Abstract
Background:
Non-small cell lung carcinoma (NSCLC) is a common malignant tumor with poor prognosis. CircRNA-100876 has been considered to be involved in NSCLC. However, the mechanism by which circRNA_100876 mediated the progression of NSCLC remains unclear.
Methods:
CCK8 assay and immunofluorescence were used to detect cell proliferation. Flow cytometry and transwell assay were performed to analyze cell apoptosis, migration and invasion, respectively. Verification of possible target for circRNA_100876 and related miR-636 were done using luciferase assay. In addition, western blot was performed to detect the protein expressions in NSCLC cells.
Results:
Silencing of circRNA_100876 notably inhibited the proliferation of NSCLC cells. Moreover, downregulation of circRNA_100876 significantly induce the apoptosis of NSCLC cells via mediation of apoptosis-related proteins. In addition, silencing of circRNA_100876 significantly inhibited migration and invasion of NSCLC cells. MiR-636 was the downstream target of circRNA_100876. Meanwhile, RET was the direct target of miR-636. Finally, circRNA_100876 shRNA2 notably suppressed the progression of NSCLC through PI3K/Akt signaling.
Conclusion:
CircRNA_100876 knockdown notably suppressed the progression of NSCLC through regulation of miR-636/RET axis, which may serve as a potential target for treatment of NSCLC.
Keywords: NSCLC, circRNA_100876, miR-636, RET
Introduction
According to the global cancer statistics report in 2018, lung cancer ranks first among the malignant tumors with high incidence and mortality all over the world.1 Lung cancers can be generally divided into 2 groups, non-small cell lung carcinoma (NSCLC) and small cell lung carcinoma (SCLC), based on the difference in morphology of tumor cells.2 Most lung cancers (80-85%) belong to NSCLC in which the predominately histological subtypes are adenocarcinoma (ADC, 40-50%) and squamous cell carcinoma (SqCC, 20-30%).3-5 In the real world, only a few patients with NSCLC can undergo surgery as they were in an early stage after diagnosis, while majority of NSCLC patients (more than 60%) have no choice but to receive conventional chemotherapy and radiotherapy with poor prognosis.2 Currently, targeted therapy as well as immunotherapy have become promisingly strategies for advanced NSCLC. The utilize of biomarkers that indicated aberrant internal environment of human beings have raised a multitude of concerns as well.2
Circular RNAs (circRNAs) are a class of endogenous single-stranded RNAs that can form a covalently closed loop without polarity end of 5’ and 3’ compared with linear RNAs, and thus are much more stable and hardly to be degraded.6 Furthermore, studies have revealed that circRNAs are rich in the binding sites of microRNA (miRNA). For instance, in one particular circRNA, scientists have found more than 70 binding sites for miR-7.7,8 As a result of the above evidence, when compared with linear RNAs, circRNAs can be considered as a more effective sponges of miRNAs, small non-coding RNAs that could interfere translation of message RNAs (mRNA), and involved in various biological processes.9,10 The relation of circRNAs dysregulation through circRNA-miRNA-mRNA axis has been widely studied while circRNAs have also been proved to play an important role especially in carcinogenesis and tumor development in recent studies.11-16 Meanwhile, Yao et al indicated that circRNA_100876 was closely associated with prognosis of NSCLC.17 Based on this recent finding, we aimed to investigate the biological function of circRNA_100876 in NSCLC cells and its underlying mechanism in regulating the development of NSCLC.
Materials and Methods
Cell Culture
Two human NSCLC cell lines (A549 and NCI-H23) and 293 T cell lines were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) with 10% FBS (Thermo Fischer Scientific), 1% penicillin and streptomycin (Thermo Fisher Scientific) at 37°C, 5% CO2.
Cell Transfection
Lentiviral expressing short-hairpin RNA (shRNA1 or shRNA2) directed target circRNA_100876 and one nontargeting sequence (negative control) were obtained from Hanbio Biotechnology Co., Ltd (Shanghai, China). Next, circRNA_100876 shRNA1 or shRNA2 was packaged into lentiviruses. Then the lentiviral vector DNAs were then transfected into 293 T cells including lenti-circRNA_100876 shRNAs and negative control (NC). After transfection, the cells were incubated at 32°C, and then the supernatant was collected. After that, supernatants of 2 circRNA_100876 shRNAs and negative control were filtered into particles. Finally, NSCLC cells were infected with lentiviral particles according to the manufactures’ protocol. After 48 h of incubation, stable NSCLC cells were then selected by puromycin (2.5 μg/mL, Sigma Aldrich, St. Louis, MO, USA). qRT-PCR assay were used to verify the efficiency of transfection.
For miR-636 transfection, A549 or NCI-H23 cells were transfected with miR-636 agonist, miR-636 antagonist or NC by Lipofectamine 2000 according to the previous reference.18 MiR-636 agonist, miR-636 antagonist and negative control RNAs were purchased from GenePharma (Shanghai, China). The efficiency of transfection was detected by qRT-PCR.
Quantitative Real-Time-PCR
CCK-8 Assay
Cell counting kit -8 (CCK-8) (Dojindo, Kumamoto, Japan) assay was used to detect cell proliferation. A549 or NCI-H23 cells were transfected with NC or circRNA_100876 shRNA2 for 0, 24, 48, and 72 h, respectively. According to the manufacturer’s protocol, 10 µL CCK8 reagent was added in each well and later cells were incubated for 2 hours before analyzing their absorbance at 450 nm in the dark.
Immunofluorescence
NSCLC cells were seeded in 24-well plates overnight. Then, cells were treated nothing (Blank), negative control (NC) or circRNA_100876 shRNA2 for 72 h. Next, cells were blocked with 10% goat serum for 30 min at room temperature and then incubated with anti-Ki67 antibody (Abcam; 1:1000) at 4°C overnight, followed by incubation with goat anti-rabbit IgG (Abcam; 1:5000) at 37°C for 1 h. Then, the nuclei were stained with DAPI (Beyotime, Shanghai, China) for 5 min. Finally, cells were were observed under a fluorescence microscope (Olympus CX23, Tokyo, Japan).
Cell Apoptosis Analysis
NSCLC cells were trypsinized, washed with phosphate buffered saline and resuspended in Annexin V Binding Buffer, followed by staining with 5 µl FITC and 5 µl propidium (PI) in the system for 15 min. Cells were analyzed using flow cytometer (BD, Franklin Lake, NJ, USA) to test the cell apoptosis rate. Western blot analysis
Total protein was isolated from cell lysates by using RIPA buffer, and quantified by BCA protein assay kit (Beyotime, Shanghai, China). Proteins were resolved on 10% SDS-PAGE, then transferred to PVDF (Bio-Rad) membranes. After blocking, the membranes were incubated with primary antibodies at 4°C overnight, then incubated with secondary anti-rabbit antibody (Abcam; 1:5000) at room temperature for 1 h. Membranes were scanned by using an Odyssey Imaging System and analyzed with Odyssey v2.0 software (LICOR Biosciences, Lincoln, NE, USA). Then, the primary antibodies used in this study as follows: anti-cleaved caspase 3 (1:1000), anti-Bcl-2 (1:1000), anti-MMP2 (1:1000), anti-RET (1:1000), anti-p-Akt (1:1000), anti-cleaved PARP (1:1000), anti-Akt (1:1000), anti-p-ERK (1:1000), anti-ERK (1:1000) and anti-β-actin (1:1000).
Measurement of Mitochondrial Membrane Potential (MMP) (ΔΨm)
Changes in mitochondrial membrane potential (MMP) were measured with JC-1 (5,5’,6,6’-Tetrachloro-1,1’,3,3’-tetraethyl-imidacarbocyanine) staining on the FACSan flow cytometer. Briefly, cells were plated into 6-well plates (3 × 10 5 cells/well) overnight. Cells were harvested, washed twice with PBS, and then resuspended in 0.5 Ml of complete medium containing 2 μM JC-1 at 37°C for 20 min. Finally, the data were analyzed using the flow cytometer in the FL1 and FL2 channels.
Transwell Assay
Briefly, 24-well Transwell plates (Corning, New York, NY, USA) were used for cell invasion and migration detection. For the cell migration assay, 2 × 10 5 A549 cells were seeded into the upper chambers of the 24-well plates in 200 µL of serum-free RPMI 1640 medium supplemented with 0.2% bovine serum albumin. The lower chambers contained RPMI 1640 medium supplemented with 1% FBS. After 24 h of incubation at 37°C, the non-migrating cells were gently removed from the upper side of each chamber with a cotton swab, while the cells that had migrated were fixed with 95% alcohol for 10 min and stained with 1% crystal violet (Sigma, Grand Island, NY, USA) for 5 min. Finally, images were captured, and the cells were counted under an inverted light microscope (Olympus) at 400x magnification.
For the invasion assay, the upper chambers of the 24-well plates were pretreated with 50 µL of Matrigel (12.5 mg/L), and the wells were pretreated with Matrigel (BD Biosciences, Franklin Lake, NJ, USA). Then, A549 cells (1 × 10 6 cells/ml) in FBS-free medium were seeded into the upper chambers. The lower chambers contained RPMI 1640 medium supplemented with 1% FBS. The cells were incubated at 37°C for 24 h, and cells that had attached to the underside of the membrane were fixed and stained with a 0.1% crystal violet solution. Finally, images were captured, and the number of invading cells was counted under a microscope at 400x magnification.
Dual Luciferase Activity Assay
The partial squences of circRNA_100876 and 3′-UTR of RET containing the putative binding sites of miR-636 were synthetized and obtained from Sangon Biotech (Shanghai, China), then were cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vectors (Promega, Madison, WI, USA) to construct wild-type reporter vectors circRNA_100876 (WT) and RET (WT), respectively. The mutant circRNA_100876 sequences and 3′-UTR of RET sequences containing the putative binding sites of miR-636 were performed by Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA, USA) and then cloned into pmirGLO vectors respectively, to construct mutant-type reporter vectors circRNA_100876 (MUT) and RET (MUT). The circRNA_100876 (WT) or circRNA_100876 (MUT) were transfected into 293 T cells together with control, vector-control (NC) or miR-636 agonist using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Similarly, the RET (WT) or RET (MUT) was transfected into 293 T cells together with control, vector-control (NC) or miR-636 agonist. The relative luciferase activity was analyzed by the Dual-Glo Luciferase Assay System (Promega).
Statistical Analysis
All data were shown in the formation of mean ± standard deviation (SD) using GraphPad Prism 7 software (La Jolla, CA, USA). One-way ANOVA followed by tukey’ test was used to make comparison between groups. Significant differences were considered when P value was less than 0.05.
Results
Silencing of circRNA_100876 Significantly Restrained Proliferation of NSCLC Cells
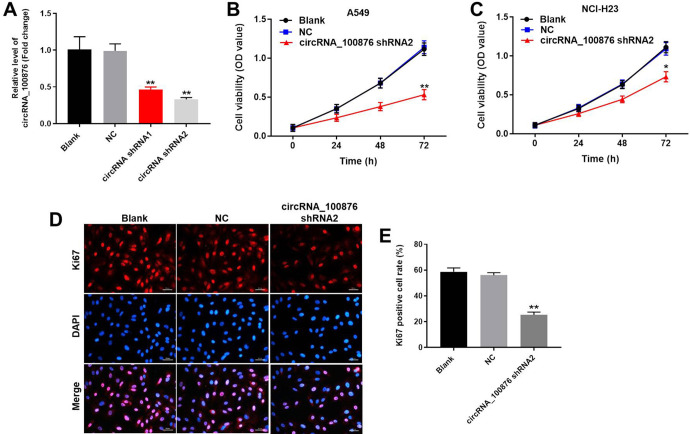
In order to evaluate the functional role of circRNA_100876 in tumorigenesis of NSCLC, A549 cells and NCI-H23 cells were transfected with circRNA_100876 shRNA. As showed in Figure 1A, the expression of circRNA_100876 in NSCLC cells was significantly inhibited by downregulation of circRNA_100876. This data confirmed that circRNA_100876 shRNA was stably transfected into NSCLC cells. Moreover, circRNA_100876 shRNA2 exhibited more significant changes. Based on this result, we used circRNA_100876 shRNA2 in following experiments. Next, to verify the effect of circRNA_100876 on proliferation of NSCLC cells, CCK-8 assay was performed. The results indicated that silencing of circRNA_100876 notably inhibited the cell growth of NSCLC (Figure 1B and 1C). Since A549 cells exhibited more obvious changes on cell proliferation, it was selected of use in following experiments. In addition, ki-67 positive rate of NSCLC cells was notably decreased by circRNA_100876 silencing (Figure 1D and 1E). Consequently, it can be considered that downregulation of circRNA_100876 could restrain cell proliferation of NSCLC.
Figure 1.
Silencing of circRNA_100876 significantly restrained proliferation of NSCLC cells. (A) NSCLC cells were transfected with circRNA_100876 shRNA1, circRNA_100876 shRNA2 or negative control (NC) respectively for 48 hours. Then, the transfection efficiency was measured using qRT-PCR. (B) A549 cells were transfected with circRNA_100876 shRNA2 or NC for 0, 24, 48 and 72 h, respectively. Then, cell viability was tested using CCK8 assay. (C) NCI-H23 cells were infected with circRNA_100876 shRNA2 or NC for 0, 24, 48 and 72 h, respectively. Then, cell viability was analyzed using CCK8 assay. (D, E) NSCLC cells were infected with circRNA_100876 shRNA2 or NC for 48 hours. Ki-67 staining assay was used to detect the cell proliferation Red immunofluorescence indicates Ki-67. Blue immunofluorescence indicates DAPI. *P < 0.05. **P < 0.01 compared to control.
CircRNA_100876 Knockdown Induced Apoptosis of NSCLC Cells
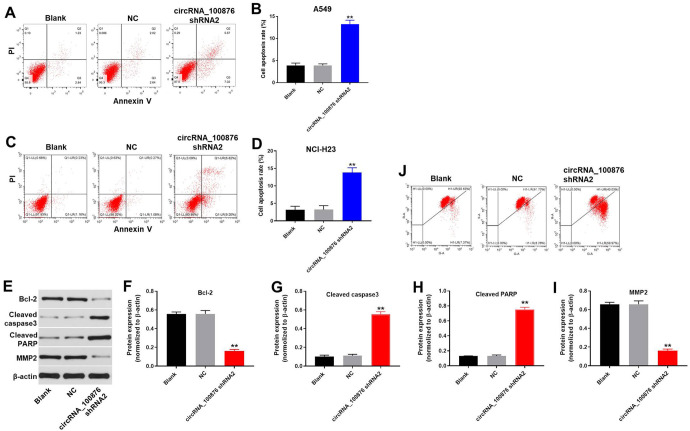
Flow cytometric assay was used to test cell apoptosis. As indicated in Figure 2A-2D, downregulation of circRNA_100876 notably induced the apoptosis of NSCLC cells. In addition, western blot was applied to evaluate protein expressions in NSCLC cells. Results suggested that the expressions of Bcl-2 and MMP-2 in NSCLC cells were greatly decreased by circRNA_100876 shRNA2. In contrast, downregulation of circRNA_100876 obviously increased the protein levels of cleaved caspase 3 and cleaved PARP (Figure 2E-2I). Moreover, silencing of circRNA_100876 notably induced mitochondrial injury in NSCLC cells (Figure 2 J). All these data had suggested that silencing of circRNA_100876 could induce apoptosis of NSCLC cells.
Figure 2.
CircRNA_100876 knockdown induced apoptosis of A549 cells. A549 cells were transfected with circRNA_100876 shRNA2 or NC for 72 h. (A, B, C, D) Cells were double stained with Annexin V-PI to distinguish apoptotic cells from normal cells using flow cytometry. (E) Protein expression of Bcl-2, cleaved PARP and cleaved caspase 3 in A549 cells were determined using western blot. (F) Relative expression of Bcl-2 was quantified via normalizing to β-actin. (G) Relative expression of cleaved caspase 3 was quantified via normalizing to β-actin. (H) Relative expression of cleaved PARP was quantified via normalizing to β-actin. (I) Relative expression of MMP2 was quantified via normalizing to β-actin. (J) Changes in mitochondrial membrane potential (MMP) were measured with JC-1 staining on the FACSan flow cytometer. **P < 0.01 compared to control.
Silencing circRNA_100876 Greatly Inhibited Migration and Invasion of NSCLC Cells
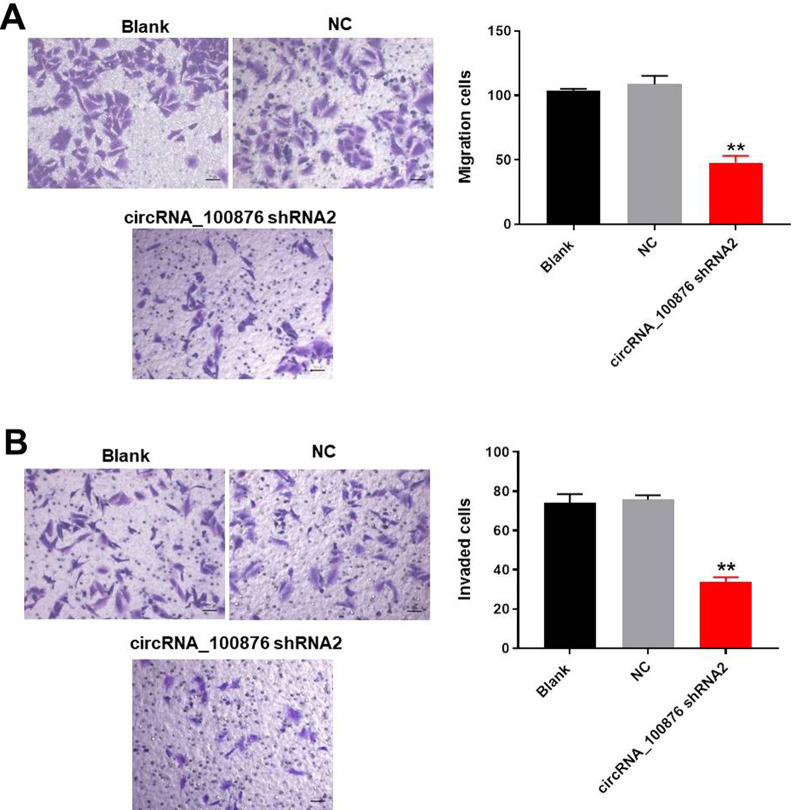
For the purpose of circRNA_100876 on cell migration and invasion of NSCLC, transwell assay was used. As demonstrated in Figure 3A and B, cirRNA_100876 shRNA2 significantly inhibited migration and invasion of A549 cells. Taken together, downregulation of circRNA_100876 could notably inhibit the progression of NSCLC in vitro.
Figure 3.
Silencing of circRNA_100876 greatly inhibited migration and invasion of NSCLC cells. A549 cells were transfected with circRNA_100876 shRNA2 or NC for 24 h. (A) Transwell migration assay was used to analyze the migration ability of NSCLC cells. (B) Transwell invasion assay was used to analyze the invasion ability of NSCLC cells. **P < 0.01 vs. NC group.
MiR-636 was a Downstream Target of circRNA_100876
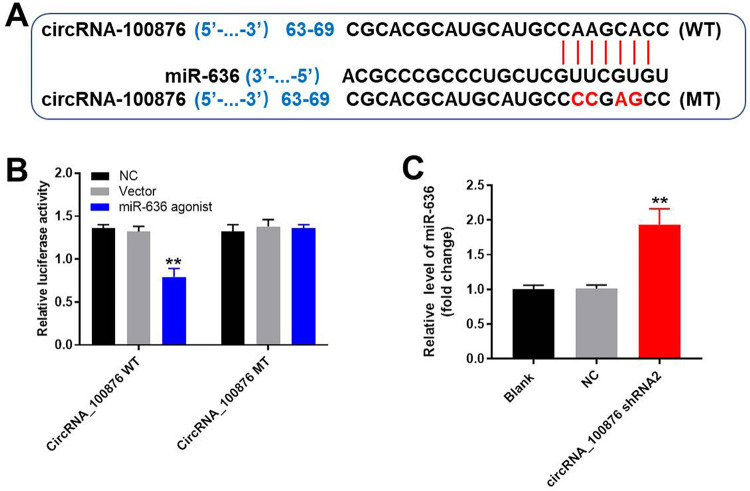
According to the prediction from the database, CircInteractome, luciferase activity assay was used to verify the relationship between miR-636 and circRNA_100876. As revealed in Figure 4A and 4B, miR-636 might be the downstream target of circRNA_100876. In addition, the result of qRT-PCR indicated that the expression of miR-636 in NSCLC cells was greatly enhanced in the presence of circRNA_100876 shRNA2 (Figure 4C). Based on these data, it can be concluded that miR-636 was a downstream target of circRNA_100876.
Figure 4.
MiR-636 was a downstream target for circRNA_100876. (A) Gene structure of wild type circRNA_100876 that indicated the predicted site which targeted miR-636 in its 3’UTR as well as the gene structure of mutant type circRNA_100876. (B) Luciferase activity was measured after co-transfecting with circRNA_100876 plasmid and miR-636 agonist. (C) A549 cells were transfected with circRNA_100876 shRNA2 or NC for 24 h. Relative expression of miR-636 in NSCLC cells was measured using qRT-PCR. **P < 0.01 compared to control.
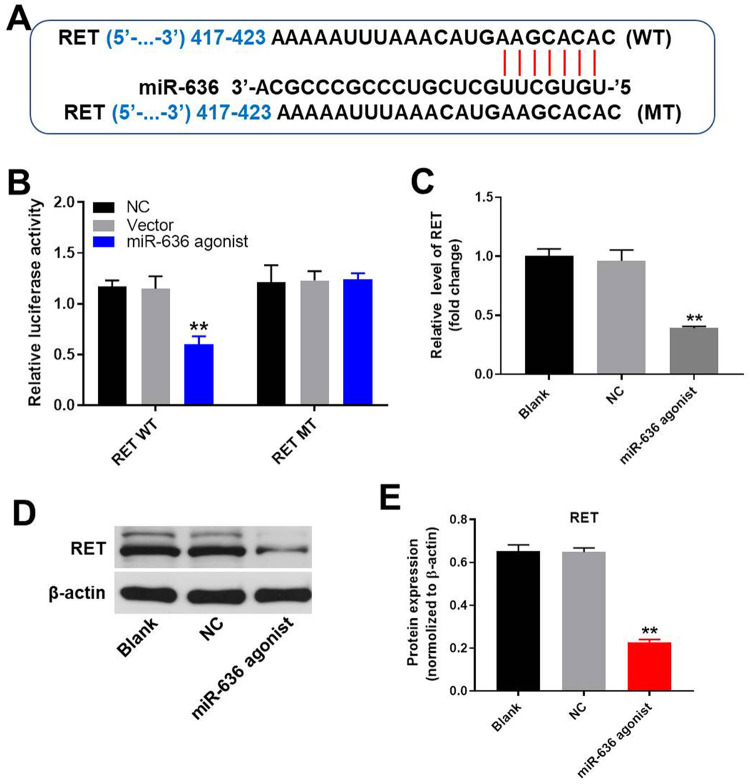
RET Was a Direct Target of miR-636
To further explore the target of miR-636, targetscan and dual luciferase report assay were performed. As indicated in Figure 5A and B, RET might be the direct target of miR-636. Moreover, the expression of RET in NSCLC cells was greatly decreased in the presence of miR-636 agonist (Figure 5C). In addition, the result of western blot revealed that the expression of RET in NSCLC cells was significantly decreased by overexpression of miR-636 antagonist, further indicating that RET was the direct target of miR-636 (Figure 5D and E). All these results showed that circRNA_100876 mediated the progression of NSCLC via regulation of miR-636/RET axis.
Figure 5.
RET was the direct target for miR-636. (A) Gene structure of wild type RET that indicated the predicted site which targeted miR-636 in its 3’UTR as well as the gene structure of mutant type RET. (B) Luciferase activity was measured after co-transfecting with RET plasmid and miR-636 agonist. (C) A549 cells were transfected with NC or miR-636 agonist for 24 h. Relative expression of RET in A549 cells was measured by qRT-PCR. (D) The protein level of RET in A549 cells was determined by western blot. (E) The relative expression of RET protein was quantified via normalizing to β-actin. **P < 0.01 compared to control.
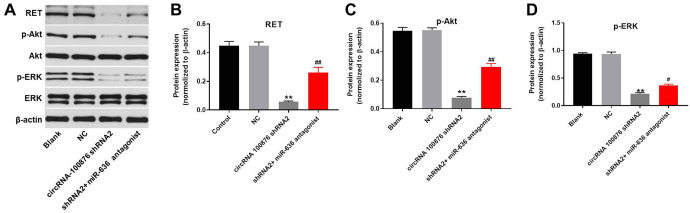
Downregulation of circ_100876 Inhibited the Progression of NSCLC via Inactivation PI3K/Akt Signaling Pathway
To further explore the mechanism by which circRNA_100876 modulated the progression of NSCLC, western blot was performed. As revealed in Figure 6A and B, the expression of RET in NSCLC cells was significantly decreased by knockdown of circRNA_100876, which was partially rescued by miR-636 antagonist. Moreover, downregulation of circRNA_100876 significantly inhibited the expression of p-Akt and p-ERK in NSCLC cells. However, the inhibitory effect of circRNA_100876 shRNA2 on these 2 proteins was significantly reversed by miR-636 downregulation (Figure 6A, C, and D). Altogether, circRNA_100876 promoted the tumorigenesis of NSCLC via activation of PI3K/Akt signaling pathway.
Figure 6.
Downregulation of circRNA_100876 inhibited the progression of NSCLC via inactivation of PI3K/Akt signaling pathway. A549 cells were transfected with circRNA_100876 shRNA2 or/and miR-636 antagonist for 72 h. Then, (A) the protein level of RET, p-Akt, Akt, p-ERK, ERK in A549 cells were measured using western blot. (B, C, D) The relative protein expressions were quantified via normalizing to β-actin. **P < 0.01 compared to control. #P < 0.05, ##P < 0.01 compared to circRNA_100876 shRNA2.
Discussion
NSCLC accounts for 80-85% of lung cancer, most NSCLC patients were found to be in the late stage with a low 5-year survival rate.3,4,19 The development and progress of NSCLC are associated with numerous complex signaling pathways,20 thus, biomarkers that involved in these complex signaling pathways are of great importance in subclassification of NSCLC as well as disease surveillance.20-22 CircRNAs are kinds of small endogenous RNAs related with carcinogenesis and tumor progression, which have been studied for the potential of being the prognosis tools.11-16 Yao et al., found that the expression level of a specific circRNA, circRNA_100876, was significantly elevated in pathological tissues of NSCLC patients, which was also correlated to metastasis of lymph node and tumor staging.17 As a result of it, we studied the functional role of circRNA_100876 and demonstrated its effect on cell proliferation, apoptosis and migration as well as its internal mechanism in NSCLC cells.
Our results have proved that silencing circRNA_100876 could restrain the development of NSCLC tumor cells in aspect of cell proliferation, migration and invasion. In addition, the apoptotic rate of NSCLC cells was significantly increased after downregulating circRNA_100876. Since the above considerable evidence had indicated the functional role circRNA_100876, we further investigated its targets and tried to build a reliable relationship between circRNA_100876 and related miRNA. According to our results, miR-636 was the downstream target for circRNA_100876 that could mediate the expression of RET which could activate carcinogenic events.23 Therefore, an axis of circRNA_100876 and miR-636 and RET could be constructed to better elucidate the role of circRNA_100876 in NSCLC development.
Actually, some studies had reported the functional role that circRNA_100876 played in other cancer diseases as well. For instance, in addition to the lung cancer, Cao and colleagues had found that poor prognosis in esophageal squamous cell carcinoma is related to overexpression of circRNA_100876.24 Yang’s research revealed that circRNA_100876 could promote proliferation and metastasis of breast cancer cells and further confirmed the internal role of its target miRNA, microRNA-361-3p.25 Last but not least, silencing circRNA_100876 could also suppress development of osteosarcoma cancer cells via acting as the sponge form of miRNA-136.26
On the basis of these former studies as well as the results shown in this paper, we could come to the conclusion that circRNA_100876 is a potential biomarker for disease prognosis, as upregulation of circRNA_100876 might indicate possible disease progress. In addition, the internal mechanism of circRNA_100876 was verified, which could modulate several kinds of miRNAs. Our study had further discovered a downstream target, RET, for miR-636 that involved in the progress of NSCLC.
As for the gene which encodes the receptor-tyrosine kinase RET (Rearranged during transfection), has been reported to serve as the actionable drivers of oncogenesis via rearrangement and mutation.23,27 Several multikinase inhibitors which could restrain the expression of RET have confirmed their therapeutic efficacy in RET-rearranged NSCLCs.28-31 However, due to the competitive relationship between RET and other non-RET kinases that resulting in sub-optimal pharmacokinetic properties and toxicities problems,32-34 current advancements could not satisfy with the treatment needs. Therefore, more molecular drugs that specific for RET-related lung diseases are needed. As shown in this paper, circRNA_100876 could sponge miR-636 to modulate the expression of RET. Thus, it is possible to further investigate its specificity for RET. If inhibiting the expression level of circRNA_100876 can only restrain the expression of RET and have no influence on other non-RET kinases, circRNA_100876 could not only be used as a biomarker for indicating aberrant internal environment, but its inhibitors could also become promisingly therapeutic target specific for RET-rearranged NSCLCs. Meanwhile, according to previous references,35,36 downregulation of RET may lead to inactivation of PI3K/Akt pathway. In addition, downregulation of PI3K/Akt can result in cell apoptosis.37,38 Thus, it can be concluded that knockdown of circ100876 induced apoptosis of NSCLC cells via indirectly targeting RET.
In general, our data revealed that inhibition of circRNA_100876 could suppress the progression of NSCLC via sponging miR-636, which may serve as a new target for treatment of NSCLC.
Footnotes
Authors’ Note: Jianxiang Song and Woda Shi contributed equally. Our study does not include animal or human studies.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Wencai Wang  https://orcid.org/0000-0002-1193-8702
https://orcid.org/0000-0002-1193-8702
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Osmani L, Askin F, Gabrielson E, Li QK, Current WHO. guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52(pt 1):103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 4. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. 2015;10(9):1240–1242. [DOI] [PubMed] [Google Scholar]
- 5. Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. [DOI] [PubMed] [Google Scholar]
- 6. Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64(3):607–613. [DOI] [PubMed] [Google Scholar]
- 7. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- 8. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- 9. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding XC, Weiler J, Grosshans H. Regulating the regulators: mechanisms controlling the maturation of microRNAs. Trends Biotechnol. 2009;27(1):27–36. [DOI] [PubMed] [Google Scholar]
- 11. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xuan L, Qu L, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8(2):932–939. [PMC free article] [PubMed] [Google Scholar]
- 14. Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 15. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. [DOI] [PubMed] [Google Scholar]
- 16. Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. [DOI] [PubMed] [Google Scholar]
- 18. Zhang S, Liu C, Zhang X. Mitochondrial damage mediated by miR-1 overexpression in cancer stem cells. Mol Ther Nucleic Acids. 2019;18:938–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(suppl 5): v1–v27. [DOI] [PubMed] [Google Scholar]
- 20. Zupa A, Improta G, Silvestri A, et al. A pilot characterization of human lung NSCLC by protein pathway activation mapping. J Thorac Oncol. 2012;7(12):1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buhrens RI, Amelung JT, Reymond MA, Beshay M. Protein expression in human non-small cell lung cancer: a systematic database. Pathobiology. 2009;76(6):277–285. [DOI] [PubMed] [Google Scholar]
- 22. Yanagisawa K, Shyr Y, Xu BJ, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362(9382):433–439. [DOI] [PubMed] [Google Scholar]
- 23. Eng C. RET proto-oncogene in the development of human cancer. J Clin Oncol. 1999;17(1):380–393. [DOI] [PubMed] [Google Scholar]
- 24. Cao S, Chen G, Yan L, Li L, Huang X. Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:7385–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang CY, Zhang FX, He JN, Wang SQ. CircRNA_100876 promote proliferation and metastasis of breast cancer cells through adsorbing microRNA-361-3p in a sponge form. Eur Rev Med Pharmacol Sci. 2019;23(16):6962–6970. [DOI] [PubMed] [Google Scholar]
- 26. Jin J, Chen A, Qiu W, et al. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. J Cell Biochem. 2019;120(9):15678–15687. [DOI] [PubMed] [Google Scholar]
- 27. Drilon A, Hu ZI, Lai GGY, Tan DSW. Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat Rev Clin Oncol. 2018;15(3):151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol. 2016;17(12):1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee SH, Lee JK, Ahn MJ, et al. Vandetanib in pretreated patients with advanced non-small cell lung cancer-harboring RET rearrangement: a phase II clinical trial. Ann Oncol. 2017;28(2):292–297. [DOI] [PubMed] [Google Scholar]
- 30. Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med. 2017;5(1):42–50. [DOI] [PubMed] [Google Scholar]
- 31. Velcheti V, Hida T, Reckamp KL, Yang JC, Tamura T. Phase 2 study of lenvatinib in patients with RET fusion-positive adenocarcinoma of the lung. Eur J Cancer. 2017;72(1): S178. [DOI] [PubMed] [Google Scholar]
- 32. Yakes FM, Chen J, Tan J, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011;10(12):2298–2308. [DOI] [PubMed] [Google Scholar]
- 33. Carlomagno F, Vitagliano D, Guida T, et al. ZD6474, an orally available inhibitor of KDR tyrosine kinase activity, efficiently blocks oncogenic RET kinases. Cancer Res. 2002;62(24):7284–7290. [PubMed] [Google Scholar]
- 34. Wedge SR, Ogilvie DJ, Dukes M, et al. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res. 2002;62(16):4645–4655. [PubMed] [Google Scholar]
- 35. Subramonian D, Phanhthilath N, Rinehardt H, et al. Regorafenib is effective against neuroblastoma in vitro and in vivo and inhibits the RAS/MAPK, PI3K/Akt/mTOR and Fos/Jun pathways. Br J Cancer. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alqahtani T, Kumarasamy VM, Huczynski A, Sun D. Salinomycin and its derivatives as potent RET transcriptional inhibitors for the treatment of medullary thyroid carcinoma. Int J Oncol. 2020;56(1):348–358. [DOI] [PubMed] [Google Scholar]
- 37. Yuan L, Qu Y, Li Q, et al. Protective effect of astaxanthin against La2O3 nanoparticles induced neurotoxicity by activating PI3K/AKT/Nrf-2 signaling in mice. Food Chem Toxicol. 2020; 144: 111582. [DOI] [PubMed] [Google Scholar]
- 38. Yao X, Tu Y, Xu Y, Guo Y, Yao F, Zhang X. Endoplasmic reticulum stress-induced exosomal miR-27a-3p promotes immune escape in breast cancer via regulating PD-L1 expression in macrophages. J Cell Mol Med. 2020. 10.1111/jcmm.15367. [DOI] [PMC free article] [PubMed] [Google Scholar]