Abstract
Selective recruitment and concentration of signaling proteins within membraneless compartments is a ubiquitous mechanism for subcellular organization1–3. The dynamic flow of molecules into and out of these compartments occurs on faster timescales than for membrane-enclosed organelles, presenting a possible mechanism to control spatial patterning within cells. Here, we combined single-molecule tracking and super-resolution microscopy, light-induced subcellular localization, reaction-diffusion modeling, and a spatially-resolved promoter activation assay to study signal exchange in and out of the 200 nm cytoplasmic PopZ microdomain at the cell pole of the asymmetrically dividing bacterium Caulobacter crescentus4–8. Two phospho-signaling proteins, the transmembrane histidine kinase CckA and the cytoplasmic phosphotransferase ChpT, provide the only phosphate source for the cell fate-determining transcription factor CtrA (Fig. 1a)9–18. We found that all three proteins exhibit restricted rates of entry into and escape from the microdomain and enhanced phospho-signaling within, leading to a submicron gradient of activated CtrA~P19 that is stable and sublinear. Entry into the microdomain is selective for cytosolic proteins and requires a binding pathway to PopZ. Our work demonstrates how nanoscale protein assemblies can modulate signal propagation with fine spatial-resolution, and that in Caulobacter, this modulation serves to reinforce asymmetry and differential cell fate of the two daughter cells.
Keywords: bacterial membraneless microdomains, polarity, phospho-signaling pathways, single-molecule tracking, reaction-diffusion modeling
In the asymmetrically dividing bacterium Caulobacter crescentus, the cytoplasmic, intrinsically disordered, multivalent, negatively-charged protein PopZ establishes a space-filling ~100–200 nm microdomain at the cell poles. The PopZ microdomain forms a curved interface against the cytoplasmic membrane and a membraneless interface with the cytoplasm5–7,20–22. While the poles of the stalked and swarmer daughter cells rapidly remodel following cytokinesis, the compositions and functions of the two poles are distinct even prior to division8. In the predivisional cell, CckA acts as a kinase exclusively at the new pole while acting as a phosphatase elsewhere18 (Fig. 1a), suggesting that the activity of the signaling pathway leading to an activated CtrA~P transcription factor might also be polarized. Indeed, another function of CtrA~P, the inhibition of DNA replication, was shown to be stronger at the new pole than the old pole of predivisional cells, supporting a mathematical model that showed a gradient of CtrA~P will form given phosphotransfer rates faster than diffusion19. However, a biophysical mechanism by which the multistep CtrA phosphotransfer pathway could accomplish such rapid biochemical rates relative to ChpT and CtrA diffusion has yet been demonstrated.
Figure 1: The phospho-signaling cascade that activates a cell fate transcription factor is sequestered within the PopZ microdomain.
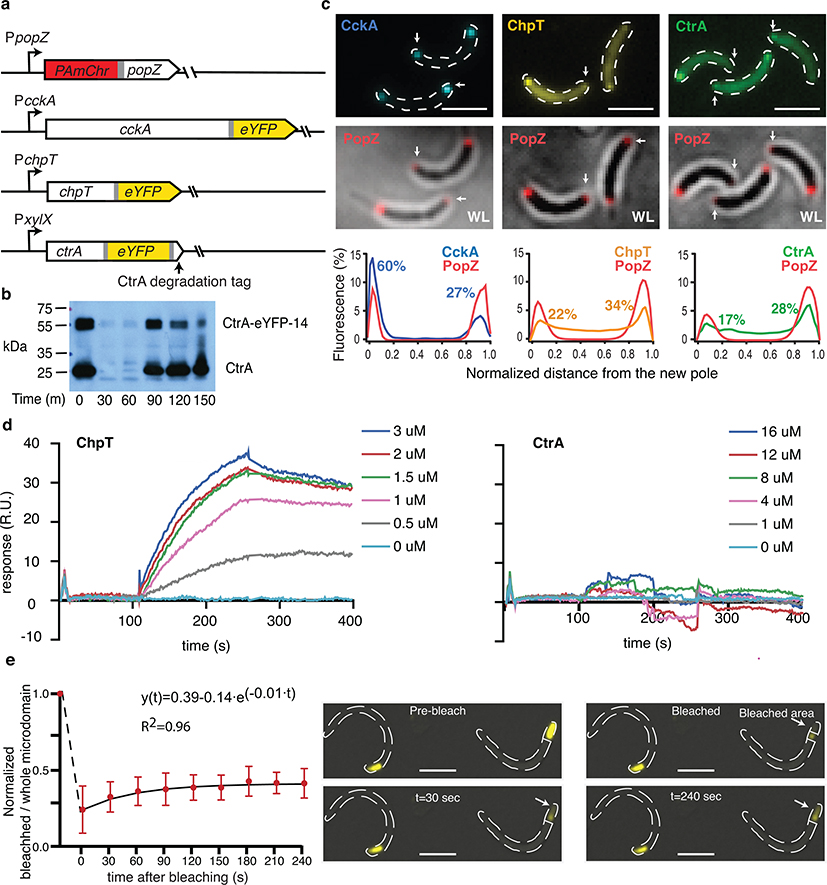
a. Diagram of signal flow and subcellular localization of the CtrA/CckA/ChpT phospho-signaling pathway relative to PopZ. In this report, we provide evidence for a gradient of CtrA~P in the predivisional cell emanating from the new cell pole, as shown in green. b. Average single-molecule distributions of PAmCherry-PopZ, CckA-eYFP, ChpT-eYFP, and CtrA-eYFP-14 at the new poles of predivisional cells (n = 27, 13, 27, and 60 poles respectively) registered within the same coordinates using PopZ as a landmark. Percentages: fraction at pole in diffraction-limited images (Extended Data Fig. 1c). c. Average CckA and PopZ polar distributions using 3D localization data from n = 29 old poles (2006 and 5282 localizations respectively). Slices (200 nm) are shown to emphasize the radial CckA distribution of on the membrane. Plot: the radial distribution of CckA and PopZ from the PopZ centroid with volume-normalized density (errorbars: 95% CI of resampled localizations). d-f. Exemplary 3D single-molecule tracks (time-coded connected dots) relative to super-resolution reconstructions of PopZ (yellow-orange) (Methods). d. Perspective views of CckA molecule motion inside and outside the pole. e. ChpT slowing upon polar entry (left), two views of apparent ChpT membrane-associated motion within the PopZ microdomain (right). f. CtrA slowing upon polar entry (left) and traversing the polar microdomain before escape (right). g. Three-dimensional Mean Square Displacement (3D MSD) curves for CckA tracks within selected cellular regions. h. Log-log MSD plots of CtrA (green) and ChpT (orange) motion along the Caulobacter cell axis, calculated separately in the cell body and poles. Blue line: MSD for simulated free diffusion with D = 1.8 μm2/s (line offset for clarity) (Extended Data Fig. 4b–d). Dotted lines: theoretical limits of observable MSD values within the pole. i. Survival distributions of labeled ChpT and CtrA molecules that either escape from the pole or photobleach. Distributions from N = 434 (77.1% bleaching) and 1149 (80.9% bleaching) events respectively. Blue line: survival distribution for simulated molecules freely diffusing with D = 1.8 μm2/s. Fits accounting for bleaching yielded similar true dwell times (~132 ms) for ChpT and CtrA (dashed line). Shaded areas: 95% confidence intervals calculated from bootstrap analysis. All scale bars: 200 nm.
In this work, we asked whether the dynamics of recruitment and turnover of the CtrA pathway proteins within the PopZ microdomain regulate phosphotransfer efficiency and facilitate a stable spatial gradient of CtrA~P. For the cytosolic ChpT phosphotransfer protein to interact with CckA kinase at the membrane of the new cell pole, it must pass through the space-filling PopZ microdomain. ChpT~P must then phosphorylate CtrA to complete the signaling pathway. Thus, critical questions are if (and how) the PopZ microdomain influences access of ChpT to the CckA kinase, where ChpT~P encounters CtrA, and ultimately, what spatial distribution of CtrA~P is established as it emerges from the new pole microdomain.
We first measured polar co-localization of the members of the CtrA activation pathway with respect to the PopZ microdomain by imaging strains expressing PAmCherry-PopZ alongside eYFP fusions of CckA, ChpT, and CtrA in Caulobacter (Extended Data Fig. 1a). Consistent with previous studies6,13, diffraction-limited microscopy showed that CckA co-localized with PopZ, with 60% of the population residing at the new pole (Extended Data Fig. 1c). We further found that both ChpT-eYFP and a CtrA-eYFP-14 sandwich fusion of CtrA, which mimics the wildtype CtrA cell-cycle degradation profile (Extended Data Fig. 1b), were recruited roughly proportionally to the number of PopZ molecules at each pole (Extended Data Fig. 1c). Surface plasmon resonance experiments showed that ChpT binds directly to PopZ, while CtrA binds to ChpT but not to PopZ (Extended Data Fig. 1d)7,15. These results suggest that ChpT is recruited primarily by PopZ while CtrA is concentrated within the pole by its interaction with pole-localized ChpT (Fig. 2f). To study the dynamics of CckA, ChpT and CtrA within the nanoscale space of the poles, we employed single-molecule tracking combined with super-resolution imaging of PopZ. Using PopZ as a landmark, we generated protein distributions within a shared “polar coordinate system” by averaging localizations from dozens of cells (Fig. 1b, Extended Data Fig. 3a–c). Each of the CtrA pathway members was concentrated and appeared uniformly distributed within the < 200 nm region of the PopZ microdomain, with concentration dropping off sharply away from PopZ.
Figure 2: Entry into the PopZ microdomain is selective and is regulated by binding.
a. Overlay of 92 total 3D single-molecule tracks of fPIF-eYFP shown relative to a PAmCherry-PopZ super-resolution reconstruction in a representative predivisional cell (consistent for the 16 cells analyzed). Scale bar 200 nm. b. Observed polar recruitment of ChpT-eYFP, CtrA-eYFP-14, fPIF-eYFP, and eYFP, quantified by counting single-molecule localizations that appeared within the super-resolved reconstruction of PopZ for (41, 89, 16, 17) cells. For comparison, the relative proportion of localizations in the cell body and in the new and old poles was simulated for molecules freely diffusing throughout the cell volume (Fig. 2b and Extended Data Fig. 4b–d). Errorbars: 95% CI magnitude (statistics based on measurement of 2750, 6289, 4616, 1153 total localizations, respectively). c. Schematic of optogenetic localization experiments using the blue-light-controlled iLID/SspB system. Diffraction-limited imaging indicates RFP-iLID (green) is reversibly bound to PopZ and enters the microdomain (red) only upon activation by blue light. Scale bar: 500 nm. d. Fluorescence intensity profiles of RFP-iLID before (black) and immediately after activation (blue), relative to SspB-CFP-PopZ (red). Shown is mean ± s.d.; n equals 250 cells. e. Fraction of RFP-iLID in the PopZ microdomain as a function of time before and after a 5 s exposure to blue light. The fraction of RFP molecules detected within the PopZ microdomain when not fused to the iLID system is shown in gray. Shown is mean ± s.d.; n equals 130 cells. f. Binding pathways that enable entry into the PopZ microdomain.
Since CckA is integrated into the curved inner membrane, we used an engineered point spread function (the DH-PSF)22 to localize and follow the motion of CckA-eYFP molecules in three dimensions (3D) (Extended Data Fig. 2a–c). At the old pole, where the PopZ microdomain fills a larger volume, we resolved the hemispherical cap of CckA on the membrane area adjacent to PopZ (Fig. 1c, Extended Data Fig. 3a,b). We found that at the old pole, CckA extended uniformly across the membrane alongside the cytoplasmic PopZ microdomain with area Aold = 0.094±0.008 μm2, while CckA at the new pole was confined to an area Anew = 0.021 ± 0.002 μm2 (Supplementary Note 1.4). In previous work we have shown that CckA autokinase activity per molecule varies based on local concentration while phosphatase activity is density-independent18. Using our single-molecule and intensity profiling measurements, we calculated CckA concentrations at the new pole, old pole, and cell body to be ~10,000, ~1,000, and ~9 molecules/μm2, respectively (Supplementary Note 1.4). The high CckA concentration at the smaller new pole microdomain is close to the previously determined CckA concentration on liposomes in vitro18 that leads to maximum autokinase activity. In contrast, the concentration of CckA at the old pole would lead to autokinase activity an order of magnitude lower than that at the new pole, and the concentration away from the poles would lead to virtually no autokinase activity. While the new pole-specific factor DivL is responsible for localization of CckA to the new pole and allosteric modulation of CckA kinase activity23–25, our direct measurement of CckA distributions suggests that the difference in CckA density between the two microdomains adds another layer of regulation to ensure asymmetric signaling.
Next, we characterized the effect of the PopZ microdomain on client protein diffusion by analyzing individual single-molecule trajectories. For CckA-eYFP, 3D tracking let us accurately trace the motion of molecules diffusing over the curved membrane at and away from the cell poles (Fig. 1d). Using mean-squared-displacement (MSD) analysis, we found that CckA diffused most rapidly in the cell body, 2-fold slower in the old pole, and 4-fold slower in the new pole (D = 0.0082 ±0.0020, 0.0040 ± 0.0014, and 0.0022 ± 0.0013 μm2/s, respectively) (Fig. 1g). These reductions in diffusion did not result from differences in pole vs. midcell geometry: the old and new poles have similar shapes4 yet exhibit asymmetric slowing, and in Monte Carlo simulations of free motion over the 3D cell surface, less than 8% of the observed reduction in diffusion could be ascribed to pole geometry (Extended Data Fig. 2b). Rather, the reduction in polar diffusivity we observed in vivo likely results in part from the binding of CckA7 and ChpT (Extended Data Fig. 1d) to PopZ, with additional slowing at the new pole induced by CckA-specific localization factors7,23 and/or the enhanced formation of higher-order CckA assemblies at high concentration26. Fluorescence recovery after photobleaching (FRAP) experiments and simulations showed that CckA was able to freely exit and enter the polar microdomain on the minutes timescale, implying that such binding events are transient (Extended Data Fig. 2d–i). CckA motion was likely not affected by the reorganization of PopZ molecules, as FRAP measurements showed slow intra-pole redistribution of PopZ-eYFP within enlarged microdomains (20% within 4 minutes) (Extended Data Fig. 1e). The finding that polar PopZ is effectively static is consistent with previous single-molecule imaging indicating that PopZ is captured for extended periods at the poles5.
Single-molecule imaging revealed that both ChpT-eYFP and CtrA-eYFP-14 were dramatically slowed within the nanoscale region defined by PopZ (Fig. 1e,f, Supplementary Movie 1). We observed multiple (<5% of all tracks with 10+ localizations) ChpT-eYFP (but not CtrA-eYFP-14) trajectories that exhibited motion only in the plane of the polar membrane (Fig. 1e), suggestive of binding to CckA at the membrane. We used MSD analysis along the long axis of the cell to quantify the diffusivity of ChpT and CtrA (Fig. 1h). Both proteins exhibited anomalous diffusion (i.e., sub-Brownian scaling with time) within the cytoplasm, with an approximate short-time Brownian diffusion coefficient D ≈ 1.8 μm2/s. Within the pole, ChpT and CtrA were slowed more than an order of magnitude, to D ≈ 0.1 μm2/s and D ≈ 0.01 μm2/s respectively (Extended Data Fig. 4e–g, Supplementary Note 1.5). (For both proteins, diffusivities inside swarmer and stalked poles were statistically indistinguishable and were pooled to increase estimate precision.) To determine the rate of protein turnover at the poles, we examined the duration of excursions within the microdomain before molecules returned to the cell body (Fig 1i). This distribution was heavy-tailed, similar between old and new poles, and confounded by contributions from photobleaching in addition to true exit events. To obtain an approximate lower bound for dwell time within the poles, we pooled old and new-pole distributions, and fit short-time behavior and corrected for bleaching events by approximating with exponential statistics (Supplementary Note 1.7). We found that ChpT and CtrA exited from the microdomain with a dwell time of at least 132±39 ms and 132±28 ms (95% CI), respectively (Fig. 1i). In all cases, molecules immediately increased in diffusivity upon returning to the cell body (Fig. 1f; ChpT not shown). This rate of exit was three-fold slower than in simulations of free diffusion (Extended Data Fig. 4a–c), indicating that the PopZ microdomain acts to slow protein exit from the poles.
The control of client protein composition in the PopZ microdomain is central to polar differentiation in Caulobacter17. Ribosomes and DNA do not enter the PopZ microdomain6,20, suggesting that the semipermeable PopZ volume at the pole mechanically excludes macromolecules above a certain pore size. From our measurements of polar volume (Fig. 1c) and estimates of protein copy number (Supplementary Note 2.1), we expect the PopZ microdomain to be highly porous, with only ~20% of its volume filled by PopZ molecules (Supplementary Note 1.4). Hypothesizing that percolation through such a porous matrix would also nonspecifically slow proteins at the pole, we performed single-molecule tracking of heterologous proteins that do not bind Caulobacter localization factors. We selected eYFP, and eYFP fused to a 100 amino acid fragment of the A. thaliana PIF6 protein, which we term fPIF27. eYFP is 0.5 times and fPIF-eYFP is 0.75 times the mass of ChpT-eYFP and CtrA-eYFP-14 (Supplementary Table 1), as reflected by their faster diffusion in the cell body (Extended Data Fig. 4f), implying their entry would not be restricted. Yet surprisingly, both eYFP and fPIF-eYFP explored the 3D volume of the cell up to the edge of the PopZ microdomain without entering it for even the length of a 20 ms camera frame (Fig. 2a, Extended Data Fig. 4d–e). This exclusion effect was specific to cytoplasmic proteins: membrane-associated proteins appeared free to explore the pole (Extended Data Fig. 5a–c).
We used optogenetics to ask whether binding to a microdomain-localized protein is sufficient to drive entry. Using the iLID-SspB light-induced dimerization system28 we found that in the absence of photostimulation, a red fluorescent protein (RFP) fused to iLID was excluded from microdomains containing overexpressed SspB-PopZ molecules, yet entered immediately upon photostimulation of iLID-SspB dimerization (Fig. 2c–e). The time RFP-iLID spent in the SspB-PopZ microdomain following photostimulation was comparable to the 20-second half-life of binding between the photostimulated iLID and SspB in vitro28, indicating that proteins were free to leave the microdomain after iLID-SspB binding was lost. Thus, despite being similar in size and charge to proteins that do enter (Supplementary Table 1), RFP-iLID molecules were only recruited into the microdomain when provided tight binding interactions with a component of the microdomain. It was previously shown that overexpression of PopZ molecules leads to their preferential accumulation at the old pole and to an expansion of the PopZ microdomain as shown by fluorescence-imaging and cryo-electron tomography5,6. Upon PopZ overexpression we observed a shift in CckA, ChpT, and CtrA from the new pole to the old pole (Extended Data Fig. 8a,b), as well as reduced expression of CtrA-regulated genes (Extended Data Fig. 8c). It had also been shown that in this background eYFP can enter the microdomain6. Further research is required to determine if overexpression of PopZ results in local changes in PopZ concentrations, concentration of client proteins, and/or structural organization of the microdomain, and whether changes in these parameters affect selective entry and signal transduction.
The dense co-localization, slow diffusion, and antagonistic activity of CckA, ChpT, and CtrA between the new and old pole microdomains9,11–13,18,19,29 is reminiscent of previous theoretical work showing the conditions under which spatial separation of opposing enzymes can lead to a gradient in protein activity30. Earlier calculations have shown that a linear gradient of CtrA~P will form given phosphotransfer rates faster than diffusion 19. As data on ChpT and CtrA diffusivities and localization (Fig. 1e,h) were not previously available, we explored in silico whether our experimental observations that these proteins are slowed and concentrated within the polar microdomain would be consistent with gradient formation. Using experimentally defined diffusivities and biochemical affinities/reaction rates inside and outside of the PopZ microdomains in the predivisional cell prior to compartmentalization, we constructed a complete reaction-diffusion model of the CtrA~P activation pathway (Extended Data Fig. 6a–d and 7a–c, Supplementary Table 7, Supplementary Note 2). We found that at steady state each step of the phosphotransfer cascade resulted in a greater spread of phospho-signal from the new pole, with 85% of total CckA~P, 30% of ChpT~P, and 12% of CtrA~P present within the new pole microdomain (Fig. 3a, left). The remaining fractions of ChpT~P and CtrA~P formed extended gradients into the cytoplasm away from the new pole, with CtrA~P showing the largest range of cytoplasmic concentrations (more than 4-fold over the length of the cell). Consistent with previous models19, we observed that weakening the affinity of ChpT for the new pole microdomain led to a shallower gradient across the cell (Fig. 3a, middle, Extended Data Fig. 7a–c, Supplementary Note 2.6). We further found that weakening the affinity of both ChpT and CtrA for the microdomain led to a near-complete loss of the gradient (Fig. 3a, middle). Sensitivity analysis demonstrated that this coordinated phosphotransfer at the new pole is a critical factor for gradient formation due to the low efficiency of CckA auto-kinase activity18 and the rapid diffusion of ChpT outside the microdomain (Extended Data Fig. 7c). Consistent with this view, we found that the vast majority of phosphotransfer events take place within the microdomain (Fig. 3a, right). Collectively, our simulations showed that the elevated concentrations of all three proteins within the microdomain increase the probability of intermolecular binding and phosphate transfer events via mass action. This result is in agreement with previous models of the function of the chromosome partitioning protein ParA21 in which high ParA concentrations, and thus increased rates of homodimerization within the PopZ microdomain, is critical for chromosome segregation.
Figure 3: Dynamics within the PopZ microdomain modulate the gradient of active CtrA~P.
a. Reaction-diffusion simulation of CtrA activation. Left panel: In silico steady-state distributions of phosphorylated CckA, ChpT, and CtrA for the wild type conditions derived from single-molecule diffusion coefficients (Fig. 1) and biochemical data (Supplementary Table 6). (Phospho-agnostic distributions in Extended Data Fig. 7b). Middle panel: In silico perturbation of the ChpT and CtrA polar binding coefficients affects phospho-gradient steepness. The binding coefficients of ChpT and CtrA to the PopZ microdomain at the new pole were either kept at their wild-type values (filled boxes) or set to 0 (empty boxes). Right panel: Phosphotransfer events from CckA to ChpT and ChpT to CtrA occur exclusively within the new pole microdomain in reaction-diffusion simulations using wild-type parameters (upper images) while an in-silico mutant with 3-fold lower binding affinity of ChpT to the PopZ microdomain results in phosphotransfer events dispersed across the cell (lower images).
b. RT-qPCR of mRNA from an eyfp gene integrated at one of 13 loci on the chromosome controlled by either the xylose-inducible promoter Pxyl or by the CtrA~P activated promoter P350. Data reported for predivisional cells where DNA replication is partially completed. No copy-number adjustment was applied. Left: (top, left) A cartoon of a predivisional cell showing the positions of the 13 loci relative to the chromosome origin (to scale and indicated by the purple gradient shading). (top, right) A diagram showing the signaling pathway for the CtrA~P-controlled promoters and for the phospho-mimetic CtrA(D51E)-controlled promoters that are independent of the signaling cascade. (bottom) A graph showing the distance of each of the 13 chromosomal loci from the new pole microdomain. Right: eyfp transcription was controlled via P350 induced by WT CtrA (red) or by the phospho-mimetic ctrA(D51E) (black) or via xylose/XylX acting on Pxyl (gray). For each condition, normalized transcription levels are shown for the 13 loci sorted by their distance from the new pole microdomain (13, 1, 12, 2, 3, 4, 11, 5, 6, 10, 7, 9, 8, Supplementary Table 5). Shown is mean ± s.d.; n equals three biologically independent experiments repeated with similar results. Additional controls for loci 1, 3, 7, and 8 (highlighted by four dots below the corresponding transcription levels) are found in Extended Data Fig. 9a–c.
We next turned to obtaining experimental verification of a gradient of CtrA~P along the long axis of the cell that was predicted by the in silico modeling of both Chen et al19 and this study, in which we included the ChpT phosphotransferase sequestered to the microdomain as a critical factor for gradient formation (Fig 3a). Accordingly, we designed a promoter activation assay as a proxy for measuring the CtrA~P spatial profile (design considerations described in Supplementary Note 3.1). As a readout of local CtrA~P levels, we measured mRNA levels of an eyfp gene driven by the CtrA~P-regulated P350 promoter (from the CCNA_00350 operon) integrated at 13 different sites along Caulobacter’s single circular chromosome (loci L1-L13) (Fig. 3b, Supplementary Table 4). Unlike most CtrA~P regulated promoters, whose activation is regulated by multiple factors31–33, P350 is activated by CtrA~P alone and is one of the few promoters that is sensitive to local levels of CtrA~P34 (Supplementary Note 3.2). In Caulobacter, the genomic position of a given chromosomal locus reflects its spatial position within the cell35, which allowed us to sort L1-L13 by their distance from the new pole (Fig. 3b, left) (Methods 14 and Supplementary Table 5). In addition, as the half-life of eyfp mRNA is on the order of 3.5 minutes (Supplementary Table 6), measurements of eyfp mRNA levels in synchronized cells report time-dependent transcriptional activity. We used RT-qPCR36 to measure reporter eyfp mRNA transcribed from each chromosomal locus in synchronized predivisional cells before compartmentalization (Fig. 3b, right). Plotting the amount of mRNA transcript as a function of locus position, we observed a smooth decline in P350-driven transcription moving away from the new pole, reaching a total decrease of 75% for loci at midcell. This decline is consistent with our model predictions for the case where both ChpT and CtrA are sequestered within the PopZ microdomain (Fig. 3, Extended Data Fig. 9d). To confirm this decrease was not due to copy-number effects from differential replication time along the chromosome, we replaced P350 with the constitutive PxylX promoter integrated at the same 13 sites and observed a close to uniform profile with at most ±20% uncorrelated variation in mRNA levels (Fig. 3b). To test whether the CtrA~P activity gradient stems from polar activation, we decoupled CtrA from the phosphotransfer pathway at the cell pole by deleting the native ctrA gene and replacing it with a plasmid-borne, xylose-inducible phosphomimetic mutant, ctrA(D51E)37 (Fig. 3b). In this background, position-specific effects on transcription from the CtrA-activated P350 promoter were lost. To confirm the abrogation of the gradient was not due to changing the levels of total CtrA protein, we sampled transcription levels for a subset of loci with the P350 promoter using xylose-induced wildtype CtrA under the same conditions used as for cells bearing the ctrA (D51E) phosphomimetic mutant. The position-specific effects were recovered when WT CtrA replaced the phosphomimetic mutant in the same plasmid-borne background (Extended Data Fig. 9c), demonstrating that the CtrA~P gradient depends on phospho-signaling from the pole but not CtrA copy number. Because CtrA availability is also strongly regulated at the level of localized proteolysis at the opposite cell pole, the cell can compensate for gradient loss and remain viable, but not healthy37. Unlike WT CtrA, high expression levels of ctrA(D51E) are required for the cells to be viable38. In the absence of a gradient in the ctrA (D51E) strain, proteolysis still compensates for this by preferentially skewing the phosphomimetic CtrA to the swarmer pole37,39,40. The cell cannot function normally if both gradient and proteolysis are compromised (in a strain bearing ctrA(D51E)Δ3Ω37). To obtain high-throughput confirmation of the observed gradient, we designed an adaptation of the TRIP (Thousands of Reports Integrated in Parallel) assay41 for Caulobacter, which we term BacTRIP (Methods 15 and Supplementary Note 4). Using the native P350 promoter, as well as an engineered variant that more strongly recruits CtrA~P (P350_AT), and a variant that is not activated by CtrA~P (P350_MT) as a control, we recapitulated the gradient that we had detected at 13 defined loci (Extended Data Fig. 9c). The BacTRIP protocol used over 200 randomly sampled genomic positions using each of the three promoters (Extended Data Fig. 10b). Importantly, the P350_MT promoter did not show any spatial pattern.
Caulobacter asymmetric cell division yields daughter cells that exhibit different genetic readouts despite having identical genomes (Fig. 1a)42. Differences in gene expression profiles stem from the spatial and temporal availability of CtrA~P, multiple other DNA binding proteins 31,43–48, as well as the methylation state of a subset of cell cycle-regulated promoters33,49,50. Here, we have focused on the asymmetric inheritance of CtrA~P transcription factor, which controls the expression of over 100 cycle-regulated genes51,52 and is critical for many swarmer cell-specific functions, including the inhibition of replication initiation53. We empirically demonstrated that the CtrA~P distribution decreases rapidly away from the new pole in predivisional cells, forming a stable sublinear CtrA~P gradient (Fig. 3b). We further showed by in silico simulation that this effect depends upon dense concentration and increased dwell time of the CckA, ChpT and CtrA signaling proteins at the pole (Fig. 3a). Finally, using single-molecule tracking we ascertained that CckA, ChpT, and CtrA indeed exhibit these properties within the PopZ microdomain (Fig. 1). Together, selective sequestration and increased dwell time of signaling proteins in the polar microdomain generates a skewed distribution of CtrA~P in the predivisional cell that would result in the capture of the majority of CtrA~P within the swarmer cell following compartmentalization and asymmetric division, ensuring robust daughter cell fate determination (Fig. 1a, Fig. 4). Hence, we posit that the function of the CtrA~P gradient is to support the robust distribution of CtrA~P to the progeny swarmer cell, which is reinforced by the selective proteolysis of CtrA in the stalked portion of the predivisional cell14,39,40,54.
Figure 4. Coordinated signaling within a membraneless microdomain at the cell pole facilitates asymmetric patterning of the CtrA~P global regulator.
(left, top). Selective entry of client proteins is permitted via direct interaction with PopZ or by interaction with a PopZ client (indirect recruitment), whereas non-client proteins cannot enter. Proteins localized to PopZ are concentrated in the microdomain due to extended dwell times relative to the cell body (Fig. 1). (left, bottom). A localized signaling pathway. Increased concentration of CckA at the cell pole, in vitro or in vivo, results in a local increase in CckA kinase activity18,66, accompanied by the accumulation of ChpT and CtrA within the microdomain, and thus the establishment of a local source of CtrA~P. The numbers indicate stages of the phospho-transfer pathway. 1) Auto-phosphorylation of CckA, 2) complex formation and transfer of phosphate from CckA~P to ChpT, 3) complex formation and transfer of phosphate from ChpT~P to CtrA. (right) CckA acts as a kinase while accumulated at the new cell pole (blue bar) while its phosphatase activity dominates when it is diffusely localized (blue dashes) in the rest of the cell. This localized source of kinase signaling flow, in conjunction with slowed CtrA diffusion in the microdomain, ultimately yields a sublinear gradient of CtrA~P activity across the long axis of the cell (Fig. 3) that primes each daughter cell for distinct cell fate. The theta structures represent the partially replicated chromosome in the predivisional cell. The portion of the predivisional cell that will become the progeny swarmer cell has a single circular chromosome whose replication is inhibited by CtrA~P bound to the origin, while the portion that will become the progeny stalked cell has a chromosome that is able to immediately initiate DNA replication in the absence of CtrA~P. Green shading along the chromosome represents a gradient of CtrA~P in the predivisional cell which serves to reinforce the biased accumulation of CtrA~P in the progeny swarmer cell upon asymmetric division. In the progeny swarmer cell, CtrA~P, along with multiple accessory factors, orchestrates the expression of swarmer cell-specific genes.
Like their eukaryotic counterparts, bacterial cells spatially organize their functions using different types of encapsulated subcellular compartments1,55–58. The PopZ microdomain of Caulobacter is formed by intrinsically disordered proteins7 and exemplifies a spatial organization strategy that shares key qualities with biomolecular condensates studied in eukaryotes2. In the WT cell, the PopZ microdomain forms a dome at the cell pole with the base of the dome forming a membraneless interface with the cytoplasm, yielding a large but porous space-filling volume (Supplementary Note 1.4). The microdomain constrains the membrane and cytoplasmic proteins of the CtrA activation pathway. The activated CtrA~P output of this pathway dynamically exchanges with the cytosol on the sub-second timescale (Fig. 1). These properties are ideal for a subcellular signaling hub that concentrates and coordinates specific signaling pathways driving a CtrA~P gradient in predivisional cells (Fig. 4). The mechanism by which this gradient is formed, switching diffusive states within spatially defined regions at the poles, is reminiscent of a general diffusion-switching mechanism shown to play a key role in development across eukaryotic species, where transient binding to membranes or protein anchors spatially concentrates morphogens59. Recent work has shown that biomolecular condensates also concentrate, transiently sequester, and slow the diffusion of signaling proteins in a wide range of eukaryotic systems60–63, and that enforcement of long dwell times within condensates is critical in multi-step pathways such as actin assembly64,65.
Methods
1. Plasmid construction
All vectors were constructed by Gibson assembly67. Supplementary Table 2 provides a description of all plasmids used in the study and Supplementary Table 4 lists the sequences of the primer sets used.
pKL400, pKL402–3, pKL518, pKL520, pKL523–27, pKL534–5, pKL545 (set A)
The template vector pXVENN-268 was linearized by PCR using primer set 1. The 1000 base-pair regions used to integrate PXylX::eyfp at loci L1-L13 were amplified by PCR from the Caulobacter genome as indicated in Supplementary Table 5.
pKL410, pKL415–6, pKL432–5, pKL506, pKL508, pKL511, and pKL531–3
The 13 vectors from set A were each linearized by PCR using primer set 2. The P350 promoter region (coordinates 365185 to 365305) was amplified by PCR from the Caulobacter genome using primer set 3.
pKL412, pKL420–22
Vectors pKL400-pKL403 were each linearized by PCR using primer set 4. The PpilA promoter region (coordinates 3198402 to 3198522) was amplified by PCR from the Caulobacter genome using primer set 5.
pKL229
pYFPC-1 was amplified with primer set 6. chpT c-term sequence was amplified from the Caulobacter genome using primer set 7.
pKL397
pVYFPN-1 was linearized by PCR with primer set 8. pif sequence was amplified by PCR from pHR SFFVp YFP-PIF-SOS2cat plasmid69 (Addgene plasmid #50851) using primer set 9.
pKL483
pVCFPC-2 was linearized by PCR with primer set 10. popZ sequence was amplified by PCR from the Caulobacter genome using primer set 11. SspB-micro was amplified from pQE-80LMBP-SspB-micro plasmid28 (Addgene plasmid #60410) using primer set 12.
pKL480
pXCHYN-1 was linearized by PCR with primer set 13. iLiD was amplified by PCR from pLL7.0:Venus-iLID-Mito plasmid28 (Addgene plasmid #60413) using primer set 14.
pKL575
pVYFPC-1 was linearized by PCR with primer set 15. TM-PAS from FixL was amplified by PCR from the Caulobacter genome using primer set 16.
pKL460: pBVMCS-6 was linearized by PCR with primer set 17. ctrA sequence was amplified by PCR from the Caulobacter genome using primer set 18.
pKL464
pKL460 was linearized by PCR with primer set 19. eYFP sequence was amplified by PCR from pVYFPC-1 using primer set 20.
Steps to construct of a barcoded plasmid for transposon delivery are detailed in Methods section 15.
2. Caulobacter sample preparation and synchrony
All Caulobacter strains used in this study (Supplementary Table 3) were derived from the synchronizable wild-type strain CB15N70. Caulobacter cells were grown in PYE (peptone-yeast extract) media70, M2 minimal medium71, or in minimal media supplemented with 0.2% d-glucose (M2G). Cells were grown in M2G media in log phase for at least 12 hours before synchronizing the cultures72,73. When appropriate, growth media was supplemented with the following antibiotics: 0.5 μg/mL gentamicin, 5 μg/mL kanamycin, 1 μg/mL oxytetracycline, 25 μg/mL spectinomycin, and/or 1 μg/mL chloramphenicol. Expression of specific genes was induced by addition of vanillate (pH 7.5) or d-xylose at the indicated concentrations and for the indicated time periods before imaging. Generalized transduction was performed with phage ϕCr3071.
For both diffraction-limited and single-molecule imaging experiments, strains were imaged at mid-exponential growth phase (OD600 = 0.3–0.5). Low induction was used to express fluorescently-tagged proteins (0.06–0.09% xylose and/or 100–150 uM vanillate, for 60–90 minutes prior to imaging, and for 60 minutes prior to synchronization, when performed). 1 μL of cells was added to 1.5% agarose/M2G pads and imaged on 22×22 mm2 no. 1.5 glass coverslips (in single-molecule experiments, Fisher 12–541-B or VWR 48366–227 cleaned by Ar plasma etching).
For single-molecule experiments, cells were washed 2–3x in M2G at room temperature prior to imaging, and fluorescent polystyrene beads (100 nm 540/560 FluoSpheres, Life Technologies) were added to provide sparse bright fiducials. To isolate predivisional cells expressing ChpT-eYFP or CtrA-eYFP-14, cells were allowed to progress through the cell cycle at 28°C for 50 minutes. Predivisional cells from synchronies and identified within mixed populations showed similar protein diffusion and localization statistics. Only predivisional cells with clearly identifiable old and new poles (based on detecting visible stalks in brightfield images or relative number of PAmCherry-PopZ localizations between the poles) were used for quantitative analysis.
3. Analysis of diffraction-limited protein distributions
Image analysis code was written in Matlab version 9.2 with the Control System, Curve Fitting, Image Processing, Optimization, Signal Processing and Statistics toolboxes. Cell outlines were identified from the binary image of intensity-thresholded brightfield images. Cells were selected for further analysis with length and width filters, preventing analysis of e.g. overlapping cells. Fluorescence images were registered to their corresponding brightfield images by cross-correlation. A local cellular coordinate system was determined for each cell by first fitting a spline to the cell outline, then defining the cell axis as the midpoint of this outline. The poles of the cells were defined as the points of the cell outline with maximum curvature. Cells were orientated from new pole to old pole based on PopZ focus size, with larger focus assigned to the old pole. Fluorescent profile for each cell was calculated as total signal within segments along the local cellular coordinate system following background substation.
4. Surface plasmon resonance binding assays
The PopZ protein was expressed and purified as described previously21,74. CtrA and ChpT were expressed and purified as described previously15. Surface plasmon resonance experiments were performed on a BIACORE 3000 biosensor system (GE Healthcare) at 25°C. PopZ was covalently immobilized on the surface of a CM5 biosensor chip (GE Healthcare) by amine coupling chemistry using N-hydroxysuccinimide (NHS) and N′-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) according to the manufacturer’s instructions. To investigate binding of either ChpT or CtrA to the immobilized PopZ, the proteins were dialyzed against Kinase Buffer (50mM Hepes, pH 8, 200mM KCl, 1mM DTT), diluted in Kinase Buffer, and injected over the PopZ surface for 2.5 minutes at different concentrations at a flow rate of 30 μL/min. For each experiment at least 5 different concentrations of ChpT or CtrA were injected over each experimental and control flow cell. Dissociation was allowed to occur at the same flow rate for 300 sec. followed by Running Buffer alone at a flow rate of 100 μL /min to allow the baseline to stabilize. All data were corrected for unspecific binding by subtracting the signal measured in a control cell lacking immobilized ligand. Data analysis was performed using the BIAevaluation software 4.1 (GE Healthcare).
5. Single-molecule microscopy
Two-color, 3D single-molecule imaging with eYFP and PAmCherry labels was performed using a microscope setup and protocols similar to previously published studies21,22 with minor modifications. More detailed discussion on the imaging workflow and analysis pipeline (including single-molecule localization and downstream processing) is available in Supplementary Note 1.
We employed an inverted microscope (Nikon Diaphot 200) with an oil-immersion, super-corrected objective (Olympus PLAPON60XOSC, 60X/NA1.4). The sample was illuminated with an activation wavelength of 405 nm (Coherent, OBIS 50 mW), and with detection lasers at 514 nm (Coherent, Sapphire 514–100 CW) and 561 nm (Coherent, Sapphire 561–100 CW diode-pumped solid state lasers in an epifluorescence (wide-field) configuration, with typical irradiances at the sample of 0.1–10 W/cm2, 0.1–0.5 kW/cm2, and 0.5–3 kW/cm2 respectively. Fluorescence signal was isolated by the microscope dichroic mirror (Chroma, zt440/514/561rpc), followed by a 514 nm long pass filter (Semrock, LP02–514RE), 561 nm notch filter (Semrock, NF03–561E) and dual band pass filter (Semrock, FF01–523/610). Light was collected with a f = 400 mm tube lens, split with a 560 nm dichroic (Semrock, FF560-FDi01) and directed through two 4f imaging systems each using f = 120 mm lenses to create a 2.54 mm diameter Fourier transform of the image in each spectral channel. Quartz-relief phase masks for each emission wavelength22 (equivalent models purchasable from Double-Helix Optics, LLC) were placed at the Fourier plane with high-repeatability magnetic mounts to generate the double-helix PSF for 3D imaging, and were removed for 2D imaging. After passing through the 4f systems, images were collected on two separate regions of an EMCCD camera (Andor Ixon DU-897E).
In all cases, eYFP labels were used for single-molecule tracking (using the transient dark states of eYFP) and PAmCherry labels were used for super-resolution imaging of PopZ (using sparse photoactivation). For CckA/PopZ imaging, data was taken with an interleaved-illumination sequence of frames with 100 ms integration times. For 3D imaging with the DH-PSF, our choice of illumination powers and integration times yielded similar localization precisions for CckA-eYFP (first-third quartile ranges of 31–41 nm lateral precision, 46–62 nm axial) and PAmCherry-PopZ (29–40 nm lateral, 44–61 nm axial) estimated from a calibration surface including total detected photons and background22. The power and integration time used for tracking cytoplasmic proteins (ChpT, CtrA, fPIF, free eYFP) resulted in somewhat higher localization precisions (38–45 nm lateral, 58–68 nm axial for ChpT-eYFP, with similar results for other proteins). 2D-only imaging of ChpT-eYFP and CtrA-eYFP-14 gave localization precisions of ~24 nm as estimated by MSD analysis. These values were used for quantitative analysis of motion along the cell axis. Exemplary tracks (e.g. Fig. 1 d–f) were selected for their clear qualitative demonstration of diffusive molecular behaviors. While these tracks were longer than typical (e.g., for CckA, only 2% of tracks lasted the full 4.0 seconds duration exhibited by the pole-localized track in Fig. 1d), their qualitative behaviors were replicated for the (40, 41, 89) cells used in analysis of each labeled protein (CckA, ChpT, CtrA). Apparent membrane-associated motion of ChpT molecules was unambiguous, but infrequent (3 out of 127 sufficiently long tracks analyzed, found in separate cells).
6. Single-molecule data analysis
All analysis was performed using custom scripts written in Matlab (The Mathworks, Inc., Natick, MA) except as noted. For 3D imaging using the double-helix PSF, we used a modified form of the Easy-DHPSF analysis suite available at https://sourceforge.net/projects/easy-dhpsf/75. Multicolor registration in 3D was performed using a locally weighted quadratic function optimized by least-squares error22 using a nanohole array to generate field-dependent calibrations and to acquire control points each day of experiments76. 1000s of control points were used to generate each registration function, with rms target registration errors of 9–18 nm. For 2D imaging, we used ThunderSTORM77 to localize single molecules from raw data. Multicolor registration in 2D was performed using the fitgeotrans function of Matlab with an affine transformation using dozens of control points generated with fluorescent beads, providing rms fiducial registration errors of < 5 nm. Localizations were filtered to avoid false positives by using PSF parameters including interlobe distance and size (for 3D imaging with the DH-PSF) and total amplitude (for both 2D and 3D imaging), and overlapping localizations were not analyzed. Drift was removed by localizing fiducials in each frame. Fiducial localization precision in each frame was generally < 5 nm and was reduced further by low-pass filtering with a boxcar or wavelet22 filter. Fiducials in different parts of the field of view gave the same correction within 10 nm, confirming most drift resulted from rigid sample translations; we used the average of the fiducials’ motion when appropriate.
7. Photobleaching experiments
All photobleaching experiments were performed using an LSM710 line scanning confocal microscope (Zeiss) employing a NA 1.4 oil-immersion objective. To measure PopZ ensemble dynamics, mVenus-PopZ was expressed from a high copy plasmid (AP323, Supplementary Table 3). Imaging was performed with 514 nm power = 1% and 86 nm pixel size. Photobleaching was performed at full 514 nm power in a 2×2 pixel region.
CckA measurements were performed with labeled CckA-eYFP as a full replacement and the marker StpX-mChy as an inducible merodiploid copy (THM 176, Supplementary Table 3). Imaging of CckA-eYFP was performed at 43 μW power at the objective back aperture, 0.7 μs dwell time per 90 nm pixel collected with a 1 Airy Unit pinhole and 800 V PMT gain. CckA-eYFP at the old poles of predivisional cells (determined using the StpX-mChy label, Extended Data Fig. 2h) was photobleached with high-intensity 514 nm laser light (490 μW at the objective back aperture, ~100 μs dwell time per targeted pixel, bleached regions masking the pole: > 1,000 larger dose than used for imaging) after which cells were imaged for many frames at set intervals without further targeted photobleaching. Corrections for incidental photobleaching during imaging are discussed in Supplementary Note 1.8.
8. Brownian motion simulations
We performed simulations of single-particle diffusion using home-built Matlab (The Mathworks, Natick, MA) scripts. We generated normally distributed displacements with mean 0 and variance 2DΔt in each dimension to simulate Brownian diffusion in either 2D (membrane protein motion) or 3D (cytoplasmic protein motion), using time steps Δt that resulted in typical displacements 6DΔt < 10 nm. In both cases, we adapted trajectories for free motion to the geometry of a 3D cylindrical cell of length l and radius r with hemispherical caps of radius r. For membrane protein motion, projection of free 2D diffusion was performed in different ways for motion of molecules within the cylindrical body vs. in the hemispherical caps. For membrane protein motion within the cylindrical body, we projected 2D motion from a 2D sheet with periodic boundary conditions onto the cylinder. For motion within the hemisphere, we projected each displacement from its 2D manifold onto the 3D surface, ensuring that the magnitude of displacements was not changed. For cytoplasmic protein motion, we modeled the confining boundary of the cell envelope using either absorptive or reflective boundary conditions. In both cases we achieved similar results; in the simulations present in Fig. 1 and Extended Data Fig. 4, we use absorbing boundary conditions. For computational convenience, we simulated both behaviors similarly: 3D motion that went beyond the surface of the cell was reflected at the tangent plane of the surface, and the reflected motion was scaled either by a factor of 1.0 (full reflection) or 0.003 (absorption, avoiding problematic numerical behavior when molecules appeared within a very small distance from the cell surface). To represent the experimental conditions, we binned the finely sampled trajectories into either 20 ms or 100 ms points to represent the effect of motion blur, then added isotropic Gaussian localization errors at each observed position of 35 nm for membrane protein trajectories or 40 nm for cytoplasmic protein trajectories.
9. Optogenetics assay
Cells expressing pVCFPC-2 PvanA::SspBmicro-CFP-PopZ, pXCHYN-1 PxylX::mCherry-iLID, and pBX-4 PopZ (strain KL6119) were grown in M2G in the dark and induced with 0.5 mM vanillate (pH 7.5) and 0.3% d-xylose for 4 hours prior to imaging. Images were collected using a Leica DMi8 S microscope equipped with a Hamamatsu C9100 EM‐CCD camera, a 100x oil-immersion objective (1.63 NA), and a SPECTRA X light engine (Lumencor). Excitation illumination was passed through a 470/20 band pass filter and delivered to cells for 1 second. The emission bandpass filters were 440/20 (CFP) and 575/25 (mCherry). To avoid stimulating the iLID system while finding the correct focus, we added a red filter after the illumination lamp (572/28 bandpass filter, Chroma).
10. A spatiotemporal model of CtrA activation pathway
We defined the spatiotemporal evolution of protein concentrations as a set of ordinary differential equations. The predivisional cell was described using one spatial dimension with the variable 0≤x≤L corresponding to the distance from the new pole. The cell cytoplasm and the two microdomains (i.e. at the new and old poles) were defined as three distinct environments. Kinetic and thermodynamic parameters were derived from experimental observations (Supplementary Note 2). For generalizable model setup, calculation, and analysis we developed a diffusion-reaction library in python 2.7.10 using Numpy, Scipy, and Matplotlib packages.
Concentrations of each molecular species (Ci) in the system were modeled as a function of position along the long axis of the cell (x), and time (t). The governing partial differential equation (PDE) for each species is:
where the first term includes all first order reactions operating on species Ci at rate αk, such as (de)phosphorylation and unbinding. The second term describes all second order reactions (such as binding) between species Ci and Cj with rate βk. The third term models the auto-kinase activity of CckA as a Hill function18 (CckA autokinase activity above), where S is the set of indexes of CckA species (e.g. CckA, CckA~P, CckA-ChpT), H is the half-maximal velocity and n is the hill coefficient. The last term defines diffusion, where D(x) is the diffusion coefficient as a function of cellular microdomain environment:
where DNP, DB, and DOP are diffusion coefficients at the new pole, cell body, and the old pole. NP and OP are the boundary positions for the new pole and the old pole microdomains (NP=0.2 and OP=3.75). To avoid unphysical behavior resulting from a sharp step function in diffusivity, we smoothed D(x) with smoothing coefficient s1.
The solution to these equations was approximated as a set of ordinary differential equations (ODEs). The spatial dimension of a 4 μm cell is discretized into n = 80 bins, each representing a 0.05 μm section of the cell. Each species is broken into n sub-species, one per bin. The simulation advances in 0.1 second steps. We use the second-order center difference formula to approximate the Laplacian operator and convert each PDE into a set of ordinary differential equations (ODEs):
where Di is a discretization of D(x). We ensure flux into and out of the bins is balanced by matching flux at the boundaries.
11. Sensitivity analysis
To measure sensitivity (Extended Data Fig. 7a–b), we varied the value of each of the 17 critical parameters in Table S6 over 3 orders of magnitude from its established literature value, across multiple combinations of all other parameters, and calculated the averaged impact this change had on the system. Simultaneously, we assessed whether small changes in other parameters could negate the effect caused by the large change in the parameter of interest. Parameters were deemed sensitive if alterations to their values irreparably altered CtrA phospho-gradient properties, indicating the importance of that parameter.
12. RT-qPCR gene expression
The effects of gene position on the transcription rate of the eyfp gene driven by various promoters were determined by measuring eyfp mRNA levels with RT-qPCR. To obtain RNA samples, 0.5 mL of cells with OD600 = 0.3–0.5 were centrifuged for 3 minutes at 15,000 r.p.m. in a benchtop centrifuge at 4°C temperature. Cell pellets were immediately flash frozen in liquid nitrogen. The pellets were resuspended in TRIzol (Ambion) and extracted using Phase Lock Gel-Heavy tubes (5 PRIME), and RNA was purified using the RNA Clean and Concentrator-5 kit (Zymo Research). Genomic DNA was eliminated by treating the samples twice with 1 MBU Baseline-ZERO DNAse (Epicentre) at 37°C for 20 minutes. RNA was recollected using Zymo-Spin columns and subsequently reverse transcribed using the SuperScript III reverse transcriptase kit (Invitrogen). Following reverse transcription, remaining RNA was degraded via RNAse H treatment, and the cDNA was diluted tenfold before beginning qPCR.
Expression levels were determined using on an Applied Biosystems 7500 Fast Real-Time PCR system, using 7500 Software v 2.0.1. The 15 mL qPCR reaction contained 2 mL of cDNA, 7.5 mL of SYBR Green Dye master mix and 5.5 mL of primer mix. The primer mix contained the forward and reverse primers to form ~100 base-pairs amplicons in the genes of interest, at a final primer concentration in the reaction of 230 nM. Expression measurements were then made by comparing cycle-threshold (CT) of the amplicons of interest to an internal standard amplicon in rho, a house-keeping gene that is insensitive to cellular concentrations or activity of CtrA. As a negative control, we verified the removal of genomic DNA template contamination by measuring CT of RNA samples not treated with reverse transcriptase. We additionally measured amplification of a genomic DNA standard curve to verify that the Rho and eYFP amplicons formed unique products and with amplification efficiencies within 10% of one another: we measured amplification efficiencies of 96.86% and 95.85%, respectively. Data was analyzed using the delta–delta-CT method78 or the normalized ratio method79. Final gene expression measurements represent the average and s.d. of three biological replicates, each composed of at least two technical replicates.
13. Modeling an array of CtrA~P gradients and their effect on transcription of a CtrA- regulated gene
CtrA~P binds multiple promoters cooperatively80,81. Because CtrA~P binds P350 as a dimer32, it is likely to exhibit cooperative activity when promoting the transcription of 350 mRNA as well. Accordingly, we modeled the transcriptional response from P350 as a Hill function,
where [mRNAi(t)] is the concentration of eyfp mRNA integrated at site (i=1–13) as a function of time. The first two terms model synthesis of eyfp mRNA from each chromosome. ks is the maximum transcription rate of the eyfp gene, K1 is the binding constant between CtrA~P and the 350 promoter, and n is the Hill coefficient describing the degree of cooperativity. xi1 is the spatial position of the eyfp gene in the original chromosome while xi2 is the spatial position of the gene on the newly replicating chromosome. The last term models first order degradation of eyfp mRNA with rate constant kd. The value of the Hill coefficient, n, was fitted to match in vivo measurements of CtrA~P levels in the cell and time resolved RNAseq measurements of P350 promoter activity, with ks, K1, and kd defined by published biochemical measurements (Supplementary Table 6).
To generate Extended Data Fig. 9d, the transcription-modeling equation was used at the predivisional time point in a quasi-steady-state approximation along with the time-dependent revealing of the underlying DNA loci with parameter values as described in Supplementary Table 5. The CtrA~P gradient was modeled using different Kds between ChpT and PopZ (Extended Data Fig. 7b).
14. Approximate gene position along the long axis of the cell
We used previously published data on the spatial organization of the Caulobacter genome and the rate of chromosome duplication to estimate gene positions over the course of the cell cycle82. The length of the cell, defined as the pole-to-pole distance along the contour of the cell medial axis, increases exponentially in rich media with a time constant of 125±8 minutes, starting at 1.6±0.2 μm for swarmer cells and reaching 4.3±0.3 μm prior to division83,84
where l(t) is the length of the cell as a function of the cell cycle time (0 ≤t ≤140 min), l0 is the swarmer cell length at the beginning of the cell cycle (l0 = 1.6), α = 0.85 and β = 0.0078 min−1.
Caulobacter possesses a single circular chromosome. DNA replication initiates at a unique origin of replication (Cori) and proceeds bidirectionally85,86. In the swarmer cell and in the stalked cell before chromosome replication, Cori is positioned near the old cell pole, where it is physically tethered to the PopZ microdomain via the ParB protein5,6,21 (Extended Data Fig. 9a, cell cycle schematic). Loci along the circular chromosome occupy well-defined and reproducible spatial coordinates, with position along the cell axis proportional to their distance from Cori on the chromosome82,87,88. The rate of chromosome duplication by the replisome is constant over the 90 minutes total replication time89. The newly replicated DNA segments are immediately and rapidly directed to their final subcellular positions following replication82 (Extended Data Fig. 9a, cell cycle schematic). We used the following two equations to approximate the spatial position of each chromosomal locus (bp) on the original chromosome, (Porig(bp)), and its newly replicated copy (Preplicated(bp,t)) during DNA replication (minutes 20 to 110 in the cell cycle), along the cell axis relative to the old pole. Because the spatial organization of the old chromosome does not change, Porig(bp) is independent of time,
where lpole is the diameter of the old pole microdomain, from which the chromosome is excluded, and M is the base-pair coordinate of the terminus region, which is positioned approximately halfway along the circular chromosome.
By contrast, the positions of loci on the new chromosome are defined by the time-dependent position of the new pole,
where rt(bp) is the “replication time” at which the chromosomal locus bp is duplicated (20 ≤ rt(bp) ≤ 110 minutes).
15. BacTRIP: High-throughput measurements of gene expression as a function of gene position
To measure gene expression as a function of gene position we adapted a high throughput assay named TRIP for Caulobacter cells. TRIP is a protocol for analyzing large numbers of reporters integrated in parallel41. In short, a barcoded operon was integrated into hundreds of positions on the chromosome using transposon mutagenesis. High-throughput sequencing was then used to extract both the barcoded mRNA and the genomic position of each integrated barcoded operon.
15.1. Creating a barcoded plasmid for transposon delivery
We took the following steps to make a barcoded plasmid for transposon delivery (Extended Data Fig. 10a). To construct the pTripTn5 delivery vector for transposon mutagenesis, the promoter and coding region of Tn5 transposase with its 5’ ME and 3’ ME motif were amplified from pXMCS2-Tn5Pxyl (LS5298) and inserted into a KpnI-SacI linearized pMCS2 vector to generate pMCS2-Transposase. A synthesized multiple cloning site (EcoRI and SacI) and a transcription terminator from the Caulobacter metK gene were inserted to EcoRI-SacI digested pMCS2-Transposase via Gibson Assembly67 to generate pTripTn5-TR. We chose the terminator of metK because it is short and displays clear transcriptional termination in RNAseq data90. The original EcoRI and SacI sites were removed during assembly so that the resulting plasmid harbors only one EcoRI or SacI site from the synthesized multiple cloning site. A DNA fragment containing a promoter sequence followed by the eyfp gene was synthesized using IDT gBlocks. Three versions of the CCNA_00350 promoter (P350) were used: wildtype P350 (WT350) with a noncanonical CtrA binding site TCAA-N7-TTAA, P350 with the consensus TTAA-N7-TTAA CtrA binding site (AT350)81,91, and a P350 with a mutated CtrA binding site TCAC-N7-TCAC, lacking the TTAA motif, that does not bind CtrA (MT350)81,91 (Extended Data Fig. 9b). An 18-bp random barcode was added between the last amino acid and the stop codon of eyfp gene by amplification, generating the intact fragment of barcoded-integrating reporter (BIR). The BIR fragment was inserted into EcoRI-SacI digested pTripTn5-TR to generate pTripTn5-PWT350::eyfp. pTripTn5-PAT350::eyfp and pTripTn5- PMT350::eyfp were constructed similarly using a Q5 site-directed mutagenesis kit (NEB). The library of reporter plasmids was then introduced into Caulobacter cells through electroporation. We collected approximately 33500, 30500, and 193000 E. coli colonies from promoter constructs pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp, respectively.
15.2. Construction of Caulobacter Tn5 transposon libraries
The transposon plasmid libraries (pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp) were introduced into Caulobacter NA1000 through electroporation. The transformants were collected on PYE agar plates containing 5 ug/ml Kanamycin. We collected 11800, 14100, and 9500 Caulobacter colonies for pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp transformed into NA1000.
15.3. Generation of mapping data
To create a sequencing library for genomic-mapping data, genomic DNA was extracted from all cells in the Caulobacter Tn5 transposon library and digested with SalI or NcoI. The SalI cut site is found once on the transposable element and 3622 times across the Caulobacter genome. To eliminate products that do not contain the transposable element (e.g., cut twice outside the transposable element), we circulated the digestion products (Extended Data Fig. 10a) by self-ligation reaction in the presence of T4 DNA ligase 4 °C. The ligation product was purified using Monarch PCR & DNA Cleanup Kit (NEB). A PCR amplification was conducted with the mapping primer pair (Supplementary Table 8) using the purified ligation product as a template. The purified PCR product was used to prepare the sequencing library using Nextera XT DNA Library Preparation Kit (Illumina). The library was sequenced by Illumina 2×300bp MiSeq Reagent Kit v3.
15.4. Generation of transcription data
To create a sequencing library for transcription data, swarmer cells from the Tn5 transposon library were synchronized and released into M2G media and allowed to proceed through the cell cycle. The early predivisional cells were collected at the 90-minute time point post synchronization and snap-frozen. The total RNA was isolated and reverse-transcribed to cDNA using Qiagen RNeasy Mini Kit and QuantiTect Reverse Transcription Kit, respectively. The eyfp gene and its corresponding barcode were amplified for 24 cycles using the counting primer pair N70X-R1barcode and S50Y-R2YFP, where X and Y represent Illumina primer indexes (Supplementary Table 8). The PCR products were gel-purified and pulled for direct sequencing by Illumina HiSeq 4000.
15.5. Generation of normalization data
To create a sequencing library for normalization data, genomic DNA was extracted from the same sample collected for the transcription data, diluted and served as a template for 20 cycles of amplification of the barcoded region using the counting primer pair (Supplementary Table 8). The PCR products were gel-purified and pulled for direct sequencing by Illumina Hi-Seq.
15.6. Data analysis
To extract the genomic integration position for each barcode, we analyzed the mapping data as follows. We selected mapping reads that satisfied the following conditions: 1) the SacI recognition sequence was detected, 2) the FASTQ read quality for each of the 18 base pairs of the barcode sequence was at least 24 (corresponding to a base call accuracy of 99%), and 3) there was a segment of at least 40 base-pairs of genomic DNA. The genomic DNA fragments within this set of reads were then aligned to the Caulobacter chromosome using BLAST92. We detected 406, 467, and 630 integration sites for pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp, respectively.
To count the number of transcripts per barcode, we analyzed three replicates of the transcription data as follows. We selected transcript reads that satisfied the following conditions: as above, 1) the SacI recognition sequence was detected and 2) FASTQ read quality for each of the 18 base pairs of the barcode sequence was at least 24, differing by the criterion 3) that the sequence of the last 5 amino acids of the eyfp gene were detected. We found 152M, 203M, and 86M transcripts that satisfied our conditions for pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp, respectively. In all three cases we found close to 25,000 distinct barcodes.
To quantitatively map transcriptional activity at specific genomic loci, we found the intersection of reads that were both well-mapped and quantitated by transcript yield. This number was primarily limited by the low yield of mapping data, and we obtained a total of 161, 156, and 148 barcodes falling into this class for pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp. Of these, for pTripTn5- PWT350::eyfp we obtained an average of 87,966 reads per barcode (standard deviation: 9,843). For pTripTn5- PAT350::eyfp we obtained an average of 125,144 reads per barcode (standard deviation: 23,1333). For pTripTn5- PMT350::eyfp we obtained an average of 35,002 reads per barcode (standard deviation: 6,479).
To count the number of cells contributing to the total mRNA yielded for each mapped barcode, we analyzed two replicates of the normalization data using the same approach we took to analyze the transcription data. We then normalized the transcript data by dividing the total number of transcript reads per barcode by the number of cells containing that barcode, allowing us to directly compare transcriptional activity between barcodes.
We found the largest abundance for eyfp mRNA when driven by the consensus PAT350 promoter, and the lowest abundance for eyfp mRNA when driven by the mutated PMT350 promoter. We further observed a gradient in eyfp mRNA levels as a function of distance from the new pole microdomain in both the PAT350 and the PAT350 datasets, with a sharper gradient for PAT350. The gradient was eliminated when eyfp was driven by the PMT350 promoter (Extended Data Fig. 10b).
Extended Data
Extended Data Fig. 1: Diffraction-limited profiles of CckA-eYFP, ChpT-eYFP, CtrA-eYFP-14 localization, ChpT/PopZ interaction, and PopZ-eYFP dynamics.
a. Fluorescent constructs used to image the co-localization of CckA, ChpT, and CtrA with respect to PopZ (cf. Supplementary Table 1). pamcherry-popZ, cckA-eyfp and chpT-eyfp are integrated at the native promotor/locus. CtrA-eYFP-14 (sandwich fusion with C-terminal degradation tag) is driven by a PxylX promoter and is expressed from a high copy plasmid. b. CtrA-eYFP-14 has similar cell cycle behavior as CtrA. Shown is a time-resolved western blot of CtrA protein expressed from CtrA promoter and CtrA-eYFP-14 protein expressed from a vanillate-regulated promoter probed with anti-CtrA antibody. The KL6039 strain was induced with 1mM vanillate for 1.5 hours prior to induction and during recovery. Data are representative of three biological replicates. c. Diffraction-limited images of predivisional Caulobacter cells expressing CckA-eYFP, ChpT-eYFP, or CtrA-eYFP-14 (top row) and PAmCherry-PopZ (middle row); new poles marked with a white arrow. Scale bars: 2 μm. Fluorescence intensity profiles, including percentage of CckA, ChpT, or CtrA signal at new and old poles, shown along normalized cell length (n = 55, 129, 143 cells respectively). d. Purified WT ChpT binds directly to PopZ, as measured by surface plasmon resonance (left), while purified CtrA does not (right). n equals three biologically independent samples. e. Recovery following targeted photobleaching of a portion of an extended PopZ microdomain. ΔpopZ cells expressing popZ-eyfp from a high copy plasmid were imaged for many frames of laser scanning confocal microscopy following targeted photobleaching with high-intensity 514 nm laser light. (left) The fraction of signal in the bleached region compared to signal in the entire microdomain is shown in red, with exponential fit in black. Shown is the mean ± s.d.; n equal to 6 cells. (right) Representative field of view from the PopZ FRAP experiment. A bleached cell and its recovery dynamics is shown next to an unbleached cell. Scale bars: 2 μm.
Extended Data Fig. 2: CckA dynamics within the poles.
a-c, simulations of membrane motion (cf. Supplementary Notes 1.9). a. A single Brownian trajectory on the cell surface. The 2D projection of the trajectory exhibits erroneously low diffusivity at the poles and equator of the cell, where 3D information is lost. b. Left: mean-squared-displacement analysis of 3D diffusion accurately infers the diffusion coefficient of motion on the cell membrane, while restriction to 2D measurements induces systematic errors. Right: comparison of D values estimated from subtrajectories inside and outside the poles. Relative D underestimation inside the poles was ≤ 8% for the measured values of polar D for CckA-eYFP (0.02–0.04 μm2/s). c. Experimental approach of 3D tracking using the DH-PSF. Left, 4 second CckA-eYFP trajectory overlaid on brightfield image of a Caulobacter cell. Middle, translation and rotation in the double-helix PSF is used to detect 2D and 3D motion. Right, the corresponding 3D trajectory along the circumference of the cell. Scale bar: 500 nm. d. Caulobacter cells expressing CckA-eYFP imaged with confocal microscopy following targeted photobleaching. Signal at the old and new pole regions is defined as the fraction of total cell fluorescence (upper plot, errorbars show ± 95% CI). After 10 minutes, the initial ratio of old to new pole signal is restored (lower plot). Average of n = 6 cells. Scale bar: 2 μm. e. Fluorescence decay in cells with photobleaching only from imaging, (n = 9, cyan), cells that were uniformly bleached with the photobleaching laser (n = 3, red), and with targeted photobleaching at the old poles (n = 6, magenta) (same statistics as d.). f-g. data shown in d. if using a correction factor from “unbleached” or “completely bleached” cells. Regardless of the correction factor used, new poles exhibit loss and old poles exhibit recovery. Errorbars: 95% CI. h. Representative images of cells showing the StpX-mChy marker used to identify stalks, i.e. the old pole, and the conditions necessary for inclusion as “predivisional,” as consistently observed in all cells. Scale bar: 2 μm. i. Predicted concentrations from reaction-diffusion simulations of CckA motion with fast and slow unbinding to the microdomain overlaid with data of (d). CckA molecules at the old pole were “bleached” by marking the molecules as dark CckA molecules.
Extended Data Fig. 3: Super-resolved distribution of PopZ, CckA, ChpT, and CtrA within the poles.
a. Single-molecule distributions of CckA and PopZ localizations averaged from old poles (n = 29 poles) and new poles (n = 13) of predivisional cells. Blue arrows show the measured diameters of CckA. Scale bar: 200 nm. b. Projection of all single-molecule localizations of CckA and PopZ in the new pole, shown facing down the cell axis (531 and 477 localizations, respectively). Top and right: 1D histogram of CckA and PopZ with 25 nm bins. Centre: mean. The half widths at half maximum (HWHM) are calculated for CckA from the standard deviations of molecule positions along the Y and Z axes, with magnitudes shown by blue arrows (error: 95% CI from localization resampling). c. 1D profiles and 2D histograms (25 nm bin size) of CtrA, ChpT, and PopZ localizations averaged with respect to PopZ (60, 27, and 27 cells respectively for each reconstruction). Scale bar: 200 nm.
Extended Data Fig. 4: Selective polar permeability and motion of cytoplasmic proteins.
a-c. Simulations of freely diffusing cytoplasmic molecules with D = 1.8 μm2/s are observed in the poles (Supplementary Notes 2.9). A step size of 0.5 ms was used, and to match experimental conditions, trajectories were reconstructed at 20 ms sampling rate (mimicking motion blur) and 40 nm localization error. a. The XY components of a single 3D trajectory, corresponding to localizations of a single freely-diffusing molecule. b. The histogram of localizations from 1,000 simulated 2-second trajectories, with poles defined as extending200 and 250 nm away from the tip (consistent with definitions used for experimental data) marked. c. 1D profile of histogram shown in (b). Approximately 4.1% and 5.5% of localizations would be expected to appear within the new and old poles (orange), respectively, if molecular motion were unobstructed. d. White light image and 2D histogram (40 nm bin size) of fPIF-eYFP and PAmCherry-PopZ single-molecule localizations. Scale bar: 2 μm. Consistent in 16 cells analyzed (Fig. 2b). e. White light image and 2D histogram (40 nm bin size) of eYFP and PAmCherry-PopZ single-molecule localizations. Scale bar: 2 μm. Consistent in 17 cells analyzed (Fig. 2b). f. 1D MSD values in the body of predivisional Caulobacter cells as shown in Fig. 1h, including eYFP and fPIF. 95% CI shown. g. Fitting the MSD data to an equation for anomalous diffusion was not able to precisely specify D and α due to the short track durations available. Points: fits values of D and α generated from 200 resamples of CtrA and ChpT tracks either from the cell body or from the poles. Blue lines mark combinations of D and α that would yield apparent diffusion coefficients Dapp for a given short-time approximation, calculated as Dapp = Dτ1−α for Dapp = 1.8 μm2/s and τ = 20 ms (the frame integration time, right line) or τ = 100 ms (the maximum time included in the fit, left line). The trajectories measured are consistent with a short-time value of Dapp = 1.8 μm2/s and suggest that α < 0.7. In contrast, diffusion within the poles appeared relatively Brownian, with 0.75 ≤ α ≤ 0.9. The MSD curve of ChpT at the poles cannot be accurately fit due to confinement effects.
Extended Data Fig. 5: Correlative imaging of a membrane-bound, pole-insensitive construct and PopZ.
a. Diffraction-limited epifluorescence data of the FixL-PAS-eYFP construct and PAmCherryPopZ. Overlap of PopZ and FixL-PAS-eYFP at the poles was consistently observed in 20 cells. Scale bar: 2 μm b. Brightfield image and 2-color histogram of single-molecule localizations of FixL-PAS-eYFP and PAmCherry-PopZ. Scale bar: 2 μm. c. A selection of five single-molecule tracks of the upper cell in b., shown relative to the PAmCherry-PopZ super-resolution reconstruction (red density). Detailed single-molecule analysis showed consistent entry into the membrane proximal to PopZ for the 4 cells analyzed. Scale bar: 200 nm.
Extended Data Fig. 6: Setup of the reaction-diffusion simulation of CtrA activation pathway.
a. A simplified schematic of the reaction and diffusion events used in our CtrA activation model. Dotted red arrows indicate CckA, ChpT, and CckA binding to either the new pole (NP) or the old pole (OP) species. The width of the arrow represents the strength of binding, with thicker arrows for tighter binding. The colored edges out of CckA (blue), ChpT (orange), and CtrA (green) boxes indicate diffusion inside the pole and in the body of the cell. The length of the arrow represents the rate of diffusion, with shorter arrows representing slower diffusion. Phosphotransfer reactions are shown in black. Each arrow is labeled with a reaction number from 1 to 39. Information on each reaction is found in Supplementary Table 7. b. A complete schematic of the phosphotransfer reactions including binding and transfer events. Reaction numbers match the simplified schematic in (a). c. Concentrations of each molecular species in the system were modeled as a function of position along the long axis of the cell and time. The governing partial differential equation (PDE) for each species was approximated as a set of ordinary differential equations (ODEs). The spatial dimension of a 4 μm cell was discretized into n = 80 bins, each representing a 0.05 μm Δx section of the cell. Each species is broken into n sub-species, one per bin. The simulation advances in 0.1 second steps. We use the second-order center difference formula to approximate the Laplacian operator and convert each PDE into a set of ODEs (Equation S7). d. Simulation results with wildtype parameters (Fig. 3a) showing steady state protein concentrations for CckA (blue), ChpT (orange), and CtrA (green) as a function of distance from the new pole in the predivisional cell.
Extended Data Fig. 7: Reaction-diffusion simulation of the CtrA activation pathway.
a. Global analysis of the effect of diffusion coefficients and reaction rates on CtrA~P amplitude and distribution. Sensitivity of CtrA~P amplitude (upper heat map) and CtrA~P distribution (lower heat map) to changes in (i) the rate of CckA, ChpT, and CtrA binding to either the new pole or old pole microdomains, (ii) diffusion coefficients of either CckA, ChpT, or CtrA, and (iii) phosphotransfer rates. Each heat map rectangle specifies one parameter (e.g. top/left rectangle specifies sensitivity to changes in the binding rate between CckA and the new pole microdomain). The color of the rectangle indicates the normalized sensitivity value (from low sensitivity (blue) to high sensitivity (red)) to changes in parameter value. b. Detailed results for changes in 8 parameters. For each parameter the fraction of total CtrA~P (top) and distribution of CtrA~P along the long axis of the cell (bottom) is shown for steady state. Gray arrows signify the increasing value of the parameter. c. Contribution of the rate of phosphotrasfer from CckA to ChpT on the distribution of CtrA~P. We simulated the distribution of CtrA~P as a function of three different phosphorylation rates of CckA by ChpT: 10/s forward and 1/s back (blue), 100/s forward and 10/s back (green), and 100/s forward and 1/s back (brown). We varied the binding affinity between ChpT and PopZ between 0 and 10μM, as well as the auto-phosphorylation rate of CckA: slow by setting CckA kHalf to 150 (left) as well as fast by setting the kHalf to 5 (right).
Extended Data Fig. 8: Diffraction-limited profiles of CckA-eYFP, ChpT-eYFP, CtrA-eYFP-14 localization and expression of CtrA~P-regulated genes in PopZ overexpression background.
a. Overexpression of PopZ does not affect CckA and ChpT abundance but does result in a reduction in CtrA abundance. Mixed population western blot of CckA-eYFP protein expressed from the cckA promoter, ChpT-eYFP expressed from the chpT promoter, and CtrA-14-eYFP protein expressed from a vanillate-regulated promoter. CckA-eYFP and ChpT-eYFP were probed with anti-YFP antibody while CtrA-14-eYFP was probed with anti-CtrA antibody. The protein abundance was measured in both wildtype (I) and PopZ overexpression (II) conditions. n equals three biologically independent experiments repeated with similar results. b. Diffraction-limited images of predivisional Caulobacter cells expressing CckA-eYFP, ChpT-eYFP, or CtrA-eYFP-14 (top row) and PAmCherry-PopZ (middle row) in a PopZ overexpression background; new poles marked with a white arrow. Scale bars: 2 μm. The percentages of total fluorescent signal at the new and old poles are indicated in blue, orange, and green for CckA, ChpT, and CtrA. Fluorescence intensity profiles shown along normalized cell length (n = 45, 52, 61 cells respectively). c. Overexpression of PopZ leads to a reduction in the expression of CtrA regulated genes. RT-qPCR of mRNA levels of five CtrA regulated genes at the 90-minute time point. Transcription was measured for wildtype cells (left) as well as cells overexpressing PopZ (right). Shown is the mean ± s.d.; n equals three biologically independent experiments repeated with similar results.
Extended Data Fig. 9: Spatially and temporally resolved transcriptional activity.
a. Time-resolved RT-qPCR of mRNA from an eyfp gene under the control of the CtrA-regulated P350 promoter integrated at four loci of the chromosome (L1, L3, L7, and L8, cf. Fig. 3). Shown is mean ± s.d.; n = 3. Below plot: Replication times of these loci during the cell cycle, and cell cycle schematic of loci/dividing cells. Chromosome: black ovals. Red areas indicate polar PopZ microdomain, green tone indicates CtrA~P levels. Transcription is normalized to the five-minutes time point of L1. b. Time-resolved RT-qPCR of eyfp expressed from PxylX::eyfp at chromosomal loci L1 and L8 demonstrates a copy number effect. The observed increase at L1 and L8 represents the effect of chromosome replication, as xylose concentration and promotor activity are not expected to vary over time. Shown is mean ± s.d.; n = 3. c. eyfp transcription measured in cells at the 90 minute cell cycle time point at L1, L3, L7, and L8 in five genetic backgrounds. In the first three backgrounds, eyfp expression is controlled by the CtrA~P-regulated P350 promoter in the presence of: native wildtype ctrA (green), the phosphomimetic ctrA(D51E) driven by PxylX in a delta ctrA background (black), or ctrA driven by PxylX in a delta ctrA background (red). In the remaining backgrounds, eyfp expression is driven by the CtrA~P-regulated promoter PpilA in the presence of native wildtype ctrA (blue), and the xylose-driven promoter PxylX in the presence of xylose (gray). The wildtype ctrA, the phosphomimetic ctrA D51E, and eyfp driven by PxylX (green, black, and gray) are a subset of the data presented in Fig. 3b. The position-specific transcriptional output present for the CtrA~P-regulated P350 promoter in native conditions is lost when the sole CtrA copy is xylose-induced phosphomimetic CtrA(D51E) (black) but is recovered with xylose-inducible wildtype ctrA (red). Shown is mean +s.d.; n = 3. d. Simulated CtrA~P distribution profiles and their effect on transcription from P350. (left), possible CtrA~P profiles simulated with altered binding coefficient value between ChpT and PopZ at the new pole (cf. Extended Data Fig. 7a). Profile 4 (light brown) reflects wildtype parameters (Fig. 3a). (right) calculated transcript levels from P350 for each CtrA~P profile (Methods 13). Profile 4 was generated using wild-type parameters and best matched the experimental data (black, Fig. 3b).
Extended Data Fig. 10: High throughput assay to probe gene expression as a function of genomic position.
a. Graphical representation of the BacTRIP method. (I) A library of reporter plasmids generates a library of barcoded Caulobacter loci (purple, green, and red). Light gray: Tn5 mediated transposon. This includes a 5’ mosaic end (ME) followed by a SalI site, kanamycin resistance cassette (KanR), the origin of replication (ori), a promoter (gray), a gene of interest (yellow), a transcribed barcode (colors), a stop codon, the metK gene terminator from Caulobacter, and the 3’ ME. Transformation of the reporter plasmid library into Caulobacter introduces at most one barcode into each cell at a random locus (light brown box). Each barcoded strain reports local transcriptional activity at its locus. (II) DNA. Genomic DNA is used both to determine the integration locus (left) and the number of times each barcode was integrated (right). To determine the locus of each integrated barcode we digested the extracted DNA with SalI and used reverse PCR with mapping primers (Supplementary Table 8) to amplify fragments with an ori element for sequencing. To determine the number of times each barcode was integrated we amplified and sequenced the extracted DNA with counting primers (Supplementary Table 8). (III) RNA. We produced cDNA for sequencing from extracted, reverse-transcribed mRNA. The resulting quantity of inferred mRNA abundance was normalized by total copies of the barcode present in the library. b. BacTRIP results. We used three different CtrA binding sites to probe the wildtype CCNA_00350 promoter (PWT350), an engineered promoter in which we replaced the CtrA binding motif (red) of P350 with a consensus motif (PAT350), as well as a mutant site that is not recognized by CtrA (PMT350). We synchronized the three populations of cells and preformed BacTRIP on predivisional cells at the 90 minute time point. The bar graphs show abundance of 161, 156, and 148 barcoded eyfp as a function of locus distance from the new pole for pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp.
Supplementary Material
Supplementary Movie 1. Single-molecule tracks of CtrA-eYFP-14 Exemplary raw data used to produce single-molecule trajectories of CtrA-eYFP-14. First track: entry to the new pole. (Data plotted in Fig. 1f, left.) Second track: exit from the old pole. As noted in Methods, while not all single molecules used to produce tracks persisted this long before photobleaching, these results (reversible slowing of proteins within the poles) were consistently observed in 89 cells for CtrA-eYFP-14.
Acknowledgments
We thank Justin W. Kern for help with plasmid design and construction, Michael D. Melfi for providing a RT-qPCR protocol, Alberto Lovell and Michael R. Eckart at the Stanford PAN Facility for support in using MySeq and RT-qPCR and for running SPR experiments, Jared M. Schrader for sharing unpublished data on mRNA half-life in Caulobacter, Andrew Olson and the Stanford Neuroscience Microscopy Service (supported by NIH NS069375) for providing equipment and assistance for photobleaching experiments, and Allison H. Squires for critical feedback on the manuscript. We also thank Harley H. McAdams for helpful discussions on modeling of signal transduction, and all members of the Shapiro and Moerner labs for helpful discussions throughout the project. We acknowledge support from the Gordon and Betty Moore Function GBMF 2550.03 to Life Sciences Research Foundation [to K.L.], the Weizmann Institute of Science National Postdoctoral Award Program for Advancing Women in Science [to K.L.], and from NIGMS of the National Institutes of Health under award numbers T32GM007276 [to T.H.M], R01-GM086196 [to W.E.M. and L.S.], R35-GM118067 [to W.E.M.], and R35-GM118071 [to L.S.]. L.S. is a Chan Zuckerberg Biohub Investigator.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests statement
The authors declare no competing interests.
Data Availability
BacTRIP sequencing data for the mapping, transcription, and normalization datasets have been uploaded to the NCBI Sequence Read Archive (SRA) at https://www.ncbi.nlm.nih.gov/sra/ under accession no. PRJNA595753. Lists of plasmids and strains generated as part of this study are available in Supplementary Tables 2 and 3. A complete list of parameters used for modeling the CtrA activation pathway is available in Supplementary Table 7. Source data for most panels in the main and extended data figures is available online in Source Data. The remainder of the data that support the findings of this study are available from the corresponding authors upon request.
Code Availability
The code that supports the findings of this study, including analysis of single-molecule tracks, reaction-diffusion model of CtrA activation pathway and analysis of BacTRIP data are available from the corresponding author upon request.
References
- 1.Shapiro L, McAdams HH & Losick R Why and how bacteria localize proteins. Science 326, 1225–1228, doi: 10.1126/science.1175685 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298, doi: 10.1038/nrm.2017.7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberti S The wisdom of crowds: regulating cell function through condensed states of living matter. J Cell Sci 130, 2789–2796, doi: 10.1242/jcs.200295 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Bowman GR et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol 76, 173–189, doi: 10.1111/j.1365-2958.2010.07088.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman GR et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134, 945–955, doi: 10.1016/j.cell.2008.07.015 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebersbach G, Briegel A, Jensen GJ & Jacobs-Wagner C A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134, 956–968, doi: 10.1016/j.cell.2008.07.016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes JA et al. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc Natl Acad Sci U S A 113, 12490–12495, doi: 10.1073/pnas.1602380113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berge M & Viollier PH End-in-Sight: Cell Polarization by the Polygamic Organizer PopZ. Trends Microbiol 26, 363–375, doi: 10.1016/j.tim.2017.11.007 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Jacobs C, Ausmees N, Cordwell SJ, Shapiro L & Laub MT Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol Microbiol 47, 1279–1290 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Biondi EG et al. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444, 899–904, doi: 10.1038/nature05321 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Werner JN et al. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci U S A 106, 7858–7863, doi: 10.1073/pnas.0901781106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelastro PS, Sliusarenko O & Jacobs-Wagner C Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J Bacteriol 192, 539–552, doi: 10.1128/JB.00985-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iniesta AA, Hillson NJ & Shapiro L Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc Natl Acad Sci U S A 107, 7012–7017, doi: 10.1073/pnas.1001767107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsokos CG & Laub MT Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol 15, 744–750, doi: 10.1016/j.mib.2012.10.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blair JA et al. Branched signal wiring of an essential bacterial cell-cycle phosphotransfer protein. Structure 21, 1590–1601, doi: 10.1016/j.str.2013.06.024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lori C et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523, 236–239, doi: 10.1038/nature14473 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Lasker K, Mann TH & Shapiro L An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr Opin Microbiol 33, 131–139, doi: 10.1016/j.mib.2016.06.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann TH, Seth Childers W, Blair JA, Eckart MR & Shapiro L A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nature communications 7, 11454, doi: 10.1038/ncomms11454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YE et al. Spatial gradient of protein phosphorylation underlies replicative asymmetry in a bacterium. Proc Natl Acad Sci U S A 108, 1052–1057, doi: 10.1073/pnas.1015397108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowman GR et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol 76, 173–189, doi: 10.1111/j.1365-2958.2010.07088.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ptacin JL et al. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci U S A 111, E2046–2055, doi: 10.1073/pnas.1405188111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gahlmann A et al. Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions. Nano letters 13, 987–993, doi: 10.1021/nl304071h (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iniesta AA, Hillson NJ & Shapiro L Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc Natl Acad Sci U S A 107, 7012–7017, doi: 10.1073/pnas.1001767107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsokos CG, Perchuk BS & Laub MT A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell 20, 329–341, doi: 10.1016/j.devcel.2011.01.007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childers WS et al. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS biology 12, e1001979, doi: 10.1371/journal.pbio.1001979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann TH & Shapiro L Integration of cell cycle signals by multi-PAS domain kinases. Proc Natl Acad Sci U S A, doi: 10.1073/pnas.1808543115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levskaya A, Weiner OD, Lim WA & Voigt CA Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001, doi: 10.1038/nature08446 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guntas G et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci U S A 112, 112–117, doi: 10.1073/pnas.1417910112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubey BN et al. Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking. Science advances 2, e1600823, doi: 10.1126/sciadv.1600823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown GC & Kholodenko BN Spatial gradients of cellular phospho-proteins. FEBS Lett 457, 452–454 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Panis G, Murray SR & Viollier PH Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS microbiology reviews 39, 120–133, doi: 10.1093/femsre/fuu002 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Zhou B et al. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS genetics 11, e1004831, doi: 10.1371/journal.pgen.1004831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collier J, McAdams HH & Shapiro L A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci U S A 104, 17111–17116, doi: 10.1073/pnas.0708112104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narayanan S, Kumar L & Radhakrishnan SK Sensory domain of the cell cycle kinase CckA regulates the differential DNA binding of the master regulator CtrA in Caulobacter crescentus. Biochim Biophys Acta Gene Regul Mech 1861, 952–961, doi: 10.1016/j.bbagrm.2018.08.006 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Viollier PH et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A 101, 9257–9262, doi: 10.1073/pnas.0402606101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heid CA, Stevens J, Livak KJ & Williams PM Real time quantitative PCR. Genome Res 6, 986–994 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Domian IJ, Quon KC & Shapiro L Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90, 415–424 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Quon KC, Marczynski GT & Shapiro L Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84, 83–93 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH & Shapiro L A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A 103, 10935–10940, doi: 10.1073/pnas.0604554103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gora KG et al. Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol Microbiol 87, 1277–1289, doi: 10.1111/mmi.12166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akhtar W et al. Using TRIP for genome-wide position effect analysis in cultured cells. Nature protocols 9, 1255–1281, doi: 10.1038/nprot.2014.072 (2014). [DOI] [PubMed] [Google Scholar]
- 42.McAdams HH & Shapiro L System-level design of bacterial cell cycle control. FEBS Lett 583, 3984–3991, doi: 10.1016/j.febslet.2009.09.030 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biondi EG et al. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol 59, 386–401, doi: 10.1111/j.1365-2958.2005.04970.x (2006). [DOI] [PubMed] [Google Scholar]
- 44.Fumeaux C et al. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nature communications 5, 4081, doi: 10.1038/ncomms5081 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan MH, Kozdon JB, Shen X, Shapiro L & McAdams HH An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation. Proc Natl Acad Sci U S A 107, 18985–18990, doi: 10.1073/pnas.1014395107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gora KG et al. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell 39, 455–467, doi: 10.1016/j.molcel.2010.06.024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haakonsen DL, Yuan AH & Laub MT The bacterial cell cycle regulator GcrA is a sigma70 cofactor that drives gene expression from a subset of methylated promoters. Genes Dev 29, 2272–2286, doi: 10.1101/gad.270660.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janakiraman B, Mignolet J, Narayanan S, Viollier PH & Radhakrishnan SK In-phase oscillation of global regulons is orchestrated by a pole-specific organizer. Proc Natl Acad Sci U S A 113, 12550–12555, doi: 10.1073/pnas.1610723113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reisenauer A & Shapiro L DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. Embo J 21, 4969–4977 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez D, Kozdon JB, McAdams HH, Shapiro L & Collier J The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res 42, 3720–3735, doi: 10.1093/nar/gkt1352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laub MT, McAdams HH, Feldblyum T, Fraser CM & Shapiro L Global analysis of the genetic network controlling a bacterial cell cycle. Science 290, 2144–2148 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Laub MT, Chen SL, Shapiro L & McAdams HH Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A 99, 4632–4637, doi: 10.1073/pnas.062065699 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quon KC, Yang B, Domian IJ, Shapiro L & Marczynski GT Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A 95, 120–125 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jenal U The role of proteolysis in the Caulobacter crescentus cell cycle and development. Res Microbiol 160, 687–695, doi: 10.1016/j.resmic.2009.09.006 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Wodarz A Establishing cell polarity in development. Nat Cell Biol 4, E39–44, doi: 10.1038/ncb0202-e39 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Misteli T Beyond the sequence: cellular organization of genome function. Cell 128, 787–800, doi: 10.1016/j.cell.2007.01.028 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Rudner DZ & Losick R Protein subcellular localization in bacteria. Cold Spring Harbor perspectives in biology 2, a000307, doi: 10.1101/cshperspect.a000307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Surovtsev IV & Jacobs-Wagner C Subcellular Organization: A Critical Feature of Bacterial Cell Replication. Cell 172, 1271–1293, doi: 10.1016/j.cell.2018.01.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Folkmann AW & Seydoux G Single-molecule study reveals the frenetic lives of proteins in gradients. Proc Natl Acad Sci U S A 115, 9336–9338, doi: 10.1073/pnas.1812248115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L et al. Real-time imaging of Huntingtin aggregates diverting target search and gene transcription. Elife 5, doi: 10.7554/eLife.17056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bianchi F et al. Steric exclusion and protein conformation determine the localization of plasma membrane transporters. Nature communications 9, 501, doi: 10.1038/s41467-018-02864-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu H et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323, doi: 10.1038/s41586-018-0174-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X et al. RIM and RIM-BP Form Presynaptic Active-Zone-like Condensates via Phase Separation. Mol Cell 73, 971–984 e975, doi: 10.1016/j.molcel.2018.12.007 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Case LB, Ditlev JA & Rosen MK Regulation of Transmembrane Signaling by Phase Separation. Annu Rev Biophys 48, 465–494, doi: 10.1146/annurev-biophys-052118-115534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang WYC et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103, doi: 10.1126/science.aau5721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mann TH & Shapiro L Integration of cell cycle signals by multi-PAS domain kinases. Proc Natl Acad Sci U S A 115, E7166–E7173, doi: 10.1073/pnas.1808543115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345, doi: 10.1038/nmeth.1318 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Thanbichler M, Iniesta AA & Shapiro L A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res 35, e137, doi: 10.1093/nar/gkm818 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toettcher JE, Weiner OD & Lim WA Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434, doi: 10.1016/j.cell.2013.11.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poindexter JS Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev 28, 231–295 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ely B Genetics of Caulobacter crescentus. Methods Enzymol 204, 372–384 (1991). [DOI] [PubMed] [Google Scholar]
- 72.Tsai JW & Alley MR Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol 183, 5001–5007 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schrader JM & Shapiro L Synchronization of Caulobacter crescentus for investigation of the bacterial cell cycle. J Vis Exp, doi: 10.3791/52633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez AM et al. A Localized Complex of Two Protein Oligomers Controls the Orientation of Cell Polarity. MBio 8, doi: 10.1128/mBio.02238-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lew MD, Backlund MP & Moerner WE Rotational mobility of single molecules affects localization accuracy in super-resolution fluorescence microscopy. Nano letters 13, 3967–3972, doi: 10.1021/nl304359p (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Diezmann A, Lee MY, Lew MD & Moerner WE Correcting field-dependent aberrations with nanoscale accuracy in three-dimensional single-molecule localization microscopy. Optica 2, 985–993, doi: 10.1364/OPTICA.2.000985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ovesny M, Krizek P, Borkovec J, Svindrych Z & Hagen GM ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390, doi: 10.1093/bioinformatics/btu202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmittgen TD & Livak KJ Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Pfaffl MW A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reisenauer A, Kahng LS, McCollum S & Shapiro L Bacterial DNA methylation: a cell cycle regulator? J Bacteriol 181, 5135–5139 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Siam R & Marczynski GT Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. Embo J 19, 1138–1147, doi: 10.1093/emboj/19.5.1138 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Viollier PH et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A 101, 9257–9262, doi: 10.1073/pnas.0402606101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iyer-Biswas S et al. Scaling laws governing stochastic growth and division of single bacterial cells. Proc Natl Acad Sci U S A 111, 15912–15917, doi: 10.1073/pnas.1403232111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright CS et al. Intergenerational continuity of cell shape dynamics in Caulobacter crescentus. Scientific reports 5, 9155, doi: 10.1038/srep09155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brassinga AK & Marczynski GT Replication intermediate analysis confirms that chromosomal replication origin initiates from an unusual intergenic region in Caulobacter crescentus. Nucleic Acids Res 29, 4441–4451 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dingwall A & Shapiro L Rate, origin, and bidirectionality of Caulobacter chromosome replication as determined by pulsed-field gel electrophoresis. Proc Natl Acad Sci U S A 86, 119–123 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Umbarger MA et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell 44, 252–264, doi: 10.1016/j.molcel.2011.09.010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le TB, Imakaev MV, Mirny LA & Laub MT High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734, doi: 10.1126/science.1242059 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kozdon JB et al. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc Natl Acad Sci U S A 110, E4658–4667, doi: 10.1073/pnas.1319315110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schrader JM et al. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS genetics 10, e1004463, doi: 10.1371/journal.pgen.1004463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spencer W, Siam R, Ouimet MC, Bastedo DP & Marczynski GT CtrA, a global response regulator, uses a distinct second category of weak DNA binding sites for cell cycle transcription control in Caulobacter crescentus. J Bacteriol 191, 5458–5470, doi: 10.1128/JB.00355-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J Mol Biol 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1. Single-molecule tracks of CtrA-eYFP-14 Exemplary raw data used to produce single-molecule trajectories of CtrA-eYFP-14. First track: entry to the new pole. (Data plotted in Fig. 1f, left.) Second track: exit from the old pole. As noted in Methods, while not all single molecules used to produce tracks persisted this long before photobleaching, these results (reversible slowing of proteins within the poles) were consistently observed in 89 cells for CtrA-eYFP-14.