Abstract
Background
The immobile nature of plants means that they can be frequently confronted by various biotic and abiotic stresses during their lifecycle. Among the various abiotic stresses, water stress, temperature extremities, salinity, and heavy metal toxicity are the major abiotic stresses challenging overall plant growth. Plants have evolved complex molecular mechanisms to adapt under the given abiotic stresses. Long non-coding RNAs (lncRNAs)—a diverse class of RNAs that contain > 200 nucleotides(nt)—play an essential role in plant adaptation to various abiotic stresses.
Results
LncRNAs play a significant role as ‘biological regulators’ for various developmental processes and biotic and abiotic stress responses in animals and plants at the transcription, post-transcription, and epigenetic level, targeting various stress-responsive mRNAs, regulatory gene(s) encoding transcription factors, and numerous microRNAs (miRNAs) that regulate the expression of different genes. However, the mechanistic role of lncRNAs at the molecular level, and possible target gene(s) contributing to plant abiotic stress response and adaptation, remain largely unknown. Here, we review various types of lncRNAs found in different plant species, with a focus on understanding the complex molecular mechanisms that contribute to abiotic stress tolerance in plants. We start by discussing the biogenesis, type and function, phylogenetic relationships, and sequence conservation of lncRNAs. Next, we review the role of lncRNAs controlling various abiotic stresses, including drought, heat, cold, heavy metal toxicity, and nutrient deficiency, with relevant examples from various plant species. Lastly, we briefly discuss the various lncRNA databases and the role of bioinformatics for predicting the structural and functional annotation of novel lncRNAs.
Conclusions
Understanding the intricate molecular mechanisms of stress-responsive lncRNAs is in its infancy. The availability of a comprehensive atlas of lncRNAs across whole genomes in crop plants, coupled with a comprehensive understanding of the complex molecular mechanisms that regulate various abiotic stress responses, will enable us to use lncRNAs as potential biomarkers for tailoring abiotic stress-tolerant plants in the future.
Keywords: Abiotic stresses, Long non-coding RNAs, Gene regulation, Target mimicry
Background
The immobile nature of plants means that they can be frequently confronted by various biotic and abiotic stresses during their lifecycle. Plants have evolved several complex mechanisms to recognize various stress factors, generate appropriate signaling pathways, and respond accordingly by reprogramming the expression of multiple genes at the transcriptional, post-transcriptional, and epigenome level to adapt under harsh environment conditions [1, 2]. The research community has successfully identified several complex mechanisms that plants use at the genetic, physiological, biochemical, and molecular levels to maintain ‘cellular homeostasis’ under unfavorable environments [2, 3]. The discovery of miRNAs (21–24 nt)—a novel class of non-coding RNAs (ncRNAs)—and their regulatory mechanisms for controlling genes involved in various developmental, biological, and stress responses has advanced our understanding of gene regulation in plants [4, 5]. The technical innovations of genome sequencing, especially next-generation sequencing, RNA-sequencing (RNA-seq), and advanced bioinformatics tools, have improved the functional elucidation of various genes at the transcription, post-transcription, post-translation, and epigenetic level [6]. These innovations have enabled the discovery of novel ncRNAs, including lncRNAs, and their role in regulating various biological processes, development, and stress responses in mammals and plants (for details, see [7, 8]). LncRNAs are a diverse class of RNAs, and the largest class acting as ‘biological regulators’ that control transcriptional regulation and genome imprinting [9, 10]. Numerous noteworthy instances of lncRNAs regulating plant development, disease resistance, nutrient acquisition, and other biological processes through chromatin remodeling, histone modification, pri-mRNA alternative splicing, or acting as ‘target mimicry’ have been recorded [11–15]. However, few studies have undertaken genome-wide exploration of lncRNAs, their complex regulatory molecular mechanisms, or functional annotation [16]. Here, we explain the types and functions of lncRNAs and update the roles of various lncRNAs, their target gene(s), and the complex operational molecular mechanisms involved in acclimating plants to the challenging environments of various abiotic stresses.
Biogenesis, type, and functions of lncRNAs
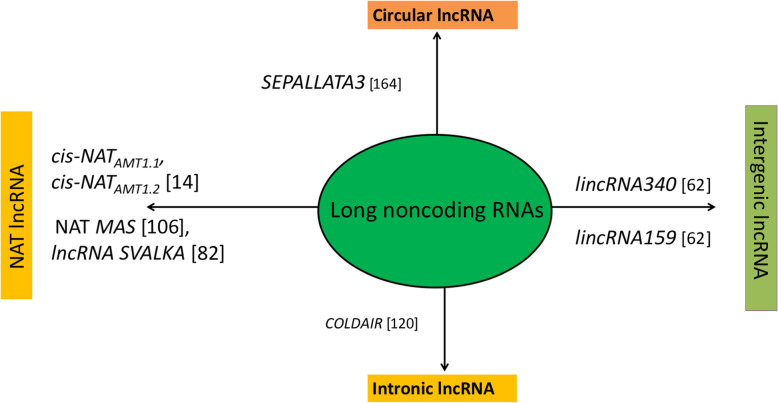
Among the various classes of ncRNAs, lncRNAs are a heterogeneous class of RNA transcripts > 200 nt that are incapable of coding proteins, act as ‘riboregulators,’ are located in the nucleus or cytoplasm, and are transcribed by RNA polymerase II or III and polymerase IV/V [17–19]. Pol IV lncRNAs serve as precursors for small interfering RNAs (siRNAs) [19]. Pol V-dependent lncRNAs assist in modulating the local chromatin loop [20], are transcribed from either strand of the protein-coding locus, may or may not have 5´ cap and poly-adenylation at 3´ tail, and are expressed in a ‘tissue-specific’ manner [21–23]. LncRNAs can be broadly classified as (i) long intergenic ncRNAs (lincRNAs), (ii) intronic ncRNAs (incRNAs), (iii) natural antisense transcripts (NATs), and (iv) circular long non-coding RNAs (circRNAs) based on their location and neighboring protein-coding genes [22, 24–26]. LincRNAs originate from intergenic regions, featuring weakly spliced, polyadenylated tissue-specific expression, and execute trans (distant gene) regulatory function [27–29], while incRNAs are transcribed from intronic regions. NATs originate from complementary DNA strands of sense coding regions [26] and feature cis- and trans-regulatory action [30]. However, circRNAs are in low abundance, originate from the ‘back-splicing reaction of internal exons in pre-mRNA’ [29, 31], feature a covalently closed structure, and display higher sequence conservation than linear lncRNAs [29, 32]. Various types of lncRNAs and their possible biogenesis are illustrated in Figs. 1 and 2. They act functionally as ‘decoy’ or ‘sponge molecules,’ ‘signal molecules,’ ‘backbone molecules,’ and ‘guide molecules’ [25, 34, 35]. Moreover, lncRNAs can be precursors of miRNAs and siRNAs, regulate alternative splicing of pre-mRNAs, and serve as endogenous target mimics (eTM) competing for various miRNAs [20, 36, 37].
Fig. 1.
Various types of lncRNAs with suitable examples in various plants [164]
Fig. 2.
Biogenesis of various lncRNAs. LincRNAs originate from intergenic regions, while incRNAs are transcribed from intronic regions. NATs originate from complementary DNA strands of sense coding regions [26], while circRNAs originate from a ‘back-splicing reaction of internal exons in pre-mRNA’ [29, 31]. The figure is modified from Wang and Chekanova [33] and Wu et al. [29]
Sequence conservation, diversity and phylogenetic features of plant lncRNAs
The highly evolved nature of lncRNAs has resulted in lower sequence conservation across plant and animal species and, thus, fewer phylogenetic relationships [38, 39]. Marques and Ponting [40] reported that < 2% of lncRNAs in Arabidopsis thaliana were evolutionarily conserved across the plant kingdom, which explains the rapid evolution of lncRNA sequences. Conservation analysis of lncRNAs from five monocot and five dicot species demonstrated high sequence conservation at the intra-species and sub-species level [41]. At the interspecific level, lncRNAs remain highly diverged at the nucleotide level and have shown a diverse regulatory role [41, 42]. Mohammadin et al. [43] also supported positional sequence conservation of lncRNAs in Aethionema arabicum and Tarenaya hassleriana at the nucleotide level using a phylogenomics approach. Likewise, Golicz et al. [44] confirmed the sequence homology of four lncRNAs in soybean, chickpea, and Medicago truncatula. Despite sequence dissimilarity, lncRNAs were similar in terms of their low expression capability, short length, and fewer exons and splice variants across numerous plant species, including Arabidopsis, cucumber, maize, chickpea, and soybean [43–47]. Likewise, the conserved function of lncRNAs in both animal and plant species has been investigated [38]. The growing database of lncRNAs and comparative genomics analyses could provide new impetus into the functional conservation of lncRNA genes and their modes of action and function across various plant species [38].
lncRNAs controlling drought stress tolerance
Globally, episodes of drought stress-related events are increasing due to the erratic pattern of rainfall, which affects plant growth and poses a serious challenge for global food security [48]. Plants have a variety of physiological, biochemical, and complex molecular networks, including cascades of various signal transduction pathways, to adapt under drought stress [49]. Advances in molecular biology have uncovered the underlying gene(s)/QTLs and various complex regulatory gene networks and molecular signaling cascades controlling the drought stress response in plants [48, 50]. Subsequently, the discovery of drought-responsive miRNAs and their candidate target genes in various plants has shed light on the molecular mechanisms involved in drought stress adaptation (see [51]). Likewise, emerging evidence has revealed a participatory role of lncRNAs in response to drought stress in plants, capitalizing on the co-expression network based on lncRNAs, miRNAs and protein-coding genes, and transcription factors [52–54]. Notable instances of drought-responsive lncRNAs have been reported in various plant species—six in Arabidopsis [55], 504 in Populus spp. [56], 98 in rice [57], 664 in maize [58], 19 in foxtail millet [59], 185 in cassava [60], and 1597 in switchgrass [52]. LncRNAs could affect the drought stress response by recruiting complex mechanisms based on eTM, antisense transcription-mediated modulation, chromatin modulation, or directly regulating the transcription of various drought-responsive genes [60–63]. Deep sequencing of foxtail millet provided an opportunity to explore 584 lncRNAs [59], of which 17 lincRNAs and two NAT lncRNAs exhibited differential expression under drought stress. Concurrently, the authors found 20 similar lincRNAs and one NAT lncRNA responding to drought stress in sorghum [59]. Only one drought-responsive lncRNA in foxtail millet exhibited sequence co-linearity with the drought-responsive lncRNA in sorghum, demonstrating the low conserved nature of lncRNAs [59]. In Populus trichocarpa, a systematic RNA-seq analysis explored a comprehensive landscape of > 2500 lncRNAs [56], of which 504 were drought-responsive. Functional validation of eight drought-responsive lncRNAs from the 504 drought-responsive lncRNAs using RT-qPCR revealed the up-regulation of six lincRNAs and down-regulation of two lincRNAs under water stress. To survey drought-responsive lncRNAs in the cassava genome, strand-specific RNA-seq data served to identify a set of 318 lncRNAs and 153 NAT lncRNAs responding to cold and drought stress [60]. Of the 51 drought-specific differentially expressed lncRNAs (DElncRNAs), 40 showed up-regulatory action under drought stress. Functional validation of selected lincRNAs using qRT-PCR revealed the up-regulation of lincRNA101, lincRNA391, and lincRNA356 and down-regulation of lincRNA64, lincRNA350, lincRNA182, and lincRNA392 under drought stress. Furthermore, relying on the target mimic mechanism increased the expression of lincRNA340 under drought, which reduced the activity of target miR169 and ultimately increased NUCLEAR FACTOR Y (NF-Y) gene expression [60] see Fig. 3]. Ding et al. [53] recovered 124 DElncRNAs under drought stress in cassava, of which 11 worked as target mimics for miR156, miR164, miR169, and miR172. Functional validation revealed that TCONS_00068353 lncRNA acted as a target mimic for miR156k and miR172c that control various abiotic stress-responsive genes, while TCONS_00060863 and TCONS_00097416 lncRNAs participated in the ABA and ethylene signaling pathways, respectively, under drought stress [53].
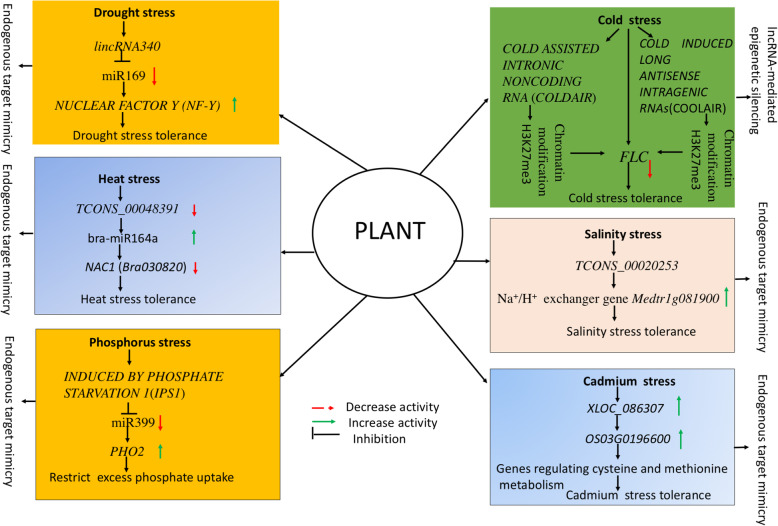
Fig. 3.
LncRNA module controlling environmental abiotic stress response in plants. Relying on the target mimic mechanism increased the expression of lincRNA340 under drought, which reduced the activity of target miR169 and ultimately increased NUCLEAR FACTOR Y (NF-Y) gene expression [60]. The lncRNAs TCONS_00048391 and TCONS_00010856 acted as endogenous target mimics for bra-miR164a, which regulates the heat stress response [64]. Repression of the FLC locus during the early onset of cold stress is controlled by COLD INDUCED LONG ANTISENSE INTRAGENIC RNAs (COOLAIR), an alternatively spliced NAT lncRNA transcribed from the antisense orientation of FLC gene by chromatin modification (reducing active histone mark H3K36me3 and enhancing repressive histone mark H3K27me3) of the FLC locus during vernalization [65]. Under salinity stress TCONS_00020253 up-regulate expression of Na+/H+ exchanger gene Medtr1g081900 in roots [68]. Inductive mechanism of lncRNA INDUCED BY PHOSPHATE STARVATION 1(IPS1) works as an eTM or decoy for miR399 and regulates the target PHO2 gene expression and phosphate homeostasis in Arabidopsis (Franco-Zorrilla et al. [11] and Bari et al. [69]). Significant up-regulatory activity of lncXLOC_086307 suggests that XLOC_086307 likely participates in Cd response processes in rice by controlling the cysteine-rich peptide metabolism-related gene OS03G0196600 [88]
Considering the regulatory mechanism of NAT lncRNA, 98 drought-responsive lncRNAs were recovered in rice using RNA-seq analysis, along with two important drought-responsive lncRNAs NAT Os02g0250700–01 (targeting late embryogenesis abundant protein gene) and NAT Os02g0180800–01 (targeting cinnamoyl-CoA reductase gene) [57]. The expression of these two lncRNAs and their corresponding target genes remained inversely correlated. A study on genome-wide drought-responsive lncRNAs in maize identified 1535 lncRNAs at various developmental stages [54]. The lncRNAs captured at the R1 stage (silking stage) had a critical role in drought stress tolerance. Furthermore, the V-ATPase encoding gene (vpp4) was unearthed as a possible target gene for lncRNAMSTRG.6838.1; vpp4 and the identified lncRNA may work as cis-acting pairs.
Apart from acting as eTM or NAT, lncRNAs could regulate the transcription of various drought stress-responsive genes [52, 62]. The possible role of lncRNAs in regulating drought stress tolerance has been explored in Arabidopsis, with the identification of a novel lncRNA DROUGHT INDUCED lncRNA (DRIR) localized in the nucleus, containing a 755 nt long lincRNA that controls several drought stress-responsive genes, including ABA-signaling genes (ABI5, P5CS1, RD29A, and RD29B), aquaporin genes (NIP1, TIP4), annexin gene (ANNAT7), FUCOSYL TRANSFERASE4 (FUT4) gene, and transcription factor genes (NAC3, WARKY8) at the transcription level [37]. The drirD(T-DNA insertion mutant) and DRIR-overexpressing Arabidopsis lines had higher drought tolerance than wild-type seedlings, as revealed in the higher-fold expression of these genes. Thus, the lncRNA DRIR conferred water-deficit stress tolerance by serving as a positive regulator.
Likewise, lncRNAs regulating various drought-responsive regulatory genes participating in ethylene and ABA synthesis and signaling, calcium signaling, starch and sucrose synthesis, and various metabolic processes have been reported in rice [63], switchgrass (Panicum virgatum L.) [52], P. betulifolia [70], cassava [53, 71], and Cleistogenes songorica [62] (see Table 1). Of the 441 DElncRNAs identified in switchgrass under drought stress imposed at various growth stages, lncRNAs XLOC_053020, XLOC_014465, and XLOC_033252 controlling ABA synthesis, XLOC_074836 contributing to ethylene signaling, and XLOC_005809 involved in trehalose phosphate synthase metabolism were up-regulated, suggesting their significant role in drought- stress tolerance [52]. Various lncRNAs and their possible target gene(s) and working mechanisms contributing to drought stress and other abiotic stress responses in various crops have been identified (see Table 2). Collectively, the various lncRNAs play a role in controlling drought stress by acting as target mimics for various miRNAs that control the expression of various drought-responsive target genes or transcription factors through up- or down-regulation. These emerging lncRNAs could act as a regulatory hub for controlling various drought-responsive hormonal signaling pathways at the transcription, post-transcription, and epigenome level.
Table 1.
Various types of lncRNAs that control abiotic stress responses in plants
| Name of stress | Crop | No. of lncRNAs identified | Number of lncRNAs expressed under stress | Platform and technique used for lncRNAs identification and their function | Function | Reference |
|---|---|---|---|---|---|---|
| Drought | Foxtail millet | 19 lncRNAs | 19 | IlluminaHiSeq 2000,qRT-PCR | Control drought stress response | [59] |
| Drought | Populus trichocarpa | 2542 lincRNAs | 504 | HiSeq™ 2000, RT-qPCR | Drought- stress response | [56] |
| Drought | Rice | 98 lncRNAs | 98 | Illumina HiSeq 2500, qRT-PCR | Regulatory role in drought response | [57] |
| Drought | Arabidopsis | DROUGHT INDUCED lncRNA (DRIR) | DROUGHT INDUCED lncRNA (DRIR) | HiSeq 2000, RT-qPCR | Participate in regulating set of drought responsive genes | [37] |
| Drought | Rice | 3714 | 21 | RT-qPCR, PLncPRO | Differentially expressed under drought stress | [72] |
| Drought | Wheat | – | 59,110 | Illumina HiSeq. 2000, qRT-PCR | Differential expression under drought stress response | [73] |
| Drought and cold | Cassava | 682 lncRNAs | 318 | HiSeq 2500,qRT-PCR, CNCI, CPC, | Hormone signal transduction, sucrose metabolism pathway etc. | [60] |
| Drought | Pyrus betulifolia | 14,478 | 251 | Illumina HiSeq 4000, CNCI, CPC, qRT-PCR | Various metabolic processes | [70] |
| Drought | Panicum virgatum L | 16,551 novel lncRNAs | 1597 | HiSeq2500, qRT-PCR | Regulating drought-stress response | [52] |
| Drought | Maize | 3488 | 1535 | Illumina HiSeq 2500, qRT-PCR | Oxidoreductase activity, water binding, and electron carrier activity | [54] |
| Drought | Cleistogenes songorica | 3397 lncRNAs | 468 | HiSeq2500, CPC, CNCI, CPATqRT-PCR | Regulating drought-stress response | [62] |
| Drought | Cassava | 833 | 124 | Hiseq 4000, qRT-PCR, CNCI, CPC, | Cell-related metabolism, Calvin cycle, hormone metabolism etc. | [53] |
| Drought | Cassava | 1405 | 185 | qRT-PCR | Melatonin responsive controlling drought-stress response | [74] |
| Drought | Cassava | 1379 | 194 | qRT-PCR | ABA signaling regulation | [71] |
| Heat stress | Wheat | 125 putative | 77 | Solexa sequencing technology wheat Affymetrix GeneChip, qRT-PCR | Heat responsive | [75] |
| Heat stress | Brassica rapa ssp. chinensis | 4594 putative lncRNAs | 1686 | Illumina Hiseq. 2500, qRT-PCR CPC,CNCI | Differential expression of these RNA suggested involvement of various phytohormones in heat stress tolerance. | [64] |
| Heat stress and drought | Brassica juncea | 7613 putative lncRNAs | 1614 | qRT-PCR | Associated with enzymatic and non-enzymatic antioxidants under drought and heat stress | [76] |
| Cold and heat | Chinease cabbage | 10,001 | 2236 | Illumina HiSeq™ 2000 qRT-PCR, CPC | Total of 67 and 192 target genes for cold and heat were regulated | [77] |
| Cold stress | Banana | 12,462 lncRNAs | 20 | Illumina HiSeqTM 4000, qPCR, CPC | Cold stress response | [78] |
| Cold stress | Arabidopsis | 379 | 135 | Illumina HiSeq 2500, RT-qPCR | Cold or freezing acclimation | [79] |
| Cold stress | Arabidopsis | SVALKA | SVALKA | Repress CBF1 expression and freezing tolerance | [80] | |
| Cold stress | Grapevine | 2088 | 466 | HiSeq 2500, qRT-PCR, CNCI, CPC, | Related to cold stress response | [81] |
| Cold stress | Chinese cabbage | 2088 | 549 | Illumina HiSeqTM 2000, qPCR | Controlling vernalization | [82] |
| Cold stress | Rice | 1485 lncRNAs | 566 | Illumina HiSeq 2500 platform, qRT-PCR | Controlling cold stress response | [83] |
| Cold stress | Medicago | |||||
| truncatula | 24,368 unique lncRNAs | 983 and 1288 | Illumina HiSeq 4000,Q-PCR | Controlling cold stress response | [84] | |
| Salinity | Arabidopsis | DROUGHT INDUCED lncRNA (DRIR) | DROUGHT INDUCED lncRNA (DRIR) | HiSeq 2000, RT-qPCR | Participate in regulating set of salinity responsive genes | [37] |
| Salinity and drought | Chickpea | 3457 | 13 | RT-qPCR, PLncPRO | Differentially expressed under drought and salinity stress | [72] |
| Salinity | Barley | CNT0018772 and CNT0031477 | 2 | qPCR | Both up- and down- regulatory role in salinity stress | [85] |
| Salinity | Cotton | 1117 unique lncRNAs | 44 | Illumina HiSeq 4000, RT-qPCR | Controls salinity stress genes | [86] |
| Salinity and boron | Maize | 48,345 | 1710 | Illumina MiSeq, RT-qPCR, AgriGO | Nicotianamine biosynthetic and metabolic processes, gene regulation | [87] |
| Salinity | Poplar | 10,646 and 10,531 lncRNAs | 8592 and 3425 | HiSeq 2500 | Regulating osmotin 34, NHX7, RARE-COLD-INDUCIBLE 2B, and WRKY 33 genes | |
| Cadmium stress | Rice | 3558 | 69 lncRNAs were up-regulated and 75 lncRNAs were down-regulated | Illumina HiSeq 2000,CPC, RT-qPCR | Genes related to phtosynthetic pathways are involved in response to Cd stress | [88] |
| and salinity | Wheat | 44,698 | 2064 and 2278 | Regulatory roles in numerous biological processes | [89] | |
| Ca2+-channel blocker | Wheat | 6309 | 177 | HiSeqTM2000, qRT-PCR | Affects various biological processes | [90] |
| Oxidative stress | Rice | 7000 lncRNAs | Hiseq2000, DEGSeq | Down-regulated poly adenylation lncRNAs participate in abiotic stress tolerance | [91] | |
| Waterlogging | Maize | 6099 | 3190 | Illumina HisSeq 4000, qRT-PCR | Metabolic pathways, such as glycolysis and methionine metabolism in response to water logging | [92] |
| Phosphate starvation | Arabidopsis | 1212 novel lncRNAs | 309 | Illumina Hiseq 2000/2500, | Phosphate starvation signaling and regulation | [93] |
| qRT-PCR | Cell wall organization and photosynthesis | |||||
| Phosphate deficiency | Medicago truncatula | 10, 785 | 358 and 224 | Illumina Hiseq2000, qRT-PCRCPC,CNCI | Involved in various signal transduction, chemical detoxification | [61] |
| Phosphorus use efficiency | barley | 188 and 209 | – | Illumina sequencing, qRT-PCR | Related to phosphate starvation | [94] |
| Nitrogen deficiency | Poplar | 388 | 126 | Low nutrition adaptation | [95] | |
| Nitrogen deficiency | Maize | 7245 | 637 | Illumina HiSeq™2500,CPC,qPCR | Nitrogen metabolism, oxidative phosphorylation | [96] |
| Nitrogen deficiency | Rice | 2588 novel putative lncRNA | 2588 | Illumina HiSeq 2500, qRT-PCR | Regulatory role in N-starvation- response | [14] |
| Nitrogen deficiency | Barley | 498 lncRNAs | 56 | Illumina Hiseq Xten platform | Regulatory role in N-starvation- response | [97] |
| qPCR | [98] | |||||
| Boron deficiency | Poncirus | 2101 unique lncRNAs | Illumina HiSeq X Ten platform | Regulatory role in B-starvation response | ||
| trifoliata | qRT-PCR | |||||
| Low nutrient deficiency | Arabidopsis | 60 differentially expressed lincRNAs | 60 differentially expressed lincRNAs | HiSeq2000TM, qRT-PCR | Controlling various nutrient response | [99] |
| CPC=Coding Potential Calculator | ||||||
| CNCI=Coding-Non-Coding Index | ||||||
| CPAT = Coding Potential Assessment Tool | ||||||
Table 2.
Function of various lncRNAs regulating various abiotic stress in plants
| Stress | Crop | Genotype | LncRNA | Target gene | Regulatory mechanism | Reference |
|---|---|---|---|---|---|---|
| Drought | Populus | Nisqually 1 | lincRNA20 and lincRNA2752 | – | Control drought stress by regulating | [56] |
| trichocarpa | lincRNA2962 and lincRNA1039 | ptc-miR476 and ptc-miR169 through eTM | ||||
| LincRNA3241 | ||||||
| Drought | Rice | Oryza sativa cv. Ilmi | NAT Os02g0250700–01 | Os02g0250600–01 | Regulate drought by NAT lncRNAs | [57] |
| NAT Os02g0180800–01 | (late embryogenesis | |||||
| abundant protein) | ||||||
| Os02g0180700–01 | ||||||
| (cinnamoyl-CoA | ||||||
| reductase) | ||||||
| Drought | Rice | DXWR | Up-regulated lncRNAs MSTRG69391 | Transcription factor, calmodulin | Regulate biological processes in | [63] |
| MSTRG41712 and MSTRG68635 and | HSP genes, mitochondrial carrier | response to drought stress | ||||
| down regulated lncRNAs MSTRG65848 | protein gene etc | |||||
| MSTRG27834 and MSTRG46301 | ||||||
| Drought | Cassava | TMS60444 | lincRNA340 | NUCLEAR FACTOR Y (NF-Y) | By targeting miR169 based on target mimicry | [60] |
| Drought | Wheat | Kiziltan and TR39477 | c70772_g2_i1 and c90557_g1_i1 | c69036_g1_i1 and | Drought stress is regulated by | [73] |
| TTD-22 | c9653_g1_i2 | lncRNA-miRNA-mRNA networks | ||||
| Drought | Panicum | Alamo | XLOC_053020 | Pavir.Ia01153 | Regulation of genes related | [52] |
| virgatum L | XLOC_014465 | Pavir.Bb00347 | to ethylene synthesis | |||
| XLOC_033252 | Pavir.Eb01847 | and signaling, ABA synthesis and signaling, | ||||
| XLOC_090250, XLOC_016922, | Pavir.J23169 and | starch and sucrose biosynthesis gene | ||||
| and XLOC_067866 | Pavir.Ca01179 | |||||
| XLOC_074836 | Pavir.J04626 | |||||
| XLOC_008122 | Pavir.J10665 | |||||
| XLOC_081155 | Pavir.Ba00729 | |||||
| XLOC_005809 | Pavir.Ab03141 | |||||
| Drought | Cassava | Ku50 | TCONS_00060863, TCONS_00068353 | CYP707A1 | Genes involved in ABA catabolism, | [53] |
| TCONS_00097416, TCONS_00069665, | CSLD5, ERL1, SPCH, | ethylene signaling. | ||||
| LAX2, HDG11,SCR | Also regulates gene by targeting miR156, | |||||
| TCONS_00040721 | GRF1 and HB51,DOX1 | miR164, miR169, and miR172 | ||||
| Drought | Cleistogenes | MSTRG.43964.1 | Genes related to | By regulating miRNA166, miRNA164, miRNA393, and miRNA397a/b and acting as endogenous target mimics | [62] | |
| songorica | MSTRG.4400.2 | abscisic acid (ABA) | ||||
| signalling pathway, | ||||||
| MSTRG.42613.1 | Genes related to starch | |||||
| MSTRG.25585.13 | and sucrose metabolism | |||||
| Drought | Maize | B73 | lncRNA MSTRG6838.1 | V-ATPase encoding gene, | lncRNA regulating transcriptional | [54] |
| vpp4 | regulation by cis- and trans-acting modes | |||||
| Drought | Cassava | TCONS_00129136, TCONS_00122745 | [71] | |||
| TCONS_00088201,TCONS_00067612 | ||||||
| Drought | Cassava | TCONS_00003360, TCONS_00015102 | Calcium signaling, ABA and | [74] | ||
| TCONS_00149293 | ethylene metabolism | |||||
| Drought | Brassica napus | Q2 and Qinyou8 | XLOC_042431, XLOC_071559, | BnaC06g05090D | IAA, Cytokinin and ABA signalling | [100] |
| XLOC_ 095305, XLOC_100682, | BnaA01g17750D | alpha-trehalose-phosphate synthase | ||||
| XLOC_019521 and XLOC_ 042894 | BnaC07g44670D | |||||
| XLOC_075476 and XLOC_074677, | BnaC02g25020D, BnaC02g25150D, | |||||
| XLOC_074677, XLOC_ 093758 | BnaC02g25200D | |||||
| XLOC_044363 and XLOC_076449 | ||||||
| XLOC_052298 | ||||||
| Heat | Wheat | TAM107 | TahlnRNA27, TalnRNA5, | – | Histone acetylation of TalnRNA5 | [75] |
| Chinease spring | TahlnRNA12, TalnRNA21 | – | ||||
| TahlnRNA23 and TahlnRNA29 | ||||||
| Heat | Chinese cabbage | GHA and XK | TCONS_00048391 | NAC1 (Bra030820) | By targeting bra-miR164a based on | [64] |
| TCONS_00010856 | Bra021232 | target mimicry mechanism | ||||
| TCONS_00004594 | ||||||
| Heat | Cucumis sativus | Improved Jinchun 2 | TCONS_00031790, TCONS_00014332, | – | Interact with miR9748 plant hormone signal | [101] |
| TCONS_00014717,TCONS_00005674 | transduction pathways | |||||
| Heat and | Brassica | TCONS_00051908 | – | By acting as targets and eTMs for the miRNAs | [76] | |
| drought | juncea | TCONS_00088973 | ||||
| Cold | Cassava | TMS60444 | lincRNA159 | NAM, ATAF1/2, | Regulate cold tolerance targeting miRNA164 | [60] |
| CUC2 | based on target mimicry mechanism | |||||
| Cold | Arabidopsis | Col-0 | SVALKA | CBF1 | SVK represses CBF1 and | [80] |
| increase cold acclimation | ||||||
| Cold | Arabidopsis | Col-0 | COLDWRAP | FLC | COLDWRAP reinforc estable | [102] |
| repression of FLC under cold stress | ||||||
| Cold | Arabidopsis | Col-0 | TAS1a | – | By alternative spicing of lncRNA | [79] |
| Cold | Arabidopsis | Col-0 | MAS | MAF4 gene | Histone modification and role of | [103] |
| NAT-lncRNAs regulating gene expression | ||||||
| Cold | Brachypodium | – | BdCOOLAIR1,BdCOOLAIR2 | BdODDSOC1, | BdCOOLAIR transcript represses | [104] |
| distachyon | BdODDSOC2 | function of BdODDSOC gene | [104] | |||
| Cold | Grapevine | Cabernet Sauvignon | VIT_203s0017n00360 | Upregulation of the following target | up and down regulation of the target genes | [81] |
| VIT_207s0031n00070 | genes VIT_216s0100g00380 | |||||
| VIT_201s0011n00530. | VIT_208s0058g00960 | |||||
| IT_209s0002n00340 | VIT_215s0046g02110 | |||||
| VIT_213s0158n00020 | VIT_202s0025g01280 | |||||
| VIT_213s0067n00110 | VIT_200s0246g00150 | |||||
| VIT_200s0225n00020 | VIT_202s0154g00610 | |||||
| Cold stress | Chinease cabbage | RJKB-T24 | MSTRG.4795, MSTRG.18513, | BrFLC and BrMAF genes | Epigenetic modification at BrFLC2as locus, | [82] |
| MSTRG21908, | related to vernalization | epigenetic modification at Bra024350 and | ||||
| MSTRG.259, MSTRG.491 | Bra031888, Bra024351 and Bra031884 loci | |||||
| MSTRG.17153 | ||||||
| Cold stress | Medicago truncatula | Jemalong A17 | lncRNA MtCIR1 | MtCBF genes | Targeting MtCBF genes | [84] |
| Salinity | Medicago truncatula | Jemalong A17 | lncRNA TCONS_00097188, | Medtr6g006990, | By regulating various genes | [68] |
| Salinity | TCONS_00046739, | cytochrome P450 | related to ROS activity, | |||
| Salinity | TCONS_00100258 | Transmembrane proteins gene | secondary messenger molecules, | |||
| and TCONS_00118328 | ||||||
| salinity | TCONS_00047650 | Medtr3g069280, | carbonic anhydrase gene etc. | |||
| Salinity | lncRNA TCONS_00020253 | Medtr1g081900 and | ||||
| Salinity | TCONS_00116877 | Medtr7g094600 | ||||
| Salinity | Arabidosis | – | DRIR | ANNAT7,NAC3 and | Affecting fucosyltransferase or | [37] |
| WRKY8 | NAC3 transcription factor | |||||
| Salinity | Cotton | SN91–11 | lnc_388,lnc_883, | Gh_A09G1182, | Targeting Gh_A09G1182, | [86] |
| Gh_D03G0339 genes | ||||||
| lnc_973 and lnc_253 | and Gh_D03G0339 genes | Regulating ghr-miR399 and ghr-156e by eTM | ||||
| Salinity | Poplar | P. euphratica | Peu_00167161, Pal_00184400 | HKT1 | – | |
| P.alba var. pyramidalis | Pal_00132209 | fucosyltransferase or NAC3 | ||||
| Salinity | Cotton | SN91–11 | lncRNA973 | SOD, CAT, POD and P5CS, | lncRNA973 regulate the | [105] |
| RBOHB, RBOHD, NHX7 | ghr-miR399 and its target gene GhPHO2 | |||||
| MYB5, WRKY46, ERF62, NAC29 | ||||||
| Salinity | Pistacia vera L. | Ghazvini and Sarakhs | 5 NAT-lncRNAs | CERK1, LEA, Laccase genes | NAT-lncRNA regulate ATPase, | [106] |
| TF genes, genes related to | cation transporter, kinase | |||||
| hormone signaling pathways | and UDP-glycosyltransferases genes | |||||
| Water | Maize | B73 | TCONS_00177501 | Zm00001d029280 | Assist in water logging tolerance | [92] |
| logging | TCONS_00124833 | Zm00001d012263 | ||||
| TCONS_00105920 | Zm00001d015618 | |||||
| logging | TCONS_00092298 | Zm00001d018819 | ||||
| Cadmium | Rice | DX142 | XLOC_086307 | OS03G0196600 | Genes regulating cysteine and | [88] |
| cysteine-rich peptide | methionine metabolism and | |||||
| XLOC_086119 and XLOC_066284 | metabolism-related gene | carotenoid biosynthesis | ||||
| carotenoid biosynthesis | ||||||
| Phosphate | Arabidopsis | XLOC_020833, XLOC_001691 | PHR1 | Regulating phosphate homeostasis | [93] | |
| starvation | and XLOC_013661 | by targeting miR399 | ||||
| Phosphate | Medicago | Jemalong A17 | PDIL1,PDIL2 and PDIL3 | MtPHO2 | [61] | |
| starvation | truncatula | Medtr1g074930 | ||||
| Nitrogen | Rice | Nipponbare | cis-NATAMT1.1,cis-NATAMT1.2 | AMT1.1and AMT1.2 | Regulate nitrogen use efficiency | [14] |
| starvation | ||||||
| Nitrogen | Arabidopsis | trans-acting siRNA3 (TAS3) | Nitrate transporter 2, NRT2.4 | Maintains cellular N homeostasis by multiple | [99] | |
| starvation | SEC14p-like protein/Patellin-4 | tasiRNAs targeting ARF2, 3, and 4 | ||||
| Regulatory component | ||||||
| of ABA receptor 3 | ||||||
| PPC3 | ||||||
| Nitrogen | Barley | Liuzhutouzidamai | lnc00090 and lnc000248 | – | lnc00090 and lnc000248 target mimics for | [97] |
| starvation | for hvu-miR399 | |||||
| Boron | Poncirus trifoliata | – | XLOC_002224 | Ciclev10008338m | Targetting Ciclev10008338m gene | [98] |
| deficiecy | ||||||
lncRNAs controlling heat stress tolerance
Heat stress is a major abiotic stress that significantly affects plant growth, physiology, metabolic activity, development, and yield performance [2]. With the current rise in global temperatures, changes in plant phenology and adaptation processes are negatively affecting crop yield, which is challenging global food security [107]. Plants recruit a variety of mechanisms, including adaptive, biochemical, and molecular, to cope with heat stress [2, 108]. Plants produce different phytohormones, heat shock proteins (HSPs)/chaperones, antioxidant enzymes, and metabolites that play a critical role in adjusting to heat stress [108, 109]. At the molecular level, the activation of regulatory pathways plays a role in plant adaptation to heat stress [2]. There is evidence for miRNAs regulating the heat stress response in various plants [110]. The accumulating evidence for lncRNAs acting as an important molecular regulatory layer offers insight into the regulatory mechanism of the heat stress response in crop plants. To explore the role of lncRNAs in conferring a heat stress response, 54 putative heat stress-responsive lncRNAs were identified in wheat using the wheat Affymetrix Gene Chip-based microarray and Solexa sequencing [75]. Among the identified lncRNA transcripts, four and 26 were precursors of miRNAs (viz., miR2004, Ta-miR2010, miR2066) and siRNAs, respectively. Up-regulation of lncRNAs TahlnRNA27 and TalnRNA5 and their corresponding miR2010 and miR2004 was confirmed by qRT-PCR analysis, indicating their significant role in the heat stress response in wheat. However, the heat stress response remained tissue-specific/dependent with TalnRNA5 displaying relatively higher expression in seed tissue than other tissues [75]. Most lncRNAs are weak in sequence conservation; their expression varies from tissue to tissue, developmental stages, and even species to species [65]. Tissue/development-specific expression of lncRNAs has been reported in maize [54, 87] and cassava [53] under drought stress, and species-specific expression was noted for Populus euphratica and Populus alba var. pyramidalis under salinity stress.
A plethora of differentially expressed lncRNAs and their corresponding protein-coding heat stress-responsive target genes and miRNAs have been identified in various crops [64, 77]. Wang et al. [64] explored the up- and down-regulation of lncRNAs and differentially expressed genes (DEGs) involved in the brassinosteroid, ABA, auxin, jasmonic acid, salicylic acid, and ethylene hormone signaling pathways, and DEGs encoding various heat shock proteins across the whole genome, using strand-specific RNA-seq in Brassica rapa under heat stress. Among the three identified heat-responsive DElncRNAs, differential expression of lncRNATCONS_00004594 downstream at the protein-coding gene Bra021232 via qRT-PCR suggested its cis-regulatory expression [64]. Further, lncRNAs TCONS_00048391 and TCONS_00010856 acted as endogenous target mimics for bra-miR164a, which regulates the heat stress response. Consequently, under heat stress, up-regulation of bra-miR164a and down-regulation of lncRNATCONS_00048391 and the target Bra030820 (NAC1) gene rendered heat tolerance in ‘XK’ variety ([64], see Fig. 3). Likewise, the binding of lincRNA159 with conserved miR164 decreased the expression of three miR164-targeted NAC genes (NAM, ATAF1/2, CUC2) in cassava under cold stress [60]. Similarly, drought-responsive lncRNA MSTRG.42613.1 was identified as the target mimic of conserved miRNA164 regulating drought stress in C. songorica [62]. In the future, manipulation of the overexpression or knockout of lncRNAs targeting genes controlling the heat stress response could help us to engineer heat-tolerant crop plants.
lncRNAs controlling cold stress tolerance
Low-temperature stress is an important abiotic stress that challenges plant growth and yield [2, 111]. Plants orchestrate several complex regulatory gene networks of C-repeat binding factors (CBFs) and cold regulated genes (COR) [112] and myriad of novel regulatory miRNAs [110] that enable them to acclimate to cold stress. Advances in genetic and genomic approaches have elucidated several QTLs and probable candidate genes contributing to cold tolerance in plants [111]. Likewise, there is emerging evidence of lncRNAs that regulate the cold stress response in plants [65, 80, 103]. The emerging role of lncRNAs regulating cold acclimation is documented in Arabidopsis [65, 82], cassava [60], Brassica rapa [15, 82], banana, grapevine [81], and Brachypodium [104].
Vernalization is a well-established phenomenon in plant species adapted to cold climates, which prevents flowering during vegetative growth in winter and allows flowering during the reproductive phase under favorable conditions in spring [113]. In Arabidopsis, FLOWERING LOCUS C (FLC) is a well-known regulatory locus that controls flowering time epigenetically [114]. FLC also acts as a suppressor of flowering during cold in Arabidopsis [115]. In this context, the participatory role of lncRNAs in inhibiting expression of the FLC locus by vernalization under cold stress through Polycomb-mediated epigenetic regulation is a well-established mechanism for controlling cold acclimation in Arabidopsis [65, 67, 116]. Repression of the FLC locus during the early onset of cold stress is controlled by COLD INDUCED LONG ANTISENSE INTRAGENIC RNAs (COOLAIR), an alternatively spliced NAT lncRNA transcribed from the antisense orientation of FLC gene by chromatin modification (reducing active histone mark H3K36me3 and enhancing repressive histone mark H3K27me3) of the FLC locus during vernalization [65–67]. Interestingly, Castaings et al. [117] demonstrated the evolutionarily conserved role of class I antisense COOLAIR that controls FLC repression during vernalization in Arabidopsis thaliana, Arabis lyrata, and Arabis alpina species.
Likewise, COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR) [116], transcribed from intron1 of the FLC gene, recruits the Polycomb Repressive Complex 2 (PRC2) that helps in chromatin modification (increase H3K27me3) of the FLC locus and thus represses expression of the FLC locus (see Fig. 3). Subsequently, Kim et al. suggested that “Polycomb-binding lncRNA, COLDWRAP” could further cooperate in the stable repression of the FLC locus during vernalization in Arabidopsis.
Recently, Kindergren et al. [80] advanced our understanding of the cold acclimation mechanism in Arabidopsis by illustrating the novel role of SVALKA and cryptic antisense CBF1 (asCBF1) lncRNAs induced by cold stress. These lncRNAs regulate cold acclimation by suppressing transcription of the CBF1gene by RNA polymerase II (RNAPII) collision derived from lncRNAs SVALKA and asCBF1. Likewise, to explore the role of lncRNAs controlling the cold stress response in Arabidopsis, strand-specific RNA-sequencing (ssRNA-seq) identified 4050 NAT lncRNAs and 2460 lincRNAs as cold-responsive lncRNAs [103]. Among these, the authors substantiated the novel role of MAS (NAT lncRNA_2962), a cis-acting NAT lncRNA induced under cold stress, which activated transcription of the corresponding cold-responsive MADS AFFECTING FLOWERING 4 (MAF4), an FLC family member, by involving WDR5a complex that deposits H3K4me3 at MAF4 gene for its activation. Thus, the activated gene eventually suppresses flowering under cold tress. Likewise, in Brassica rapa, three FLC paralogs that act as a floral repressor during vernalization have been reported [118, 119]. The involvement of NATs at the FLC2 locus of Brassica rapa under cold stress has been reported [120]. RNA-seq driven transcriptome analysis of control and cold-treated leaves of Brassica rapa identified 2088 lncRNAs [82], of which three BrFLC loci contributed to cold stress regulation—only BrFLC2, harboring NAT BrFLC2as (MSTRG.2765), had homology to the COOLAIR transcript of Arabidopsis thaliana and displayed up-regulation under cold stress [82]. Functionally, COOLAIR acts as “cis-NAT with respect to the AtFLC locus” [67]; however, the action of BrFLC2as as cis- or trans-acting mode needs further investigation. Likewise, considering the role of the MAF gene, the Bra024350 locus (homologous to AtMAF1)—with a NAT known as MSTRG.14523—was down-regulated under cold stress. However, the Bra024351 locus (homologous to AtMAF4)—with a NAT known as MSTRG.14524—was not down-regulated under cold stress in Brassica rapa, suggesting that the working mechanism of the lncRNAs mentioned above differed from the lncRNAs involved in vernalization in Arabidopsis thaliana [82]. Furthermore, among the plethora of differentially expressed lincRNAs, NAT lncRNAs identified lncRNAs MSTRG.4795, MSTRG.18513, and MSTRG21908 as up-regulated and MSTRG.259, MSTRG.491, and MSTRG.17153 as down-regulated under cold stress imposed at various stages in Brassica rapa [82].
A genome-wide survey for cold-responsive lncRNAs in grapevine using RNA-seq analysis recovered 284 novel up-regulated lncRNAs, 182 novel down-regulated lncRNAs, 242 DElncRNAs targeting 326 protein-coding genes, and various stress-responsive genes including CBF4 transcription factor genes, late embryogenesis abundant protein genes, and WRKY transcription factor genes [81]. Functional validation of selected lncRNAs through qRT-PCR confirmed up-regulation of lncRNAs VIT_200s0179n00030, VIT_207s0141n00070, and VIT_207s0005n0048 and down-regulation of VIT_201s0010n00070, VIT_208s0007n00270, and VIT_209s0002n00020, suggesting their important role in regulating cold stress tolerance in grapevine [81]. In cassava, to unveil cold and drought-responsive lncRNAs genome-wide, 318 lncRNAs were captured [60]. Considering their contributory role in cold stress tolerance, functional validation of lincRNA419, 207, and 234 revealed their up-regulated activity under cold stress. To decipher the regulatory network of miRNAs, lncRNAs, and the stress-responsive gene controlling cold tolerance, lincRNA159 acting as target mimic for miR164 decreased the expression of NAC genes under cold stress [60]. Apart from these mechanisms, alternative splicing (AS) of lncRNAs and pri-miRNAs could participate in controlling the cold stress response in Arabidopsis [79]. Of the 135 lncRNAs identified with cold-dependent differential expression and differential alternative splicing, induction of TAS1a lncRNA regulated by AS under cold stress was uncovered in Arabidopsis. The unspliced intron-containing transcript AT2G27400.1 produced from TAS1a contained “miR173binding site and tasiRNAs generation site” while the spliced transcript AT2G27400_ID1 remained intronless. Given the decrease in temperature, the AT2G27400_ID1 transcript decreased rapidly in the first 6 h after cold treatment, whereas unspliced AT2G27400.1 increased in the first 3 h. Subsequently, it declined over the next 12 h [79]. Thus, AS of lncRNAs plays an important role in regulating cold stress tolerance. LncRNAs could regulate cold tolerance through chromatin modulation/remodeling, AS mechanisms, and transcriptional regulation of genes contributing to cold tolerance. Further understanding of the working mechanism of lncRNAs controlling cold stress may provide opportunities for engineering cold-tolerant crops.
lncRNAs as new players in plant acclimation under salinity stress
The indiscriminate practice of excessively irrigating farmland and the rapid depletion of groundwater are major factors associated with the increase in salinity-related problems worldwide [121]. Globally, 45 Mha of irrigated land and 32 Mha of hardy land are challenged by salinity stress [122, 123]. Thus, soil salinization remains an increasing constraint to global food production. Under salinity stress, plants suffer from an excessive load of toxic ions, which reduces plant growth and development and grain yield [124].
Plants have evolved several cellular and physiological mechanisms to adapt to salinity stress (see [124]). At the molecular level, a plethora of ion transporter proteins encoded by gene(s)/QTLs and other regulatory genes play a crucial role in controlling salinity stress in various plants (see [121, 124]). Likewise, evidence of regulatory roles of lncRNAs enabling plants to tolerate salinity stress has advanced our understanding of the molecular mechanisms controlling the salinity stress response in plants [37, 55].
To elucidate the functional role of lncRNAs in Medicago truncatula under salinity stress and osmotic stress, several lncRNAs have been identified, including TCONS_00046739 (regulating cytochrome P450 in roots), TCONS_00097188 (regulating photosynthesis by up-regulating Medtr6g006990 gene), TCONS_00047650 (up-regulating expression of the Medtr3g069280 gene encoding phosphatidylinositol-specific phospholipase C), TCONS_00116877 (up-regulating the Medtr7g094600 gene encoding glutathione peroxidase in roots), and TCONS_00020253 (up-regulating expression of Na+/H+ exchanger gene Medtr1g081900 in roots) ([68], see Fig. 3). Likewise, the presence of lncRNA DRIR regulates higher expression of P5CS1, RD29A, RD29B, AtrbohB, FUT4, ANNAT7, and NAC3 genes that confer salinity stress tolerance in the drirD mutant and DRIR-overexpressing lines in Arabidopsis [37]. In cotton, deep transcriptome sequencing of salt-treated leaf tissue facilitated the identification of 44 differentially expressed lincRNAs from 1117 unique lncRNAs [86]. Functional validation of selected lincRNAs via RT-qPCR revealed the up-regulatory role of lnc_388 on cis-acting target leucine-rich repeat 8 (Gh_A09G1182) gene and lnc_883lncRNA targeting on Gh_D03G0339MS_channel protein-coding gene under salinity stress (Table 2). The authors also confirmed the role of lncRNAs lnc_973 and lnc_253 acting as target mimics for ghr-miR399 and ghr-156e under salinity stress [86]. Likewise, 1710 lncRNAs that were responsive to combined salinity and boron stress were explored in the Lluteño landrace of maize using deep transcriptome analysis of leaf and root tissue-derived RNA libraries [87]. Interestingly, a comparison of the genome sequences of three popular maize cultivars (B73, Mo17, and Palomero) and the Lluteño landrace identified the presence of 955 conserved lncRNA transcripts; however, 755 were exclusive to the Lluteño landrace, which may explain its salinity and boron stress tolerance [87]. To gain insight into the salinity and boron response of lncRNAs, functional validation of 12 trans-NAT lncRNAs from 848 differentially expressed trans-NAT lncRNAs suggested their significant role in controlling various stress regulatory gene expression, including combined salinity and boron stress and the nicotinamide metabolic process [87]. Thus, the identified lncRNAs conferred salinity stress tolerance by controlling oxidative stress through modulating genes encoding for antioxidant enzymes and regulating various Na+/H+ exchanger genes and other regulatory genes related to salinity stress.
lncRNAs regulating nutrient deficiency in plants
Nutrient acquisition from soil remains one of the essential physiological processes for regulating plant growth and development [125]. Several molecular mechanisms, including many nutrient transporters, are actively involved in plant nutrient homeostasis [126, 127]. Among the various non-coding regulatory RNAs, evidence of miRNAs and lncRNAs regulating nutrient acquisition has been found in various plants [11, 14, 61].
Among the major nutrients, phosphorus (P) serves as a fundamentally important element contributing to plant growth and development; it also acts as a P source for ATP production [128]. The availability of soil inorganic phosphate (Pi) to plants is constrained by several factors that limit overall plant growth and development [129]. Little information is available on the complex regulatory network of P homeostasis in plants [127, 130]. Several molecular and biochemical mechanisms are activated by plants to improve soil inorganic phosphate availability and increase phosphorus use efficiency (PUE) [127, 129, 130]. In this context, the role of miRNAs controlling phosphate availability has been reported in various plants [110]. Likewise, the emerging role of lncRNAs regulating phosphate content in plants is well-established in Arabidopsis [11, 93], rice [14, 131], and the model legume Medicago truncatula [61]. The working mechanism of miR399 and its target gene PHOSPHATE2 (PHO2) is well-recognized for regulating phosphate content in Arabidopsis [132, 133]. Subsequently, Franco-Zorrilla et al. [11] revealed the inductive mechanism of lncRNA INDUCED BY PHOSPHATE STARVATION 1(IPS1) that works as an eTM or decoy for miR399 and regulates the target PHO2 gene expression and phosphate homeostasis in Arabidopsis. Thus, given the abundance of phosphate, lncRNA IPS1 binds to miR399 and prevents it from acting on target gene PHO2, which presumably abolishes the functional role of phosphate transporters by the ubiquitination pathway, thereby restricting root uptake of excessive phosphate ([69], see Fig. 3). Under phosphate-deficient conditions, the PHO2 gene is suppressed as miR399 degrades the transcript of PHO2 and eventually allows phosphate transporters to accumulate phosphate [132].
Furthermore, the phosphate regulation mechanism—based on the “PHR1–miR399–PHO2” pathway in association with phosphate deficiency-responsive lncRNAPDIL1, a paralog of Mt4—has been demonstrated in Medicago truncatula [61]. The authors established a negative regulatory role of the lncRNAsPDIL2 and PDIL3 controlling the expression of the phosphate transporter gene Medtr1g074930. Likewise, the working mechanism of cis-NATPHO1;2 lncRNA functioning as a translational enhancer of the PHO1;2 gene for phosphate homeostasis has been reported in rice [131].
Like P, nitrogen (N) is an essential nutrient for plant growth and development, and also serves as an N source for amino acids, ATP, and N metabolism in plants [134]. Several QTLs in various crops of agricultural importance reportedly improve nitrogen use efficiency (NUE) [134]. Advances in functional genomics approaches have identified several regulatory gene(s) and transporter genes controlling NUE in crop plants [135]. However, the entire molecular mechanism of N assimilation is not understood in plants [136]. State-of-the-art deep transcriptome sequencing via RNA-seq has further advanced our understanding of N-responsive lncRNAs contributions to N homeostasis in plants. Numerous N-responsive lncRNAs have been uncovered in various plant species viz., rice, maize, poplar [14, 95, 96]. The operating mechanism of lncRNAs cis-NATAMT1.1 and cis-NATAMT1.2, targeting the AMT1 gene for N homeostasis, is well-recognized in rice [14]. A study on lncRNAs in the Arabidopsis genome under various nutrient-deficient conditions uncovered the role of trans-acting siRNA3 (TAS3) as an important lincRNA targeting the nitrate transporter 2 gene, thereby regulating N transport in N-starved environments [99].
Among the various micronutrients, boron (B) is an essential micronutrient for plant growth and development, membrane integrity, and cell wall synthesis [137–139]. Genome-wide exploration of lncRNA regulating B deficiency response in Poncirus trifoliata through strand-specific deep transcriptome analysis detected 2101 unique lncRNAs [98]. Further, expression profiling analysis identified 729 up-regulated and 721 down-regulated lncRNAs under B deficiency stress. Functional validation of selected lncRNAs shed light on the target genes involved in the calcium signaling and plant hormone signal transduction pathways under B deficiency stress in Poncirus trifoliata [98].
The above findings have laid the foundation for future in-depth research on the regulatory role of various lncRNAs controlling nutrient deficiency in plants.
Role of lncRNAs under heavy metal toxicity
The outcome of rapid industrialization, application of heavy doses of chemical fertilizers, and indiscriminate contamination of heavy metals in irrigation water and arable land have posed a serious challenge for crop yields and human health [140], particularly cadmium. To minimize heavy metals moving from the soil into plants, plants use several regulatory molecular mechanisms [140]—lncRNAs may play a crucial role in controlling the uptake of heavy metals into the plant system.
RNA-seq profiling identified 301 cadmium-responsive lncRNAs in Brassica napus, of which 67 were eTMs for 36 Cd-responsive miRNAs [141]. Functional validation of TCONS_00091906, TCONS_00033487, and TCONS_00097191 lncRNA under Cd stress using qRT-PCR analysis indicated their significant role as target mimicry for EL628609, TC182597, and TC203372 mRNAs involved in Cd uptake and detoxification [141]. Likewise, Chen et al. [88] undertook a genome-wide survey of lncRNAs using RNA deep transcriptome sequencing that provided evidence of both up- and down-regulation of lncRNAs involved in the Cd response. Furthermore, functional analysis of DElncRNA provided insight into the role of lncRNAs regulating target genes associated with cysteine and methionine metabolism under Cd stress (see Fig. 3). Considering the mounting evidence of arsenic (As) toxicity in rice, Tang et al. [142] provided novel insights into As-responsive lncRNAs along with other non-coding RNAs regulating the As toxicity response in rice. However, the mechanisms involved in the regulatory role of lncRNAs controlling heavy metals is unknown and needs further research.
Database and web-based resources of lncRNAs
Advances in functional genomics, especially RNA-seq analysis, have enabled the discovery of novel lncRNAs that regulate various biological processes, including stress responses. However, the accurate prediction of lncRNAs, their structure, genomic content, conservation, and functional annotation remains a challenge (see [8]). To address these shortcomings, several web-based resources and databases have been developed, viz., NONCODE provides the comprehensive biological functions of lncRNAs [143–145], PLNlncRbase contains information on 1187 plant lncRNAs from more than 40 species [146], and Plant Long non-coding RNA Database (PLncDB) offers information on 6480 lncRNAs in Arabidopsis [147]. Likewise, the Plant Natural Antisense Transcripts Database (PlantNATsDB) provides information on plant NATs controlling various physiological and development processes [148], Plant ncRNA Database (PNRD) maintains records of 25,739 non-coding RNAs including lncRNAs [149], CANTATAdb maintains 45,117 lncRNAs from 10 plant species [16], CANTATAdb 2.0. annotates plant lncRNAs [155] and PLncPRO provides information on abiotic stress-responsive lncRNAs in rice and chickpea [72]. A detailed list of plant lncRNA databases is in Table 3. Several important tools, such as CPPred [158], REPTree [159], Pfamscan [160], COME [161], PLIT [156], and CPC2 [162], are available to distinguish lncRNAs from mRNAs. Advances in bioinformatics tools and new algorithms could further boost our efforts in discovering novel lncRNAs and their accurate functional annotations.
Table 3.
Databases of various lncRNAs obtained in plant species
| Name | Characteristics | lncRNA and details | References | Link |
|---|---|---|---|---|
| PLncDB | It provides comprehensive data on Arabidopsis lncRNAs | Arabidopsis lncRNAs | [147] | http://chualab.rockefeller.edu/gbrowse2/homepage.html |
| PLNlncRbase | Detailed information on experimentally identified plant lncRNAs | Supply information on 1187 plant lncRNAs in | [146] | http://bioinformatics.ahau.edu.cn/PLNlncRbase/ |
| 43 plant species | ||||
| PNRD | It provides information on different types of ncRNAs | 150 plant species | [149] | http://structuralbiology.cau.edu.cn/PNRD |
| CANTATAdb | Used for annotation of identified lncRNAs | Covers information on lnc RNA on 10 plant species | [16] | http://cantata.amu.edu.pl, http://yeti.amu.edu.pl/CANTATA/ |
| GREENC | Used for annotate lncRNAs | Annotation of more than 120,000 lncRNAs associated to 37 plant species could be done | [150] | http://greenc.sciencedesigners.com/ |
| PLncPRO | Used for prediction of lncRNAs in plants and used fornvestigating abiotic stress responsive lncRNAs in rice and chickpea | 3714 and 3457 lncRNAs in rice and chickpea for drought and salinity | [72] | http://ccbb.jnu.ac.in/plncpro. |
| PlaNC-TE | Provide insights about the relationship between ncRNA and TEs in plants | Information on overlapping of ncRNA and transposon elements from 40 plant genomes | [151] | http://planc-te.cp.utfpr.edu.br |
| EVLncRNAs | It contains lncRNA information on various species including plant | 1543 lncRNAs from 77 species and also 428 plant lncRNAs from 44 plant species | [152, 153] | http://biophy.dzu.edu.cn/EVLncRNAs. |
| CRISPRlnc | Database for validated CRISPR/Cas9 sgRNAs for lncRNAs from variousspecies including plants | 305 lncRNAs and 2102 validated sgRNAs on eight species including plant | [154] | http://www.crisprlnc.org or http://crisprlnc.xtbg.ac.cn |
| CANTATAdb 2.0 | It provides information on annotation of plant lncRNAs | Covers information on lnc RNA on 39 plant species | [155] | http://cantata.amu.edu.pl, http://yeti.amu.edu.pl/CANTATA/ |
| PLIT | Used for investigating of plant lncRNAs from RNA seq data. | Provides information on lncRNA from 8 plant species | [156] | |
| PLncDB | Detail information on plant lncRNAs | Provides plant lincRNAs and lncNATs information | [157] | – |
Conclusion
The rapidly increasing number of plant lncRNAs and their multifaceted regulatory roles in governing various biological processes is becoming a hotspot in biological research [8, 12]. However, genome-wide discovery, characterization, and functional annotation of lncRNAs remain limited in plant species. The increasing availability of reference genome sequences of crop plants could offer opportunities to explore various lncRNAs and their sequence similarity and ‘functional conservation’ using comparative genome analysis [38]. Further, in-depth transcriptome sequencing, rapid advances in computational biology, and increasing databases for lncRNAs and efficient methods/tools could assist in the prediction of accurate lncRNAs and functional annotation of novel lncRNAs. The paucity of mutants corresponding to lncRNAs is another challenge for functional analysis of novel lncRNAs [17]. In this context, CRISPR/Cas9 engineered mutation in novel abiotic stress-responsive lncRNAs could shed light on the function of lncRNAs, and thus help in the design of abiotic stress-tolerant crop plants [163]. The availability of a comprehensive atlas of lncRNAs across whole genomes in crop plants, coupled with a comprehensive understanding of the complex molecular mechanisms that regulate various abiotic stress responses, will enable us to use lncRNAs as potential biomarkers for tailoring abiotic stress-tolerant plants in the future.
Acknowledgments
RJ, MK, MZ acknowledge support from the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China.
Abbreviations
- lncRNA
Long non-coding RNA
- miRNA
MicroRNA
- ncRNAs
Non-coding RNAs
- RNA-seq
RNA-sequencing
- siRNAs
Small interfering RNAs
- eTM
Endogenous target mimics
- lincRNAs
Long intergenic lncRNAs
- NAT
Natural anti-transcript
- DElncRNAs
Differentially expressed lncRNAs
- vpp4
V-ATPase encoding gene
- DRIR
DROUGHT INDUCED lncRNA
- DEGs
Differentially expressed genes
- CBFs
C-repeat binding factors
- COR
Cold regulated genes
- FLC
FLOWERING LOCUS C
- COOLAIR
COLD INDUCED LONG ANTISENSE INTRAGENIC RNAs
- COLDAIR
COLD ASSISTED INTRONIC NONCODING RNA
- RNAPII
RNA polymerase II
- P
Phosphorus
- N
Nitrogen
- B
Boron
- IPS1
INDUCED BY PHOSPHATE STARVATION 1
- NUE
Nitrogen use efficiency
Authors’ contributions
UCJ conceived the idea and wrote the MS. HN, RJ, NM, MK. MZ, KHMS edited the MS. UCJ acknowledges support from the Indian Council of Agricultural Research (ICAR), New Delhi, India. The authors have read and approved the manuscript.
Funding
No fund is required for writing this article.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Uday Chand Jha, Email: uday_gene@yahoo.co.in.
Kadambot H. M. Siddique, Email: kadambot.siddique@uwa.edu.au
References
- 1.He M, He CQ, Ding NZ. Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front Plant Sci. 2018;9:1771. doi: 10.3389/fpls.2018.01771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Li XM, Li HX, Chong K. Crop improvement through temperature resilience. Annu Rev Plant Biol. 2019;70:753–780. doi: 10.1146/annurev-arplant-050718-100016. [DOI] [PubMed] [Google Scholar]
- 3.Kollist H, Zandalinas SI, Sengupta S, Nuhkat M, Kangasjärvi J, Mittler R. Rapid responses to abiotic stress: priming the landscape for the signal transduction network. Trends Plant Sci. 2018;24:25–37. doi: 10.1016/j.tplants.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Swarup R. Denyer T. miRNAs in plant development. Annu Plant Rev. 2019;2 10.1002/9781119312994.apr0649.
- 5.Wani SH, Kumar V, Khare T, Tripathi P, Shah T, Ramakrishna C, et al. miRNA applications for engineering abiotic stress tolerance in plants. Biologia. 2020; 10.2478/s11756-019-00397-7.
- 6.Li J, Liu C, Sun C, et al. Plant stress RNA-seq nexus: a stress-specific transcriptome database in plant cells. BMC Genomics. 2018;19:966. doi: 10.1186/s12864-018-5367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y, Zhang Y, Chen X, Chen Y. Plant noncoding RNAs: hidden players in development and stress responses. Annu Rev Cell Dev Biol. 2019;35:407–431. doi: 10.1146/annurev-cellbio-100818-125218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budak H, Kaya SB, Cagirici HB. Long non-coding RNA in plants in the era of reference sequences. Front Plant Sci. 2020;11:276. doi: 10.3389/fpls.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Böhmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chekanova JA. Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol. 2015;27:207–216. doi: 10.1016/j.pbi.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 12.Nejat N, Mantri N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit Rev Biotechnol. 2018;38:93–105. doi: 10.1080/07388551.2017.1312270. [DOI] [PubMed] [Google Scholar]
- 13.Sun Z, Huang K, Han Z, et al. Genome-wide identification of Arabidopsis long noncoding RNAs in response to the blue light. Sci Rep. 2020;10:6229. doi: 10.1038/s41598-020-63187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin SY, Jeong JS, Lim JY, Kim T, Park JH, Kim JK, et al. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genomics. 2018;19:532. doi: 10.1186/s12864-018-4897-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corona-Gomez JA, Garcia-Lopez IJ, Stadler PF, Fernandez-Valverde SL. Splicing conservation signals in plant long non-coding RNAs. RNA. 2020; 10.1261/rna.074393.119. [DOI] [PMC free article] [PubMed]
- 16.Szcześniak MW, Rosikiewicz W, Makałowska I. CANTATAdb: a collection of plant long non-coding RNAs. Plant Cell Physiol. 2016;l57:e8. doi: 10.1093/pcp/pcv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Hao L, Li D, Zhu L, Hu S. Long non-coding RNAs and their biological roles in plants. Genomics Proteomics Bioinformatics. 2015;13:137–147. doi: 10.1016/j.gpb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, et al. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell. 2014;55:383–396. doi: 10.1016/j.molcel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Herr AJ, Jensen MB, Dalmay T. Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 20.Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M. Battles and hijacks: noncoding transcription in plants. Trends Plant Sci. 2015;20:362–371. doi: 10.1016/j.tplants.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Laurent GS, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 24.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 25.Sun X, Zheng H, Sui N. Regulation mechanism of long noncoding RNA in plant response to stress. Biochem Biophys Res Commun. 2018;503:402–407. doi: 10.1016/j.bbrc.2018.07.072. [DOI] [PubMed] [Google Scholar]
- 26.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Chung PJ, Liu J, Jang IC, Kean MJ. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014;24:444–453. doi: 10.1101/gr.165555.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017;33:540–552. doi: 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence–mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Chu Q, Bai P, Zhu X, Zhang X, Mao L, et al. Characteristics of plant circular RNAs. Brief Bioinform. 2018; 10.1093/bib/bby111. [DOI] [PubMed]
- 33.Wang HV, Chekanova JA. Long noncoding RNAs in plants. Adv Exp Med Biol. 2017;1008:133–154. doi: 10.1007/978-981-10-5203-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang KC, Chang HY. Molecular mechanisms of long non-coding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang N, Cui J, Shi Y, et al. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic Res. 2019;6:28. doi: 10.1038/s41438-018-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin T, Zhao HY, Cui P, Albesher N, Xiong LM. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017;175:1321–1336. doi: 10.1104/pp.17.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 39.Simopoulos CMA, Weretilnyk EA, Golding GB. Molecular traits of long non-protein coding RNAs from diverse plant species show little evidence of phylogenetic relationships. G3 (Bethesda). 2019;9:2511–2520. doi: 10.1534/g3.119.400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng P, Liu S, Nie X, Weining S, Wu L. Conservation analysis of long non-coding RNAs in plants. Sci China Life Sci. 2018;61:190–198. doi: 10.1007/s11427-017-9174-9. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Bai X, Luo W, Feng Y, Shao X, Bai Q, et al. Genome-wide identification of long noncoding RNAs and their responses to salt stress in two closely related poplars. Front Genet. 2019;10:777. doi: 10.3389/fgene.2019.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohammadin S, Edger PP, Pires JC, Schranz ME. Positionally-conserved but sequence-diverged: identification of long non-coding RNAs in the Brassicaceae and Cleomaceae. BMC Plant Biol. 2015;15:217. doi: 10.1186/s12870-015-0603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Golicz AA, Singh MB, Bhalla PL. The Long intergenic noncoding RNA (LincRNA) landscape of the soybean genome. Plant Physiol. 2018;176:2133–2147. doi: 10.1104/pp.17.01657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014;15:R40. doi: 10.1186/gb-2014-15-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao Z, Fan C, Cheng T, Su Y, Wei Q, Li G. Genome-wide identification, characterization and evolutionary analysis of long intergenic noncoding RNAs in cucumber. PLoS One. 2015;10:e0121800. doi: 10.1371/journal.pone.0121800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khemka N, Singh VK, Garg R, Jain M. Genome-wide analysis of long intergenic non-coding RNAs in chickpea and their potential role in flower development. Sci Rep. 2016;6:33297. doi: 10.1038/srep33297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jha UC, Bohra A, Nayyar H. Advances in “omics” approaches to tackle drought stress in grain legumes. Plant Breed. 2019;139:1–26. doi: 10.1111/pbr.12761. [DOI] [Google Scholar]
- 49.Lamaoui M, Jemo M, Datla R, Bekkaoui F. Heat and drought stresses in crops and approaches for their mitigation. Front Chem. 2018;6:26. doi: 10.3389/fchem.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta PK, Balyan HS, Gahlaut V. QTL analysis for drought tolerance in wheat: present status and future possibilities. Agronomy. 2017;7:5. doi: 10.3390/agronomy7010005. [DOI] [Google Scholar]
- 51.Ferdous J, Hussain SS, Shi BJ. Role of microRNAs in plant drought tolerance. Plant Biotechnol J. 2015;13:293–305. doi: 10.1111/pbi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang C, Tang G, Peng X, Sun F, Liu S, Xi Y. Long non-coding RNAs of switchgrass (Panicum virgatum L.) in multiple dehydration stresses. BMC Plant Biol. 2018;18:79. doi: 10.1186/s12870-018-1288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding Z, Tie W, Fu L, Yan Y, Liu G, Yan W, et al. Strand-specific RNA-seq based identification and functional prediction of drought-responsive lncRNAs in cassava. BMC Genomics. 2019;20:214. doi: 10.1186/s12864-019-5585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pang J, Zhang X, Ma X, Zhao J. Spatio-temporal transcriptional dynamics of maize long non-coding RNAs responsive to drought stress. Genes (Basel). 2019;10:138. doi: 10.3390/genes10020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben Amor B, Wirth S, Merchan F, Laporte P, d'Aubenton-Carafa Y, Hirsch J, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shuai P, Liang D, Tang S, Zhang Z, Ye CY, Su Y, et al. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot. 2014;65:4975–4983. doi: 10.1093/jxb/eru256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung PJ, Jung H, Jeong DH, Ha SH, Choi YD, Kim JK. Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genomics. 2016;17:563. doi: 10.1186/s12864-016-2997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang W, Han Z, Guo Q, Liu Y, Zheng Y, Wu F, et al. Identification of maize long non-coding RNAs responsive to drought stress. PLoS One. 2014;9:e98958. doi: 10.1371/journal.pone.0098958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi X, Xie S, Liu Y, Yi F, Yu J. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol Biol. 2013;83:459–473. doi: 10.1007/s11103-013-0104-6. [DOI] [PubMed] [Google Scholar]
- 60.Li S, Yu X, Cheng Z, Zhao P, He Y, Wang W, et al. Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci Rep. 2017;7:45981. doi: 10.1038/srep45981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang T, Zhao M, Zhang X, Liu M, Yang C, Chen Y, et al. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J Exp Bot. 2017;68:5937–5948. doi: 10.1093/jxb/erx384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan Q, Wu F, Yan Z, Li J, Ma T, Zhang Y, et al. Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery. BMC Plant Biol. 2019;19:23. doi: 10.1186/s12870-018-1626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weidong Q, Hongping C, et al. Systematic characterization of long non-coding RNAs and their responses to drought stress in Dongxiang wild rice. Ric Sci. 2020;27:21–31. doi: 10.1016/j.rsci.2019.12.003. [DOI] [Google Scholar]
- 64.Wang A, Hu J, Gao C, Chen G, Wang B, Lin C, et al. Genome-wide analysis of long noncoding RNAs unveils the regulatory roles in the heat tolerance of Chinese cabbage (Brassica rapa ssp. chinensis) Sci Rep. 2019;9:5002. doi: 10.1038/s41598-019-41428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 66.Sun QW, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Csorba T, Questa JI, Sun Q, Dean C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci U S A. 2014;111:16160–16165. doi: 10.1073/pnas.1419030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang TZ, Liu M, Zhao MG, Chen R, Zhang WH. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015;15:131. doi: 10.1186/s12870-015-0530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bari R, Datt Pant B, Stitt M, Scheible WR. PHO2, micro- RNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J, Lin J, Kan J, Wang H, Li X, Yang Q, et al. Genome-wide identification and functional prediction of novel drought-responsive lncRNAs in Pyrus betulifolia. Genes (Basel). 2018;9:311. doi: 10.3390/genes9060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu C, Ding Z, Chen M, Yang G, et al. Identification and functional prediction of lncRNAs in response to PEG and ABA treatment in cassava. Environ Exp Bot. 2019;166:103809. doi: 10.1016/j.envexpbot.2019.103809. [DOI] [Google Scholar]
- 72.Singh U, Khemka N, Rajkumar MS, Garg R, Jain M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017;45:e183. doi: 10.1093/nar/gkx866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cagirici HB, Alptekin B, Budak H. RNA sequencing and co-expressed long non-coding RNA in modern and wild wheats. Sci Rep. 2017;7:10670. doi: 10.1038/s41598-017-11170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding Z, Wu C, Tie W, Yan Y, He G, Hu W. Strand-specific RNA-seq based identification and functional prediction of lncRNAs in response to melatonin and simulated drought stresses in cassava. Plant Physiol Biochem. 2019;140:96–104. doi: 10.1016/j.plaphy.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 75.Xin M, Wang Y, Yao Y, Song N, Hu Z, Qin D, et al. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011;11:61. doi: 10.1186/1471-2229-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhatia G, Singh A, Verma D, Sharma S, Singh K. Genome-wide investigation of regulatory roles of lncRNAs in response to heat and drought stress in Brassica juncea (Indian mustard) Environ Exp Bot. 2019;171:103922. doi: 10.1016/j.envexpbot.2019.103922. [DOI] [Google Scholar]
- 77.Song XM, Liu GF, Huang ZN, Duan WK, Tan HW, Li Y, et al. Temperature expression patterns of genes and their co expression with LncRNAs revealed by RNA-Seq in non-heading Chinese cabbage. BMC Genomics. 2016;17:297. doi: 10.1186/s12864-016-2625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu W, Cheng C, Lin Y, XuHan X, Lai Z. Genome-wide identification and characterization of mRNAs and lncRNAs involved in cold stress in the wild banana (Musa itinerans) PLoS One. 2018;13:e0200002. doi: 10.1371/journal.pone.0200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calixto CPG, Tzioutziou NA, James AB, Hornyik C, Guo W, Zhang R, et al. Cold-dependent expression and alternative splicing of arabidopsis long non-coding RNAs. Front Plant Sci. 2019;10:235. doi: 10.3389/fpls.2019.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kindgren P, Ard R, Ivanov M, Marquardt S. Transcriptional read-through of the long noncoding RNA SVALKA governs plant cold acclimation. Nat Commun. 2018;9:4561. doi: 10.1038/s41467-018-07010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang P, Dai L, Ai J, Wang Y, Ren F. Identification and functional prediction of cold-related long noncoding RNA (lncRNA) in grapevine. Sci Rep. 2019;9:6638. doi: 10.1038/s41598-019-43269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shea DJ, Nishida N, Takada S, Itabashi E, Takahashi S, Akter A, et al. Long noncoding RNAs in Brassica rapa L following vernalization. Sci Rep. 2019;9:9302. doi: 10.1038/s41598-019-45650-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leng Y, Sun J, Wang J, Liu H, Zheng H, et al. Genome-wide lncRNAs identification and association analysis for cold-responsive genes at the booting stage in rice (Oryza sativa L.) plant. Genome. 2020;13:e20020. doi: 10.1002/tpg2.20020. [DOI] [PubMed] [Google Scholar]
- 84.Zhao M, Wang T, Sun T, Yu X, Tian R, Zhang WH. Identification of tissue-specific and cold- responsive lncRNAs in Medicago truncatula by high-throughput RNA sequencing. BMC Plant Biol. 2020;20:99. doi: 10.1186/s12870-020-2301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karlik E, Gözükırmızı N. Evaluation of barley lncRNAs expression analysis in salinity stress. Russ J Genet. 2018;54:198–220. doi: 10.1134/S1022795418020096. [DOI] [Google Scholar]
- 86.Deng F, Zhang X, Wang W, Yuan R, Shen F. Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol. 2018;18:23. doi: 10.1186/s12870-018-1238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huanca-Mamani W, Arias-Carrasco R, Cárdenas-Ninasivincha S, Rojas-Herrera M, Sepúlveda-Hermosilla G, Caris-Maldonado JC, et al. Long non-coding RNAs responsive to salt and boron stress in the hyper-arid Lluteño maize from Atacama desert. Genes. 2018;9:170. doi: 10.3390/genes9030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L, Shi S, Jiang N, Khanzada H, Wassan GM, Zhu C, et al. Genome-wide analysis of long non-coding RNAs affecting roots development at an early stage in the rice response to cadmium stress. BMC Genomics. 2018;19:460. doi: 10.1186/s12864-018-4807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shumayla SS, Taneja M, Tyagi S, Singh K, Upadhyay SK. Survey of high throughput RNA-Seq data reveals potential roles for lncRNAs during development and stress response in bread wheat. Front Plant Sci. 2017;8:1019. doi: 10.3389/fpls.2017.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma K, Shi W, Xu M, Liu J, Zhang F. Genome-wide identification and characterization of long non-coding RNA in wheat roots in response to Ca2+ channel blocker. Front Plant Sci. 2018;9:244. doi: 10.3389/fpls.2018.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan J, Li J, Yang Y, Tan C, Zhu Y, Hu L, et al. Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa. Plant J. 2018;93:814–827. doi: 10.1111/tpj.13804. [DOI] [PubMed] [Google Scholar]
- 92.Yu F, Tan Z, Fang T, Tang K, Liang K, Qiu F. A comprehensive transcriptomics analysis reveals long non-coding RNA to be involved in the key metabolic pathway in response to waterlogging stress in maize. Genes. 2020;11:267. doi: 10.3390/genes11030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan J, Zhang Y, Dong J, Sun Y, Lim BL, Liu D, et al. Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics. 2016;17:655. doi: 10.1186/s12864-016-2929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ren P, Meng Y, Li B, Ma X, Si E, Lai Y, et al. Molecular mechanisms of acclimatization to phosphorus starvation and recovery underlying full-length transcriptome profiling in barley (Hordeum vulgare L.) Front Plant Sci. 2018;9:500. doi: 10.3389/fpls.2018.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen M, Wang C, Bao H, Chen H, Wang Y. Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol Genet Genomics. 2016;291:1663–1680. doi: 10.1007/s00438-016-1210-3. [DOI] [PubMed] [Google Scholar]
- 96.Lv Y, Liang Z, Ge M, Qi W, Zhang T, Lin F, et al. Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.) BMC Genomics. 2016;17:350. doi: 10.1186/s12864-016-2650-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Z, Jiang Q, Jiang P, et al. Novel low-nitrogen stress-responsive long non-coding RNAs (lncRNA) in barley landrace B968 (Liuzhutouzidamai) at seedling stage. BMC Plant Biol. 2020;20:142. doi: 10.1186/s12870-020-02350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou GF, Zhang LP, Li BX, et al. Genome-wide identification of long non-coding RNA in trifoliate Orange (Poncirus trifoliata (L.) Raf) leaves in response to boron deficiency. Int J Mol Sci. 2019;2:5419. doi: 10.3390/ijms20215419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fukuda M, Nishida S, Kakei Y, Shimada Y, Fujiwara T. Genome-wide analysis of long intergenic noncoding RNAs responding to low-nutrient conditions in Arabidopsis thaliana - possible involvement of trans-acting siRNA3 in response to low nitrogen. Plant Cell Physiol. 2019;60:1961–1973. doi: 10.1093/pcp/pcz048. [DOI] [PubMed] [Google Scholar]
- 100.Tan X, Li S, Hu L, Zhang C. Genome-wide analysis of long non-coding RNAs (lncRNAs) in two contrasting rapeseed (Brassica napus L.) genotypes subjected to drought stress and re- watering. BMC Plant Biol. 2020;20:81. doi: 10.1186/s12870-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He X, Guo S, Wang Y, Wang L, Shu S, Sun J. Systematic identification and analysis of heat-stress-responsive lncRNAs, circRNAs and miRNAs with associated co-expression and ceRNA networks in cucumber (Cucumis sativus L.) Physiol Plant. 2020;168:736–754. doi: 10.1111/ppl.12997. [DOI] [PubMed] [Google Scholar]
- 102.Kim DH, Sung S. Vernalization-triggered intragenic chromatin-loop formation by long noncoding RNAs. Dev Cell. 2017;40:302–12.e4. doi: 10.1016/j.devcel.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao X, Li J, Lian B, Gu H, Li Y, Qi Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun. 2018;9:5056. doi: 10.1038/s41467-018-07500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiao F, Pahwa K, Manning M, Dichy N, Geuten K. Cold induced antisense transcription of FLOWERING LOCUS C in distant grasses. Front Plant Sci. 2019;10:72. doi: 10.3389/fpls.2019.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang X, et al. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019;19:459. doi: 10.1186/s12870-019-2088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jannesar M, Seyedi SM, Moazzam JM, et al. A genome-wide identification, characterization and functional analysis of salt-related long non-coding RNAs in non-model plant Pistacia vera L. using transcriptome high throughput sequencing. Sci Rep. 2020;10:5585. doi: 10.1038/s41598-020-62108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Piao S, Liu Q, Chen A, Janssens I, Fu Y, Dai J, et al. Plant phenology and global climate change: current progresses and challenges. Glob Chang Biol. 2019;25:1922–1940. doi: 10.1111/gcb.14619. [DOI] [PubMed] [Google Scholar]
- 108.Janni M, Gullì M, et al. Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J Exp Bot. 2020; 10.1093/jxb/eraa034. [DOI] [PMC free article] [PubMed]
- 109.Jha UC, Bohra A, Jha R, Parida S. Integrated ‘omics’ approaches to sustain major global grain legume productivity under heat stress. Plant Breed. 2017;136:437–459. doi: 10.1111/pbr.12489. [DOI] [Google Scholar]
- 110.Song X, Li Y, Cao X, Qi Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol. 2019;70:489–525. doi: 10.1146/annurev-arplant-050718-100334. [DOI] [PubMed] [Google Scholar]
- 111.Jha UC, Bohra AB, Jha R. Breeding approaches and genomics technologies to increase crop yield under low-temperature stress. Plant Cell Rep. 2017;36:1–35. doi: 10.1007/s00299-016-2073-0. [DOI] [PubMed] [Google Scholar]
- 112.Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- 113.Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 114.Whittaker C, Dean C. The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu Rev Cell Dev Biol. 2017;33:555–575. doi: 10.1146/annurev-cellbio-100616-060546. [DOI] [PubMed] [Google Scholar]
- 115.Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 116.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 117.Castaings L, Bergonzi S, Albani MC, Kemi U, Savolainen O, Coupland G. Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat Commun. 2014;5:4457. doi: 10.1038/ncomms5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Itabashi E, Osabe K, Fujimoto R, Kakizaki T. Epigenetic regulation of agronomical traits in Brassicaceae. Plant Cell Rep. 2018;37:87–101. doi: 10.1007/s00299-017-2223-z. [DOI] [PubMed] [Google Scholar]
- 119.Shea DJ, Itabashi E, Takada S, Fukai E, et al. The role of FLOWERING LOCUS C in vernalization of brassica: the importance of vernalization research in the face of climate change. Crop Pasture Sci. 2018;69:30–39. doi: 10.1071/CP16468. [DOI] [Google Scholar]
- 120.Li X, Zhang S, Bai J, He Y. Tuning growth cycles of Brassica crops via natural antisense transcripts of BrFLC. Plant Biotechnol J. 2016;14:905–914. doi: 10.1111/pbi.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jha UC, Bohra A, Jha R, Parida SK. Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 2019;38:255–277. doi: 10.1007/s00299-019-02374-5. [DOI] [PubMed] [Google Scholar]
- 122.Zelm EV, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020;71:403–433. doi: 10.1146/annurev-arplant-050718-100005. [DOI] [PubMed] [Google Scholar]
- 123.FAO . FAO land and plant nutrition management service. Rome: Food and Agriculture Organization of the United Nations; 2015. [Google Scholar]
- 124.Ismail AM, Horie T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol. 2017;68:19.1–19.30. doi: 10.1146/annurev-arplant-042916-040936. [DOI] [PubMed] [Google Scholar]
- 125.Islam MS. Sensing and uptake of nitrogen in rice plant: a molecular view. Ric Sci. 2019;26:343–355. doi: 10.1016/j.rsci.2018.12.007. [DOI] [Google Scholar]
- 126.Tegeder M, Masclaux-Daubresse C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2017;217:35–53. doi: 10.1111/nph.14876. [DOI] [PubMed] [Google Scholar]
- 127.Wang Z, Zheng Z, Song L, Liu D. Functional characterization of Arabidopsis PHL4 in plant response to phosphate starvation. Front Plant Sci. 2018;9:1432. doi: 10.3389/fpls.2018.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Adnan M, Zahir S, Fahad S, Arif M, Mukhtar A, Imtiaz AK, et al. Phosphate-solubilizing bacteria nullify the antagonistic effect of soil calcification on bioavailability of phosphorus in alkaline soils. Sci Rep. 2018;8:4339. doi: 10.1038/s41598-018-22653-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Puga MI, Rojas-Triana M, de Lorenzo L, Leyva A, Rubio V, Paz-Ares J. Novel signals in the regulation of pi starvation responses in plants: facts and promises. Curr Opin Plant Biol. 2017;39:40–49. doi: 10.1016/j.pbi.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 130.Wang F, Deng M, Xu J, Zhu X, Mao C. Molecular mechanism of phosphate transport and signaling in higher plants. Semin Cell Dev Biol. 2018;74:114–122. doi: 10.1016/j.semcdb.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 131.Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell. 2013;25:4166–4182. doi: 10.1105/tpc.113.116251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang TK, Han CL, Lin SI, et al. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell. 2013;25:4044–4060. doi: 10.1105/tpc.113.115998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hawkesford MJ, Griffiths S. Exploiting genetic variation in nitrogen use efficiency for cereal crop improvement. Curr Opin Plant Biol. 2019;49:35–42. doi: 10.1016/j.pbi.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Curci PL, AieseCigliano R, Zuluaga DL, Janni M, Sanseverino W, Sonnante G. Transcriptomic response of durum wheat to nitrogen starvation. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-01377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Simons M, Saha R, Guillard L, Clement G, Armengaud P, Canas R, et al. Nitrogen-use efficiency in maize (Zea mays L.): from ‘omics’ studies to metabolic modelling. J Exp Bot. 2014;65:5657–5671. doi: 10.1093/jxb/eru227. [DOI] [PubMed] [Google Scholar]
- 137.Cakmak I, Römheld V. Boron deficiency-induced impairments of cellular functions in plants. Plant and Soil. 1997;193:71–83. doi: 10.1023/A:1004259808322. [DOI] [Google Scholar]
- 138.Camacho-Cristóbal JJ, Anzellotti D, González-Fontes A. Changes in phenolic metabolism of tobacco plants during short-term boron deficiency. Plant Physiol Biochem. 2002;40:997–1002. doi: 10.1016/S0981-9428(02)01463-8. [DOI] [Google Scholar]
- 139.Camacho-Cristóbal JJ, González-Fontes A. Boron deficiency decreases plasmalemma H+-ATPase expression and nitrate uptake and promotes ammonium assimilation into asparagine in tobacco roots. Planta. 2007;226:443–51. [DOI] [PubMed]
- 140.Ali H, Khan E, Ilahi I. Environmental chemistry and ecotoxicology of hazardous heavy metals: environmental persistence, toxicity, and bioaccumulation. J Chem. 2019;6730305:1–14. [Google Scholar]
- 141.Feng SJ, Zhang XD, Liu XS, Tan SK, Chu SS, Meng JG, et al. Characterization of long non-coding RNAs involved in cadmium toxic response in Brassica napus. RSC Adv. 2016;6:82157. doi: 10.1039/C6RA05459E. [DOI] [Google Scholar]
- 142.Tang Z, Xu M, Ito H, Cai J, Ma X, Qin J, Yu D, Meng Y. Deciphering the non-coding RNA-level response to arsenic stress in rice. Plant Signal Behav. 2019;14:1629268. doi: 10.1080/15592324.2019.1629268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liao P, Li S, Cui X, Zheng Y. A comprehensive review of web-based resources of non-coding RNAs for plant science research. Int J Biol Sci. 2018;14:819–832. doi: 10.7150/ijbs.24593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu C, Bai B, Skogerbo G, Cai L, Deng W, Zhang Y, et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao Y, Hui L, Fang S, et al. NONCODE 2016: an informative and valuable data source of long noncoding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xuan H, Zhang L, Liu X, et al. PLNlncRbase: a resource for experimentally identified lncRNAs in plants. Gene. 2015;573:328–332. doi: 10.1016/j.gene.2015.07.069. [DOI] [PubMed] [Google Scholar]
- 147.Jin J, Liu J, Wang H, et al. PLncDB: plant long noncoding RNA database. Bioinformatics. 2013;29:1068–1071. doi: 10.1093/bioinformatics/btt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen D, Yuan C, Zhang J, Zhang Z, Bai L, Meng Y, et al. PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res. 2012. 2012;40:D1187–D1193. doi: 10.1093/nar/gkr823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yi X, Zhang Z, Ling Y, et al. PNRD: a plant non-coding RNA database. Nucleic Acids Res. 2015;43:S982–9. Szcześniak MW, Rosikiewicz W, Makałowska I. CANTATAdb: a collection of plant long non-coding RNAs. Plant Cell Physiol. 2016;57:e8.
- 150.PaytuvíGallart A, Hermoso Pulido A, Anzar Martínez de Lagrán I, Sanseverino W, Aiese Cigliano R. GREENC: a wiki-based database of plant lncRNAs. Nucleic AcidRes. 2016;44:D1161–D1166. doi: 10.1093/nar/gkv1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pedro DLF, Lorenzetti APR, Domingues DS, Paschoal AR. PlaNC-TE: a comprehensive knowledgebase of noncoding RNAs and transposable elements in plants. Database (Oxford). 2018;2018:1–7. doi: 10.1093/database/bay078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhou B, Zhao H, Yu J, Guo C, Dou X, Song F, et al. EVLncRNAs: a manually curated database for long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 2018;46:D100–D105. doi: 10.1093/nar/gkx677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhou B, Zhao H, Yu J, Guo C, Dou X, Song F, et al. Experimentally validated plant lncRNAs in EVLncRNAs database. Methods Mol Biol. 1933;2019:431–437. doi: 10.1007/978-1-4939-9045-0_27. [DOI] [PubMed] [Google Scholar]
- 154.Chen W, Zhang G, Li J, Zhang X, Huang S, Xiang S, et al. CRISPRlnc: a manually curated database of validated sgRNAs for lncRNAs. Nucleic Acids Res. 2019;47:D63–D68. doi: 10.1093/nar/gky904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Szcześniak MW, Bryzghalov O, Ciomborowska-Basheer J, Makałowska I. CANTATAdb 2.0: expanding the collection of plant long noncoding RNAs. Methods Mol Biol. 2019;1933:415–429. doi: 10.1007/978-1-4939-9045-0_26. [DOI] [PubMed] [Google Scholar]
- 156.Deshpande S, Shuttleworth J, Yang J, Taramonli S, England M. PLIT: an alignment-free computational tool for identification of long non-coding RNAs in plant transcriptomic datasets. Comput Biol Med. 2019;105:169–181. doi: 10.1016/j.compbiomed.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 157.Sun HX, Chua NH. Bioinformatics approaches to studying plant long noncoding RNAs (lncRNAs): identification and functional interpretation of lncRNAs from RNA-Seq data sets. Methods Mol Biol. 2019;1933:197–205. doi: 10.1007/978-1-4939-9045-0_11. [DOI] [PubMed] [Google Scholar]
- 158.Tong X, Liu S. CPPred: coding potential prediction based on the global description of RNA sequence. Nucleic Acids Res. 2019;47:e43. doi: 10.1093/nar/gkz087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Negri TDC, Alves WAL, Bugatti PH, Saito PTM, Domingues DS, Paschoal AR. Pattern recognition analysis on long noncoding RNAs: a tool for prediction in plants. Brief Bioinform. 2018;20:682–689. doi: 10.1093/bib/bby034. [DOI] [PubMed] [Google Scholar]
- 160.Mistry J, Bateman A, Finn RD. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007;8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hu L, Xu Z, Hu B, Lu ZJ. COME: a robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Res. 2017;45:e2. doi: 10.1093/nar/gkw798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu D, Mewalal R, Hu R, et al. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic Res. 2017;4:17031. doi: 10.1038/hortres.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.