from Escherichia coli (EcSSB) for R-loop formation activity (Fig. 3). No activity was observed in a broad range of EcSSB protein concentrations under standard R-loop formation conditions. We also tested two other conditions, in which EcSSB protein showed the strongest annealing activity, such as a buffer with pH 5.5 or the presence of 2 mm spermidine (pH 7.0) (25). However, at any of the tested conditions, EcSSB did not show R-loop formation activity. The optimal RPA concentration for R-loop formation was one RPA heterotrimer per 15 nt and 30 nt of RNA for human and yeast orthologs, respectively (Fig. S10). Human RPA produces R-loops over a broad range of Mg2+ concentration, peaking near 1 mm, whereas yeast RPA shows a sharper peak of Mg2+ dependence with a maximal R-loop yield at 2 mm (Fig. S11). We also found that RAD52 or RAD51 recombinase, which efficiently promoted D-loop formation, did not promote R-loop formation (Fig. 4). Recently, we reported that RAD52 promotes inverse strand exchange between linear dsDNA and homologous RNA. Here, we tested whether human RAD52 can promote R-loop formation through the “inverse” strand exchange mechanism. RAD52 at different concentrations was mixed with supercoiled pUC19 dsDNA (9.3 nm (molecules) or 50 μm (nt)) first, followed by the addition of RNA (93 nm (molecules) or 4.5 μm (nt)) (Fig. S12). We found that under these conditions, R-loops can be formed, but their yield even under optimal RAD52 concentration (200 nm) was very low (0.6%). These results indicate that RAD52 promotes inverse RNA strand exchange in proximity to dsDNA break ends. Thus, R-loop formation appeared to be a unique activity of RPA among tested proteins.
Figure 3.
Human and yeast RPA promote R-loop formation. A, human RPA (HsRPA) (200 nm) and yeast RPA (ScRPA) (100 nm), but not E. coli SSB (EcSSB) (270 nm), promote R-loop formation between a 48-mer RNA (no. 501; 3 μm, nt) and pUC19 dsDNA (67.2 μm, nt). The R-loops were analyzed by electrophoresis in a 1% agarose gel. B, data from A presented as a graph. C, kinetics of R-loop formation by ScRPA (100 nm) between 48-mer RNA (no. 501; 3 μm, nt) and pUC19 dsDNA (67.2 μm, nt) analyzed by electrophoresis in a 1% agarose gel. D, data from C presented as a graph. Error bars, S.E.
Figure 4.
Human RAD52 and RAD51 promote formation of D-loops, but not R-loops. A, scheme of D/R-loop formation. *, 32P label. B, kinetics of RAD52-promoted D- and R-loop formation. RAD52 (450 nm) was preincubated with a 48-mer ssDNA (no. 211; 3 μm, nt) or RNA (no. 501; 3 μm, nt) of the same sequence. The reactions were initiated by the addition of supercoiled pUC19 dsDNA (50 μm, nt), and the products were analyzed by electrophoresis in a 1% agarose gel. 60sp, RAD52-independent (spontaneous) D/R-loop formation after 60 min of reaction. C, data from B shown as a graph. D, kinetics of RAD51-promoted D- and R-loop formation. RAD51 (1 μm) was preincubated with a 48-mer ssDNA (no. 211; 3 μm, nt) or RNA (no. 501; 3 μm, nt). The reactions were initiated by the addition of supercoiled pUC19 dsDNA (50 μm, nt). The products were analyzed by electrophoresis in a 1% agarose gel. 60sp, RAD51-independent (spontaneous) D/R-loop formation after 60 min of incubation. E, data from D shown as a graph.
Reconstitution of DNA replication restart using R-loops
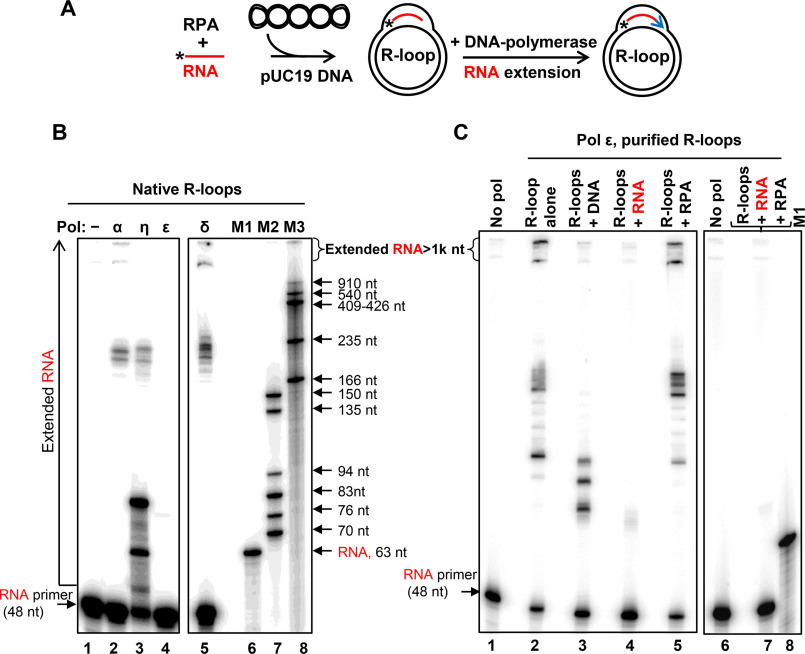
Previously, it was suggested by Kogoma (16) that R-loops may be used to initiate the restart of DNA replication stalled at DNA damage sites. The ability of RPA to form R-loop may be especially relevant to this hypothesis because of a strong and well-documented RPA association with DNA replication. Thus, RPA was initially discovered in human cell extracts as a component essential for SV40 DNA replication in vitro (26–28). Here, we wanted to test whether human DNA polymerases pol δ, pol α, and pol ε and the translesion polymerase pol η can use R-loops for initiation of DNA replication. Pol η was shown to promote DNA synthesis from homologous recombination intermediates (D-loops) (29). In our experiments, DNA polymerases were directly added to the R-loops generated by RPA with 32P-labeled RNA (no. 501) and pUC19 DNA in the presence of four dNTPs (Fig. 5A). RNA extension by DNA polymerases was visualized by electrophoresis in denaturing polyacrylamide gels. We found that pol η was the most efficient in utilizing R-loops for initiation of DNA synthesis, but most of its products were short, ≤83 nt, as could be expected for translesion DNA polymerases (30) (Fig. 5B, lane 3). Pol α and pol δ (in the presence of RFC and PCNA) were less efficient but generated longer DNA products, ∼235 nt (approximate size of the largest R-loops that can form on pUC19 supercoiled dsDNA) and even ≥1000 nt (due to the synthesis-driven strand displacement) (31). In contrast, pol ε could not efficiently use native R-loops to initiate DNA synthesis (Fig. 5B, lane 4); even the addition of RFC and PCNA did not stimulate this reaction.4
Figure 5.
In vitro reconstitution of DNA synthesis restart from R-loops. A, experimental scheme. *, 32P label at 5′-end of RNA (48 nt of no. 501). Blue arrow, extension of RNA by DNA polymerases. B, R-loops (3 nm) were generated in pUC19 using RPA. RNA extension in R-loops was carried out using DNA pol α (50 nm), η (38 nm), ε (50 nm), or pol δ (0.5 nm). The products of RNA extension were analyzed by electrophoresis in 8% polyacrylamide denaturing gels. In control (lane 1), DNA polymerases were omitted. 32P-labeled markers are shown in lanes 6–8. C, effect of RNA, ssDNA, RPA, and RPA-RNA on DNA synthesis by pol ε. RNA extension by pol ε (50 nm) was carried out using deproteinized and purified R-loops (1 nm) (lane 2). The R-loops were premixed with ssDNA (no. 2; 3 μm, nt) (lane 3), RNA (no. 517; 3 μm, nt) (lane 4), RPA (5 nm) (lane 5), or a mixture of RPA (200 nm) and RNA (no. 517; 3 μm, nt) (lane 7) prior to pol ε addition. In control (lanes 1 and 6), pol ε was substituted with storage buffer.
To investigate whether the poor ability of pol ε to use native R-loops for DNA extension is intrinsic or is caused by an inhibitory effect of RPA present in the reaction, we deproteinized and purified R-loops. In this case, pol ε and other tested DNA polymerases efficiently extended RNA (Fig. 5C and Fig. S13). Moreover, the reactions were not affected significantly when RPA was added back to purified R-loops at a concentration that was sufficient to cover the displaced ssDNA strand in R-loops at a stoichiometry of 1 trimer per 15 nt. However, when free RNA (no. 517; 3 μm, nt) or RPA-RNA complexes were added to R-loops, we found a strong inhibition of pol ε (Fig. 5C, lanes 4 and 7). Thus, the presence of RPA-RNA complexes inhibited activity of pol ε in the reconstitution experiments with nondeproteinized R-loops. Pol ε was also sensitive to free ssDNA (no. 2; 3 μm, nt), albeit to a lesser degree (Fig. 5C, lane 3). Strong inhibition of pol ε with RNA (Fig. 5C, lane 4) was unexpected and requires further investigation. Among other tested polymerases, only pol η showed some mild sensitivity to free ssDNA and RNA (Fig. S13, compare the RNA primer uptake in lane 7 with those in lanes 8 and 9), and none of them showed significant sensitivity to RPA under tested conditions (Fig. S13, lanes 5, 10, and 18). Thus, RPA-generated R-loops can be used for initiation of DNA synthesis by human DNA polymerases: pol α, pol η, pol δ, and pol ε.
Discussion
In this study, we identified novel unanticipated activities of RPA: high-affinity binding to RNA and formation of R-loops between RNA and homologous supercoiled dsDNA. We show that human DNA polymerases α, δ, ε, and η can utilize RPA-generated R-loops for initiation of DNA synthesis in vitro, supporting a previously proposed role of R-loops in DNA replication restart (16, 17).
Whereas binding of RPA to RNA was demonstrated in early studies, the RPA-binding affinity for RNA was underestimated. The affinity of RPA for RNA was measured indirectly using RNA as a nonlabeled poly(rA) and poly(rIC) competitor against labeled ssDNA in the filter-binding assay (23). As we show here, poly(rA) is an exceptionally poor competitor against ssDNA, not adequately representing RNA with mixed-base composition. Poly(rIC) readily forms double-stranded structures to which RPA is known to bind poorly. Recently, using fluorescence anisotropy, the KD values for RPA binding to ssDNA and RNA were determined as 4 and 15.2 nm, respectively (32). Based on these data, one could conclude that RPA binds to RNA with only 3–4-fold lower affinity than to ssDNA. However, the RNA and ssDNA concentrations (5 nm) in this study were too close to the reported KD values, especially for ssDNA, making the accuracy of these measurements problematic. In addition, the length and the structure of this RNA substrate were not reported in the paper. Moreover, in other experiments, the poly(dU) sequences were referred to as “RNA,” making the need for the substrate description even stronger. In contrast, our data show that RPA binds to RNA with high affinity (KD ≈ 100 pm for a 48-mer), about 500-fold stronger than to dsDNA but still 30–60-fold weaker than to ssDNA.
The high affinity of RPA to RNA in vitro may suggest that RPA binds RNA also in vivo. Recent proteomics studies support this proposal. Thus, RPA has been identified among RNA-interacting proteins in mammalian (33, 34) and yeast cells (35). Bonasio's group (33), by protein-RNA photocross-linking and quantitative MS, identified RPA among the proteins that interact with RNA regardless of its polyadenylation status in the nuclei of mouse embryonic stem cells. The RNA-cross-linked peptide 263VYYFSK268 was mapped in the DNA-binding domain A of RPA70. Mendell's group (34) identified RPA among the proteins that interact with long noncoding RNA NORAD. In that study, biotinylated RNA fragments of NORAD were incubated with human HTT-116 whole-cell lysates, and the proteins that bind to these fragments were eluted and identified by MS. RPA70, RPA32, and RPA14 subunits were among the proteins that specifically bound NORAD RNA. Parker's group (35), by UV cross-linking proteins to mRNAs, identified RPA among the proteins that directly interact with mRNA in vivo. mRNA-protein complexes were then purified under denaturing conditions using oligo(dT) columns, and the RNA–bound proteins were analyzed by LC–MS/MS (35). ScRFA1 subunit (ortholog of HsRPA70) was identified among the mRNA-bound proteins. The biological role of RPA-RNA interactions remains to be understood. RPA may protect RNA from RNases or recruit proteins that are involved in RNA metabolism. A putative role of RPA in mRNA nuclear export was reported (36). Additional studies are needed to further characterize RPA-RNA interactions in vivo.
Even though RNA is abundant in the cell, RPA binding to RNA may not necessarily interfere with its well-established functions in DNA metabolism that require RPA binding to ssDNA. It is likely that RPA will transfer from RNA to ssDNA generated during DNA replication stress or damage due to its 60-fold higher affinity for ssDNA. The dynamic nature of RPA binding was demonstrated for ssDNA; RPA can migrate along the ssDNA axis and transfer from one polynucleotide to another (22, 37–39).
R-loop formation promoted by RPA is a unique type of strand exchange, as it is initiated by a complex that RPA forms with RNA. In contrast, all other known types of strand exchange, both forward and inverse, are initiated by a recombinase-DNA complex. For instance, E.coli RecA promotes formation of R-loops or RNA-DNA heteroduplexes in inverse RNA strand exchange by assembling an active complex on dsDNA, which then engages free RNA (17, 40). Similarly, RAD52 promotes formation of RNA-DNA heteroduplex in inverse RNA strand exchange by forming an active complex with dsDNA (41). R-loop formation was reported for ICP8, the herpes simplex virus type-1 ssDNA-binding protein (42). However, this reaction occurred only with the alkali-denatured form of plasmid dsDNA through the annealing mechanism. Thus, RPA appeared to be the first known protein that promotes formation of bona fide R-loops by forming an active complex with RNA and by promoting invasion of RNA into covalently closed duplex DNA.
Although the mechanism of R-loop formation by RPA remains to be investigated, several assumptions can be made. During the initial step of R-loop formation, RPA acts in a complex with RNA due to its ∼500-fold higher affinity for RNA compared with the plasmid dsDNA. Moreover, the optimal amount of RPA required for R-loop formation corresponds to its stoichiometric coverage of RNA, but not dsDNA (Fig. S10). Next, RPA-RNA complex needs to engage dsDNA in the homology search process. The RPA trimer has at least four DNA-binding domains (3, 22), which could potentially provide binding space to both RNA and dsDNA, juxtaposing them for RNA:DNA pairing. Binding of dsDNA by the RPA-RNA complex should be by necessity weak to allow multiple association-dissociation steps during the homology probing. After homology is found and initial R-loops are formed, RPA may not remain bound to the newly formed RNA-DNA heteroduplex but be transferred to the displaced ssDNA strand, to which it has much higher affinity (Fig. 1). This RPA binding to the displaced ssDNA strand may help to stabilize and further expand the R-loop. A relatively weaker RPA binding to RNA compared with ssDNA may favor its R-loop formation activity as opposed to D-loop formation. Because of a strong binding to RPA, ssDNA may occupy all available binding space, preventing dsDNA binding that is needed for formation of D-loops.
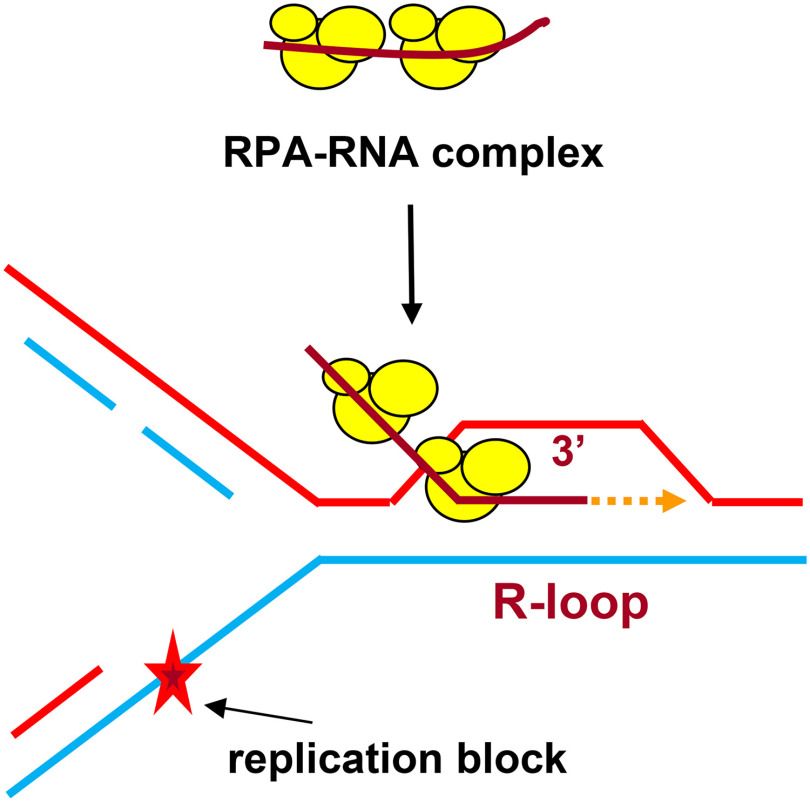
It is highly plausible that R-loop formation by RPA is not the only mechanism that exists in the cell. Because we did not find such activity in EcSSB, we assume that E. coli may use other mechanisms that remain to be identified. RecA-mediated inverse RNA strand exchange for R-loop formation was previously proposed to be one of these mechanisms (17). We also cannot exclude the possibility that some auxiliary proteins are required for stimulation of EcSSB R-loop formation activity. Recent data indicate that R-loops are a common structure in genomes of humans and other species (12, 13). The biological role of R-loops is currently under intense investigation. It was found that R-loops are essential for repair of DNA double-strand breaks in actively transcribed genome regions through transcription-coupled homologous recombination (6, 14, 15) or non-homologous end joining (43). It was proposed by Kogoma (16) that R-loops may serve as a primer to restart DNA replication stalled at DNA lesions (Fig. 6). The R-loop formation activity of RPA may be especially relevant to replication restart because of a strong RPA association with DNA replication (26–28). The RPA32 subunit was directly UV cross-linked with the RNA strand of the nascent RNA-DNA primer during SV40 replication in nuclei of monkey CV-1 cells (44). It was demonstrated that RPA interacts with pol α, RFC, and pol δ (45, 46). RPA stabilizes a complex between short RNA primer and pol α and then coordinates loading of RFC, PCNA, and pol δ to initiate DNA synthesis. We found that all tested human DNA polymerases, pol α, pol δ, pol ε, and pol η, can utilize RPA-generated R-loops for initiation of DNA synthesis in vitro. These in vitro reconstitution experiments further support Kogoma's hypothesis and suggest the mechanisms of genome maintenance that depend on RPA and RNA.
Figure 6.
Proposed RPA-dependent DNA replication restart initiated at R-loop. RPA promotes formation of the R-loop that serves as a primer for a DNA polymerase during replication restart.
Experimental procedures
Proteins, DNA, and RNA
Human RPA, RAD51, and RAD52 were purified as described (47–49). E. coli SSB protein was purchased from Affimetrix Inc. Human DNA polymerases pol η, the catalytic core of pol α p180(335–1257), the FLAG-tagged four-subunit pol δ, RFC, and PCNA were purified as described (50–53). The catalytic core of human pol ε p261(1–1172)exo− was purified according to Ref. 54 with the following modifications: His tag was placed on the N terminus before a SUMO tag and removed by SUMO protease after the first purification step, which included the nickel ion affinity chromatography. The oligodeoxyribonucleotides (Table S1) were purchased from IDT Inc. and further purified by electrophoresis in polyacrylamide gels containing 50% urea (55). HPLC-purified oligoribonucleotides were purchased from IDT Inc. All experiments with RNA were carried out in the presence of 1× Ambion RNAsecure RNase inactivation reagent. Double-stranded oligonucleotides were prepared by annealing of equimolar (molecules) amounts of complementary oligonucleotides (55). When indicated, oligonucleotides were 5-end–labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Supercoiled pUC19 plasmid dsDNA was prepared by a method that did not involve DNA denaturation (56) with modifications. Briefly, E. coli host cells were treated with lysozyme and lysed with Triton X-100. The lysate was cleared by centrifuged at 4 °C for 30 min at 40,000 × g. The cleared lysate was mixed with ethidium bromide to 700 µg/ml and CsCl (0.59 g/1 ml of cleared lysate) and loaded at the top of CsCl (1.58 g/ml in water) solution that filled the bottom half of the centrifuge tube. The samples were centrifuged in an angle rotor for 24 h at 200,000 × g at the ambient temperature. Isolated supercoiled plasmid DNA was further purified by 3× butanol extractions followed by gel filtration on a Sephacryl S-500 column. Supercoiled pHSG299 plasmid dsDNA purified by CsCl-ethidium bromide gradient centrifugation was purchased from Takara Bio Inc. pHSG299 is a derivative of pUC19 plasmid in which an ampicillin-resistant gene was replaced with a kanamycin-resistant gene. DNA and RNA concentrations are expressed in moles of molecules or, when indicated, in moles of nucleotides.
RPA binding to RNA, ssDNA, dsDNA, and RNA-DNA hybrid using EMSA
20-µl mixtures contained human RPA at the indicated concentrations, 25 mm Tris·acetate (pH 7.5), 10 mm KCl (added with the protein stock), 2 mm DTT, 1 mm magnesium acetate, 100 μg/ml BSA, and 32P-labeled 48-mer nucleic acid substrates: RNA (no. 501; 5 pm, molecules), ssDNA (no. 211; 0.5 pm, molecules), dsDNA (nos. 211/212; 3 nm, molecules), or RNA-DNA hybrid (nos. 501/212; 3 nm, molecules). The mixtures were incubated for 15 min at 37 °C and then placed on ice. Each sample was supplemented with 3 µl of 50% glycerol and loaded onto a 6% polyacrylamide (29:1) gel in 0.25× TBE buffer (22.5 mm Tris, 22.5 mm borate, and 0.25 mm EDTA, pH 8.3). Bromphenol blue was added only in the sample containing 32P-labeled probe without RPA. Electrophoresis was carried out at 13 V/cm for 1 h at room temperature. The gels were dried on Amersham Biosciences Hybond-N+ membrane (GE Healthcare) and analyzed using a Typhoon FLA 7000 biomolecular imager. The KD and Bmax values were obtained by fitting the data to a one-site binding hyperbola in GraphPad Prism 5.0. Bmax values were 1.20 ± 0.07, 1.11 ± 0.08, 1.20 ± 0.07, and 1.10 ± 0.02 for RNA (no. 501), ssDNA (no. 211), dsDNA (nos. 211/212), and RNA-DNA hybrid (nos. 501/212), respectively.
RPA binding to RNA or ssDNA in the presence of competitors
RPA (20 pm) was incubated with a 32P-labeled 48-mer ssDNA (no. 211; 5 pm (molecules) or 240 pm (nt)) that was premixed with various indicated amounts of nonlabeled RNA (no. 501) or ssDNA (no. 211) for 15 min at 37 °C. In other experiments, RPA at the indicated concentrations was incubated with 32P-labeled RNA (no. 501; 5 pm (molecules) or 240 pm (nt)) or 32P-labeled ssDNA (no. 211; 5 pm (molecules) or 240 pm (nt)) that was premixed with various indicated amounts of pHSG299 supercoiled plasmid dsDNA for 15 min at 37 °C. The nonhomologous pHSG299 dsDNA competitor was used to avoid possible DNA/RNA pairing that might interfere with the RPA-binding measurements. RPA-[32P]RNA and RPA-[32P]DNA complexes were analyzed by EMSA as described above.
D-loop and R-loop formation
Human RPA (200 nm) was incubated with 32P-labeled RNA (3 μm, nt) or ssDNA (3 μm, nt) in buffer A containing 25 mm Tris·acetate (pH 7.5), 10 mm KCl (added with the protein stock), 2 mm DTT, 1 mm magnesium acetate, and 100 μg/ml BSA for 15 min at 37 °C. The reactions were initiated by the addition of supercoiled pUC19 dsDNA (67.2 μm, nt). Aliquots (10 µl) were withdrawn at the indicated time points and deproteinized by incubation with 1% SDS, 1.6 mg/ml proteinase K, 6% glycerol, and 0.01% bromphenol blue for 15 min at 37 °C. Samples were analyzed by electrophoresis in 1% agarose gels in TAE buffer (40 mm Tris·acetate, pH 8.0, and 1 mm EDTA). Electrophoresis was carried out at 5 V/cm for 1.5 h at room temperature. The gels were dried and analyzed as described above for EMSA. The D-loop/R-loop yield was expressed as a percentage of the input plasmid DNA. For ScRPA and EcSSB, the R-loop formation was carried out as described above, except that magnesium acetate concentration was 2 mm, and the protein concentrations were 100 and 270 nm, respectively.
For human RAD52, the R/D-loop formation reactions were carried out as described for RPA, except that RAD52 (450 nm) was used instead of RPA, magnesium acetate was 0.3 mm, and supercoiled pUC19 dsDNA was 50 μm (nt). For human RAD51, the R/D-loop formation reactions were carried out as described for RPA, except that 1 mm ATP was included in the reaction mixture, 1 mm CaCl2 was used instead of magnesium acetate, and the concentrations of RAD51 and supercoiled pUC19 dsDNA were 1 μm and 50 μm (nt), respectively.
Cleavage of the R-loops with EcoRI restriction endonuclease
RPA-promoted R-loop formation was carried out for 3 h at 37 °C, as described above. Then 1.5 µl of 50 mm magnesium acetate and 0.5 µl (10 units) of EcoRI restriction endonuclease were added to 10 µl of the reaction mixture, and incubation was continued for another 15 min. The samples were deproteinized and analyzed by electrophoresis in 1% agarose gels. The gels were dried and analyzed as described above for the R-loop formation. The dried gels were then rehydrated, stained with ethidium bromide (2 µg/ml in water) for 30 min at room temperature, destained for 30 min in a large volume of water, and subjected to image analysis using an AlphaImager 3400 gel documentation station.
Treatment of the R-loops with RNase H
RPA-promoted R-loop formation was carried out for 3 h at 37 °C, as described above. Then 1 µl of 10× RNase H reaction buffer and 1 µl (5 units) of RNase H (New England Biolabs) were added to 8 µl of the reaction mixture, and incubation was continued for another 30 min. The samples were deproteinized and analyzed by electrophoresis in 1% agarose gels. The gels were dried and autoradiographed and analyzed using a Typhoon FLA 7000 biomolecular imager as described above for EMSA. The dried gels were then rehydrated by soaking in water, detached from the Amersham Biosciences Hybond-N+ membrane, stained with ethidium bromide, and analyzed as described above for EcoRI cleavage of R-loops.
Reconstitution of DNA synthesis restart using R-loops
RPA-promoted R-loop formation between pUC19 and 32P-labeled 48-mer RNA (no. 501) was carried out in buffer containing 25 mm Tris·acetate, pH 7.5, 1 mm magnesium acetate, 100 μm each of four dNTPs, 250 µg/ml BSA, and 10 mm DTT for 3 h at 37 °C. Then KCl was added to a final concentration of 40 mm. To initiate DNA synthesis from R-loops (3 nm, molecules), 9-µl aliquots were mixed with DNA pol α (50 nm), pol η (38 nm), or pol ε (50 nm) and incubated for 30 min at 37 °C. The addition of the DNA polymerases increased final KCl concentration to 60 mm.
For RNA extension by pol δ, RPA-promoted R-loop formation was performed in standard buffer A for 3 h at 37 °C. Reconstitution reactions (10 µl) contained R-loops (3 nm, molecules), RFC (8 nm), PCNA (48 nm), pol δ (0.5 nm), 30 mm Tris·acetate, pH 7.5, 5 mm magnesium acetate, 100 μm each of four dNTPs, 1 mm ATP, 250 µg/ml BSA, 10 mm DTT, 60 mm KCl. R-loops were preincubated with RCF and PCNA for 5 min at 37 °C, and then pol δ was added, and incubation was continued for another 30 min.
All DNA polymerization reactions were terminated by adding 15 µl of 99.9% formamide, containing 0.1% bromphenol blue. The samples were heated for 4 min at 80 °C, and the products of RNA extension were analyzed by electrophoresis in an 8% denaturing PAGE (19:1), containing 50% urea. The migration markers M1–M3 were 63-nt RNA, 70–150-nt ssDNA oligonucleotides, and 166-910-nt denatured DdeI restriction fragments of pUC19, respectively. After electrophoresis, the gels were fixed in 10% glacial acetic acid and 10% ethanol for 20 min at room temperature, dried, and analyzed using a Typhoon FLA 7000 biomolecular imager.
RNA extension by DNA polymerases using deproteinized R-loops
RPA-promoted R-loop formation was performed in buffer A for 3 h at 37 °C. The R-loops were deproteinized by treatment with proteinase K (1 mg/ml) and 0.8% SDS for 30 min at 37 °C, and then 1 mm EDTA, pH 8.0, was added to chelate magnesium ions. The deproteinized R-loops were purified by passing twice through S-400 spin columns (GE Healthcare) equilibrated with 25 mm Tris-HCl (pH 7.5) and 25 mm KCl. The purified R-loops were supplemented with 1 mm of magnesium acetate and kept at −20 °C.
Reactions (10 µl) were initiated by adding DNA pol α (50 nm), pol η (4 nm), or pol ε (50 nm) to deproteinized R-loops (1 nm) in 30 mm Tris-HCl (pH 7.5), 1 mm magnesium acetate, 100 μm each of four dNTPs, 250 µg/ml BSA, 10 mm DTT, 60 mm KCl and carried out for 30 min at 37 °C. For DNA pol δ, the reactions (10 µl) were carried out in 30 mm Tris·acetate, pH 7.5, 5 mm magnesium acetate, 100 μm each of four dNTPs, 1 mm ATP, 250 µg/ml BSA, 10 mm DTT, and 60 mm KCl. Deproteinized R-loops (1 nm, molecules) were preincubated with RCF (2 nm) and PCNA (10 nm) for 5 min at 37 °C, and then pol δ (0.5 nm) was added, followed by incubation for another 30 min.
Quantification and statistical analysis
For statistical analysis, GraphPad Prism 5 software was used. In vitro experiments were repeated at least three times; S.E. values are presented on the graphs.
Data availability
All data are contained within the article and supporting information.
Supplementary Material
Acknowledgments
We thank Patrick Sung (University of Texas Health Science Center at San Antonio) and Stephen Kowalczykowski (University of California, Davis) for providing yeast RPA and all members of the Mazin laboratory for comments and discussion.
This article contains supporting information.
Author contributions—O. M. M. and A. V. M. conceptualization; O. M. M. data curation; O. M. M. investigation; O. M. M., L. Y. K., A. G. B., T. H. T., F. A. K., and A. V. M. methodology; O. M. M. writing-original draft; O. M. M. and A. V. M. writing-review and editing; S. S. software; S. S. and A. V. M. formal analysis; L. Y. K., A. G. B., T. H. T., and F. A. K. resources; A. V. M. supervision; A. V. M. funding acquisition; A. V. M. project administration.
Funding and additional information—This work was supported by NCI and NIGMS, National Institutes of Health, Grants R01 CA188347, P30CA056036, and R01 GM136717 and by a Drexel-Coulter Program Award (to A. M.) and by NIGMS, National Institutes of Health, Grants R35 GM127085 (to T. T.) and R01 GM095758 (to F. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
O. M. Mazina and A. V. Mazin, unpublished results.
- RPA
- replication protein A
- EMSA
- electrophoretic mobility shift assay
- nt
- nucleotide(s)
- pol
- polymerase
- RFC
- replication factor C
- PCNA
- proliferating cell nuclear antigen
- SUMO
- small ubiquitin-like modifier.
References
- 1. Haring S. J., Mason A. C., Binz S. K., and Wold M. S. (2008) Cellular functions of human RPA1: multiple roles of domains in replication, repair, and checkpoints. J. Biol. Chem. 283, 19095–19111 10.1074/jbc.M800881200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wold M. S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 10.1146/annurev.biochem.66.1.61 [DOI] [PubMed] [Google Scholar]
- 3. Chen R., and Wold M. S. (2014) Replication protein A: single-stranded DNA's first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays 36, 1156–1161 10.1002/bies.201400107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zou Y., Liu Y., Wu X., and Shell S. M. (2006) Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 208, 267–273 10.1002/jcp.20622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borgstahl G. E., Brader K., Mosel A., Liu S., Kremmer E., Goettsch K. A., Kolar C., Nasheuer H. P., and Oakley G. G. (2014) Interplay of DNA damage and cell cycle signaling at the level of human replication protein A. DNA Repair (Amst.) 21, 12–23 10.1016/j.dnarep.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yasuhara T., Kato R., Hagiwara Y., Shiotani B., Yamauchi M., Nakada S., Shibata A., and Miyagawa K. (2018) Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 175, 558–570.e11 10.1016/j.cell.2018.08.056 [DOI] [PubMed] [Google Scholar]
- 7. Wei L., Nakajima S., Bohm S., Bernstein K. A., Shen Z., Tsang M., Levine A. S., and Lan L. (2015) DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 112, E3495–E3504 10.1073/pnas.1507105112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nguyen H. D., Yadav T., Giri S., Saez B., Graubert T. A., and Zou L. (2017) Functions of replication protein A as a sensor of R loops and a regulator of RNase H1. Mol. Cell 65, 832–847.e34 10.1016/j.molcel.2017.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stork C. T., Bocek M., Crossley M. P., Sollier J., Sanz L. A., Chédin F., Swigut T., and Cimprich K. A. (2016) Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5, e17548 10.7554/eLife.17548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lang K. S., Hall A. N., Merrikh C. N., Ragheb M., Tabakh H., Pollock A. J., Woodward J. J., Dreifus J. E., and Merrikh H. (2017) Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 170, 787–799.e18 10.1016/j.cell.2017.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tresini M., Warmerdam D. O., Kolovos P., Snijder L., Vrouwe M. G., Demmers J. A., van I. W. F., Grosveld F. G., Medema R. H., Hoeijmakers J. H., Mullenders L. H., Vermeulen W., and Marteijn J. A. (2015) The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58 10.1038/nature14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanz L. A., Hartono S. R., Lim Y. W., Steyaert S., Rajpurkar A., Ginno P. A., Xu X., and Chédin F. (2016) Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 10.1016/j.molcel.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chédin F. (2016) Nascent connections: R-loops and chromatin patterning. Trends Genet. 32, 828–838 10.1016/j.tig.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marnef A., Cohen S., and Legube G. (2017) Transcription-coupled DNA double-strand break repair: active genes need special care. J. Mol. Biol. 429, 1277–1288 10.1016/j.jmb.2017.03.024 [DOI] [PubMed] [Google Scholar]
- 15. Wei L., Levine A. S., and Lan L. (2016) Transcription-coupled homologous recombination after oxidative damage. DNA Repair (Amst.) 44, 76–80 10.1016/j.dnarep.2016.05.009 [DOI] [PubMed] [Google Scholar]
- 16. Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238 10.1128/.61.2.212-238.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaitsev E. N., and Kowalczykowski S. C. (2000) A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev. 14, 740–749 [PMC free article] [PubMed] [Google Scholar]
- 18. Itoh T., and Tomizawa J. (1980) Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. U. S. A. 77, 2450–2454 10.1073/pnas.77.5.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stuckey R., García-Rodríguez N., Aguilera A., and Wellinger R. E. (2015) Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proc. Natl. Acad. Sci. U. S. A. 112, 5779–5784 10.1073/pnas.1501769112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aguilera A., and García-Muse T. (2012) R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 21. Santos-Pereira J. M., and Aguilera A. (2015) R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- 22. Pokhrel N., Caldwell C. C., Corless E. I., Tillison E. A., Tibbs J., Jocic N., Tabei S. M. A., Wold M. S., Spies M., and Antony E. (2019) Dynamics and selective remodeling of the DNA-binding domains of RPA. Nat. Struct. Mol. Biol. 26, 129–136 10.1038/s41594-018-0181-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim C., Snyder R. O., and Wold M. S. (1992) Binding properties of replication protein A from human and yeast cells. Mol. Cell Biol. 12, 3050–3059 10.1128/mcb.12.7.3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bugreev D. V., Hanaoka F., and Mazin A. V. (2007) Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 14, 746–753 10.1038/nsmb1268 [DOI] [PubMed] [Google Scholar]
- 25. Christiansen C., and Baldwin R. L. (1977) Catalysis of DNA reassociation by the Escherichia coli DNA binding protein: a polyamine-dependent reaction. J. Mol. Biol. 115, 441–454 10.1016/0022-2836(77)90164-4 [DOI] [PubMed] [Google Scholar]
- 26. Wold M. S., and Kelly T. (1988) Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. U. S. A. 85, 2523–2527 10.1073/pnas.85.8.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fairman M. P., and Stillman B. (1988) Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 7, 1211–1218 10.1002/j.1460-2075.1988.tb02933.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wobbe C. R., Weissbach L., Borowiec J. A., Dean F. B., Murakami Y., Bullock P., and Hurwitz J. (1987) Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc. Natl. Acad. Sci. U. S. A. 84, 1834–1838 10.1073/pnas.84.7.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McIlwraith M. J., Mcllwraith M. J., Vaisman A., Liu Y., Fanning E., Woodgate R., and West S. C. (2005) Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 20, 783–792 10.1016/j.molcel.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 30. Plosky B. S., and Woodgate R. (2004) Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr. Opin. Genet. Dev. 14, 113–119 10.1016/j.gde.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 31. Stith C. M., Sterling J., Resnick M. A., Gordenin D. A., and Burgers P. M. (2008) Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 283, 34129–34140 10.1074/jbc.M806668200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Q. M., Yang Y. T., Wang Y. R., Gao B., Xi X. G., and Hou X. M. (2019) Human replication protein A induces dynamic changes in single-stranded DNA and RNA structures. J. Biol. Chem. 294, 13915–13927 10.1074/jbc.RA119.009737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He C., Sidoli S., Warneford-Thomson R., Tatomer D. C., Wilusz J. E., Garcia B. A., and Bonasio R. (2016) High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell 64, 416–430 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee S., Kopp F., Chang T. C., Sataluri A., Chen B., Sivakumar S., Yu H., Xie Y., and Mendell J. T. (2016) Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 10.1016/j.cell.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitchell S. F., Jain S., She M., and Parker R. (2013) Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20, 127–133 10.1038/nsmb.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen C. C., Lee J. C., and Chang M. C. (2012) 4E-BP3 regulates eIF4E-mediated nuclear mRNA export and interacts with replication protein A2. FEBS Lett. 586, 2260–2266 10.1016/j.febslet.2012.05.059 [DOI] [PubMed] [Google Scholar]
- 37. Chen R., Subramanyam S., Elcock A. H., Spies M., and Wold M. S. (2016) Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Res. 44, 5758–5772 10.1093/nar/gkw339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nguyen B., Sokoloski J., Galletto R., Elson E. L., Wold M. S., and Lohman T. M. (2014) Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 426, 3246–3261 10.1016/j.jmb.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gibb B., Ye L. F., Gergoudis S. C., Kwon Y., Niu H., Sung P., and Greene E. C. (2014) Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS ONE 9, e87922 10.1371/journal.pone.0087922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kasahara M., Clikeman J. A., Bates D. B., and Kogoma T. (2000) RecA protein-dependent R-loop formation in vitro. Genes Dev. 14, 360–365 [PMC free article] [PubMed] [Google Scholar]
- 41. Mazina O. M., Keskin H., Hanamshet K., Storici F., and Mazin A. V. (2017) Rad52 inverse strand exchange drives RNA-templated DNA double-strand break repair. Mol. Cell 67, 19–29.e13 10.1016/j.molcel.2017.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boehmer P. E. (2004) RNA binding and R-loop formation by the herpes simplex virus type-1 single-stranded DNA-binding protein (ICP8). Nucleic Acids Res. 32, 4576–4584 10.1093/nar/gkh797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chakraborty A., Tapryal N., Venkova T., Horikoshi N., Pandita R. K., Sarker A. H., Sarkar P. S., Pandita T. K., and Hazra T. K. (2016) Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 7, 13049 10.1038/ncomms13049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mass G., Nethanel T., and Kaufmann G. (1998) The middle subunit of replication protein A contacts growing RNA-DNA primers in replicating simian virus 40 chromosomes. Mol. Cell Biol. 18, 6399–6407 10.1128/mcb.18.11.6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mo J., Liu L., Leon A., Mazloum N., and Lee M. Y. (2000) Evidence that DNA polymerase delta isolated by immunoaffinity chromatography exhibits high-molecular weight characteristics and is associated with the KIAA0039 protein and RPA. Biochemistry 39, 7245–7254 10.1021/bi0000871 [DOI] [PubMed] [Google Scholar]
- 46. Yuzhakov A., Kelman Z., Hurwitz J., and O'Donnell M. (1999) Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 18, 6189–6199 10.1093/emboj/18.21.6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Henricksen L. A., Umbricht C. B., and Wold M. S. (1994) Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 48. Sigurdsson S., Trujillo K., Song B., Stratton S., and Sung P. (2001) Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 276, 8798–8806 10.1074/jbc.M010011200 [DOI] [PubMed] [Google Scholar]
- 49. Singleton M. R., Wentzell L. M., Liu Y., West S. C., and Wigley D. B. (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. U. S. A. 99, 13492–13497 10.1073/pnas.212449899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kadyrov F. A., Genschel J., Fang Y., Penland E., Edelmann W., and Modrich P. (2009) A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 106, 8495–8500 10.1073/pnas.0903654106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masutani C., Kusumoto R., Iwai S., and Hanaoka F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 19, 3100–3109 10.1093/emboj/19.12.3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baranovskiy A. G., Babayeva N. D., Suwa Y., Gu J., Pavlov Y. I., and Tahirov T. H. (2014) Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 42, 14013–14021 10.1093/nar/gku1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rodriges Blanko E., Kadyrova L. Y., and Kadyrov F. A. (2016) DNA mismatch repair interacts with CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition. J. Biol. Chem. 291, 9203–9217 10.1074/jbc.M115.713271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zahurancik W. J., Klein S. J., and Suo Z. (2013) Kinetic mechanism of DNA polymerization catalyzed by human DNA polymerase ε. Biochemistry 52, 7041–7049 10.1021/bi400803v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rossi M. J., Mazina O. M., Bugreev D. V., and Mazin A. V. (2010) Analyzing the branch migration activities of eukaryotic proteins. Methods 51, 336–346 10.1016/j.ymeth.2010.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clewell D. B., and Helinski D. R. (1969) Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc. Natl. Acad. Sci. U.S.A. 62, 1159–1166 10.1073/pnas.62.4.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting information.