Abstract
Photoaffinity labeling (PAL) is one of the upcoming and powerful tools in the field of molecular recognition. It includes the determination of dynamic parameters, such as the identification and localization of the target protein and the site of drug binding. In this study, a photoaffinity-labeled probe for full-length human immunodeficiency virus-1 integrase (HIV-1 IN) capture was designed and synthesized, following the structure of the FDA-approved drug Raltegravir. This photoprobe was found to retain the HIV IN inhibitory potential in comparison with its parent molecule and demonstrates the ability to label the HIV-1 IN protein. Putative photoprobe/inhibitor binding sites near the catalytic site were then identified after protein digestion coupled to mass and molecular modeling analyses.
Keywords: HIV-1 integrase inhibitors, Photoaffinity labeling, Raltegravir, Molecular recognition
A prerequisite for structure-based drug design (SBDD) is an understanding of the principles of molecular recognition in protein–ligand complexes.1,2 Experimental and computational techniques such as X-ray crystallography, biochemical assays, and molecular modeling are well-validated tools in providing structural details at the molecular and atomic levels.1,3 Each of these methods possesses peculiar characteristics with advantages and limitations, and thus the management of complementary data constitutes a major goal for a better understanding of biochemical and drug binding mechanisms.
Photoaffinity labeling (PAL), that is, the “capture” of proteins by small molecules via photoactivation-mediated irreversible cross-linking, sequentially analyzed by mass spectrometry, represents a powerful method for identifying sites of molecular interactions.4 PAL is based on the ability of some functional groups (photophores) to form reactive species that are able to create covalent bonds if irradiated with particular wavelengths.5−9
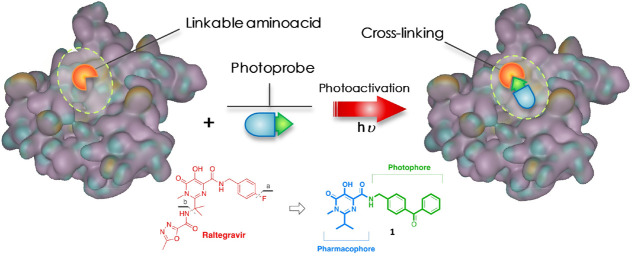
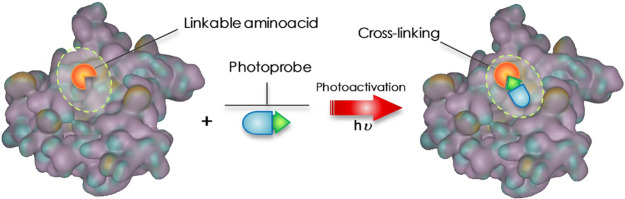
In PAL, a photophore is incorporated into the structure of a ligand capable of interacting with a particular target. The thus-obtained photoprobe will selectively establish interactions with its putative binding site and, following a procedure called “photoactivation”, will form irreversible covalent bonds (photolinkage) with adjacent amino acid residues (Figure 1).5,6
Figure 1.
Schematic concept of cross-linking by the photoprobe with a “linkable” amino acid.
The choice of the photophore is a critical point for the PAL. This chemical entity must satisfy various criteria such as sufficient affinity for the target, reduced dimensions, chemical stability, photoactivation at a wavelength that does not compromise the structural integrity of the target, as well as the ability to easily form a single covalent bond. Moreover, an optimal activity of the photoprobe (i.e., within the same order of magnitude as the inhibitor used as a model) is required for a successful PAL experiment. However, in some cases, it has been possible to use a photoprobe with markedly lower activities than the “ancestor compounds”, and compounds with as much as 100 times lower activity can still be useful.
Among the various photoreactive groups available for photoprobe generation, compounds containing a benzophenone chemotype demonstrate some advantages in the study of protein structures and their interactions.10 By exploiting the highly electrophilic character of the carbonyl in the excited state, they are able to also react with generally inert bonds. Moreover, the greater chemical stability under normal conditions compared with other photophores such as azides and diaziridines, allows benzophenone derivatives to be manipulated, even in the presence of ambient light, without running the risk of triggering protein degradation processes.
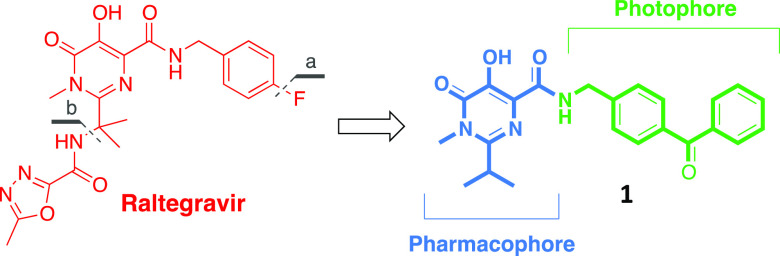
As a suitable model for a PAL investigation, we chose the enzyme HIV-1 integrase (HIV-1 IN), which is a key viral enzyme in HIV replication that catalyzes the insertion of retro-transcribed viral cDNA into the host genome to form a stable provirus.11−13 Because of its role in the HIV cell cycle and considering that it does not have any host cellular homologue, HIV-1 IN has been validated as a target for antiretroviral drug development.14,15 First-generation HIV-1 IN inhibitors such as Raltegravir (RAL, Chart 1) and Elvitegravir and the second-generation inhibitor Dolutegravir, followed by Cabotegravir and Bictegravir, evolved from the β-diketo acid (DKA) pharmacophore.15−17
Chart 1. Structures of the Drug Raltegravir and the Designed Photoprobe.
All of the developed inhibitors predominantly inhibit the strand transfer (ST) step by acting as a chelating functional group, which is required to chelate two Mg2+ metal ions present in the active site of the catalytic core domain (CCD).18−22 Although the addition of an HIV-1 IN inhibitor to existing components of antiretroviral therapy has improved the outcome of highly active antiretroviral therapy (HAART) by potential synergism, there is still a significant need to acquire new and more reliable information detailing the interaction between IN inhibitors and the binding site, possibly by exploring the mechanistic behavior under a “dynamic” condition.
Recently, we and others have reported the incorporation of photoreactive groups into DKA-based HIV-1 IN inhibitors, and these agents can be used to identify the DKA binding site on the IN protein.23−25 Nevertheless, because of the relative lability of the 2-hydroxy-4-oxobut-2-enoic motif, typical DKAs were found to be unsuitable for the advanced stage of PAL analyses. Again, the PAL approach and mass spectrometry analysis were successfully used to identify a unique HIV-1 IN inhibitor-binding site for a series of coumarin-containing IN inhibitors.26 Therefore, although many structural X-ray crystallographic structures are available to elucidate the interaction between HIV-1 IN inhibitors within the active site,27−29 further insights could be useful to clarify the binding mode of HIV-1 IN inhibitors through the determination of dynamic parameters as well as to identify other sites of action.14,15,17,30−32
In this scenario, we sought to design and synthesize a novel photoaffinity probe (1, Chart 1), structurally related to the drug RAL, that could effectively label the viral full-length HIV-1 IN, which could be further used for extensive PAL studies.
After the preparation of the photoprobe, Western blotting (WB) analyses coupled to mass spectrometry were performed to provide an effective linkage of 1 to the target protein. Then, protein digestion, mass experiments, and computational modeling, that is, docking and molecular dynamics, helped us to rationalize the behavior of the putative probe-HIV-1 IN interactions.
Specifically, considering the greater chemical stability, a photoactivable benzophenone group as a photophore was introduced in the carboxamide side chain of the pyrimidine scaffold in RAL, in place of the p-F-benzyl moiety. Moreover, for molecular simplification purposes, the heterocyclic substituent was removed from the isopropyl substituent (Chart 1a,b, respectively).
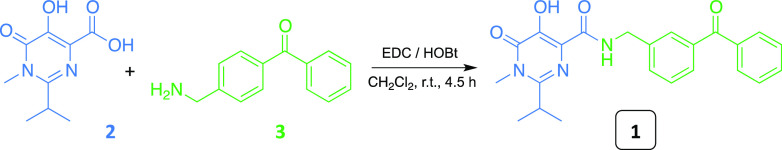
The preparation of the desired photoprobe 1 was achieved by following the synthetic approach previously reported by us for the preparation of the RAL-based analogue HL2,33,34 with some modifications. In particular, 1 was obtained by amidation of the “synthons” 2 and 3 in the presence of N-(3-dimethylpropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and 1-hydroxybenzotriazole hydrate (HOBt) in dichloromethane (CH2Cl2) at room temperature (Scheme 1).
Scheme 1. Preparation of Photoprobe 1.
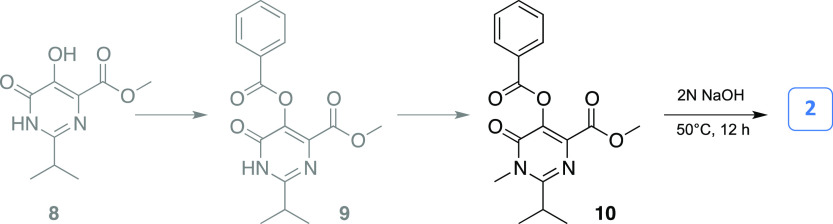
The key synthones 2 and 3 were synthesized by two separate synthetic routes, as outlined in Scheme 2 and in the Supporting Information.
Scheme 2. Synthetic Route for the Preparation of the Synthon 2.
In brief, N-methyl precursor 10 was obtained in good yield by sequential protection of the hydroxyl function in position five of dihydroxypyrimidine derivative 8 by reaction with benzoic anhydride in pyridine, with the formation of the intermediate 9, which was selectively methylated at the N1 with methyl iodide and Cs2CO3 in anhydrous tetrahydrofuran (THF) (Scheme 2; see the Supporting Information for experimental details).33 Alkaline hydrolysis of ester 10 gave dihydropyrimidine carboxylic acid 2.
The preparation of (commercially available) aminodiphenylketone 3 was accomplished by starting from 4-methylbenzophenone 11 (Scheme 3 in the Supporting Information), following a procedure previously reported by us.33 All compounds have been characterized by means of NMR, IR, and mass spectrometry.
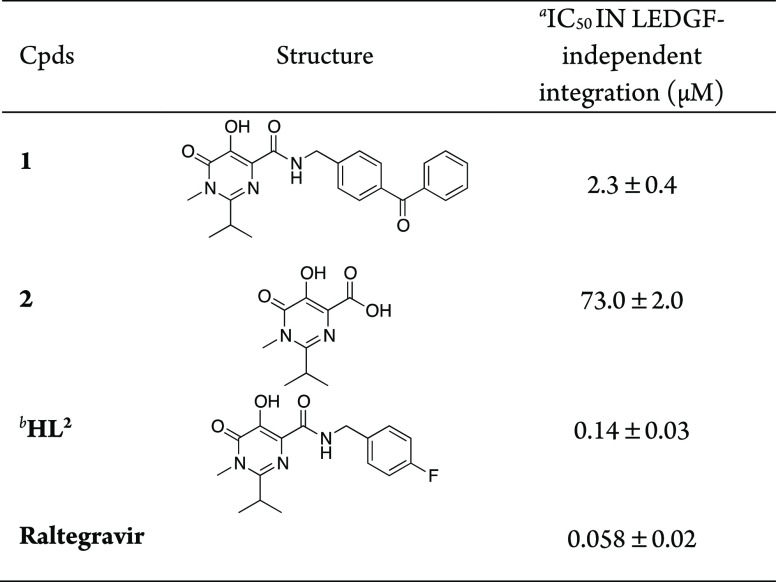
Next, photoprobe 1 was tested for its ability to inhibit the overall HIV-1 IN integration in in vitro assays, employing purified enzyme and using RAL and HL2 as reference compounds (Table 1). To define the minimal chemical features of 1 required for inhibitory activity, synthon 2, without the benzyl moiety, was used as a control.
Table 1. In Vitro HIV-1 Integrase Inhibition Assay.
Compound concentration required to inhibit the HIV-1 IN catalytic activities by 50% in the absence of LEDGF.
Data from ref (33).
The results indicated that photoprobe 1 was able to inhibit the HIV-1 IN catalytic activity, with an IC50 value of 2.3 ± 0.4 μM, although it resulted in being 16 times less potent than HL2 and ∼40 times less effective with respect to RAL (IC50s = 2.3 ± 0.4 μM vs IC50 = 0.058 ± 0.02 for 1 and RAL, respectively). Furthermore, acid intermediate 2 (which did not bear any aromatic substituent in position four) seems to retain a residual inhibitory activity (IC50 = 73.0 ± 2.0). These results confirm that the nature of the substituents in the carboxamide side chain of the pyrimidine ring significantly influences the potency of the pharmacophoric scaffold. However, probe 1 retains an activity in the low micromolar range, thus making it suitable to be used for microlevel PAL analysis.
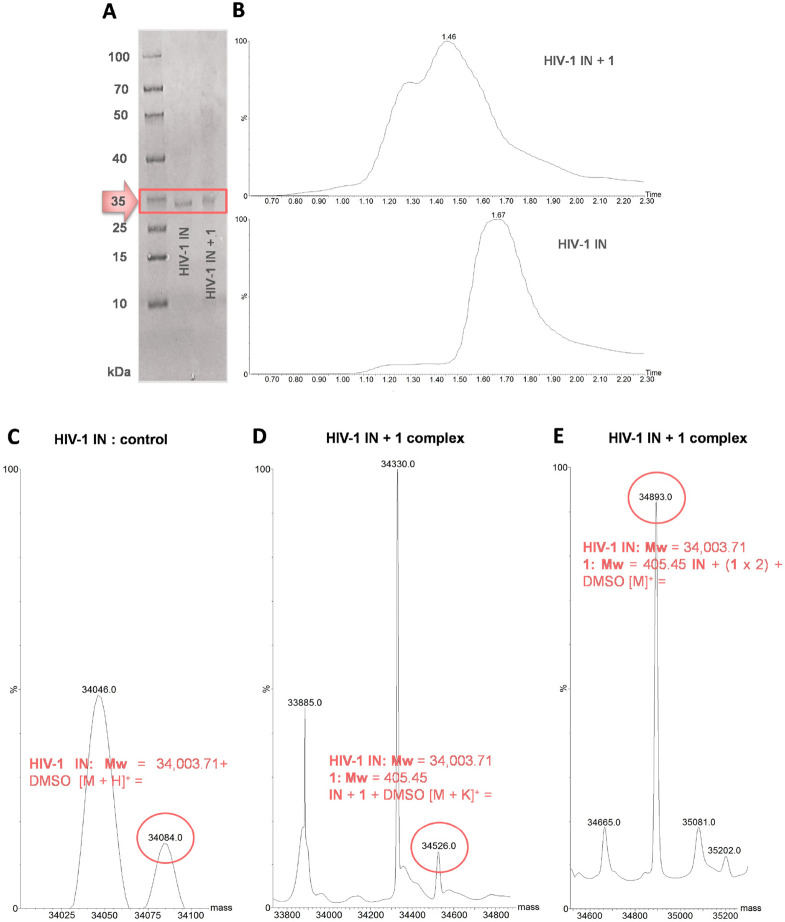
Next, 1 was submitted to a cross-linking experiment, where it was illuminated at 360 nm (UV cross-linking) in the presence of full-length HIV-1 IN enzyme (see the amino acid sequence in the Supporting Information), then subjected to polyacrylamide gel electrophoresis (SDS-PAGE gel). Exposure of the gel to UV transillumination showed clear fluorescence bands at the expected region (i.e., ∼35 kDa), corresponding to labeled IN protein, further confirmed by staining. Figure 2A shows a representative picture of gel electrophoresis analysis demonstrating the capture of HIV-1 IN by the photoprobe, thus supporting the ability of 1 to irreversibly conjugate the enzyme protein.
Figure 2.
(A) Results of the gel electrophoresis analysis of the capture of HIV-1 IN by photoprobe 1. Lines show both the HIV-1 IN alone and the HIV-1 IN + photoprobe 1 mixture after UV irradiation at 360 nm. (B) LC–MS total-ion current (TIC) traces showing a distinct shift in the retention time between the HIV-1 IN alone (bottom) and the protein after activation (top) with the probe in molar excess. (C) ESI-MS spectrum expansion of the HIV-1 IN free protein. Spectrum of the HIV-1 IN + 1 complex incubated in (D) 1:1.5 and (E) 1:5 protein/photoprobe ratio after irradiation.
After capture of the protein, the enzyme–protoprobe (HIV-1 IN + 1) complex was directly investigated by mass spectrometry (MS) experiments (see LC−MS traces of free and labeled proteins in Figure 2B). Considering the molecular weight of the free HIV-1 IN full protein (MS spectrum of HIV-1 IN alone is shown in Figure 2C), the analyses of the ESI-MS spectra of the incubated photoreacted sample revealed that the molecular weight shift would correspond to one (Figure 2D) or two molecules (Figure 2E) of photoprobe 1. It is worth noting that the stoichiometric ratio of the HIV-1 IN full protein to the photoprobe should be 1:1 or 1:2 or more (other peaks could be revealed), depending on the protein/photoprobe ratio before photoactivation. An excess of probe (i.e., from 1:1.5 to 1:5 ratio) can lead to multiple labeling.
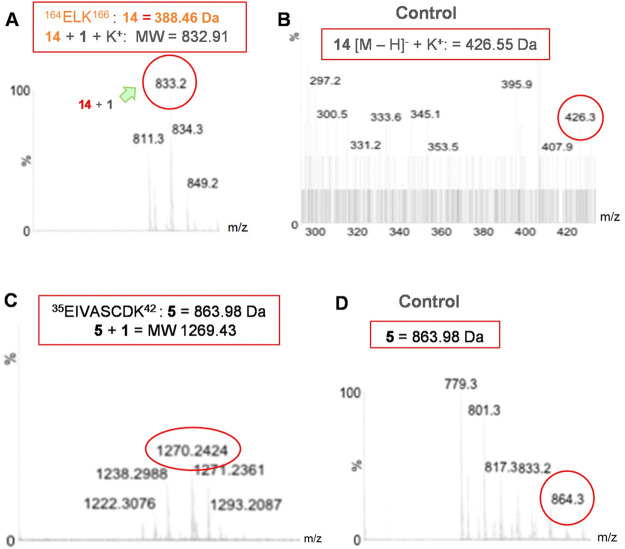
To investigate which amino acid pattern could be involved in photolinking/interaction by a photoprobe/inhibitor with the protein, the HIV-1 IN + 1 complexes were submitted to tryptic digestion, and the total digest and obtained fractions were analyzed by MS experiments. From the predicted peptide fragments suggested by trypsic digestion (see the Supporting Information), we identified 164ELK166 (fragment 14) as a potential peptide cross-linked to photoprobe 1 (Figure 3A–D). Curiously, in addition to 14, another peptide, that is, 42EIVASCDK49 (fragment 5), can be found after incubation with a molar excess of probe 1.
Figure 3.
MS spectra of (A) fragment 164ELK166 (157ELK159 in regular HIV-1 IN) linked to photoprobe 1, (B) its respective free peptide, (C) 157ELK159, 35EIVASCDK42 + 1, and (D) the unlabeled peptide from the total tryptic digest of labeled HIV-1 IN + 1 conjugated.
Finally, keeping in mind the indications provided from mass analyses, we envisaged to computationally explore the topological space of the enzyme accessible to probe 1 on the catalytic site.
Specifically, whereas the identified peptide 14 was located in the CCD, in the region 50–212, fragment 5 was positioned in the N-terminal domains (NTDs). Therefore, we focused our exploration on the catalytic site, close to key catalytic amino acid E152. For clarification purposes, the fragment 164ELK166 that resulted from the digested full protein corresponds to 157ELK159 of the regular HIV IN sequence. (See the Supporting Information.) According to mass results, 1 would bind to the peptide 157ELK159, located near the catalytic site. Interestingly, along with other residues, this peptide bears the K159, which is known for having an important role in DNA binding with the enzyme and may affect catalytic activity.35 Additionally, we realized that the other peptide, that is, 35EIVASCDK42 (i.e., 42EIVASCDK49 in our digested protein), found after incubation with a molar excess of probe 1, can be located within the NTD and would include an amino acid, that is, Cys40, involved in the zinc finger motif.
We then performed docking and molecular dynamics studies to predict the interaction of photoprobe 1 in the IN core domain (PDB: 1QS4).36
Docking results indicated that the amino acid residues involved in the binding were as follows: Ser119, Glu92, His67, Thr66, Lys159, Lys156, Glu152, Gly149, and Gln48. In particular, the benzophenone moiety of compound 1 occupied an area close to the backbone amino acid residues K156 and K159. Collectively, the disposition of this probe within this amino acid pocket is coherent with the established binding mode of this class of HIV-1 IN inhibitors. The interaction of 1 on the surface of the HIV-1 IN core domain is shown in Figure 4A.
Figure 4.
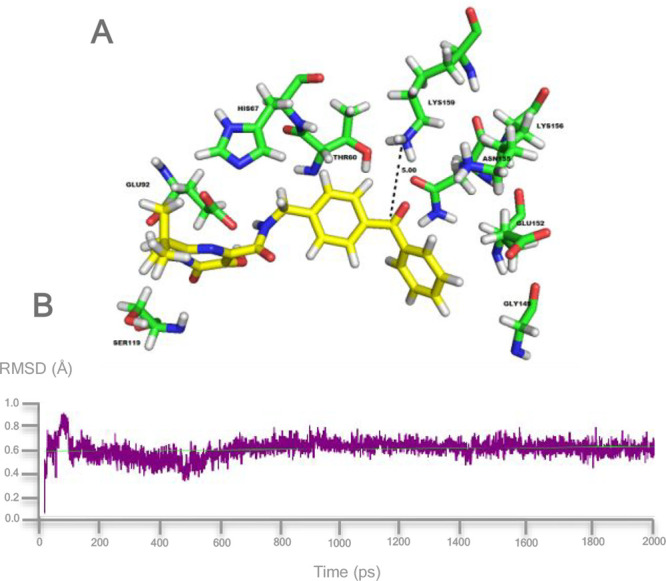
(A) Binding mode for photoprobe 1 within the HIV-1 IN catalytic site after MD simulation for 2 ns. The residues interacting with probe 1 are shown. The distance between the Lys159 and the carbonyl group of 1 is also highlighted. (B) RMSD from the starting protein structure by MD simulations..
MD simulations revealed that the pharmacophoric group on probe 1 maintained the key interaction responsible for the inhibitory activity (Figure 4B).
The RMSD plot disclosed that a stable complex was formed between the photoprobe and HIV IN, where the carbonyl of the benzophenone (photophoric moiety) and the K159 of the protein backbone aligned in the vicinity of a distance of ∼5 Å after the simulation time period of 2 ns calculations (Figure 4B). This finding suggests the formation of a covalent linkage between the backbone peptide 157ELK159 and the benzophenone moiety of 1 upon photoactivation.
In this study, we report on the design and synthesis and a preliminary experimental validation of a RAL-based photoaffinity probe for full-length HIV-1 IN capture to be used in extensive PAL analysis. The developed photoprobe 1 retained IN inhibition potency against purified enzyme with respect to the reference compounds. The analysis of the photoactivation experiment as well as data obtained from trypsin digestion coupled to mass experiments seems to prove an efficient capture of HIV-1 IN by the probe, which would establish a photolinkage in the proximity of the enzyme active site, as also supported by computational modeling. Work is in progress to plan tandem HPLC-MS for a complete PAL procedure.
Acknowledgments
We are grateful to the Schrödinger Team for the opportunity to evaluate a trial of the latest version of software Maestro.
Glossary
Abbreviations
- HIV-1
human immunodeficiency virus-1
- IN
integrase
- PAL
photoaffinity labeling
- HAART
highly active antiretroviral therapy
- CCD
catalytic core domain
- RAL
raltegravir
- WB
Western blotting
- SBDD
structure-based drug design
- DKA
β-diketo acid
- cDNA
complementary deoxyribonucleic acid
- ST
strand transfer
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- LEDGF
lens epithelium-derived growth factor
- RMSD
root-mean-square deviation
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00009.
Experimental methods; synthesis and characterization details of photoprobe 1; NMR and mass spectra of 1; preparation of the synthons 2 and 3 and their intermediates 6–10, 12, and 13; expression and purification of recombinant IN; HIV-1 IN HTRF inhibition assays; photoaffinity labeling of HIV-1 IN; SDS-PAGE experiments; trypsic digestion of HIV-1 IN; site of cleavage and peptides generated by tryptic digestion; alignment of the HIV-1 IN sequences; and molecular modeling (PDF)
Author Contributions
This work was done through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Neamati N.; Barchi J. J. Jr. New paradigms in drug design and discovery. Curr. Top. Med. Chem. 2002, 2, 211–227. 10.2174/1568026023394227. [DOI] [PubMed] [Google Scholar]
- Hughes J. P.; Rees S.; Kalindjian S. B.; Philpott K. L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. R.; Porebski P. J.; Chruszcz M.; Minor W. X-ray crystallography: Assessment and validation of protein-small molecule complexes for drug discovery. Expert Opin. Drug Discovery 2011, 6, 771–782. 10.1517/17460441.2011.585154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.; Collins I. Photoaffinity labeling in target- and binding-site identification. Future Med. Chem. 2015, 7, 159–183. 10.4155/fmc.14.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman G.; Prestwich G. D. Using photolabile ligands in drug discovery and development. Trends Biotechnol. 2000, 18, 64–77. 10.1016/S0167-7799(99)01402-X. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y.; Sadakane Y. Photoaffinity labeling in drug discovery and developments: chemical gateway for entering proteomic frontier. Curr. Top. Med. Chem. 2002, 2, 271–288. 10.2174/1568026023394182. [DOI] [PubMed] [Google Scholar]
- Robinette D.; Neamati N.; Tomer K. B.; Borchers C. H. Photoaffinity labeling combined with mass spectrometric approaches as a tool for structural proteomics. Expert Rev. Proteomics 2006, 3, 399–408. 10.1586/14789450.3.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakane Y.; Hatanaka Y. Photochemical fishing approaches for identifying target proteins and elucidating the structure of a ligand-binding region using carbene-generating photoreactive probes. Anal. Sci. 2006, 22, 209–218. 10.2116/analsci.22.209. [DOI] [PubMed] [Google Scholar]
- Brunner J. New photolabeling and crosslinking methods. Annu. Rev. Biochem. 1993, 62, 483–514. 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- Dorman G.; Prestwich G. D. Benzophenone photophores in biochemistry. Biochemistry 1994, 33, 5661–5673. 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- Hazuda D. J. HIV Integrase as a Target for Antiretroviral Therapy. Curr. Opin. HIV AIDS 2012, 7, 383–389. 10.1097/COH.0b013e3283567309. [DOI] [PubMed] [Google Scholar]
- Pommier Y.; Johnson A. A.; Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discovery 2005, 4, 236–248. 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- Li X.; Krishnan L.; Cherepanov P.; Engelman A. Structural biology of retroviral DNA integration. Virology 2011, 411, 194–205. 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito F.; Tramontano E. Past and future. Current drugs targeting HIV-1 integrase and reverse transcriptase-associated ribonuclease H activity: single and dual active site inhibitors. Antivir. Chem. Chemother. 2014, 23, 129–144. 10.3851/IMP2690. [DOI] [PubMed] [Google Scholar]
- Li Y.; Xuan S.; Feng Y.; Yan A. Targeting HIV-1 integrase with strand transfer inhibitors. Drug Discovery Today 2015, 20 (4), 435–449. 10.1016/j.drudis.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Hazuda D. J.; Felock P.; Witmer M.; Wolfe A.; Stillmock K.; Grobler J. A.; Espeseth A.; Gabryelski L.; Schleif W.; Blau C.; Miller M. D. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 2000, 287, 646–650. 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- Choi E.; Mallareddy J. R.; Lu D.; Kolluru S. Recent advances in the discovery of small-molecule inhibitors of HIV-1 integrase. Future Science OA 2018, 4 (9), FSO338. 10.4155/fsoa-2018-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D.; Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: Specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 1339–1343. 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neamati N.Role of Metals in HIV-1 Integrase Inhibitor Design. In HIV-1 Integrase: Mechanism of Action and Inhibitor Design; Wiley Series in Drug Discovery and Development; Wiley: Hoboken, NJ, 2011. [Google Scholar]
- Rogolino D.; Carcelli M.; Sechi M.; Neamati N. Viral enzymes containing magnesium: metal binding as a successful strategy in drug design. Coord. Chem. Rev. 2012, 256, 3063–3086. 10.1016/j.ccr.2012.07.006. [DOI] [Google Scholar]
- Lesbats P.; Engelman A. N.; Cherepanov P. Retroviral DNA Integration. Chem. Rev. 2016, 116, 12730–12757. 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler J. A.; Stillmock K.; Hu B.; Witmer M.; Felock P.; Espeseth A. S.; Wolfe A.; Egbertson M.; Bourgeois M.; Melamed J.; Wai J. S.; Young S.; Vacca J.; Hazuda D. J. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 6661–6666. 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Marchand C.; Pommier Y.; Burke T. R. Jr. Design and Synthesis of Photoactivatable Aryl Diketo Acid-Containing HIV-1 Integrase Inhibitors as Potential Affinity Probes. Bioorg. Med. Chem. Lett. 2004, 14, 1205–2007. 10.1016/j.bmcl.2003.12.064. [DOI] [PubMed] [Google Scholar]
- Zhao X. Z.; Semenova E. A.; Liao C.; Nicklaus M.; Pommier Y.; Burke T. R. Jr. Biotinylated biphenyl ketone-containing 2,4-dioxobutanoic acids designed as HIV-1 integrase photoaffinity ligands. Bioorg. Med. Chem. 2006, 14, 7816–7825. 10.1016/j.bmc.2006.07.064. [DOI] [PubMed] [Google Scholar]
- Sechi M.; Carta F.; Sannia L.; Dallocchio R.; Dessì A.; Al-Safi R. I.; Neamati N. Design, synthesis, molecular modeling and anti-HIV 1 integrase activity of a series of photoactivatable diketo acid-containing inhibitors as affinity probes. Antiviral Res. 2009, 81, 267–276. 10.1016/j.antiviral.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Al-Mawsawi L. Q.; Fikkert V.; Dayam R.; Witvrouw M.; Burke T. R.; Borchers C. H.; Neamati N. Discovery of a small-molecule HIV-1 integrase inhibitor-binding site. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 10080–10085. 10.1073/pnas.0511254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G. N.; Hare S.; Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 2010, 468, 326–329. 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S.; Gupta S. S.; Valkov E.; Engelman A.; Cherepanov P. Retroviral Intasome Assembly and Inhibition of DNA Strand Transfer. Nature 2010, 464, 232–236. 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S.; Vos A. M.; Clayton R. F.; Thuring J. W.; Cummings M. D.; Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 20057–20062. 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcelli M.; Rogolino D.; Gatti A.; Pala N.; Corona A.; Caredda A.; Tramontano E.; Pannecouque C.; Naesens L.; Esposito F. Chelation motifs affecting metal-dependent viral enzymes: N’-acylhydrazone ligands as dual target inhibitors of HIV-1 integrase and reverse transcriptase ribonuclease H domain. Front. Microbiol. 2017, 8, 1–10. 10.3389/fmicb.2017.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona A.; Di Leva F. S.; Rigogliuso G.; Pescatori L.; Madia V. N.; Subra F.; Delelis O.; Esposito F.; Cadeddu M.; Costi R.; Cosconati S.; Novellino E.; di Santo R.; Tramontano E. New insights into the interaction between pyrrolyl diketoacids and HIV-1 integrase active site and comparison with RNase H. Antiviral Res. 2016, 134, 236–243. 10.1016/j.antiviral.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Esposito F.; Sechi M.; Pala N.; Sanna A.; Koneru P. C.; Kvaratskhelia M.; Naesens N.; Corona A.; Grandi N.; di Santo R.; D’Amore V. M.; Di Leva F. S.; Novellino N.; Cosconati S.; Tramontano E. Discovery of dihydroxyindole-2-carboxylic acid derivatives as dual allosteric HIV-1 Integrase and Reverse Transcriptase associated Ribonuclease H inhibitors. Antiviral Res. 2020, 174, 104671. 10.1016/j.antiviral.2019.104671. [DOI] [PubMed] [Google Scholar]
- Bacchi A.; Carcelli M.; Compari C.; Fisicaro E.; Pala N.; Rispoli G.; Rogolino D.; Sanchez T. W.; Sechi M.; Sinisi V.; Neamati N. Investigating the role of metal chelation in HIV-1 integrase strand transfer inhibitors. J. Med. Chem. 2011, 54, 8407–8420. 10.1021/jm200851g. [DOI] [PubMed] [Google Scholar]
- Gardelli C.; Nizi E.; Muraglia E.; Crescenzi B.; Ferrara M.; Orvieto F.; Pace P.; Pescatore G.; Poma M.; Rico Ferreira M. d. R.; Scarpelli R.; Homnick C. F.; Ikemoto N.; Alfieri A.; Verdirame M.; Bonelli F.; Gonzalez Paz O.; Taliani M.; Monteagudo E.; Pesci S.; Laufer R.; Felock P.; Stillmock K. A.; Hazuda D.; Rowley M.; Summa V. Discovery and synthesis of HIV integrase inhibitors: development of potent and orally bioavailable N-methyl pyrimidones. J. Med. Chem. 2007, 50, 4953–4975. 10.1021/jm0704705. [DOI] [PubMed] [Google Scholar]
- Krishnan L.; Li X.; Naraharisetty H. L.; Hare S.; Cherepanov P.; Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 15910–15915. 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgur Y.; Craigie R.; Cohen G. H.; Fujiwara T.; Yoshinaga T.; Fujishita T.; Sugimoto H.; Endo T.; Murai H.; Davies D. R. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 13040–13043. 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.