Sharp et al. investigate cell cycle control of chromatin-associated RNP complexes. They find that Aurora-B and SAF-A control the redistribution of RNAs away from chromosomes in mitosis. This early mitotic chromosomal remodeling process is required for the fidelity of chromosome segregation.
Abstract
During mitosis, the genome is transformed from a decondensed, transcriptionally active state to a highly condensed, transcriptionally inactive state. Mitotic chromosome reorganization is marked by the general attenuation of transcription on chromosome arms, yet how the cell regulates nuclear and chromatin-associated RNAs after chromosome condensation and nuclear envelope breakdown is unknown. SAF-A/hnRNPU is an abundant nuclear protein with RNA-to-DNA tethering activity, coordinated by two spatially distinct nucleic acid–binding domains. Here we show that RNA is evicted from prophase chromosomes through Aurora-B–dependent phosphorylation of the SAF-A DNA-binding domain; failure to execute this pathway leads to accumulation of SAF-A–RNA complexes on mitotic chromosomes, defects in metaphase chromosome alignment, and elevated rates of chromosome missegregation in anaphase. This work reveals a role for Aurora-B in removing chromatin-associated RNAs during prophase and demonstrates that Aurora-B–dependent relocalization of SAF-A during cell division contributes to the fidelity of chromosome segregation.
Introduction
During cell division, chromosomes are dramatically restructured to facilitate segregation to daughter cells. At the onset of mitosis, most nuclear transcription ceases through a block of transcription initiation, causing a runoff of actively elongating RNA polymerase (Akoulitchev and Reinberg, 1998; Liang et al., 2015; Palozola et al., 2017; Prescott and Bender, 1962; Segil et al., 1996). During the same period, interphase chromosome architecture is erased through the removal of the majority of cohesin complexes from euchromatin and the loading of condensin complexes (Gibcus et al., 2018). This coordinated exchange of cohesin and condensin complexes leads to dramatic chromosome condensation and the characteristic X shape of mitotic chromosomes (Haarhuis et al., 2014). Collectively, each of these genome remodeling pathways contributes to the accurate segregation of chromosomes during anaphase.
Aurora-B is a master regulatory kinase that controls mitotic chromosome segregation and cytokinesis. Aurora-B is a member of the “chromosome passenger complex” (CPC) that dynamically changes localization throughout mitosis. The CPC localizes throughout the chromosomes at prophase, concentrates at the inner centromere region at metaphase, and transfers from chromosomes to the spindle midzone during anaphase (Carmena et al., 2012). During prophase, the CPC, Plk1, and Cdk1 phosphorylate the cohesin-associated proteins Sororin and SA2, which allows the cohesin release factor WAPL to open cohesin rings and strip cohesin from euchromatic regions (Haarhuis et al., 2014). In addition to phosphorylation of cohesin complexes, Aurora-B phosphorylates histone H3 during prophase to release HP1 from chromatin (Fischle et al., 2005; Hirota et al., 2005). However, the full spectrum of mitotic Aurora-B substrates and functions is not currently known.
During interphase, a large proportion of the genome is transcribed into RNA. Most mRNAs are spliced, capped, polyadenylated, and exported from the nucleus. However, a small fraction of fully processed mRNAs are retained in the nucleus (Bahar Halpern et al., 2015). Many nuclear transcripts do not code for proteins but are part of a diverse group of functional noncoding RNAs (Djebali et al., 2012). Noncoding RNAs influence gene expression by many mechanisms, including interacting with both transcriptional activators and repressors, promoting 3D genome organization, coating specific chromosomal domains, promoting DNA replication origin usage, and silencing an entire chromosome (Wang and Chang, 2011). Because noncoding RNA function has been studied primarily during interphase, it is not clear how genome organization promoted by nuclear RNAs is regulated during mitosis when chromosomes are restructured.
The prototypical nuclear noncoding RNA is the XIST RNA. This transcript is expressed from the inactive X chromosome in female cells and coats the Xi chromosome in cis to silence most genes (Galupa and Heard, 2015). XIST RNA is tethered to the Xi chromosome during interphase through a combination of factors, including Ciz1 (Ridings-Figueroa et al., 2017; Sunwoo et al., 2017), heterogeneous nuclear RNP K (hnRNP-K; Colognori et al., 2019; Pintacuda et al., 2017), and hnRNP-U/SAF-A (Hasegawa et al., 2010). Interestingly, XIST RNA is removed from the mitotic Xi chromosome in an Aurora-B–dependent manner, but the molecular mechanism is not known (Hall et al., 2009). Additionally, it is not known if other nuclear RNAs are also removed from chromosomes during mitosis.
SAF-A is a highly abundant nuclear protein originally identified as a factor that binds with high affinity to scaffold attachment regions of chromosomes and as a protein that binds to heterogeneous nuclear RNA (Fackelmayer and Richter, 1994; Kiledjian and Dreyfuss, 1992). Indeed, SAF-A has been shown to interact with hundreds of RNAs, and recent mass spectrometry studies have shown that all cellular SAF-A is complexed with RNA (Caudron-Herger et al., 2019; Huelga et al., 2012; Xiao et al., 2012). SAF-A contains a modular domain architecture with an N-terminal SAP DNA-binding domain, a central AAA+ domain, and a C-terminal RGG-type RNA-binding domain. SAF-A is important for restricting XIST RNA localization to the Xi chromosome territory during interphase through a mechanism requiring the SAP and Arginine/Glycine-rich (RGG) domains. The presence of two spatially and functionally distinct nucleic acid–binding domains suggested that SAF-A directly tethers XIST RNA to chromatin (Hasegawa et al., 2010). Subsequently, SAF-A has been implicated in a wide variety of nuclear RNA regulatory processes: forming interchromosomal connections through interactions with the FIRRE noncoding RNA (Hacisuleyman et al., 2014), mRNA splicing (Ye et al., 2015), and decompaction of euchromatic DNA through interaction with RNA (Nozawa et al., 2017). Notably, all described SAF-A functions occur in the interphase nucleus, and many of these processes are reversed during early mitosis. We therefore tested whether SAF-A is regulated during mitosis or if SAF-A–mediated chromosomal structures are remodeled during chromosome condensation.
In this work, we investigate the molecular mechanisms that regulate nuclear RNA localization during mitosis. We find that nuclear RNAs are removed from the surface of prophase chromosomes in an Aurora-B– and SAF-A–dependent manner. We show that Aurora-B phosphorylates SAF-A at two sites in the SAP domain to release SAF-A–RNA complexes from chromatin during mitosis. Additionally, we find that nonphosphorylatable SAF-A phenocopies the genome-wide retention of RNA on mitotic chromosomes observed in Aurora-B–inhibited cells. Finally, we demonstrate that cells expressing nonphosphorylatable SAF-A experience a high frequency of chromosome misalignment in metaphase and chromosome segregation errors in anaphase. Our results show that removal of SAF-A complexed to nuclear RNAs from chromatin is a key aspect of mitotic chromosomal restructuring and is essential for accurate chromosome segregation.
Results
SAF-A–RNA complexes undergo dynamic interactions with chromatin during the cell cycle
SAF-A is involved in several processes central to interphase nuclear function, including chromatin-bound RNA localization, interchromosomal interactions, and DNA decondensation. Because these processes are all downregulated or reversed to allow for chromosome condensation and individualization during early mitosis, we hypothesized that SAF-A interactions with DNA or RNA are regulated in a cell cycle–dependent manner.
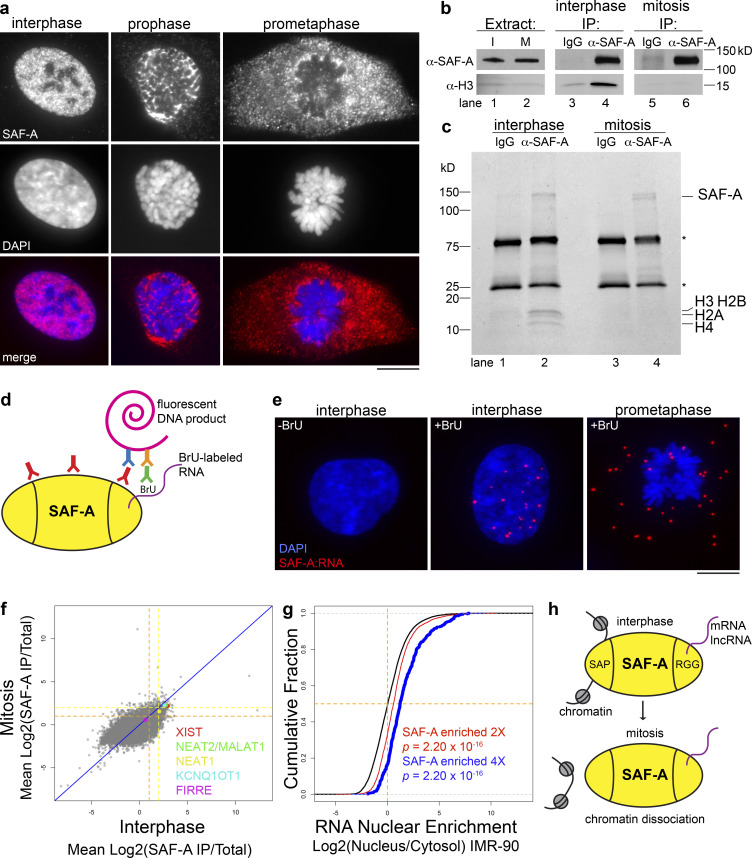
To test for SAF-A–chromatin interactions, we monitored SAF-A localization across the cell cycle in human, diploid RPE-1 cells (Fig. 1 a). Interphase cells showed strong nuclear staining of SAF-A as previously described (Dreyfuss et al., 1984; Hasegawa et al., 2010). In mitotic cells, we observed that SAF-A was dramatically cleared from chromatin in prophase, and that SAF-A staining of chromatin was absent by prometaphase. The exclusion of SAF-A from mitotic chromosomes was observed with two different antibodies to detect the native protein (Fig. 1 a and data not shown), in SAF-A–GFP–transfected cells imaged under both live and fixed conditions (see Fig. 4 d, Fig. 7 c, and Fig. S2 e), and in cells expressing a SAF-A–AID–mCherry allele knocked into the endogenous locus (see Fig. S2 d).
Figure 1.
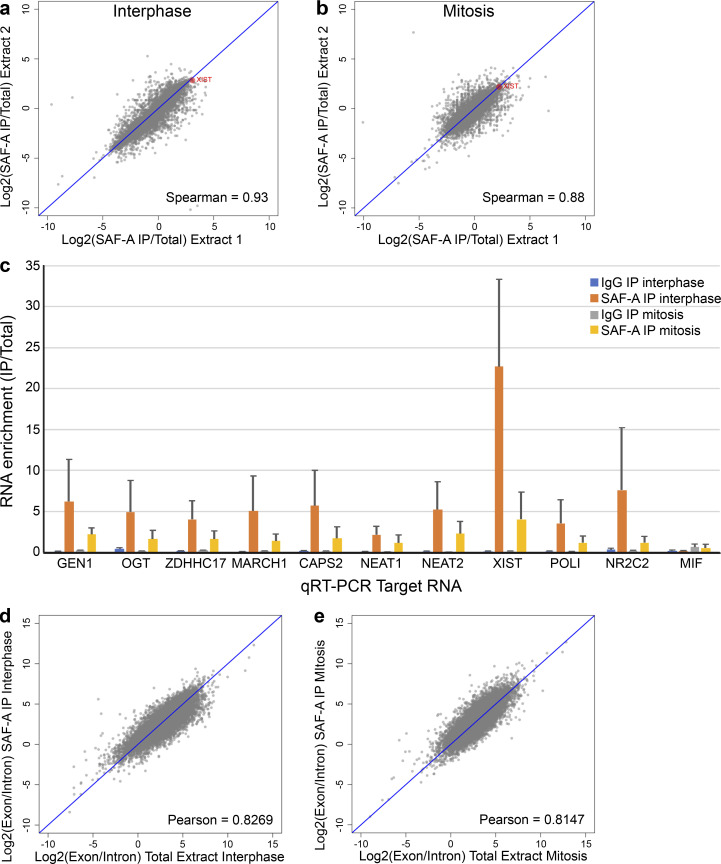
SAF-A–RNA complexes undergo dynamic interactions with chromatin during the cell cycle. (a) IF of SAF-A in RPE-1 cells relative to chromosomes (DAPI). Scale bar = 10 µm. (b) Western blot analysis of SAF-A and histone H3 levels in cell extracts (lanes 1 and 2) and IPs (lanes 3–6) from cells arrested in interphase (I) or mitosis (M). Mock IPs included mouse IgG (lanes 3 and 5); α-SAF-A IPs (lanes 4 and 6) were performed with a mouse mAb specific for SAF-A. (c) Coomassie-stained gel analysis of IgG IPs (lanes 1 and 3) and α-SAF-A IPs (lanes 2 and 4). IgG is denoted by the asterisks. (d) The PLA was modified to detect interactions between SAF-A and RNA. RPE-1 cells were labeled with BrU and incubated with primary antibodies to SAF-A and BrU. When the antigens are <150 nm apart, secondary antibodies conjugated to oligos will generate a fluorescent DNA product. (e) SAF-A–RNA PLA interactions detected in RPE-1 cells incubated with or without BrU to label RNA. (f) RIP-seq was performed to identify SAF-A–interacting RNAs in interphase and mitotic RPE-1 cell extracts. To determine which RNAs associated with SAF-A in both interphase and mitosis, the average RNA enrichment in mitosis was plotted against RNA enrichment in interphase. Average enrichment values were determined from two independent biological replicates; all RNAs greater than or equal to twofold enriched are gated by the dashed orange line, whereas all RNAs greater than or equal to fourfold enriched are gated by the dashed yellow line. (g) Nuclear retention of SAF-A–interacting RNAs. SAF-A–interacting RNAs were compared with the nuclear/cytosolic distribution determined for each expressed RNA. SAF-A–interacting RNAs are shown in blue (fourfold enriched) and red (twofold enriched); total RNA expression is shown in black. A rightward shift on the x axis indicates a higher degree of nuclear retention for SAF-A–interacting RNAs relative to the total RNA population. P values were calculated using a Wilcoxon rank-sum test. (h) Summary of SAF-A interactions with chromatin and RNA during interphase and mitosis.
Figure 4.
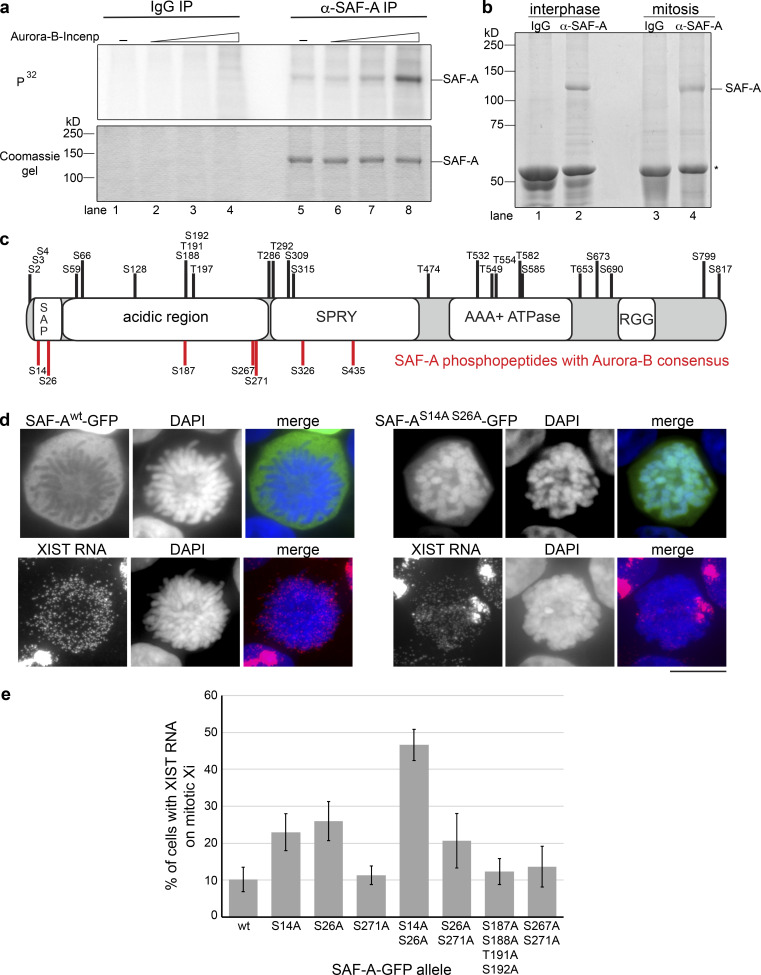
Aurora-B phosphorylates SAF-A on residues S14 and S26 of the SAP domain. (a) IP kinase assay. Control IgG IPs (lanes 1–4) and α-SAF-A IPs (lanes 5–8) were incubated with purified Aurora-B–Incenp complex. Phosphorylation of SAF-A was detected using 32P-labeled ATP (upper panel), and SAF-A was detected using Coomassie blue (lower panel). Aurora-B–Incenp complex was present at 0 nM (lanes 1 and 5), 1 nM (lanes 2 and 6), 10 nM (lanes 3 and 7), or 100 nM (lanes 4 and 8). (b) SAF-A was immunopurified from interphase and mitotic cell extracts for mass spectrometric identification of phosphopeptides. IgG heavy chain is denoted by the asterisk. (c) Phosphorylated serines and threonines were mapped relative to the domain structure of SAF-A as described in the text. Serines depicted in red fit the Aurora-B consensus sequence (R/K-X-S/T or R/K-X-S/T); serines and threonines depicted in black do not have the Aurora-B consensus motif. (d) GFP IF for wild-type and phosphomutant versions of SAF-A–GFP expressed in HEK293T cells, combined with XIST RNA FISH. Scale bar = 10 µm. (e) Quantitation of the percentage of cells expressing either wild-type or phosphomutant SAF-A–GFP showing retention of XIST RNA on the mitotic Xi. 100 cells were scored for each transfection; the average and SD of multiple independent transfections are shown. Error bars represent the SD. For SAF-Awt–GFP and SAF-AS14A S26A–GFP, n = 5; n = 3 for all other alleles tested.
Figure 7.
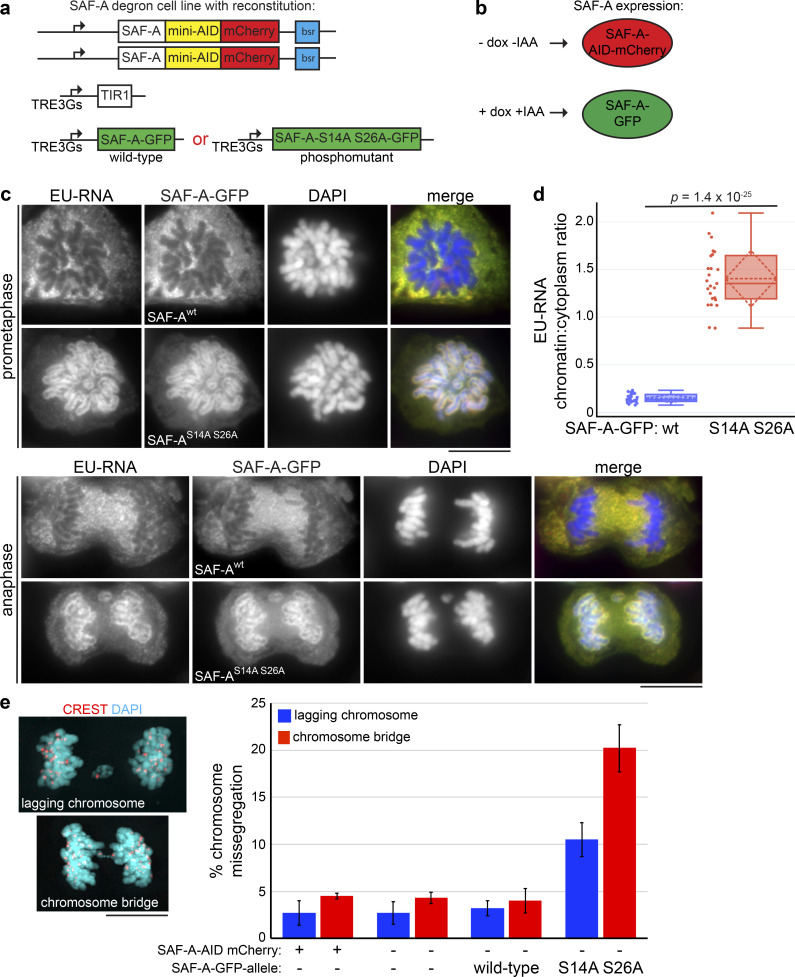
Dynamic localization of SAF-A promotes normal chromosome segregation in anaphase. (a) SAF-A–AID–mCherry degron cells were transduced with lentivirus encoding either SAF-Awt–GFP or the phosphomutant SAF-AS14A S26A–GFP. (b) In this scheme, untreated cells express only SAF-A–AID–mCherry. Addition of doxycycline and auxin (IAA) induces SAF-A–GFP alleles while also causing degradation of SAF-A–AID–mCherry protein through induction of TIR1. (c) The SAF-A degron cell line was reconstituted with either SAF-Awt–GFP or SAF-AS14 S26–GFP. Cells were labeled with EU to detect RNA localization and immunostained for GFP to detect SAF-A–GFP alleles. Examples of cells in prometaphase and late anaphase are shown. (d) Quantitation of EU-RNA localization in SAF-A–GFP-reconstituted cell lines, expressed as a ratio of chromatin-associated RNA versus cytoplasmic RNA. The mean and SD are represented by the horizontal and diamond-shaped dashed lines, respectively. SAF-Awt–GFP, n = 25; SAF-AS14A S26A, n = 25. The P value was calculated using a Student’s t test. (e) Chromosome missegregation during anaphase was measured in cells with and without SAF-A–AID–mCherry depletion and in cells reconstituted with SAF-Awt–GFP or SAF-AS14 S26–GFP. Three biological replicates were performed with 200 anaphases scored in each experiment for the incidence of lagging chromosomes and chromosome bridge formation. Error bars indicate SD. An example of each type of aberrant anaphase is depicted beside the graph with CREST immunostaining to mark kinetochores. Scale bar = 10 µm.
Figure S2.
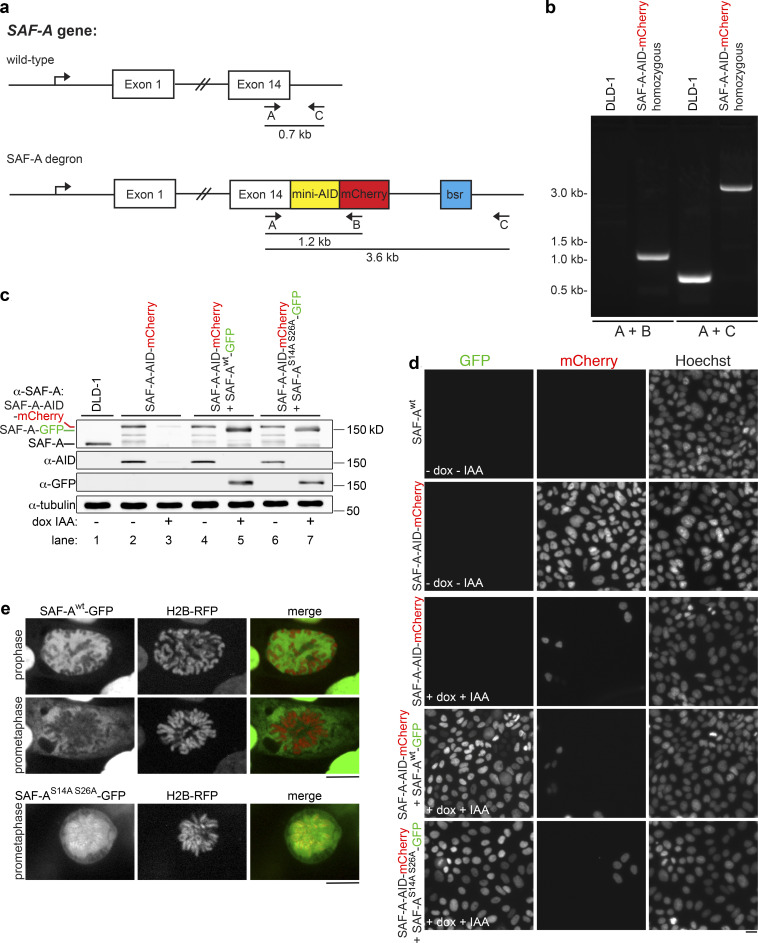
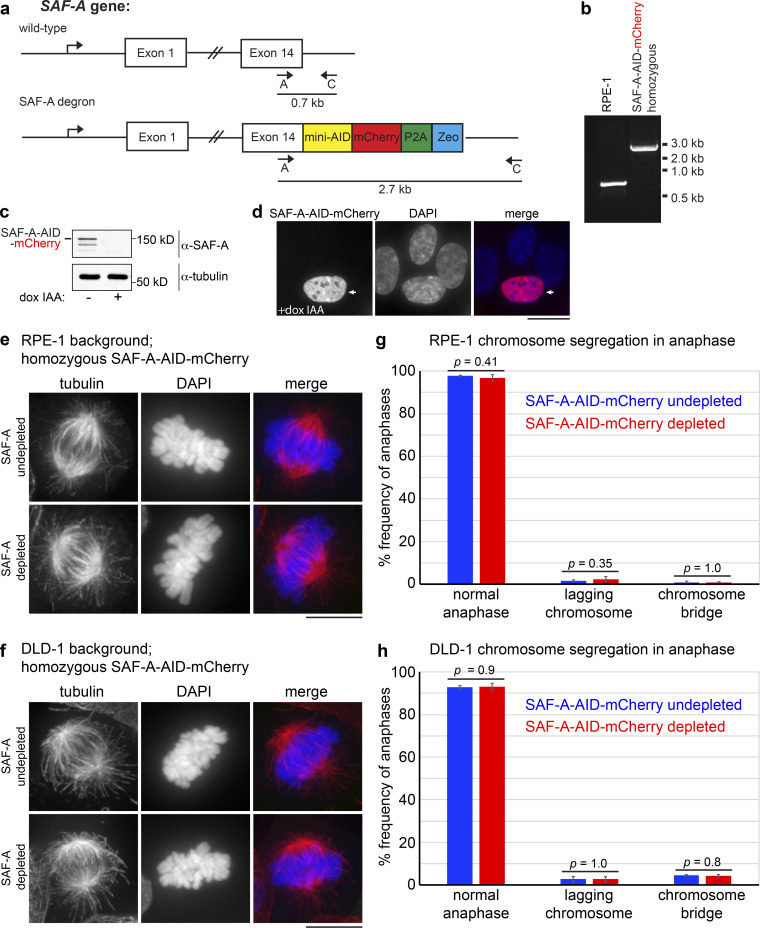
SAF-A depletion and reconstitution. (a) The endogenous SAF-A locus was targeted for recombination in diploid DLD-1 cells, using CRISPR to introduce a C-terminal tag encoding both the minimal auxin-inducible degron sequence and mCherry. Clonal isolates were screened for homozygous SAF-A–AID–mCherry recombinants using the indicated combinations of primers A, B, and C. (b) PCR analysis of genomic DNA from wild-type untransfected DLD-1 cells and the homozygous SAF-A–AID–mCherry clone used for depletion and reconstitution experiments. (c) Western blot analysis of SAF-A alleles in DLD-1 cells (lane 1), SAF-A–AID–mCherry cells (lanes 2 and 3), SAF-A–AID–mCherry + SAF-Awt–GFP cells (lanes 4 and 5), and SAF-AS14A S26A–GFP cells (lanes 6 and 7). SAF-A–AID–mCherry cells expressed the recombinant tagged protein at levels comparable to the wild-type untagged protein (lanes 1 and 2); addition of doxycycline and auxin to cell cultures resulted in extensive depletion of SAF-A–AID–mCherry (lane 3). The exclusive protein expression states represented in Fig. 7 a were confirmed for all tagged cell lines (lanes 2–7; compare − and + dox IAA), as depicted in the α-SAF-A, α-AID, and α-GFP blots. The relative mobility of all SAF-A protein species is indicated on the α-SAF-A blot. We note that all AID-tagged cell lines showed some degree of minor degradation bands. (d) Live-cell imaging of cell lines with and without doxycycline and auxin treatment. SAF-A–AID–mCherry was extensively depleted in all cell lines treated with doxycycline and auxin (+dox +IAA). In addition, induction of SAF-A–GFP allele expression was observed throughout cell populations containing the wild-type or phosphomutant SAF-A–GFP alleles. Scale bar = 20 µm. (e) Live-cell imaging of SAF-Awt–GFP cells and SAF-AS14A S26A–GFP cells. SAF-A localization in live cells is identical to the SAF-A localization patterns observed in fixed cell populations. Scale bar = 10 µm.
Recent analysis of chromatin-bound transcription factors has suggested that formaldehyde fixation can alter localization properties of some transcription factors (Teves et al., 2018). To confirm that the exclusion of SAF-A from chromatin reflected a change in its physical association with chromatin during mitosis, we immunoprecipitated SAF-A from interphase and mitotic cell extracts prepared in the absence of formaldehyde cross-linking, and tested for the presence of the core histones in both control and α-SAF-A eluates (Fig. 1 b). Histone H3 was enriched in interphase α-SAF-A immunoprecipitations (IPs) relative to control IPs using IgG, but was absent in mitotic α-SAF-A IPs. Furthermore, Coomassie-stained gel analysis of control and α-SAF-A IPs confirmed the presence of all four core histones in interphase but not mitotic α-SAF-A IPs. In concordance with our live imaging (see Fig. S2 e), these data demonstrate that SAF-A is removed from chromosomes in mitosis and is not a fixation artifact.
Previous work has shown that SAF-A directly binds to hundreds of RNAs (Huelga et al., 2012; Xiao et al., 2012), but has not addressed whether SAF-A RNA binding is cell cycle regulated. To determine whether differential chromatin association during the cell cycle influences SAF-A interactions with RNA, we modified the proximity ligation assay (PLA) to detect SAF-A–RNA complexes in asynchronous cell populations (Fig. 1 d). In this scheme, all RNA is labeled with 5-bromouridine (BrU) before immunostaining with BrU and SAF-A primary antibodies. PLA detection of SAF-A–RNA interactions is then achieved through DNA polymerase amplification of oligonucleotide sequences conjugated to secondary antibodies. In interphase cells, SAF-A–RNA interactions were exclusively nuclear, whereas in mitotic cells, SAF-A–RNA interactions were dispersed throughout the entire cell (Fig. 1 e). Therefore, SAF-A maintains RNA interactions throughout the cell cycle, even after removal from chromosomes during mitosis.
To identify SAF-A–interacting RNAs across the cell cycle, we immunoaffinity purified native SAF-A–RNA complexes from interphase and mitotic RPE-1 cell extracts and performed high-throughput RNA sequencing (RNA-seq; Fig. 1 f). Sequencing reads from two independent biological replicates showed a high degree of correlation (Fig. S1, a and b), demonstrating the reproducibility of our sequencing data. Overall, we identified 1,761 transcripts enriched greater than or equal to twofold in SAF-A IPs from both interphase and mitotic cell extracts, representing 13% of all expressed RNAs. Quantitative RT-PCR (qRT-PCR) confirmed the enrichment of specific transcripts in SAF-A IPs for all RNAs tested (Fig. S1 c). We noted that several prominent long noncoding RNAs (lncRNAs), such as XIST, NEAT2/MALAT1, and KCNQ1OT1, were enriched with SAF-A across the cell cycle; however, the majority of SAF-A–interacting transcripts were mature, fully spliced mRNAs (Fig. S1, c–e). SAF-A–associated RNAs were well correlated in interphase and mitosis (Spearman ρ = 0.69), confirming that SAF-A–RNA interactions are stable throughout the cell cycle.
Figure S1.
RIP-seq was performed to identify SAF-A–enriched RNAs in interphase and mitotic cell extracts; cDNA reads present in SAF-A IPs were aligned to the human genome. To determine the reproducibility of two biological RIP-seq experiments, we plotted RNA enrichment in SAF-A IPs in the first experiment against RNA enrichment in the second experiment. (a and b) Correlation plots for RIP-seq data from interphase extracts (a) and mitotic extracts (b). XIST RNA is highlighted in red. The Spearman correlation value is depicted on each plot. (c) qRT-PCR of SAF-A–interacting RNAs identified by RIP-seq. All SAF-A–interacting RNAs tested showed significant enrichment in SAF-A IPs relative to their abundance in control IPs. MIF was used as a negative control because this RNA was not enriched in SAF-A IPs. SAF-A enrichment values represent the average obtained from two independent biological replicates; error bars represent the SD. (d and e) To determine whether SAF-A primarily interacted with mature or nascent RNA transcripts, we plotted the ratio of exon reads and intron reads for each transcript in the total extract (x axis) and SAF-A IPs (y axis). Correlation plots are shown for RIP-seq data from interphase extracts (d) and mitotic extracts (e). Each point represents the average of two biological replicates. The Pearson correlation value is shown on each plot.
To determine if SAF-A–associated RNAs are preferentially retained in the nucleus, we compared our interphase RNA-seq data with previous data examining relative nuclear enrichment of all RNAs in IMR-90 cells (Fig. 1 g; Djebali et al, 2012). Indeed, we found a highly statistically significant enrichment of nuclear-retained RNAs present in the SAF-A–interacting RNA population, and that a higher SAF-A RNA enrichment was correlated with a higher nucleus/cytosol localization ratio (Fig. 1 g, see twofold versus fourfold population). Taken together, our data show that SAF-A–RNA complexes are stable throughout the cell cycle, that SAF-A associates with hundreds of nuclear-retained mRNAs and lncRNAs during interphase, and that mitotic removal of SAF-A during prophase releases SAF-A–RNA complexes from condensing chromosomes (Fig. 1 h).
Aurora-B triggers relocalization of SAF-A–RNA complexes in early mitosis
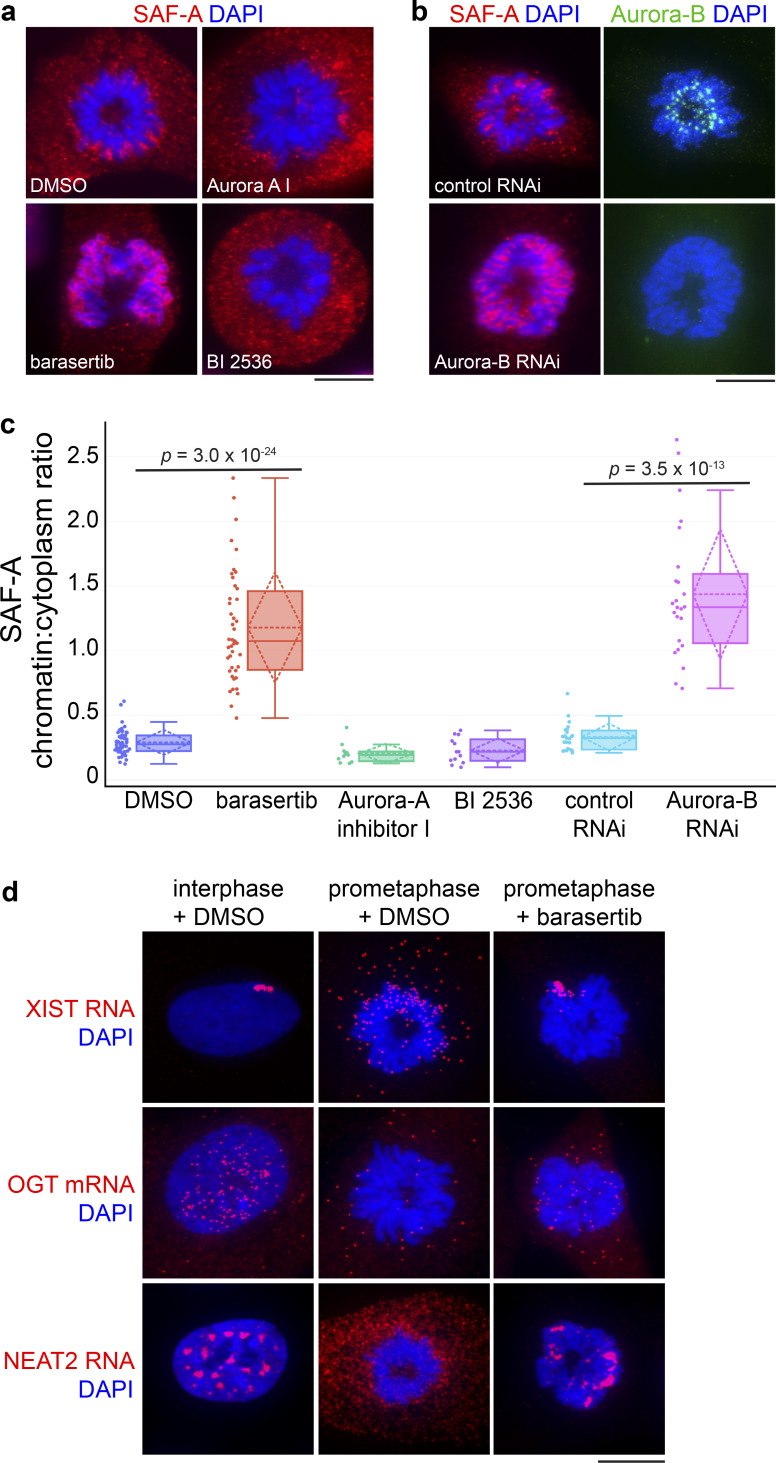
Our data suggested there is a signal during prophase that triggers SAF-A removal from chromatin, prompting us to query whether inhibition of the Aurora-A, PLK1, or Aurora-B kinases that function during early mitosis would alter SAF-A localization in prometaphase. Whereas mitotic cells treated with DMSO showed the normal SAF-A chromatin exclusion pattern, treatment of cells with the Aurora-B selective inhibitor barasertib caused SAF-A enrichment on prometaphase chromosomes (Fig. 2, a and c). Similar results were observed using siRNAs to deplete Aurora-B (Fig. 2, b and c). In contrast, Plk1 or Aurora-A inhibition had no discernable effect on SAF-A localization (Fig. 2 c).
Figure 2.
Aurora-B triggers relocalization of SAF-A–RNA complexes in early mitosis. (a) SAF-A IF in RPE-1 cells incubated with either DMSO or mitotic inhibitors to Aurora-B (barasertib), Aurora-A (Aurora-A inhibitor I), and Plk1 (BI 2536). Scale bar = 10 µm. (b) RPE-1 cells were cultured in the presence of either control siRNAs or Aurora-B siRNAs. Cells were immunostained for SAF-A and Aurora-B. (c) Image quantitation of mitotic SAF-A localization under the indicated treatments, expressed as a ratio of chromatin-localized SAF-A versus cytoplasmic SAF-A. For each box plot, the median is represented by the horizontal solid line within the shaded box; the mean and SD are represented by the horizontal and diamond-shaped dashed lines, respectively. Data points representing individual mitotic figures are rendered beside each box plot in circles. DMSO, n = 49; barasertib, n = 43; Aurora-A inhibitor 1, n = 11; BI 2536, n = 13; control siRNA, n = 23; Aurora-B siRNA, n = 24. P values were calculated using a Student’s t test. (d) RNA FISH for the SAF-A–interacting XIST, OGT, and NEAT2 RNAs was performed in RPE-1 cells incubated with DMSO or barasertib.
To test whether Aurora-B inhibition caused SAF-A–interacting RNAs to be retained on chromatin, we treated cells with barasertib and performed FISH to detect three RNAs significantly enriched in SAF-A IPs: the XIST and NEAT2/MALAT1 lncRNAs and O-linked N-Acetylglucosamine transferase (OGT) mRNA (Fig. 2 d). In interphase, all three RNAs showed prominent nuclear retention, either as a chromosome-sized focus in the case of XIST (Brown et al., 1992) or as multiple nuclear foci in the case of OGT and NEAT2. We note that, in contrast to the lncRNAs, OGT mRNA was also present as small granular cytoplasmic particles, consistent with it being a substrate for translation. In prometaphase cells treated with DMSO, XIST RNA was dispersed as small cytoplasmic foci; in contrast, prometaphase cells treated with barasertib showed an abnormal accumulation of XIST RNA on the mitotic Xi chromosome, consistent with previous observations (Clemson et al., 1996; Hall et al., 2009). A similar trend was observed for OGT and NEAT2 localization: whereas prometaphase cells showed a general trend of nuclear RNA disaggregation and cytoplasmic dispersal, Aurora-B inhibition resulted in markedly increased overlap of OGT and NEAT2 RNAs with mitotic chromosomes. Together, these data suggest that the Aurora-B kinase is responsible for the removal of SAF-A–RNA complexes from prophase chromatin.
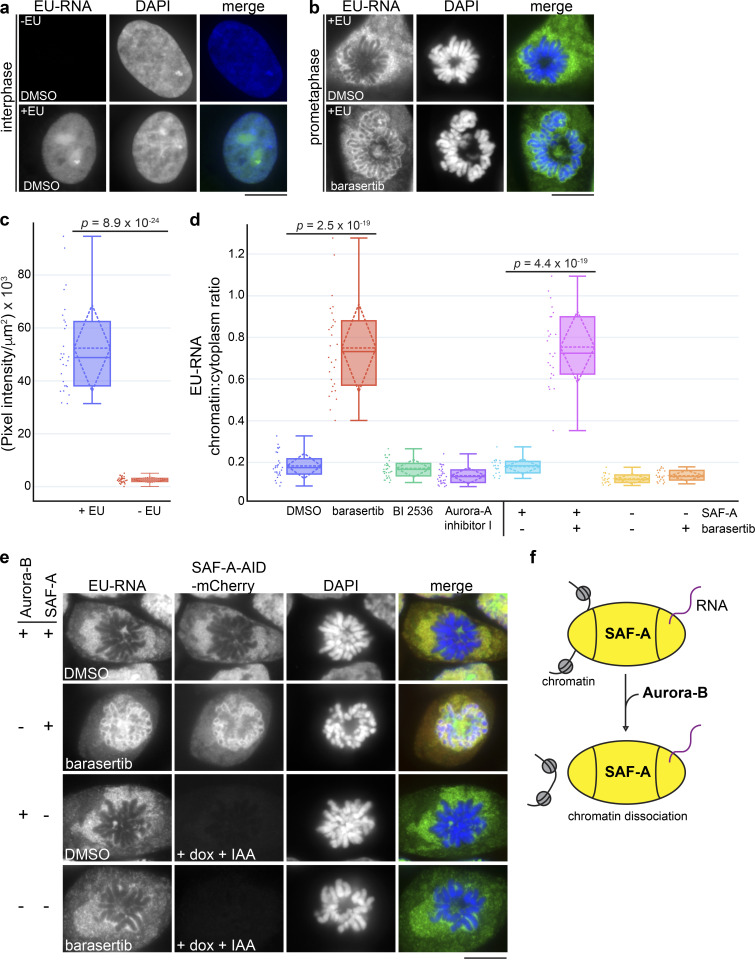
To determine whether Aurora-B has a global role in regulating RNA localization, we labeled cells with a 3-h pulse of 5-ethynyl uridine (EU; Jao and Salic, 2008) and monitored total RNA localization in prometaphase cells with or without Aurora-B inhibition (Fig. 3). In control cell populations, we observed that EU-labeled RNA was substantially enriched in the nucleus relative to the cytoplasm, as previously observed (Fig. 3, a and c; Jao and Salic, 2008; Johnson et al., 2017; Palozola et al., 2017). However, RNA was excluded from condensed prophase and prometaphase chromosomes (Fig. 3 b). Strikingly, barasertib-treated cells showed RNA retention on the chromosome surface, reminiscent of the SAF-A localization pattern in Aurora-B–inhibited cells (Fig. 3, b and d). Therefore, Aurora-B releases a large population of nuclear/chromosomal RNAs from chromosomes in prometaphase.
Figure 3.
Aurora-B and SAF-A together define a pathway for removing RNP complexes from mitotic chromosomes. (a and b) EU-labeled RNA was detected in interphase and prometaphase DLD-1 cells treated with DMSO or barasertib. Chromosomes were stained with DAPI. Scale bar = 10 µm. (c) Quantitative image analysis demonstrated a 21-fold increase in fluorescence intensity measured in +EU-labeled cell populations (n = 29) relative to control cells (n = 31). (d) Quantitation of mitotic EU-RNA localization, expressed as a ratio of chromatin-localized RNA versus cytoplasmic RNA. Cells were treated with DMSO or mitotic inhibitors as shown. DMSO, n = 29; barasertib, n = 28; BI 2536, n = 34; Aurora-A inhibitor 1, n = 31. P values were calculated using a Student’s t test. (e) EU-RNA was detected in DLD-1 cells homozygous for a SAF-A–AID–mCherry degron allele. Cells were treated with either DMSO or barasertib, with or without SAF-A depletion (+dox +IAA) to determine the epistatic relationship of Aurora-B and SAF-A. Quantitation of mitotic EU-RNA localization in SAF-A degron cells under all conditions is shown in d. SAF-A undepleted + DMSO, n = 20; SAF-A undepleted + barasertib, n = 25; SAF-A depleted + DMSO, n = 20; SAF-A depleted + barasertib, n = 22. (f) Model for Aurora-B–dependent regulation of SAF-A–RNA chromatin association.
If Aurora-B primarily targets SAF-A to remove RNAs from chromatin in mitosis, we hypothesized that SAF-A ablation should result in RNA release from chromatin in Aurora-B–inhibited cells. To test this, we constructed a human diploid DLD-1 cell line with both copies of endogenous SAF-A fused to an auxin-inducible degron sequence and mCherry (SAF-A–AID–mCherry; Fig. S2, a and b; Holland et al., 2012; Natsume et al., 2016); a doxycycline-inducible TIR1 E3 ligase was also integrated to enable auxin-mediated degradation of SAF-A–AID–mCherry. Control experiments demonstrated that SAF-A–AID–mCherry was homozygous, was expressed at normal levels, showed a localization pattern identical to the wild-type protein, and could be depleted within 24 h of treatment with doxycycline and auxin (indole-3-acetic acid [IAA]; Fig. S2, c and d). We then tested for EU-RNA localization under all combinations of SAF-A–AID–mCherry depletion and Aurora-B inhibition. In cells expressing SAF-A–AID–mCherry, EU-RNA was retained on chromatin in the presence of barasertib, as was observed in wild-type cells (Fig. 3 e). However, in SAF-A–depleted cells, Aurora-B inhibition no longer caused EU-RNA retention on chromosomes, demonstrating that Aurora-B is epistatic to SAF-A (Fig. 3, d and e). We conclude that Aurora-B and SAF-A together define a pathway that releases RNA from chromosomes en masse during early mitosis (Fig. 3 f).
Phosphorylation of SAF-A by Aurora-B controls chromatin association
To test whether Aurora-B promotes chromosomal removal of SAF-A through direct phosphorylation, we performed a kinase assay using recombinant Aurora-B–Incenp complex (Bolton et al., 2002; Rosasco-Nitcher et al., 2008) and immunoaffinity-captured SAF-A (Fig. 4 a). We observed dose-dependent phosphorylation of SAF-A in the presence of the active Aurora-B–Incenp complex. A low level of SAF-A phosphorylation was present in reactions lacking Aurora-B–Incenp complex, possibly due to a trace amount of kinase activity copurifying with SAF-A (Fig. 4 A, lane 5). In contrast, there were no specific phosphorylation events observed in IgG control IPs. Therefore, the Aurora-B–Incenp complex can directly phosphorylate SAF-A.
To identify phosphorylation sites on SAF-A in vivo, we isolated SAF-A from interphase and mitotic cell extracts and used mass spectrometry to identify modified peptides (Fig. 4 b and Table S1). We pooled our phosphopeptide data with that from several phosphoproteomic studies (https://www.phosphosite.org/homeAction.action) and mapped serine and threonine phosphorylation sites relative to SAF-A domain structure (Fig. 4 c and Table S1). Potential Aurora-B sites were then identified on the basis of whether the proximal amino acids fit the Aurora-B consensus sequence (K/R-S/T or K/R-X-S/T; Alexander et al., 2011; Cheeseman et al., 2002; Hengeveld et al., 2012; Kettenbach et al., 2011). All seven of the putative Aurora-B sites were located in the N-terminal half of SAF-A, with two positioned in the SAP domain required for DNA binding (S14, S26), three positioned in a low-complexity acidic domain (S187, S267, S271), and two located within the SPRY domain (S326, S435). Phosphorylated SAF-A S14, S26, S59, S187, and S271 were overrepresented in mitosis (Table S1; Douglas et al., 2015; Kettenbach et al., 2011; Olsen et al., 2010).
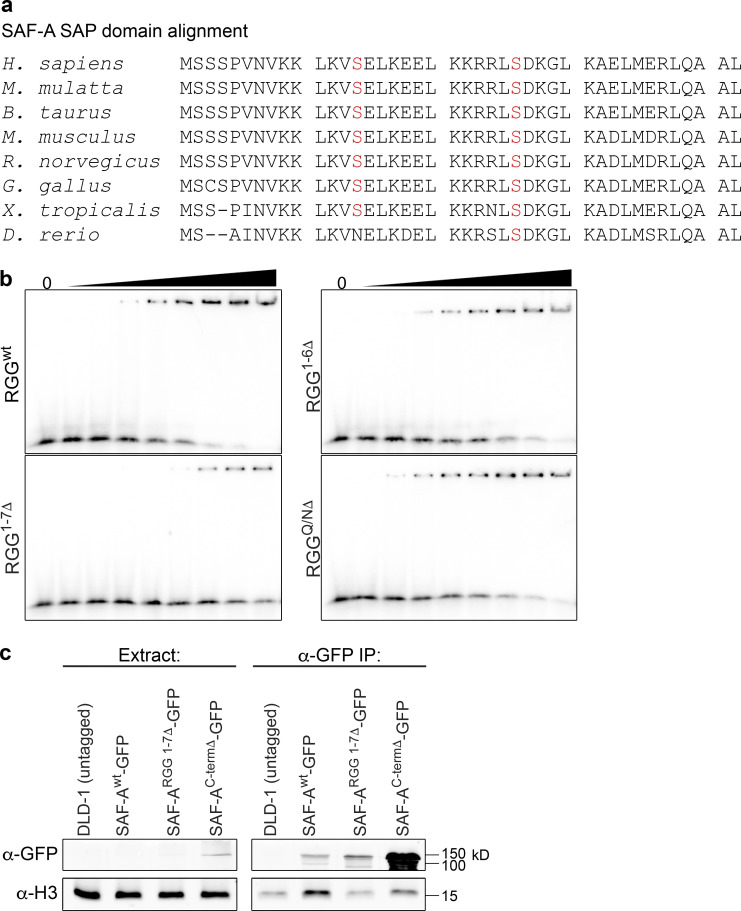
To determine if the predicted Aurora-B phosphorylation sites regulate SAF-A–RNA localization during mitosis, we constructed a series of phosphomutants with single or multiple alanine substitutions in the SAP domain and acidic domain and analyzed mitotic HEK293T cells expressing wild-type or phosphomutant SAF-A–GFP for XIST RNA localization (Fig. 4, d and e). Cells expressing wild-type SAF-A–GFP showed normal localization of the tagged protein and XIST RNA: both components showed nuclear localization during interphase and exclusion from mitotic chromosomes. In contrast, we found that mutation of S14A and S26A, either alone or in combination, significantly increased the number of cells showing ectopic retention of SAF-A on mitotic chromosomes and focal XIST RNA staining on the Xi during prometaphase (Fig. 4 d). In particular, prometaphase cells expressing double-mutant SAF-AS14A S26A–GFP had a 50% frequency of cells with focal, chromatin-bound XIST RNA staining, nearly a fivefold increase relative to the SAF-Awt–GFP control (Fig. 4 e). Expression of either SAF-AS14A–GFP or SAF-AS26A–GFP caused a less dramatic yet significant effect, with both causing a greater than twofold increase in prometaphase cells with focal XIST localization. Furthermore, cells expressing SAF-AS26A S271A–GFP had an XIST RNA localization pattern similar to SAF-AS26A–GFP, demonstrating that the cumulative effects of mutating two phosphorylatable serines was specific to the SAF-A SAP domain residues S14 and S26. The other SAF-A phosphomutants analyzed had no effect on XIST RNA localization distinguishable from SAF-Awt–GFP. An alignment of SAF-A from multiple species showed conservation of S14 and S26 residues throughout the vertebrate lineage (Fig. S4 a). On the basis of these data, we conclude that S14 and S26 regulate mitotic SAF-A localization.
Figure S4.
Analysis of SAF-A nucleic acid binding domains. (a) Alignment of the SAF-A SAP domain indicates strong conservation of serines 14 and 26 throughout vertebrates, except in zebrafish SAF-A, which retains only the second serine. (b) RNA-binding mutants have reduced chromatin interactions. For each experimental replicate (n = 2), RNA EMSA reactions were performed concurrently for the GFP–SAF-A RNA-binding domain constructs depicted in Fig. 8 a. Constructs were RGGwt, RGG1-6Δ, RGG1-7Δ, and RGGQ/NΔ. Quantitation of RNA-binding data are shown in Fig. 8, c and d. (c) To test whether SAF-A RNA-binding mutations still interacted with interphase chromatin, we immunoprecipitated SAF-Awt–GFP, SAF-ARGG1-7Δ–GFP, or SAF-AC-termΔ–GFP and examined eluates for the presence of histone H3. Untagged DLD-1 cells were used as a control. To compare chromatin interactions, histone H3 coprecipitation was measured as a ratio of histone H3/SAF-A and is shown in Fig. 8 f.
Phosphorylation of the SAP domain reduces SAF-A binding to DNA
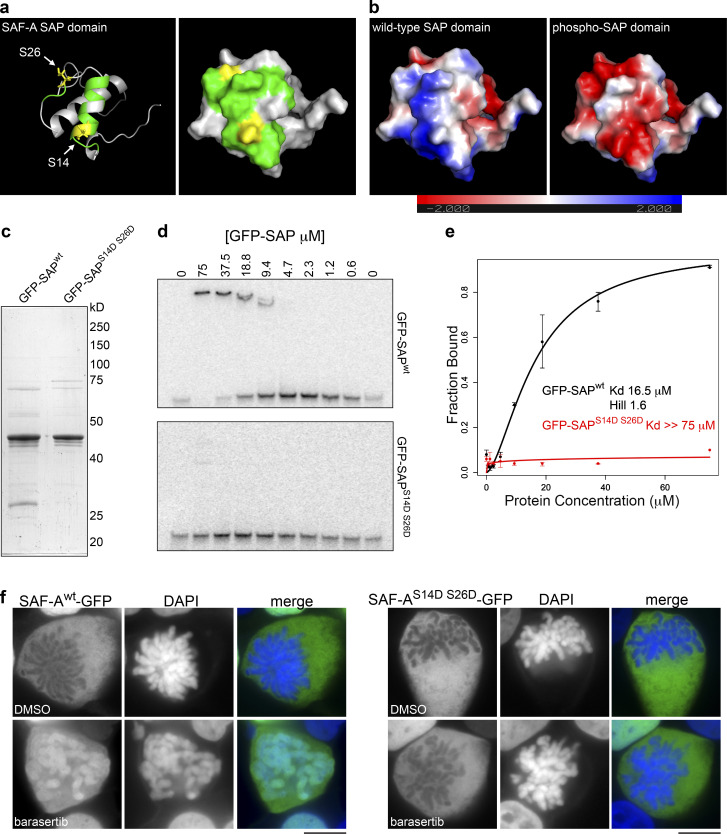
Previous work has demonstrated that SAF-A can bind directly to AT-rich DNA in vitro and that the N-terminal SAP domain is a DNA-binding domain (Fackelmayer et al., 1994; Göhring et al., 1997; Kipp et al., 2000; Romig et al., 1992). To gain molecular insight into how phosphorylation of residues S14 and S26 on SAF-A could block DNA binding, we performed molecular modeling of the SAF-A SAP domain using I-TASSER software, which uses a combination of ab initio modeling and refinement based on known structures in the Protein Data Bank (Fig. 5 a; Yang et al., 2015). I-TASSER identified several close sequence and structural homologues of SAF-A, all of which featured a SAP domain with a helix-turn-helix structural motif. Using this model, we found that S14 is positioned within the first helix, whereas S26 is positioned in a loop between the first and second helices (depicted in yellow in Fig. 5 a).
Figure 5.
Aurora-B phosphorylation of the SAP domain reduces SAF-A binding to DNA. (a) Molecular modeling of the SAP domain of SAF-A, rendered as a ribbon model (left) and as a space-filling model (right). Residues S14 and S26 are highlighted in yellow; residues predicted to define the DNA-binding interface are colored in green. (b) Electrostatic surface charge potential was determined for the unphosphorylated SAP domain (left) or the SAP domain phosphorylated on residues S14 and S26 (right). The color gradient inset shows the charge distribution as a continuum between red (more negative) and blue (more positive). (c) Purification of recombinant wild-type (GFP-SAPwt) and phosphomimetic (GFP-SAPS14D S26D) SAP domains of SAF-A from E. coli. (d) DNA EMSAs for GFP-SAPwt and GFP-SAPS14D S26D with an AT-rich DNA template. (e) The fraction of bound DNA template versus the protein concentration of GFP-SAPwt or GFP-SAPS14D S26D was plotted to determine the dissociation constant, Kd, and the Hill coefficient. Error bars represent the SD of n = 2 replicates. (f) Wild-type and phosphomimetic alleles of SAF-A–GFP were expressed in HEK293T cells in the presence of either DMSO or barasertib. SAF-Awt–GFP and SAF-AS14D S26D–GFP were visualized by GFP immunostaining. Scale bar = 10 µm.
We used the nuclear magnetic resonance structure of the SAP domain–containing protein PIAS1 to map the residues contacting DNA onto the equivalent positions in the SAF-A model (depicted in green in Fig. 5 a; Okubo et al., 2004), and observed that both S14 and S26 were positioned on the same molecular surface of the SAP domain as the residues predicted to contact DNA. To test whether phosphorylation of residues S14 and S26 altered the electrostatic surface potential of the SAP domain, we calculated the surface charge for both the unphosphorylated and phospho-S14 S26 models of the SAF-A SAP domain. We found that the unphosphorylated SAF-A SAP domain has an overall net positive charge (Fig. 5 b, blue) and that this charge is dramatically reversed by phosphorylation of S14 and S26 (Fig. 5 b, red). These data suggest that charge repulsion between the phospho-SAP domain and the DNA phosphate backbone is a likely mechanism for the release of SAF-A from chromatin after phosphorylation by Aurora-B.
To directly test if altered surface charge potential of the SAF-A SAP domain affects DNA binding, we reconstituted DNA binding by the SAF-A SAP domain in vitro using GFP-SAPwt and GFP-SAPS14D S26D expressed and purified from Escherichia coli (Fig. 5 c). We then employed a DNA electrophoretic mobility shift assay (EMSA) using a 50-bp AT-rich SAR sequence (Okubo et al., 2004) to measure the DNA binding affinity of each protein (Fig. 5, D and E). The SAPwt domain readily bound the SAR DNA with an apparent Kd of 16.5 µM. In contrast, the SAPS14D S16D domain displayed a dramatically reduced DNA binding, such that we were able to detect only minimal DNA-binding activity at the highest protein concentration tested (Kd > 75 µM). We conclude that phosphorylation of the SAP domain of SAF-A changes the surface charge of the DNA-binding domain and causes dramatically reduced DNA binding.
The reduced DNA-binding affinity of the phosphomimetic SAP domain in vitro suggested that a phosphomimetic SAF-A allele would show reduced chromatin affinity in vivo. We therefore compared the localization of SAF-Awt–GFP with SAF-AS14D S26D–GFP, with and without barasertib treatment (Fig. 5 f). In barasertib-treated cell populations, 95% of cells showed SAF-Awt–GFP retained on prometaphase chromosomes compared with only 3% of cells treated with DMSO. In contrast, the localization of SAF-AS14D S26D–GFP was completely excluded from mitotic chromosomes with or without barasertib treatment, thus demonstrating that the Aurora-B inhibitor acts through SAF-A S14 and S26. Taken together, our mutant analysis data indicate that Aurora-B phosphorylates SAF-A on residues S14 and S26 in the DNA-binding SAP domain in vivo and that phosphorylation of these two residues acts additively to dissociate SAF-A–RNA complexes from chromatin during early mitosis.
Evidence that monomeric SAF-A mediates RNA–DNA tethering
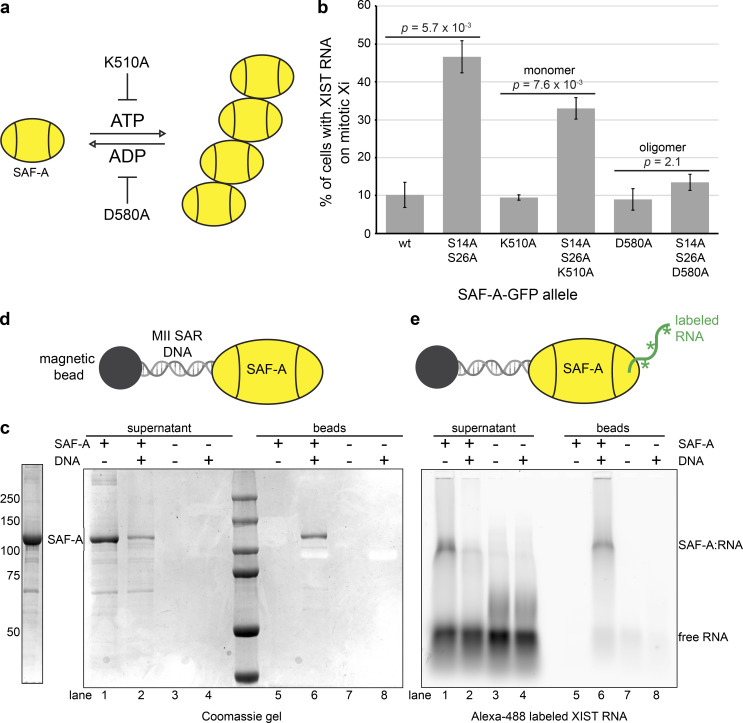
The domain structure of SAF-A includes a central AAA+-type ATPase domain that controls protein oligomerization through ATP binding and hydrolysis (Nozawa et al., 2017). Consequently, mutations in specific Walker box residues can be used to manipulate the SAF-A oligomeric state (Fig. 6 a). SAF-A exists as a monomer in its most abundant cellular form, yet it has not been determined whether the monomeric or oligomeric form of SAF-A contributes to RNA localization. XIST RNA FISH in mitotic cells expressing the SAF-A phosphomutant allowed direct visualization of SAF-A–RNA–chromatin interactions in vivo. To determine whether SAF-A mediates XIST RNA localization as a monomer or SAF-A oligomer, we generated SAF-AK510A–GFP (monomeric) and SAF-AD580A–GFP (oligomeric) alleles, either alone or in combination with the SAF-AS14A S26A–GFP phosphomutant, and monitored mitotic XIST RNA localization in transfected cell populations (Fig. 6 b). Interestingly, we found that the monomeric SAF-A phosphomutant (SAF-AS14A S26A K510A–GFP) anchored XIST RNA to mitotic chromatin nearly as efficiently as the SAF-AS14A S26A–GFP allele. In contrast, the oligomeric SAF-A phosphomutant (SAF-AS14A S26A D580A) was indistinguishable from the wild-type allele. These data suggested that the monomeric form of SAF-A is sufficient to tether RNA to chromosomes.
Figure 6.
Monomeric SAF-A is sufficient for RNA tethering. (a) Model of ATP-dependent oligomerization cycle of AAA+-type ATPases. The K510A mutation in the SAF-A Walker A box inhibits ATP binding and protein oligomerization, whereas the D580A mutation in the Walker B box inhibits ATP hydrolysis, trapping SAF-A in the oligomeric state. (b) Quantitation of the percentage of cells showing XIST RNA retention on the mitotic Xi in HEK293T cells transfected with SAF-A–GFP alleles. 100 cells were scored for each transfection; the average and SD of multiple independent transfections are shown. For SAF-Awt–GFP and SAF-AS14A S26A–GFP, n = 5; n = 2 for all other alleles tested. P values were calculated using a Student’s t test. (c) Purification of full-length monomeric SAF-A from Sf9 cells. (d) Capture of SAF-A on DNA-coated beads. The SAF-A–interacting DNA sequence from the MII SAR element was conjugated to magnetic beads and used to capture SAF-A. Coomassie gel staining was used to examine the presence of unbound SAF-A in reaction supernatants (lanes 1–4) and SAF-A binding to DNA-coated beads (lanes 5–8). (e) RNA-tethering assay. SAF-A complexed with an FITC-labeled XIST RNA fragment (lanes 5–8) was incubated with DNA beads. FITC fluorescence was used to image SAF-A–RNA complexes in reaction supernatants (lanes 1–4) and retention of RNA with SAF-A bound to DNA beads (lanes 5–8). Asterisks indicate FITC-labeled nucleotides.
To test this idea, we devised an in vitro RNA–DNA tethering assay and queried whether monomeric SAF-A could bridge interactions between DNA-conjugated beads and RNA. We expressed and purified full-length monomeric SAF-A protein (Fig. 6 c) and used a nuclear scaffold DNA sequence coupled to magnetic beads to capture SAF-A in the absence of ATP to block oligomerization of SAF-A (MII SAR DNA; Fackelmayer et al., 1994). SAF-A bound efficiently to DNA-conjugated beads, without detectable binding to control beads (Fig. 6 d). To assay whether SAF-A bound to DNA could simultaneously bind RNA, we incubated SAF-A complexed with a fragment of the XIST RNA with DNA beads (Huelga et al., 2012) and found that XIST RNA was retained specifically on SAF-A–DNA beads (Fig. 6 e). These data support the idea that SAF-A has intrinsic RNA–DNA tethering activity in the absence of protein oligomerization or accessory proteins. Given that the abundance of SAF-A is estimated to be 2 × 106 molecules per cell (Fackelmayer and Richter, 1994), our data suggest that numerous DNA–SAF-A–RNA bridges are formed in the interphase nucleus.
Dynamic localization of SAF-A promotes normal chromosome segregation in mitosis
The data suggested that removal of SAF-A–RNA complexes from chromatin is a normal feature of prometaphase, prompting us to test whether the role of Aurora-B in determining mitotic localization of SAF-A is important for normal chromosome transmission. For this purpose, we used lentiviruses to integrate tet-inducible versions of SAF-Awt–GFP or SAF-AS14A S26A–GFP into the human cell line engineered with the tagged, degradable SAF-A–AID–mCherry (Fig. 7 a and Fig. S2). To simultaneously deplete SAF-A–AID–mCherry and reconstitute SAF-A function with the GFP-tagged alleles, cells were first treated with doxycycline for 24 h to induce SAF-A–GFP and TIR1 and then treated with both doxycycline and auxin for another 24 h to induce degradation of SAF-A–AID–mCherry before analysis (Fig. 7 b).
To determine whether the GFP-tagged SAF-A alleles showed localization properties predicted by our data, we visualized GFP immunofluorescence (IF) together with RNA localization (Fig. 7 c). Inspection of individual mitotic figures revealed that the SAF-Awt–GFP cell line showed exclusion of the protein and EU-RNA from chromosomes, comparable to endogenous RNA and SAF-A localization in untreated wild-type cells. Conversely, cells expressing the unphosphorylatable mutant SAF-AS14A S26A–GFP showed retention of SAF-A and RNA on chromosomes, in a pattern that persisted throughout mitosis. Quantitation of EU-RNA confirmed these observations because cells expressing the SAF-A phosphomutant had 9.6-fold more chromosome-associated RNA than SAF-Awt–GFP cells (Fig. 7 d). These data confirm that the SAF-A allele reconstitution assay recapitulates the phenotypes predicted by chemical inhibition data (Figs. 2 and 3) and transient transfection (Fig. 4). Furthermore, the data strongly argue that Aurora-B causes relocalization of nuclear and chromosomal RNAs during mitosis through targeting S14 and S26 in the SAF-A SAP domain.
To test whether mitotic RNA localization impacts chromosome inheritance in anaphase, we stained SAF-Awt–GFP and SAF-AS14A S26A–GFP cells with CREST antibodies that label kinetochores and scored anaphases in asynchronous cell populations (Fig. 7 e). We observed that wild-type DLD-1, undepleted SAF-A–AID–mCherry cells, depleted SAF-A–AID–mCherry cells, and cells expressing SAF-A–GFP all displayed a comparable low frequency of aberrant anaphases (Fig. 7 e). In the case of SAF-A–AID–mCherry cells, these findings were confirmed in both DLD-1 and RPE-1 cell backgrounds (Fig. S3).
Figure S3.
Normal spindle morphology and chromosome segregation in cells depleted for SAF-A. Recent work reported that SAF-A RNAi leads to increased chromosome segregation defects in RPE-1 cells (Nozawa et al., 2017). (a) We constructed an SAF-A–AID–mCherry degron line in the RPE-1 background, using a strategy similar to that used for the DLD-1 SAF-A degron line. (b) RPE-1 cells expressing SAF-A–AID–mCherry at the endogenous locus were confirmed by PCR. (c) Addition of doxycycline and IAA to SAF-A–AID–mCherry RPE-1 cultures led to depletion of SAF-A, as demonstrated by Western blot analysis. (d) Imaging of SAF-A–AID–mCherry RPE-1 cells also showed extensive depletion of SAF-A upon addition of doxycycline and IAA. An arrow depicts an undepleted cell for comparison. (e and f) IF staining of tubulin revealed normal spindle morphology in SAF-A–AID–mCherry cultures in both RPE-1 and DLD-1 cell backgrounds, with or without SAF-A depletion. (g and h) We monitored chromosome segregation rates in SAF-A–AID–mCherry cultures, with and without SAF-A depletion in both RPE-1 and DLD-1 cell backgrounds. Both cell lines showed normal chromosome segregation rates with or without SAF-A depletion. Chromosome segregation in anaphase was scored in 200 cells for each experimental condition in three independent experiments. P values were calculated using a Student’s t test and are included on the bar graphs to indicate that there were no statistically significant differences between SAF-A–undepleted and SAF-A–depleted cell populations. It is possible that the mitotic defects previously reported in SAF-A RNAi experiments could result from off-target RNAi effects rather than from SAF-A depletion. Scale bar = 10 µm.
In contrast to control cell populations, cells expressing SAF-AS14A S26A–GFP showed a significant increase in the rate of lagging chromosomes present at the midzone (3.3-fold increase; P = 2.9 × 10−3) and the rate of anaphase chromosome bridge formation (fivefold increase; P = 6.0 × 10−4). We conclude that the proper mitotic localization of SAF-A–RNA complexes is important for normal chromosome segregation in anaphase.
Coordination between SAF-A RNA and DNA binding
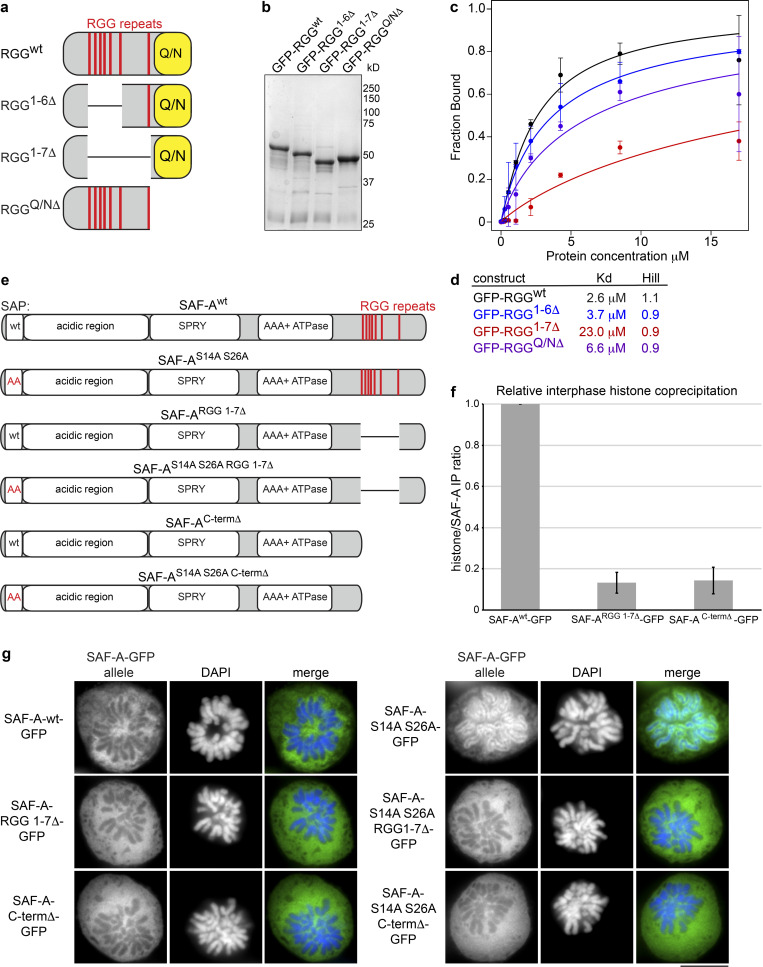
SAF-AS14A S26A–GFP cells have an abnormal accumulation of SAF-A and RNA on mitotic chromatin. To determine whether the chromosome segregation defects in SAF-AS14A S26A–GFP cells were attributable to the ectopic retention of SAF-A or RNA on mitotic chromosomes, we sought to identify a mutation that would allow SAF-A chromatin binding but block RNA binding. Previous work demonstrated that SAF-A RNA-binding activity maps to the C-terminal RGG domain (Kiledjian and Dreyfuss, 1992), which has recently been confirmed in vitro (Ozdilek et al., 2017). However, the quantitative contributions of various RGG motifs to RNA binding are not currently known. The SAF-A C-terminal RGG domain contains a central cluster of six perfect RGG repeats and a seventh perfect repeat C-terminal to the central RGG cluster, followed by a Q/N-rich C terminus (Fig. 8 a). To determine the quantitative contributions of each of these sequence features to RNA binding, we examined the binding of purified recombinant RGG domain deletions to a fragment of the XIST RNA using RNA EMSA (Fig. 8, a–c; and Fig. S4 b). The full-length RGG domain bound to RNA with an apparent affinity of 2.6 µM, consistent with a recent study (Ozdilek et al., 2017). Deletion of the C-terminal Q/N-rich domain resulted in a very modest decrease in RNA binding, consistent with previous work (Kiledjian and Dreyfuss, 1992). Interestingly, deletion of the central cluster of six RGG repeats resulted in very little change in RNA binding, but deletion of all seven RGG repeats resulted in a ninefold reduction in RNA binding (Fig. 8, c and d). We conclude that deletion of all perfect RGG repeats is necessary to dramatically reduce RNA-binding activity of SAF-A and that the C-terminal Q/N-rich domain does not contribute to RNA-binding activity.
Figure 8.
Interdependency of SAF-A RNA and DNA binding. (a) Cartoon depicting the SAF-A RGG domain and deletions analyzed by EMSA. RGG repeats are indicated by a red line; the Q/N-rich domain is indicated by a yellow box. (b) Coomassie-stained gel of purified, recombinant RGG domain proteins used for RNA EMSA reactions. (c) Plot depicting fraction of RNA bound versus protein concentration for various SAF-A RGG proteins. Error bars are the SDs of two experiments. (d) Kd and Hill coefficients were calculated for each RGG construct. (e) Cartoon depicting SAF-A–GFP constructs used to reconstitute DLD-1 SAF-A–AID–mCherry cells. (f) SAF-A–GFP constructs with or without RNA-binding domain mutations were immunoprecipitated from interphase cells. To compare chromatin interactions, we used quantitative Western blot analysis to calculate the ratio of SAF-A complexed with histone H3 (Fig. S4 c). The bar graph depicts the histone H3/SAF-A ratio after normalization of SAF-Awt–GFP; the average and SD were calculated from two independent experiments. (g) GFP IF was used to monitor chromatin localization of the indicated SAF-A constructs (from e) in prometaphase cells. Scale bar = 10 µm.
To determine how RNA binding contributes to SAF-A chromatin binding in vivo, we constructed cell lines expressing tet-inducible SAF-ARGG1-7Δ–GFP and SAF-ACtermΔ–GFP in the SAF-A–AID–mCherry background to deplete endogenous SAF-A (Fig. 8 e). First, we tested for chromatin binding in interphase cells by comparing relative levels of SAF-Awt–GFP coimmunoprecipitation with histone H3 (Fig. 8 f and Fig. S4 c). Both SAF-ARGG1-7Δ–GFP and SAF-ACtermΔ–GFP showed a greater than sevenfold reduction in histone H3 interaction relative to SAF-Awt–GFP, indicating that SAF-A RNA-binding mutations also have reduced chromatin binding during interphase.
To assay for mitotic SAF-A chromatin binding, we combined the nonphosphorylatable SAP domain mutation with the RNA-binding mutations to generate cell lines expressing SAF-AS14A S26A RGG1-7Δ–GFP and SAF-AS14A S26A CtermΔ–GFP (Fig. 8 e). We then tested for mitotic chromatin binding by examining SAF-A–GFP allele IF in prometaphase figures. Interestingly, both SAF-A mutations that prevented RNA binding failed to retain SAF-A on mitotic chromosomes (Fig. 8 g), again suggesting that RNA binding by SAF-A is required for chromatin binding in vivo. On the basis of these data, we speculate that it may not be possible to genetically separate chromatin and RNA-binding functions by SAF-A.
Retention of SAF-A–RNPs on mitotic chromosomes causes metaphase alignment defects
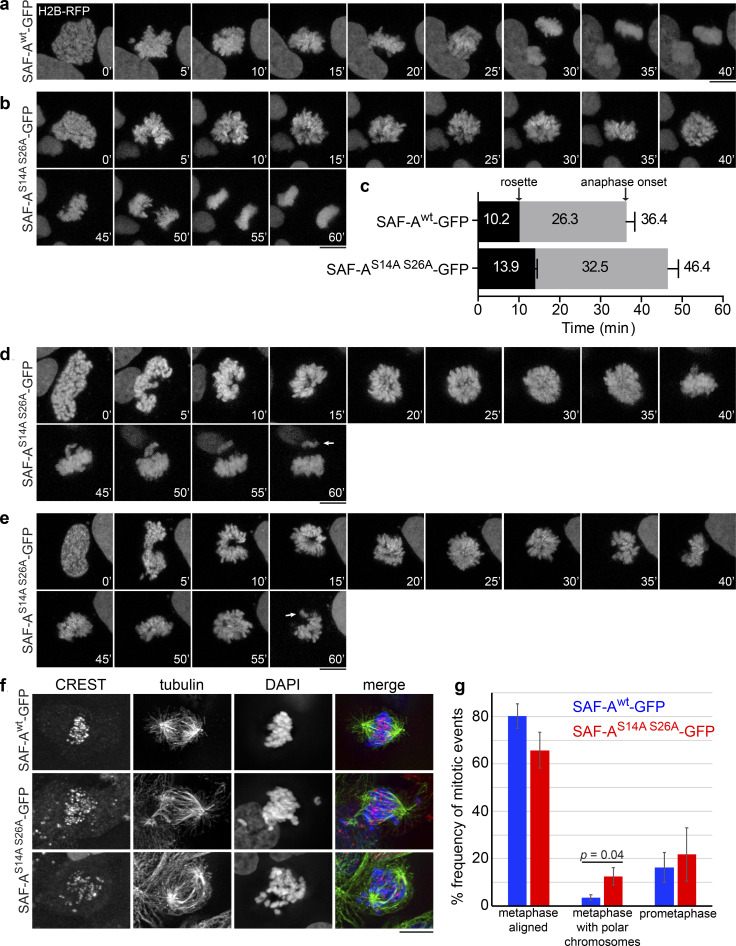
To understand how retention of SAF-A–RNP complexes on mitotic chromosomes causes anaphase segregation defects, we integrated H2B-RFP into SAF-A allele replacement cell lines and examined chromosome behavior in living cells during mitosis. Cells expressing SAF-Awt–GFP excluded SAF-A from chromosomes during prophase (Fig. S2 e) and progressed normally from prophase to anaphase onset in 36.4 min, on average (Fig. 9 a). In contrast, cells expressing SAF-AS14A S26A–GFP retained SAF-A on mitotic chromosomes and exhibited defective mitotic progression (Fig. 9, b–e). In the three examples shown, SAF-AS14A S26A–GFP cells exhibited a significant delay in progression through prometaphase and did not achieve the compact metaphase plate observed in SAF-Awt–GFP cells. In one class of events (Fig. 9 b; n = 14 for both genotypes), SAF-AS14A S26A–GFP cells were able to complete anaphase within 60 min, extending the length from prophase to anaphase onset to an average of 46.4 min (Fig. 9, b and c). In a second class of events, SAF-AS14A S26A–GFP cells displayed poor resolution of individual chromosomes after nuclear envelope breakdown (NEBD; Fig. 9, d and e, at 5-min time point; n = 14 of 28 total movies), a phenotype that was never observed in SAF-Awt–GFP cell populations. In addition, we could observe misalignment of individual chromosomes (Fig. 9, d and e, arrows). We conclude that retention of SAF-A–RNPs on mitotic chromosomes leads to delays in early stages of mitosis and chromosome alignment defects, resulting in an overall delay in completion of mitosis.
Figure 9.
Retention of SAF-A RNPs leads to chromosome alignment defects. (a and b) Live-cell imaging of chromosome segregation in SAF-Awt and SAF-AS14A S26A cells expressing H2B-RFP. (c) Quantitation of the times from NEBD to rosette formation, and from prometaphase to anaphase onset, in cells that completed anaphase within the 60 min of imaging (n = 14 for both genotypes). Error bars depict the SD. P values were as follows: t = 0 to rosette, P < 10−4; rosette to anaphase onset, P = 7.5 × 10−2 (NS); t = 0 to anaphase onset, P = 8.6 × 10−3. (d and e) Examples of poorly resolved chromosomes in SAF-AS14A S26A cells just after NEBD (n = 14 of 28 total movies). Two examples are shown, depicting an extended prometaphase with chromosome misalignments (arrows). (f) Examples of spindles and chromosome alignment in SAF-Awt and SAF-AS14A S26A cells arrested in metaphase with MG132. Two examples of SAF-AS14A S26A cells with polar chromosomes are shown. (g) Quantitation of metaphase chromosome alignment in MG132-arrested cells. For each genotype, >200 cells were scored in two independent experiments. SAF-AS14A S26A cells showed an average 3.5-fold increase in metaphase cells with polar chromosomes. Error bars represent the SD (n = 2). P values were calculated using a Student’s t test. Scale bar = 10 µm.
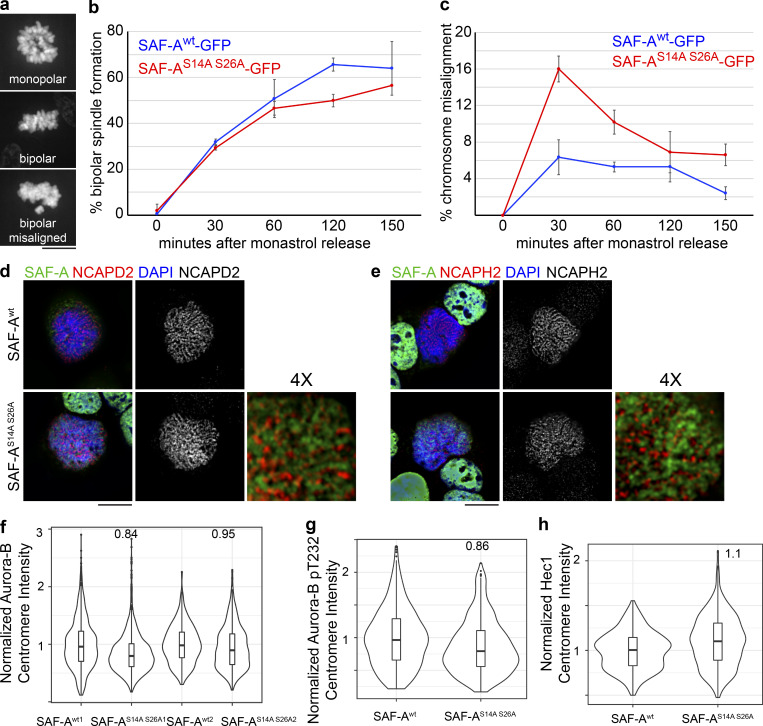
We tested whether the prometaphase delay in SAF-AS14A S26A–GFP cells reflected a problem with correcting kinetochore–microtubule attachment errors (Lampson et al., 2004). Cell populations were treated with monastrol to arrest cells with monopolar spindles and then released into media containing MG132 to arrest bipolar spindles that formed after monastrol washout (Fig. S5, a–c). We found that SAF-Awt and SAF-AS14A S26A–GFP cells formed bipolar spindles at very similar rates following monastrol washout (Fig. S5 b), suggesting normal spindle assembly kinetics in SAF-AS14A S26A–GFP cells. However, at early time points after monastrol release, we observed that SAF-AS14A S26A–GFP cells showed a 2.5-fold increase in chromosome misalignment, indicating a transient defect in kinetochore–microtubule error correction (Fig. S5 c).
Figure S5.
Cells expressing phosphomutant SAF-A show a transient defect in kinetochore–microtubule error correction but show normal localization of mitotic factors. (a–c) To test for correction of kinetochore–microtubule attachment errors, SAF-Awt–GFP and SAF-AS14A S26A–GFP cells were treated with monastrol and subsequently released into media containing MG132. For each time point in the experiment, 100–300 cells were scored. (a) Examples of each chromosome alignment category scored after monastrol washout. (b) Quantitation of bipolar spindle assembly following monastrol washout. Error bars are the SD values from two experiments. (c) Quantitation of chromosome alignment following monastrol washout. We observed a 2.5-fold increase in chromosome misalignment in SAF-AS14A S26A–GFP cells 30 min after monastrol release. Error bars are the SD from two experiments. (d) Localization of condensin I subunit NCAPD2 in SAF-Awt–GFP and SAF-AS14A S26A–GFP cells. 4× magnification was produced using Adobe Photoshop. (e) Localization of condensin II subunit NCAPH2 in SAF-Awt–GFP and SAF-AS14A S26A–GFP cells. 4× magnification was produced in Photoshop. (f) Violin plot depicting normalized Aurora-B centromere intensity in SAF-Awt–GFP and SAF-AS14A S26A–GFP cells. For replicate 1, SAF-Awt1 (n = 415 kinetochores), SAF-AS14A S26A1 (n = 704 kinetochores). In replicate 2, SAF-Awt2 (n = 1,447 kinetochores), SAF-AS14A S26A2 (n = 1,927 kinetochores). (g) Violin plot depicting normalized autophosphorylated Aurora-B (pT232) centromere intensity in SAF-Awt–GFP and SAF-AS14A S26A–GFP cells. For SAF-Awt, n = 535 kinetochores; for SAF-AS14A S26A, n = 646 kinetochores. (h) Violin plot depicting normalized Hec1 kinetochore intensity in SAF-Awt–GFP and SAF-AS14A S26A–GFP cells. For SAF-Awt, n = 617 kinetochores; for SAF-AS14A S26A, n = 644 kinetochores. Scale bar = 10 µm.
We then tested whether SAF-AS14A S26A–GFP cells have higher rates of chromosome misalignment by monitoring cells arrested in metaphase with MG132. SAF-AS14A S26A–GFP cells exhibited a 3.5-fold increase in unaligned, polar chromosomes in metaphase-arrested cells (Fig. 9, f and g). Spindle assembly defects were not observed in any genotype (Fig. 9 f and Fig. S4, d–g). We examined several markers of early mitotic chromatin and kinetochores in SAF-A S14A S26A–GFP cells, including condensins I and II, Aurora-B, and Hec1, but found no gross changes in localization (Fig. S5, d–h). Taken together, these data show that the ectopic retention of SAF-A–RNP complexes on chromatin causes defective metaphase chromosome alignment in a manner that is independent of spindle assembly, condensin localization, Aurora-B localization/activation, or Hec1 recruitment to kinetochores.
Mitotic phosphorylation of SAF-A promotes chromosome alignment through centromere protein E (CENP-E) and chromokinesins
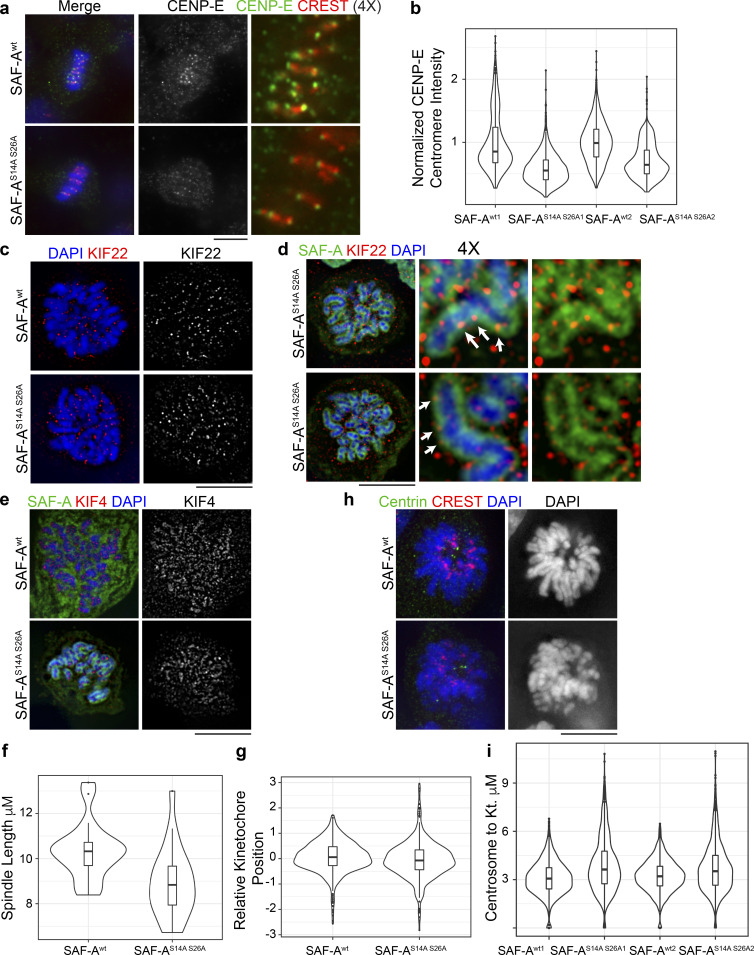
Metaphase chromosome alignment is achieved through the combined action of several chromokinesins and kinetochore-bound motor proteins (Maiato et al., 2017). To gain insight into the molecular causes of chromosome misalignment observed in SAF-AS14A S26A cells, we examined kinetochore and chromosomal markers by IF. CENP-E is a kinetochore-localized kinesin motor important for metaphase chromosome alignment (Putkey et al., 2002) through establishment of lateral kinetochore–microtubule attachments (Kapoor et al., 2006). CENP-E staining at kinetochores was significantly reduced in SAF-AS14A S26A cells (Fig. 10, a and b), suggesting that a lack of normal CENP-E levels at kinetochores could contribute to chromosome alignment defects.
Figure 10.
Mitotic retention of SAF-A RNPs affects CENP-E and chromokinesins. (a) Localization of CENP-E and CREST in SAF-Awt– and SAF-AS14A S26A–expressing cells. (b) Violin plots depicting normalized kinetochore fluorescence of CENP-E. For replicate 1, SAF-Awt1 (n = 761 kinetochores), SAF-AS14A S26A1 (n = 1,192 kinetochores); P = 2.2 × 10−16. In replicate 2, SAF-Awt2 (n = 1,285 kinetochores), SAF-AS14A S26A2 (n = 1,170 kinetochores); P = 2.2 × 10−16. (c) Localization of KIF22 in SAF-Awt– and SAF-AS14A S26A–expressing cells. (d) Colocalization of KIF22 with SAF-AS14A S26A at the chromosome periphery (arrows) in two different cells. Magnified insets were produced using Adobe Photoshop. (e) Localization of KIF4 in SAF-Awt– and SAF-AS14A S26A–expressing cells. (f) Violin plots of spindle length in SAF-Awt– and SAF-AS14A S26A–expressing cells (SAF-Awt [n = 30], SAF-AS14A S26A [n = 30]; P = 8.0 × 10−4). (g) Violin plot depicting the relative position of kinetochores between the spindle poles in SAF-Awt– and SAF-AS14A S26A–expressing cells (SAF-Awt [n = 998 kinetochores]; SAF-AS14A S26A [n = 736 kinetochores]; P = 6 × 10−2). The distance of each kinetochore to both spindle poles was measured, and the ratio of distances between the two poles was calculated. The plot depicts the log2 value of the ratio of the distance between both poles; thus, a value of 0 is equidistant to both poles. (h) SAF-Awt– and SAF-AS14A S26A–expressing cells were treated with monastrol and stained for centrin and CREST. SAF-AS14A S26A cells frequently showed disorganized chromosome orientation within monopolar spindles compared with SAF-Awt cells. (i) Quantitation of centrosome-to-kinetochore distance in SAF-Awt and SAF-AS14A S26A cells. For replicate 1, SAF-Awt1 (n = 1,462 kinetochores), SAF-AS14A S26A1 (n = 1,290 kinetochores); P = 2.2 × 10−16. In replicate 2, SAF-Awt2 (n = 1,295 kinetochores), SAF-AS14A S26A2 (n = 1,166 kinetochores); P = 2.2 × 10−12. For all violin plots, P values were calculated using a Wilcoxon rank-sum test. Scale bar = 10 µm.
Chromosomes congress to the metaphase plate through the combined action of the chromokinesins KIF22 and KIF4 (Levesque and Compton, 2001; Stumpff et al., 2012; Wandke et al., 2012). We therefore tested for altered chromosomal localization of KIF22 or KIF4 in SAF-AS14A S26A cells. KIF22 localization was unchanged in SAF-AS14A S26A cells compared with cells expressing SAF-Awt (Fig. 10 c). Interestingly, KIF22 colocalized with SAF-AS14A S26A at the chromosome periphery (Fig. 10 d), suggesting that chromosomal SAF-A–RNA complexes could alter KIF22 activity. KIF4 localization was also not affected in SAF-AS14A S26A cells. Consistent with previous work, KIF4 localized to the chromosome axis (Samejima et al., 2012) and did not colocalize with SAF-AS14A S26A (Fig. 10 e).
KIF22 and KIF4 play different roles in ensuring metaphase chromosome alignment (Samejima et al., 2012; Stumpff et al., 2012; Wandke et al., 2012); the respective roles of each of these chromokinesins can be revealed by examining spindle length, kinetochore positioning, and the chromosome positioning relative to the centrosomes in monastrol-treated monopolar spindles (Stumpff et al., 2012; Wandke et al., 2012). We found that spindle length was significantly decreased in SAF-AS14A S26A cells (8.91 ± 1.5 µm versus 10.4 ± 1.2 µm in SAF-Awt; P = 8.0 × 10−3) on a scale comparable to loss of KIF22 function (Fig. 10 f; Wandke et al., 2012). To determine the relative position of the kinetochore within the spindle, we calculated the distance from each kinetochore to both spindle poles and examined the ratio between these distances. Using this measure, a ratio near 0 (log2[1]) indicates a kinetochore positioned in the middle of the spindle. Kinetochore alignment in SAF-AS14A S26A cells was not significantly different from that in SAF-Awt cells, but it did show more outliers because of polar chromosomes (Fig. 10 g). This result is also consistent with loss of KIF22 function (Stumpff et al., 2012).
Chromokinesins KIF22 and KIF4 also influence the kinetochore-to-centrosome distance in monopolar spindles (Stumpff et al., 2012). As a second test to determine whether chromokinesin function was compromised in SAF-AS14A S26A cells, we examined chromosome positioning in monastrol-treated cells. SAF-Awt cells exhibited well-formed, rosette-shaped monopolar spindles with a regular distance of chromosomes to kinetochores and chromosome arms oriented away from the centrosomes (Fig. 10 h). In contrast, SAF-AS14A S26A–expressing cells exhibited a significantly increased distance of kinetochores to centrosomes (Fig. 10 i; 0.65 ± 0.21µm increase; P = 2.2 × 10−16), similar to the increase observed in KIF4-depleted cells (Stumpff et al., 2012). In addition, SAF-AS14A S26A–expressing cells exhibited poorly ordered rosette structures and frequently contained chromosomes where the arms were not oriented away from the centrosomes. We conclude that ectopic retention of SAF-A–RNA complexes on mitotic chromosomes leads to defects in metaphase chromosome congression through interfering with normal CENP-E localization and chromokinesin function.
Discussion
In this work, we show that Aurora-B phosphorylates the SAF-A SAP domain to remove nuclear RNAs from the surface of all chromosomes during prophase. Furthermore, we demonstrate that removal of mitotic SAF-A–RNP complexes from the chromosome surface is important for accurate chromosome segregation during anaphase. This study identifies the first pathway to control the global removal of RNPs from the surface of chromatin during mitosis. Our work therefore has identified a previously unrecognized chromosomal remodeling process that is temporally correlated with other genome restructuring events during early mitosis.
During prophase, chromosome structure is dramatically remodeled through the coordinated action of several pathways that impact chromosome condensation, sister chromatid resolution, and transcriptional silencing. Recent work using Hi-C methods demonstrated that interphase chromosomal structures such as topologically associated domains and A/B compartments are removed from chromosomes during the first 15 min of mitosis and are replaced by a structure of nested loop domains orchestrated by the condensin I and II complexes (Gibcus et al., 2018; Naumova et al., 2013; Walther et al., 2018). During interphase, topologically associated domains are maintained by the combined action of the cohesin complex and the CCCTC-binding factor, which together facilitate interactions between enhancer and promoter regions (Kagey et al., 2010; Nora et al., 2017; Rao et al., 2017). Quantitative proteomics of chromosomal proteins during mitosis showed that the cohesin complex is removed from chromosome arms at the same time as condensin complexes begin to associate with chromosomes (Gibcus et al., 2018; Ohta et al., 2010). Mitotic transcriptional silencing is synchronous with mitotic chromosomal remodeling and occurs through the phosphorylation of transcription initiation factors and runoff of elongating RNA polymerase II, which is facilitated by prophase cohesin removal (Akoulitchev and Reinberg, 1998; Liang et al., 2015; Perea-Resa et al., 2020; Segil et al., 1996). Our work shows that in addition to these chromosome remodeling events, chromosome-associated RNP complexes are removed during early mitosis. Thus, during prophase, multiple different pathways function in parallel to erase interphase genome structure and condense chromosomes in preparation for chromosome segregation.
In early mitosis, Aurora-B regulates chromosome structure through cohesin removal, condensin I loading, and heterochromatin dissociation (Fischle et al., 2005; Giet and Glover, 2001; Hirota et al., 2005; Lipp et al., 2007; Losada et al., 2002). We now show that phosphorylation of SAF-A by Aurora-B also contributes to chromosomal remodeling by releasing chromatin-bound RNAs. Interestingly, Aurora-B phosphorylates the pluripotency transcription factor Oct4 in embryonic stem cells to promote chromatin release during mitosis (Shin et al., 2016). Collectively, these observations demonstrate that Aurora-B controls many different pathways important for restructuring interphase chromatin during early mitosis. The fact that Aurora-B triggers the release of chromatin-bound RNAs and core transcription factors suggests that Aurora-B may be a key factor responsible for resetting the transcriptional program as cells pass through mitosis. Passage through mitosis is a key step mediating transcriptional reprogramming and cell fate transitions (Egli et al., 2008; Soufi and Dalton, 2016), and our work suggests that phosphorylation of SAF-A may be a key component of this process. Recent work has demonstrated that there are examples of RNAs that maintain association with chromatin during mitosis, such as nascent transcripts at mitotically expressed genes (Palozola et al., 2017) or noncoding RNAs at centromeric loci (Chan et al., 2012). Whether these RNAs function independently of SAF-A or whether a phosphatase protects SAF-A from Aurora-B activity at these loci is a question for future studies.
SAF-A has been implicated in several processes that control interphase genome structure, all of which are mediated by SAF-A interactions with RNA. For example, SAF-A impacts inactive X chromosome structure through the control of XIST RNA–Xi localization (Hasegawa et al., 2010; Helbig and Fackelmayer, 2003; Pullirsch et al., 2010). SAF-A is also required for nuclear localization of the FIRRE RNA, which promotes interchromosomal interactions between a subset of autosomes (Hacisuleyman et al., 2014). In this study, we identify 1,800 additional RNAs that associate with SAF-A throughout the cell cycle, therefore significantly expanding the repertoire of known SAF-A–RNA interactions. Furthermore, because we could visualize SAF-A–RNA complexes bound to chromosomes in total RNA labeling experiments, we hypothesize that many of the SAF-A–RNA complexes we have identified are anchored to chromatin in a fashion similar to the XIST and FIRRE RNAs. Indeed, on the basis of cytological evidence (Fig. 3), it appears that most or all of Aurora-B–regulated, chromatin-associated RNA localization depends on SAF-A function.
In addition to its role in RNA localization, SAF-A promotes decondensation of expressed regions of the interphase genome by interacting with nuclear RNAs (Fan et al., 2018; Nozawa et al., 2017). Interestingly, the decondensation function of SAF-A is linked to SAF-A oligomerization through the AAA+ domain and RNA-binding activity, but it is independent of the DNA-binding domain (Nozawa et al., 2017). Because this study did not explicitly address RNA localization, it was not clear how the SAF-A chromatin decondensation and RNA-tethering activities were related. We now show that RNA–DNA tethering by SAF-A is performed by the monomeric form of the protein and that mutations that trap SAF-A in an oligomeric state cannot tether RNA to DNA. Additionally, we show that recombinant monomeric SAF-A is alone sufficient to tether RNA to DNA. Furthermore, because all cellular SAF-A is complexed with RNA (Caudron-Herger et al., 2019), these results suggest that at least two distinct populations of SAF-A–RNA complexes are present in cells: a monomeric population that tethers RNA to DNA and an oligomeric population that decondenses transcriptionally active chromatin. We show that failure to remove the RNA-tethering form of SAF-A from chromosomes during mitosis results in chromosome segregation defects. It is currently not clear how oligomerization of SAF-A is controlled during mitosis, and this will require further investigation. In addition, we note that the interactions of both types of SAF-A oligomers with chromatin are likely to require RNA binding, but it is currently unclear how the two different populations are specified.
Our findings show that failure to execute the normal removal of SAF-A–RNPs from mitotic chromosomes led to significant defects in metaphase alignment. We hypothesize that the ectopic retention of SAF-A–RNPs causes physical changes to the surface of the chromatin fiber, impacting the function of several complexes involved in mitotic chromosome function. For example, we found that the amount of CENP-E present at kinetochores was significantly decreased when SAF-A RNPs were present on chromosome arms. CENP-E has been reported to be a direct RNA-binding protein (He et al., 2016), which suggests that the inappropriate presence of RNPs on chromatin could negatively regulate its localization to the kinetochore. In addition, we observed that chromatin-bound SAF-A–RNPs led to spindle and chromosome defects consistent with defects in the function of the chromokinesins KIF22 and KIF4 in a manner independent of their overall localization.
Our live imaging data revealed that SAF-AS14A S26A cells show defects in chromosome individualization during early mitosis in a stochastic manner. Defects in chromosome individualization could be caused by persistent interactions between chromosomes in trans, which has been shown to be controlled at some loci by SAF-A (Hacisuleyman et al., 2014). Although the localization of condensin complexes is not altered in SAF-AS14A S26A cells, the presence of chromatin-bound RNPs may reduce the processivity of condensin complexes. Furthermore, it is possible that the presence of SAF-A–RNPs on the surface of chromosomes could change the mechanical properties of the chromatin fiber, which has been shown to be important for chromosome alignment (Jaqaman et al., 2010). Additional work will be necessary to determine the full import through which removal of chromatin-bound RNPs contributes to normal chromosome segregation.
Materials and methods
Cell culture
hTERT-RPE-1 cells were a gift from Brian Chadwick (Florida State University, Tallahassee, FL; CRL-4000; American Type Culture Collection [ATCC]) and were cultured in DMEM/F-12 supplemented with 10% FBS (HyClone Laboratories), penicillin/streptomycin, 2 mM glutamine, and 7.5% sodium bicarbonate. HEK293T cells were a gift from Paul Kaufman (University of Massachusetts Medical School, Worcester, MA; 12022001; MilliporeSigma) and were cultured as recommended by the ATCC. DLD-1 cells (CCL-221; ATCC) were a gift from Andrew Holland (Johns Hopkins School of Medicine, Baltimore, MD). Cell lines were authenticated by microscopic observation of cell morphology and confirmation of the presence and number of Xi chromosomes, and they were determined to be free of viral infection or mycoplasma contamination after testing by Charles River Laboratories. Cell lines in the DLD-1 background were grown on collagen-coated glass coverslips before imaging (A1048301; Thermo Fisher Scientific). All other cell lines were cultured on uncoated glass coverslips.
Cell synchronization and drug treatment
Cells were synchronized in interphase by incubating cells in 2 mM thymidine (T-1895; MilliporeSigma) for 24 h. For synchronization in mitosis, cells were arrested first in a single thymidine block, then washed twice in PBS and released into thymidine-free media for 6 h, followed by an overnight incubation in 50 µM S-trityl-L-cysteine (93450; Fluka). Cells were harvested the next morning by mitotic shake-off. To arrest cells with monopolar spindles in the error correction assay (Lampson et al., 2004), cells were treated with 100 µM monastrol for 3 h, washed three times in PBS, and then released into media containing 5 µM MG132. Cells were then assayed at 30-min intervals after monastrol release. To arrest cells with bipolar spindles, cells were treated with 5 µM MG132 for 2–3 h. For experiments with kinase inhibitors, cells were incubated in the presence of the drugs for 3–4 h at 37°C before fixation.
Barasertib (Mortlock et al., 2007; AZD1152-HQPA, Selleck Chemicals) and Aurora-A inhibitor I (Aliagas-Martin et al., 2009; S1451; Selleck Chemicals) were each used at 1 µM. BI 2536 (Lénárt et al., 2007; S1109; Selleck Chemicals) was used at 100 nM. Monastrol (M8515; MilliporeSigma) was used at 100 µM. MG132 (S2619; Selleck Chemicals) was used at 5 µM. An equivalent volume of DMSO alone was added to control cells. All drug stocks were prepared as 10 mM in DMSO and frozen until use.
RNA labeling and detection
Total cellular RNA was labeled for 3 h with EU (E10345; Thermo Fisher Scientific) or BrU (850187; MilliporeSigma). EU-RNA was detected with Alexa Fluor 488 azide or Alexa Fluor 594 azide using the Click-iT RNA Imaging Kit (C10329 and C10330; Thermo Fisher Scientific) according to the manufacturer’s instructions. When EU detection was combined with SAF-A IF, we performed the SAF-A IF first, followed by a 10-min fixation with 2% PFA in PBS. Coverslips were then processed for EU-RNA detection as described above. SAF-A:RNA complexes were detected in BrU-labeled cell populations using the rabbit α-SAF-A and mouse α-BrU antibodies in conjunction with the Duolink PLA kit (DUO92102; MilliporeSigma; Cy3-compatible fluorochrome).
IF and FISH
Cells were washed briefly in PBS and fixed for 10 min in PBS containing 4% PFA (Electron Microscopy Sciences) at room temperature. Cells were washed with room temperature PBS and then gently extracted in PBS containing 0.5% Triton X-100 for 20 min. In the course of this study, we compared several fixation methods and found these conditions best for preserving mitotic chromosome structure as well as SAF-A staining and RNA localization. In particular, we found that extracting the cells in the presence of hypotonic cytoskeletal buffer caused extensive swelling of chromatin and suboptimal imaging of mitotic stages. These conditions were used throughout the study (SAF-A, RNA FISH, EU-RNA, GFP, CENP-E, and Hec1), except as noted below.
For NCAPD2 and NCAPH2 IF, cells were washed twice with PBS, prepermeabilized with PBS + 0.1% Triton X-100 for 1 min at room temperature, fixed with PBS + 4% PFA for 10 min at room temperature, and then washed twice with PBS. For KIF22 IF, cells were washed twice with PBS and then simultaneously fixed and permeabilized with PBS + 4% PFA + 0.1% Triton X-100 for 10 min at room temperature, followed by two washes in PBS.
For Aurora-B and phospho–Aurora-B IF, cells were incubated with 1 µM nocodazole for 3 h, collected by mitotic shake-off, and then cytospun onto charged slides (500 µl of cells at 0.25 × 106 cells/ml; 1,500 rpm for 5 min) using a Shandon Cytospin device. Cells were fixed in PBS + 4% PFA for 10 min at room temperature and permeabilized with PBS 0.1% Triton X-100 for 10 min. To image spindle microtubules in SAF-A–AID–mCherry cells, cells were simultaneously fixed and permeabilized for 10 min in 4% PFA, 100 mM Pipes, pH 6.8 (KOH), 10 mM EGTA, 1 mM MgCl2, and 0.2% Triton X-100.
After fixation, cells were blocked briefly in PBS containing 1% RNase-free BSA (Ambion UltraPure BSA, AM2616; Thermo Fisher Scientific) and 0.2% Tween. Cells were incubated with the primary antibody at 37°C and washed three times in PBS + 0.2% Tween. Secondary antibody incubations and washes were performed similarly. Chromosomes were stained with 40 ng/ml DAPI. In experiments in which FISH was performed after immunostaining, RNAsin (Promega) was added to the blocking buffer and antibody incubations at 400 U/ml, and DAPI staining was omitted. Prior to FISH, cells were fixed for 10 min in 2% PFA to preserve antibody–antigen interactions throughout the FISH protocol.
For FISH, cells were fixed as described above and dehydrated sequentially in 70%, 85%, 95%, and 100% ethanol. The XIST RNA probe was directly labeled with Cy3-dCTP (PA53021; GE Healthcare) using Klenow (BioPrime DNA labeling system; Invitrogen) and a plasmid template with human XIST cDNA 1–16,474 described previously (Xiao et al., 2007). Probes for the NEAT2 and OGT RNAs were labeled similarly, using a PCR product amplified from cDNA as the template. Cells were hybridized with the probe overnight in a humid chamber at 37°C in 50% formamide, 2× SSC, and 10% dextran sulfate. Coverslips were washed twice with 50% formamide/2× SSC, once with 2× SSC, and three times with 1× SSC. All washes were performed at 39°C. DAPI was included in the second 1× SSC wash at 40 ng/ml. Coverslips were mounted onto slides with VECTASHIELD mounting medium (Vector Laboratories).
IP
After synchronization, cells were washed in PBS and incubated for 30 min at 4°C in ice-cold lysis buffer (1 ml per 15-cm plate) containing 25 mM Tris, pH 7.4, 150 mM KCl, 5 mM EDTA, 5 mM MgCl2, 1% NP-40, 0.5 mM DTT, and protease inhibitors (Roche). Cells were collected from the plate and passed several times through a syringe with a 22-gauge needle. Lysates were centrifuged for 30 min at 4°C (∼22,000 ×g) to remove insoluble material. The protein concentration of the extract was determined using a Bradford assay. Cell extracts were normalized to between 1–2 mg/ml and incubated with 6 µg SAF-A antibody per ml of extract for 1.5–2 h at 4°C. Control IPs included 6 µg rabbit or mouse IgG (Jackson ImmunoResearch) per milliliter of extract, matched to the host species of SAF-A antibody used in experimental IPs. Immune complexes were collected 1.5–2 h at 4°C, with preequilibrated Dynabeads (Thermo Fisher Scientific; 4.5 µl Dynabeads per μg of antibody) conjugated to either protein A or protein G, according to whether the antibody to SAF-A was generated in rabbit or mouse, respectively. The slurry of beads bound to immune complexes was washed three times in 1 ml lysis buffer before elution.
For immunoaffinity purification of SAF-A for mass spectrometry, we used a rabbit polyclonal antibody to SAF-A and included protein phosphatase inhibitor cocktails I and III (MilliporeSigma) in cell extracts. Samples were submitted for analysis to the Taplin Biological Mass Spectrometry Facility (http://taplin.med.harvard.edu/). Phosphopeptides identified in this study for both interphase and mitotic SAF-A are detailed in Table S1. The schematic of SAF-A phosphopeptides mapped relative to domain structure was assembled from this study (Kettenbach et al., 2011; Olsen et al., 2010; https://www.phosphosite.org/homeAction.action and references therein).
To investigate SAF-A–associated RNAs (RNA IP sequencing [RIP-seq]), we performed IP as described above, except that a mouse mAb to SAF-A was used. RNAsin (Promega) was added to the IP at 40 U/ml. Samples were eluted in TRIzol reagent (Thermo Fisher Scientific) for RNA purification before library production. The coprecipitation of native SAF-A with chromatin was also performed with the mouse mAb to SAF-A (Fig. 1, b and c). In SAF-A–GFP–tagged cell lines (Fig. 8 and Fig. S5 b), immune complexes were collected with GFP-Trap magnetic agarose beads (GTMA020; Bulldog Bio). Samples were treated as described above, except that immune complexes were formed during an overnight incubation at 4°C.
qRT-PCR
Superscript III reverse transcription (Invitrogen) was used to synthesize cDNA at 50°C according to the manufacturer’s instructions. For quantitative real-time PCR, we used 0.05 vol of a cDNA reaction to program a PCR using iQ SYBR Green Supermix (Bio-Rad Laboratories). PCRs were amplified on a CFX96 Real-Time System (Bio-Rad Laboratories) and analyzed with the CFX software package.
Antibodies
Primary antibodies used in this study were as follows: rabbit mouse α-SAF-A (ab10297; Abcam), rabbit α-SAF-A (ab20666; Abcam), rabbit α-histone H3 (ab1791; Abcam), mouse α-histone H3-S10P (ab14955; Abcam), mouse α-tubulin (DM1A, T6199; MilliporeSigma), mouse α-GFP (ab1218; Abcam), mouse α-Aurora-B (ab3609; Abcam; 611082; BD Transduction Laboratories), mouse α-mini-AID (M214-3; MBL International), rabbit α-NCAPD2 (HPA036947; MilliporeSigma), rabbit α-NCAPH2 (PA564393; Invitrogen), rabbit α-KIF22 (222187; Abcam), mouse α-Hec1 (3613; Abcam), rabbit α-Aurora-B pT232 (600-401-677; Rockland Immunochemicals), human α-CREST (HCT-100; ImmunoVision), mouse α-KIF4 (sc-365141; Santa Cruz Biotechnology), rabbit α-Centrin (ab101332; Abcam), rabbit α-CENP-E (AKIN04; Cytoskeleton), and a mouse antibody with reactivity to BrU (Paulsen et al., 2014; 555627; BD Biosciences). SAF-A–AID–mCherry protein was detected using an alpaca α-RFP antibody conjugated to Atto-594 (RBA594; Bulldog Bio). Tet-inducible SAF-A–GFP alleles were detected using alpaca α-GFP conjugated to Atto-488 (GBA488; Bulldog Bio).
Secondary antibodies used in this study were as follows: donkey α-rabbit Cy3 (Jackson ImmunoResearch), goat α-rabbit Cy3 (Jackson ImmunoResearch), goat α-mouse Alexa Fluor 488 (Jackson ImmunoResearch), goat α-mouse Cy3 (Jackson ImmunoResearch), goat α-human Alexa Fluor 647 (Jackson ImmunoResearch), and goat α-mouse Alexa Fluor 647 (Thermo Fisher Scientific).
Transfections
Full-length, wild-type SAF-A was cloned into pEGFP-N1 (Clontech Laboratories) and confirmed by DNA sequencing. The resulting plasmid encoding SAF-A (pMB918) was used as a template for site-directed mutagenesis to generate phosphomutant and phosphomimetic alleles of SAF-A. Plasmids used for transfections were as follows: pMB918 (SAF-Awt–GFP), pMB935 (SAF-AS14A–GFP), pMB936 (SAF-AS26A–GFP), pMB1003 (SAF-AS14A S26A–GFP), pMB992 (SAF-AS271A–GFP), pMB1004 (SAF-AS26A S271A–GFP), pMB932 (SAF-AS267A S271A–GFP), pMB931 (SAF-AS187A S188A T191A S192A–GFP), pMB1013 (SAF-AS14D S26D–GFP), and pMB1014 (SAF-AS14E S26E–GFP).
HEK293T cells were plated onto coverslips the day before transfection at a density of 0.3 × 106 cells per well. Cells were transfected for 48 h with wild-type or mutant versions of SAF-A–GFP using 2.5 µg plasmid DNA and 2.0 µl Lipofectamine 3000 (Invitrogen). Multiple independent transfections (n ≥ 2–5) were analyzed to assess the frequency of the phenotypes reported in Figs. 4, 5, and 6.
To deplete Aurora-B by RNAi, RPE-1 cells were transfected using RNAiMAX (Thermo Fisher Scientific) and incubated for 48 h before processing coverslips for FISH and immunostaining. Cells were transfected using nontargeting siRNA (D-001210-01-05, 5′-UAGCGACUAAACACAUCAA-3′; Dharmacon) or a pool of siRNAs specific for Aurora-B (M-003326-08-0005, 5′-CAGAAGAGCUGCACAUUUG-3′, 5′-CCAAACUGCUCAGGCAUAA-3′, 5′-ACGCGGCACUUCACAAUUG-3′, 5′-UGGGACACCCGACAUCUUA-3′; Dharmacon). Transfections were performed in 24-well format, using 30 pmol siRNA and 1.5 µl Lipofectamine RNAiMAX per well.
Microscopy
Cells were visualized on an Olympus microscope (BX61) equipped with a disc-scanning unit spinning disc confocal attachment. Images were captured as 3D optical stacks (0.2 µm per slice) using a charge-coupled device camera (ORCA-ER, C4742-80; Hamamatsu Photonics) and MetaMorph software (Molecular Devices). A 100× oil-immersion lens (NA 1.4) or a 60× oil-immersion lens (NA 1.42) was used for all images. Coverslips were mounted using VECTASHIELD mounting medium (H-1000; Vector Laboratories) and visualized under Olympus immersion oil. Images were captured at room temperature. Image stacks were processed and analyzed using the MetaMorph software package. Some images were further processed using Autoquant constrained, iterative deconvolution in MetaMorph.
For live-cell imaging, we used SAF-A degron cells expressing either SAF-Awt–GFP or SAF-AS14A S26A–GFP. A lentivirus encoding H2B-RFP was integrated to image chromosomes. Cells were grown in 3-cm glass-bottomed plates in RPMI 1640 medium and were imaged under temperature- and CO2-controlled conditions using a LiveCell Stage Top Incubation System (BioVision Technologies). To deplete endogenous SAF-A and simultaneously induce SAF-A–GFP alleles, cells were treated with doxycycline and IAA as described below. For high-resolution images (Fig. S2 e), a spinning disk confocal microscope (Ultraview Confocal Scanner ERS; PerkinElmer) with a charge-coupled device camera (ORCA-ER; Hamamatsu Photonics) was employed. Images were acquired as 3D optical stacks (0.8 µm per slice) using an Apochromat total internal reflection microscopy 60× oil immersion objective (NA 1.45) and Volocity software (PerkinElmer). For time-lapse imaging (Fig. 9, a–e), cells were imaged with a Nikon A1R confocal microscope (Nikon Instruments Inc.), and pictures were acquired every 5 min for 1 h using the Galvano Scanner Detector. Images were acquired as 3D optical stacks (0.8 µm per slice) using an Apochromat total internal reflection microscopy 60× oil immersion objective (NA 1.49) and NIS-Elements software (Nikon Instruments Inc.). All live images were processed using Fiji software.
For image quantitation, we acquired image series for each experimental condition using equivalent exposure times. In each optical stack, we identified a single slice representing the midpoint of the chromosome rosette. We measured total EU-RNA or SAF-A fluorescence by subtracting background fluorescence, thresholding the image, marking the cell boundary, and then summing pixel intensity over the selected area. To measure chromatin overlap of EU-RNA and SAF-A, DAPI images were deconvolved and used to select the chromatin area by thresholding. We then summed pixel intensity of EU-RNA or SAF-A within the area overlapping chromatin. The chromosome-localized fluorescent signal was subtracted from the total intensity to obtain the cytoplasmic fluorescence. EU-RNA or SAF-A localization was then expressed as a ratio of chromosome overlap/cytoplasmic localization.
For quantitation of Aurora-B, Hec1, and CENP-E centromere intensity, we acquired images of ≥20 cells for each genotype. We then identified kinetochores by CREST signal. Antigen intensity was quantitated by manually drawing a circle around the centromere region in ImageJ. Kinetochore intensity plots were prepared for each biological replicate using R. To calculate the distance between kinetochores and centrosomes, we identified the position between two centrosomes and then calculated the 3D distance from that point to all kinetochores using the PointPicker plugin in ImageJ.
Aurora-B kinase assay
The Aurora-B–Incenp complex was expressed and purified from E. coli as described previously (Jambhekar et al., 2014). SAF-A was immunoaffinity purified from interphase cells using the α-SAF-A rabbit polyclonal antibody. Kinase assays were performed as described previously (Bolton et al., 2002; Rosasco-Nitcher et al., 2008).
Molecular modeling
The DNA-binding SAP domain of SAF-A (residues 1–62) was submitted to the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/; Roy et al., 2010; Yang et al., 2015) to identify structural homologues present in the Protein Data Bank. We chose the highest ranked of five models for further analysis. Phosphoserines were modeled onto the SAF-A SAP domain model using Vienna-PTM 2.0 (http://vienna-ptm.univie.ac.at; Margreitter et al., 2013; Petrov et al., 2013). Molecular models of wild-type and phospho-SAF-A were rendered in PyMOL (https://pymol.org/2/). Electrostatic surface potential and surface charge calculations were rendered in PyMOL using the Adaptive Poisson-Boltzmann Solver plugin (http://www.poissonboltzmann.org/).
SAF-A SAP domain purification and DNA EMSA
The wild-type SAP domain of SAF-A was amplified by PCR and cloned into a modified pET30a vector containing GFP using In-Fusion cloning (pMB1018; Clontech Laboratories). SAPS14D S26D (pMB1028) was created using site-directed mutagenesis. Clones were verified by sequencing. Proteins were expressed in BL21 Rosetta 2 cells overnight at 18°C and were lysed using a French press in PBS containing 300 mM NaCl and EDTA-free protease inhibitors. Lysates were cleared by centrifugation at 25,000 ×g for 30 min at 4°C. Cleared lysates were bound to Ni-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) for 1 h at 4°C. Beads were washed with ∼100 column volumes of PBS containing 300 mM NaCl and 10 mM imidazole. Proteins were eluted using PBS containing 500 mM imidazole. Eluted proteins were immediately loaded onto a Superdex S200 column equilibrated with 50 mM Tris, pH 7.5, and 150 mM NaCl. Peak fractions were pooled, adjusted to 10% glycerol, and stored at −80°C.
The DNA template for EMSA was an AT-rich SAR sequence (Okubo et al., 2004). 5′-AATTCAGAAAATAATAAAATAAAACTAGCTATTTTATATTTTTTC-3′ was ordered as DNA oligos from Integrated DNA Technologies. Oligos were annealed and end labeled with γ-32P-ATP using PNK. PNK labeling reactions were purified using a G50 spin column, then 1 µg of labeled DNA was gel purified from a 10% native PAGE gel. After elution, labeled DNA was ethanol precipitated and quantitated by using a NanoDrop system. Purified DNA duplex was adjusted to 50 nM and stored at −20°C.
DNA EMSA reactions contained the indicated protein concentrations (Fig. 5 d), 5 nM annealed SAR oligos, in a buffer containing 50 mM Tris, pH 7.5, 50 mM NaCl, 1 mM EDTA, and 4% glycerol. All components were mixed and incubated at room temperature for 30 min. Reactions were separated in a 10% native PAGE gel in 1× Tris-borate-EDTA run at 120 V for 1 h. Gels were dried and exposed to a phosphor screen overnight. Binding reactions were performed in duplicate. The fraction of the SAF-A SAP domain bound to the double-stranded DNA SAR template was calculated using ImageJ, and binding curves were fit to the data using R.
Expression and purification of SAF-A in Sf9 cells
Full-length SAF-A was cloned into pFB-HTA using InPhusion cloning (pMB1105). Sf9 cells were grown to a density of 106 cells per ml and infected with an MOI ∼5 from a P2 viral stock. Infected cells were grown for 48 h after infection. Infected cells were collected by centrifugation at 1,000 ×g for 15 min. Cells were washed once with ice-cold PBS and collected by centrifugation as above. Cell pellets (∼109 cells) were resuspended in 25 ml PBS (400 mM NaCl final concentration), 1× protease inhibitors (Roche Complete Mini), Benzonase (50 U; MilliporeSigma), and 1% CHAPS. Resuspended cells were lysed by 10 passages of a tight-pestle Dounce homogenizer. Cells were centrifuged at 40,000 ×g for 15 min at 4°C. A volume of 0.8 ml Ni-NTA (Qiagen) beads was added to the soluble lysate and incubated in batch at 4°C for 1 h. Beads were collected and washed in column format with ∼50 column volumes of PBS. Protein was eluted with 1 ml PBS + 50 mM imidazole. Eluted protein was diluted 2:3 with cold water. Eluted protein was applied directly to a HiTrap heparin column on a fast protein liquid chromatography instrument (Bio-Rad Laboratories). Bound protein was eluted with an NaCl gradient from 100 to 1,000 mM NaCl. SAF-A was eluted in a single peak from the heparin column. Peak fractions from the heparin column were loaded onto a Sephacryl S200 column run in 50 mM Tris, pH 8.0, 100 mM NaCl, and 0.5 mM EDTA. Peak fractions were collected and quantified by Bradford assay. The final yield was 1.5 mg of purified SAF-A from 1.5 liters of cultured Sf9 cells.
DNA bead-tethering assays
An 875-bp fragment of the MII SAR region (hg38 chr20:41104718-41105592) was amplified from human genomic DNA and cloned into pCR2.1 using TOPO cloning (pMB1156). This fragment was amplified from the plasmid using M13F-biotin and M13R. Purified DNA was coupled to M280 streptavidin Dynabeads as described elsewhere (Hannak and Heald, 2006) A 935-nt fragment from exon 1 of human XIST RNA (hg38 chrX:73845035-73845969) was prepared by in vitro transcription incorporating FITC-UTP. Transcribed RNA was purified by LiCl2 precipitation.
Tethering reactions were performed in 25-µl reaction volumes. We first prepared protein–RNA complexes by incubating purified SAF-A with XIST RNA. Reactions contained 100 nM XIST RNA, 2 µM SAF-A, 0.1 mg/ml yeast tRNA, 0.1% CHAPS in 50 mM Tris, pH 8.0, 100 mM NaCl, and 1 mM DTT. Reactions were incubated for 30 min at room temperature, then added to 15 µl DNA (or empty streptavidin beads) and incubated for 30 min at room temperature with occasional flicking. Beads were washed three times, 1 ml per wash, in 50 mM Tris, pH 8.0, 100 mM NaCl, and 0.1% CHAPS, switching tubes once between washes 2 and 3. Protein–RNA–DNA complexes were eluted with 1× SDS sample buffer by incubating at 80°C for 5 min. Half the eluted samples were run on agarose and acrylamide gels for RNA and protein analyses.
SAF-A RGG expression and RNA EMSA
Full-length SAF-A RGG was cloned into pMB809 to create an N-terminal fusion with His-GFP-3XHA-PreScission Protease (pMB1211). Deletion mutants encompassing the central RGG cluster (pMB1241), full RGG cluster (pMB1240), or C-terminus (pMB1234) were created using site-directed mutagenesis using NEB Q5 polymerase.
Plasmids were transformed into BL21 Rosetta 2 cells. Single colonies were used to inoculate a 50-ml overnight culture in Luria broth + kanamycin. The next morning, 1 liter of Terrific Broth was inoculated with the overnight culture and grown to an OD ∼0.6 at 37°C. IPTG was added to 0.1 mM for 4 h at 37°C. Cells were collected by centrifugation and stored at −20°C. Cell pellets were thawed in PBS (500 mM NaCl), 500 mM arginine, 10 mM imidazole, and 1× protease inhibitors. Cells were lysed by a single passage through a French press (1,500 psi), Benzonase was added, and cells were centrifuged at 45,000 ×g for 20 min at 4°C. Cleared lysate was added to 0.5 ml Ni-NTA beads and bound at 4°C for 1 h. Beads were collected by centrifugation and washed in column format (∼100 column volume) using lysis buffer. Protein was eluted from the column using PBS + 500 mM arginine + 500 mM imidazole. The entire elution was immediately applied to a Superdex S200 column equilibrated in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 500 mM arginine. Peak fractions were collected and concentrated using Microcon spin columns. We found that the isolated SAF-A RGG domain was extremely aggregation prone and that 500 mM arginine in all buffers was necessary to stabilize this protein, as has been observed for other aggregation-prone proteins (Boke et al., 2016).
A fragment of human XIST (hg38 chrX:73845035-73845969) was internally labeled with 32P-UTP by in vitro transcription. Transcribed RNA was gel purified from a urea PAGE gel. EMSA reactions were performed in the following buffer: 25 mM Tris, pH 7.5, 25 mM NaCl, 1 mM EDTA, 4% glycerol, 0.1% CHAPS, 0.1 mg/ml yeast tRNA, and 0.1 nM XIST RNA. Radiolabeled XIST RNA was denatured by heating in water at 65°C for 10 min, then cooled on ice for 2 min. All proteins were diluted into Sephadex S200 column buffer containing 500 mM arginine. Diluted proteins were mixed with reaction mixture and incubated at room temperature for 30 min. All EMSA reactions contained 250 mM arginine after addition to diluted proteins. Reactions were separated on a 7.5% PAGE gel in 1× Tris-borate-EDTA for 60 min at 90 V. Gels were then dried and exposed to phosphor screens overnight. The screen was developed using a GE Typhoon imager. The fraction of bound RNA was calculated using ImageJ; Kd values were calculated using R. EMSA reactions were performed for all four proteins at the same time, and a duplicate experiment was conducted on a different day.
SAF-A RIP-seq and bioinformatics
RNA associated with SAF-A was immunoprecipitated as described above and purified in TRIzol reagent (Invitrogen). Any potential contaminating DNA was removed by DNase digestion (RQ-1; Promega); RNA was subsequently purified on a column (RNA Clean & Concentrator-25; Zymo Research). rRNA sequences were removed using a Ribo-Zero Gold rRNA removal kit (Illumina), and the rRNA-free eluate was again column purified (RNA Clean & Concentrator-5; Zymo Research). RNA libraries were generated using the NEBNext Ultra RNA Directional Library (New England Biolabs), PAGE purified, and sequenced using an Illumina HiSeq 2000 platform.
Reads from RNA-seq libraries were aligned to the human genome builds hg19 and hg38 using TopHat (Trapnell et al., 2012). Reads were counted against UCSC Genome Browser gene annotations (for hg38). To analyze differential gene expression, we first filtered our RNA-seq data to eliminate any genes that had a fragments per kilobase of transcript per million mapped reads (FPKM) of 0 in any of the eight sequenced libraries. All plots were created using R. To identify coding and noncoding genes in the UCSC Genome Browser genome annotation, we extracted a representative RefSeq transcript for each gene annotation. We categorized genes as being an mRNA if the representative transcript had a name starting with “NM_,” and noncoding RNA genes had the format “NR_.” Comparisons of noncoding RNA overrepresentation were calculated using Fisher’s exact test in R. All sequences and summary tables generated by high-throughput sequencing have been deposited in the Gene Expression Omnibus database under accession no. GSE87200.
To compare SAF-A RNA-seq data with nuclear retained RNAs (Djebali et al., 2012), we downloaded datasets of whole-cell, nucleus, and cytoplasm RNAs from IMR-90 cells, an untransformed, female diploid primary cell line (SRR534291, SRR534292, SRR524299, SRR534300, SRR534301, and SRR534302). Reads were aligned to hg38 using TopHat and counted against UCSC Genome Browser gene models using Cufflinks as described above. Read counts were filtered for genes with a FPKM >0 and combined with SAF-A IP-sequencing data.
Statistics
Two or more independent biological replicates were performed for all experiments described in this study, with exceptions as follows. Identification of SAF-A phosphopeptides by mass spectrometry was performed once and pooled with published data as described above (Fig. 4, b and c). Molecular modeling of the SAF-A SAP domain was also performed once (Fig. 5, a and b). Image quantitation of phospho-Aurora-B, Hec1, spindle length, and relative kinetochore position were each performed once (Fig. 10 and Fig. S5). For statistical analysis of quantitative data, the central tendency is represented by the mean, and variation is represented by the SD. In all figures (except Fig. 10 and Fig. S5), P values were calculated using a two-sided Student’s t test specifying equal variance of the two samples; the number of individual data points (n) measured for each comparison is specified in the figure legends. In Fig. 10 and Fig. S5, P values were calculated using a Wilcoxon rank-sum test. Correlation statistics for RNA-seq experiments were calculated using Spearman’s correlation coefficient in R. Graphs were generated using Excel, Plotly, and R software.
Data availability
All sequences and summary tables generated by high-throughput sequencing have been deposited in the Gene Expression Omnibus database under accession no. GSE87200. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ghuhkaawdtkxfwf&acc=GSE87200
Construction of cell lines expressing degron-tagged SAF-A
We targeted the endogenous SAF-A locus for C-terminal tagging using the CRISPR-Cas9 system as previously described (Natsume et al., 2016; Ran et al., 2013). In brief, we used an online CRISPR Design Tool to identify a PAM sequence near the SAF-A stop codon and to design a corresponding guide RNA for cloning into the BbsI sites of pX330-U6-chimeric_BB-CBh-hSpCas9 (42230; Addgene). The resulting construct, pMB1050, was used in conjunction with the targeting construct to modify the SAF-A locus.
To generate the SAF-A targeting construct, we fused 200-bp homology arms to a cassette encoding the minimal auxin-inducible degron sequence fused to the mCherry tag and the blasticidin-resistance gene. The 5′ homology arm corresponds to human chromosome 1, nt 244854433–244854462. The 3′ homology arm corresponds to human chromosome 1, nt 244854250–244854449. The tagging cassette was derived from plasmid pMK294 (mAID-mCherry2-Bsr, 72832; Addgene). All elements of the targeting cassette were ligated together in the pBluescript KS+ vector (In-Fusion; Takara Bio) to make pMB1116.
DLD-1 cells were cotransfected with pMB1050 and pMB1116 using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific). After 72 h, cells were selected for 6 d with 5 µg/ml blasticidin S (Thermo Fisher Scientific). Individual clones were obtained by FACS sorting mCherry-positive cells at 1 cell/well of a 96-well plate. Cell lines were then screened by genomic DNA PCR, Western blot analysis, and live-cell imaging to identify positive clones expressing SAF-A–AID–mCherry (Fig. S2).
SAF-A–AID–mCherry cells in the DLD-1 background were transfected with a plasmid encoding codon-optimized, tet-inducible OsTIR1 (pMK243, 72835; Addgene). Cells were selected with 0.5 mg/ml Geneticin (Thermo Fisher Scientific); individual drug-resistant colonies were picked for expansion using cloning cylinders. Positive clones were first screened by RT-PCR to identify lines that express TIR1 after doxycycline treatment and were then tested for inducible degradation of SAF-A–AID–mCherry. We observed that SAF-A degron cells required 24 h of treatment with 1 µg/ml doxycycline (D3447; MilliporeSigma) and 500 µM auxin (IAA, I-5148; MilliporeSigma) for optimal depletion of SAF-A–AID–mCherry.
To generate homozygous SAF-A–AID knock-in cells in the RPE-1 cell background (Fig. S3, a–d), we modified our homology-directed repair (HDR) cassette to create a P2A fusion between mAID-mCherry and zeocin resistance (pMB1201). We then used PCR with 5′ biotin–modified oligos to PCR amplify this repair cassette. The HDR PCR product was cotransfected with the SAF-A single-guide RNA plasmid (pMB1050) into RPE-1 cells using nucleofection. Briefly, 2 µg of the HDR cassette and single-guide RNA plasmid were nucleofected into 106 cells. Cells recovered for 7 h, followed by addition of pfithrin-α (30 µM; Komarov et al., 1999), pfithrin-µ (10 µM), or no drug for 3 d. Cells were then split and selected for zeocin resistance, in the absence of pifithrin-µ or pifithrin-α, for 2 wk. Single-cell colonies were isolated and analyzed for homozygous mAID-mCherry insertion by Western blot analysis and PCR. We isolated three homozygous clones in cells treated with pifithrin-α. Both untreated cells and cells treated with pifithrin-µ yielded only heterozygous clones. Homozygous SAF-A–mAID–mCherry RPE-1 cells were then transduced with lentivirus expressing tet-inducible OsTir1 and neomycin resistance. Cells were selected for neomycin-resistant colonies for 2 wk. Inducible SAF-A degradation was monitored by fluorescence and Western blot analysis.
SAF-A–AID–mCherry depletion and reconstitution
GFP-tagged SAF-A alleles were cloned into the lentiviral expression vector pLVX-TetOne-puro (Takara Bio) using In-Fusion cloning. Lentiviral plasmids used for reconstitution were pMB1103 (SAF-Awt–GFP), pMB1109 (SAF-AS14A S26A–GFP), pMB1246 (SAF-ARGG 1–7Δ–GFP), pMB1248 (SAF-AS14A S26A RGG 1–7Δ–GFP), pMB1110 (SAF-AC-termΔ–GFP), and pMB1119 (SAF-AS14A S26A C-termΔ–GFP). Lentiviral plasmids were cotransfected with packaging plasmids pLP1, pLP2, and pVSV/G into HEK293T cells using Lipofectamine 2000 (Thermo Fisher Scientific). Lentivirus-containing supernatant was collected at 72 h and used to transduce SAF-A–AID–mCherry cells in the presence of RPMI media supplemented with 10% FBS and 6 µg/ml polybrene (H-9268; MilliporeSigma). After 48 h, cells were passaged before FACS sorting of GFP-positive cells. All cells were gated similarly for GFP expression. Individual clones were then tested by Western blot analysis and live-cell imaging to confirm SAF-A–GFP expression levels. In experiments in which SAF-A function was reconstituted with GFP-tagged inducible proteins, cells were first treated with 1 µg/ml doxycycline for 24 h to induce TIR1 and SAF-A–GFP, followed by another 24-h incubation in doxycycline and 500 µM IAA to degrade SAF-A–AID–mCherry.
Online supplemental material
Fig. S1 shows statistical analysis of SAF-A RNA-seq data and independent validation of SAF-A–interacting RNAs. Fig. S2 shows control experiments to validate SAF-A depletion and rescue in DLD-1 SAF-A–AID–mCherry cell lines. Fig. S3 includes control experiments to validate the RPE-1 SAF-A–AID–mCherry cell line and shows that depletion of SAF-A results in normal spindle assembly and chromosome segregation rates. Fig. S4 shows SAF-A SAP domain alignments, SAF-A RNA EMSA gels, and SAF-A RGG mutant interactions with chromatin. Fig. S5 shows analysis of spindle formation and error correction in SAF-A phosphomutant cells and IF analysis of mitotic regulators in SAF-A phosphomutant cells. Table S1 lists SAF-A phosphopeptides identified in this study. Table S2 contains RNA-seq data from SAF-A IPs in interphase and mitosis.
Supplementary Material
lists SAF-A phosphopeptides identified in interphase and mitosis from purification shown in Fig. 4 b.
contains RIP-seq data for SAF-A IPs in interphase and mitosis.
Acknowledgments
We thank Brian Chadwick, Iain Cheeseman (Whitehead Institute for Biomedical Research and Massachusetts Institute of Technology, Cambridge, MA), Andrew Holland, and Paul Kaufman for sharing plasmids and cell lines. We are indebted to Jesse Cochrane and Radhika Subramanian for advice on performing the molecular modeling of SAF-A. We thank Iain Cheeseman, Paul Kaufman, Jeannie Lee, Barbara Panning, and Radhika Subramanian for insightful discussion and comments on the manuscript.
Live-cell imaging was performed in the Massachusetts General Hospital Microscopy Core of the Program in Membrane Biology, which is partially supported by a Center for the Study of Inflammatory Bowel Disease grant (DK043351) and a Boston Area Diabetes and Endocrinology Research Center award (DK057521). The Nikon A1R confocal system is supported by National Institutes of Health grant 1S10 RR031563-01. This work was supported by a grant from the National Institutes of Health to M.D. Blower (GM122893).
The authors declare no competing financial interests.
Author contributions: Conceptualization, J.A. Sharp and M.D. Blower; Methodology, J.A. Sharp and M.D. Blower; Software, M.D. Blower; Validation, J.A. Sharp and W. Wang; Formal Analysis, J.A. Sharp, C. Perea-Resa, and M.D. Blower; Investigation, J.A. Sharp, C. Perea-Resa, W. Wang, and M.D. Blower; Data Curation, M.D. Blower; Writing – Original Draft, J.A. Sharp and M.D. Blower; Writing – Review and Editing, J.A. Sharp and M.D. Blower; Visualization, J.A. Sharp; Supervision, J.A. Sharp and M.D. Blower; Funding Acquisition, M.D. Blower.
References
- Akoulitchev S., and Reinberg D.. 1998. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12:3541–3550. 10.1101/gad.12.22.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J., Lim D., Joughin B.A., Hegemann B., Hutchins J.R., Ehrenberger T., Ivins F., Sessa F., Hudecz O., Nigg E.A., et al. 2011. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4:ra42 10.1126/scisignal.2001796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliagas-Martin I., Burdick D., Corson L., Dotson J., Drummond J., Fields C., Huang O.W., Hunsaker T., Kleinheinz T., Krueger E., et al. 2009. A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B. J. Med. Chem. 52:3300–3307. 10.1021/jm9000314 [DOI] [PubMed] [Google Scholar]
- Bahar Halpern K., Caspi I., Lemze D., Levy M., Landen S., Elinav E., Ulitsky I., and Itzkovitz S.. 2015. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 13:2653–2662. 10.1016/j.celrep.2015.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke E., Ruer M., Wühr M., Coughlin M., Lemaitre R., Gygi S.P., Alberti S., Drechsel D., Hyman A.A., and Mitchison T.J.. 2016. Amyloid-like self-assembly of a cellular compartment. Cell. 166:637–650. 10.1016/j.cell.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M.A., Lan W., Powers S.E., McCleland M.L., Kuang J., and Stukenberg P.T.. 2002. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell. 13:3064–3077. 10.1091/mbc.e02-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.J., Hendrich B.D., Rupert J.L., Lafrenière R.G., Xing Y., Lawrence J., and Willard H.F.. 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 71:527–542. 10.1016/0092-8674(92)90520-M [DOI] [PubMed] [Google Scholar]
- Carmena M., Wheelock M., Funabiki H., and Earnshaw W.C.. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13:789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron-Herger M., Rusin S.F., Adamo M.E., Seiler J., Schmid V.K., Barreau E., Kettenbach A.N., and Diederichs S.. 2019. R-DeeP: proteome-wide and quantitative identification of RNA-dependent proteins by density gradient ultracentrifugation. Mol. Cell. 75:184–199.e10. 10.1016/j.molcel.2019.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F.L., Marshall O.J., Saffery R., Kim B.W., Earle E., Choo K.H., and Wong L.H.. 2012. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. USA. 109:1979–1984. 10.1073/pnas.1108705109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R. III, Chan C.S., Drubin D.G., and Barnes G.. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Clemson C.M., McNeil J.A., Willard H.F., and Lawrence J.B.. 1996. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132:259–275. 10.1083/jcb.132.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognori D., Sunwoo H., Kriz A.J., Wang C.Y., and Lee J.T.. 2019. Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell. 74:101–117.e10. 10.1016/j.molcel.2019.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. 2012. Landscape of transcription in human cells. Nature. 489:101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P., Ye R., Morrice N., Britton S., Trinkle-Mulcahy L., and Lees-Miller S.P.. 2015. Phosphorylation of SAF-A/hnRNP-U serine 59 by Polo-like kinase 1 is required for mitosis. Mol. Cell. Biol. 35:2699–2713. 10.1128/MCB.01312-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Choi Y.D., and Adam S.A.. 1984. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol. Cell. Biol. 4:1104–1114. 10.1128/MCB.4.6.1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Birkhoff G., and Eggan K.. 2008. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat. Rev. Mol. Cell Biol. 9:505–516. 10.1038/nrm2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackelmayer F.O., and Richter A.. 1994. Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry. 33:10416–10422. 10.1021/bi00200a024 [DOI] [PubMed] [Google Scholar]
- Fackelmayer F.O., Dahm K., Renz A., Ramsperger U., and Richter A.. 1994. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur. J. Biochem. 221:749–757. 10.1111/j.1432-1033.1994.tb18788.x [DOI] [PubMed] [Google Scholar]
- Fan H., Lv P., Huo X., Wu J., Wang Q., Cheng L., Liu Y., Tang Q.Q., Zhang L., Zhang F., et al. 2018. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res. 28:192–202. 10.1101/gr.224576.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Tseng B.S., Dormann H.L., Ueberheide B.M., Garcia B.A., Shabanowitz J., Hunt D.F., Funabiki H., and Allis C.D.. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 438:1116–1122. 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- Galupa R., and Heard E.. 2015. X-chromosome inactivation: new insights into cis and trans regulation. Curr. Opin. Genet. Dev. 31:57–66. 10.1016/j.gde.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Gibcus J.H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J., Kanemaki M.T., Xie L., Paulson J.R., Earnshaw W.C., et al. 2018. A pathway for mitotic chromosome formation. Science. 359 eaao6135 10.1126/science.aao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., and Glover D.M.. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682. 10.1083/jcb.152.4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhring F., Schwab B.L., Nicotera P., Leist M., and Fackelmayer F.O.. 1997. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 16:7361–7371. 10.1093/emboj/16.24.7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis J.H., Elbatsh A.M., and Rowland B.D.. 2014. Cohesin and its regulation: on the logic of X-shaped chromosomes. Dev. Cell. 31:7–18. 10.1016/j.devcel.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R., et al. 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21:198–206. 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.L., Byron M., Pageau G., and Lawrence J.B.. 2009. AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J. Cell Biol. 186:491–507. 10.1083/jcb.200811143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E., and Heald R.. 2006. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 1:2305–2314. 10.1038/nprot.2006.396 [DOI] [PubMed] [Google Scholar]
- Hasegawa Y., Brockdorff N., Kawano S., Tsutui K., Tsutui K., and Nakagawa S.. 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell. 19:469–476. 10.1016/j.devcel.2010.08.006 [DOI] [PubMed] [Google Scholar]
- He C., Sidoli S., Warneford-Thomson R., Tatomer D.C., Wilusz J.E., Garcia B.A., and Bonasio R.. 2016. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell. 64:416–430. 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig R., and Fackelmayer F.O.. 2003. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma. 112:173–182. 10.1007/s00412-003-0258-0 [DOI] [PubMed] [Google Scholar]
- Hengeveld R.C., Hertz N.T., Vromans M.J., Zhang C., Burlingame A.L., Shokat K.M., and Lens S.M.. 2012. Development of a chemical genetic approach for human aurora B kinase identifies novel substrates of the chromosomal passenger complex. Mol. Cell. Proteomics. 11:47–59. 10.1074/mcp.M111.013912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Lipp J.J., Toh B.H., and Peters J.M.. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 438:1176–1180. 10.1038/nature04254 [DOI] [PubMed] [Google Scholar]
- Holland A.J., Fachinetti D., Han J.S., and Cleveland D.W.. 2012. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 109:E3350–E3357. 10.1073/pnas.1216880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelga S.C., Vu A.Q., Arnold J.D., Liang T.Y., Liu P.P., Yan B.Y., Donohue J.P., Shiue L., Hoon S., Brenner S., et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 1:167–178. 10.1016/j.celrep.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A., Emerman A.B., Schweidenback C.T., and Blower M.D.. 2014. RNA stimulates Aurora B kinase activity during mitosis. PLoS One. 9 e100748 10.1371/journal.pone.0100748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C.Y., and Salic A.. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA. 105:15779–15784. 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., King E.M., Amaro A.C., Winter J.R., Dorn J.F., Elliott H.L., McHedlishvili N., McClelland S.E., Porter I.M., Posch M., et al. 2010. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 188:665–679. 10.1083/jcb.200909005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.L., Yewdell W.T., Bell J.C., McNulty S.M., Duda Z., O’Neill R.J., Sullivan B.A., and Straight A.F.. 2017. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife. 6 e25299 10.7554/eLife.25299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 467:430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach A.N., Schweppe D.K., Faherty B.K., Pechenick D., Pletnev A.A., and Gerber S.A.. 2011. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4:rs5 10.1126/scisignal.2001497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M., and Dreyfuss G.. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 11:2655–2664. 10.1002/j.1460-2075.1992.tb05331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipp M., Göhring F., Ostendorp T., van Drunen C.M., van Driel R., Przybylski M., and Fackelmayer F.O.. 2000. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 20:7480–7489. 10.1128/MCB.20.20.7480-7489.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarov P.G., Komarova E.A., Kondratov R.V., Christov-Tselkov K., Coon J.S., Chernov M.V., and Gudkov A.V.. 1999. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 285:1733–1737. 10.1126/science.285.5434.1733 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., and Kapoor T.M.. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237. 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J.J., Hoffmann M., Rettig W.J., Kraut N., and Peters J.M.. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17:304–315. 10.1016/j.cub.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Levesque A.A., and Compton D.A.. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154:1135–1146. 10.1083/jcb.200106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K., Woodfin A.R., Slaughter B.D., Unruh J.R., Box A.C., Rickels R.A., Gao X., Haug J.S., Jaspersen S.L., and Shilatifard A.. 2015. Mitotic transcriptional activation: clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol. Cell. 60:435–445. 10.1016/j.molcel.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp J.J., Hirota T., Poser I., and Peters J.M.. 2007. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J. Cell Sci. 120:1245–1255. 10.1242/jcs.03425 [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano M., and Hirano T.. 2002. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16:3004–3016. 10.1101/gad.249202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Gomes A.M., Sousa F., and Barisic M.. 2017. Mechanisms of chromosome congression during mitosis. Biology (Basel). 6 E13 10.3390/biology6010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margreitter C., Petrov D., and Zagrovic B.. 2013. Vienna-PTM web server: a toolkit for MD simulations of protein post-translational modifications. Nucleic Acids Res. 41(Web Server issue, W1). W422-6 10.1093/nar/gkt416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock A.A., Foote K.M., Heron N.M., Jung F.H., Pasquet G., Lohmann J.J., Warin N., Renaud F., De Savi C., Roberts N.J., et al. 2007. Discovery, synthesis, and in vivo activity of a new class of pyrazoloquinazolines as selective inhibitors of aurora B kinase. J. Med. Chem. 50:2213–2224. 10.1021/jm061335f [DOI] [PubMed] [Google Scholar]
- Natsume T., Kiyomitsu T., Saga Y., and Kanemaki M.T.. 2016. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15:210–218. 10.1016/j.celrep.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Naumova N., Imakaev M., Fudenberg G., Zhan Y., Lajoie B.R., Mirny L.A., and Dekker J.. 2013. Organization of the mitotic chromosome. Science. 342:948–953. 10.1126/science.1236083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E.P., Goloborodko A., Valton A.L., Gibcus J.H., Uebersohn A., Abdennur N., Dekker J., Mirny L.A., and Bruneau B.G.. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 169:930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R.-S., Boteva L., Soares D.C., Naughton C., Dun A.R., Buckle A., Ramsahoye B., Bruton P.C., Saleeb R.S., Arnedo M., et al. 2017. SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell. 169:1214–1227.e18. 10.1016/j.cell.2017.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Bukowski-Wills J.C., Sanchez-Pulido L., Alves F.L., Wood L., Chen Z.A., Platani M., Fischer L., Hudson D.F., Ponting C.P., et al. 2010. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 142:810–821. 10.1016/j.cell.2010.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo S., Hara F., Tsuchida Y., Shimotakahara S., Suzuki S., Hatanaka H., Yokoyama S., Tanaka H., Yasuda H., and Shindo H.. 2004. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 279:31455–31461. 10.1074/jbc.M403561200 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Ozdilek B.A., Thompson V.F., Ahmed N.S., White C.I., Batey R.T., and Schwartz J.C.. 2017. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 45:7984–7996. 10.1093/nar/gkx460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palozola K.C., Donahue G., Liu H., Grant G.R., Becker J.S., Cote A., Yu H., Raj A., and Zaret K.S.. 2017. Mitotic transcription and waves of gene reactivation during mitotic exit. Science. 358:119–122. 10.1126/science.aal4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M.T., Veloso A., Prasad J., Bedi K., Ljungman E.A., Magnuson B., Wilson T.E., and Ljungman M.. 2014. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods. 67:45–54. 10.1016/j.ymeth.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C., Bury L., Cheeseman I.M., and Blower M.D.. 2020. Cohesin removal reprograms gene expression upon mitotic entry. Mol. Cell. 78:127–140.e7. 10.1016/j.molcel.2020.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D., Margreitter C., Grandits M., Oostenbrink C., and Zagrovic B.. 2013. A systematic framework for molecular dynamics simulations of protein post-translational modifications. PLOS Comput. Biol. 9 e1003154 10.1371/journal.pcbi.1003154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G., Wei G., Roustan C., Kirmizitas B.A., Solcan N., Cerase A., Castello A., Mohammed S., Moindrot B., Nesterova T.B., et al. 2017. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell. 68:955–969.e10. 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott D.M., and Bender M.A.. 1962. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26:260–268. 10.1016/0014-4827(62)90176-3 [DOI] [PubMed] [Google Scholar]
- Pullirsch D., Härtel R., Kishimoto H., Leeb M., Steiner G., and Wutz A.. 2010. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development. 137:935–943. 10.1242/dev.035956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey F.R., Cramer T., Morphew M.K., Silk A.D., Johnson R.S., McIntosh J.R., and Cleveland D.W.. 2002. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell. 3:351–365. 10.1016/S1534-5807(02)00255-1 [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D., et al. 2017. Cohesin loss eliminates all loop domains. Cell. 171:305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridings-Figueroa R., Stewart E.R., Nesterova T.B., Coker H., Pintacuda G., Godwin J., Wilson R., Haslam A., Lilley F., Ruigrok R., et al. 2017. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 31:876–888. 10.1101/gad.295907.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romig H., Fackelmayer F.O., Renz A., Ramsperger U., and Richter A.. 1992. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 11:3431–3440. 10.1002/j.1460-2075.1992.tb05422.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., and Stukenberg P.T.. 2008. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 319:469–472. 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., and Zhang Y.. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K., Samejima I., Vagnarelli P., Ogawa H., Vargiu G., Kelly D.A., de Lima Alves F., Kerr A., Green L.C., Hudson D.F., et al. 2012. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J. Cell Biol. 199:755–770. 10.1083/jcb.201202155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N., Guermah M., Hoffmann A., Roeder R.G., and Heintz N.. 1996. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10:2389–2400. 10.1101/gad.10.19.2389 [DOI] [PubMed] [Google Scholar]
- Shin J., Kim T.W., Kim H., Kim H.J., Suh M.Y., Lee S., Lee H.T., Kwak S., Lee S.E., Lee J.H., et al. 2016. Aurkb/PP1-mediated resetting of Oct4 during the cell cycle determines the identity of embryonic stem cells. eLife. 5 e10877 10.7554/eLife.10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., and Dalton S.. 2016. Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development. 143:4301–4311. 10.1242/dev.142075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., Wagenbach M., Franck A., Asbury C.L., and Wordeman L.. 2012. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell. 22:1017–1029. 10.1016/j.devcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H., Colognori D., Froberg J.E., Jeon Y., and Lee J.T.. 2017. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. USA. 114:10654–10659. 10.1073/pnas.1711206114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves S.S., An L., Bhargava-Shah A., Xie L., Darzacq X., and Tjian R.. 2018. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife. 7 e35621 10.7554/eLife.35621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther N., Hossain M.J., Politi A.Z., Koch B., Kueblbeck M., Ødegård-Fougner Ø., Lampe M., and Ellenberg J.. 2018. A quantitative map of human Condensins provides new insights into mitotic chromosome architecture. J. Cell Biol. 217:2309–2328. 10.1083/jcb.201801048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H., et al. 2012. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 198:847–863. 10.1083/jcb.201110060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.C., and Chang H.Y.. 2011. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 43:904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Sharp J.A., Kawahara M., Davalos A.R., Difilippantonio M.J., Hu Y., Li W., Cao L., Buetow K., Ried T., et al. 2007. The XIST noncoding RNA functions independently of BRCA1 in X inactivation. Cell. 128:977–989. 10.1016/j.cell.2007.01.034 [DOI] [PubMed] [Google Scholar]
- Xiao R., Tang P., Yang B., Huang J., Zhou Y., Shao C., Li H., Sun H., Zhang Y., and Fu X.D.. 2012. Nuclear matrix factor hnRNP U/SAF-A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Mol. Cell. 45:656–668. 10.1016/j.molcel.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y.. 2015. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 12:7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Beetz N., O’Keeffe S., Tapia J.C., Macpherson L., Chen W.V., Bassel-Duby R., Olson E.N., and Maniatis T.. 2015. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc. Natl. Acad. Sci. USA. 112:E3020–E3029. 10.1073/pnas.1508461112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists SAF-A phosphopeptides identified in interphase and mitosis from purification shown in Fig. 4 b.
contains RIP-seq data for SAF-A IPs in interphase and mitosis.
Data Availability Statement
All sequences and summary tables generated by high-throughput sequencing have been deposited in the Gene Expression Omnibus database under accession no. GSE87200. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ghuhkaawdtkxfwf&acc=GSE87200