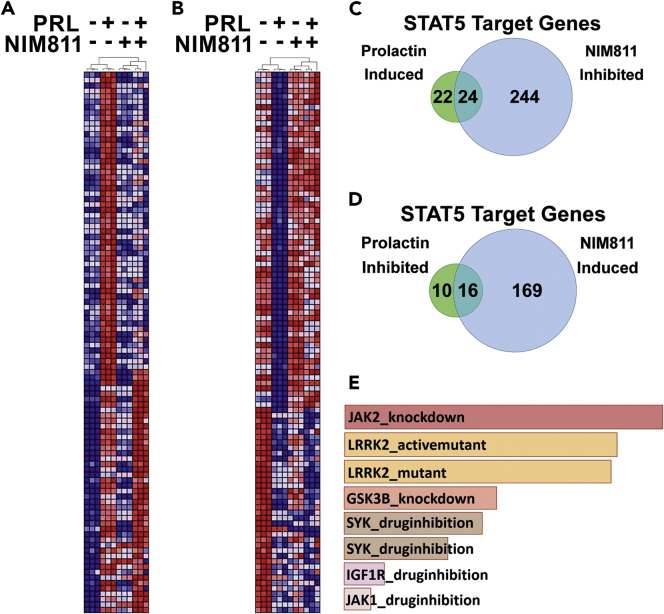
Figure 4.
NIM811 Opposes Prolactin-Induced Gene Expression Globally and at Stat5 Target Genes
(A and B) Heatmaps depict hierarchical clustering of the top 100 PRL-induced (A) and PRL-inhibited (B) genes (represented by rows) identified by microarray analysis. Samples are represented by columns and cluster with biological replicates according to the treatment conditions indicated above each dendrogram. Red and blue represent high and low gene expression values, respectively.
(C and D) Venn diagrams quantify the Stat5 target genes defined as those previously reported by Kang et al. (2014a) from the microarray analysis that are significantly induced (C) or inhibited (D) by PRL (fold change>1.2) and that are also significantly inhibited (C) or induced (D) by NIM811. Differential expression between treatment conditions was assessed by moderated t test adjusted for multiple hypotheses by the Benjamini and Hochberg method. The false discovery rate (FDR) was controlled so that only those probe sets with q < 0.01 were deemed significant.
(E) Bar graph depicts the highest scoring gene set enrichments when NIM811-inhibited genes (fold change>1.2) are compared with gene signatures from kinase perturbation experiments deposited in the Gene Expression Omnibus (GEO). The gene signatures with the highest concordance with NIM811-inhibited gene set are listed in descending order from top to bottom (# 1–8) based on p value ranking. (1) Jak2_knockdown_192_GSES4645, p < 3.18 × 10−12; (2) LRRK2_activemutant_159_GSE36321, p < 3.50 × 10−12; (3) LRRK2_mutant_GDS4401, p < 5.24 × 10−12; (4) GSK3β_knockdown_206_GDS4305, p < 2.27 × 10−9; (5) SYK_druginhibition_153_GSE34176, p < 5.22 × 10−9; (6) SYK_druginhibition_154_GSE34176, p < 1.23 × 10−8; (7) IGF1R_druginhibition_46_GSE14024, p < 1.62 × 10−7; and (8) JAK1_druginhibition_166_GSE38335, p < 3.79 × 10−7.