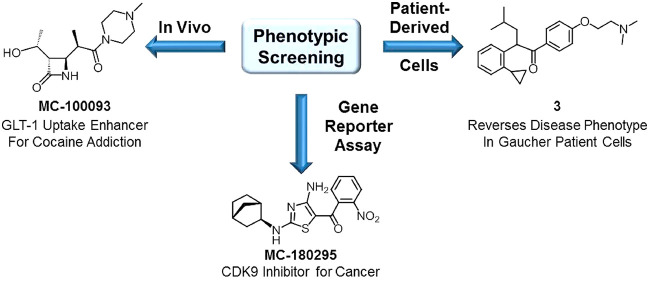
Abstract
Prior to genetic mapping, the majority of drug discovery efforts involved phenotypic screening, wherein compounds were screened in either in vitro or in vivo models thought to mimic the disease state of interest. While never completely abandoning phenotypic approaches, the labor intensive nature of such tests encouraged the pharmaceutical industry to move away from them in favor of target-based drug discovery, which facilitated throughput and allowed for the efficient screening of large numbers of compounds. However, a consequence of reliance on target-based screening was an increased number of failures in clinical trials due to poor correlation between novel mechanistic targets and the actual disease state. As a result, the field has seen a recent resurrection in phenotypic drug discovery approaches. In this work, we highlight some recent phenotypic projects from our industrial past and in our current academic drug discovery environment that have provided encouraging results.
Keywords: CDK9, Cell-based assays, Drug discovery, GLT-1, Gaucher disease, Phenotypic
Ever since Paul Ehrlich’s side chain theory heralded the birth of the field of medicinal chemistry in 1901,1,2 drug discovery has followed two general strategies: phenotypic and target-based. Prior to the advancements in genomics and molecular biology realized in the last two decades of the 20th century, almost all drug discovery involved testing compounds against some living system to determine their effects on a phenotype thought to be associated with the disease state of interest. But such efforts were relatively low throughput and labor intensive, and in the 1980s, the pharmaceutical industry began to abandon phenotypic screening in favor of a more efficient, higher throughput method, namely, the use of biochemical molecular target-based screening. Target-based screening became even more appealing at the end of the 20th century when advances made it possible to screen compounds in miniature assays run in multiwell plates, with the laborious tasks performed by robots. Such high-throughput screening methodology now made it possible to obtain pharmacological data on thousands of compounds in a relatively short period of time. The advantages of target-based screening included simplicity, lower cost compared to phenotypic screening, and the ease with which structure–activity relationship (SAR) information could be generated,3 and Pharma embraced this efficiency.
By 2005, the results of the Human Genome Project (HGP) suggested that there could be over 3000 druggable proteins, only a small fraction of which had been exploited by drug discovery efforts.4,5 This realization sparked drug discovery research on a multitude of novel mechanistic targets, the majority of which was driven by target-based approaches. Yet a decade later, it had become clear that relatively few novel therapies had been identified as a result of the HGP and the target-based discovery approaches derived from the information that the HGP had provided.6,7 Two influential papers by Swinney and Anthony in 2011 and Swinney in 2013 provided clear analyses that showed that the majority of first-in-class drugs identified from 1999 to 2008 had come from phenotypic drug discovery approaches.8,9
That being said, the pharmaceutical industry never completely abandoned phenotypic drug discovery. Much of the successful antibiotic drug discovery to date started from a phenotypic approach such as inhibition of bacterial growth in culture or functional inhibition of cell wall synthesis (without knowing the actual mechanistic target). Some oncology research has involved screening compounds phenotypically (e.g., cytostatic/cytotoxic effects on cancer cells in culture),10 as has CNS drug discovery, which often relies on in vivo behavioral assays11 to the point that efforts have been made to build upon early strategies involving fruit flies and develop high throughput platforms for phenotypic in vivo CNS drug discovery using zebrafish and planarians.12 However, the application of new technologies such as artificial intelligence, inducible stem cells, patient-derived cells, and tissue modeling has changed perception and rekindled interest in phenotypic approaches.11,13−16 The complexity that was once thought to be a disadvantage to drug discovery is becoming a stimulus for tackling challenging therapeutic targets of high unmet medical need.
Wyeth Research (the company that the authors worked for prior to its acquisition by Pfizer in 2009) developed and marketed a number of drugs that were discovered using phenotypic approaches. Perhaps the earliest example of this comes from its over-the-counter portfolio. While working at the Institutum Divi Thomae,17 George Sperti developed an ointment (later called Sperti ointment) that promoted wound healing and healthy cell growth, although the actual efficacy of the ointment has been debated for several decades.18,19 The primary ingredients of the ointment were live yeast cell derivative (LYCD, later referred to as “biodynes”) and shark liver oil. Rights to the ointment were acquired in 1935 by American Home Products20 (which later renamed itself to become Wyeth Research), and the ointment was repositioned for the treatment of hemorrhoids under the now famous name “Preparation H”. To this day, the exact mechanism(s) of action for LYCD is poorly understood at best,21 and the live yeast component of Preparation H has since been replaced with drugs of known mechanism (e.g., phenylephrine), but the ointment serves to remind researchers that there was a time when all drug discovery was done phenotypically.
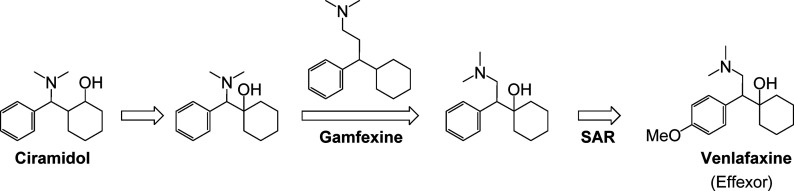
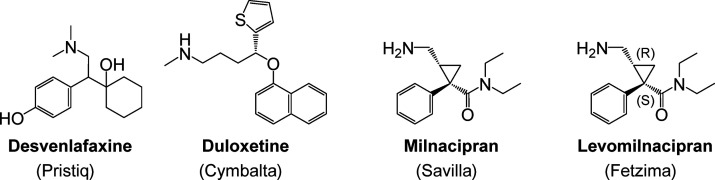
A second example is the block buster antidepressant drug venlafaxine (Effexor). Venlafaxine was identified as an antidepressant via screening in three in vivo animal models of depression.22 The structural origin of venlafaxine came out of a desire to eliminate the chiral complexity of ciramidol, an opiate-based analgesic agent identified several years earlier (Figure 1).23 Structural modifications that eliminated two of the three chiral centers of ciramidol resulted in a chemical scaffold that resembled that of gamfexine,24 a compound known to have antidepressant activity. Venlafaxine’s antidepressant activity was identified through in vivo screening. Its mechanisms of action (inhibition of serotonin and norepinephrine reuptake) were defined retrospectively after the antidepressant activity had been discovered. Venlafaxine (FDA approval date 199325) was a first-in-class serotonin–norepinephrine reuptake inhibitor (SNRI) that was developed and marketed as the racemic mixture. Its favorable antidepressant profile in a broad range of patients (including patients refractory to other antidepressant agents) prompted the later discovery of other SNRIs (Figure 2) such as desvenlafaxine (Pristiq, FDA approval date 200826), duloxetine (Cymbalta, FDA approval date 200427), milnacipran (Savilla, FDA approval date 200928), and levomilnacipran (Fetzima, FDA approval date 201329).
Figure 1.
Structural evolution of venlafaxine (Effexor).
Figure 2.
Marketed serotonin–norepinephrine reuptake inhibitor antidepressants that followed venlafaxine.
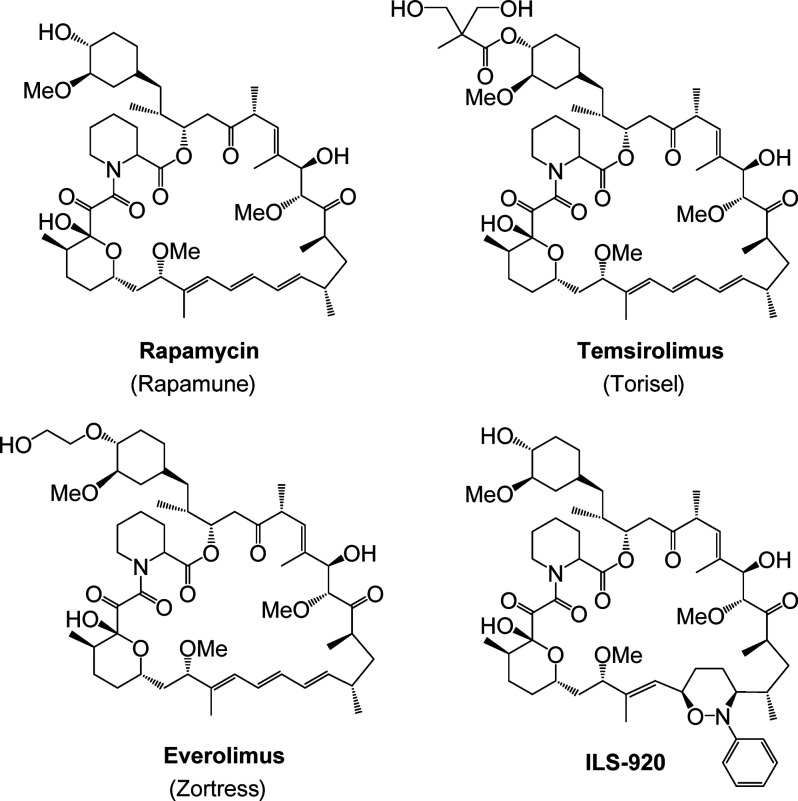
A prime example from Wyeth’s drug development history of a productive phenotypic screening campaign is their macrocyclic immunophilins (Figure 3) and the first-in-class macrolide immunosuppressant agent rapamycin (sirolimus, Rapamune). Rapamycin was discovered in soil samples taken from Easter Island (Rapa Nui) as a product produced by the bacteria Streptomyces hygroscopicus.30 Its original antifungal activity was identified through phenotypic screening against samples of fungus in culture. Rapamycin’s development as an antifungal drug was later abandoned in favor of its potent immunosuppressant activity, again discovered phenotypically in vivo.31 It was not until 199132 that the actual mechanistic target for the immunosuppressant activity of rapamycin was discovered, which turned out to be a pair of previously uncharacterized proteins dubbed mTORs (mammalian targets of rapamycin). This discovery was cemented when the crystal structure of the ternary complex of rapamycin, mTOR (referred to as FRAP at the time), and FKBP12 was solved and published in 1996 (Figure 4).33 However, the impact of phenotypic screening within this chemical scaffold did not stop there. Results from the National Cancer Institute’s NCI60 cancer panel had demonstrated that rapamycin demonstrated cytostatic activity in a number of tumor cell lines.34 While rapamycin was never developed as an anticancer drug, that phenotypic discovery ultimately led to the development and marketing of other structurally similar macrolides (Figure 3), such as Wyeth’s temsirolimus (Torisel) and Novartis’ everolimus (Zortress, Afinitor). And a campaign to eliminate the immunosuppressive activity (assessed phenotypically on CD4+ T cells in culture) led to the discovery of ILS-920, a neuroprotectant and neuroregenerative agent identified through screening in rat cortical neurons.35 The mechanistic targets for these activities were later discovered to be FKBP52 and L-type calcium channels.35
Figure 3.
Structures of macrocyclic immunophilins.
Figure 4.
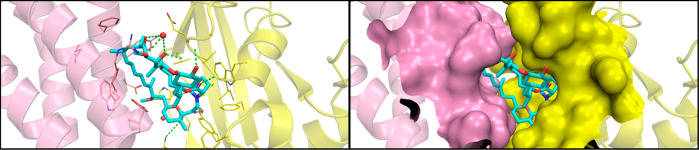
Model of the tertiary complex between FKBP12, mTOR, and rapamycin. The complex structure (rapamycin and mTOR taken from PDB 4DRJ; FKBP12 taken from PDB 1C9H) was prepared with Protein Preparation Wizard (Schrodinger, New York, NY) to add missing hydrogen atoms, side chains, and loops, correct bond orders, and remove water molecules beyond 5 Å of the ligand.
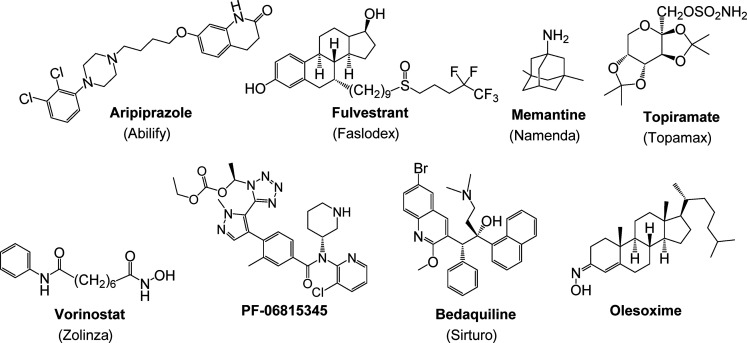
Wyeth, of course, was not the only company who continued to embrace phenotypic drug discovery approaches. Notable examples (Figure 5) cited by the 2011 Swinney and Anthony paper8 include Otsuka/BristolMyers Squibb’s aripiprazole (Abilify), AstraZeneca’s fulvestrant (Faslodex), Forrest Laboratories’ memantine (Namenda), and Merck’s vorinostat (Zolinza). Following the realization made clear by the Swinney papers8,9 and others that followed,10,16,36−38 there has been something of a resurrection of phenotypic drug discovery in the industrial setting. A resurgence in interest in phenotypic approaches is being seen in the area of neurodegenerative disease drug discovery following decades of disappointing clinical results with drugs derived from target-based approaches for Alzheimer’s disease.39 And there have been success stories for projects pursued through phenotypic screening. For example, the beneficial impact of proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitory antibodies on treating hypercholesterolemia spurred significant interest in small molecule inhibitors.40 Despite the availability of X-ray crystal structures of both PCSK9 itself and the interaction of PCSK9 with the low density lipoprotein receptor (LDLR), the one small molecule to advance to Phase II clinical trials to date is Pfizer’s PF-06815345 (Figure 5),41 which was derived from optimization of R-IMPP, a small molecule antisecretagogue that was found to inhibit the translation of PCSK9 protein via a high-throughput phenotypic screen.42 Phenotypic screening of various chemical scaffolds resulted in the identification of Sirturo (bedaquiline), the first new antituberculosis drug approved in over 40 years (Figure 5).43 Examination of a compound originally prepared as a synthetic intermediate in a series of in vivo epilepsy models ultimately led to the discovery of topiramate (Topamax, Figure 5), an antiepileptic agent and neurostabilizer that was later found to possess a complex collection of antiseizure and antimigrane mechanisms.16,44 However, despite the potential for greater translational accuracy to the disease state, phenotypic drug discovery is not without its challenges and disappointments, as discussed in a recent perspective by Haasen et al.45 For example, Trophos’s olesoxime (TRO19622, Figure 5) was discovered via a phenotypic screen measuring neuronal survival. The company and compound were acquired by Roche in 2015. However, clinical trials for the compound in Alzheimer’s disease and spinal muscular atrophy have produced disappointing results to date.46
Figure 5.
Marketed drugs and advanced clinical candidates discovered through phenotypic drug discovery.
We and others have published numerous comparative perspectives on drug discovery in the academic versus the industrial setting.47−52 One aspect of academic drug discovery that some perceive as an advantage is the ability to more readily embrace higher risk projects and approaches (as long as funding can be identified to support them). One of the higher risk approaches that some academic drug discovery groups have incorporated into their research is phenotypic screening, especially for diseases that have few or no options for treatment.53 Since establishing the Moulder Center for Drug Discovery Research within Temple University School of Pharmacy in 2009, the research team has incorporated phenotypic screening approaches into its drug discovery strategy in order to pursue new drugs for diseases that have little or no options for treatment. Below we describe three phenotypic screening-based projects that have resulted in intriguing results that are currently being positioned for possible development.
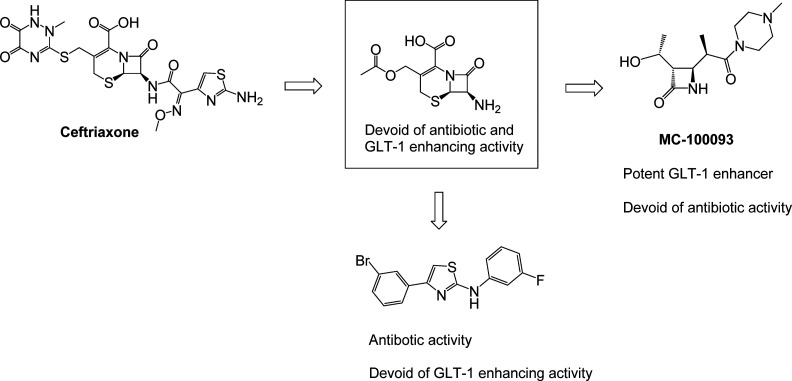
Addiction to cocaine remains an unmet medical need for which there is no FDA-approved treatment. While many cocaine users are able to sustain periods of abstinence from cocaine use, the majority of them eventually relapse when exposed to cocaine-usage associated cues, stress, or using the drug again.54−56 In 2005, Rothstein and co-workers used a phenotypic screen to examine a library of 1040 FDA-approved drugs and nutritional supplements to identify compounds that could enhance the expression of the glutamate transporter GLT-1 (EAAT2) as part of program looking for neuroprotective agents.57 Active hits included a number of β-lactam antibiotics, the most potent of which was ceftriaxone (Figure 6). It was later discovered that ceftriaxone, by virtue of its ability to enhance GLT-1 expression, was able to attenuate the reinstatement of cocaine seeking that is primed by either cues or cocaine administration.58,59 However, there are limitations associated with using ceftriaxone subchronically as a drug for preventing cocaine relapse, including its lack of oral bioavailability, its poor CNS penetration (1–2%),60 and the risk for developing bacterial resistance. To overcome these issues we embarked on a phenotypically driven SAR campaign to identify β-lactam analogs that retained the ability to enhance expression of GLT-1 while eliminating antibiotic activity and increasing oral and CNS bioavailability (Figure 6). Removing the side chains of ceftriaxone eliminated both antibiotic activity and GLT-1 enhancing effects. Eliminating the carboxylic acid group and the 6-membered ring (while retaining the β-lactam moiety) led to our lead molecule MC-100093. These structural modifications resulted in a molecule that possesses oral and CNS bioavailability and enhanced GLT-1 expression potency compared to ceftriaxone. The detailed profile of MC-100093 will be presented in a forthcoming publication (Knackstedt, Rothstein, Abou-Gharbia et al., submitted). While the exact mechanistic target of β-lactam-based GLT-1 enhancers like ceftriaxone and MC-100093 has yet to be identified, some insight is available from studies using ceftriaxone. Ceftriaxone has been shown to stimulate GLT-1 mRNA and protein expression in several cellular and animal models.61,62 At least part of this effect is thought to occur via the interaction of NF-κB with the GLT-1 promoter region.63 However, other biochemical pathways such as Akt,64 GPR30,65 and perhaps even brain region-specific epigenetic mechanisms66 may also be involved in the GLT-1 enhancing effects of ceftriaxone. Detailed safety studies have not been performed with MC-100093, but ceftriaxone was shown to be safe when given at high doses in Phase III clinical trials examining its efficacy in Amyotropic Lateral Sclerosis.67 In addition to its efficacy in cocaine reinstatement, preliminary studies have also demonstrated that MC-100093 reverses neurodegenerative phenotype in an animal model of cerebral palsy.68,69 Interestingly, we were also able to retain some of the antibiotic activity while eliminating the GLT-1 enhancing effects of the ceftriaxone scaffold in the absence of the β-lactam group by manipulating the thiazole substituent.70
Figure 6.
SAR studies driven by phenotypic screening identified MC-100093, an orally bioavailable β-lactam-based glutamate uptake enhancer with no antibiotic activity.
The use of patient derived cells in phenotypic assays has great potential to discover new, breakthrough therapeutics and offers an alternative to target based strategies, which have failed to deliver effective treatments.71,72 We recently employed a high throughput phenotypic screen to identify a new series of small molecules that reverse disease phenotype in Gaucher patient-derived fibroblasts.73 Gaucher disease is an autosomal recessive disorder that results from mutations in the GBA1 gene encoding the lysosomal enzyme β-glucocerebrosidase (Gcase). Gcase deficiency or loss of Gcase activity results in accumulation of its substrates glucosylceramide and glucosylsphingosine, which in turn leads to lysosomal dysfunction and, ultimately, cell death. Clinically, Gaucher disease is divided into three types. Type 1 is restricted to the periphery, while Types 2 and 3 have CNS involvement. Type 2 Gaucher disease is the most severe. Patients with Type 2 Gaucher disease suffer rapid and progressive neurodegeneration during the first years of life. Some success has been achieved in treating Type 1 Gaucher disease through enzyme replacement therapy with recombinant Gcase (Cerezyme, imiglucarase) and substrate reduction therapy using small molecule inhibitors of the upstream enzyme glucosylceramide synthase (Zavesca (miglustat); Cerdaltga (eliglustat)). However, none of these therapeutic options are effective in treating Types 2 and 3 Gaucher disease because the drugs do not penetrate the CNS.
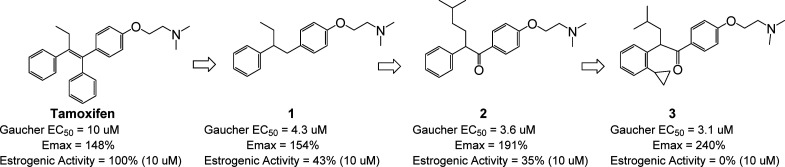
To address this unmet medical need we chose an alternative, more holistic phenotypic approach. Disrupted calcium homeostasis is thought to play a role in a number of lysosomal storage disorders, including Type 2 Gaucher disease.74,75 Evidence for this association comes from both neuronal cellular models of Gaucher disease76,77 and from brains taken post-mortem from Type 2 Gaucher disease patients.78 We discovered that Type 2 Gaucher disease patient derived fibroblasts (L444P/L444P genotype) display reduced calcium release from acidic stores in response to Gly-Phe-β-naphthylamide (GPN) compared to normal cells. GPN is a cathepsin C substrate, which, upon hydrolysis, causes osmotic lysis and calcium release from lysosomal acidic stores into the cytoplasm.79 On the basis of this phenotypic difference, we developed a high throughput fluorescence-based assay to screen for compounds with activity to reverse the lysosomal calcium release deficit found in patient-derived cells.80 A screen of 1200 known FDA approved drugs (Prestwick Chemical Library) was performed.73 One of the hits identified was tamoxifen (Figure 7), a selective estrogen receptor modulator used to treat estrogen-sensitive breast cancer.
Figure 7.
SAR around the screening hit tamoxifen led to compound 3, which reversed the diseased phenotype in Gaucher disease patient-derived cells and displayed reduced estrogenic activity.
Tamoxifen displayed modest functional potency (EC50 = 10 μM with 148% activation over the basal response to GPN-induced calcium release measured in patient-derived cells in the absence of compound treatment). By contrast, tamoxifen induced a maximum 28% activation over basal GPN-induced calcium release in normal (wild-type) cells. The drug-like properties of tamoxifen made it a good starting point for hit-to-lead activities. However, its potent antiestrogenic activity would be considered unacceptable for chronic therapy, especially in the young patient population affected by Type 2 Gaucher disease. Our structure–activity and structure–property studies (Figure 7)73 therefore focused on improving potency and efficacy within the tamoxifen chemical scaffold, reducing estrogenic activity, and identifying novel intellectual property. Since the exact molecular target was not known, we carried out a pharmacophore-driven campaign. SAR around tamoxifen itself identified compound 1, which retained potency and efficacy in the GPN-induced calcium release assay but showed reduced estrogenic potency compared to tamoxifen. Hypothesizing that the optimal pharmacophore preferred the aryl ring in a nonplanar conformation (planarity is stabilized by the extended conjugation of the tamoxifen scaffold), we introduced a carbonyl group and a modified alkyl substituent into the system to give compound 2, which was more potent and efficacious than tamoxifen or compound 1. Further conformational restriction through introduction of a 2-substituent on the distal aryl ring provided compound 3, which showed enhanced efficacy and a lack of estrogenic activity at concentrations up to 10 μM.
One of the key factors for success in phenotypic drug discovery is the relevance of the assay to the disease state.81 Reporter-linked assays have been used as drug discovery screens for several years.82 These assays, which can be easily miniaturized to high throughput platforms, have been engineered to link expression of a gene with a measurable readout to a variety of biological activities, including signal transduction, gene expression, and epigenetic modification. Several reporter systems have been utilized successfully, the two most prominent being (arguably) luciferase and green fluorescent protein (GFP). Reporter gene assays have become a valuable screening tool in drug discovery targeting cancer.10,83 Assays can be targeted to monitor specific pathways, which sometimes makes deconvolution of the mechanism of action of screening hits somewhat easier.
We have a long-standing collaboration with the group of Dr. J. P. Issa (formerly of the Fels Institute for Cancer Research and Molecular Biology, Lewis Katz School of Medicine at Temple University, presently located at the Coriell Institute for Medical Research). Issa and colleagues developed a high throughput gene reporter-based assay to screen for novel compounds that restore epigenetic control to tumor suppressor genes that have been inactivated in cells that have become cancerous.84 The screen uses the well-established YB5 cell-based system, which is derived from the human colon cancer cell line SW48.85,86 YB5 contains a single insertion of the cytomegalovirus promotor driving GFP expression that is epigenetically silenced through DNA methylation and histone acetylation. In the YB5 system, GFP expression behaves similarly to endogenous tumor suppressor genes that have been epigenetically silenced. The screen was used to successfully identify a novel epigenetic mechanism, namely calcium signaling through calcium-calmodulin kinase.86
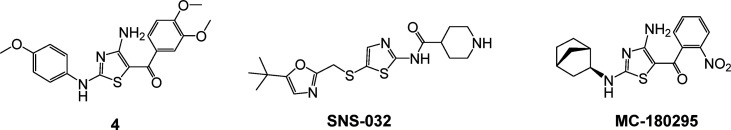
Screening of the TimTec NDL-3000 library (consisting of 3040 seminatural compounds, derived from natural or synthetic “natural-like” small molecule derivatives) identified aminothiazole hit 4 (Figure 8) as an epigenetic reactivator of silenced tumor suppressor genes.87 Compound 4 showed no activity against the main known regulators of epigenetic silencing, DNA methyl transferase and histone deacetylase. Connectivity mapping88 using RNA sequencing (RNA-seq) revealed that a number of compounds possessing pan-cyclin-dependent-kinase (CDK) inhibitory activity shared a similar transcriptional profile. Interestingly, one of these CDK inhibitors (SNS-032, Figure 8) also possessed an aminothiazole moiety in its chemical structure. Detailed profiling of the pan-CDK inhibitors and compound 4 revealed that the compounds that were most effective at gene reactivation in the YB5 cell system had the lowest IC50 values for CDK9. An SAR campaign using the YB5 system as the primary screen ultimately identified MC-180295 (Figure 8) as the most potent and efficacious analog within the series. Further deconvolution finally revealed MC-180295 to be one of the most selective CDK9 inhibitors reported to date (22.5-fold selective over the nearest CDK target, CDK4, Table 1) with an IC50 value against CDK9 of 5 nM. This was, to our knowledge, the first report of epigenetic modulatory activity for CDK9. MC-180295 displayed broad anticancer activity in vitro. In vivo, treatment with the compound deceased tumor burden and increased survival time in NSG mice inoculated with SW48 cells. In addition, CDK9 inhibition also sensitized cells to checkpoint inhibition induced by an anti-PD-1 antibody in vivo.
Figure 8.
Structures of SNS-032, compound 4, and MC-180295.
Table 1. Inhibitory Activity for MC-180295 against Cyclin-Dependent Kinasesa.
| target | CDK1/cyclin B | CDK2/cyclin A | CDK2/cyclin E | CDK3/cyclin E | CDK4/cyclin D | CDK5/P35 | CDK5/P25 | CdK8/cyclin D3 | CDK7/CycH/MAT1 | CDK9/cyclin T1 |
| IC50, nM | 138 | 233 | 367 | 399 | 112 | 159 | 186 | 712 | 555 | 5 |
MC-180295 displayed 22.5-fold selectivity for CDK9 versus its next closest target, CDK4.87
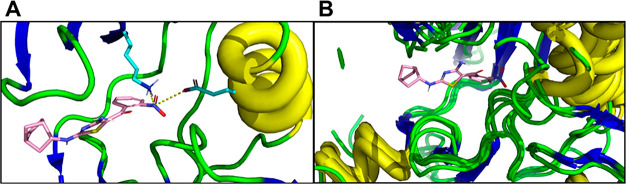
Molecular modeling studies (Figure 9) provide a hypothesis for the selectivity that MC-180295 possesses for CDK9 over other CDKs. MC-180295 was docked to the ATP binding site of CDK1, CDK2, CDK5, CDK6, and CDK7 and CDK9. The model suggests that MC-180295 binds to the hinge region of all seven proteins in a similar conformation. However, the nitro group of MC-180295 appears to make hydrogen bonds to Lys48 on the N-lobe and Glu66 on the αC helix of CDK9 that are not present in the models for CDKs 1, 2, 5, 6, and 7. These additional binding interactions may be responsible for the enhanced affinity that MC-180295 shows for CDK9.
Figure 9.
Docking of MC-180295 with ATP binding site of cyclin-dependent kinases. Crystal structures of the protein kinases were downloaded from RCSB Protein Data Bank (CDK1, 5LQF; CDK2, 1HCK; CDK5, 1UNH; CDK6, 2EUF; CDK7, 1UA2; CDK9, 3BLQ) and prepared with Protein Preparation Wizard (Schrodinger, New York NY). Receptor Grid Preparation was used to create the docking sites. Standard Precision was employed for the docking step. Other protein kinases were aligned using CDK9 as a reference frame, and the coordinate of the docking pose was copied to have the same binding model for all kinases. (A) MC-180295 docked to ATP binding site of CDK9. The nitro group engages Lys48 and Glu66. (B) MC-180295 docked to CDK1, CDK2, CDK5, CDK6, and CDK7. The nitro group does not make significant binding interactions with the proteins.
Phenotypic screening appears to have risen to take a place among the tools used by drug discovery scientists in their continuing effort to identify new treatments for unmet medical needs. The approach has been resuscitated, in part, by advancements in technology that enable the design of meaningful high throughput assays and facilitate deconvolution of the mechanism(s) of action for the hits that are identified. Today’s phenotypic screening approaches are encouraging researchers to tackle disease states and biochemical mechanisms that were once thought of as “undruggable”. But they are only part of the solution to what many perceive to be a problem in drug discovery, namely, lower productivity now that the “low hanging fruit” has been picked from the drug discovery orchard. Numerous perspectives have been written in the past two decades detailing thoughts on the issues impacting the industry. Opinions on how to fix the problem vary, but what is clear is that the model moving forward needs to change and it is changing. Like the principal of Yin and Yang, phenotypic screening is part of a balance, part of a complementary collection of approaches and technologies that is being embraced in an effort to ensure that we continue to address the unmet medical challenges of today and tomorrow.89
Acknowledgments
Part of the work described in this manuscript was supported by the National Institutes of Health under Award Number R01DA037270-01 to M.A.
Glossary
Abbreviations
- ATP
adenosine triphosphate
- CD4
cluster of differentiation-4
- CDK
cyclin-dependent kinase
- CycH
cyclin H
- CNS
central nervous system
- EAAT2
excitatory amino acid transporter-2
- FDA
Food and Drug Administration
- FKBP
FK binding protein
- Gcase
β-glucocerebrosidase
- GFP
green fluorescent protein
- GLT-1
glutamate transporter-1
- GPN
Gly-Phe-β-naphthylamide
- HGP
Human Genome Project
- LDLR
low density lipoprotein receptor
- LYCD
live yeast cellular derivative
- MAT1
ménage à trois 1
- mTOR
mammalian target of rapamycin
- RNA-seq
RNA sequencing
- SNRI
serotonin–norepinephrine reuptake inhibitor
- PCSK9
proprotein convertase subtilisin/kexin type 9
- SAR
structure–activity relationship
The authors declare no competing financial interest.
References
- Valent P.; Groner B.; Schumacher U.; Superti-Furga G.; Busslinger M.; Kralovics R.; Zielinski C.; Penninger J. M.; Kerjaschki D.; Stingl G.; Smolen J. S.; Valenta R.; Lassmann H.; Kovar H. L.; Jager U.; Kornek G.; Muller M.; Sorgel G. Paul Ehrlich (1854–1915) and his contibutions to the foundation and birth of translational medicine. J. Innate Immun. 2016, 8, 111–120. 10.1159/000443526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich P. Die Seitenkettentheorie und ihre gegner. Munchner Med. Wochenschr. 1901, 2121–2124. [Google Scholar]
- Croston G. E. The utility of target-based discovery. Expert Opin. Drug Discovery 2017, 12, 427–429. 10.1080/17460441.2017.1308351. [DOI] [PubMed] [Google Scholar]
- Hopkins A. L.; Groom C. R. The druggable genome. Nat. Rev. Drug Discovery 2002, 1, 727–730. 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Russ A.; Lampel S. The druggable genome: an update. Drug Discovery Today 2005, 10, 1607–1610. 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- Wu-Pong S.The human genome project and drug development. In Biopharmaceutical Drug Design and Development, 2nd ed.; Wu-Pong S., Rojanasakul Y., Eds.; Humana Press: New York, NY, 2008; pp 15–30. [Google Scholar]
- Edwards A. M.; Isserlin R.; Bader G. D.; Frye S. V.; Willson T. M.; Yu F. H. Too many roads not taken. Nature 2011, 470, 163–165. 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- Swinney D. C.; Anthony J. How were new medicines discovered?. Nat. Rev. Drug Discovery 2011, 10, 507–519. 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Swinney D. C. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharmacol. Ther. 2013, 93, 299–301. 10.1038/clpt.2012.236. [DOI] [PubMed] [Google Scholar]
- Moffat J. G.; Rudolph J.; Bailey D. Phenotypic screening in cancer drug discovery – past, present and future. Nat. Rev. Drug Discovery 2014, 13, 588–602. 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- Williams C.; Fecke W.. The impact of phenotypic and functional screening in CNS drug discovery. Drug Target Review, 2015, https://www.drugtargetreview.com/article/3361/the-impact-of-phenotypic-and-functional-screening-in-cns-drug-discovery-2/ (accessed May 5, 2020).
- Henry J.; Wlodkowic D. Towards high-throughput chemobehavioural phenomics in neuropsychiatric drug discovery. Mar. Drugs 2019, 17, 340. 10.3390/md17060340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulner N.; Danckaert A.; Ihm J.; Shum D.; Shorte S. L. Next-generation phenotypic screening n early drug discovery for infectious diseases. Trends Parasitol. 2019, 35, 559–570. 10.1016/j.pt.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Wagner B. K. The resurgence of phenotypic screening in drug discovery and development. Expert Opin. Drug Discovery 2016, 11, 121–125. 10.1517/17460441.2016.1122589. [DOI] [PubMed] [Google Scholar]
- Szabo M.; Akusjarvi S. S.; Saxena A.; Liu J.; Chandrasekar Janebjer G.; Kitambi S. S. Cell and small animal models for phenotypic drug discovery. Drug Des., Dev. Ther. 2017, 11, 1957–1967. 10.2147/DDDT.S129447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanoff B. E. Phenotypic assessment and the discovery of topiramate. ACS Med. Chem. Lett. 2016, 7, 662–665. 10.1021/acsmedchemlett.6b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. M. Biodynes. A miracle of wound healing and tissue repair – the life cell’s secret. Sci. Am. 1943, 168, 14–16. 10.1038/scientificamerican0143-14. [DOI] [Google Scholar]
- Hirshfeld J. W.; Pilling M. A.; Maun M. F. The use of bio-dyne ointment for burns. J. Am. Med. Assoc. 1943, 123, 476. 10.1001/jama.1943.82840430003007a. [DOI] [Google Scholar]
- Crowe M. J.; McNeill R. B.; Schlemm D. J.; Greenhalgh D. G.; Keller S. J. Topical application of yeast extract accelerates the wound healing of diabetic mice. J. Burn Care Rehabil. 1999, 20, 155–162. 10.1097/00004630-199903000-00032. [DOI] [PubMed] [Google Scholar]
- Funding Universe, Wyeth: Company History, http://www.fundinguniverse.com/company-histories/wyeth-history/, accessed 12/11/2019.
- Gruenstein E. I.; Schlemm D. J.; Bethi M.; Keller S. J. The early signaling pathway of live yeast cell derivative in THP-1 monocytes. Cell Calcium 2018, 73, 112–120. 10.1016/j.ceca.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Yardley J. P.; Husbands G. E. M.; Stack G.; Butch J.; Bicksler J.; Moyer J. A.; Muth E. A.; Andree T.; Fletcher H. III; James M. N. G.; Sielecki A. R. 2-Phenyl-2-(1-hydroxycycloalkyl)ethylamine derivatives: Synthesis and antidepressant activity. J. Med. Chem. 1990, 33, 2899–2905. 10.1021/jm00172a035. [DOI] [PubMed] [Google Scholar]
- Yardley J. P.; Fletcher H. III; Russell P. B. A potent benzylamine analgesic: (−) cis-2-(Y-dimethylamino-m-hydroxybenzyl) cyclohexanol. Experientia 1978, 34, 1124–1125. 10.1007/BF01922905. [DOI] [PubMed] [Google Scholar]
- Biel J.Antidepressants, hallucinogens and stimulants, In Annual Reports in Medicinal Chemistry; Cain C. K., Ed.; Academic Press: New York, NY, 1967; Vol. 2, pp 11–23. [Google Scholar]
- Drugs@FDA: FDA-Approved Drugs 1993, US Food & Drug Administration website, approved 12/28/1993, https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=reportsSearch.process&rptName=2&reportSelectMonth=12&reportSelectYear=1993&nav, accessed 02/04/2020.
- Pristiq Drug Approval Package, US Food & Drug Administration website, approved 02/29/2008, https://www.accessdata.fda.gov/drugsatfda_docs/nda/2008/021992s000TOC.cfm, accessed 02/04/2020.
- Cymbalta Drug Approval Package, US Food & Drug Administration website, approved 08/03/2004, https://www.accessdata.fda.gov/drugsatfda_docs/nda/2004/021427_s000_Cymbalta.cfm, accessed 02/04/2020.
- Savella Drug Approval Package, US Food & Drug Administration website, approved 01/14/2009, https://www.accessdata.fda.gov/drugsatfda_docs/nda/2009/022256s000TOC.cfm, accessed 02/04/2020.
- Fetzima Drug Approval Package, US Food & Drug Administration website, approved 07/25/2013, https://www.accessdata.fda.gov/drugsatfda_docs/nda/2013/204168Orig1s000TOC.cfm, accessed 02/04/2020.
- Vezina C.; Kudelski A.; Sehgal S. N. Rapamycin (AY-22989). A new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principal. J. Antibiot. 1975, 28, 721–726. 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- Martel R. R.; Klicius J.; Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can. J. Physiol. Pharmacol. 1977, 55, 48–51. 10.1139/y77-007. [DOI] [PubMed] [Google Scholar]
- Heitman J.; Movva N. R.; Hall M. N. Target for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Choi J.; Chen J.; Schreiber S. L.; Clardy J. Structure of the FKBP12-rapamycin complex interacting with binding domain of human FRAP. Science 1996, 273, 239–242. 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Faivre S.; Kroemer G.; Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat. Rev. Drug Discovery 2006, 5, 671–688. 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- Ruan B.; Pong K.; Jow F.; Bowlby M.; Crozier R. A.; Liu D.; Liang S.; Chen Y.; Mercado M. L.; Feng X.; Bennett F.; von Schack D.; McDonald L.; Zaleska M. M.; Wood A.; Reinhart P. H.; Magolda R. L.; Skotnicki J.; Pangalos M. N.; Koehn F. E.; Carter G. T.; Abou-Gharbia M.; Graziani E. I. Binding of rapamycin analogs to calcium channels and FKBP52 contributes to their neuroprotective activities. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 33–38. 10.1073/pnas.0710424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney D. C.; Xia S. The discovery of medicines for rare diseases. Future Med. Chem. 2014, 6, 987–1002. 10.4155/fmc.14.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B. K.; Schreiber S. L. The power of sophisticated phenotypic screening and modern mechanism-of-action methods. Cell Chem. Biol. 2016, 23, 3–9. 10.1016/j.chembiol.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ali H. The evolution of drug discovery: from phenotypes to gargets, and back. MedChemComm 2016, 7, 788–798. 10.1039/C6MD00129G. [DOI] [Google Scholar]
- Brown D. G.; Wobst H. J. Opportunities and challenges in phenotypic screening for neurodegenerative disease research. J. Med. Chem. 2019, 10.1021/acs.jmedchem.9b00797. [DOI] [PubMed] [Google Scholar]
- Xu S.; Luo S.; Zhu Z.; Xu J. Small molecules as inhibitors of PCSK9: current status and future challenges. Eur. J. Med. Chem. 2019, 162, 212–133. 10.1016/j.ejmech.2018.11.011. [DOI] [PubMed] [Google Scholar]
- Lintner N. G.; McClure K. F.; Petersen D.; Londregan A. T.; Piotrowski D. W.; Wei L.; Xiao J.; Bolt M.; Loria P. M.; Maguire B.; Geoghegan K. F.; Huang A.; Rolph T.; Liras S.; Doudna J. A.; Dullea R. G.; Cate J. H. D. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol. 2017, 15, e2001882 10.1371/journal.pbio.2001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen D. N.; Hawkins J.; Ruangsiriluk W.; Stevens K. A.; Maguire B. A.; O’Connell T. N.; Rocke B. N.; Boehm M.; Ruggeri R. B.; Rolph T.; Hepworth D.; Loria P. M.; Carpino P. A. A small-molecule anti-secretagogue of PCSK9 targets the 80S ribosome to inhibit PCSK9 protein translation. Cell Chem. Biol. 2016, 23, 1362–1371. 10.1016/j.chembiol.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Andries K.; Verhasselt P.; Guillemont J.; Gohlmann W. H.; Neefs J.-M.; Winkler H.; Van Gestel J.; Timmerman P.; Zhu M.; Lee E.; Williams P.; de Chaffoy D.; Huitric E.; Hoffner S.; Cambau E.; Truffot-Pernot C.; Lounis N.; Jarlier v. A diaryl quinolone drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005, 307, 223–227. 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- Maryanoff B. E. Pharmaceutical “gold” from neurostabilizing agents: topiramate and successor molecules. J. Med. Chem. 2009, 52, 3431–3440. 10.1021/jm900141j. [DOI] [PubMed] [Google Scholar]
- Haasen D.; Schopfer U.; Antczak C.; Guy C.; Fuchs F.; Selzer P. How phenotypic screening influenced drug discovery: Lessons from five years of practice. Assay Drug Dev. Technol. 2017, 15, 239–246. 10.1089/adt.2017.796. [DOI] [PubMed] [Google Scholar]
- Weber J. J.; Clemensson L. E.; Schioth H. B.; Nguyen H. P. Olesoxime in neurodegenerative diseases: scrutinizing a promising drug candidate. Biochem. Pharmacol. 2019, 168, 305–318. 10.1016/j.bcp.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Abou-Gharbia M.; Blass B. E.; Childers W. E.. Academic drug discovery centers: Key players in the future of the pharmaceutical industry. In Comprehensive Medicinal Chemistry III; Chackalamannil S., Rotella D. P., Ward S. E., Eds.; Elsevier Limited: Oxford, UK, 2017; Vol. 1, pp 1–10. [Google Scholar]
- Everett J. R. Academic drug discovery: Current status and prospects. Expert Opin. Drug Discovery 2015, 10, 937–944. 10.1517/17460441.2015.1059816. [DOI] [PubMed] [Google Scholar]
- Huryn D. M. Drug discovery in an academic setting: Playing to the strengths. ACS Med. Chem. Lett. 2013, 4, 313–315. 10.1021/ml400012g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T.; Imai R.; Ono S. The current status of drug discovery and development as originated in United States Academia: The influence of industrial and academic collaboration on drug discovery. Clin. Transl., Sci. 2018, 11, 597–606. 10.1111/cts.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier J. S. Academia and industry: Allocating credit for discovery and development of new therapies. J. Clin. Invest. 2019, 129, 2172–2174. 10.1172/JCI129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow J. C. A medicinal chemist’s perspective on transitioning from industry to academic drug discovery. ACS Med. Chem. Lett. 2019, 10, 687–689. 10.1021/acsmedchemlett.9b00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle M. The academic pill: How academia contributes to curing diseases. SLAS Disc. 2019, 24, 203–212. 10.1177/2472555218824280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J. R.; Alterman A. I.; Mulvaney F. D.; Koppenhaver J. M. Predicting proximal factors in cocaine relapse and near miss episodes: Clinical and theoretical implications. Drug Alcohol Depend. 1999, 56, 67–78. 10.1016/S0376-8716(99)00013-7. [DOI] [PubMed] [Google Scholar]
- Sinha R. The role of stress in addiction relapse. Curr. Psychiatry Rep. 2007, 9, 388–395. 10.1007/s11920-007-0050-6. [DOI] [PubMed] [Google Scholar]
- Nestler E. J. The neurobiology of cocaine addiction. Sci. Pract. Perspect. 2005, 3, 4–10. 10.1151/spp05314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein J. D.; Patel S.; Regan M. R.; Haenggeli C.; Huang Y. H.; Bergles D. E.; Jin L.; Hoberg M. D.; Vidensky S.; Chung D. S.; Toan S. V.; Bruijn L. I.; Su Z.-Z.; Gupta P.; Fisher P. B. R-Lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature 2005, 433, 73–77. 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- Sari Y.; Smith K. D.; Ali P. K.; Rebec G. V. Upregulation of GLT1 attenuates cue-induced reinstatement of cocaine-seeking behavior in rats. J. Neurosci. 2009, 29, 9239–9243. 10.1523/JNEUROSCI.1746-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knackstedt L. A.; Melendez R. I.; Kalivas P. W. Ceftriaxone restores glutamate homeostasis and prevents relapse to cocaine seeking. Biol. Psychiatry 2010, 67, 81–84. 10.1016/j.biopsych.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau R.; Sorgel F.; Eiffert H. Penetration of drugs through the blood-cerebrospinal fluid/blood-brain barrier for treatment of central nervous system infections. Clin. Microbiol. Rev. 2010, 23, 858–883. 10.1128/CMR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein J. D.; Van Kammen M.; Levey A. I.; Martin L. J.; Kuncl R. W. Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann. Neurol. 1995, 38, 73–84. 10.1002/ana.410380114. [DOI] [PubMed] [Google Scholar]
- Thone-Reineke C.; Neumann C.; Namsolleck P.; Schmerbach K.; Krikov M.; Schefe J. H.; Lucht K.; Hortnagl H.; Godes M.; Muller S.; Rumschussel K.; Funke-Kaiser H.; Villringer A.; Steckelings U. M.; Unger T. the u-lactam antibiotic, ceftriaxone, dramatically improves survival, increases glutamate uptake and induces neurotrophins in stroke. J. Hypertens. 2008, 26, 2426–2435. 10.1097/HJH.0b013e328313e403. [DOI] [PubMed] [Google Scholar]
- Lee S. G.; Su Z. Z.; Emdad L.; Gupta P.; Sarkar D.; Borjabad A.; Volsky D. J.; Fisher P. B. Mechanism of ceftriaxone induction of excitatory amino acid transporter-2 expression and glutamate uptake in primary human astrocytes. J. Biol. Chem. 2008, 283, 13116–13123. 10.1074/jbc.M707697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-B.; Toan S. V.; Zelenaia O.; Watson D. J.; Wolfe J. H.; Rothstein J. D.; Robinson M. B. Regulation of astrocytic glutamate transporter expression by Akt: evidence for a selective transcriptional effect on the GLT-1/EAAT2 subtype. J. Neurochem. 2006, 97, 759–771. 10.1111/j.1471-4159.2006.03743.x. [DOI] [PubMed] [Google Scholar]
- Lee E.; Sidoryk-Wegrzynowicz M.; Wang N.; Webb A.; Son D.-S.; Lee K.; Aschner M. GPR30 regulates glutamate transporter GLT-1 expresson in rat primary astrocytes. J. Biol. Chem. 2012, 287, 26817–26828. 10.1074/jbc.M112.341867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic T.; Holsboer F.; Rein T.; Zschocke J. The CpG island shore of the GLT-1 gene acts as a methylation-sensitive sensitive enhancer. Glia 2012, 60, 1345–1355. 10.1002/glia.22353. [DOI] [PubMed] [Google Scholar]
- Cudkowicz M. E.; Titus S.; Kearney M.; Yu H.; Sherman A.; Schoenfeld D.; Hayden D.; Shui A.; Brooks B.; Conwit R.; Felsenstein D.; Greenblatt D. J.; Keroack M.; Kissel J. T.; Miller R.; Rosenfeld J.; Rothstein J.; Simpson E.; Tolkoff-Rubin N.; Zinman L.; Shefner J. M. Efficacy and safety of ceftriaxone for ayltrophic lateral sclerosis: results of a multi-stage, randomized, double-blind, placebo-controlled, Phase 3 study. Lancet Neurol. 2014, 13, 1083–1091. 10.1016/S1474-4422(14)70222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feather D.; McCormack N.; Dykstra H.; Childers W.; Blass B.; Rawls S.; Ramirez S.; Ferguson T.. Therapeutic intervention as treatments to cerebral palsy, Program and Abstract Book 2014 Mid-Atlantic Pharmacology Society Annual Meeting, Philadelphia, PA; American Society for Pharmacology and Experimental Therapeutics, 2014; p 7, Poster #1. [Google Scholar]
- Feather-Schussler D.; Ferguson T. A battery of motor tests in a neonatal mouse model of cerebral palsy. J. Visualized Exp. 2016, e53569. 10.3791/53569. [DOI] [PMC free article] [PubMed] [Google Scholar]; https://www.jove.com/video/53569/a-battery-of-motor-tests-in-a-neonatal-mouse-model-of-cerebral-palsy (accessed May 5, 2020).
- Annadurai S.; Martinez R.; Canney D. J.; Eidem T.; Dunman T.; Abou-Gharbia M. Design and synthesis of 2-aminothiazole based antimicrobials targeting MRSA. Bioorg. Med. Chem. Lett. 2012, 22, 7719–7725. 10.1016/j.bmcl.2012.09.095. [DOI] [PubMed] [Google Scholar]
- Swinney D. C. The contribution of mechanistic understanding to phenotypic screening for first-in-class medicines phenotypic assays in drug discovery. J. Biomol. Screening 2013, 18, 1186–1192. 10.1177/1087057113501199. [DOI] [PubMed] [Google Scholar]
- Zheng W.; Thorne N.; McKew J. C. Phenotypic screens as a renewed approach for drug discovery. Drug Discovery Today 2013, 18, 1067–1073. 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers W.; Fan R.; Martinez R.; Colussi D. J.; Melenski E.; Liu Y.; Gordon J.; Abou-Gharbia M.; Jacobson M. A. Novel compounds that reverse the disease phenotype in Type 2 Gaucher disease patient-derived cells. Bioorg. Med. Chem. Lett. 2020, 30, 126806. 10.1016/j.bmcl.2019.126806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselyov K.; Yamaguchi S.; Lyons C. W.; Muallem S. Aberrant Ca2+ handling in lysosomal storage disorders. Cell Calcium 2010, 47, 103–111. 10.1016/j.ceca.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans E.; Platt F. M. Lysosomal Ca2+ homeostasis: role in pathogenesis of lysosomal storage disorders. Cell Calcium 2011, 50, 200–205. 10.1016/j.ceca.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Korkotian E.; Schwarz A.; Pelled D.; Schwarzmann G.; Segal M.; Futerman A. H. Elevation of intracellular glucosylceramide levels results in increase in endoplasmic reticulum density and in functional caldium stores in cultured neurons. J. Biol. Chem. 1999, 274, 21673–21678. 10.1074/jbc.274.31.21673. [DOI] [PubMed] [Google Scholar]
- Lloyd-Evans E.; Pelled D.; Riebeling C.; Bodennec J.; de-Morgan A.; Waller H.; Schiffmann R.; Futerman A. H. Glucosylceramide and bucosylsphingosine modulate calcium mobilization from brain microsomes via different mechanisms. J. Biol. Chem. 2003, 278, 23594–23599. 10.1074/jbc.M300212200. [DOI] [PubMed] [Google Scholar]
- Pelled D.; Trajkovic-Bodennec S.; Lloyd-Evans E.; Sidransky E.; Schiffmann R.; Futerman A. H. Enhanced calcium release in the acute neuronopathic form of Gaucher disease. Neurobiol. Dis. 2005, 18, 83–88. 10.1016/j.nbd.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Berg T. O.; Stromhaug E.; Lovdal T.; Seglen P. O.; Berg T. Use of glycyl-L-phenylalanine-2-naphthylamide, a lysosome-disrupting cathepsin C substrate to distinguish between lysosomes and prelysosomal endocytic vacuoles. Biochem. J. 1994, 300, 229–236. 10.1042/bj3000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A manuscript on the Gaucher patient-derived cell HIS is in preparation. A similar assay protocol employing Tay-Sachs patient-derived cells can be found in:; Colussi D. J.; Jacobson M. A. Patient-derived phenotypic high-throughput assay to identify small molecules restoring lysosomal function in Tay Sachs Disease. SLAS Discovery 2019, 24, 295–303. 10.1177/2472555218814538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent F.; Loria P.; Pregel M.; Stanton R.; Kitching L.; Nocka K.; Doyonnas R.; Steppan C.; Gilbert A.; Schroeter T.; Peakman M.-C. Developing predictive assays: The phenotypic screening “rule of 3. Sci. Transl. Med. 2015, 7, 293ps15. 10.1126/scitranslmed.aab1201. [DOI] [PubMed] [Google Scholar]
- Welch G.; Damoiseaux R.; Loren M.. Making it all work: Functional genomics and reporter gene assays. In Reporter Gene Assays: Methods in Molecular Biology; Damoiseaux R., Hasson S., Eds.; Humana Press, New York, NY, 2018; Vol. 1755, pp 89–105. [DOI] [PubMed] [Google Scholar]
- Ediriweera M. K.; Tennekoon K. H.; Samarakoon S. R. In vitro assays and techniques utilized in anticancer drug discovery. J. Appl. Toxicol. 2019, 39, 38–71. 10.1002/jat.3658. [DOI] [PubMed] [Google Scholar]
- Raynal N. J.-M.; Lee J. T.; Wang Y.; Beaudry A.; Madireddi P.; Garriga J.; Malouf G. G.; Dumont S.; Dettman E. J.; Gharibyan V.; Ahmed S.; Chung W.; Childers W. E.; Abou-Gharbia M.; Henry R. A.; Andrews A. J.; Jelinek J.; Cui Y.; Baylin S. B.; Gill D. L.; Issa J.-P. J. Targeting calcium signaling induces reactivation of tumor suppressor genes in cancer. Cancer Res. 2016, 76, 1494–1505. 10.1158/0008-5472.CAN-14-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si J.; Boumber Y. A.; Shu J.; Qin T.; Ahmed S.; He R.; Jelinek J.; Issa J.-P. J. Chromatin remodeling is required for gene reactivation after decitabine-mediated DNA hypomethylation. Cancer Res. 2010, 70, 6968–6970. 10.1158/0008-5472.CAN-09-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal N. J-M.; Si J.; Taby R. F.; Gharibyan V.; Ahmed S.; Jelinek J.; Estecio M. R. H.; Issa J.-P. J. DNA methylation does not stably lock gene expression but instead serves as a molecular mark for gene silencing memory. Cancer Res. 2012, 72, 1170–1181. 10.1158/0008-5472.CAN-11-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Pandey S.; Travers M.; Sun H.; Morton G.; Madzo J.; Chung W.; Khowsathit J.; Perez-Leal O.; Barrero C. A.; Merali C.; Okamoto Y.; Sato T.; Pan J.; Garriga J.; Bhanu N. V.; Simithy J.; Patel B.; Huang J.; Raynal N. J.-M.; Garcia B. A.; Jacobson M. A.; Kadoch C.; Merali S.; Zhang Y.; Childers W.; Abou-Gharbia M.; Karanicolas J.; Baylin S. B.; Zahnow C. A.; Jelinek J.; Grana X.; Issa J.-P. J. Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell 2018, 175, 1244–1258. 10.1016/j.cell.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.; Crawford E. D.; Peck D.; Modell J. W.; Blatt I. C.; Wrobel M. J.; Lerner J.; Brunet J. P.; Subramanian A.; Ross K. N.; Reich M.; Hieronymus H.; Wei G.; Armstrong S. A.; Haggarty S. J.; Clemons P. A.; Wei R.; Carr S. A.; Lander E. S.; Golub T. R. The Connectivity Map: using gene-expression signatures to connect small molecules, genes and disease. Science 2006, 313, 1929–1935. 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Vaidya A.; Roy A.; Chaguturu R. How to rekindle drug discovery process through integrative therapeutic targeting. Expert Opin. Drug Discovery 2018, 13, 893–898. 10.1080/17460441.2018.1514010. [DOI] [PMC free article] [PubMed] [Google Scholar]