Summary
Specific recording, labeling, and spatiotemporal manipulating neurons are essential for neuroscience research. In this study, we developed a tripartite spatiotemporal gene induction system in C. elegans, which is based on the knockout of two transcriptional terminators (stops in short) by two different recombinases FLP and CRE. The recombinase sites (loxP and FRT) flanked stops after a ubiquitous promoter terminate transcription of target genes. FLP and CRE, induced by two promoters of overlapping expression, remove the stops (subsequent FLP/CRE-out). The system provides an "AND" gate strategy for specific gene expression in single types of cell(s). Combined with an inducible promoter or element, the system can control the spatiotemporal expression of genes in defined cell types, especially in cells or tissues lacking a specific promoter. This tripartite FLP/CRE-out gene expression system is a simple, labor- and cost-saving toolbox for cell type-specific and inducible gene expression in C. elegans.
Subject Areas: Molecular Biology, Neuroscience, Genetic Engineering
Graphical Abstract
Highlights
-
•
A tripartite FLP/CRE-out spatiotemporal gene expression system in C. elegans
-
•
Eight promoter pairs with the targeting of a single-type neuron(s) were identified
-
•
The system is easy to use and cost and labor saving
Molecular Biology; Neuroscience; Genetic Engineering
Introduction
Genetic methods are widely used to study functions and neurosignal integration of neural circuits in the nervous system (Dymecki and Kim, 2007; Luo et al., 2008, 2018). Spatiotemporally specific (i.e., cell type-specific and inducible) expression of genes is required to identify neuron type, measure the activity of genetically defined neurons, analyze function(s) of the tested gene(s) in neurons, construct neuronal circuit diagrams, and dissect the process of signal integration in neuronal circuits. The genes expressed include the tested genes of living organisms and those encoding indicators, markers, and proteins used in optogenetic and chemogenetic manipulation of neurons. Owing to the conservation of genes in the animal genera, experimentally tractable animal models are generally used to study the relationships among genes, proteins, and neural circuits that underlie behaviors. Caenorhabditis elegans (C. elegans) is a prevalent animal model for studying conserved molecular and neural bases of the nervous system. The limited specific promoters of single/paired neuron(s) hinder the pace and veracity of neuroscience studies, not only in C. elegans but also in other species (Dymecki and Kim, 2007; Luo et al., 2008, 2018).
For spatial and/or temporal gene expression in C. elegans, several strategies have been developed. Bipartite interaction systems of spatial gene expression provide cell type-specific gene expression, which employ two promoters of intersectional expression patterns to drive two interactional parts of a molecule (Chelur and Chalfie, 2007; Zhang et al., 2004). A recently developed strategy of native and tissue-specific fluorescence (NATF) combined with genome editing and split-GFP allows cell-specific fluorescence labeling of native proteins in C. elegans (He et al., 2019). Another method used cell-specific rescue of heat shock factor-1 (hsf-1) mutants to regulate spatial and temporal transgene expression in C. elegans (Bacaj and Shaham, 2007).
The bipartite FLP-out or CRE-out systems induce temporal or spatiotemporal gene expression in C. elegans. It is based on the knockout of a transcriptional terminator (stop) by recombinase FLP (flipase) or CRE (cyclization recombination) derived from the yeast Saccharomyces cerevisiae and bacteriophage P1, respectively (Buchholz et al., 1998; Dymecki, 1996; Golic and Lindquist, 1989; Hoess et al., 1984; Raymond and Soriano, 2007; Sauer and Henderson, 1988). FLP-out and CRE-out are the processes of recombinase-catalyzed intramolecular excision of spacer DNA that lies between tandemly oriented FRT (FLP recognition target) and LoxP (locus of crossing over in P1) sites (Davis et al., 2008; Guo et al., 2015; Hubbard, 2014; Lopez-Cruz et al., 2019; Macosko et al., 2009; Schmitt et al., 2012; Voutev and Hubbard, 2008; Wang et al., 2017). However, these systems need an inducible promoter combined with a cell type-specific promoter for spatiotemporal gene expression. Moreover, bipartite systems of CRE-out of mCherry were designed for lineage tracing with varied fluorescence in C. elegans (Ruijtenberg and van den Heuvel, 2015). Other bipartite expression systems were also developed to achieve spatiotemporal transgene control in C. elegans, such as GAL4-UAS (cGAL) system (Wang et al., 2017), split intein based split cGAL system (Wang et al., 2018), Q system together with split Q system (Wei et al., 2012), and recently the tetracycline-dependent ribozyme switch system (Wurmthaler et al., 2019). An interesting strategy combing heat shock promoter hsp-16.2 with infrared laser illumination (Churgin et al., 2013, 2014; Singhal and Shaham, 2017) was also implemented for spatiotemporal gene induction. Nevertheless, a feasible, reliable, labor- and cost-saving toolbox for cell type-specific and inducible expression of genes of interest in C. elegans is still needed.
Here, a tripartite spatiotemporal gene expression system was developed, which works on dual knockout of two distinct recombinase sites flanked stops by two different recombinases FLP and CRE (and subsequent FLP/CRE-out). Vectors used in the system were constructed by the flexible Three-Fragment Multisite Gateway method. This tripartite FLP/CRE-out gene expression system not only offers highly refined spatial and temporal control of gene expression, but is also feasible and labor and cost saving.
Results
Construction and Work Principle of the Tripartite FLP/CRE-out Gene Expression System
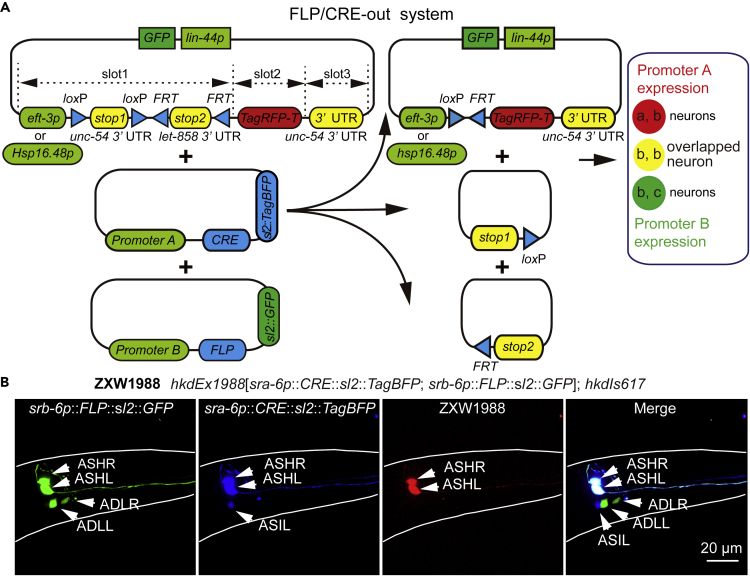
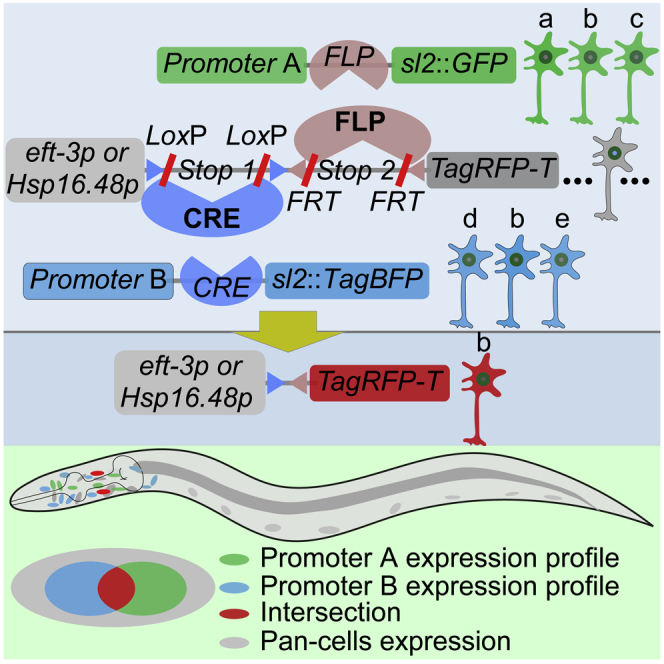
Recombining-out transcriptional terminator with recombinase enables spatial, temporal, or spatiotemporal gene expressions (Davis et al., 2008; Guo et al., 2015; Hubbard, 2014; Lopez-Cruz et al., 2019; Macosko et al., 2009; Schmitt et al., 2012; Voutev and Hubbard, 2008; Wang et al., 2017). However, these bipartite systems have challenges providing spatial and temporal gene expression in the neuron(s) or tissue lacking a specific promoter. Therefore, to induce spatiotemporal gene expression in C. elegans, we developed a tripartite gene induction system (Figure 1A). The system is composed of a core vector for the expression of a tested gene and two recombinase-encoding vectors. The core vector contains eft-3 (strong pan-expression) or hsp-16.48 promoter (heat shock inducible), two different tandem recombinase-sites-flanked transcriptional terminators (stops), and a gene of interest. For an easy change of promoters and genes, we used the Three-Fragment Multisite Gateway method (Magnani et al., 2006) to construct these three vectors (Figures S1A–S1C), using the Invitrogen system from Thermo Fisher Scientific (Thermo Fisher Scientific, MA, USA). To construct the core vector, we used the following sequences: eft-3 or hsp-16.48 promoter with loxP-flanked stop1 and FRT-flanked stop2 as slot1 (promoter::d-stops, in short); the gene of interest as slot2; and unc-54 3′ UTR as slot3. The loxP and FRT are recombination sites of CRE and FLP. The transcriptional terminators stop1 and stop2, which block the transcription of the target gene, consist of unc-54 3′ UTR and let-858 3′ UTR. Recombining-out two stops by recombinase CRE and FLP driven by two distinct promoters with overlapping expression in a type of cell(s), enabled gene transcription in the defined cell(s). CRE and FLP used in the FLP/CRE-out system derived from a nucleolus-transport-enhanced Cre (nCre) of yeast S. cerevisiae (Hoess et al., 1984; Sauer and Henderson, 1988) and mouse codon-optimized FLP (FLPo) (Raymond and Soriano, 2007). We made further modifications to improve their expression and referred to as CRE and FLP. This system provides an "AND" gate strategy for intersectional targeting of a single cell type, i.e., controls gene expression in a type of cell(s) that lacks a specific promoter (Figure 1A).
Figure 1.
A Tripartite Spatiotemporal Gene Expression Control System based on Dual Knocking Out Two Transcriptional Terminators
(A) Design and working principle of the tripartite FLP/CRE-out-mediated gene expression control system. The system consists of three vectors: a core vector and two plasmids encoding CRE and FLP recombinases. The core plasmid consists of sequences of a strong pan-expression eft-3 or a promoter of the hsp-16 family, two transcription terminators flanked by loxP or FRT recombination sites, a target gene and lin-44p::GFP.
(B) Confocal fluorescence images of a ZXW1988 transgenic worm. Different fluorescent channels show expression profile of green, srb-6p in ASHs and ADLs; blue, sra-6p in ASHs and ASIs; and red channels, TagRFP-T in ASHs. The merged image (right) indicated the co-localization of the three fluorescence colors. Scale bar, 20 μm.
The FLP/CRE-out Gene Expression System Is Efficient to Label and Identify a Single Type of Neurons
Spatiotemporally manipulating and recording of neuronal activity is vital for studies of neuronal circuits and circuital neurosignal integration. The FLP/CRE-out system should be a convenient tool for the studies. Neuron-type-specific promoters or promoter pairs are essential for the FLP/CRE-out system. In C. elegans, many neurons, especially sensory neurons, have been identified to possess specific promoters or promoter pairs. However, numerous neurons, especially interneurons, still lack known specific promoters or promoter pairs. Identifying promoter pairs for these neurons will facilitate research in these cells. For easy identification of promoter pairs, we constructed a chromosomally integrated C. elegans transgene ZXW617 strain using a standard UV/trimethylpsoralen (UV/TMP) integration method (Yandell et al., 1994). Its genotype is hkdIs617[eft-3p::d-stops::TagRFP-T::unc-54 3′ UTR::lin-44p::GFP]. Co-injection of FLP and CRE plasmids with a promoter pair of a single neuron-type intersectional expression into the transgenic worms should enable single neuron-specific fluorescent labeling. As expected, the hkdIs617[sra-6p::CRE::sl2::TagBFP; srb-6p::FLP::sl2::GFP] transgene displayed ASHs-specific TagRFP-T labeling (Figure 1B). The expression patterns of sra-6p and srb-6p were ASHs/ASIs and ASHs/ADLs with an intersection of ASH neurons. To identify whether each of the two stops is sufficient to prevent the expression of the downstream reporter gene, the strains of hkdEx2000[sra-6p::CRE::sl2::TagBFP]; hkdIs617 and hkdEx2001[srb-6p::FLP::sl2::GFP]; hkdIs617 were generated, respectively. As shown in Figures S2A and S2B, no red fluorescent protein was detected; however, blue and green channels showed the normal expression of CRE and FLP, respectively. This result illustrates that each of the two stops was able to prevent the expression of the downstream reporter gene.
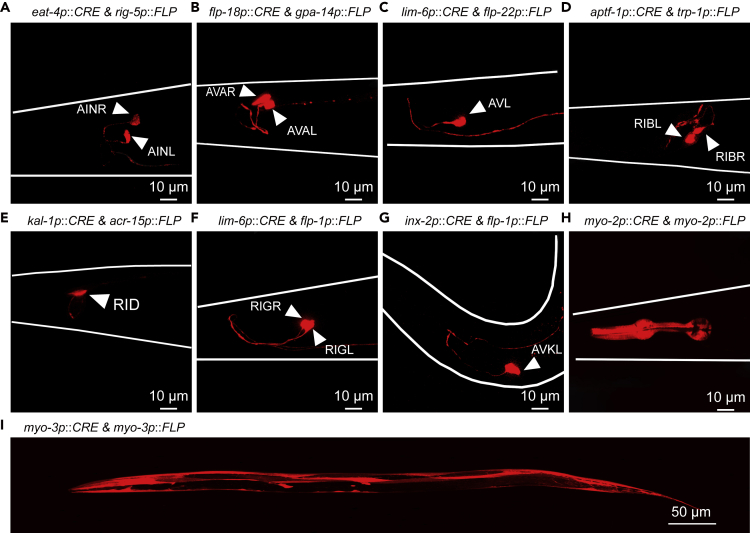
Next, we used the integrated strain ZXW617 to identify various promoter pairs for intersectional targeting of single-neuron type. We successfully identified eight promoter pairs with a single neuron-type intersection. Shown in Figures 2A–2G and Table S1, the following promoter pairs enabled TagRFP-T expression in single types of neuron(s): eat-4p & rig-5p in AINs (paired L/R), flp-18p & gpa-14p in AVAs (paired L/R), lim-6p & flp-22p in AVL (single), aptf-1p & trp-1p in RIBs (paired L/R), kal-1p & acr-15p in RID (single), lim-6p & flp-1p in RIGs (paired L/R), and inx-2p & flp-1p in AVKs (paired L/R). An FLP-out approach, which used single-copy transgenic strains that stably express an optimized version of FLP in specific tissues or by heat induction, reached 100% recombination efficiencies in several cell types such as muscles, intestine, and serotonin-producing neurons (Munoz-Jimenez et al., 2017). Compared with this system, the FLP/CRE-out system offers efficiencies of 100% in muscle cells and 53%–86% in neurons. The differential efficiencies in neurons may be caused by the varied expression of the recombinases. The ZXW617 strain may be an array of TagRFP-T. In the array, both local excision and excision between different integrated plasmids should be possible. Thus, the array could be reduced to just a single plasmid over time, since every recombination event leaves behind a LoxP or FRT site that can recombine again. After some observations, we observed no obvious variability of TagRFP-T fluorescence in different worms, no gradual dimming of the fluorescence over time, and even brighter fluorescence in older animals.
Figure 2.
Identifying Promoter Pairs of Spatial Targeting Using Integrated ZXW617 Worms
(A–G) Confocal fluorescence images of TagRFP-T in AIN (A, paired L/R), AVA (B, paired L/R), AVL (C, single), RIB (D, paired L/R), RID (E, single), RIG (F, paired L/R), and AVK (G, paired L/R) neurons, respectively. Neuron-specific TagRFP-T expression was controlled by the FLP/CRE-out system with the indicated vectors of CRE and FLP. "Paired L/R″ and "single" indicate the type of neuron(s). Scale bar, 10 μm.
(H and I) Confocal fluorescence images of TagRFP-T in pharyngeal muscle cells (H) and body wall musculature (I). Scale bars, 10 μm (H) and 50 μm (I).
Since the core plasmid used a strong pan-expression eft-3 promoter to drive target gene expression, the FLP/CRE-out system should be able to control specific gene expression in cells/tissues other than neurons. We thus set out to induce TagRFP-T expression in pharyngeal muscle and body wall musculature. As shown in Figures 2H and 2I, two types of muscle cells showed specific TagRFP-T labeling. In summary, a tripartite cell-specific gene expression system based on recombining-out two different stops was developed. We also have identified eight promoter pairs with intersectional targeting of a single-type neuron(s).
The Highly Refined Spatial and Temporal Control System of Gene Expression Based on FLP/CRE-out
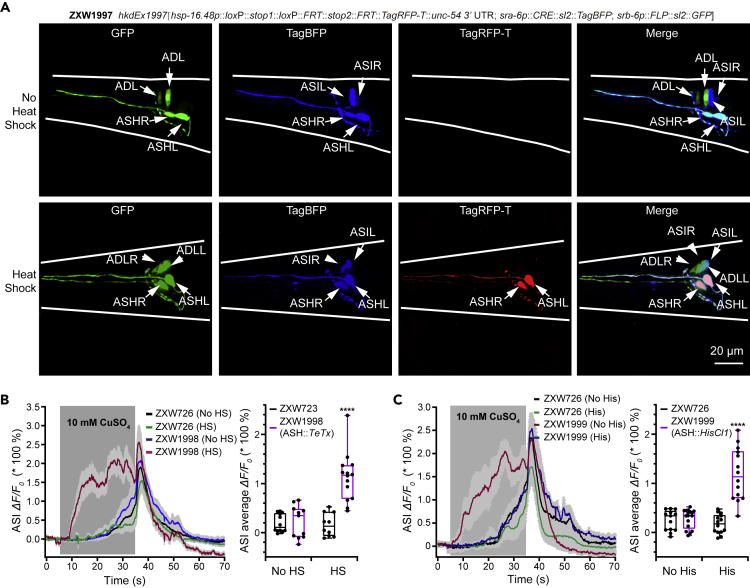
The FLP/CRE-out system is reliable for inducing cell type-specific gene expression. Replacement of eft-3 promoter in the core vector with an inducible element or promoter should empower the trinary system to control both spatial and temporal gene expression. Heat shock promoters were widely used to induce temporal gene expression in C. elegans (Bacaj and Shaham, 2007; Churgin et al., 2013, 2014; Davis et al., 2008; Hubbard, 2014; Jones et al., 1989; Monsalve et al., 2019; Munoz-Jimenez et al., 2017; Shim et al., 2003; Singhal and Shaham, 2017; Stringham et al., 1992; Voutev and Hubbard, 2008; Wang et al., 2018). So, we replaced eft-3p with hsp16.48p in the "A" entry clone to construct an hsp-16.48p::d-stops::TagRFP-T vector (Figure S3B). To examine the inducibility and neuron-specificity of TagRFP-T expression, we tested gene induction in ASH neurons. We generated a transgenic strain ZXW1997 of genotype hkdEx1997[hsp-16.48p::loxP::stop1::loxP::FRT::stop2::FRT::TagRFP-T::unc-54 3′ UTR; sra-6p::CRE::sl2::TagBFP; srb-6p::FLP::sl2::GFP]. As expected, no obvious red fluorescence in ASH neurons was detected in heat shock-untreated worms. In contrast, bright red fluorescence in ASHs was observed in heat shock (2 h heat shock at 34°C and 12 h post-treatment culture under standard 20°C) treated worms (Figure 3A).
Figure 3.
Inducible and Cell Type-Specific Expression of Target Genes Controlled by FLP/CRE-Out System and Uses in Genetic and Chemogenetic Manipulation of Single-Type Neurons
(A) Confocal fluorescence images of GFP, TagBFP, and TagRFP-T before and after the treatment of heat shock; 12 h after the 2-h heat shock treatment, a single pair of ASH neurons were specifically labeled by red fluorescence. The expression patterns of sra-6p and srb-6p were ASHs/ASIs and ASHs/ADLs, respectively. Scale bar, 20 μm.
(B) Changes in Ca2+ transients (in curves in the left panel, and box-and-whisker in the right panel) in response to CuSO4 solution (10 mM) in ASI neurons in worms of the indicated genotypes. No HS, without heat shock treatment; HS, with heat shock treatment. The genotypes of ZXW726 and ZXW1998 are hkdEx726[gpa-4p::R-GECO1; lin-44p::GFP] and hkdEx1998[hsp16.48p::loxP::stop1::loxP::FRT::stop2::FRT::TeTx; sra-6p::CRE::sl2::TagBFP; srb-6p::FLP::sl2::GFP; gpa-4p::R-GECO1], respectively.
(C) Changes in the Ca2+ signals (in curves in the left panel, and box-and-whisker in the right panel) in ASI neurons in response to CuSO4 solution (10 mM) in worms of the indicated genotypes. Histamine of 10 mM was used to activate the HisCl1 channel. The genotype of ZXW1999 is hkdEx1999[eft-3p::loxP::stop1::loxP::FRT::stop2::FRT::HisCl1; sra-6p::CRE::sl2::TagBFP; srb-6p::FLP::sl2::GFP; gpa-4p::R-GECO1]. No His, without histamine treatment; His, with histamine treatment. Data of Ca2+ signals were expressed as the means ± SEM as indicated by solid traces ± gray shading and box plots with each dot representing those in each individual tested worm. The significance of statistical difference was analyzed by two-way ANOVA analysis with post hoc test of Tukey's multiple comparison correction and indicated as ∗∗∗∗p < 0.0001.
Spatial and temporal manipulation of neurons is essential for neuroscience studies. Interruption of neurotransmitter release in a tested neuron(s) with the light chain of tetanus toxin (TeTx) and optogenetic and chemogenetic manipulation is commonly used (Boyden et al., 2005; Guo et al., 2015; Kim et al., 2017; Luo et al., 2008, 2018; Macosko et al., 2009; Pokala et al., 2014; Wang et al., 2016; Yizhar et al., 2011). One primary goal of our study is to develop a toolbox for spatiotemporal manipulation of neurons. TeTx, a specific synaptobrevin protease that blocks vesicle fusion with the plasma membrane and thus neurotransmitter release (Schiavo et al., 1992), has been successfully used to inhibit chemical synaptic transmission of tested neurons in C. elegans (Guo et al., 2015; Liu et al., 2019; Macosko et al., 2009; Wang et al., 2016). Continual inhibition of neurotransmission by TeTx, beginning from embryonic periods, may interfere with the development of the nervous system. Therefore, the inducible expression of the toxin might be an appropriate solution.
In C. elegans nociception and avoidance of copper ion, ASIs and ASHs reciprocally inhibit. The inhibition of ASHs augments the activity in ASIs (Guo et al., 2015). Here we used this neuronal interaction to probe further spatiotemporal gene induction of the FLP/CRE-out system. We blocked ASH neurotransmission with inducible expression of TeTx, chemogenetically inhibited ASHs, and examined changes in ASI Ca2+ transients evoked by Cu2+. We generated ZXW1998 transgenic strain of genotype hkdEx1998[hsp-16.48p::d-stops::TeTx; sra-6p::CRE::sl2::TagBFP; srb-6p::FLP::sl2::GFP; gpa-4p::R-GECO1]. Expectedly, heat-shock treatment (34°C for 2 h) of the transgenic worms caused a significant increase in ASI Ca2+ signals evoked by 10 mM CuSO4. In contrast, the heat shock-untreated ZXW1998 and heat shock-treated hkdEx726[gpa-4p::R-GECO1; lin-44p::GFP] worms (Figure 3B) did not display an obvious change in Cu2+-elicited Ca2+ signals in ASIs. This result indicated a successful temporal induction of TeTx in ASHs.
The Drosophila histamine-gated chloride channel subunit 1 (HisCl1) and administration of exogenous histamine are used to inhibit neurons acutely in C. elegans (Guo et al., 2015; Pokala et al., 2014). Next, we chemogenetically inhibited ASHs and examined ASI Ca2+-responses to the Cu2+ treatment. We generated ZXW1999 transgenic strain of genotype hkdEx1999[eft-3p::d-stops::HisCl1; sra-6p::CRE::sl2::TagBFP; srb-6p::FLP::sl2::GFP; gpa-4p::R-GECO1]. As shown in Figure 3C, the administration of 10 mM exogenous histamine for 10 min activates HisCl1 channel and thus acutely inactivates ASHs, resulting in a significant increase in Cu2+-evoked Ca2+ signals in ASIs. In comparison, the HisCl1 expression (in histamine-untreated ZXW1999) or the treatment of histamine (in ZXW726) alone did not significantly change the Cu2+-evoked Ca2+ transients in ASIs. In summary, the above experiments corroborate the feasibility and efficiency of the tripartite FLP/CRE-out system of spatiotemporal gene expression.
Discussion
In the present study, we develop a tripartite FLP/CRE-out gene induction method/system for spatiotemporal control of transgene expression in single/single type of neuron(s) and other cells or tissues in C. elegans. In addition, the system provides an "AND" gate strategy for the spatial expression of a gene. The advantages of the FLP/CRE-out method are as follows. (1) Ability to spatiotemporally control gene expression in the neuron(s) or other cells that lack a specific promoter. Especially this system is useful for optogenetic and chemogenetic manipulation of single types of neuron(s). (2) Simple and cost saving. Changes of promoter pairs, which direct CRE and FLP expression, are needed only for expressing a target gene in varied types of neurons and other cells. Meanwhile, the change of a target gene in the core vector is needed to control the spatiotemporal expression of different genes in defined cell types. Replacing promoters and genes in the FLP/CRE-out system is easy by the use of the MultiSite gateway method. All plasmids and worms used in this study are available to the scientific community. The integrated worm strain ZXW617 of genotype hkdIs617[eft-3p::d-stops::TagRFP-T::unc-54 3′ UTR::lin-44p::GFP] is a useful tool to identify new promoter pairs for spatial control of gene expression in a single type of neuron(s). Using this worm strain, we identified eight promoter pairs for controlling spatial gene expression in the neuron(s) lacking a known specific promoter or promoter pair. Our work lays an important foundation for studies in these neurons. Importantly, ∼6,000 promoters in the C. elegans promoterome (Dupuy et al., 2004) are available for constructing recombinase vectors by the three fragment gateway method. Moreover, single-copy transgenic strains that stably express an optimized version of FLP in specific tissues or by heat induction (Munoz-Jimenez et al., 2017) are useful for the FLP/CRE-out system. (3) The strong pan-expression eft-3p and hsp-16.48p promoters in the core gene expression plasmid of the FLP/CRE-out system lay a foundation for the use of the system in cells and tissues other than neurons in C. elegans. (4) In some bipartite systems, the effector transgene is placed downstream of a tissue-specific promoter; therefore, it can only be expressed as long as the promoter is active. Compared with these bipartite induction systems, the use of eft-3 promoter in the FLP/CRE-out system offers the possibility of prolonged and continuous expression of the effector transgene.
Multiple methods have been developed for controlling spatial or spatiotemporal gene expression in C. elegans. Bipartite systems, such as FLP-out and CRE-out systems, are not able to induce spatial and temporal gene expression in a single type of neuron(s) and other cells lacking a specific promoter (Davis et al., 2008; Guo et al., 2015; Hubbard, 2014; Lopez-Cruz et al., 2019; Macosko et al., 2009; Schmitt et al., 2012; Voutev and Hubbard, 2008; Wang et al., 2017), HSF-1 system (Bacaj and Shaham, 2007), GAL4-UAS system (Wang et al., 2017), QF-GR system (Monsalve et al., 2019). Moreover, the tripartite Q system and split Q system (Wei et al., 2012) empower to control spatiotemporal gene expression. However, this system lacked tight regulation. QS and QF have to be expressed constitutively, which require a minimum of 6 h for quinic acid to alleviate QS repression and allow transgene induction, whereas complete restoration of transgene expression takes 24 h (Monsalve et al., 2019; Nance and Frokjaer-Jensen, 2019). Tripartite split cGAL confers spatiotemporal control of transgene expression but is likely to introduce a temporal delay, compared with a direct heat shock promoter::gene fusion (Wang et al., 2018). The dynamic range of the tetracycline-dependent ribozyme switch system (with maximal gene 3.75-fold ± 0.85-fold induction) is weaker compared with the classical use of heat shock promoters (Wurmthaler et al., 2019). To temporally and spatially control gene expression and protein activity/localization are essential in modern biology and even modern medicine. Given the advantages of the FLP/CRE-out-mediated gene control system, this system expands the genetic toolbox for C. elegans.
Limitations of the Study
All of the transgenic strains except ZXW617 were not integrated. The ZXW617 strain was generated by a standard ultraviolet/trimethylpsoralen (UV/TMP) integration method, not by single-copy insertion. The strain may be an array of TagRFP-T.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Zheng-Xing Wu (ibbwuzx@mail.hust.edu.cn).
Materials Availability
All data generated or analyzed in this study are included in this published article (and its Supplemental Information files). The strains and plasmids will be deposited at CGC (https://cgc.umn.edu/) and Addgene (https://www.addgene.org/), respectively.
Data and Code Availability
The published article includes all data generated or analyzed during this study.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Caenorhabditis Genetic Center (CGC) and National BioResource Project (NBRP) for the worm strains used in this study, Dr. B.F. Liu for the support in the fabrication of microfluidic devices, Dr. C. Bargmann for the plasmids contenting of HisCl1 cDNA, and Dr. Peter Simms for revising the manuscript. This work was supported by grants from the National Science Foundation of China (31471034) and Fundamental Research Funds for the Central Universities (2016YXZD062).
Author Contributions
Z.-X.W. supervised the project. M.-H.G., T.-H.W., and W.W. performed the majority of the plasmid construction and imaging experiments. Z.-X.W., M.-H.G., and U.A.-S. wrote the manuscript with help from all of the other authors.
Declaration of Interests
The authors declare no competing interests.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101567.
Supplemental Information
References
- Bacaj T., Shaham S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics. 2007;176:2651–2655. doi: 10.1534/genetics.107.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E.S., Zhang F., Bamberg E., Nagel G., Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Buchholz F., Angrand P.O., Stewart A.F. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- Chelur D.S., Chalfie M. Targeted cell killing by reconstituted caspases. Proc. Natl. Acad. Sci. USA. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churgin M.A., He L., Murray J.I., Fang-Yen C. Efficient single-cell transgene induction in Caenorhabditis elegans using a pulsed infrared laser. G3 (Bethesda) 2013;3:1827–1832. doi: 10.1534/g3.113.007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churgin M.A., He L., Murray J.I., Fang-Yen C. Construction of a system for single-cell transgene induction in Caenorhabditis elegans using a pulsed infrared laser. Methods. 2014;68:431–436. doi: 10.1016/j.ymeth.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.W., Morton J.J., Carroll D., Jorgensen E.M. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy D., Li Q.R., Deplancke B., Boxem M., Hao T., Lamesch P., Sequerra R., Bosak S., Doucette-Stamm L., Hope I.A. A first version of the Caenorhabditis elegans promoterome. Genome Res. 2004;14:2169–2175. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki S.M. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc. Natl. Acad. Sci. U S A. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki S.M., Kim J.C. Molecular neuroanatomy's "Three Gs": a primer. Neuron. 2007;54:17–34. doi: 10.1016/j.neuron.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic K.G., Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Guo M., Wu T.H., Song Y.X., Ge M.H., Su C.M., Niu W.P., Li L.L., Xu Z.J., Ge C.L., Al-Mhanawi M.T.H. Reciprocal inhibition between sensory ASH and ASI neurons modulates nociception and avoidance in Caenorhabditis elegans. Nat. Commun. 2015;6:e5655. doi: 10.1038/ncomms6655. [DOI] [PubMed] [Google Scholar]
- He S., Cuentas-Condori A., Miller D.M., 3rd NATF (Native and Tissue-Specific Fluorescence): a strategy for bright, tissue-specific GFP labeling of native proteins in Caenorhabditis elegans. Genetics. 2019;212:387–395. doi: 10.1534/genetics.119.302063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R., Abremski K., Sternberg N. The nature of the interaction of the P1 recombinase Cre with the recombining site loxP. Cold Spring Harb. Symp. Quant. Biol. 1984;49:761–768. doi: 10.1101/sqb.1984.049.01.086. [DOI] [PubMed] [Google Scholar]
- Hubbard E.J. FLP/FRT and Cre/lox recombination technology in C. elegans. Methods. 2014;68:417–424. doi: 10.1016/j.ymeth.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D., Dixon D.K., Graham R.W., Candido E.P. Differential regulation of closely related members of the hsp16 gene family in Caenorhabditis elegans. DNA. 1989;8:481–490. doi: 10.1089/dna.1.1989.8.481. [DOI] [PubMed] [Google Scholar]
- Kim C.K., Adhikari A., Deisseroth K. Integration of optogenetics with complementary methodologies in systems neuroscience. Nat. Rev. Neurosci. 2017;18:222–235. doi: 10.1038/nrn.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Qin L.W., Li R., Zhang C., Al-Sheikh U., Wu Z.X. Reciprocal modulation of 5-HT and octopamine regulates pumping via feedforward and feedback circuits in C. elegans. Proc. Natl. Acad. Sci. U S A. 2019;116:7107–7112. doi: 10.1073/pnas.1819261116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cruz A., Sordillo A., Pokala N., Liu Q., McGrath P.T., Bargmann C.I. Parallel multimodal circuits control an innate foraging behavior. Neuron. 2019;102:407–419.e408. doi: 10.1016/j.neuron.2019.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Callaway E.M., Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Callaway E.M., Svoboda K. Genetic dissection of neural circuits: a decade of progress. Neuron. 2018;98:256–281. doi: 10.1016/j.neuron.2018.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Pokala N., Feinberg E.H., Chalasani S.H., Butcher R.A., Clardy J., Bargmann C.I. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E., Bartling L., Hake S. From Gateway to Multisite Gateway in one recombination event. BMC Mol. Biol. 2006;7:46. doi: 10.1186/1471-2199-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve G.C., Yamamoto K.R., Ward J.D. A new tool for inducible gene expression in Caenorhabditis elegans. Genetics. 2019;211:419–430. doi: 10.1534/genetics.118.301705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jimenez C., Ayuso C., Dobrzynska A., Torres-Mendez A., Ruiz P.C., Askjaer P. An efficient FLP-based toolkit for spatiotemporal control of gene expression in Caenorhabditis elegans. Genetics. 2017;206:1763–1778. doi: 10.1534/genetics.117.201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., Frokjaer-Jensen C. The Caenorhabditis elegans transgenic toolbox. Genetics. 2019;212:959–990. doi: 10.1534/genetics.119.301506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokala N., Liu Q., Gordus A., Bargmann C.I. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc. Natl. Acad. Sci. U S A. 2014;111:2770–2775. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C.S., Soriano P. High-efficiency FLP and ΦC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S., van den Heuvel S. G1/S Inhibitors and the SWI/SNF complex control cell-cycle exit during muscle differentiation. Cell. 2015;162:300–313. doi: 10.1016/j.cell.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Sauer B., Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U S A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G., Benfenati F., Poulain B., Rossetto O., Polverino de Laureto P., DasGupta B.R., Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schmitt C., Schultheis C., Pokala N., Husson S.J., Liewald J.F., Bargmann C.I., Gottschalk A. Specific expression of channelrhodopsin-2 in single neurons of Caenorhabditis elegans. PLoS One. 2012;7:e43164. doi: 10.1371/journal.pone.0043164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J., Im S.H., Lee J. Tissue-specific expression, heat inducibility, and biological roles of two hsp16 genes in Caenorhabditis elegans. FEBS Lett. 2003;537:139–145. doi: 10.1016/s0014-5793(03)00111-x. [DOI] [PubMed] [Google Scholar]
- Singhal A., Shaham S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat. Commun. 2017;8:14100. doi: 10.1038/ncomms14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham E.G., Dixon D.K., Jones D., Candido E.P. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R., Hubbard E.J. A "FLP-Out" system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu J., Gharib S., Chai C.M., Schwarz E.M., Pokala N., Sternberg P.W. cGAL, a temperature-robust GAL4-UAS system for Caenorhabditis elegans. Nat. Methods. 2017;14:145–148. doi: 10.1038/nmeth.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu J., Yuet K.P., Hill A.J., Sternberg P.W. Split cGAL, an intersectional strategy using a split intein for refined spatiotemporal transgene control in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 2018;115:3900–3905. doi: 10.1073/pnas.1720063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Qin L.W., Wu T.H., Ge C.L., Wu Y.Q., Zhang Q., Song Y.X., Chen Y.H., Ge M.H., Wu J.J. cGMP signalling mediates water sensation (hydrosensation) and hydrotaxis in Caenorhabditis elegans. Sci. Rep. 2016;6:19779. doi: 10.1038/srep19779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Potter C.J., Luo L., Shen K. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nat. Methods. 2012;9:391–395. doi: 10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmthaler L.A., Sack M., Gense K., Hartig J.S., Gamerdinger M. A tetracycline-dependent ribozyme switch allows conditional induction of gene expression in Caenorhabditis elegans. Nat. Commun. 2019;10:491. doi: 10.1038/s41467-019-08412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M.D., Edgar L.G., Wood W.B. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 1994;91:1381–1385. doi: 10.1073/pnas.91.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O., Fenno L.E., Davidson T.J., Mogri M., Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang S., Ma C., Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study.