ABSTRACT
Background
It is unclear how high fructose consumption induces disparate metabolic responses in genetically diverse mouse strains.
Objective
We aimed to investigate whether the gut microbiota contributes to differential metabolic responses to fructose.
Methods
Eight-week-old male C57BL/6J (B6), DBA/2J (DBA), and FVB/NJ (FVB) mice were given 8% fructose solution or regular water (control) for 12 wk. The gut microbiota composition in cecum and feces was analyzed using 16S ribosomal DNA sequencing, and permutational multivariate ANOVA (PERMANOVA) was used to compare community across mouse strains, treatments, and time points. Microbiota abundance was correlated with metabolic phenotypes and host gene expression in hypothalamus, liver, and adipose tissues using Biweight midcorrelation. To test the causal role of the gut microbiota in determining fructose response, we conducted fecal transplants from B6 to DBA mice and vice versa for 4 wk, as well as gavaged antibiotic-treated DBA mice with Akkermansia for 9 wk, accompanied with or without fructose treatment.
Results
Compared with B6 and FVB, DBA mice had significantly higher Firmicutes to Bacteroidetes ratio and lower baseline abundance of Akkermansia and S24–7 (P < 0.05), accompanied by metabolic dysregulation after fructose consumption. Fructose altered specific microbial taxa in individual mouse strains, such as a 7.27-fold increase in Akkermansia in B6 and 0.374-fold change in Rikenellaceae in DBA (false discovery rate <5%), which demonstrated strain-specific correlations with host metabolic and transcriptomic phenotypes. Fecal transplant experiments indicated that B6 microbes conferred resistance to fructose-induced weight gain in DBA mice (F = 43.1, P < 0.001), and Akkermansia colonization abrogated the fructose-induced weight gain (F = 17.8, P < 0.001) and glycemic dysfunctions (F = 11.8, P = 0.004) in DBA mice.
Conclusions
Our findings support that differential microbiota composition between mouse strains is partially responsible for host metabolic sensitivity to fructose, and that Akkermansia is a key bacterium that confers resistance to fructose-induced metabolic dysregulation.
Keywords: gut microbiota, fructose, metabolic syndrome, fecal transplant, Akkermansia, microbiota-host interaction, gene by diet interaction
Introduction
The drastic increase in fructose consumption over the past few decades has been paralleled by the rising prevalence of metabolic syndrome and diabetes (1). Our recent systems nutrigenomics study unraveled the impact of fructose on transcriptome, epigenome, and gene–gene interactions in the hypothalamus, which is the main regulator of metabolism (2). Additionally, studies on liver and adipose tissues have also revealed fructose-induced alterations in genes involved in several aspects of systemic metabolism (3–5). Interestingly, genetically diverse mouse strains demonstrate disparate metabolic responses to fructose consumption, a finding reproducible across multiple studies, including ours (5, 6). However, the causal mechanisms underlying the interindividual differences in metabolic responses to fructose have yet to be elucidated.
The gut microbiome is emerging as an important modulator of metabolism, obesity, and other metabolic disorders (7). Its dynamic nature, ease of manipulation, and response to dietary intervention have made the gut microbiome a suitable therapeutic target to mitigate metabolic syndrome (8). Diet influences trillions of gut microorganisms as early as 1 d post–dietary intervention (9). The gut microbiome has been suggested to contribute to phenotypic diversity in different mouse strains fed a high-fat, high-sucrose diet (10, 11). It is plausible that the gut microbiome also plays a role in determining interindividual variability in metabolic responses to fructose consumption.
Dietary fructose is mainly absorbed in the small intestine, where it can be extensively metabolized into glucose and organic acids. However, an excessive amount of fructose that is not absorbed in the small intestine due to malabsorption or high intake can undergo colonic bacterial fermentation, resulting in the production of metabolites such as SCFAs (12). Fructose also has the potential to modify the microbiota community and its normal function. For instance, fructose metabolites can shape the gut environment and be an energy source for the gut microbiota (13). Fructose can also suppress gut bacterial colonization by silencing a colonization factor in a commensal bacterium (14).
Here, we investigated gut microbiota as a potential link between fructose consumption and differential metabolic phenotypes in mice with diverse genetic backgrounds. We tested our paradigm in 3 mouse strains, namely C57BL/6J (B6), DBA/2J (DBA), and FVB/NJ (FVB), which have contrasting metabolic susceptibility to high-caloric diets (5, 6, 11).
Methods
Animals and study design
To investigate the role of the gut microbiota in host responses to fructose consumption, we examined microbial composition in the context of differential metabolic and transcriptomic responses in multiple mouse strains (Supplemental Figure 1). Seven-week-old male mice (20–25 g) from 3 inbred strains, namely B6, DBA, and FVB, were obtained from the Jackson Laboratory (Bar Harbor, ME) and housed in a pathogen-free barrier facility with a 12-h light/dark cycle at the University of California, Los Angeles. Mice were fed Lab Rodent Diet 5001, containing 234 g/kg protein, 45 g/kg fat, 499 g/kg carbohydrate, and 2.89 kcal/g metabolizable energy (LabDiet). After a 1-wk acclimation period, mice from each strain were randomly divided into 2 groups. One group was provided with regular water (control group, n = 8–10 mice/strain) and the other group was given 8% (wt/vol) fructose (3.75 kcal/g energy; NOW Real Food) dissolved in regular water (fructose group, n = 10–12 mice/strain) for 12 wk ad libitum. Food and drink intakes were monitored daily on a per-cage basis. Distinct alterations in body weight, fat mass, plasma lipids, glycemic traits, intraperitoneal glucose-tolerance test (IPGTT), and the transcriptome of liver, adipose, and hypothalamus in response to fructose consumption across mouse strains were observed and previously published (5). Gut microbiota compositions of cecal and fecal samples were analyzed. Fructose-responsive microbiota were correlated with metabolic phenotypes and host gene expression to prioritize microbial taxa that may contribute to differential host fructose responses. Last, fecal microbiota transplant (FMT) and Akkermansia muciniphila (AM) colonization experiments were conducted to test the causal role of gut microbiota in determining mouse strain–specific fructose responses. The study was performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals. All experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of California, Los Angeles.
Fecal and cecal microbiota analysis
Feces were collected at 1, 2, 4, and 12 wk of fructose treatment, and cecal contents were collected at the end of fructose treatment (12 wk). Samples were snap-frozen and then stored at −80°C until DNA isolation. Microbial DNA was isolated from samples using the MO BIO PowerSoil®-htp 96 Well Soil DNA Isolation Kit (MO BIO). For the fecal and cecal samples, the same lot isolation kit was used; for the fecal transplant samples a different lot kit was used. The V4 region (15, 16) of the 16S ribosomal DNA (rDNA) gene was amplified in triplicate with barcoded primers (515f and 806r) (17). PCR products were quantified with a Quant-iTTM PicoGreen® dsDNA Assay Kit (ThermoFisher Scientific), and all samples from a specific experiment (e.g., fecal, cecal, or fecal transplant experiment) were combined in equal amounts (∼250 ng/sample) into a single pooled submission to be purified with the UltraClean PCR® Clean-Up Kit (MO BIO). Single-end reads were generated on the Illumina HiSeq 2500 platform using 2 lanes for each pool of mixed samples to create an unbiased sequencing approach. Approximately 500 samples were sequenced per submission, and each dataset of fecal, cecal, and fecal transplant samples were sequenced in different submissions. Raw sequences were processed using QIIME to produce de-multiplexed and quality-controlled sequences. Reads were binned into operational taxonomic units (OTUs) at 97% similarity using UCLUST against the Greengenes reference database (18). Singletons, OTUs representing <0.005% total relative abundance, unsuccessfully sequenced samples, and outliers were removed. Samples were normalized to a rarefied level specific for each dataset to reduce the effect of unequal sequencing depth to result in 60,000, 106,815, and 37,614 reads per sample for fecal, cecal, and fecal transplant samples, respectively. A total of 180 fecal, 46 cecal, and 170 fecal transplant samples were used for downstream analysis.
Correlation analysis between gut microbiota and metabolic phenotypes or fructose signature genes
Correlation analysis was performed between the relative abundance or proportion of microbiota with metabolic phenotypes, including body weight, adiposity, and area under the curve (AUC) for glucose tolerance. Microbiota proportions were also correlated with differentially expressed genes in response to fructose consumption, or “fructose signature genes,” in hypothalamus, liver, and adipose tissue. The fructose signature genes correlated with bacterial abundance were classified for their biological functions in Gene Ontology (GO), REACTOME, and Kyoto Encyclopedia of Genes and Genomes (KEGG) using the Gene Set Enrichment Analysis (GSEA) tool (19).
Antibiotics treatment prior to fecal transplant or Akkermansia colonization
Six-week-old B6 or DBA recipient mice were orally gavaged with a solution of vancomycin (50 mg/kg), neomycin (100 mg/kg), and metronidazole (100 mg/kg) twice daily for 7 d. Ampicillin (1 mg/mL) was also provided ad libitum in drinking water (20). Antibiotic-treated mice were housed in sterile cages with sterile water and food throughout the experiment. Following antibiotics treatment, mice were used in either fecal transplant or Akkermansia colonization experiments, as described below.
Fecal transplant between B6 and DBA mice
Reciprocal fecal transplant between B6 and DBA mice was conducted. B6 mice receiving DBA feces were designated as B6(DBA) and DBA mice receiving B6 feces were designated as DBA(B6). To serve as control groups, DBA mice were transplanted with DBA feces and designated as DBA(DBA), and B6 mice were transplanted with B6 feces and designated as B6(B6). Fecal transplant was performed according to previous studies, with some modification (21, 22). Briefly, freshly collected feces were pooled from 4 donor mice and suspended at 40 mg/mL concentration in anaerobic PBS. The suspension (150 μL) was orally gavaged to the recipient mice for 4 wk. Total time between collecting feces and delivery of microbial contents into recipients was kept as short as possible (<15 min) to protect anaerobes. After 1 wk of fecal transplant, mice from each of the 4 transplant experiments, namely B6(DBA), B6(B6), DBA(B6), and DBA(DBA), were divided into 2 groups and treated with 8% fructose or regular water for 12 wk (n = 7–14 mice/group). Body weight and IPGTT were measured using the methods described previously (5).
Akkermansia colonization in DBA mice
Akkermansia colonization was conducted according to Olson et al. (20). Antibiotic-treated DBA mice were orally gavaged with 200 μL bacterial suspension (5 × 109 CFU/mL in anaerobic PBS) throughout the experiment. After 1 wk of bacterial gavage, DBA mice were treated with 8% fructose or regular water for 8 wk (n = 10–14/group). DBA mice receiving anaerobic PBS served as controls (n = 8–10/group). Body weight and IPGTT were measured as described previously (5).
Statistical analysis
The microbiota data were summarized into relative abundance by taxonomic level in QIIME, and communities were visualized with principal coordinates analysis (PCoA) based on the weighted UniFrac distance measure (23). Categorical groups (treatment, time, mouse strain) were confirmed to have similar multivariate homogeneity of group dispersions to allow them to be compared using the nonparametric permutational multivariate ANOVA (PERMANOVA) test with the adonis function (24). Microbial composition was analyzed at the phylum, family, and genus taxonomic levels using the Statistical Analysis of Metagenomic Profiles (STAMP) package (25).
Linear discriminant analysis (LDA) effect size (LEfSe) was used to identify taxa differentially represented between 3 mouse strains using standard parameters (P < 0.05, LDA score >2.0) (26). Using the identified features from the LEfSe analysis, we selected 6 fecal genera that showed contrasting patterns in their baseline levels between DBA and the other 2 mouse strains, B6 and FVB. To visualize the baseline differences of these 6 genera between the 3 mouse strains, boxplots were plotted using centered log-ratio (CLR) transformed values from OTU counts with the “rgr” package in R software (24). The difference between strains was assessed using a 1-factor ANOVA followed by Sidak's post hoc test. Because Turicibacter was not normally distributed, the difference between strains was analyzed by Kruskal-Wallis test followed by Dunn's test.
Taxa that differed between the fructose and regular water groups were identified using White's nonparametric T-test (27), followed by Storey's false discovery rate (FDR) estimation using relative abundance data and STAMP (28). A 1-factor ANOVA followed by Sidak's post hoc test was used to determine the difference in Firmicutes to Bacteroidetes (F/B) ratio between 3 mouse strains. F/B ratio was calculated by dividing the proportion of Firmicutes and Bacteroidetes for each sample, and then log-transformed to achieve normal distribution.
Correlation between gut microbiota and metabolic phenotypes or fructose signature genes from individual tissues was assessed using Biweight midcorrelation (bicor) (29). Statistical P values were adjusted using the Benjamini-Hochberg approach and an FDR <0.05 was considered significant.
To analyze body-weight gain and glucose tolerance data involving multiple time points in fecal transplant and Akkermansia colonization experiments, a 3-factor repeated-measures ANOVA followed by Sidak's post hoc test was used. The effects and interaction of 3 factors (microbial manipulation, fructose, time) on the metabolic phenotypes were tested.
To evaluate the effects of fructose under each unique microbiota manipulation (e.g., within Akkermansia-colonized groups or within B6 mice transplanted with DBA microbe) across multiple time points, a 2-factor repeated-measures ANOVA was used. Fructose treatment was used as the between-group factor and time was used as the within-subject factor. To assess the effect of a given microbiota manipulation under fructose treatment across multiple time points, a 2-factor repeated-measures ANOVA was used. Microbiota manipulation was used as the between-group factor and time was used as the within-subject factor. If the interaction was significant, microbiota manipulations within each time point were compared by Sidak's post hoc test. Statistical analyses were performed using STATISTICA (version 7; Statsoft, Inc.) and GraphPad Prism (version 8; GraphPad Software, Inc.). Data are expressed as means ± SEMs. P < 0.05 was considered statistically significant.
Results
Strain-specific metabolic responses to fructose treatment
We have previously reported that B6, DBA, and FVB mice demonstrated striking differences in their metabolic responses to 8% fructose treatment for 12 wk (5). DBA mice were more sensitive to fructose in terms of obesity and diabetes-related phenotypes including body weight, adiposity, and glucose intolerance. B6 mice showed significant increases while FVB mice had decreases in plasma cholesterol concentrations. These results demonstrate strong interstrain variability in fructose response in genetically divergent mouse strains. Importantly, the disparate metabolic responses were not due to differences in overall energy intake [14.6 ± 0.384, 17.9 ± 0.880, or 18.0 ± 0.403 kcal/(mouse · d) for B6, DBA, or FVB, respectively] (5). The differences in metabolic responses (DBA >B6 or FVB) were also not correlated with the amount of fructose water intake FVB [23.6 ± 1.36 mL/(mouse · d)] >B6 [8.74 ± 0.187 mL/(mouse · d)] or DBA [8.47 ± 0.390 mL/(mouse · d)]) (5). Amount of fructose intake was likely driven by differences in fructose perception and preference between the mouse strains (6, 30).
Overall effects of fructose on gut microbiota community
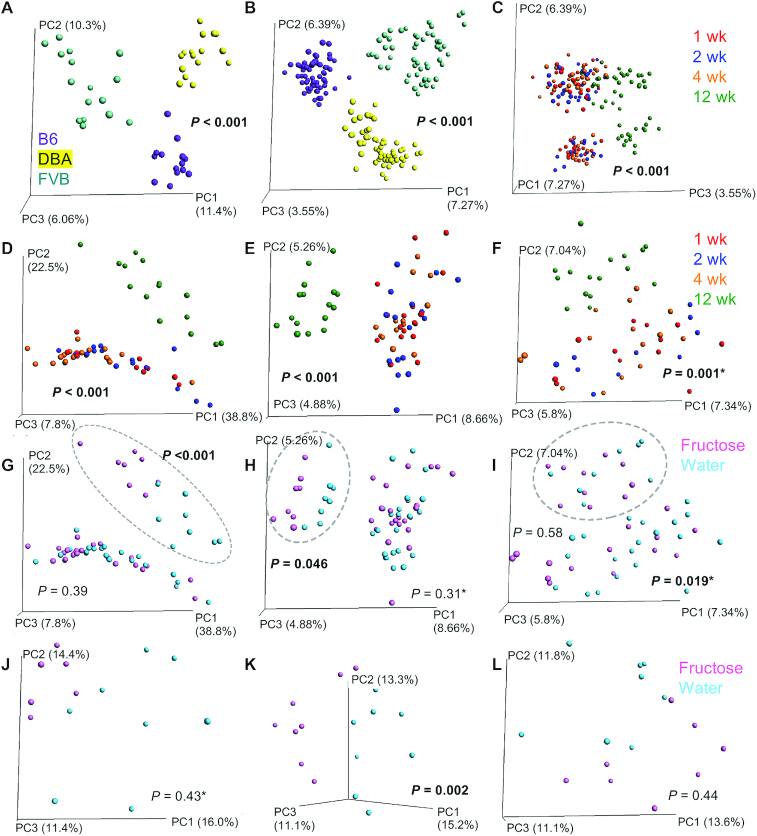
The gut microbiota in the 3 mouse strains were assessed using 16S rDNA sequencing. PCoA plots showed distinct clusters of mouse strain for both the cecal (P < 0.001 by PERMANOVA; Figure 1A) and the fecal (P < 0.001; Figure 1B) samples. Time was also a significant factor in fecal samples (P < 0.001), with week 12 separating from the earlier weeks when evaluating all mouse strains together (Figure 1C) or each mouse strain separately (Figure 1D–F).
FIGURE 1.
PCoA of cecal and fecal microbiota of B6, DBA, and FVB mice consuming fructose or regular water. (A) Cecal microbiota samples across 3 mouse strains were shown to separate by strain (A; n = 16/strain; week 12). (B, C) Fecal microbiota samples across 3 mouse strains were separated by both strain (B; n = 64/strain across 4 time points) and time (C; n = 16/time point for each strain). Panel C used the same ordination as panel B, except that PC3 was presented as the x-axis to show the relation with time. (D–F) For each mouse strain, fecal samples were colored by time points for B6 (D), DBA (E), and FVB (F) to show the time effect. (G–I) Fecal samples were colored with fructose or water treatment for B6 (G), DBA (H), and FVB (I) to show treatment effect. Samples for the 12-wk time point are shown in dotted circles, with the corresponding P values for fructose treatment effect reported. (J–L) Cecal samples were colored by the fructose or water treatment for B6 (J), DBA (K), and FVB (L). P values were generated by PERMANOVA, and significant results are presented in bold. P values with an asterisk designate that significantly different dispersions were observed, which may influence the P values reported as PERMANOVA assumes similar dispersion. B6, C57BL/6J; DBA, DBA/2J; FVB, FVB/NJ; PC, principal coordinate; PCoA, principal coordinates analysis; PERMANOVA, permutational multivariate ANOVA.
Fecal microbiota differences were seen between fructose and water groups at 12 wk in B6 (P < 0.001 by PERMANOVA; Figure 1G) and DBA (P = 0.046; Figure 1H) mice, but not in FVB mice (P = 0.58; Figure 1I). In cecum, the fructose group showed separation from controls for B6 mice (Figure 1J); however, the separation was not statistically significant with PERMANOVA (P = 0.43). This result may be influenced by the difference in multivariate spread (P = 0.038) since PERMANOVA assumes equal dispersion. Fructose treatment was a significant factor for cecal microbiota composition in DBA (P = 0.002; Figure 1K) but not for FVB (P = 0.44; Figure 1L) mice. Overall, consistent with the differential effects of fructose on metabolic phenotypes, DBA gut microbiota in both feces and cecum were sensitive to fructose treatment.
Baseline differences in gut microbiota between mouse strains show correlation with host metabolic phenotypes
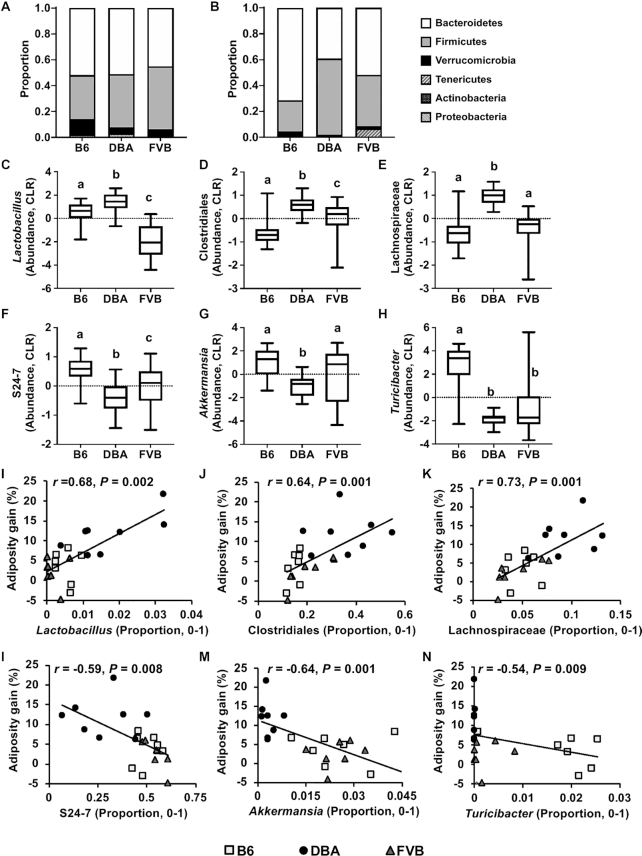
Differences in baseline microbial composition can drive distinct host responses to the same dietary manipulation (31, 32). At the phylum level, there were no significant differences in cecum Firmicutes/Bacteroidetes (F/B) ratios between the 3 mouse strains (Figure 2A). However, in fecal samples, DBA had a significantly higher F/B ratio (1.78 ± 0.169) than B6 (0.346 ± 0.024, P < 0.001 by 1-factor ANOVA) or FVB (0.898 ± 0.091, P < 0.001) (Figure 2B), agreeing with the known association of higher Firmicutes abundance with obesity (33) and the increased adiposity in DBA (5). The microbial taxa that accounted for the greatest differences between the 3 mouse strains included 21 cecal (Supplemental Figure 2A, B) and 14 fecal (Supplemental Figure 2C, D) microbial genera based on the LEfSe analysis.
FIGURE 2.
Cecal and fecal baseline microbial composition in B6, DBA, and FVB mice and correlation with adiposity gain. (A, B) Taxa bar plots of baseline cecal (A) and fecal (B) microbiota of 3 mouse strains at the phylum level. (C–H) Baseline abundance profiles for specific fecal microbiota of 3 mouse strains at the genus level. CLR values were used for plotting the abundance of each microbiota. Box-and-whiskers plots from minimum to maximum showing abundance distribution of Lactobacillus (C), unknown genus of Clostridiales (D), unknown genus of Lachnospiraceae (E), unknown genus of S24–7 (F), Akkermansia (G), and Turicibacter (H). The center line in the box denotes the median value. One-factor ANOVA followed by Sidak's post hoc test was conducted to calculate significant differences between the 3 mouse strains. Labeled means without a common letter differ, P < 0.05. n = 7–8/group. (I–N) Correlation analysis plots between microbiota baseline proportion and adiposity gain at week 12 of fructose treatment. r = Biweight midcorrelation (bicor) coefficient, P = Benjamini-Hochberg–adjusted P values. n = 7–8/group. B6, C57BL/6J; CLR, centered log-ratio; DBA, DBA/2J; FVB, FVB/NJ.
We reasoned that if any specific microbial taxon determines the differential fructose response between the mouse strains, its abundance likely shows contrasting patterns between the susceptible strain DBA and the 2 resistant strains. Of 21 cecal genera (Supplemental Figure 2B), none showed contrasting patterns between DBA and the 2 resistant strains. Of the 14 fecal genera (Supplemental Figure 2D), 6 showed distinct patterns in DBA mice compared with the resistant strains (Figure 2C–H). DBA mice showed higher CLR abundance for Lactobacillus, an unknown bacterium of the order Clostridiales, and an unknown bacterium of the family Lachnospiraceae compared with B6 and FVB. On the other hand, DBA mice had lower CLR abundance for unknown bacteria of family S24–7, Akkermansia and Turicibacter. To understand the potential role of these microbial taxa in metabolic regulation, the correlations between the proportion of these taxa and adiposity gain across all mouse strains were tested. We found that all taxa were significantly correlated with adiposity gain (P < 0.01; Figure 2I–N).
Fructose-responsive microbiota and correlation with host metabolic phenotypes
Next, we explored the differentially abundant microbiota between fructose and water groups in the 3 mouse strains at 12 wk. We observed more fructose-responsive microbiota in feces (9 taxa) compared with cecum samples (1 taxon) across mouse strains at the family level (Table 1).
TABLE 1.
Differentially abundant microbiota between fructose and water groups in cecal and fecal samples of B6, DBA, and FVB mice1
| Mouse strain and taxonomic level | Source | Fructose-responsive microbiota | Fold-change2 |
|---|---|---|---|
| DBA | |||
| Family | Cecum | Erysipelotrichaceae | 0.055* |
| B6 | |||
| Family | Feces | Rikenellaceae | 0.620* |
| Family | Feces | S24–7 | 1.19* |
| Family | Feces | Dehalobacteriaceae | 0.600* |
| Family | Feces | Lachnospiraceae | 0.590* |
| Family | Feces | Mogibacteriaceae | 0.333* |
| Family | Feces | Ruminococcaceae | 0.706* |
| Family | Feces | Turicibacteraceae | 0.348* |
| Family | Feces | Pseudomonadaceae | 3.33* |
| Family | Feces | Verrucomicrobiaceae | 7.25** |
| DBA | |||
| Family | Feces | Rikenellaceae | 0.374* |
| Family | Feces | Pseudomonadaceae | 33.3* |
| DBA | |||
| Genus | Cecum | Erysipelotrichaceae (unknown genus) | 0.004* |
| Genus | Cecum | Clostridium | 0.009** |
| Genus | Cecum | Anaerostipes | 0.025** |
| FVB | |||
| Genus | Cecum | Bifidobacterium | 16.5* |
| B6 | |||
| Genus | Feces | Dehalobacterium | 0.600* |
| Genus | Feces | Mogibacteriaceae (unknown genus) | 0.333* |
| Genus | Feces | Akkermansia | 7.27** |
| DBA | |||
| Genus | Feces | Rikenellaceae (unknown genus) | 0.374* |
| Genus | Feces | Pseudomonadaceae (unknown genus) | 60.0* |
| Genus | Feces | Pseudomonas | 28.0* |
n = 8/group/mouse strain. *FDR < 0.05, **FDR < 0.01. B6, C57BL/6J; DBA, DBA/2J; FDR, false discovery rate; FVB, FVB/NJ.
Fold-change is calculated as the ratio of the relative abundance of microbiota between fructose and water groups (fructose/water).
In feces, fructose treatment showed the highest impact in B6 mice by altering the abundance of many microbiota (9 families and 3 genera). Significant decreases were observed in 5 taxa belonging to the phylum Firmicutes. There were also significant increases in S24–7 of the phylum Bacteroidetes and Verrucomicrobiaceae of the phylum Verrucomicrobia. Within the Verrucomicrobiaceae family, Akkermansia significantly increased in B6 mice after fructose treatment. In DBA feces, fructose altered the abundance of Rikenellaceae and Pseudomonadaceae. These 2 families were also found to be fructose responsive in B6 mice; however, the response was more dramatic in DBA mice (0.374-fold and 33.3-fold changes in DBA compared with 0.620-fold and 3.33-fold changes in B6 for Rikenellaceae and Pseudomonadaceae, respectively). No fecal microbial taxa were significantly altered by fructose in FVB mice (Table 1; Supplemental Figure 3A, B). In DBA cecum, the family Erysipelotrichaceae and 2 of its genera (Clostridum and an unknown genus), and Anaerostipes were all significantly decreased by fructose. Fructose also significantly increased cecal Bifidobacterium in FVB mice, while no cecal taxa were significantly changed in B6 mice (Table 1; Supplemental Figure 3C, D).
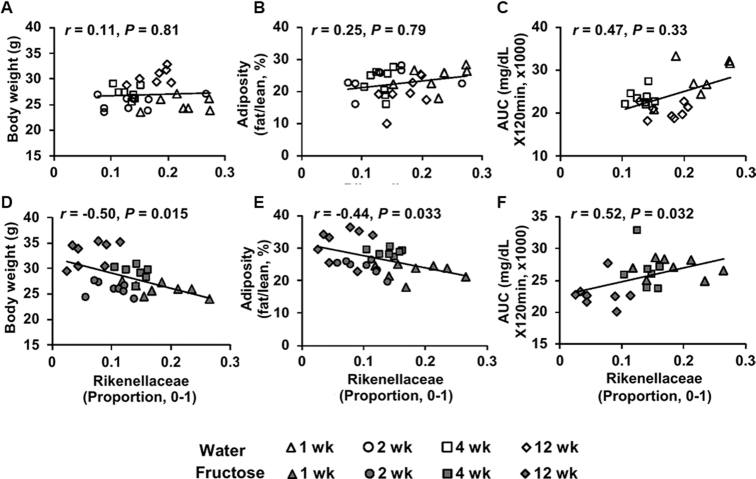
We next correlated the abundance of these fructose-responsive taxa with metabolic phenotypes in water- or fructose-treated mice. In DBA mice, cecal Erysipelotrichaceae was negatively correlated with adiposity and AUC (Supplemental Figure 4A, B). Fecal Rikenellaceae in DBA mice had negative correlations with body weight and adiposity, and a positive correlation with AUC in the fructose group, while no significant correlation was observed with these phenotypes in the water group (Figure 3). No phenotypic correlation was observed for the fructose-responsive taxa in B6 and FVB mice, which is not surprising given the weaker phenotypic alterations in these mouse strains in response to fructose consumption.
FIGURE 3.
Correlation analysis of fructose-responsive microbiota with metabolic phenotypes in DBA mice. (A–C) Correlation plots between Rikenellaceae proportion and body weight (A), adiposity (B), and glucose tolerance AUC (C) across time points in the water group. (D–F) Correlation plots between Rikenellaceae proportion and body weight (D), adiposity (E), and glucose tolerance AUC (F) across time points in the fructose group. r = Biweight midcorrelation (bicor) coefficient, P = Benjamini-Hochberg–adjusted P-values. n = 7–8/group/time point. DBA, DBA/2J.
Correlation of fructose-responsive microbiota with fructose signature genes in host metabolic tissues
We then analyzed the correlation between the abundance of fructose-responsive microbiota and the host fructose signature genes in liver, adipose tissue, and hypothalamus (5). We observed distinct correlation patterns in the 3 mouse strains: the B6 fructose-responsive taxa were correlated with only hypothalamic fructose signature genes, while the fructose-responsive taxa in DBA cecum or feces were correlated with only liver or adipose tissue signature genes, respectively (summary in Table 2; the full list of genes correlated with fructose-responsive taxa shown in Supplemental Table 1).
TABLE 2.
Correlation between fructose-responsive microbiota and host fructose signature genes in 3 metabolic tissues in B6 and DBA mice1
| Mouse strain and fructose-responsive microbiota | No. of correlated genes at FDR <0.05 | Correlation with host fructose-responsive genes | ||
|---|---|---|---|---|
| Host tissue | Top correlated genes2,3 | Overrepresented pathways3 | ||
| B6 (feces)4 | ||||
| Rikenellaceae | Hypo | 37 | Emg1, Rpl37a, Oat, Gna 13, Ctsz* | RNA metabolism, protein localization in endoplasmic reticulum, ribosome, amino acid metabolism, influenza infection |
| S24–7 | Hypo | 11 | Numbl*, Cxx1c*, Gnb2*,1500009L16Rik*, Slc25a17* | — |
| Dehalobacteriaceae | Hypo | 7 | Atp2b2*, Slc6a3*,Nefh*,Nrarp, Hint2 | — |
| Dehalobacterium | Hypo | 42 | Atp2b2*, Slc6a3*,Nefh*, Nrarp, Atg3* | Ribosome, response to alcohol, blood circulation, dopamine biosynthesis, locomotory behavior |
| Lachnospiraceae | Hypo | 2 | Maneal, Rbm39 | — |
| Mogibacteriaceae | Hypo | 54 | Mogs, Snrpe,Nrarp, Timp3*,Sgsm1 | Ribosome, metabolism of proteins, RNA, organonitrogen compound, protein targeting to membrane metabolism |
| Mogibacteriaceae (unknown genus) | Hypo | 130 | Mogs, Psmb4, Derl1, Timp3*,Sdcbp | Ribosome, metabolism of protein, RNA, protein localization to endoplasmic reticulum, signaling by robo receptors |
| Ruminococcaceae | Hypo | 12 | Trappc2l, Gnb2, Uck1, Mid1ip1, Itm2a* | — |
| Pseudomonadaceae | Hypo | 2 | Cxx1c*, Numbl* | — |
| Verrucomicrobiaceae | Hypo | 13 | Scarna2*, AW209491*,Gja1,Rps9*,Th | Ribosome, protein localization to endoplasmic reticulum, mRNA activation |
| Akkermansia | Hypo | 44 | Gja1,Rps9*,Th, Bmp7, Oxt* | Ribosome, protein localization to endoplasmic reticulum, selenoamino acid metabolism, influenza infection, signaling by robo receptors |
| DBA (cecum)4 | ||||
| Erysipelotrichaceae (unknown genus) | Liver | 5 | Gsdmd*, Wdr82*, Tmem62*, S100a10*, Pgk1* | — |
| Clostridium | Liver | 5 | Epha2*, Mad2l2*, Cyp8b1*, Ero1l*, Pus10* | — |
| Anaerostipes | Liver | 4 | Rogdi*, Cyp8b1*, Mad2l2*, Baiap2* | — |
| DBA (feces)4 | ||||
| Rikenellaceae | Adipose | 447 | Ccr1, Sema3e, Msr 1, Abhd3, Fas | RNA metabolism, response to lipid, cytokines, bacterial molecule, lipid metabolism, immune system, endocytosis |
| Rikenellaceae (unknown genus) | Adipose | 432 | Ccr1, Apobec1, Sema3e, Creb1, Fas | RNA metabolism, response to lipid, cytokines, bacterial molecule, lipid metabolism, immune system, signaling pathway |
| Pseudomonadaceae | Adipose | 77 | Sfrp4*, Msr1*, Sema3e*, Abhd3*, Fas* | — |
| Pseudomonadaceae (unknown genus) | Adipose | 209 | Creb1*, Fabp3*, Msr1*, Sema3e*, Fas* | RNA processing, response to cytokine, lipid, defense response, lipid metabolism, response to hormone |
| Pseudomonas | Adipose | 43 | Ppp1r13b*, Jmjd1c*, Msr1*, Slc25a10*, Fabp3* | — |
Fructose-responsive genes in hypothalamus (Hypo), liver, and adipose tissue. B6, C57BL/6J; DBA, DBA/2J; FDR, false discovery rate. Correlations were done at both family or genus levels. The microbiota correlated at the genus level are italicized.
Top 5 correlated genes that belong to major gene sets analyzed using GSEA (Gene Set Enrichment Analysis). Genes with positive correlation with microbiota are indicated with an asterisk.
Full lists of correlated genes are shown in Supplemental Table 1 and enriched pathways are shown in Supplemental Table 2.
Sample size, n = 4–6/group/mouse strain.
In B6, Dehalobacterium showed a positive correlation with hypothalamic genes encoding the neurotransmitter transporter Slc6a3, a notch signaling component Nrarp, and an autophagy gene Atg3. Akkermansia was correlated with several neurotransmitter-related genes, including Oxt encoding precursor of oxytocine/neurophysin 1 and Th encoding tyrosine hydroxylase. In DBA cecum, both Anaerostipes and Clostridium were positively correlated with Cyp8b1 in liver, which is responsible for bile acid synthesis (34).
In DBA feces, all fructose-responsive taxa were correlated with host signature genes of the adipose tissue, and these genes were involved in lipid metabolism, immune system, and response to lipids, cytokines, and hormones (Supplemental Table 2). Adipose genes such as Abhd3, Msr1, Ccr1, Creb1, and Fas were correlated with Rikenellaceae and Pseudomonadaceae as well as genera within these families (Table 2; Supplemental Table 2). Taken together, these correlations suggest that gut microbiota may interact with host genes in a mouse strain– and tissue-specific manner in response to fructose.
Alteration of gut microbiota modulates fructose response
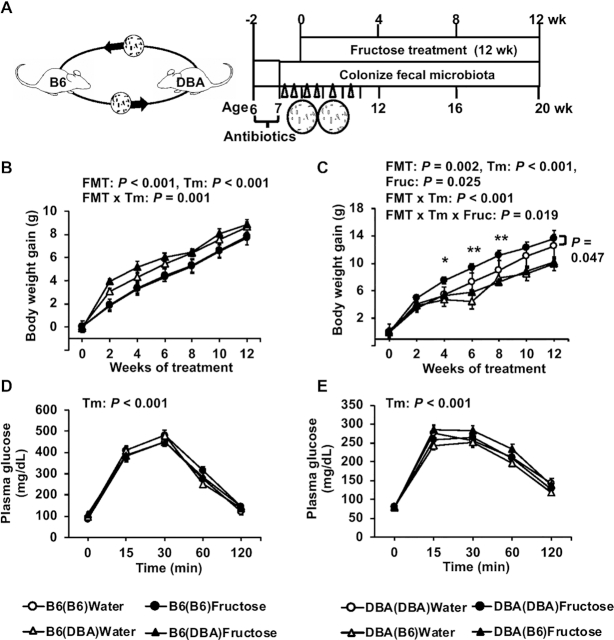
Since B6 and DBA mice showed disparate metabolic responses to fructose, we tested whether B6 microbiota confer resistance and DBA microbiota confer vulnerability to fructose effects by transplanting B6 feces to antibiotic-treated DBA mice and vice versa (Figure 4A). Using 16S rDNA sequencing, we confirmed that the recipient mice gut microbiome shifted after fecal transplant (Supplemental Figure 5A, B).
FIGURE 4.
Metabolic phenotypes post–fecal transplant in B6 and DBA mice with or without fructose consumption. (A) Schematic design of FMT. (B–E) Body-weight gain (B, C) and glucose tolerance (D, E) of recipient B6 and DBA mice, respectively, with or without 8% fructose water. Data are presented as means ± SEMs, n = 7–14/group. The P values of the main factors (FMT, fructose, time) and interactions by 3-factor repeated-measures ANOVA are shown on the top of the graph. Asterisks in panel C indicate time points at which significant differences were found between DBA(DBA) and DBA(B6) under fructose treatment based on 2-factor repeated-measures ANOVA with Sidak's post hoc test; *P < 0.05, **P < 0.01. Fructose effects within each FMT group across time points were assessed by 2-factor repeated-measures ANOVA, and significant difference between fructose and water treatments for the DBA(DBA) FMT group is indicated by a P value with a side bar (C). B6, C57BL/6J; B6(B6), B6 mice receiving B6 feces; B6(DBA), B6 mice receiving DBA feces; DBA, DBA/2J; DBA(B6), DBA mice receiving B6 feces; DBA(DBA), DBA mice receiving DBA feces; FMT, fecal microbiota transplant; Fruc, fructose; Tm, time.
When the main effects of FMT, fructose, and time were tested, there were significant FMT effects on weight gain in both B6 (P < 0.001; Figure 4B) and DBA (P = 0.002; Figure 4C) mice, but there was no effect of FMT on glucose tolerance in both mouse strains (Figure 4D, E). Overall, there was a significant fructose effect on weight gain in DBA mice (P = 0.025; Figure 4C), which was not observed in B6 mice (Figure 4B). In those fed fructose, body-weight gain in DBA mice receiving B6 bacteria [DBA(B6)] was significantly lower compared with DBA(DBA) mice at 4 wk (P = 0.028), 6 wk (P = 0.006), and 8 wk (P = 0.007). In contrast, there was no significant fructose effect in B6 mice receiving DBA bacteria [B6(DBA)] or in B6(B6) mice (Figure 4B). These results suggest that DBA microbiota failed to induce fructose sensitivity in B6 mice. On the other hand, DBA(B6) mice no longer displayed fructose-induced weight gain (fructose effect, P = 0.66) as seen in DBA(DBA) mice (fructose effect, P = 0.047; Figure 4C), supporting that B6 microbiota conferred resistance to body-weight gain upon fructose consumption.
The results from FMT experiments support a causal role of B6 microbiota in conferring fructose resistance to DBA. We next focused on prioritizing the potential microbes in B6 that may determine the fructose-resistance phenotype. Akkermansia was found to be a highly plausible candidate to explain the dampened response to fructose in B6 for the following reasons. First, Akkermansia has been previously demonstrated to carry antiobesity and insulin-sensitizing effects (32, 35, 36). The beneficial effect of Akkermansia was also previously observed in mice fed a high-fat, high-sucrose diet (37). Second, it was depleted in the vulnerable strain DBA but was highly abundant in both resistant mouse strains, B6 and FVB (Figure 2G). Third, fructose treatment caused an increase in Akkermansia in B6 mice (Table 1; Supplemental Figure 3A). Last, Akkermansia abundance in B6 is significantly correlated with a large number of hypothalamic genes such as Oxt and Th, which regulate metabolism (Table 2; Supplemental Table 1).
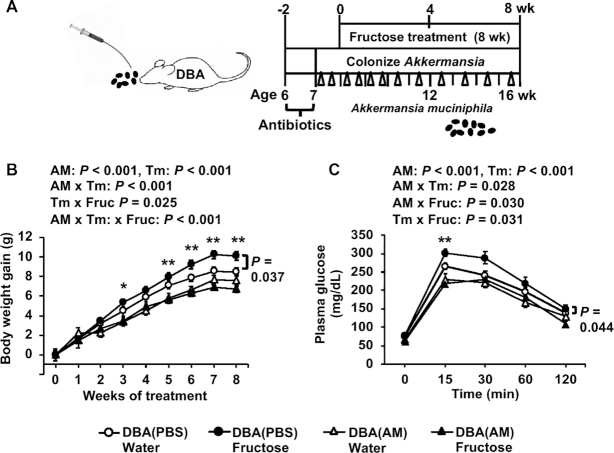
To test the role of Akkermansia in protecting against fructose-induced metabolic dysregulation as predicted above, we gavaged DBA mice with AM or PBS media along with fructose treatment for 8 wk (Figure 5A). There were significant effects of AM on both body-weight gain (P < 0.001) and glucose tolerance (P < 0.001) (Figure 5B, C). Under fructose treatment, DBA mice receiving AM had significantly lower weight gains at 3 wk (P = 0.023) and 5–8 wk (P < 0.01) compared with control mice receiving PBS. No AM effect was observed in the water treatment mice (P = 0.14). Within the AM or PBS group, fructose had significant effects on body-weight gain in the PBS control group (P = 0.037), whereas DBA mice receiving AM no longer displayed fructose-induced body-weight gain as seen in control mice (Figure 5B). Furthermore, fructose increased glucose intolerance in the PBS control group (fructose effect, P = 0.044), whereas AM treatment abrogated fructose-induced glucose intolerance in DBA mice (Figure 5C). These results support that Akkermansia confers resistance to fructose-mediated metabolic dysregulation.
FIGURE 5.
Metabolic phenotypes post–Akkermansia colonization in DBA mice with or without fructose consumption. (A) Schematic design of AM colonization. PBS serves as the control for AM. (B, C) Body-weight gain (B) and glucose tolerance (C) of recipient DBA mice with or without 8% fructose water. Data are presented as means ± SEMs, n = 8–14/group. The P values of main factors (AM, fructose, time) and interactions by 3-factor repeated-measures ANOVA are shown on the top of the graph. Asterisks indicate time points at which significant differences were found between the AM colonization and PBS control groups under fructose treatment based on 2-factor repeated-measures ANOVA with Sidak's post hoc test; *P < 0.05, **P < 0.01. Fructose effects within PBS or AM groups across time points were assessed by 2-factor repeated-measures ANOVA. A significant fructose effect on weight gain and glucose tolerance in mice receiving PBS is indicated by P values with a side bar (B and C). AM, Akkermansia muciniphila; DBA, DBA/2J; Fruc, fructose; Tm, time.
Discussion
Our previous study showed that 3 mouse strains representing a range of genetic diversity differed in their metabolic and transcriptomic responses to high-fructose treatment (5). Since gut microbiota are an important modulator of metabolic capacity (7), here we tested the hypothesis that disparate fructose responses among mouse strains were at least partially driven by the gut microbiota. Our 16S rDNA sequencing analysis revealed that baseline microbiota composition and its response to fructose varied by mouse strains. The fecal transplant data indicate that B6 mice carry microbiota that confer resistance to fructose-induced body-weight gain. We next evaluated candidate taxa to explain the dampened response to fructose in B6 mice. We prioritized Akkermansia since it is enriched in B6 compared with DBA mice, which have lower abundances. Indeed, gavaging AM to DBA mice mitigated fructose-induced obesity and glucose intolerance. These results support a causal role of gut microbiota in determining the differential metabolic responses to fructose among genetically diverse mouse strains.
As initial colonizing microbial species are important for establishing a favorable environment for bacterial growth in a particular context (38), differences in baseline microbial composition between mouse strains can lead to variations in the metabolic processes of individuals in response to diet (31, 32). We found that Akkermansia, Turicibacter, and S24–7 were lower in DBA mice but were abundant in B6 and FVB mice. On the other hand, the baseline abundance of Lactobacillus, Clostridiales, and Lachnospiraceae were higher in DBA mice. Previously, Akkermansia has been associated with obesity resistance and improved metabolic parameters in humans and mice, and beneficial effects of dietary interventions have been associated with the higher abundance of Akkermansia at baseline (32, 36). S24–7 has a protective association against diabetes, whereas Lachnospiraceae promotes pathogenesis of diabetes in non-obese diabetic (NOD) mice (39). The observed lack of protective Akkermansia and S24–7, along with the higher abundance in pathogenic Lachnospiraceae in DBA mice, agrees with the vulnerability of DBA mice to fructose-induced metabolic dysregulation. Therefore, these bacterial taxa that differ significantly at baseline between strains have the potential to regulate the differential response to fructose treatment.
In addition to differences in baseline microbes that can explain intermouse strain variability, bacteria altered by fructose may also play a role in the variability in metabolic responses to fructose. It has been suggested that fructose shifts the gut microbiota and leads to a westernized microbiome acquisition with altered metabolic capacity, resulting in development of obesity or metabolic disorders (40). In our study, we found various microbes with significantly altered abundances in DBA and B6 but not FVB, which may relate to their differential sensitivity to fructose. Cecal Erypsipelotricaceae and Anaerostipes in DBA mice decreased in abundance upon fructose consumption (Table 1; Supplemental Figure 3), and they are known as butyrate-producing bacteria (41). Butyrate promotes the intestinal barrier development, and decreased butyrate production can increase intestinal permeability (42). In both the B6 and DBA feces, we also detected decreased abundance of Rikenellaceae and increased abundance of Pseudomonadaceae following fructose treatment (Table 1). Increases in Pseudomonadaceae and Pseudomonas were previously found in obese individuals with higher insulin resistance (43). Therefore, despite the numerous differences between B6 and DBA, there were shared microbial changes in response to fructose consumption. The weaker phenotypic responses in B6 can be a result of compensatory balancing effects of higher abundances of beneficial bacteria such as Akkermansia and S24–7 in B6.
We detected more fructose-responsive microbiota in B6 mice than in DBA and FVB mice (Table 1), and interestingly, most of these B6 taxa were associated with the hypothalamic fructose signature genes (Table 2; Supplemental Table 1). Among them are genes that are important for energy homeostasis such as Nrarp, Oxt, and Th. Nrarp encodes an intracellular component of the Notch signaling pathway and regulates differentiation of mouse hypothalamic arcuate neurons responsible for feeding and energy balance. Dysregulation of this homeostatic mediator is an underlying cause of various diseases ranging from growth failure to obesity (44). Oxt encodes oxytocin, which is the anorexigenic peptide. Oxytocin maintains homeostasis in feeding-related behavior (45). The Th gene encodes tyrosine hydrogenase (TH). Hypothalamic arcuate nucleus TH neurons play a role in energy homeostasis, and silencing of TH neurons reduces body weight (46). Our previous mice study (2) reported that fructose is an inducer of both genomic and epigenomic variability in the hypothalamus and has the ability to reorganize gene networks that play a central role in metabolic regulation and neuronal processes. The current study suggests that fructose-induced transcriptomic changes in the hypothalamus of B6 mice could be partly driven by fructose-responsive gut microbiota.
In contrast to B6 mice, fructose-responsive microbiota in DBA mice were associated with lipid and inflammatory genes in the adipose tissue (Table 2; Supplemental Table 2). Abhd3 is a crucial factor for insulin resistance in adipose tissue (47); Sema3e contributes to inflammation and insulin resistance in obese mice (48); Msr1 provides protection from excessive insulin resistance in obese mice (49); Creb1 promotes expression of transcriptional factors of adipogenesis and insulin resistance in obesity (50); Fas contributes to adipose tissue inflammation, hepatic steatosis, and insulin resistance induced by obesity (51). Therefore, these adipose-tissue genes correlating with fructose-responsive bacteria in DBA are relevant to the increased adiposity and compromised insulin sensitivity seen in DBA mice. However, we acknowledge that the correlative relation observed here does not directly imply causation, and future experiments are needed to directly test the causal role of the genes as well as the bacteria implicated.
Our fecal transplant study support that B6 mice carry gut microbes that confer resistance to fructose-induced metabolic syndrome, while DBA microbiota did not significantly induce fructose sensitivity in B6 mice. We further demonstrate that Akkermansia partially mediates the protective effect of B6 microbiota. This is the first time that Akkermansia is implicated in determining fructose response. Given the recognized therapeutic potential of modulating the gut microbiota (8), probiotic treatment with Akkermansia may represent a viable approach to mitigate fructose-induced metabolic abnormalities. In addition to Akkermansia, other bacteria may also play roles in modulating the differential fructose response between individuals. Future efforts will explore the pathogenic or protective roles of other bacterial species, such as Lactobacillus, Turicibacter, or Pseudomonas.
In summary, our multistrain, multi-omics (gut microbiome, transcriptome, and phenome) integrative studies of interindividual variability in fructose-induced metabolic syndrome established a causal role of gut microbiota in modulating host responses to fructose, a significant metabolic risk in modern societies. Importantly, our study identified key microbial species that may serve as preventative or therapeutic targets for metabolic syndrome. By exploring gut–host interactions, our study also opens numerous hypotheses regarding specific microbiota–host interactions to be further elucidated in the future.
Supplementary Material
Acknowledgments
The authors thank Dr Richard C Davis for assistance in oral gavage and fecal transplant, Drs Yuqi Zhao and Zeyneb Kurt for assistance with data analysis, and Dr Sana Majid, Paul Patel, Maya Singh, Jessica Yang, and Justin Yoon for helping with animal experiments. The authors’ responsibilities were as follows—XY, EYH, ISA, and CAO: designed the research; ISA, CAO, JML, ZY, IC, GD, PC, HRB, and JD: conducted the research including animal experiments; JML, ISA, GZ, IK, and JD: analyzed data; ISA, XY, and JML: wrote the manuscript; FG-P, AJL, and all authors: contributed to manuscript revision; XY: had primary responsibility for the final content; and all authors: read and approved the final manuscript.
Notes
XY and FG-P are funded by NIH DK104363. JML was supported by NIH T32 DK007789. GD was supported by National Institute of Environmental Health Sciences NIH T32ES015457 and an American Diabetes Association Postdoctoral Fellowship 1-19-PDF-007-R. IK is supported in part by NIH DK077162, the Allan Smidt Charitable Fund, and the Factor Family Foundation Chair in Nephrology. AJL is supported by NIH HL28481 and HL30568. CAO is supported by National Institute on Aging F31AG064844. EYH is supported by Department of Defense Army Research office W911NF1710402 and is a New York Stem Cell Foundation–Robertson Investigator.
Author disclosures: The authors report no conflicts of interest.
Supplemental Figures 1–5 and Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
ISA and JML contributed equally to this work.
Abbreviations used: AM, Akkermansia muciniphila; AUC, area under the curve; B6, C57BL/6J; CLR, centered log-ratio; DBA, DBA/2J; F/B, Firmicutes to Bacteroidetes; FDR, false discovery rate; FMT, fecal microbiota transplant; FVB, FVB/NJ; IPGTT, intraperitoneal glucose-tolerance test; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; OTU, operational taxonomic unit; PCoA, principal coordinates analysis; PERMANOVA, permutational multivariate analysis of variance; rDNA, ribosomal DNA; STAMP, Statistical Analysis of Metagenomic Profiles; TH, tyrosine hydrogenase.
Contributor Information
In Sook Ahn, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Jennifer M Lang, Department of Medicine, Division of Cardiology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Christine A Olson, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Graciel Diamante, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Guanglin Zhang, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Zhe Ying, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Hyae Ran Byun, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Ingrid Cely, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Jessica Ding, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Peter Cohn, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Ira Kurtz, Department of Medicine, Division of Nephrology, University of California, Los Angeles, CA, USA; Brain Research Institute, University of California, Los Angeles, CA, USA.
Fernando Gomez-Pinilla, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA; Department of Neurosurgery, University of California, Los Angeles, CA, USA.
Aldons J Lusis, Department of Medicine, Division of Cardiology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Elaine Y Hsiao, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA.
Xia Yang, Department of Integrative Biology and Physiology, University of California, Los Angeles, CA, USA; Brain Research Institute, University of California, Los Angeles, CA, USA; Molecular Biology Institute, University of California, Los Angeles, CA, USA; Institute for Quantitative and Computational Biosciences, University of California, Los Angeles, CA, USA.
References
- 1. Basciano H, Federico L, Adeli K. Fructose, insulin resistance, and metabolic dyslipidemia. Nutr Metab. 2005;2:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meng Q, Ying Z, Noble E, Zhao Y, Agrawal R, Mikhail A, Zhuang Y, Tyagi E, Zhang Q, Lee JH et al. Systems nutrigenomics reveals brain gene networks linking metabolic and brain disorders. EBioMedicine. 2016;7:157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koo HY, Wallig MA, Chung BH, Nara TY, Cho BHS, Nakamura MT. Dietary fructose induces a wide range of genes with distinct shift in carbohydrate and lipid metabolism in fed and fasted rat liver. Biochim Biophys Acta Mol Basis Dis. 2008;1782:341–8. [DOI] [PubMed] [Google Scholar]
- 4. Li Z, Xiong C, Mo S, Tian H, Yu M, Mao T, Chen Q, Luo H, Li Q, Lu J et al. Comprehensive transcriptome analyses of the fructose-fed syrian golden hamster liver provides novel insights into lipid metabolism. PLoS One. 2016;11:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang G, Byun HR, Ying Z, Blencowe M, Zhao Y, Hong J, Shu L, Chella Krishnan K, Gomez-Pinilla F, Yang X. Differential metabolic and multi-tissue transcriptomic responses to fructose consumption among genetically diverse mice. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glendinnning J, Breinager L, Kyrillou E, Lacuna K, Rocha R, Sclafani S. Differential effects of sucrose and fructose on dietary obesity in four mouse strains. Physiol Behav. 2010;101:331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. [DOI] [PubMed] [Google Scholar]
- 8. Fändriks L. Roles of the gut in the metabolic syndrome: an overview. J Intern Med. 2017;281:319–36. [DOI] [PubMed] [Google Scholar]
- 9. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oh JH, Alexander LM, Pan M, Schueler KL, Keller MP, Attie AD, Walter J, van Pijkeren JP. Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont Lactobacillus reuteri. Cell Host Microbe. 2019;25:273–84. [DOI] [PubMed] [Google Scholar]
- 13. Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, De los Reyes-Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Townsend GE, Han W, Schwalm ND, Raghavan V, Barry NA. Dietary sugar silences a colonization factor in a mammalian gut symbiont. 2019;116:233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, Ding X, Guan R, Zhu C, Xu C, Zhu B, Zhang H, Xiong Z, Xue Y, Tu J et al. Evaluation of different 16S rRNA gene V regions for exploring bacterial diversity in a eutrophic freshwater lake. Sci Total Environ. 2018;618:1254–67. [DOI] [PubMed] [Google Scholar]
- 17. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci. 2011;108:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell. 2018;173:1728–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015;290:5647–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, Israeli D, Zmora N, Gilad S, Weinberger A et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–6. [DOI] [PubMed] [Google Scholar]
- 23. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H Vegan: Community Ecology Package. R package version 2.2-0. 2014. [Google Scholar]
- 25. Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles, Bioinformatics. 2014;30:3123–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Storey JDD, Taylor JE, Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J R Stat Soc. 2004;66:187–205. [Google Scholar]
- 29. Zheng CH, Yuan L, Sha W, Sun ZL. Gene differential coexpression analysis based on biweight correlation and maximum clique. BMC Bioinformatics. 2014;15:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reed DR, Li S, Li X, Huang L, Tordoff MG, Starling-Roney R, Taniguchi K, West DB, Ohmen JD, Beauchamp GK et al. Polymorphisms in the taste receptor gene (Tas1r3) region are associated with saccharin preference in 30 mouse strains. J Neurosci. 2004;24:938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kootte RS, Levin E, Salojärvi J, Smits LP, Hartstra AV, Udayappan SD, Hermes G, Bouter KE, Koopen AM, Holst JJ et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26:611–9. [DOI] [PubMed] [Google Scholar]
- 32. Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, Kayser BD, Levenez F, Chilloux J, Hoyles L et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–36. [DOI] [PubMed] [Google Scholar]
- 33. Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci. 2005;102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. [DOI] [PubMed] [Google Scholar]
- 35. Derrien M, van Baarlen P, Hooiveld G, Norin E, Muller M, de Vos W. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011;2:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y. Cross-talk between Akkermansiamuciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Org E, Parks BW, Joo JWJ, Emert B, Schwartzman W, Kang EY, Mehrabian M, Pan C, Knight R, Gunsalus R. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015;25:1558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7:e44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krych F, Nielsen DS, Hansen AK, Hansen CHF. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-γ level in NOD mice. Gut Microbes. 2015;6:101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Payne AN, Chassard C, Lacroix C. Gut microbial adaptation to dietary consumption of fructose, artificial sweeteners and sugar alcohols: implications for host–microbe interactions contributing to obesity. Obes Rev. 2012;13:799–809. [DOI] [PubMed] [Google Scholar]
- 41. Levine UY, Looft T, Allen HK, Stanton TB. Butyrate-producing bacteria, including mucin degraders, from the swine intestinal tract. Appl Environ Microbiol. 2013;79:3879–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peng L, Li Z-R, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moreno-Indias I, Sánchez-Alcoholado L, García-Fuentes E, Cardona F, Queipo-Ortuño MI, Tinahones FJ. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am J Transl Res. 2016;8:5672–84. [PMC free article] [PubMed] [Google Scholar]
- 44. Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, Niehrs C, Kintner C. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 2001;15:1885–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Olszewski PK, Klockars A, Schiöth HB, Levine AS. Oxytocin as feeding inhibitor: maintaining homeostasis in consummatory behavior. Pharmacol Biochem Behav. 2010;97:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang X, Van Den Pol AN. Hypothalamic arcuate nucleus tyrosine hydroxylase neurons play orexigenic role in energy homeostasis. Nat Neurosci. 2016;19:1341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xia W, Pessentheiner AR, Hofer DC, Amor M, Schreiber R, Schoiswohl G, Eichmann TO, Walenta E, Itariu B, Prager G et al. Loss of ABHD15 impairs the anti-lipolytic action of insulin by altering PDE3B stability and contributes to insulin resistance. Cell Rep. 2018;23:1948–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shimizu I, Yoshida Y, Moriya J, Nojima A, Uemura A, Kobayashi Y, Minamino T. Semaphorin3E-induced inflammation contributes to insulin resistance in dietary obesity. Cell Metab. 2013;18:491–504. [DOI] [PubMed] [Google Scholar]
- 49. Cavallari JF, Anhe FF, Foley KP, Denou E, Chan RW, Bowdish DME, Schertzer JD. Targeting macrophage scavenger receptor 1 promotes insulin resistance in obese male mice. 2018;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qi L, Saberi M, Zmuda E, Wang Y, Altarejos J, Dentin R, Hedrick S, Bandyopadhyay G, Hai T, Montminy M. Adipocyte CREB promote insulin resistance in obesity. Cell Metab. 2010;9:277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wueest S, Rapold RA, Schumann DM, Rytka JM, Schildknecht A, Nov O, Chervonsky AV, Rudich A, Schoenle EJ, Donath MY et al. Deletion of Fas in adipocytes relieves adipose tissue inflammation and hepatic manifestations of obesity in mice. J Clin Invest. 2010;120:191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.