Highlights
-
•
This study provides population-based expression pattern of DNMT1 which is an actionable target for osteosarcoma treatment.
-
•
Overexpression of DNMT1 was observed in osteosarcoma, in which levels of DNMT1 were consistent in biopsy and post-chemotherapy tissues.
-
•
We highlight synergistic effect of drug targeting DNMT1 enzyme (decitabine) and chemotherapy for the treatment of osteosarcoma.
Keywords: Osteosarcoma, DNA methylation, Immunohistochemistry, Drug combinations, Drug Synergism
Abstract
Background
Abnormality in the DNA methylation process is one of the hallmarks of cancer. Emerging evidence strongly supports the idea that defects in DNA methyl transferases (DNMTs) are involved in tumor development and progression. This alteration has major effects at the transcription level of various cancer-associated genes.
Methods
Expression profiles of DNMT1 were investigated in fresh frozen tissues, patient-derived cells, and formalin-fixed paraffin-embedded tissues using immunoblotting and immunohistochemistry analysis. We also examined an anti-tumor effect of single DNA-hypomethylating agent (decitabine) and a combination of decitabine and chemotherapy in osteosarcoma cell lines.
Results
The results showed an overexpression of DNMT1 in most cases compared to normal cells and tissue samples. DNMT1 was also expressed at the same levels in paired primary cells derived from biopsy and post-chemotherapy tissues. Expression patterns of DNMT1 were examined in 77 osteosarcoma patients of whom 82% had positive DNMT1 with an IRS score > 0. Most of the cases expressed low to moderate levels of DNMT1 (IRS range 1–8, median = 2.0). Furthermore, we found that a combination of decitabine and chemotherapy had a synergistic effect in most of the tested osteosarcoma cells at a low dose therapeutic range of decitabine.
Conclusions
Our study revealed DNMT1 expression patterns that indicated potential roles of DNMT1 in osteosarcoma transformation and progression. This finding also suggests the efficacy of a combination therapy of decitabine with chemotherapy for osteosarcoma treatment.
1. Introduction
Osteosarcoma is a primary malignant bone tumor that occurs most frequently in children and adolescents and is one of the most aggressive and hard-to-treat cancers. After chemotherapy was introduced as an adjunct to surgery for the treatment of osteosarcoma, there was a very striking increase in 5-year survival of up to 70% in patients with localized disease at diagnosis [1]. Unfortunately, the patients evolve resistance to neoadjuvant chemotherapy leading to metastasis and shorter overall survival (25–40% at 5 years) [1], [2]. Presently, no effective therapeutic options are available for the treatment of patients who respond poorly to conventional chemotherapy. A major challenge in the development of new therapeutic alternatives is the complicated etiologies of osteosarcoma, which occur as a result from their extremely complex genomic profiles. Additionally, epigenetic changes can create more intricate mechanisms in the tumorigenesis of osteosarcoma.
DNA methylation is one of the best-known epigenetic events in human cancers. Promoter hypermethylation mediates the silencing process of various genes including tumor suppressors and genes controlling the immune response and drug sensitivity [3]. This abnormality is a consequence of an overexpression of de novo DNA methyltransferases (DNMTs) early in tumor progression. Earlier work has demonstrated that DNMT3A and DNMT3B are essential for de novo DNA methylation patterns, while DNMT1 acts as a maintenance enzyme that preserves DNA methylation during DNA synthesis [4]. However, later studies support the idea that DNMT1 also has an important role in de novo DNA methylation. Overexpression of DNMT1 has been reported in various types of cancer including but not limit to breast, bladder, colon, kidney, pancreatic, gastric and esophageal squamous cell carcinoma [5], [6], [7], [8], [9], [10]. Accumulating evidence shows that cancer cells depend on DNA methylation-mediated silencing of specific genes. Overexpression of DNMT1 is associated with down-regulation of a number of tumor suppressor genes. For instance, silencing DNMT1 inhibits proliferation, metastasis and invasion of esophageal squamous cell carcinoma (ESCC) cells [11]. It has also been shown to decrease methylation of RASSF1A and DAPK promoters leading to increased expression of these tumor suppressor genes in ESCC cells and in a xenograft mouse model. Additionally, it has been demonstrated that blocking of DNMT1 expression by miR-152 reduces promoter methylation of KLF4 in pancreatic cancer cells which consequently inhibit cell growth in vitro and in animal models [6].
Unlike genetic perturbations, epigenetic changes can be reversed with agents targeting epigenetic modifications. Much research has been performed to investigate the structure and functions of DNMTs to develop more effective inhibitors that target these key enzymes. Decitabine (5-aza-2′-deoxycytidine) is one of DNMT inhibitors that has been approved for use as a single agent to treat patients with myelodysplastic syndromes (MDS) and elderly patients with acute myeloid leukemia (AML) [12]. In addition to treatment of hematological malignancies, many clinical trials have tested the efficacy of decitabine in solid tumors both as a single agent and in combination with chemotherapy [13]. The findings of high response rate and mild toxicity in individual patients highlight a potential of applying decitabine in cancer therapy.
In this study, DNMT1 expression profiles were investigated in different types of samples including fresh frozen tissues, patient-derived cells and formalin-fixed paraffin-embedded (FFPE) tissues in order to examine potential roles of DNMT1 in osteosarcoma development and progression. We tested the efficacy of decitabine both as a lone agent and also as part of a combination chemotherapy in osteosarcoma treatment.
2. Materials and methods
2.1. Patients and tissue samples
For immunohistological analysis of DNMT1, we collected formalin-fixed paraffin-embed embedded tissues (FFPE) of biopsy samples from 77 osteosarcoma patients who had been diagnosed at Maharaj Nakorn Chiang Mai Hospital, Thailand, between 2000 and 2019 and had been treated with a standard neoadjuvant regimen and/or had undergone surgery at Maharaj Nakorn Chiang Mai Hospital and were followed-up for survival analysis until 11 Jan 2020. All primary biopsy H&E slides were reviewed by a bone and soft tissue pathologist (JS).
Osteosarcoma (N = 9) and soft callus tissues were obtained either from biopsy samples or from the fracture site of donors treated at the Trauma Unit. All tissue specimens were frozen at −80 °C within 30 min of surgery and stored until used.
This study protocol has been approved by the Research Ethics Committee of the Faculty of Medicine, Chiang Mai University. All patients and/or parent provided informed consent for patient information to be stored in the hospital database. All methods were carried out in accordance with good clinical practices (GCPs) and relevant guidelines. Clinicopathological parameters, including date of diagnosis, Enneking staging, metastatic status, and percentage of tumor necrosis after chemotherapy were retrieved from hospital records and pathology reports (Table 1).
Table 1.
Characteristics of osteosarcoma patients in the study cohort (N = 77).
| Factor | Number of patients | DNMT1 expression |
P-value | |
|---|---|---|---|---|
| Mean ± SD | Median (min–max) | |||
| Age at diagnosis (years) [mean = 18.1 ± 11.4, median = 15 (range 5–73)] | ||||
| ≤15 | 42 | 2.83 ± 2.17 | 3.00 (0.00–10.32) | 0.152 |
| >15 | 35 | 2.36 ± 2.46 | 1.40 (0.00–8.00) | |
| Gender | ||||
| Male | 43 | 2.59 ± 2.13 | 2.00 (0.00–8.00) | 0.847 |
| Female | 34 | 2.66 ± 2.54 | 2.05 (0.00–10.32) | |
| Enneking stage | ||||
| IIB | 45 | 2.85 ± 2.52 | 2.10 (0.00–10.32) | 0.442 |
| III | 32 | 2.29 ± 1.94 | 2.00 (0.00–7.00) | |
| Metastasis | ||||
| No | 16 | 3.57 ± 3.20 | 3.28 (0.00–10.32) | 0.226 |
| Yes | 61 | 2.37 ± 1.96 | 2.00 (0.00–7.11) | |
| Chemoresistance | ||||
| Good responder | 8 | 4.51 ± 3.61 | 3.78 (0.00–10.32) | 0.055 |
| Poor responder | 37 | 2.07 ± 1.93 | 2.00 (0.00–7.43) | |
P-values were calculated using the Mann-Whitney U test.
2.2. Tissue extraction
Protein extraction protocol was performed according to previously report with modification [15]. Fresh frozen tissues of osteosarcoma and soft callus (50–100 mg) were cut into small pieces and crushed in liquid nitrogen using a chilled mortar and pestle. The tissue powder was then incubated in Radioimmunoprecipitation assay (RIPA) buffer containing 1% protease inhibitor cocktail with agitation on a rocking mixer at 4 °C for 30 min. The lysate was centrifuged at 12,000×g for 10 min at 4 °C. Supernatant was collected to determine protein concentration with BCA assay.
2.3. Western blot analysis
Crude proteins (10–15 μg) were separated in 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon‑P; EMD Millipore, Billerica, MA, USA). Membranes were blocked with 10% skimmed milk in TBS/T buffer (TBS, 0.1% Tween‑20) and incubated overnight with antibodies specific to DNMT1 and actin at 4 °C. Membranes were then washed with TBS/T and incubated for 1 h with secondary antibody conjugated with horseradish peroxidase at room temperature. Band intensity was determined using an ECL-Advance Western Blotting Detection kit (GE Healthcare, Chicago, IL, USA). This protocol was modified from previous report [16].
2.4. Immunohistochemistry
Immunohistochemical analysis and scoring were followed our workflow [17]. Formalin-fixed paraffin-embedded (FFPE) tissues (N = 77) were obtained from archival paraffin blocks at the Department of Pathology, Faculty of Medicine, Chiang Mai University. FFPE tissues were immunostained using the Ventana automated straining system (Ventana Medical Systems, Tucson, AZ, USA). An Ultraview Universal DAB Detection Kit (Ventana Medical Systems, Tucson, AZ, USA), an indirect biotin-free system, was used to detect primary antibodies. Antigen retrieval was performed by heating the FFPE tissue in citrate buffer (pH 6) and subsequently incubating it with anti-DNMT1 at 1:50 dilution for 32 min (sc-20701 (H-300); Santa Cruz, CA, USA). The positive controls for DNMT1 staining were colon and tonsil tissues. Nuclear staining of DNMT1 was evaluated (by PC and JS) without prior knowledge of the clinical data using a semi-quantitative immunoreactive scoring system. The percentage of immunoreactive cells was estimated and scored as follows: no staining = 0, positive staining < 10% = 1, positive staining ≥ 10 and > 33% = 2, positive staining ≥ 33% and ≤ 66% = 3, positive staining ≥ 66% = 4. Intensity of staining was scored on a scale of 0–3: no color reaction = 0, mild reaction = 1, moderate reaction = 2, and intense reaction = 3. Immunoreactive score (IRS) was derived by multiplying immunoreactive cell scores and intensity of staining scores to compute an immunoreactive score ranging from 0 to 12.
2.5. Cell culture
Primary osteoblasts used in this study were derived from a set of bone graft specimens. Primary osteosarcoma cells were extracted from chemona.ve biopsy and post-chemotherapy tissues of patients with osteosarcoma. Extraction, culturing and characterization of the primary cells were performed according to previously described protocols [14]. To characterize primary osteoblasts and patient-derived osteosarcoma cells, the expression of osteoblastic markers, including type I collagen (COLIA1), osteonectin (ON) and bone sialoprotein (IBSP) and the oncogenic marker matrix-metalloproteinase-9 (MMP-9) were determined using quantitative real-time PCR (RT-PCR) according to our previous report [14]. Osteosarcoma and osteoblastic cell lines used in this study include 143B (CRL‑8303), MG63 (CRL-1427), SaOS2 (HTB-85), and normal human osteoblast cell line, hFOB1.19 (CRL‑11372) purchased from ATCC (Manassas, VA, USA). U2OS (CLS-300364) and MNNG/HOS (CLS-300289) were from Cell Lines Service (GmbH, Eppelheim, Germany). MNNG/HOS was cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc., Boston, MA, USA). MG63 was cultured in DMEM supplemented with 10% FBS. 143B cells were cultured in DMEM supplemented with 10% FBS and 0.015 mg/ml 5‑bromo‑2′‑deoxyuridine (Merck KGaA, Darmstadt, Germany). U2OS, SaOS2, and hFOB1.19 were cultured in F‑12 Medium supplemented with 10% FBS. All cells were maintained at 37 °C in a humidified 5% CO2 incubator. All culture conditions used in this study were optimized according to our published work [15], [16], [17].
2.6. Cell viability assay and drug combination analysis
Cell viability of osteosarcoma cells treated with chemotherapy and decitabine was assessed using MTT assay according to previously report [15]. Osteosarcoma cells were seeded in 96-well tissue culture plates (5 × 103 cells/100 μl freshly prepared culture media/well) and incubated at 37 °C with 5% CO2 overnight. Osteosarcoma cells were treated with decitabine (Selleckchem, Houston, TX, USA) at concentrations of 0, 0.01, 0.1, 1, 10, and 100 μM for 72 h. The cells were further treated with clinically achievable concentrations of decitabine in combination with doxorubicin (D1515; Sigma, St. Louis, MO, USA) or cisplatin (P4394; Sigma, St. Louis, MO, USA) for 72 h. After a 72-hour incubation, the culture medium was removed, and 100 μl of fresh medium containing 5 mg/ml of MTT solution was added to each well which were then incubated for 2 h at 37 °C. The MTT-formazan crystal had been dissolved in 100 μl of dimethyl sulfoxide with vigorous mixing. Finally, a spectrophotometer was used to measure absorbance at 550 nm. IC50 values were calculated by performing nonlinear regression analysis using GraphPad Prism version 8.0 (GraphPad Software, Inc., La Jolla, CA, USA).
To evaluate the synergism of the combination drug treatment, we used a reference model named zero interaction potency (ZIP) [18]. Synergy score values were calculated using SynergyFinder software [19].
2.7. Statistical analysis
Statistical analyses were carried out using STATA version 16.0 and GraphPad Prism version 8.4.0. Survival curves were estimated using the Kaplan–Meier method together with the log-rank test to evaluate association between DNMT1 expression and overall survival of osteosarcoma patients. Cox regression of proportional hazards was applied to probe for significance at the 95% confidence interval (CI). The significance of the correlation between staining patterns of DNMT1 and clinicopathological data was determined using Mann-Whitney U test for nonparametric data. P values <0.05 were considered to be statistically significant.
3. Results
3.1. Expression of DNMT1 in osteosarcoma tissues
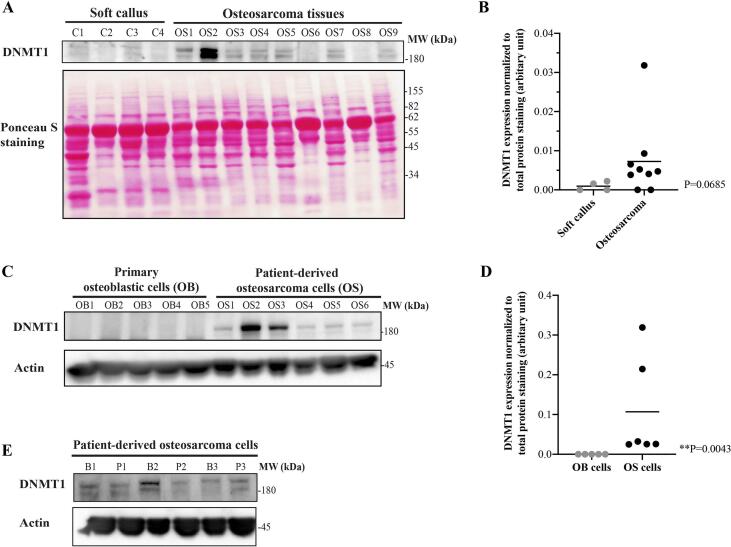
In this study, expression of DNMT1 in osteosarcoma tissues was compared to soft tissue callus which was used as a normal control. The results showed that DNMT1 was overexpressed in 78% (7/9) of osteosarcoma tissues (Fig. 1A and B). The expression level of DNMT1 was significantly higher in all patient-derived osteosarcoma cells (OS1-OS6) compared to primary osteoblastic cells (OB1-OB5) (Fig. 1C and D). We found DNMT1 was expressed in primary osteosarcoma cells derived from both biopsy and surgical tissues (post-chemotherapy) (Fig. 1E) which indicates roles of DNMT1 in primary tumors and residual tissues.
Fig. 1.
Differential expression of DNMT1 (A and B) in osteosarcoma tissues (OS1-OS9) vs soft callus tissues (C1-C4), (C and D) in patient-derived osteosarcoma cells (OS1-OS6) vs primary osteoblastic cells (OB1-OB5), and (E) in paired patient-derived cells from biopsy (B1-B3) and post-chemotherapy tissues (P1-P3).
3.2. Nuclear expression of DNMT1 in osteosarcoma patients in the study cohort
Overall, patient survival in study cohort of 77 patients ranged from 2 to 209 months after initial diagnosis with a median of 21 months. Fifty-two patients died at 2 to 102 months. Median follow-up of alive patients (n = 25) was 113 months (range 56–209 months). The 1-year and 5-year survival rates of all patients were 75% and 38%, respectively. Average age at diagnosis was 18.6 years (median = 15, range 5–73).
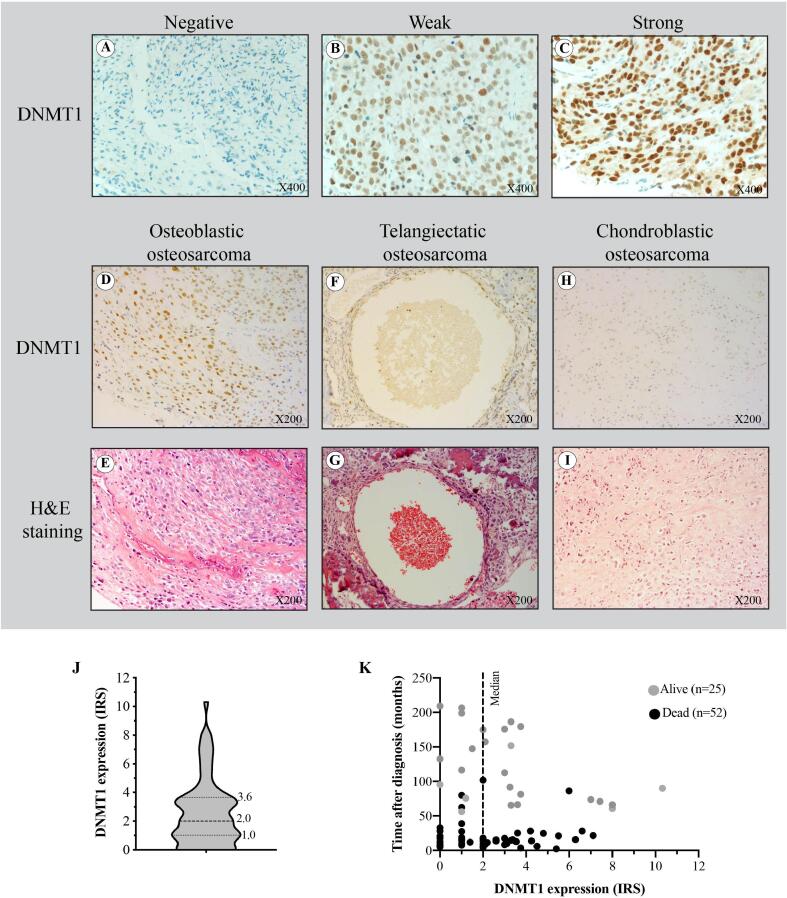
Under microscopic investigation, DNMT1 was found to be expressed predominantly in the nuclei of osteosarcoma cells. Positive nuclear immunohistochemical staining was observed to be weak (IRS 1–4), moderate (IRS 5–8), and strong (IRS 9–12) immunoreactivity in 64.9%, 15.6%, and 1.3% of cases, respectively (Fig. 2A–C). We found negative staining (IRS = 0) in 18.2% of cases. Median IRS of DNMT1 in all cases was 2.0 (Fig. 2J). Survival data and IRS of DNMT1 of individual patients is shown in Fig. 2K.
Fig. 2.
Levels of DNMT1 expression in our osteosarcoma cohort. Representative immunohistochemical staining of DNMT1 in osteosarcoma tissues (X400) showing different DNMT1 intensity; (A) negative (IRS = 0), (B) weak (IRS = 2), and (C) strong (IRS = 9) nuclei immunoreactivity of DNMT1. Representative immunohistochemical staining of DNMT1 in (D–E) osteoblastic osteosarcoma, (F–G) telangiectatic osteosarcoma as well as (H–I) chondroblastic osteosarcoma (X200). (J) The violin plot shows the median and distribution of DNMT1 expression levels in osteosarcoma patients (77 cases). (K) Survival scatter plot of individual patients.
3.3. Correlation between DNMT1 expression and clinicopathological factors
In direct comparisons between expression levels of DNMT1 and individual clinical parameters, DNMT1 expression was found to be only very weakly associated with most of those parameters (Table 1). Interestingly, expression of DNMT1 was lower in poor responders but the statistical significance was marginal (p = 0.055).
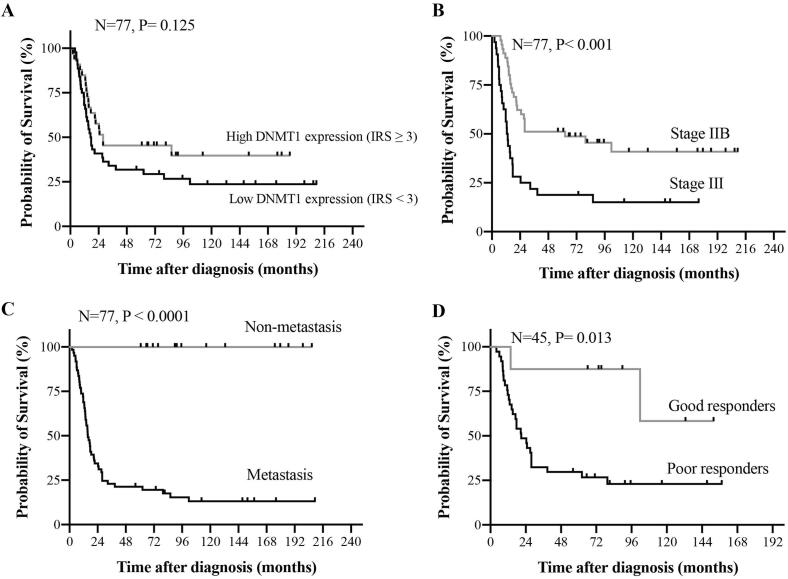
Kaplan-Meier survival analysis and log rank tests found no association between expression level of DNMT1 and survival rates of stage IIB and III osteosarcoma patients (Fig. 3). We re-computed the survival analysis using alternative cutoffs of IRS, but none reached statistical significance. Factors which were statistically significant indicators of poor prognosis included advanced Enneking stage (p = 0.001), the presence of metastasis (p < 0.001) and poor response to chemotherapy (p = 0.026), P-values calculated using the Cox regression of proportional hazards (Fig. 3 and Table 2).
Fig. 3.
Kaplan-Meier curves showing overall survival as a function of (A) DNMT1 expression in osteosarcoma patients, (B) Enneking stages, (C) metastatic status, and (D) chemotherapeutic sensitivity. P-values were calculated using the log-rank test.
Table 2.
Median survival of patients with high-grade osteosarcoma.
| Factor | Patients | Events (Death) | Median survival, months | HR (95% CI) | P-value |
|---|---|---|---|---|---|
| Age at diagnosis, years | |||||
| ≤15 | 42 | 26 | 27.8 | 1.00 | – |
| >15 | 35 | 26 | 18.5 | 1.34 (0.77–2.31) | 0.299 |
| Gender | |||||
| Male | 43 | 34 | 17.1 | 1.00 | – |
| Female | 34 | 18 | 79.6 | 0.56 (0.32–1.00) | 0.050 |
| Enneking stage | |||||
| IIB | 45 | 25 | 62.3 | 1.00 | – |
| III | 32 | 27 | 12.7 | 2.62 (1.51–4.54) | 0.001 |
| Metastasis | |||||
| No | 16 | 0 | Undefined | – | – |
| Yes | 61 | 52 | 15.6 | – | <0.001 |
| Chemoresistance | |||||
| Good responders | 8 | 2 | Undefined | 1.00 | – |
| Poor responders | 37 | 28 | 20.9 | 5.13 (1.21–21.70) | 0.026 |
| DNMT1 expression | |||||
| Low (IRS < 3) | 44 | 33 | 17.1 | 1.00 | – |
| High (IRS ≥ 3) | 33 | 19 | 27.8 | 0.64 (0.37–1.14) | 0.129 |
P-values were calculated using the Cox regression of proportional hazards; P-values <0.05 shown in bold.
3.4. The effect of decitabine on viability of osteosarcoma cells
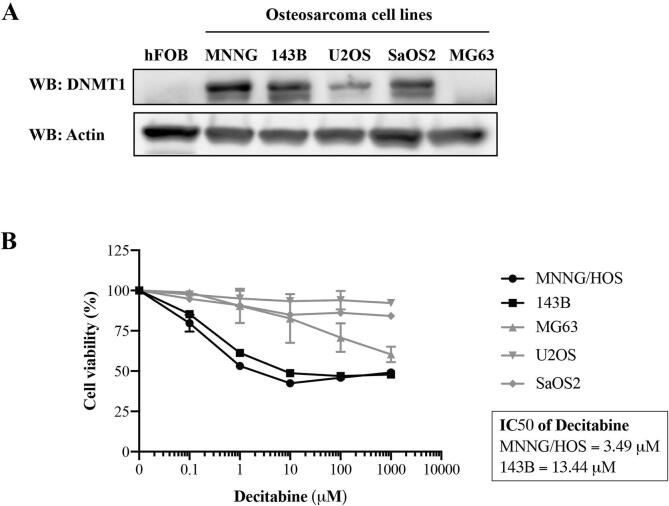
In this study, we examined the effect of decitabine, an inhibitor of DNMT1, as a single agent on cell viability of osteosarcoma cells including MNNG/HOS, 143B, U2OS, SaOS2, and MG63 cells. Immunoblotting showed that DNMT1 was overexpressed in most osteosarcoma cells but not in MG63 and hFOB 1.19 osteoblastic cells (Fig. 4A). The MTT assay demonstrated an anti-growth effect of decitabine exclusively in MNNG/HOS and 143B cells in a concentration dependent manner, whereas cell viability of the other cell lines (U2OS, SaOS2 and MG63) was not affected even at very high concentrations of decitabine (1000 μM) (Fig. 4B). The results show that the osteosarcoma cells responded to decitabine independently from their DNMT1 expression, since DNMT1-negative cells (MG63) and DNMT1-positive cells (U2OS and SaOS2) were not sensitive to decitabine treatment.
Fig. 4.
Association of DNMT1 expression levels and sensitivity to decitabine. (A) Expression levels of DNMT1 in osteoblast (hFOB 1.19) and osteosarcoma cell lines (MNNG/HOS, 143B, U2OS, SaOS2, and MG63). (B) Percentage of cell viability of osteosarcoma cell lines measured using MTT assay after treatment with decitabine at indicated concentrations for 72 h.
3.5. Synergistic effects of decitabine and chemotherapy in osteosarcoma cells
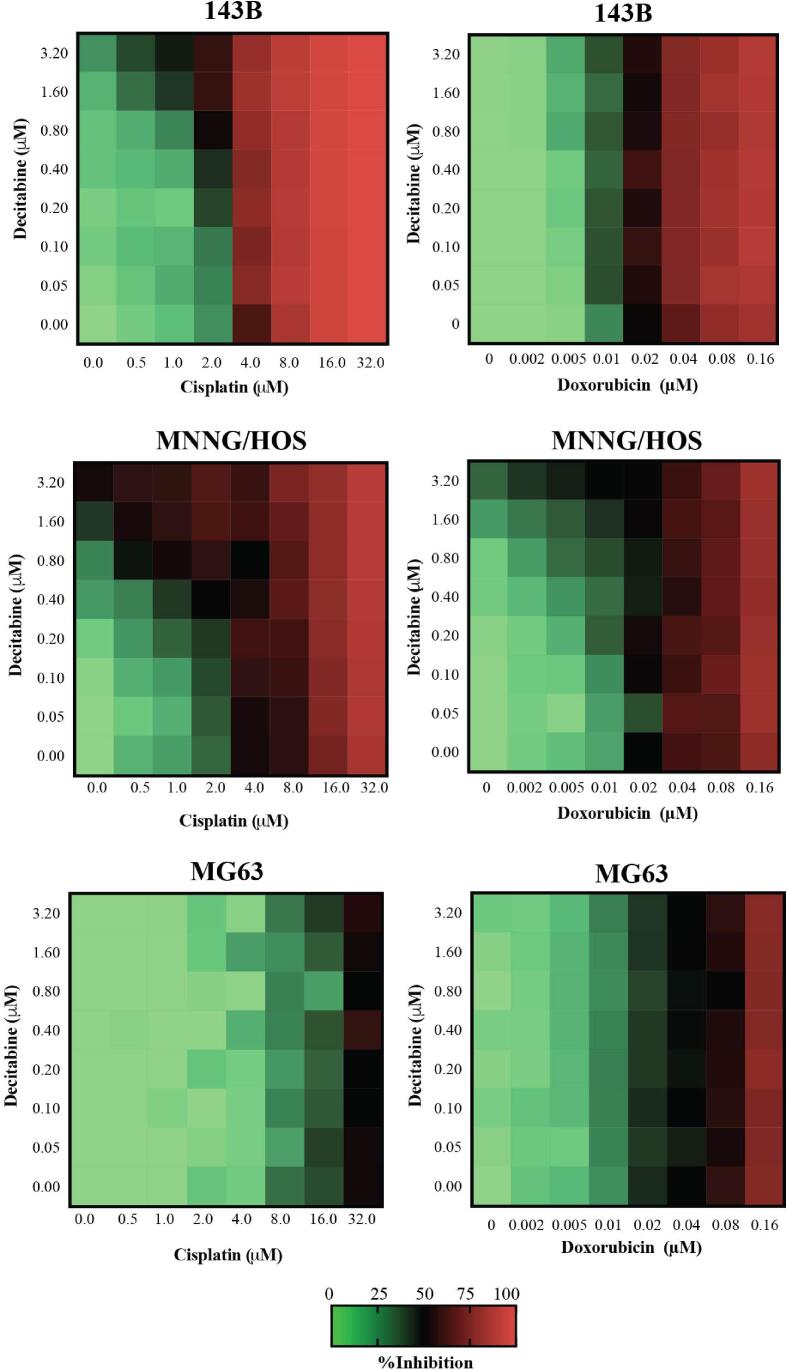
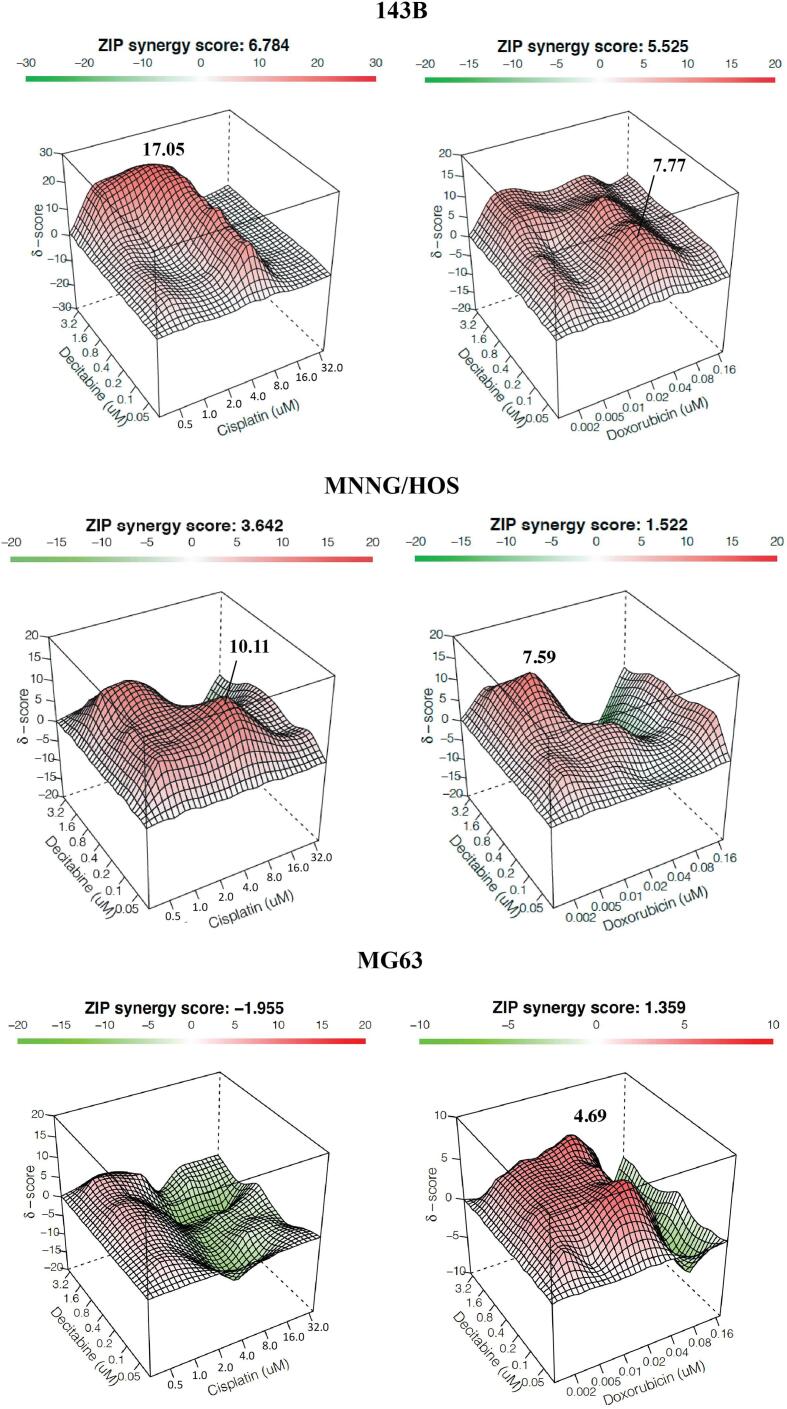
To investigate the synergistic effects of decitabine and chemotherapy, high-DNMT1-expressed osteosarcoma cells (143B and MNNG/HOS) and negative-DNMT1-expressed cells (MG63) were treated with decitabine in a concurrent combination with either doxorubicin or cisplatin. The dose–response matrix of each treatment combination was assessed (Fig. 5). We found synergistic effects (ZIP score > 0) of a combination of decitabine and cisplatin exclusively in high-DNMT1-expressed cells, but not in negative-DNMT1-expressed cells (ZIP score < 0) (Fig. 6). Interestingly, synergistic effects of decitabine plus doxorubicin were similar in all tested osteosarcoma cell lines and were not correlated with DNMT1 levels.
Fig. 5.
Percentage inhibition of decitabine in combination with a chemotherapeutic drug (cisplatin or doxorubicin) at indicated concentrations for 72 h in 143B, MNNG/HOS, and MG63 cells.
Fig. 6.
Synergistic effects of combinations of decitabine and chemotherapeutic drugs in 143B, MNNG/HOS and MG63 cells. Synergy scores were calculated using Synergyfinder software. ZIP Synergy scores > 0 indicate synergism (red regions) and scores < 0 indicate antagonism (green regions). Values in the white boxes represent the average synergy score for the region of highest synergy. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The present study demonstrated expression patterns of DNMT1 in osteosarcoma. We found that DNMT1 was overexpressed in most osteosarcoma tissues compared to soft callus (osteoblast-enriched tissues). Consistently, an upregulation of DNMT1 was observed in osteosarcoma cell lines compared to human fetal osteoblastic cells. Furthermore, DNMT1 was significantly expressed in the osteosarcoma cohort, of which 82% of the patients (63/77 cases) had positive DNMT1 expression. Interestingly, the present study also indicated that DNMT1 was expressed in all longitudinal samples including matched primary cells derived from biopsy and post-chemotherapy tissues. This implies that DNMT1 might play some role in residual cells after chemotherapy treatment. Evidence from several research studies has shown interplay between DNMT1 expression and non-coding RNA involved in regulating gene expression and biological effects of osteosarcoma cells. A study by Shi et al. demonstrated that HOTAIR (HOX transcript antisense intergenic RNA) activates DNMT1 expression and global DNA methylation levels through repressing miR-126 [20]. Thus, depletion of HOTAIR induces higher sensitivity of osteosarcoma cells to DNMT1 inhibitor through an apoptosis mechanism. Another study showed that overexpression of miR-139-5p inhibited osteosarcoma cell growth, migration and invasion and also reduced tumor growth in tumor xenografts by decreasing the level of DNMT1 [21]. A series of experiments have indicated that DNMT1 is a direct target of miR-139-5p, in which miR-139-5p controls the down-regulation of DNMT1 expression. Interestingly, it has also been found that long noncoding RNA nuclear enriched abundant transcript 1 (NEAT1) epigenetically regulates the G9a-DNMT1-Snail complex that is involved with metastasis of osteosarcoma cells [22]. Reducing NEAT1 suppresses EMT through the G9a-DNMT1-Snail axis leading to a reduction of in vitro invasive and migratory capacity and a decrease of in vivo lung metastasis.
Repurposing of approved drugs is a promising strategy for rapid development of new medicines for rare diseases or diseases that lack effective drug treatment. We previously identified new targets for the treatment of osteosarcoma which rely on protein profiles [23]. DNMT1 is included in our target list of FDA-approved drugs. In the present study, we tested the efficacy of decitabine, a DNMT inhibitor, in three osteosarcoma cell lines including highly-invasive cells (143B and MNNG/HOS) and chemo-less-sensitive cells (MG63). The results showed the low potency of decitabine as a single agent treatment. Similar to other drug repurposing studies, many individual compounds have low potency at safe therapeutic concentrations, while effective doses exceed safe levels [24]. Evaluating the efficacy of decitabine in increasing the sensitivity of chemotherapeutic drugs in other types of cancers is an example of drug combinations increasing the success rate of drug repurposing. We examined the synergistic effect of decitabine alone and in combination with conventional chemotherapy in osteosarcoma cells. The results showed that decitabine is able to sensitize osteosarcoma cells to chemotherapy drugs at low concentrations which are within safe therapeutic levels. We found that a combination of decitabine and cisplatin had a synergistic effect (higher ZIP score) exclusively in high-DNMT1-expressing cells (MNNG/HOS and 143B cells), whereas synergy was not observed in negative-DNMT1-expressing cells (MG63). We did not observe a similar synergistic effect with decitabine and doxorubicin combination treatment due to the ability of decitabine to sensitize osteosarcoma cells to doxorubicin independent of DNMT1 expression levels. Our findings suggest an association between DNMT1 levels and efficacy of decitabine as a chemosensitizer in platinum-based chemotherapy. There is accumulating evidence that decitabine has a dual dose-dependent mechanism of action [25]. At high doses, the cytotoxic effect of decitabine is mainly due to covalent trapping of DNMT enzyme into DNA. At lower and nontoxic doses, the antineoplastic action of decitabine is the result of its ability to induce DNA hypomethylation and to reactivate tumor suppressor genes. Several preclinical studies have indicated that decitabine can reverse platinum resistance in ovarian cancer cell lines and xenograft models. This chemo-sensitization is due to the re-expression of various tumor suppressor genes and development-associated transcription factors following treatment with decitabine [26], [27], [28]. In a phase II clinical trial, low-dose decitabine (10 mg/m2) was administered before carboplatin to ovarian cancer patients who had resisted to a platinum-based agents [29]. This combination regimen of decitabine effectively induced 35% of objective response rate (RR). Furthermore, they reported an average progression-free survival (PFS) of 10.2 months in which 53% of the patients had disease-free at 6 months. That study also found a positive correlation among demethylation of MLH1, RASSF1A, HOXA10, and HOXA11 genes in tumors with PFS (P < 0.05).
5. Conclusions
DNMT1 plays a pivotal role in osteosarcoma as demonstrated by DNMT1 expression patterns in osteosarcoma patients. The synergistic effect of decitabine combined with chemotherapy is worthy of further investigation to identify potential additional applications of decitabine in new regimens for the treatment of osteosarcoma.
CRediT authorship contribution statement
Parunya Chaiyawat: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing. Nutnicha Sirikaew: Investigation, Validation. Piyaporn Budprom: Investigation, Validation. Jeerawan Klangjorhor: Investigation, Validation. Areerak Phanphaisarn: Formal analysis. Pimpisa Teeyakasem: Investigation, Validation. Jongkolnee Settakorn: Supervision, Investigation. Dumnoensun Pruksakorn: Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was supported by the Faculty of medicine, Chiang Mai University, the National Science and Technology Development Agency (NSTDA), code P-18-51991, the National Research University (NRU) fund, and the Musculoskeletal Science and Translational Research Center. The authors would also like to express their sincere thanks to Dr. G. Lamar Robert, PhD, and Assoc. Prof. Dr. Chongchit Sripun Robert, PhD, for editing the English manuscript.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Parunya Chaiyawat, Email: parunya.chaiyawat@cmu.ac.th.
Jongkolnee Settakorn, Email: jsettakorn@gmail.com.
Dumnoensun Pruksakorn, Email: dumnoensun.p@cmu.ac.th.
References
- 1.Friebele J.C., Peck J., Pan X., Abdel-Rasoul M., Mayerson J.L. Osteosarcoma: a meta-analysis and review of the literature. Am. J. Orthop. (Belle Mead NJ) 2015;44(12):547–553. [PubMed] [Google Scholar]
- 2.Bielack S.S., Kempf-Bielack B., Delling G., Exner G.U., Flege S., Helmke K., Kotz R., Salzer-Kuntschik M., Werner M., Winkelmann W., Zoubek A., Jurgens H., Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20(3):776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M., Corn P.G., Baylin S.B., Herman J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 4.Subramaniam D., Thombre R., Dhar A., Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front. Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M., An S., Wang D., Ji H., Guo X., Wang Z. Activation of PAR4 upregulates p16 through Inhibition of DNMT1 and HDAC2 expression via MAPK signals in esophageal squamous cell carcinoma cells. J. Immunol. Res. 2018;2018:4735752. doi: 10.1155/2018/4735752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie V.K., Li Z., Yan Y., Jia Z., Zuo X., Ju Z., Wang J., Du J., Xie D., Xie K., Wei D. DNA-methyltransferase 1 induces dedifferentiation of pancreatic cancer cells through silencing of Kruppel-Like factor 4 expression. Clin. Cancer Res. 2017;23(18):5585–5597. doi: 10.1158/1078-0432.CCR-17-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y., Wang M., Jia X., Zhu H., Zhi Y., Yuan L. Wnt signaling pathway upregulates DNMT1 to trigger NHERF1 promoter hypermethylation in colon cancer. Oncol. Rep. 2018;40(2):1165–1173. doi: 10.3892/or.2018.6494. [DOI] [PubMed] [Google Scholar]
- 8.Ma T., Li H., Sun M., Yuan Y., Sun L.P. DNMT1 overexpression predicting gastric carcinogenesis, subsequent progression and prognosis: a meta and bioinformatic analysis. Oncotarget. 2017;8(56):96396–96408. doi: 10.18632/oncotarget.21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson K.D., Uzvolgyi E., Liang G., Talmadge C., Sumegi J., Gonzales F.A., Jones P.A. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27(11):2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girault I., Tozlu S., Lidereau R., Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin. Cancer Res. 2003;9(12):4415–4422. [PubMed] [Google Scholar]
- 11.Bai J., Zhang X., Hu K., Liu B., Wang H., Li A., Lin F., Zhang L., Sun X., Du Z., Song J. Silencing DNA methyltransferase 1 (DNMT1) inhibits proliferation, metastasis and invasion in ESCC by suppressing methylation of RASSF1A and DAPK. Oncotarget. 2016;7(28):44129–44141. doi: 10.18632/oncotarget.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welch J.S., Petti A.A., Miller C.A., Fronick C.C., O'Laughlin M., Fulton R.S., Wilson R.K., Baty J.D., Duncavage E.J., Tandon B., Lee Y.S., Wartman L.D., Uy G.L., Ghobadi A., Tomasson M.H., Pusic I., Romee R., Fehniger T.A., Stockerl-Goldstein K.E., Vij R., Oh S.T., Abboud C.N., Cashen A.F., Schroeder M.A., Jacoby M.A., Heath S.E., Luber K., Janke M.R., Hantel A., Khan N., Sukhanova M.J., Knoebel R.W., Stock W., Graubert T.A., Walter M.J., Westervelt P., Link D.C., DiPersio J.F., Ley T.J. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N. Engl. J. Med. 2016;375(21):2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie J., Liu L., Li X., Han W. Decitabine, a new star in epigenetic therapy: the clinical application and biological mechanism in solid tumors. Cancer Lett. 2014;354(1):12–20. doi: 10.1016/j.canlet.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Pruksakorn D., Teeyakasem P., Klangjorhor J., Chaiyawat P., Settakorn J., Diskul-Na-Ayudthaya P., Chokchaichamnankit D., Pothacharoen P., Srisomsap C. Overexpression of KH-type splicing regulatory protein regulates proliferation, migration, and implantation ability of osteosarcoma. Int. J. Oncol. 2016;49(3):903–912. doi: 10.3892/ijo.2016.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaiyawat P., Klangjorhor J., Settakorn J., Champattanachai V., Phanphaisarn A., Teeyakasem P., Svasti J., Pruksakorn D. Activation status of receptor tyrosine kinases as an early predictive marker of response to chemotherapy in osteosarcoma. Transl. Oncol. 2017;10(5):846–853. doi: 10.1016/j.tranon.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaiyawat P., Sungngam P., Teeyakasem P., Sirikaew N., Klangjorhor J., Settakorn J., Diskul-Na-Ayudthaya P., Chokchaichamnankit D., Srisomsap C., Svasti J., Pruksakorn D. Protein profiling of osteosarcoma tissue and soft callus unveils activation of the unfolded protein response pathway. Int. J. Oncol. 2019;54(5):1704–1718. doi: 10.3892/ijo.2019.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaiyawat P., Pruksakorn D., Pipatwattana P., Phanphaisarn A., Teeyakasem P., Klangjorhor J., Settakorn J. Endoplasmic reticulum protein 29 (ERp29) as a novel prognostic marker and tumor suppressor in osteosarcoma. J. Bone Oncol. 2019;16 doi: 10.1016/j.jbo.2019.100233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadav B., Wennerberg K., Aittokallio T., Tang J. Searching for Drug Synergy In Complex Dose-Response Landscapes Using An Interaction Potency Model. Comput. Struct. Biotechnol. J. 2015;13:504–513. doi: 10.1016/j.csbj.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ianevski A., He L., Aittokallio T., Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2017;33(15):2413–2415. doi: 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Lu H., Fan G., He M., Sun Y., Xu K., Shi F. A novel interplay between HOTAIR and DNA methylation in osteosarcoma cells indicates a new therapeutic strategy. J. Cancer Res. Clin. Oncol. 2017;143(11):2189–2200. doi: 10.1007/s00432-017-2478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y.K., Guo Y.H. MiR-139-5p suppresses osteosarcoma cell growth and invasion through regulating DNMT1. Biochem. Biophys. Res. Commun. 2018;503(2):459–466. doi: 10.1016/j.bbrc.2018.04.124. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Cheng C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am. J. Cancer Res. 2018;8(1):81–90. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Chaiyawat P., Settakorn J., Sangsin A., Teeyakasem P., Klangjorhor J., Soongkhaw A., Pruksakorn D. Exploring targeted therapy of osteosarcoma using proteomics data. Onco. Targets Ther. 2017;10:565–577. doi: 10.2147/OTT.S119993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun W., Sanderson P.E., Zheng W. Drug combination therapy increases successful drug repositioning. Drug Discov. Today. 2016;21(7):1189–1195. doi: 10.1016/j.drudis.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jabbour E., Issa J.P., Garcia-Manero G., Kantarjian H. Evolution of decitabine development: accomplishments, ongoing investigations, and future strategies. Cancer. 2008;112(11):2341–2351. doi: 10.1002/cncr.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plumb J.A., Strathdee G., Sludden J., Kaye S.B., Brown R. Reversal of drug resistance in human tumor xenografts by 2'-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60(21):6039–6044. [PubMed] [Google Scholar]
- 27.Fang F., Balch C., Schilder J., Breen T., Zhang S., Shen C., Li L., Kulesavage C., Snyder A.J., Nephew K.P., Matei D.E. A phase 1 and pharmacodynamic study of decitabine in combination with carboplatin in patients with recurrent, platinum-resistant, epithelial ovarian cancer. Cancer. 2010;116(17):4043–4053. doi: 10.1002/cncr.25204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Y. Li, W. Hu, D.Y. Shen, J.J. Kavanagh, S. Fu, Azacitidine enhances sensitivity of platinum-resistant ovarian cancer cells to carboplatin through induction of apoptosis, Am. J. Obstet. Gynecol. 200(2) (2009) 177 e1-9.r. [DOI] [PubMed]
- 29.Matei D., Fang F., Shen C., Schilder J., Arnold A., Zeng Y., Berry W.A., Huang T., Nephew K.P. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72(9):2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]