G12C inhibitors (G12C-Is) have significant but limited efficacy in KRAS-mutant malignancies, mostly due to “adaptive resistance.” Combining SHP2 inhibitor (SHP2-I) with G12C-I abrogates adaptive resistance; evokes beneficial, tumor-specific changes in the immune microenvironment; and potentiates PD-1 blockade. SHP2-I also has direct, context-dependent effects on tumor vasculature.
Abstract
KRAS is the most frequently mutated human oncogene, and KRAS inhibition has been a longtime goal. Recently, inhibitors were developed that bind KRASG12C-GDP and react with Cys-12 (G12C-Is). Using new affinity reagents to monitor KRASG12C activation and inhibitor engagement, we found that an SHP2 inhibitor (SHP2-I) increases KRAS-GDP occupancy, enhancing G12C-I efficacy. The SHP2-I abrogated RTK feedback signaling and adaptive resistance to G12C-Is in vitro, in xenografts, and in syngeneic KRASG12C-mutant pancreatic ductal adenocarcinoma (PDAC) and non–small cell lung cancer (NSCLC). SHP2-I/G12C-I combination evoked favorable but tumor site–specific changes in the immune microenvironment, decreasing myeloid suppressor cells, increasing CD8+ T cells, and sensitizing tumors to PD-1 blockade. Experiments using cells expressing inhibitor-resistant SHP2 showed that SHP2 inhibition in PDAC cells is required for PDAC regression and remodeling of the immune microenvironment but revealed direct inhibitory effects on tumor angiogenesis and vascularity. Our results demonstrate that SHP2-I/G12C-I combinations confer a substantial survival benefit in PDAC and NSCLC and identify additional potential combination strategies.
Introduction
The RAS/ERK MAPK cascade is among the most frequently affected pathways in human cancer (Ciriello et al., 2013; Kandoth et al., 2013; Zehir et al., 2017). Mutations in genes for pathway components, including receptor tyrosine kinases (RTKs), SHP2, NF1, RAS, or RAF family members or MEK1/2, cause aberrant pathway activation and oncogenesis. RAS (KRAS, HRAS, and NRAS) mutations occur in ∼20% of all human neoplasms (Prior et al., 2020). KRAS is the most-often altered RAS isoform in solid tumors; nearly all pancreatic ductal adenocarcinomas (PDACs), ∼50% of colorectal carcinomas (CRCs), and 25–30% of non–small cell lung cancers (NSCLCs) express mutant KRAS. KRAS mutations almost always (∼95%) affect codons 12, 13, or 61, markedly increase the RAS guanosine triphosphate (GTP)/RAS guanosine diphosphate (GDP) ratio and inappropriately activate effectors (Hobbs et al., 2016; Simanshu et al., 2017).
Mutant RAS was once viewed as impervious to GTPase-activating protein (GAP)–stimulated or intrinsic hydrolysis, “locked” in the GTP state, and “undruggable.” More recent analyses revealed subtle but key differences between RAS mutants in intrinsic and residual GAP-catalyzed GTPase activity, intrinsic and SOS-stimulated exchange, and effector binding (Bandaru et al., 2017; Gremer et al., 2011; Hunter et al., 2015; Johnson et al., 2017; Mazhab-Jafari et al., 2015; Smith and Ikura, 2014). Some oncogenic mutants, notably KRASG12C (G12C) and, to a lesser extent, KRASG12D (G12D), retain significant intrinsic GTPase activity. GTP hydrolysis in G12C is refractory to (and possibly inhibited by) RAS-GAP; G12D, G12A, G12R, G12V, and Q61L/Q61H retain some GAP responsiveness and also might undergo limited KRAS-GTP hydrolysis in cells.
Recent successes in developing clinical-grade G12C inhibitors (G12C-Is) emphasize the importance of these details (Hansen et al., 2018; Janes et al., 2018; Lito et al., 2016; Ostrem and Shokat, 2016; Patricelli et al., 2016). G12C-Is bind an evanescent pocket in KRAS-GDP, positioning a reactive group to couple to the mutant cysteine. Three are in phase I trials (AMG510, MRTX849, and JNJ74699157; Sheridan, 2020), and there are initial reports of efficacy in NSCLC patients with KRASG12C mutations (Canon et al., 2019; Govindan et al., 2019; Hallin et al., 2020; Lindsay and Blackhall, 2019). For these drugs to engage G12C, hydrolysis sufficient to generate RAS-GDP must occur. As G12C is GAP refractory, agents that inhibit exchange (as opposed to enhancing GAPs) could increase occupancy of the KRASG12C-GDP state and enhance the ability of G12C-Is to couple to mutant KRAS. Hence, SOS1/2 can effectively be viewed as competitors of G12C-Is (and vice versa).
SHP2, encoded by PTPN11, comprises two SH2 domains (N-SH2 and C-SH2), a catalytic protein-tyrosine phosphatase (PTP) domain, and a C-terminal domain with two tyrosine residues that, when phosphorylated, bind GRB2. In its “closed” (inactive) state, the N-SH2 occludes the PTP domain, blocking substrate access, while the PTP domain contorts the N-SH2, rendering it unable to bind phosphotyrosyl (pY) peptides (Barford and Neel, 1998; Hof et al., 1998; Ran et al., 2016). Conversely, pY-peptide binding drives SHP2 to the “open” state. RTKs, cytokine receptors, “scaffolding adapters” (e.g., GAB, IRS, and FRS proteins), or immune checkpoint receptors have bis-pY motifs that engage both SHP2 SH2 domains. This elegant “molecular switch” ensures SHP2 activation in response to appropriate signals at proper cellular locales and has been exploited to develop potent, selective, orally available allosteric SHP2 inhibitors (SHP2-Is; Chen et al., 2016; Garcia Fortanet et al., 2016; LaRochelle et al., 2017; Nichols et al., 2018; Ran et al., 2016; Wu et al., 2019). These drugs bind a previously unrecognized pocket in “closed” SHP2, acting as “molecular glue” to impede the N-SH2/loop/C-SH2 movements needed for activation (LaRochelle et al., 2018; Pádua et al., 2018; Ran et al., 2016). Four SHP2-Is are in phase I trials (TNO155, RMC4630, JAB3068, and RLY1971), and an initial efficacy signal has been reported for RMC4630 in KRAS-mutant NSCLC (Ou et al., 2020). SHP2 is required for full activation of RAS and the RAS/ERK cascade, but whether SHP2 regulates RAS exchange or RAS-GAP had been unclear. Recently, several groups, including ours, provided strong evidence that SHP2 acts upstream of SOS1/SOS2 to regulate exchange; consequently, SHP2-Is abrogate adaptive resistance to BRAF or MEK inhibitors (Ahmed et al., 2019; Fedele et al., 2018; Hao et al., 2019; Mainardi et al., 2018; Nichols et al., 2018). Recent reports (and our unpublished observations; see Results) show that KRASG12C mutant cancer cell lines treated with G12C-Is also develop adaptive resistance (Hallin et al., 2020; Lou et al., 2019; Misale et al., 2019; Ryan et al., 2020; Xue et al., 2020). These studies reported that adaptive response to G12C-Is could be minimized by combining G12C-I with RTK or SHP2-Is (Hallin et al., 2020; Lou et al., 2019; Ryan et al., 2020; Xue et al., 2020). Some of these findings were validated in human cell–derived xenografts or patient-derived xenografts (PDXs; Misale et al., 2019).
Tumors are not, however, mere collections of neoplastic cells. Rather, they resemble defective “mini-organs” with complex interactions between cancer cells and cells of the tumor microenvironment (TME), which includes resident and infiltrating immune, mesenchymal, and endothelial cells (Binnewies et al., 2018; Quail and Joyce, 2013). G12C-Is are mutant specific and thus have direct effects only on KRASG12C mutant tumor cells. Nevertheless, they could modulate the TME by altering tumor cell production of growth factors, cytokines, and chemokines (Canon et al., 2019). Most other targeted agents, including SHP2-Is, can affect RAS/ERK signaling in normal as well as neoplastic cells. SHP2 also has effects on parallel pathways (e.g., JAK/STAT signaling) and is implicated as an effector of inhibitory signaling by PD-1 and some other immune checkpoint receptors (Hui et al., 2017; Peled et al., 2018; Yokosuka et al., 2012).
A sophisticated understanding of cancer therapeutics requires delineation of tumor cell–autonomous and nonautonomous actions. Here, we report the effects of G12C-I, SHP2-I, and G12C-I/SHP2-I combinations in syngeneic KRASG12C mutant PDAC and NSCLC models. We find that G12C-I/SHP2-I efficacy derives from the combined effects on tumor cells and cells in the TME, reveal direct antiangiogenic effects of SHP2-Is, and identify additional potential combination strategies.
Results
SHP2 inhibition enhances KRASG12C inhibitor effects in PDAC and NSCLC cell lines
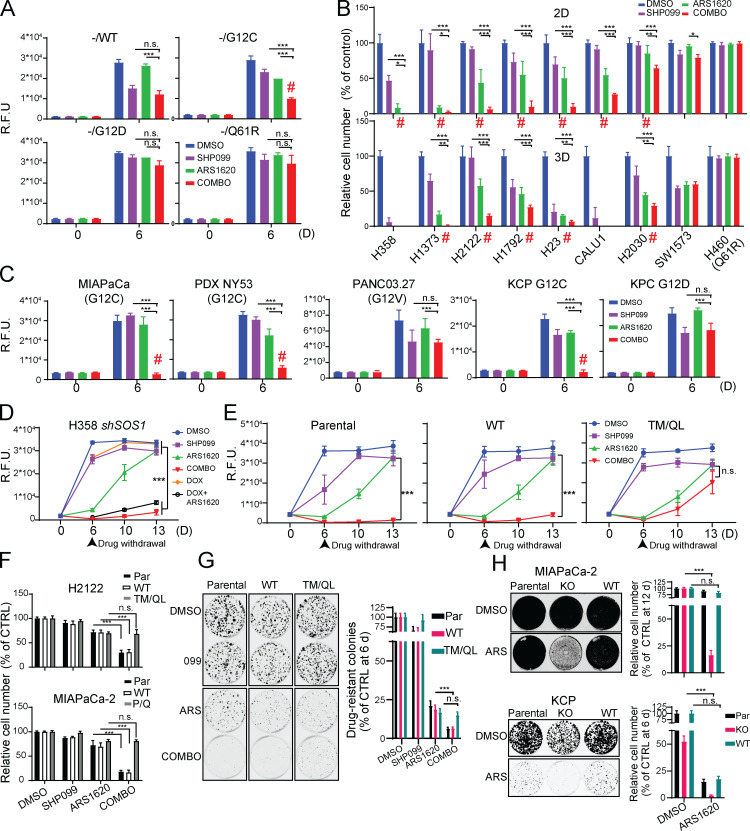
Allosteric SHP2-Is (e.g., SHP099) reduce the activation of KRAS mutants with residual GTPase activity (“cycling mutants”), most notably KRASG12C and, to a lesser extent, KRASG12D and KRASG12V (hereafter G12C, G12D, and G12V) in cancer cell lines and reconstituted “RAS-less” MEFs (Ahmed et al., 2019; Fedele et al., 2018; Hao et al., 2019; Mainardi et al., 2018; Nichols et al., 2018). As G12C is impervious to RAS-GAPs (Hunter et al., 2015), these and other data established that SHP2 acts upstream of SOS1/2. We hypothesized that SHP2 inhibition, by decreasing SOS1/2 activity, would increase occupancy of the KRASG12C-GDP state, thereby potentiating the effect of G12C-Is. We therefore assessed the effects of SHP099, the G12C-I ARS1620 (ARS), SHP099/ARS, or vehicle (DMSO) control on the proliferation of RAS-less MEFs reconstituted with KRAS mutants (Fig. 1 A). Consistent with our previous results (Fedele et al., 2018), SHP099 inhibited WT-reconstituted MEFs, whereas ARS had no effect. By contrast, SHP099 and ARS each inhibited G12C-MEFs to some extent, but SHP099/ARS had far greater efficacy. Neither SHP099 nor ARS alone or in combination significantly impaired the proliferation of G12D- or Q61R-reconstituted cells. ARS-induced adaptive resistance was comparable in Kraswt/KRASG12C and Kras−/−/KRASG12C MEFs (Fig. S1 A), suggesting a more important role for mutant KRAS in promoting adaptive resistance (see Discussion).
Figure 1.
SHP2 inhibition enhances KRASG12C inhibitor effects in PDAC and NSCLC cell lines. (A–C) Reconstituted RAS-less MEF (A), human NSCLC (B), and human or mouse PDAC (C) cell lines were treated as indicated (key at upper right). Cell viability was assessed at 6 d by PrestoBlue assay. (D) Cell viability assays on H358 NSCLC cells expressing DOX-inducible shSOS1 (shSOS), treated as indicated. Drugs were withdrawn after 6 d (arrowhead), and regrowth was quantified. (E) Cell viability assays on H358 NSCLC cells expressing SHP099-resistant PTPN11 mutant (T253M/Q257L) or WT PTPN11 (WT), treated as indicated. Drugs were withdrawn after 6 d (arrowhead), and regrowth was quantified. (F) PrestoBlue assays of H2122 and MIAPaCa-2 cells expressing SHP099-resistant PTPN11 (T253M/Q257L or P491Q respectively) or WT PTPN11 (WT) after 6-d treatment. CTRL, control; PAR, parental. (G) Colony assays (6 d) on KCP cells expressing TM/QL or WT PTPN11 (WT). (H) Colony assays on PTPN11-KO or WT-PTPN11-reconstituted MIAPaCa-2 (12 d, top) or KCP (6 d, bottom) cells. Representative results are shown from a minimum of three biological replicates per condition each with triplicate determinations for each value. Drug doses were SHP099 10 µM, ARS 10 µM, COMBO = SHP099 10 µM + ARS 10 µM. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA followed by Tukey’s multiple comparison test. Red # symbols indicate synergy of by Bliss independent analysis. n.s., not significant; R.F.U., relative fluorescence units.
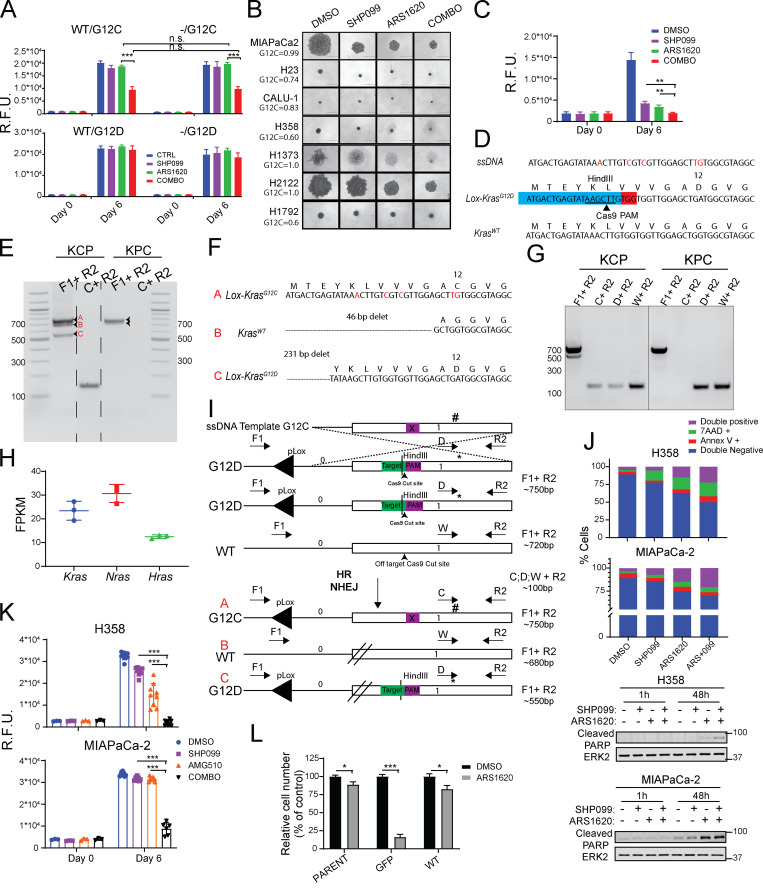
Figure S1.
SHP2 inhibition enhances KRASG12C-inhibitor effects in PDAC and NSCLC cell lines. (A) Cell viability was assessed by PrestoBlue assay at 6 d in Kraswt/KRASG12C and Kras−/−/KRASG12C MEFs. (B) Micrographs of spheroid cultures of KRASG12C-expressing cells treated as indicated for 6 d. Numbers at left indicate KRASG12C allele fraction. Scale bars, 1,000 µm. (C) Cell viability, assessed by PrestoBlue assay, at 0 and 6 d of MIAPaCa-2 spheroid cultures. (D) Schematic showing strategy used to generate the Lox-KrasG12C allele from the pancreatic KCP 1203 cells, which carry a Lox-KrasG12D allele and a pancreas-specific Cre. KCP 1203 cells were cotransfected with a vector expressing Cas9 and a Kras-targeted sgRNA, together with a ssODN template bearing the new mutation. (E) PCR products using F1 + R2 primers to discriminate between mutant Kras (∼750 bp) and WT-Kras (∼720 bp) and C1 + R2 to detect the presence of the new G12C mutation in the KCP (G12C) clone, compared with parental KPC 1203 (G12D) cells. Primer forward F1 flanks the Lox region in intron 0, primer reverse R2 anneals the end of exon 1, and forward C1 specifically anneals on exon 1 in the presence of the new generated G12C mutation. (F) Sanger sequencing of TOPO-cloned PCR products (A–C in red) from E. (G) Allele-specific PCR using forward primers WT (W), G12D (D), and G12C (C) and reverse R2 in KCP G12C clone and parental KPC 1203 (G12D) cells. (H) Fragments per kilobase of exon per million reads mapped (FPKM) for Kras alleles in KCP cells. (I) Summary of genetic events that generated the new Lox-KrasG12C allele starting from parental KCP 1203 cells. (J) Cell death after 48 h of drug treatment, quantified by flow cytometry and Annexin V/7AAD staining (top), and immunoblot for cleaved PARP in lysates from MIAPaCa-2 and H358 cells, treated as indicated (bottom). (K) Viability of MIAPaCa-2 and H358 cells, assessed by PrestoBlue assay, after 6 d of treatment with DMSO, SHP099, AMG510 (0.1 µM) or COMBO. (L) PrestoBlue assays (6 d) on parental MIAPaCa-2 cells and PTPN11-KO MIAPaCa-2 cells reconstituted with GFP or WT PTPN11 (WT). For all experiments, drug doses were SHP099 10 µM, ARS 10 µM, and AMG510 0.1 µM. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA with Tukey’s multiple comparison test. Data represent average of at least three biological replicates with triplicate determinations of each point in each replicate. n.s., not significant; HR, homologous recombination; NHEJ, non-homologous end joining; R.F.U., relative fluorescence units.
Next, we tested KRASG12C mutant NSCLC lines cultured in monolayer (2D) or spheroid (3D) conditions; 3D cultures are more dependent on the RAS/ERK pathway and more sensitive to pathway inhibition (Janes et al., 2018; Patricelli et al., 2016). Single-agent SHP099 or ARS variably inhibited 2D proliferation, and, as expected, generally had greater effects on cells in 3D (Fig. 1 B and Fig. S1 B). Again, however, SHP099/ARS was more effective than either agent alone in nearly all KRASG12C lines. In most cases, the antiproliferative effect of combining the inhibitors was synergistic (Fig. 1 B, red symbols). The KRASG12C line SW1573 failed to respond; notably, these cells express PIK3CAK111E, a known gain-of-function allele, which likely renders them KRAS-mutant independent. As expected, H460 cells, which harbor KRASQ61R, were unresponsive to either the single agent or the combination (Fig. 1 B). SHP099/ARS also had greater ability than SHP099 or ARS alone to inhibit the proliferation of KRASG12C mutant MIAPaCa-2 PDAC cells, cells derived from a KRASG12C mutant patient-derived PDAC xenografts (PDX-NY53), and mouse PDAC cells (KCP) engineered from a KRasG12D/Tp53R172H cell line (KPC) to have a single KrasG12C allele (Fig. 1 C and Fig. S1, B and C) and two inactive Kras alleles (Fig. S1, D–I). RASG12V-expressing PANC03.27 cells and parental KPC cells were inhibited to some extent by SHP099, but as expected, ARS had no effect alone nor any additional effect when combined with SHP099 (Fig. 1 C and Fig. S1, B and C). SHP099/ARS enhanced cell death (measured at 48 h of treatment) compared with either single agent (Fig. S1 J), most likely explaining its increased antiproliferative action. Some newer G12C-Is are more potent than ARS (Canon et al., 2019; Hallin et al., 2020). However, SHP099 also enhanced the effects of the clinical-grade inhibitor AMG510 (Fig. S1 K).
If SHP2 inhibition potentiates G12C-I action by lowering SOS activity, then SOS down-regulation should phenocopy the effects of SHP099. To test this possibility, we generated H358 cells expressing doxycycline (DOX)-inducible shSOS1. Indeed, ARS inhibition and SOS1 shRNA expression had similar effects to SHP099/ARS treatment (Fig. 1 D). Expression of PTPN11T253M/Q257L (TM/QL), a mutant predicted to lack SHP099 binding (Chen et al., 2016), eliminated the effects of SHP099 in combination-treated H358, H2122, and KCP cells (Fig. 1, E–G). Similarly, another drug-resistant mutant, PTPN11P491Q, rescued the effects of SHP099/ARS on MIAPaCa-2 cells (Fig. 1 F). Moreover, ARS had similar effects on PTPN11-KO MIAPaCa-2 and -KCP cells (generated by CRISPR-Cas9) as SHP099/ARS had on parental or PTPN11-reconstituted cells (Fig. 1 H and Fig. S1 L). Hence, SHP099 and ARS are “on-target” and that SHP2 inhibition improves the effect of G12C-Is in multiple KRASG12C mutant cancer cell lines, arising from two tissues.
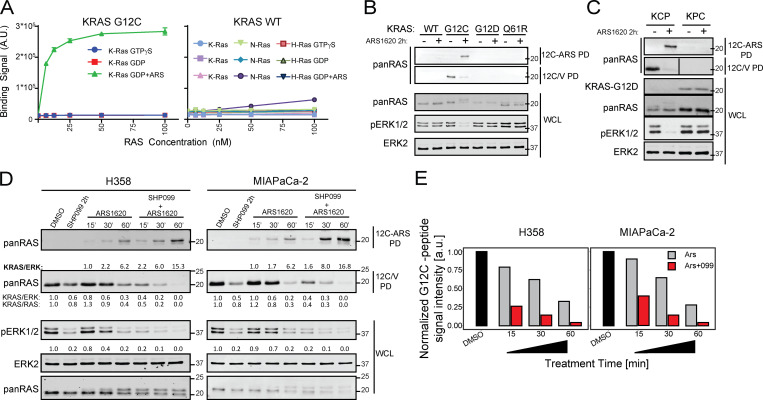
To enable direct assessment of G12C-I action, we used two novel affinity reagents in pull-down (PD) experiments. First, we employed a recently developed “monobody” (MB; 12C/V-MB) that selectively binds KRASG12C-GTP, but not KRASG12C-GDP, and to a lesser extent, KRASG12V (unpublished data). Also, using phage display, we isolated a synthetic antigen-binding fragment (Fab; 12C-ARS-Ab) that specifically recognizes ARS-adducted G12C with high affinity (Fig. 2 A and Fig. S2 A; data not shown; see Materials and methods). To validate these reagents, MEFs reconstituted with WT-KRAS or various KRAS mutants, as well as KCP and KPC mouse PDAC cells, were treated with ARS (2 h) or left untreated, and lysates were subjected to PD assays. ARS treatment lowered KRASG12C-GTP levels, as indicated by decreased RAS signals in the 12C/V-MB PDs from G12C-expressing MEFs and KCP cells, but not from the other lines. Conversely, the RAS signal was increased in 12C-ARS-Ab PDs from RAS-less MEFs reconstituted with KRASG12C, but not other KRAS mutants, as well as from KCP, but not KPC, cells (Fig. 2, B and C). Hence, the 12C/V-MB and 12C-ARS-Ab PD assays provide reciprocal information on the amount of KRASG12C-GTP and KRASG12C-ARS complexes, thereby facilitating investigation of how SHP099 potentiates ARS efficacy.
Figure 2.
SHP099 increases KRASG12C-ARS adducts. (A) 12C-ARS Fab binding to KRASG12C (left) or WT RAS isoforms (right) with/without ARS and GTPγS or GDP. (B and C) Immunoblots of cell lysates and 12C/V MB- or 12C-ARS Fab PDs from RAS-less MEFs reconstituted with the indicated mutants (B) and KCP cells (C), treated as indicated. (D) Immunoblots of cell lysates and 12C/V MB- or 12C-ARS Fab PDs from H358 and MIAPaCa-2 cells, treated as indicated. (E) ARS-adduct formation in samples from C, quantified by LC-MS/MS. ARS and SHP099 concentrations were 10 µM in all panels. Representative results are shown from a minimum of three biological replicates per condition.
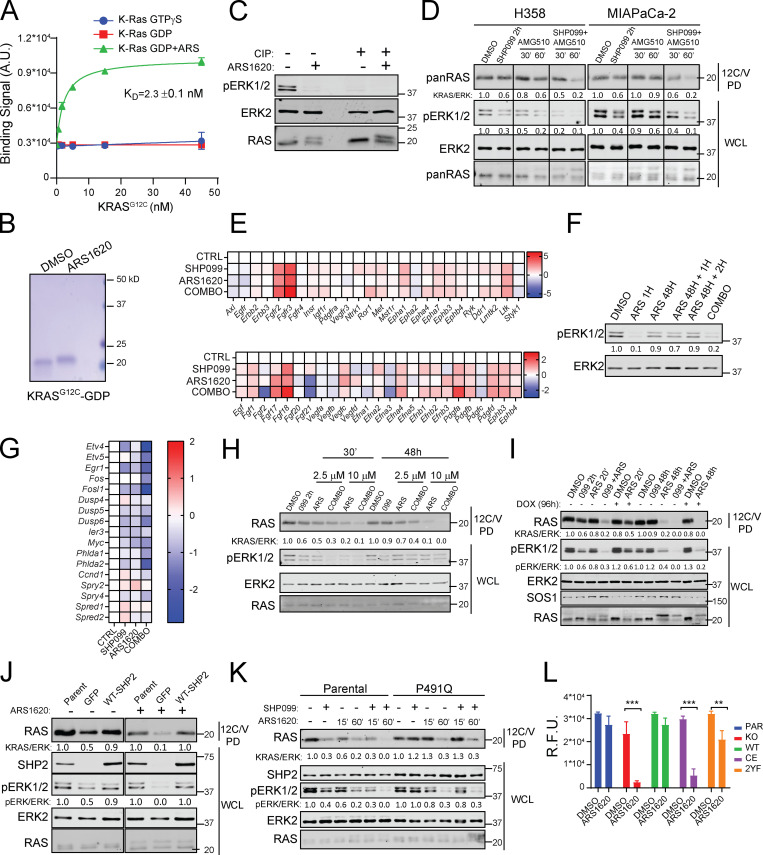
Figure S2.
SHP099 increases KRASG12C-ARS adducts and acts upstream of RAS to block G12C-I–evoked ERK pathway reactivation. (A) 12C-ARS Fab binding to KRASG12C with/without ARS and GTPγS or GDP. (B) Coomassie-stained SDS-PAGE of purified, recombinant KRASG12C, preincubated with DMSO or ARS for 2 h. (C) Immunoblots of WCLs from H358 cells, treated with ARS for 2 h or left untreated, with or without incubation with calf-intestinal phosphatase (CIP). Note that CIP treatment eliminates the pERK signal but does not affect migration of KRAS, arguing against phosphorylation as the cause of the KRAS mobility shift. (D) Immunoblots of WCLs and 12C/V MB PDs from H358 and MIAPaCa-2 cells after treatment with DMSO, SHP099, AMG510, or COMBO, as indicated. For all experiments, drug doses were: SHP099 10 µM, ARS 10 µM, AMG510 0.1 µM. (E) Time-dependent increases in RTK (top) and RTK ligand (bottom) gene expression in KCP cells treated for 48 h with DMSO (CTRL), SHP099, ARS, or COMBO, as determined by RNA-seq (colors indicate log2FC). (F) ERK reactivation, as shown by immunoblot of lysates from MIAPaCa-2 cells treated with DMSO, ARS, or ARS + SHP099 (COMBO) for the indicated times. (G) ERK-dependent gene expression in KCP cells, as assessed by RNA-seq. (H–K) Immunoblots of WCLs and 12C/V MB PDs from KCP cells (H); H358 cells expressing DOX-inducible SOS1 shRNA (shSOS), ±DOX, as indicated (I); parental MIAPaCa-2 cells or MIAPaCa-2 cells PTPN11-KO expressing GFP or reconstituted with WT-PTPN11, treated with ARS for 48 h (J); or MIAPaCa-2 cells expressing SHP099-resistant PTPN11 mutant (P491Q) or WT PTPN11 (WT) treated as described for the indicated times (K). (L) PrestoBlue assays, performed on PTPN11-KO or PTPN11-KO MIAPaCa-2 cells reconstituted with WT, C459E (CE), or Y542F + Y580F (2YF) PTPN11, after 6 d of treatment with ARS or DMSO. For all experiments, drug doses were SHP099 10 µM or ARS 10 µM. Data represent mean ± SD; significance was assessed by multiple unpaired Student’s t test (two tailed). At least two biological replicates with triplicate determinations for each point in each replicate were performed. PAR, parental; R.F.U., relative fluorescence units.
To this end, we treated H358 and MIAPaCa-2 cells with ARS alone, SHP099 alone, or SHP099/ARS for various times and performed PD assays. The inverse recovery of KRAS in 12C/V-MB and 12C-ARS Fab PDs from ARS-treated cell lysates demonstrated time-dependent formation of ARS adducts. SHP099 pretreatment accelerated ARS-adduct formation; for example, ARS engagement of KRASG12 was complete by 1 h in SHP099/ARS–treated cells compared with only ∼70% engagement in cells treated with G12C-I alone (Fig. 2 D). These events were paralleled by more efficient phosphorylated ERK (pERK) inhibition and slower mobility (in SDS-PAGE) of mutant KRAS upon SHP099/ARS treatment (Fig. 2 D and Fig. S2, B and C). Increased G12C-I engagement was also observed in experiments using 12C/V-MB PDs on lysates from AMG510- and AMG510/SHP099–treated MIAPaCa-2 or H358 cells (Fig. S2 D). We verified these findings with the current gold standard MS assay (Patricelli et al., 2016), which measures a decrease in C12–containing peptide relative to isotopic standards (G12C peptide, LVVVGACGVGK; KRAS/NRAS normalization peptide, SYGIPFIETSAK; both spiked into lysates) in tryptic digests of ARS- or SHP099-treated cell lysates; Fig. 2 E). These results, along with the known biochemical properties of G12C (retained intrinsic GTPase, GAP nonresponsive), provide additional, incontrovertible evidence that SHP2-Is impede RAS-GEF action.
SHP099 abrogates adaptive resistance to G12C-Is in vitro
MEK-I treatment of KRAS-mutant tumors fails, at least in part due to induction of genes encoding multiple RTKs and/or their ligands, which differ between tumors even of a single histotype (Hymowitz and Malek, 2018; Manchado et al., 2016; Ryan and Corcoran, 2018; Sun et al., 2014). We (Fedele et al., 2018) and others (Ahmed et al., 2019; Hao et al., 2019; Mainardi et al., 2018; Nichols et al., 2018; Ruess et al., 2018; Wong et al., 2018) reported that SHP2-Is, by blocking RAS activation evoked by MEK-I–induced RTKs/RTK ligands, prevent adaptive resistance to MEK-Is and that SHP2-I/MEK-I combinations synergistically inhibited the proliferation of multiple KRAS-mutant cancer models. We analyzed RTK and RTK ligand gene expression in ARS-treated MIAPaCa-2 and H358 cells by quantitative RT-PCR (qRT-PCR; Fig. 3 A). Several (but different) RTKs were induced by G12C-I treatment, including EGFR, FGFR3, IGFR1, MET, VEGFR1, and PDGFRA/B in MIAPaCa-2 cells and ERBB2/3, FGFR2/3, and PDGFRA/B in H358 cells. The same lines variably induced EGF, FGF2, PDGFB, PDGFC, PDGFD, and/or VEGFA/B RNA. Consequently, it is difficult, if not impossible, to design an efficient G12C-I/RTK inhibitor combination therapy. Notably, the RTK/RTK ligand genes induced by G12C-I treatment were similar, but not identical, to those evoked by MEK-I (Fig. 3 A). Analogous results were obtained in studies of ARS-treated mouse KCP cells (Fig. S2 E).
Figure 3.
SHP2-I acts upstream of RAS to abrogate G12C-I–evoked ERK–MAPK pathway reactivation. (A) Heat map showing increases in RTK/RTK ligand gene expression in MIAPaCa-2 (M) and H358 (H) cells after the indicated treatments for 48 h, determined by qRT-PCR. (B) Immunoblots of WCLs from KRASG12C-expressing cells, treated as indicated in 2D or 3D conditions. (C) ERK-dependent gene expression (ETV1, ETV4, ETV5, and DUSP6), as assessed by qRT-PCR, in KRASG12C lines treated as indicated. (D) SHP099 blocks RAS/ERK reactivation after 48-h ARS treatment of H358 and MIAPaCa-2 cells, as assessed by 12C/V MB PD. (E) Immunoblots of WCL and 12C/V MB PDs from parental or PTPN11-KO MIAPaCa-2 cells treated as indicated. (F) Immunoblots of WCL and 12C/V MB PDs from KCP cells or Ptpn11-KO KCP cells with or without reconstitution with WT-PTPN11, treated as indicated. (G) SHP2, pERK, and ERK immunoblots from MIAPaCa-2 and H358 cells ectopically expressing WT SHP2 (WT) or an SHP099-resistant mutant (P491Q or T253M/Q257L, respectively), treated as indicated. (H) Colony assays (12 d) on parental, PTPN11 KO MIAPaCa-2, or PTPN11 KO MIAPaCa-2 cells reconstituted with WT, phosphatase-inactive C459E (CE), or C-terminal tyrosine phosphorylation site-defective Y542F+Y580F (2YF) PTPN11 mutants, treated as indicated. (I) Immunoblots of WCLs and 12C/V MB PDs from parental, PTPN11-KO MIAPaCa-2, or PTPN11 KO cells reconstituted with WT, C459E (CE), or Y542F+Y580F (2YF) PTPN11, treated as indicated. All data are representative of at least two independent biological replicates. Drug doses were SHP099 10 µM and ARS 10 µM. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA followed by Tukey’s multiple comparison test. Numbers under blots indicate relative intensities, compared with untreated controls, quantified by LI-COR. A.U. and a.u., arbitrary units; ns, not significant.
To probe the mechanism of adaptive resistance to G12C-Is, we assessed the biochemical effects of each single agent and the drug combination on RAS/ERK pathway activity after brief (1 h) and longer-term (48 h) treatments. Single-agent ARS blocked ERK1/2 phosphorylation after 1 h in cells grown in 2D or 3D, but these effects were abolished after 48 h of treatment (Fig. 3 B). Addition of fresh ARS to MIAPaCa-2 cells after 48 h did not prevent pERK rebound (Fig. S2 F), indicating that loss of pathway inhibition did not reflect drug metabolism or instability. By contrast, coadministration of SHP099 prevented the ARS-induced adaptive increase in ERK phosphorylation (Fig. 3 B and Fig. S2 F). ERK-dependent gene expression can provide a better assessment of RAS/ERK pathway output than pERK levels (Pratilas et al., 2009), so we measured key ERK-dependent genes in a panel of human G12C lines (by qRT-PCR) and in KCP cells (by RNA sequencing [RNA-seq]). Compared with either single agent, SHP099/ARS more effectively suppressed ERK-dependent transcripts (Fig. 3 C and Fig. S2 G). Mutant KRAS also was reactivated after 48-h treatment with ARS (as revealed 12C/V-MB PDs); presumably, so were endogenous WT RAS isoforms. SHP099 blocked the adaptive increase in KRASG12C-GTP (Fig. 3 D and Fig. S2 H), as did SOS1 knockdown (Fig. S2 I). PTPN11 deletion had similar biochemical effects as SHP2 inhibition (Fig. 3 E), whereas reexpressing WT SHP2 restored adaptive resistance to ARS and sensitivity to SHP099 (Fig. 3 F and Fig. S2 J), showing that the effects of SHP099 were on target. The biochemical effects of SHP099 (like its effects on viability; Fig. 1, E–G) were reversed in MIAPaCa-2 cells expressing PTPN11P491Q (Fig. 3 G and Fig. S2 K) and PTPN11TM/QL-expressing H358 cells (Fig. 3 G). Hence, mutant KRAS is reactivated in G12C-I–treated cells, leading to RAS/ERK pathway reactivation, and SHP2-I, by impeding SOS1/2 activation, blocks this adaptive response.
SHP2 catalytic activity is required for RAS/ERK pathway activation by most, if not all, RTKs, but its C-terminal tyrosyl residues are essential for only some RTK signaling pathways (Araki et al., 2003; O’Reilly and Neel, 1998; Yamauchi et al., 1995). Reconstituting PTPN11-KO MIAPaCa-2 cells with WT PTPN11, but not a phosphatase-inactive mutant, PTPN11C459E (CE), restored ARS-induced adaptive resistance. SHP2 lacking both C-terminal tyrosine phosphorylation sites (PTPN11Y542F/Y580F, 2YF) partially restored adaptive resistance (Fig. 3, H and I; and Fig. S2 L). Thus, as in RTK signaling, PTP activity is essential, whereas C-terminal tyrosine residues play a modulatory role, in ARS-invoked activation of RTK signaling.
Combined SHP2/ARS inhibition is efficacious in PDAC models in vivo
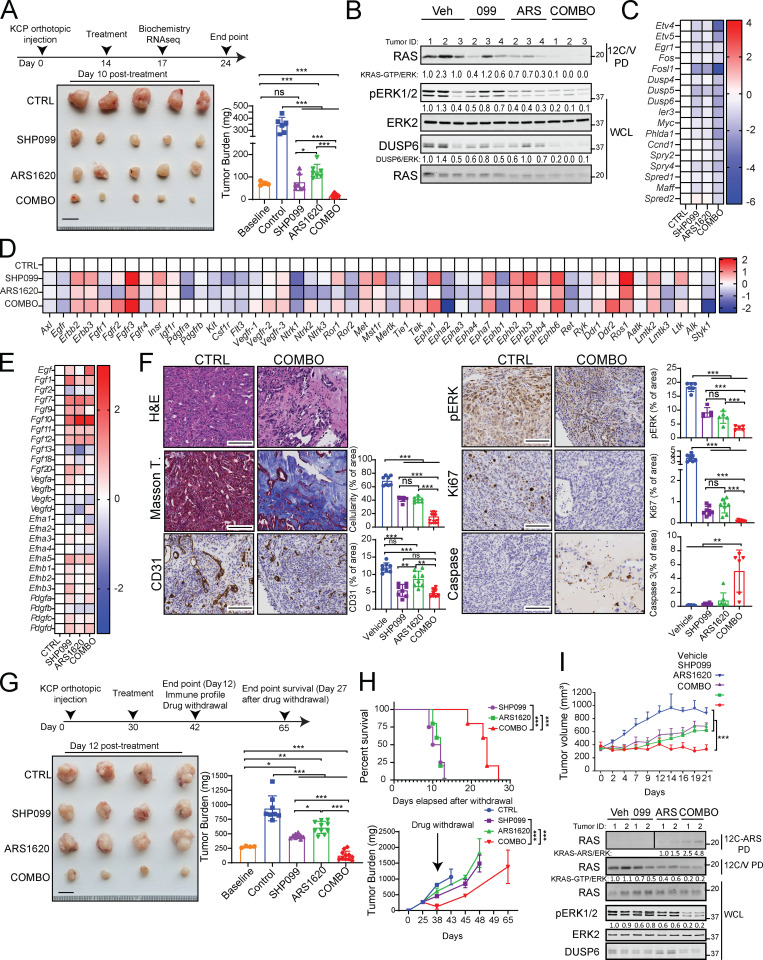
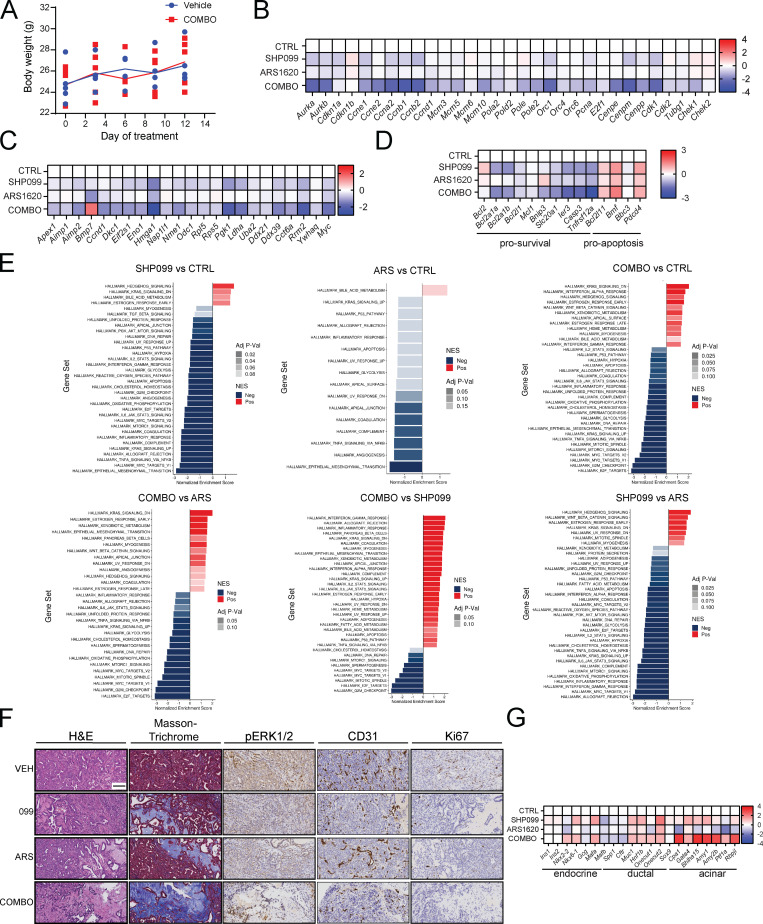
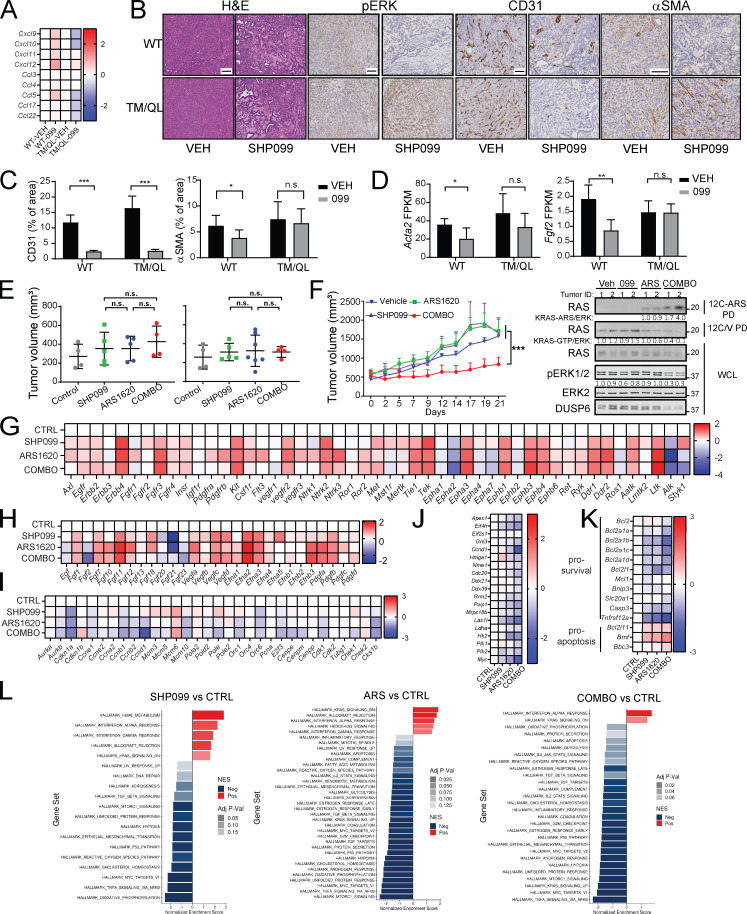
We next assessed the effects of ARS (200 mg/kg/d), SHP099 (75 mg/kg/d) or SHP099/ARS (full doses of each) on orthotopic KCP tumors in syngeneic C57BL6 mice (Fedele et al., 2018). Tumors were allowed to grow for 14 d; four mice were sacrificed to obtain baseline tumor sizes (average 100 mm3), and the rest were treated with single agent or SHP099/ARS for 3 or 10 d, respectively. After 10 d, control tumors (vehicle) had, on average, quadrupled in mass compared with the average pretreatment baseline. Single agents largely caused tumor stasis, although SHP099 treatment was more efficacious. By contrast, all tumors in the SHP099/ARS arm regressed markedly (Fig. 4 A). Treated mice showed no evident toxicity, and blood counts were normal (Fig. S3 A and data not shown). Immunoblot analysis of tumor lysates after 3 d of treatment revealed greater inhibition of KRASG12C, pERK, and the ERK-induced protein DUSP6 following combination treatment (Fig. 4 B). RNA-seq showed that compared with vehicle or either single agent, SHP2-Is/G12C-Is enhance the suppression of ERK, MYC, antiapoptotic, and cell cycle genes while increasing proapoptotic genes (Fig. 4 C and Fig. S3, B–E). In accord with the above biochemical findings, SHP099/ARS showed greater effects on KRAS signaling gene sets than either single agent (Fig. S3 E, compare agent combinations [COMBO] versus ARS and COMBO versus SHP099). Notably, single-agent ARS inhibited pERK and ERK-dependent gene expression at least as well as did SHP099, although SHP099 showed greater inhibition of KRAS pathway gene sets (Fig. S3 E, compare SHP099 versus control [CTRL], ARS versus CTRL, and SHP099 versus ARS). Nevertheless, these findings, together with the greater efficacy of SHP099 versus ARS, raised the possibility of effects of SHP2 inhibition either on tumor cells themselves or on the TME. Indeed, pathway analysis revealed effects of SHP099 on gene sets related to IL-6 JAK/STAT3 signaling, TNF-α signaling, IFN-γ response, and angiogenesis (also see below), among several others (Fig. S3 E). RNA-seq also showed that several RTKs and RTK ligands were induced by G12C-I (within 3 d of treatment, when the RNA samples were obtained); thus, adaptive resistance via RTK overactivation clearly occurs in vivo. SHP099 also induced multiple RTK/RTK ligands, but while there was substantial overlap with ARS effects, several genes were affected differently (qualitatively and quantitatively) by each agent (Fig. 4 D).
Figure 4.
Combined ARS/SHP2 inhibition is highly efficacious in PDAC models in vivo. (A) Scheme showing establishment of pancreas tumors by orthotopic injection of KCP cells into syngeneic mice, followed by treatment with vehicle (n = 6), SHP099 (n = 7), ARS (n = 7), or both drugs (COMBO; n = 9). Tumor weight was quantified in a cohort of mice at day 0 (baseline; n = 4) and in treated mice at day 10. Scale bar, 1 cm. (B) Immunoblots of KCP-derived tumor lysates showing effects of the indicated treatments on KRASG12C-GTP, pERK, and DUSP6 levels. (C–E) ERK-dependent (C), RTK (D), and RTK ligand (E) gene expression, assessed by RNA-seq, in KCP tumors treated for 3 d as in A (colors indicate log2FC, n = 3 per each group). (F) H&E, Masson Trichome, CD31, pERK, Ki67, and cleaved caspase-3 staining and quantification in KCP tumors from mice after 10 d of treatment, as indicated (n = 3 per each group). Scale bars, 100 µm. (G) Scheme showing establishment and treatment of larger KCP tumors. Tumor weight was quantified in one cohort before treatment, another cohort after 12 d of treatment, and after drug withdrawal, at day 27, as indicated. Data were pooled from two independent experiments. Scale bars, 1 cm. (H) Kaplan–Meier curves of KCP tumor-bearing mice after withdrawal of the indicated drugs (top). Tumor growth curves after withdrawal of the indicated treatment at day 12 (bottom). Data were pooled from two independent experiments. (I) Response of SQ NY53 PDXs to the indicated treatments (n = 6/group). For all experiments, doses were SHP099 (75 mg/kg body weight, daily), ARS (200 mg/kg body weight, daily), or both drugs (daily). Veh, vehicle. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA with Tukey’s multiple comparison test. For Kaplan–Meier curves, a log-rank test was used. Numbers under blots indicate relative intensities compared with untreated controls, quantified by LI-COR. ns, not significant.
Figure S3.
ARS/SHP099 combination is efficacious in PDAC model in vivo. (A) ARS/SHP099 regimen is well tolerated in KCP-derived orthotopic tumors, with no significant decrease in body weight after 12 d of treatment (n = 4). (B–D) Cell cycle (B), MYC target (C), and apoptosis (D) gene expression in KCP-derived orthotopic tumors after vehicle (n = 3), SHP099 (n = 3), ARS (n = 3), or COMBO (n = 3) treatment for 3 d, as determined by RNA-seq (colors indicate log2FC). (E) Pathway analysis using MSigDB Hallmark genes, ranked by fold change between the indicated groups. (F) H&E, Masson trichrome, CD31, pERK, and Ki67 staining of sections from KCP tumors treated for 10 d with vehicle (VEH), SHP099, ARS, or COMBO (20× magnification). Scale bar, 100 µm. (G) Pancreatic epithelial lineage–specific gene expression in control (CTRL) and treated KCP-derived orthotopic tumors, determined by RNA-seq (colors indicate log2FC, n = 3). For all experiments, drug doses were SHP099 75 mg/kg body weight (daily), ARS 200 mg/kg body weight (daily), or both drugs (daily).
H&E-stained sections of tumors from SHP099/ARS–treated mice revealed markedly increased collagenized stroma with scattered histiocytes, histiocytic giant cells, hemosiderin-laden histiocytes, and lymphocytes, compared with the effects of vehicle or single agents (Fig. 4 F and Fig. S3 F). Residual cancer cells were widely spaced with scattered glands in COMBO-treated tumors, unlike the solid sheets of malignant cells seen in control tumors. Masson trichrome staining confirmed the increase in collagen and diminished cellularity. As we reported previously (Fedele et al., 2018), SHP099 decreased KCP tumor vascularity, as shown by CD31 immunostaining. Also similar to our previous findings on SHP099/MEK-I–treated KPC tumors, residual tumor cells in SHP099/ARS–treated (but not single-agent– or vehicle-treated) mice showed ductal/acinar differentiation (Fig. S3 F). Consistent with these observations, ductal, acinar, as well as endocrine, genes were induced (Fig. S3 G), while epithelial–mesenchymal transition genes sets were decreased (Fig. S3 E, compare COMBO versus CTRL). Immunohistochemical (IHC) analysis confirmed more profound pERK inhibition in tumors from SHP099/ARS– compared with single-agent–treated mice, as well as decreased proliferation and increased apoptotic cell death (Fig. 4 F and Fig. S3 F).
To evaluate efficacy more stringently, we allowed KCP tumors to grow to 250 mm3 before initiating treatment. Single agents again inhibited tumor growth, but SHP099/ARS caused dramatic tumor regression (Fig. 4 G). When treatment was stopped after 12 d, tumors recurred in all groups, but regrowth of SHP099/ARS–treated tumors was delayed, and median survival of this cohort was more than doubled compared with single-agent–treated mice (Fig. 4 H). SHP099/ARS also was more effective than either single agent in inhibiting tumor growth (Fig. 4 I, top panel) and RAS/ERK pathway activation (Fig. 4 I, bottom panel) in a highly aggressive KRASG12C PDX model.
SHP099/ARS evokes an antitumor immune program and is potentiated by anti–PD-1
The PDAC TME features abundant immune-suppressive myeloid cells and regulatory T cells (T reg cells) and scarce cytotoxic lymphocytes (Clark et al., 2007; Vonderheide and Bayne, 2013). Consequently, these tumors are often termed immunologically “cold.” SHP2 and the RAS/ERK pathway have roles in most, if not all, TME cells, often affecting several signaling pathways. G12C-Is, owing to their mutant specificity, affect only cancer cell signaling but, by altering growth factor/cytokine/chemokine production, they could also affect the PDAC TME.
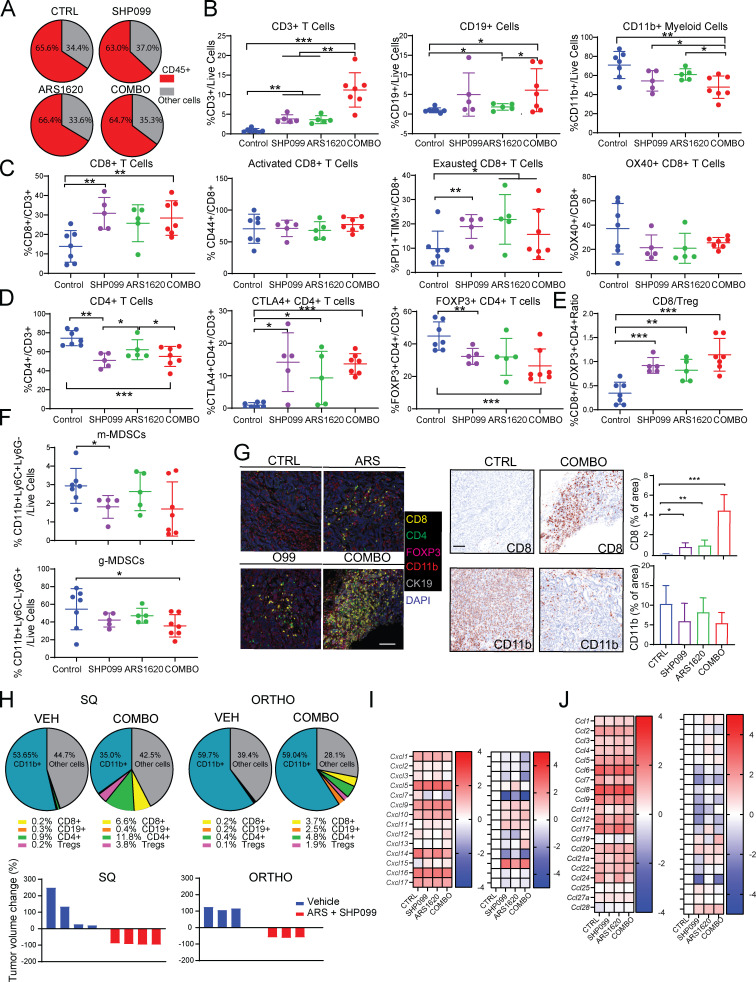
We surveyed the immune composition of the KCP TME using the tumors from Fig. 4 G. Although the percentage of CD45+ cells was unchanged in tumors from ARS-, SHP099-, and SHP099/ARS–treated mice (Fig. 5 A), the composition of the CD45+ population was altered (Fig. 5, B–F). Total T lymphocytes (percentage of CD3/live cells) were increased in single-agent– and, more substantially, SHP099/ARS–treated groups (Fig. 5 B). Each single agent also increased B lymphocytes (although only nominally for SHP099), and this increase was preserved in SHP099/ARS–treated mice. By contrast, there was a trend toward decreased total CD11b+ myeloid cells, mostly comprising granulocytic myeloid-derived suppressor cells (g-MDSCs), following SHP099 alone (P = 0.11), and a nearly 50% decrease (P = 0.045) after SHP099/ARS (Fig. 5, B and F). Subset analyses revealed that single-agent, and especially combination, treatment preferentially increased CD8 T cells (as percentage of total T cells), but these cells exhibited markers (TIM3, PD-1, and OX40) consistent with “exhaustion” (Fig. 5 C). Conversely, CD4 cells and T reg cells decreased; consequently, CD8/T reg cell ratios increased, most prominently after SHP099/ARS (Fig. 5, D and E). We also examined the spatial distribution of immune cells in the TME by multicolor immunofluorescence (IF) and IHC. Although there were regional differences, single agents significantly increased intratumor lymphocytes, but SHP099/ARS treatment evoked greater CD8 T cell immigration (Fig. 5 G).
Figure 5.
ARS/SHP099 combination provokes an antitumor immune program in syngeneic PDAC model. (A) Pie charts showing %CD45+ (immune) cells and %CD45- (cancer plus stromal) cells in KCP tumors after 12 d of treatment, as in Fig. 4. (B) Frequencies of infiltrating CD3+ T cells, CD19+ B cells, and CD11b+ myeloid cells. (C) Frequencies of infiltrating CD8+ T cells and respective subpopulations. (D) Frequencies of infiltrating CD4+ T cells and respective subpopulations. (E) Ratio of infiltrating CD8+ T cells to FOXP3+ regulatory CD4+ T cells. (F) Frequencies of infiltrating MDSCs. Data were verified in at least two independent experiments for each subset. (G) Multiplex IF/IHC analysis of KCP tumors, stained with the indicated markers and quantified (n = 3 per each group). Scale bar, 100 µm. For B–G, tumors were analyzed at day 12 after the indicated treatments. CTRL, control. (H) Pie charts showing immune cell (CD45+) composition (top) and tumor volume (bottom) in SQ versus orthotopic (ORTHO) KCP tumors, treated with vehicle (VEH) or ARS + SHP099 (COMBO) for 10 d (n = 3–4/group). Note the greater response of SQ tumors. (I and J) CXCL (I) and CCL (J) chemokine expression in KCP tumors after 3 d of treatment, as assessed by RNA-seq (colors are log10 of raw counts averages and log2FC [left], n = 3 per group [right]). Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA with Tukey’s multiple comparison test.
Single-agent AMG510 caused an ∼50-fold increase in intratumor CD3+ and CD8+ T cells in subcutaneous (SQ) xenografts of KRASG12C-engineered CT26 CRC cells (Canon et al., 2019), but T cells were more modestly increased in ARS-treated orthotopic KCP tumors (Fig. 5 H). To ask whether this difference might reflect the distinct location of the tumors, we compared the efficacy of SHP099/ARS in mice with orthotopic or SQ KCP tumors. Notably, SQ tumors showed a more robust antitumor T cell response and much greater regression (Fig. 5 H).
SHP099 and G12C-I each increased the expression of chemokine and cytokine genes that promote T cell recruitment (e.g., CXCL9-11; CCL5; Araujo et al., 2018; Chow and Luster, 2014; Lavergne et al., 2004; Nagarsheth et al., 2017; Tokunaga et al., 2018) while decreasing the expression of those that favor immune-suppressive CD11b+ myeloid cells (e.g., CXCL1-5; CCL9; Chow and Luster, 2014; Kortlever et al., 2017; Liao et al., 2019; Nagarsheth et al., 2017; Fig. 5, I and J). SHP099/ARS evoked much greater differential expression of these genes, which likely accounts, at least in part, for the more favorable immune modulatory effects of the combination. The increase in PD-1+ (potentially “exhausted”) T cells in SHP099/ARS–treated mice suggested a possible benefit of adding PD-1 blockade. Indeed, SHP099/ARS/PD-1 resulted in even greater regression than SHP099/ARS or either single agent plus anti–PD-1 (Fig. 6 A). H&E- and Masson trichrome–stained sections of tumors from SHP099/ARS/PD-1–treated mice revealed large areas of collagen scarring and only scattered residual cancer (Fig. 6 B). Residual tumors were too small for flow cytometry, so we used multiple IF to assess the immunological consequences of the “triple” combination (Fig. 6 C). Notably, many more CD8+ T cells penetrated into KCP tumors treated with the triple combination (COMBO + αPD-1) compared with SHP099/ARS (COMBO + IgG). These cells showed lower expression of PD-1, expressed the cytolytic marker granzyme B, and directly contacted tumor cells (CK19+).
Figure 6.
ARS/SHP099 efficacy is enhanced by anti–PD-1 in the PDAC model. (A) Syngeneic mice bearing KCP tumors treated with vehicle + isotype IgG (200 µg/mouse three times/week; n = 6), SHP099 (75 mg/kg body weight, daily) + anti–PD-1 (200 µg/mouse three times/week; n = 5), ARS (200 mg/kg body weight, daily) + anti–PD-1 (200 µg/mouse three times/week; n = 5), ARS+SHP099 (COMBO; daily) + isotype IgG (200 µg/mouse three times/week; n = 9), or COMBO (daily) + anti–PD-1 (200 µg/mouse three times/week; n = 9), as depicted. Tumor weights were measured at day 0 (baseline) and day 12. Rightmost panel shows expanded scale for the indicated treatments from the middle panel. (B) H&E, Masson trichrome, and Ki67 staining and quantification of sections from orthotopic KCP tumors, analyzed after treatments in A. Bottommost panel shows expanded scale for the indicated treatments from the panel above (n = 3 per group). Scale bars, 10 µm. (C) Multiplex IF analysis of KCP tumors, after 12-d treatment as indicated, stained with the indicated markers and quantified at right (n = 3 per group). Scale bar, 50 µm. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; one-way ANOVA with Tukey’s multiple comparison test.
Effects of SHP2 inhibition in PDAC cancer cells and the TME
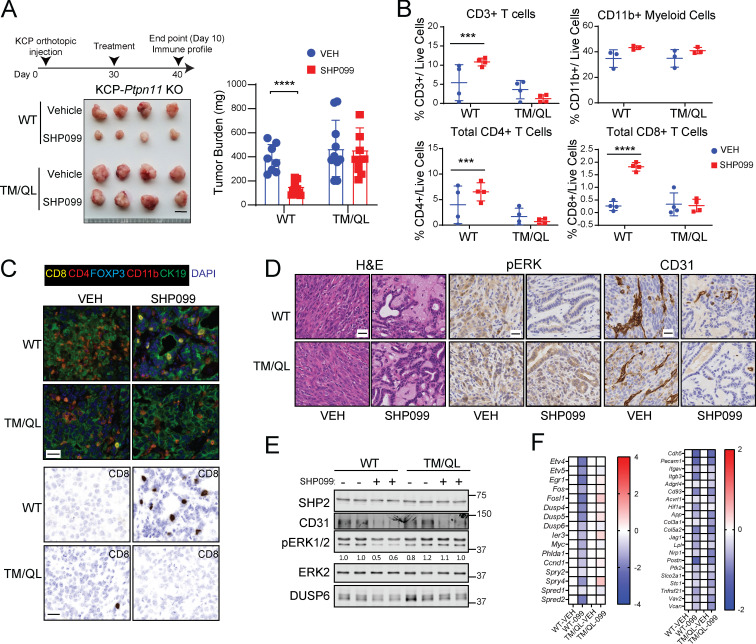
To begin to define direct effects of SHP099 on tumor cells versus indirect effects on cells in the TME, we established orthotopic tumors of Ptpn11-KO KCP cells reconstituted with PTPN11 or the SHP099-resistant mutant TM/QL. As expected, SHP099 (for 10 d) suppressed PTPN11-reconstituted KCP tumor growth, but TM/QL-reconstituted tumors failed to regress (Fig. 7 A). As in parental KCP tumors, SHP099 evoked an influx of CD8 and CD4+ T cells in PTPN11-reconstituted KCP tumors. This influx also was abrogated in TM/QL-KCP tumors (Fig. 7, B and C), most likely because TM/QL tumors failed to alter their expression of chemokines and cytokines that likely mediate the T cell infiltration observed in mice bearing parental KCP tumors or WT-reconstituted, KCP-KO tumors (Fig. S4 A). Therefore, SHP099 (and, presumably, SHP099/ARS) must alter signaling in, and induce death of, KCP cells to evoke changes in the immune microenvironment. Nevertheless, direct effects of SHP099 on immigrating immune cells still might be required for the antitumor response (see Discussion).
Figure 7.
Tumor cell–autonomous and nonautonomous effects of SHP2 inhibition in PDAC. (A) Tumors were established in syngeneic mice by orthotopic injection of Ptpn11-KO KCP cells reconstituted with WT or SHP099-resistant TM/QL mutant and treated with vehicle or SHP099 (75 mg/kg body weight, daily), as depicted. Tumor weights were measured at day 10. Scale bar, 1 cm. (B) Tumor-infiltrating immune cells from experiment in A (n = 4). (C) Multiplex IF/IHC analysis of representative tumors from A. Scale bars, 20 µm. (D) H&E, pERK, and CD31 staining of representative KCP tumors from A (n = 6). Scale bars, 10 µm. (E) Immunoblot showing CD31, pERK, and DUSP6 levels in representative tumors. (F) ERK-dependent and angiogenesis gene expression, as assessed by RNA-seq, in KCP tumors from A (n = 5; colors indicate log2FC). Data were pooled from two independent experiments. Data represent mean ± SD; ***, P < 0.001; ****, P < 0.0001; significance was assessed by multiple unpaired Student’s t test (two tailed).
Figure S4.
Tumor cell–autonomous and –nonautonomous effects of SHP2 inhibition and ARS/SHP099 efficacy in NSCLC GEMMs. (A) Expression of chemokines potentially involved in T cell immigration in tumors from Ptpn11-KO KCP cells reconstituted with WT or SHP099-resistant TM/QL mutant, treated for 10 d with vehicle (n = 5) or SHP099 (75 mg/kg body weight daily; n = 5). (B) H&E, pERK, CD31, and αSMA staining of sections from KCP tumor, established as in Fig. 7 A (n = 3/group). Scale bars, 100 µm. (C) CD31 and αSMA quantification from sections KCP tumors established as in B. (D) FKPM for Acta2 (top) and Fgf2 (bottom) in RNA from KCP tumors established as in Fig. 7 A (n = 5/group). (E) Baseline tumor volumes for KCP (left) and KC (right) mice before accrual to the indicated treatments. (F) Growth of H2122 cell–derived xenografts (left) and immunoblots (right) of tumor lysates and 12C/V MB or 12C-ARS Fab PDs from mice treated as indicated (n = 4/group). (G–J) Time-dependent expression of RTK (G), RTK ligand (H), cell cycle (I), MYC target (J), and apoptotic (K) genes in LSL-KRASG12C-Tp53R270H tumors after vehicle (n = 3), SHP099 (n = 3), ARS (n = 3), or COMBO (n = 3) treatment for 3 d, as assessed by RNA-seq (colors indicate log2FC). (L) Pathway analysis of MSigDB Hallmark genes ranked by fold change between the indicated groups. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test (two tailed). n.s., not significant; NES, normalized enrichment score.
Although SHP099 did not decrease TM/QL tumor size, histological examination revealed substantial tumor necrosis, replacement with eosinophilic material, and more duct-like epithelial architecture (Fig. 7 D). IHC confirmed clear rescue of pERK staining, and RNA-seq showed that ERK target gene expression was unaffected in TM/QL tumors from SHP099-treated mice, confirming that this mutant was, as expected, SHP099 resistant and that TM/QL tumor cells were unaffected by SHP099. Nevertheless, TM/QL-KCP tumors, like their WT-reconstituted (and parental KCP) counterparts, were markedly hypovascular, as shown by reduced CD31 staining and decreased angiogenic gene expression (Fig. 7, D–F; and Fig. S4, B and C). Hence, in addition to its effects on tumor cells, SHP099 has direct antiangiogenic actions. SHP099 also reduced the number of activated fibroblasts in KCP-WT tumors, as indicated by αSMA staining and Acta2 expression, but these effects were reversed in TM/QL-KCP tumor–bearing mice (Fig. S4, B–D). Therefore, the ability of SHP099 to modulate tumor-associated fibrosis requires inhibitor action in tumor cells, presumably to lower production of secreted factors that act on stromal fibroblasts. Intriguingly, Fgf2 levels decreased after SHP099 treatment in WT-KCP tumor–bearing mice, and levels were restored in TM/QL-KCP tumor–bearing mice (Fig. S4 D). Conceivably, FGF2 might be critical for tumor-associated fibrosis in this model.
ARS/SHP099 is also efficacious in KRASG12C NSCLC
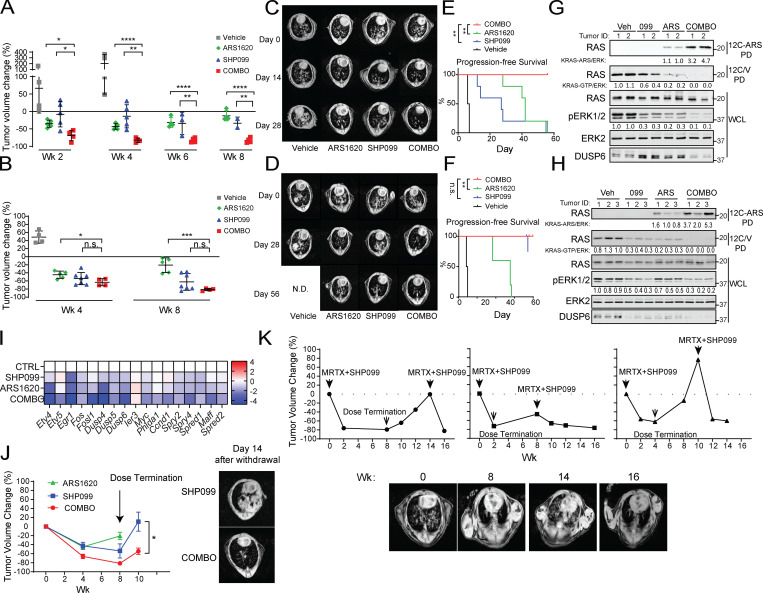
To ask if SHP099/ARS efficacy extended to other KRASG12C mutant malignancies, we monitored the effects of SHP099, ARS, and SHP099/ARS on KRASG12C (KC) and KrasG12C;Tp53R270H (KCP) NSCLC genetically engineered mouse models (GEMMs; Li et al., 2018) by serial magnetic resonance imaging (MRI). SHP099/ARS treatment induced deep responses by 2 wk and complete responses after 4 wk (Fig. 8, A–D). Remarkably, all combination-treated mice remained in remission over an 8-wk treatment period. Mice bearing KC or KCP tumors of comparable initial size (Fig. S4 E) had much shallower initial responses when treated with either single agent. SHP099-treated KC tumors remained in remission, but tumors recurred in single-agent ARS-treated mice by 8 wk on therapy, and KCP tumors recurred by 8 wk of treatment with either agent. Consequently, while ARS or SHP099 treatment resulted in a marginal survival advantage, all SHP099/ARS–treated mice remained alive and disease-free throughout the treatment period (Fig. 8, E and F). SHP099/ARS also inhibited the growth of KRASG12C-driven H2122 xenografts more effectively than either single agent (Fig. S4 F).
Figure 8.
ARS/SHP099 combination is also efficacious in NSCLC GEMMs. (A and B) Tumor volume in LSL-KRASG12C-Trp53R270H (A) and LSL-KRASG12C (B) NSCLC GEMMs, quantified by MRI, after treatment with vehicle, SHP099, ARS, or both (COMBO) at the indicated times. (C and D) Representative magnetic resonance images showing lungs from LSL-KRASG12C-Trp53R270H (C) and LSL-KRASG12C (D) NSCLC GEMMs before and after treatment, as indicated. (E and F) Kaplan–Meier curves for LSL-KRASG12C-Trp53R270H (E) and LSL-KRASG12C (F) models after the indicated treatments. (G and H) Immunoblots of lysates and 12C/V-MB or 12C-ARS Fab PDs from LSL-KRASG12C-Trp53R270H (G) and LSL-KRASG12C (H) tumors after 3 d of treatment. (I) ERK-dependent gene expression, as assessed by RNA-seq, in tumors from LSL-KRASG12C-Trp53R270H mice, treated for 3 d, as indicated (colors indicate log2FC). (J) LSL-KRASG12C tumor volume after treatment and drug withdrawal, as indicated (left); representative magnetic resonance images 14 d after drug withdrawal are shown at right. Data were pooled from two independent experiments. (K) Tumor growth curves (top) from three LSL-KRASG12C-Trp53R270H mice after MRTX1257 + SHP099 treatment, drug withdrawal, and rechallenge, as indicated. Representative magnetic resonance images (bottom) of the mouse in the left panel above at the indicated times. Doses were SHP099 75 mg/kg body weight (daily) and MRTX1257 50 mg/kg body weight (daily). Data represent mean ± SD; *, P < 0.05, **, P < 0.01, ***, P < 0.001; one-way ANOVA with Tukey’s multiple comparison test. For the curves in E and F, significance was evaluated by log-rank test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant; N.D., not determined.
Biochemical analysis revealed comparable G12C inhibition by SHP099 or ARS but substantially greater inhibition with SHP099/ARS (Fig. 8, G and H; and Fig. S4 F). Direct measurements using the 12C-ARS-Ab confirmed increased adduct formation in tumors from SHP099/ARS–treated compared with single-agent ARS–treated mice. As in the orthotopic PDAC model, RNA-seq (performed after 3 d of treatment) showed induction of several RTKs and RTK ligands by G12C-I, confirming that adaptive resistance also occurs in vivo in NSCLC (Fig. S4, G and H). Addition of SHP2-I abrogated this resistance, as shown by the greater suppression of pERK and ERK, MYC, apoptotic, and cell cycle gene expression evoked in SHP099/ARS– compared with ARS-treated (or SHP099-treated) mice (Fig. 8 I and Fig. S4, I–L).
When treatment of KC tumor–bearing mice was stopped at 8 wk, tumors recurred in SHP099- and SHP099/ARS–treated mice. As in PDAC, though, recurrence was substantially slower in the latter group, indicating substantially fewer live tumor cells (Fig. 8 J). We also tested the effects of the clinical-grade G12C-I MRTX1257 (MRTX), alone or combined with SHP099, on KC tumors. In accord with its greater potency, single-agent MRTX was more efficacious than ARS in this model. Even so, SHP099 also enhanced MRTX efficacy, as revealed by the slower tumor recurrence after drug withdrawal (Fig. S5 A). To test whether recurrent tumors remained sensitive to combination treatment, we treated KCP tumor–bearing mice as above with MRTX plus SHP099 for 8 wk and then stopped therapy for 6 wk. We then resumed treatment, and consistent with bona fide adaptive resistance, tumors regressed again to the same extent as seen after initial treatment (Fig. 8 K).
Figure S5.
ARS/SHP099 also evokes antitumor immune response in NSCLC GEMMs. (A) Quantification of tumor volumes in LSL-KRASG12C NSCLC GEMMs after treatment with vehicle (n = 5), SHP099 (75 mg/kg, daily; n = 7), ARS (200 mg/kg, daily; n = 5), ARS + SHP099 (daily; n = 4), MRTX1257 (50 mg/kg, daily; n = 2), or MRTX1257 + SHP099 (daily, n = 3) at the indicated times. (B) Pie charts showing immune cell populations in LSL-KRASG12C tumors, treated as indicated for 6 d. (C) Multiplex IF/IHC analysis of LSL-KRASG12C- and LSL-KRASG12C; Tp53R270H tumors, treated as indicated for 3 d, and stained with the indicated markers (n = 3/group). Scale bars, 100 µm. (D and E) Infiltrating myeloid cells in LSL-KRASG12C ( D) and LSL-KRASG12C-Tp53R270H (E) tumors analyzed after 6 d of treatment. Data pooled from at least two independent experiments. (F) CD31 staining of sections from LSL-KRASG12C–derived and LSL-KRASG12C-Tp53R270H–derived tumors after 3 d of treatment, as indicated (n = 3/group). Scale bars represent 100 µm and 10 µm for 10× and 40× magnification, respectively. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test (two tailed). n.s., not significant.
As in PDAC, disease control was better in SHP099- than ARS-treated KC tumor–bearing mice, despite a comparable decrease in KRASG12C-GTP and, if anything, greater suppression of ERK target genes following ARS treatment (Fig. 8, B and D). These data again suggested additional effects of SHP2-I, potentially on the TME. Indeed, flow cytometry revealed enhanced immune cell infiltration into tumors from 6-d ARS-, SHP099-, and SHP099/ARS–treated KC mice (Fig. 9 A and Fig. S5 B). Total T and B lymphocytes (as percentage of live cells) were increased by single-agent and SHP099/ARS treatment (Fig. 9 B), and there was a relative increase in CD8+ T cells in all groups (Fig. 9 C). CD44+CD62L− effector CD8+ cells were increased substantially by SHP099 or SHP099/ARS treatment, concomitant with a relative decrease in naive cells (CD44−CD62L+). However, in contrast to their effects on PDAC, ARS, SHP099, and SHP099/ARS did not evoke PD-1+TIM3+ CD8+ cells in lung tumors. Single-agent SHP099 decreased CD4+ T cell infiltration, but this decrease was mitigated by combination treatment. Nevertheless, SHP099/ARS resulted in decreased T reg cells (FOXP3+) and consequently an increased CD8/T reg ratio (Fig. 9, D and E). Similar effects were seen in KCP tumors (Fig. 9 F). Enhanced T cell infiltration into KC and KCP tumors from SHP099/ARS–treated mice was also evident by multiplex IF/IHC (Fig. S5 C).
Figure 9.
ARS/SHP099 provokes an antitumor immune program in syngeneic NSCLC model. (A) Pie chart showing the percentage of CD45+ and CD45− cells in LSL-KRASG12C tumors after 6 d of treatment, as indicated (data are presented as the average of each treatment). (B–E) Frequencies of infiltrating immune cells in LSL-KRASG12C tumors analyzed at day 6 of the indicated treatments. (F) Frequencies of indicated infiltrating immune cells in LSL-KRASG12C-Trp53R270H tumors after 6 d of the indicated treatments. Data represent mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA with Tukey’s multiple comparison test. Data were pooled from at least two independent experiments.
Concomitant with these potentially beneficial actions on tumor-associated T cells, we saw complex effects on tumor-associated myeloid cells in both NSCLC models. SHP099 (in the KC tumors) or ARS (in KCP tumors) increased total CD11b+ cells and increased the fraction of this compartment composed of macrophages (F4/80+Gr1−) in tumors of either genotype (Fig. 9, B and F; and Fig. S5, D and E). Surprisingly, combining the two agents reversed these increases, leading to a slight overall decrease in total CD11b+ cells and macrophages (Fig. 9, B and F; and Fig. S5, D and E). In contrast to a recent report on the effects of SHP2-I on KrasG12C-engineered CT26 SQ allografts, single-agent SHP099 increased tumor-associated M2 macrophages in KC tumors and tended to increase these cells in KCP tumors, although the latter was not statistically significant. ARS had the opposite effect in both models (although again, only the effects in KC tumors were statistically significant). Consequently, ARS alone or SHP099/ARS resulted in a significant increase in the M1/M2 ration in both models (Fig. S5 E). Each single agent increased the monocytic myeloid-derived suppressor cells (m-MDSC; CD11b+Ly6C+LY6G−) and g-MDSC (CD11b+Ly6C-LY6G+) populations in KCP tumors, but only SHP099 increased this population significantly in the KC model. SHP099/ARS reversed these increases (Fig. S5, D and E). Overall, SHP099/ARS diminished myeloid cell populations (M2 macrophages; myeloid-derived suppressor cells [MDSCs]) that typically have tumor-promoting effects. Table 1 summarizes the effects of each single agent and SHP099/ARS combination in the PDAC and NSCLC GEMMs.
Table 1. Drug-related changes in immune infiltration cell types in different models and studies.
| Treatment | KC GEMM | KCP GEMM | PDAC model | CT26 KRAS G12D (Quintana et al., 2020) | CT26 KRAS G12C (Canon et al., 2019) |
|---|---|---|---|---|---|
| CD3+ T cells | |||||
| SHP2-I | ↑ | NS | ↑ | ↑ | ND |
| G12C-I | ↑ | ↑ | ↑ | ND | ↑ |
| COMBO | ↑ | ↑ | ↑ | ND | ND |
| CD19+ cells | |||||
| SHP2-I | ↑ | ↑ | NS | ND | ND |
| G12C-I | ↑ | NS | ↑ | ND | ND |
| COMBO | ↑ | NS | ↑ | ND | ND |
| CD11b+ myeloid cells | |||||
| SHP2-I | ↑ | NS | NS | ↓ | ND |
| G12C-I | NS | ↑ | NS | ND | ND |
| COMBO | NS | NS | ↓ | ND | ND |
| CD8+ T cells | |||||
| SHP2-I | ↑ | ↑ | ↑ | ↑ | ND |
| G12C-I | ↑ | NS | NS | ND | ↑ |
| COMBO | ↑ | ↑ | ↑ | ND | ND |
| Activated CD8+ T cells | |||||
| SHP2-I | ↑ | ↑ | NS | ↑ | ND |
| G12C-I | NS | NS | NS | ND | ND |
| COMBO | NS | ↑ | NS | ND | ND |
| Exhausted CD8+ T cells | |||||
| SHP2-I | NS | ↑ | ↑ | NS | ND |
| G12C-I | NS | NS | ↑ | ND | ND |
| COMBO | NS | ↑ | ↑ | ND | ND |
| Effector CD8+ T cells | |||||
| SHP2-I | ↑ | ↑ | ↑ | ND | ND |
| G12C-I | NS | NS | ↑ | ND | ND |
| COMBO | ↑ | ↑ | ↑ | ND | ND |
| CD4+ T cells | |||||
| SHP2-I | ↓ | ↓ | ↓ | ↑ | ND |
| G12C-I | NS | NS | NS | ND | ↑ |
| COMBO | NS | NS | ↓ | ND | ND |
| CTLA4+ CD4+ T cells | |||||
| SHP2-I | NS | ↓ | ↑ | ND | ND |
| G12C-I | NS | NS | ↑ | ND | ND |
| COMBO | ↓ | NS | ↑ | ND | ND |
| FOXP3+ CD4+ T cells | |||||
| SHP2-I | NS | NS | ↓ | ND | ND |
| G12C-I | NS | NS | NS | ND | ND |
| COMBO | ↓ | NS | ↓ | ND | ND |
| CD8+/T reg cells | |||||
| SHP2-I | ↑ | ↑ | ↑ | NS | ND |
| G12C-I | ↑ | ↑ | ↑ | ND | ND |
| COMBO | ↑ | NS | ↑ | ND | ND |
| m-MDSCs | |||||
| SHP2-I | ↑ | ↑ | ↓ | NS | ND |
| G12C-I | NS | ↑ | NS | ND | ND |
| COMBO | NS | ↑ | NS | ND | ND |
| g-MDSCs | |||||
| SHP2-I | ↑ | NS | NS | NS | ND |
| G12C-I | NS | ↑ | NS | ND | ND |
| COMBO | NS | NS | ↓ | ND | ND |
| F4/80 macrophages | |||||
| SHP2-I | ↑ | NS | NS | ↓ | ND |
| G12C-I | ↑ | ↑ | NS | ND | ↑ |
| COMBO | NS | ↑ | NS | ND | ND |
| M1 macrophages | |||||
| SHP2-I | NS | ↓ | NS | ↑ | ND |
| G12C-I | NS | ↓ | NS | ND | ND |
| COMBO | NS | NS | NS | ND | ND |
| M2 macrophages | |||||
| SHP2-I | ↑ | NS | NS | ↓ | ND |
| G12C-I | ↓ | NS | NS | ND | ND |
| COMBO | ↓ | NS | ↓ | ND | ND |
Summary of changes (relative to vehicle control) in immune cell populations in the indicated models, treated as indicated. For KC and KCP GEMMs and the orthotopic PDAC model, the G12C-I was ARS and the SHP2-I was SHP099. For Quintana et al. (2020), the SHP2-I was RMC-4630, and for Canon et al. (2019), the G12C-I was AMG510. Only significant changes (P < 0.05) are shown.
Finally, we assessed the effects of SHP099, ARS, and SHP099/ARS on tumor-associated vasculature. In contrast to the effects of SHP099 in PDAC (Fig. 4 F and Fig. S3 F), in KC and KCP NSCLC, each single agent and SHP099/ARS increased lung tumor–associated blood vessels (Fig. S5 F). These data indicate that SHP099 (and, presumably, SHP099/ARS) might trigger a tumor cell–autonomous secretory program that promotes tumor angiogenesis specifically in the NSCLC TME (see Discussion).
Discussion
Clinically active, covalent KRASG12C inhibitors have provided the first opportunity to directly target this key oncoprotein (Canon et al., 2019; Hallin et al., 2020; Stephen et al., 2014). However, initial reports from phase I trials show tumor responses in KRASG12C tumors are partial and restricted to a subset of patients (Canon et al., 2019; Hallin et al., 2020). These results suggest that, like other targeted therapies (Ahronian and Corcoran, 2017; Konieczkowski et al., 2018; Ryan and Corcoran, 2018), G12C-Is will have limited impact as single agents due to drug resistance. Adaptive resistance (termed “rebound” by some investigators), in which the inhibited pathway is reactivated due to induction of RTKs/RTK ligand genes, is a common form of intrinsic resistance (Anderson et al., 2017; Duncan et al., 2012; Lin et al., 2015; Manchado et al., 2016; Nazarian et al., 2010; Sun et al., 2014; Zawistowski et al., 2017). The host immune system can cure some malignancies; conceivably, all cancer cures might require generation of a durable antitumor response. However, most tumors evade antitumor immunity via diverse mechanisms, and conventional chemotherapy and most targeted therapies also affect the TME. A sophisticated approach to developing curative cancer regimens requires delineating the mechanism of action of antineoplastics on cancer and TME cells and using these insights to develop complementary combinations that prevent tumor resistance. Here, by using two new affinity reagents that allow direct monitoring of KRASG12C activation and inhibition, we find that G12C-Is evoke adaptive resistance in vitro and in vivo by inducing KRASG12C reactivation. Similar to the effects of other RAS/ERK pathway inhibitors (Anderson et al., 2017; Duncan et al., 2012; Lin et al., 2015; Manchado et al., 2016; Nazarian et al., 2010; Sun et al., 2014; Zawistowski et al., 2017), G12C-Is induce RTK/RTK ligand genes, increasing RTK signaling to RAS. SHP2 inhibition, by increasing G12C-I accessibility to mutant KRAS, abrogates this resistance mechanism. Moreover, by studying their effects on KRASG12C PDAC and NSCLC GEMMs, we find that SHP2-Is and G12C-I/SHP2-I combinations have complex effects on the TME; some are mediated indirectly via effects on tumor cells, but others reflect direct SHP2-I action on tumor endothelium. Moreover, these effects are tumor genotype, tumor histotype, and tumor location dependent. These intricate details of tumor–TME, drug–tumor, and drug–TME interactions suggest additional combination approaches to further enhance G12C-I efficacy.
While this work was in progress, others reported that G12C-Is induce adaptive resistance in G12C-mutant cell lines and PDXs. In general, our findings agree with these studies, but we differ in important details. Misale et al. (2019) reported that ARS evokes adaptive resistance by activating the PI3K–AKT pathway. However, we did not observe increased activation of AKT (phosphorylated AKT) or downstream targets of the pathway (e.g., pS6) in ARS-treated H358 and MIAPaCa-2 cells or in KCP pancreas tumors from ARS-treated mice (data not shown). Others argue that adaptive resistance to G12C-Is involves up-regulation of RTK signaling and activation of WT RAS, which cannot be targeted by the inhibitor (Ryan et al., 2020), whereas Xue et al. (2020) reported that resistance arises from preexisting heterogeneity that enables some tumor cells to survive by inducing mutant KRAS to levels that exceed inhibitor targeting capacity. Like Ryan et al. (2020), we observed induction of RTKs/RTK ligand genes following ARS treatment, and similar to our previous findings on the effects of MEK-Is (Fedele et al., 2018), we saw different patterns of RTK/RTK ligands induced by G12C-I in different cell lines, even within the same histotype. We also noted qualitative and quantitative differences in RTK/RTK ligand gene induction by MEK-Is and G12C-Is (Fig. 3 A; data not shown); the reason(s) for these differences merits future study. In contrast to the previous reports, though, we did not observe altered KRAS gene expression in vitro or in vivo in response to G12C-I treatment. Instead, capitalizing on our novel 12C/V-MB PD assay, which enables specific monitoring of G12C-GTP (in the presence of normal RAS-GTP) in vitro and in vivo, we show clearly that adaptive resistance to ARS is accompanied by reactivation of KRASG12C. Although normal KRAS/other RAS isoforms might also be reactivated, KRASG12C-GTP (given its GAP resistance) should accumulate to much higher levels than other RAS-GTP species and thus should be the main effector of ERK pathway reactivation. Consistent with this conclusion, ARS induces adaptive resistance equivalently in Kraswt/KRASG12C and Kras−/−/KRASG12C MEFs (Fig. S1 A). Our novel affinity capture reagents will facilitate similar assessments of G12C activation state in other systems.
Several lines of evidence show that SHP099 was “on-target” in our experiments and hence that SHP099 effects were due to SHP2 inhibition. First, SHP099 had the expected biochemical effects on the RAS/ERK pathway in the multiple lines tested. Moreover, two different drug-resistant mutants (PTPN11P491Q, TM/QL) rescued these effects (in PDAC and NSCLC cells, respectively), whereas PTPN11 KO or SOS1 knockdown had biological and biochemical consequences similar to those of SHP099. Our results comport with and strengthen previous studies of the effects of SHP2 modulation on G12C-I action (Hallin et al., 2020; Lou et al., 2019; Misale et al., 2019; Ryan et al., 2020; Xue et al., 2020). Furthermore, our PTPN11-KO cell reconstitution experiments (Fig. 3, H and I; and Fig. S2, J and L) demonstrate that PTP activity is essential for, whereas C-terminal tyrosine residues play a modulatory role in, adaptive resistance to G12C-Is. These results align with previous studies on SHP2 action in RTK signaling (Araki et al., 2003; O'Reilly and Neel, 1998; Yamauchi et al., 1995), which found that C-terminal phosphorylation is essential for some, but not all, RTK pathways and thus provide clues into which reactivated RTKs are most critical to mediating adaptive resistance (i.e., those requiring SHP2 tyrosine phosphorylation).
Most importantly, we find that SHP2 and/or G12C inhibition in immune-competent PDAC and NSCLC GEMMs has important and complex effects both on tumor cells and on cells in the TME. Notably, similar to the effects of single-agent G12C-I or SHP2-I in the clinic (Canon et al., 2019; Hallin et al., 2020; Ou et al., 2020), ARS or SHP099 alone had limited efficacy in all of our models (Fig. 4, A, G, and H; and Fig. 8, A–H), By contrast, G12C-I/SHP2-I dramatically improved efficacy and extended survival in all models tested without evident toxicity (Fig. 4, A, G, and H; and Fig. 8, A–H). Moreover, SHP099 abrogated adaptive resistance to ARS in tumors via the same mechanism observed in vitro, as it increased occupancy of the KRASG12C-GDP state, facilitating greater ARS engagement, and thereby restored KRAS/ERK pathway inhibition; enhanced suppression of ERK, MYC, antiapoptotic, and cell cycle genes; and concomitantly induced differentiation (lineage) and proapoptotic genes.
Although each single agent altered the TME, SHP2-I/G12C-I evoked a much broader adaptive immune response in PDAC and NSCLC, featuring increased CD8+ T cell infiltration, decreased T reg cells (and consequently a higher CD8/T reg cell ratio), and increased tumor-associated B cells (Table 1). Nevertheless, the responses in the PDAC and NSCLC models diverged in several important ways. For example, the presumed immune-suppressive populations differed; CTLA4+/CD4+ T cells increased after SHP099, ARS, or SHP099/ARS treatment in PDAC (Fig. 5 D) but decreased in NSCLC (Fig. 9 D). Immune-suppressive CD11b+ myeloid subpopulations (m-MDSCs and g-MDSCs) generally decreased in PDAC (Fig. 5, B and F) but increased in response to some therapies in the NSCLC models (Fig. 9, B and F; Fig S5, D and E; and Table 1). Furthermore, SHP2-I/G12C-I–induced CD8+ T cells displayed exhaustion markers only in the PDAC model (Fig. 5 C). Notably, adding anti–PD-1 to ARS alone or SHP099/ARS enhanced antitumor immunity and conferred additional therapeutic benefit in PDAC (Fig. 6, A–C), emphasizing the pathological importance of these observations.
Gene expression analysis suggests key chemokines that likely mediate SHP2-I and G12C-I effects on the TME. Increased CXCL9-11 in response to SHP099, ARS, or SHP099/ARS treatment probably promote enhanced T cell immigration into orthotopic KCP tumors, while increased CXCL13 could evoke B cell immigration. By contrast, decreased CXCL1-3/5 and/or CCL9 could account for the altered myeloid cell populations (Fig. 5, I and J). Previous studies reported that activated KRAS enhances CCL9 and CXCL3 secretion, leading to recruitment of immunosuppressive macrophages and MDSCs. These actions were attributed to MYC activation or repression of IFN-regulatory factor 2, respectively (Kortlever et al., 2017; Liao et al., 2019). Similarly, SHP099, ARS, or SHP099/ARS treatment decreased Myc (Fig. 4 C) while inducing Irf1, Irf2, Irf7, and Irf9 (data not shown). Enhanced IFN-regulatory factor activity might also explain the observed increase in CXCL9-11 and CXCL13 (Kanda and Watanabe, 2007; Lazear et al., 2013; Muthalagu et al., 2020).
Importantly, our results differ substantially from recent studies of SQ syngeneic tumor models (Table 1). AMG510 evoked an ∼50 fold increase in intratumor CD3+ and CD8+ T cells in KrasG12C-engineered CT26 CRC allografts in BALB/c mice (Canon et al., 2019), but we observed much more modest increases in ARS-treated mice with orthotopic pancreas or autochthonous lung tumors. This discrepancy is due, at least in part, to differences in tumor location; SQ KCP tumors showed more robust T cell infiltration and larger antitumor responses than orthotopic tumors (Fig. 5 H). Our findings comport with site-dependent differences (SQ versus autochthonous) in PDAC response to other agents (Vonderheide and Bayne, 2013). The precise genetics and/or histotype of CT26 and KCP tumors, the strain in which the tumors were established (BALB/c versus C57BL/6), and/or the respective potency of ARS versus AMG also could contribute. While our work was in review, it was \reported that treatment of SQ CT26 tumors with the SHP2-I RMC-4630, by inhibiting macrophage colony-stimulating factor signaling, alters macrophage phenotype from M2 to M1 (Quintana et al., 2020). By contrast, M2 macrophages increased in KC and KCP tumors from SHP099-treated mice and were not affected by SHP2-I in our other models (Fig. S5, D and E; and Table 1). Again, this discrepancy likely reflects differences in tumor location, tumor genotype (G12D versus G12C, mutational burden), and histotype (PDAC, NSCLC versus CRC) in the two studies. Regardless, these results emphasize the need to carefully investigate all such parameters in credentialing single agents or combinations, particularly given that the initial clinical signals for G12C-Is and SHP2-Is show tumor-type–specific differences (Canon et al., 2019; Hallin et al., 2020; Ou et al., 2020).
Single SHP099 was more efficacious than ARS alone both in PDAC and NSCLC, even though ARS directly targets the driver oncogene. ARS is less potent than other G12C-Is, such as AMG-510 and MRTX1257 (Canon et al., 2019; Hallin et al., 2020). Nevertheless, inadequate potency is unlikely to explain the superior single-agent efficacy of SHP099 (compared with ARS) or the improved efficacy of SHP099/ARS combinations in vitro or in vivo. ARS was at least as effective as SHP099 in lowering KRASG12C-GTP, pERK levels, and ERK-dependent gene expression in vitro (Fig. 3 D) and in vivo (Fig. 4, B and I; Fig. 8, G and H; and Fig. S4 F). SHP099/ARS further suppressed these parameters. Furthermore, SHP099 potentiated the effects of AMG510 in vitro (Fig. S1 K) and of MRTX1257 in vivo (Fig. S5 A).
Although AMG-510 and MRTX1257 were each more efficacious than ARS or SHP099 in vitro and in vivo, our results suggest that the superior antitumor effects of SHP099 compared with ARS are the result of SHP2 actions on cells in the TME. By reconstituting Ptpn11-KO KCP cells with the drug-resistant mutant PTPN11TM/QL, we could distinguish direct effects of SHP2 inhibition in tumor and TME cells, respectively. Indeed, SHP099-resistant KCP pancreas tumors showed marked decreases in tumor vasculature in response to SHP099, similar to parental (Fedele et al., 2018) and WT-PTPN11–reconstituted (Fig. 7 D) cells. Decreased vascularity likely reduced tumor perfusion, given the histological evidence of central necrosis and decreased tumor cell density (Fig. 7 D and Fig. S4 B). Histological and gene expression analysis also suggest that TM/QL tumors partially differentiated in SHP099-treated mice, even though tumor cells themselves were unresponsive to SHP099, as confirmed by retained pERK levels and ERK-dependent gene expression. Whether these effects reflect altered production of a factor from endothelial cells (or some other, as-yet-undefined, SHP099-sensitive component of the TME) or decreased autocrine signaling by the reduced tumor cell population remains unclear. Even more intriguingly, SHP099 has context-dependent effects on tumor angiogenesis, as it evoked a moderate increase in tumor endothelium in NSCLC (Fig. S5 F). Such differences might arise from intrinsic endothelial cell heterogeneity (Aird, 2012; Jambusaria et al., 2020) and/or the marked differences in oxygen levels in PDAC and NSCLC.
By contrast, the SHP099-evoked influx of T cells and the decrease in activated fibroblasts in WT-PTPN11–expressing tumors was abrogated in TM/QL-KCP tumors, indicating that SHP2 must be inhibited within tumor cells to evoke these changes. ARS only affects mutant KRAS, so its action on cells in the TME (and by inference, any additional effects it contributes to SHP099/ARS) must reflect altered mediator production by tumor cells, a notion supported by our RNA-seq data (Fig. 5, I and J; Fig. S3 E; and Fig. S4 L). However, there might well be additional, direct effects of SHP2-I on signaling pathways in TME cells. Mice that express drug-resistant SHP2 in specific cells in the TME are required to address such issues; such studies are underway in our laboratory.
In summary, G12C-I/SHP2-I efficacy derives from their combined actions on tumor cells and cells in the TME and is tumor genotype and histotype dependent. There also are direct antiangiogenic effects of SHP2-I and G12C-I/SHP2-I that, when combined with PD-1 blockade, can further improve therapeutic outcomes in KRASG12C tumors. In NSCLC models, mice remain in remission for up to 8 wk of continuous combination therapy. Nevertheless, after treatment cessation, PDAC and NSCLC relapse. Our results also suggest additional rational combinations that might enhance efficacy and effect cure (e.g., SHP2-I/ G12C-I + anti-CTLA4 or OX40 agonism in PDAC; SHP2-I/ G12C-I + g-MDSC–targeted therapy in NSCLC). Future studies will be directed toward achieving this critical goal.
Materials and methods
Cell lines and reagents
MIAPaCa-2, Panc03.27, CALU-1, H23, H358, H2030, H1373, SW1573, and H460 cells were from laboratory stocks, obtained as described previously (Fedele et al., 2018). H1792 and H2122 cells were obtained from Dr. Thales Papagiannakopulos (NYU School of Medicine, New York, NY). NYU 59 primary low-passage human pancreatic cancer PDX-derived cells were from Dr. Diane Simeone (NYU School of Medicine) and were generated as described previously (Fedele et al., 2018). KPC 1203 cells were the gift of Dr. Dafna Bar-Sagi (NYU School of Medicine) and were derived from a pancreatic tumor in an LSL-KrasG12D/Tp53R172H/+; Pdx1-Cre (KPC) mouse on C57BL/6 background, as described previously (Hingorani et al., 2005). Immortalized RAS-less (Nras−/−; Hras−/−, Krasf/f, CreERTam) MEFs were provided by the National Cancer Institute RAS Initiative at the Frederick National Laboratory for Cancer Research under a material transfer agreement.
All cells were grown in 5% CO2 at 37C° under media conditions described by the vendor or the source laboratory; details are available from C. Fedele upon request. Cells were tested routinely (every 3 mo) for mycoplasma contamination by PCR (Young et al., 2010) and genotyped by STR analysis at IDEXX Bioresearch. SHP099 was purchased from Wuxi. ARS and AMG510 were purchased from Selleckchem. MRTX1257 was provided by Mirati Therapeutics under a collaborative agreement.
Plasmids and virus production
Human SHP2 cDNA was cloned into pMSCV-IRES-GFP, pCW57.1, and PLX304. pCW57.1 and PLX304 were gifts from David Root (Broad Institute, Cambridge, MA; Addgene; plasmids 41393 and 25890). Mutations were introduced by using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). Sequences encoding Kras and SHP2 sgRNAs were cloned into the BbsI site of pX458 (a gift from Feng Zhang [Broad Institute, Cambridge, MA]; Addgene; plasmid 48138). The following oligonucleotides were used for cloning sgRNAs into pX458: mKras G12D forward, 5′-CACCGAATGACTGAGTATAAGCTTG-3′; mKras G12D reverse, 5′-AAACCAAGCTTATACTCAGTCATTC-3′; mPtpn11 forward, 5′-CACCGAAAACTGCCATCGACTCCTC-3′; mPtpn11 reverse, 5′-AAACGAGGAGTCGATGGCAGTTTTC-3′; PTPN11 forward, 5′-CACCGGATTACTATGACCTGTATGG-3′; PTPN11 reverse, 5′-AAACCCATACAGGTCATAGTAATCC-3′. The pTRIPZ shSOS1 construct was a gift from Dr. Dafna Bar-Sagi (NYU School of Medicine, New York, NY).
Viruses were produced by cotransfecting HEK293T cells with lentiviral or retroviral constructs and packaging vectors (pVSV-G + pvPac for retroviruses; pVSV-G+ dR8.91 for lentiviruses). 48 h later, culture media were passed through a 0.45-mm filter and viral supernatants, supplemented with 8 µg/ml polybrene (Sigma), were used to infect 70% confluent cells in 6-well dishes for 16 h at 37°C. Stable pools were selected either by using the appropriate antibiotic or by FACS for EGFP.
G12C targeting
KPC 1203 cells (2 × 105) were cotransfected with 2 μg of the Cas9/sgRNA vector PX458 and 4 μl of single-stranded donor oligonucleotide (ssODN) homology-directed repair template (20 μM) using Xtreme gene (Roche). 2 d following transfection, GFP+ cells were purified by FACS, and single cells were seeded into a 96-well plate. Clones were screened by immunoblotting with RASG12D-specific Rabbit mAb D8H7 (Cell Signaling; 14429). Further characterization of the only clone (1/96) that had lost KRASG12D expression was performed by analyzing genomic DNA. The region flanking Kras exon 1 was amplified by PCR, and the product was subcloned into the Zero Blunt TOPO vector (Thermo Fisher Scientific; K287540), followed by Sanger sequencing of the insert using M13 primers. The following oligos were used: mKras intron 0 forward (F1), 5′-GTCTTTCCCCAGCACAGTGC-3′; mKras WT site-specific forward (W), 5′-ACTTGTGGTGGTTGGAGCTGG-3′; mKras G12D site-specific forward (D), 5′-GCTTGTGGTGGTTGGAGCTGA-3′; mKras G12C site-specific forward (C), 5′-ACTTGTCGTCGTTGGAGCTTG-3′; mKras intron 1 reverse (R2), 5′-CCTTTACAAGCGCACGCAGACTGTAGAGC-3′; ssODN homology-directed repair template, 5′-CACACAAAGGTGAGTGTTAAAATATTGATAAAGTTTTTGATAATCTTGTGTGAGACATGTTCTAATTTAGTTGTATTTTATTATTTTTATTGTAAGGCCTGCTGAAAATGACTGAGTATAAACTTGTCGTCGTTGGAGCTTGTGGCGTAGGCAAGAGCGCCTTGACGATACAGCTAATTCAGAATCACTTTGTGGATGAG-3′.
RAS nucleotide exchange
Purified RAS proteins used in binding experiments were prepared by diluting stock protein (typically containing 20–250 µM RAS) 25-fold with 20 mM Tris-Cl buffer, pH 7.5, containing 5 mM EDTA, 0.1 mM dithiothreitol, and 1 mM (final concentration) of nucleotide (GDP or GTPγS). Samples were incubated at 30°C for 30 min. MgCl2 was then added to a final concentration of 20 mM, and the solution was incubated on ice for at least 5 min before use.
Selection of phage-displayed antibody fragments against 12C-ARS
General procedures for the development of Fabs against purified protein targets have been described previously (Fellouse et al., 2007). Four rounds of phage display library selection with biotinylated KRASG12C-GDP+ARS at 100 nM, 100 nM, 50 nM, and 20 nM were performed. The first round recovered clones that bound to KRASG12C-GDP+ARS; the second round recovered clones that bound to KRASG12C-GDP+ARS, previously precleared with KRASG12C-GDP; the third round recovered clones that bound to KRASG12C-GDP+ARS, previously precleared with KRASG12C-GTP. The final round recovered clones that bound to KRASG12C-GDP+ARS, previously precleared with KRASG12C-GDP. Phage captured on beads were eluted in 100 µl of 0.1 M Gly-HCl, pH 2.1, and immediately neutralized with 35 µl of 1 M Tris-Cl, pH 8. Recovered clones were analyzed by phage ELISA and DNA sequencing, as described previously (Fellouse et al., 2007).
Expression, purification, and characterization of recombinant Fabs
Phage display vectors were converted into Fab expression vectors that contain a substrate tag for the biotin ligase BirA (Avidity; AviTag) at the C terminus of the heavy chain. Fabs were expressed in Escherichia coli strain 55244 (American Type Culture Collection) and were purified by protein G affinity chromatography, followed by cation exchange chromatography, as described previously (Fellouse et al., 2007). Purified Fabs were biotinylated in vitro using purified BirA. Approximately 2–5 mg purified Fabs were obtained routinely from a 1-liter bacterial culture. SDS-PAGE showed that Fabs were >90% pure. Fab binding to targets was assessed by a bead binding assay, as described previously (Hattori et al., 2016). Briefly, biotinylated Fabs were immobilized on Dynabeads M280 streptavidin, and then excess biotin-binding sites on streptavidin were blocked with biotin. Biotinylated RAS proteins were titrated into the solution containing Fab-immobilized beads. After washing, biotinylated RAS proteins bound to Fabs on the beads were detected with neutravidin Dylight650. The median signal intensity in the Dylight650 channel for the 75th–95th percentile population was taken as representative. The 12C-ARS Fab showed the highest specificity among Fabs tested, so this antibody was used for further analyses.
The DNA sequence of 12C-ARS Fab, light chain is 5′-GATATCCAGATGACCCAGTCCCCGAGCTCCCTGTCCGCCTCTGTGGGCGATAGGGTCACCATCACCTGCCGTGCCAGTCAGTCCGTGTCCAGCGCTGTAGCCTGGTATCAACAGAAACCAGGAAAAGCTCCGAAGCTTCTGATTTACTCGGCATCCAGCCTCTACTCTGGAGTCCCTTCTCGCTTCTCTGGTAGCCGTTCCGGGACGGATTTCACTCTGACCATCAGCAGTCTGCAGCCGGAAGACTTCGCAACTTATTACTGTCAGCAAGACTGGTACTTCCCGATCACGTTCGGACAGGGTACCAAGGTGGAGATCAAACGAACTGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATTCACAGTTGAAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAAAAACATAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAGGGGAGAGTGT-3′.
The DNA sequence of 12C-ARS Fab, heavy chain is 5′-GAGGTTCAGCTGGTGGAGTCTGGCGGTGGCCTGGTGCAGCCAGGGGGCTCACTCCGTTTGTCCTGTGCAGCTTCTGGCTTCACTTTCTCTTCTTATTATATACACTGGGTGCGTCAGGCCCCGGGTAAGGGCCTGGAATGGGTTGCATCTATTTCTCCTTCTTCTGGCTCTACTTATTATGCCGATAGCGTCAAGGGCCGTTTCACTATAAGCGCAGACACATCCAAAAACACAGCCTACCTACAAATGAACAGCTTAAGAGCTGAGGACACTGCCGTCTATTATTGTGCTCGCTACGGTGGTCGTTCTTACTGGCAGAAACAGGACTCTTACTTCTACCAGCATGGTTTGGACTACTGGGGTCAAGGAACCCTGGTCACCGTCTCCTCGGCCTCCACCAAGGGTCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTCGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACA-3′.
Proliferation assays
Cells (500–2,000/well) were seeded into 96-well plates. Following incubation with DMSO, 10 µM ARS, 10 µM SHP099, or both drugs, cell viability (n = 3) was assayed at different times using the PrestoBlue cytotoxicity assay (Thermo Fisher Scientific) according to the manufacturer’s protocol. Media (including drugs) were refreshed every 48 h. Briefly, 10 µl PrestoBlue reagent was added to each well, and after 2 h, fluorescence was measured on a multiplate reader, with an excitation wavelength of 530 nm and an emission wavelength of 590 nm. Data were corrected for PrestoBlue background fluorescence in media alone. All data represent at least two biological independent experiments in which technical triplicates were performed.
Clonogenic assays
Cells (100–2,000) were seeded in 6-well plates 1 d before treatment with DMSO, 10 µM ARS, 10 µM SH099, or both drugs; allowed to grow until they formed colonies (7–14 d); rinsed twice with PBS to remove floating cells; fixed in 4% formaldehyde in PBS (vol/vol) for 10–15 min; and stained in 0.1% crystal violet/10% ethanol for 20 min. Staining solution was aspirated, and colonies were washed with water three times, air-dried, and visualized with an Odyssey Imaging System (LI-COR). Results were quantified by using the ImageJ Colony Area PlugIn (Guzmán et al., 2014). At least three biological replicates were performed.
Immunoblotting
Whole-cell lysates (WCLs) were generated in modified radioimmunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% NP-40, and 0.1% SDS, without sodium deoxycholate), supplemented with protease (40 µg/ml PMSF, 2 µg/ml antipain, 2 µg/ml pepstatin A, 20 µg/ml leupeptin, and 20 µg/ml aprotinin) and phosphatase (10 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate, and 10 mM sodium pyrophosphate) inhibitors. After clarification of debris by a microfuge, samples were quantified with the DC Protein Assay Kit (Bio-Rad). Total lysate protein was resolved by standard SDS-PAGE and transferred in 1× transfer buffer and 15% methanol. Membranes were incubated with their respective primary and secondary antibodies labeled with IRDye (680 and 800 nm) and then visualized using the LI-COR system. Antibodies against phospho-p42/44 MAPK (rabbit polyclonal; #9101; 1:1,000) were obtained from Cell Signaling. Monoclonal pan-RAS antibody (clone Ab-3; OP40-100UG; 1:1,000) was obtained from Millipore. Rabbit polyclonal antibodies against SHP2 (sc-280; 1:1,000) and mouse monoclonal ERK-2 (D2: sc-1647; 1:1,000) were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-SOS1 (MA5-17234) was purchased from Invitrogen. Rabbit monoclonal anti-DUSP6 antibody EPR129Y was obtained from Abcam (#ab76310).
KRASG12C activity measurements
Cells cultured in 6-well plates were treated as described in the figures with G12C-I (ARS, AMG510, and/or SHP099). Cells were lysed by incubating them in GTPase lysis buffer (25 mM Tris-Cl, pH7.2, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, and 5% glycerol supplemented with protease inhibitors and phosphatase inhibitors on ice for 15 min immediately before analysis. After centrifugation for 15 min at 15,000 g, supernatants were collected and incubated with streptavidin agarose resin (Thermo Fisher Scientific) for 1 h at 4°C, followed by a brief centrifugation, to decrease nonspecific binding to the resin. Precleared lysates were incubated with biotinylated 12C/V-MB or 12C-ARS-Fab bound to streptavidin agarose for 1.5 h at 4°C while rotating. Agarose beads were then washed twice with GTPase lysis buffer, boiled in 1x SDS-PAGE sample buffer, and subjected to immunoblotting with a pan-RAS antibody (Millipore).
LC-MS/MS Assay for ARS binding to KRASG12C
Cells (5 × 105) were treated with the indicated compounds for the times listed and subsequently washed twice with PBS and prepared for protein extraction and liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis, as described previously (Patricelli et al., 2016). LC-MS/MS was performed at the PCC Proteomics Shared Resource at NYU School of Medicine.
Histology and IHC
H&E, Masson trichome staining, and IHC were performed by the PCC Experimental Pathology Shared Resource at NYU School of Medicine. IHC for pERK (Cell Signaling; 4370), CD31 (Cell Signaling; D8V9E), cleaved caspase-3 (Cell Signaling; D3E9), Ki67 (Spring Biosciences; SP6), and αSMA (Abcam; ab5694) was performed on sections from paraformaldehyde-fixed tumors. Quantification was performed by ImageJ as described before (Fedele et al., 2018).
Xenografts
All animal experiments were approved by, and conducted in accordance with the procedures of, the Institutional Animal Care and Use Committees at New York University School of Medicine (protocol no. 170602). NY53 and H2122 xenografts were established by SQ injection of 5 × 106 cells in 50% Matrigel (Corning) into the right flanks of 8- to 10-wk-old nu/nu mice (Charles River; #088). Each treatment group contained 8–10 mice. When tumors reached 100–500 mm3, as measured by calipers (size = length × width2 × 0.5), mice were randomized to four groups (10 mice/group) for each model and treated with vehicle, SHP099, ARS, or SHP099/ARS. Investigators were not blinded to group allocation. The following oral gavage dosing regimens were employed: ARS (200 mg/kg, daily), SHP099 (75 mg/kg daily), or ARS 200 mg/kg daily, SHP099 75 mg/kg daily. SHP099 was resuspended in 0.6% methylcellulose, 0.5% Tween80 in 0.9% saline. ARS was dissolved 1% N-methyl-2-pyrrolidone + 19% polyethylene glycol 400 (PEG400) + 80% (10% hydroxypropyl in water). Caliper and weight measurements were performed every other day and continued until termination of the experiments.
Orthotopic PDAC model
KCP cells were generated as described above. Cells (105) were suspended in Matrigel, implanted into the pancreata of 6- to 8-wk-old syngeneic mice (C57BL/6), and allowed to establish for 14–30 d before beginning treatment. Mice were randomized to four groups (10 mice/group) for each model, and treated with vehicle, SHP099, ARS, or SHP099/ARS. Investigators were not blinded to group allocation. Vehicle, ARS (200 mg/kg daily), SHP099 (75 mg/kg daily), ARS 200 mg/kg daily, or SHP099 75 mg/kg daily was administered for the indicated time, and mice were euthanized. Where indicated, α-PD-1 antibody (200 µg; Bio X Cell; RMP1-14) was used. Dosing was repeated every 3 d for the duration of the experiment. Control mice were injected with PBS or isotype control antibody (Bio X Cell; clone LTF-2).
NSCLC GEMM studies
KRASG12C or KRASG12C;Tp53R270H mice (mixed background; Li et al., 2018) were monitored by MRI for tumor development after intranasal induction with adeno-Cre (2.5 × 106 PFU). Tumor-bearing mice were dosed with vehicle, ARS (200 mg/kg daily), SHP099 (75 mg/kg daily), ARS 200 mg/kg daily, or SHP099 75 mg/kg daily and monitored by MRI every 2 wk. In some experiments, MRTX1257 (50 mg/kg daily) was used in place of ARS. Mice were randomized as described above.
Flow cytometry
Tumors were minced, chopped, and digested in DMEM containing 2.0 mg/ml collagenase IV (GIBCO), 1.0 mg/ml hyaluronidase (Worthington), 0.1% soybean trypsin inhibitor, and 50 U/ml DNase I (STEMCELL Technologies) at 37°C for 1 h. Single-cell suspensions were obtained by passage through a strainer (70 µm), washed in FACS buffer (PBS with 5% FBS), incubated with LIVE/DEAD Fixable Zombie Yellow Fixable Viability Kit (BioLegend; 423104) for 30 min, blocked with anti-CD16/32 (BioLegend; clone 93) for 5 min on ice, and then incubated with fluorophore-conjugated antibodies on ice for 45 min. For detection of intracellular markers, FOXP3 Fixation/Permeabilization Buffer Set (BioLegend) was used, according to the manufacturer’s instructions. Antibodies for flow cytometry were CD45 BUV395 #564279, CD4 BUV737 #564933,Ki-67 Alexa700 #50-112-4690, CD11c BUV737 #564986 from Fisher; CD3e BV421 #100336, CD8a BV570 #100740, CD44 APC/Cy7 #103028, PD-1 BV711 #135231, CTLA4 APC #106310, OX40 PerCP/Cy5.5 #119425, Tim3 BV785 #119725, Foxp3 PE #126404,CD19 BV650 #115540, CD11b BV421 #101236, Ly6C BV570 #128030, Ly6G BV711 #108431, F4/80 APC/Cy7 #123118, Dx5 FITC #108906, CD19 PerCP5.5 #115534, CD69 PE-CF594 #104536, Ly6C APC/Cy7 #128026, CD45 BV605 #103140, CD8 BV711 #100748, CD3 AF700 #100216, PD-1 APC/Cy7 #135224, IA/IE FITC #107616, Ly6G PerCP5.5 #127616, CD8 PE # 100708, F4/80 PE-CF594 #123146, CD44 BV421 #103039, Ki-67 BV421 #652411, PD-L1 PECy7 #124314, CD62L APC #104412, CD4 BV785 #100453, CD11b BV711 #101242, CD11c BV785 #117336 from BioLegend; CTLA4 PE #12-1522-83, Tim3 PE/Cy7 #25-5870-82, and Foxp3 APC #17-5773-82 from eBioscience. For quantifying apoptosis, cells were stained by using the PE Annexin V Apoptosis Detection Kit I according to manufacturer’s protocol (BD). Flow cytometry was performed on an LSR II flow cytometer (BD) at the PCC Precision Immunology Shared Resource at NYU School of Medicine and analyzed by using FlowJo software (BD).
RNA extraction and sequencing
RNA was extracted from frozen tumors using the miRNeasy Mini Kit (Qiagen), according to the manufacturer’s instructions. RNA-seq was performed by the PCC Genome Technology Center Shared Resource. Libraries were prepared by using the Illumina TruSeq Stranded Total RNA Sample Preparation Kit and sequenced on an Illumina NovaSeq 6000 using 150-bp paired-end reads. Sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. The average number of read pairs/sample was 35.4 million. Data were processed by the Perlmutter Cancer Center Applied Bioinformatics Laboratories shared resource.
qRT-PCR
Total cellular RNA was isolated by using the Qiagen RNeasy kit. cDNA was generated by using the SuperScript IV First Strand Synthesis System (Invitrogen) for RT-PCR. qRT-PCR was performed with Fast SYBR Green Master Mix (Applied Biosystems), following the manufacturer’s protocol, in 384-well format in C1000 Touch Thermal Cycler (Bio-Rad). Differential gene expression analysis was performed with CFX Manager (Bio-Rad) and normalized to GAPDH expression. Primers used are listed elsewhere (Fedele et al., 2018).
Bliss analysis
Potential drug synergy was assessed by Bliss analysis as Yab,P = Ya + Yb − YaYb, where Ya stands for percentage inhibition of drug a and Yb stands for percentage inhibition of drug b (Zhao et al., 2014). Synergistic effects were defined as the percentage of observed effect greater than Yab,P.
Statistical analysis
Data are expressed as mean ± SD. Significance was assessed using Welch’s t test or one-way ANOVA, as appropriate. Survival rates were analyzed by log-rank test. Statistical analyses were performed in Prism 8 (GraphPad Software). Significance was set at P = 0.05.
Data availability
RNA-seq data have been deposited in the Gene Expression Omnibus database under the accession code GSE149815. All other data supporting the findings of this study are available within the article, in the supplemental information files, or from the corresponding author upon request.
Online supplemental material
Fig. S1 shows the effects of SHP2-I and KRASG12C inhibitor as single agents or in combination on PDAC and NSCLC cell lines. Fig. S2 provides additional information on how SHP099 increases KRASG12C-ARS adducts and acts upstream of KRAS to block G12C-I–evoked ERK pathway reactivation. Fig. S3 provides additional information on ARS/SHP099 combination effects in PDAC. Fig. S4 dissects the tumor cell–autonomous and nonautonomous effects of SHP2 inhibition and provides further information on ARS/SHP099 efficacy in NSCLC GEMMs. Fig. S5 describes the antitumor immune response evoked by ARS/SHP099 treatments in NSCLC GEMMs.
Acknowledgments
We thank Drs. Thales Papagiannakopoulos (NYU School of Medicine, New York, NY) and Diane Simeone (NYU School of Medicine, New York, NY) for sharing cell lines; Drs. Dafna Bar-Sagi (NYU School of Medicine, New York, NY), David Root (Broad Institute, Cambridge, MA), and Feng Zhang (Broad Institute, Cambridge, MA) for plasmids; and the PCC Experimental Pathology, Precision Immunology, Genome Technology Center, Applied Bioinformatics Laboratories and Proteomics Laboratory shared resources (National Institutes of Health grant P30CA016087) for technical support. We thank Dr. Jamie Christensen (Mirati Therapuetics, San Diego, CA) for generously providing MRTX1257 and for helpful discussions. We also thank Drs. Toshiyuki Araki, Kiyomi Araki, Abhishek Bhardwaj, Jayu Jen, and Shuang Zhang, and Ms. Angel Sing and Wei Wei (Neel lab) for advice and discussion on this project.
This work was supported by National Institutes of Health grants CA49152 (B.G. Neel), CA219670 (K-K. Wong), CA194864 (S. Koide), American Cancer Society grant 132652-PF-18-180-01-TBE (K.W. Teng), and Cancer Center Core Grant P30 CA016087 and a Mirati Therapeutics sponsored research agreement (K-K. Wong).
Author contributions: Conception and design: C. Fedele, S. Li, K.W. Teng, K-K. Wong, S. Koide, A. Koide, and B.G. Neel; acquisition of data: C. Fedele, S. Li, K.W. Teng, C.J.R. Foster, D. Peng, H. Ran, T. Hattori, A. Koide, and Y. Wang; analysis and interpretation of data: C. Fedele, S. Li, K.W. Teng, C.J.R. Foster, D. Peng, H. Ran, T. Hattori, A. Koide, H. Ran, P. Mita, M.J. Geer, K.H. Tang, I. Dolgalev, U. Ozerdem, S. Koide, K.-K. Wong, and B.G. Neel; administrative, technical, or material support: C. Fedele, S. Li, K.W. Teng, C.J.R. Foster, D. Peng, H. Ran, T. Hattori, A. Koide, H. Ran, P. Mita, M.J. Geer, K.H. Tang, I. Dolgalev, U. Ozerdem, J. Deng, T. Chen, J. Leinwand, W. Wang, B. Diskin, and G. Miller; writing (original draft): C. Fedele and B.G. Neel; writing (review and editing): C. Fedele, S. Li, K-K. Wong, and B.G. Neel; study supervision: C. Fedele, K-K. Wong, and B.G. Neel; funding acquisition: S. Koide, K-K. Wong, and B.G. Neel.
References
- Ahmed T.A., Adamopoulos C., Karoulia Z., Wu X., Sachidanandam R., Aaronson S.A., and Poulikakos P.I.. 2019. SHP2 Drives Adaptive Resistance to ERK Signaling Inhibition in Molecularly Defined Subsets of ERK-Dependent Tumors. Cell Rep. 26:65–78.e5. 10.1016/j.celrep.2018.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahronian L.G., and Corcoran R.B.. 2017. Strategies for monitoring and combating resistance to combination kinase inhibitors for cancer therapy. Genome Med. 9:37 10.1186/s13073-017-0431-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird W.C. 2012. Endothelial cell heterogeneity. Cold Spring Harb. Perspect. Med. 2 a006429 10.1101/cshperspect.a006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G.R., Winter P.S., Lin K.H., Nussbaum D.P., Cakir M., Stein E.M., Soderquist R.S., Crawford L., Leeds J.C., Newcomb R., et al. 2017. A Landscape of Therapeutic Cooperativity in KRAS Mutant Cancers Reveals Principles for Controlling Tumor Evolution. Cell Rep. 20:999–1015. 10.1016/j.celrep.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T., Nawa H., and Neel B.G.. 2003. Tyrosyl phosphorylation of Shp2 is required for normal ERK activation in response to some, but not all, growth factors. J. Biol. Chem. 278:41677–41684. 10.1074/jbc.M306461200 [DOI] [PubMed] [Google Scholar]
- Araujo J.M., Gomez A.C., Aguilar A., Salgado R., Balko J.M., Bravo L., Doimi F., Bretel D., Morante Z., Flores C., et al. 2018. Effect of CCL5 expression in the recruitment of immune cells in triple negative breast cancer. Sci. Rep. 8:4899 10.1038/s41598-018-23099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaru P., Shah N.H., Bhattacharyya M., Barton J.P., Kondo Y., Cofsky J.C., Gee C.L., Chakraborty A.K., Kortemme T., Ranganathan R., et al. 2017. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife. 6 e27810 10.7554/eLife.27810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D., and Neel B.G.. 1998. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 6:249–254. 10.1016/S0969-2126(98)00027-6 [DOI] [PubMed] [Google Scholar]
- Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., et al. 2018. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24:541–550. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., et al. 2019. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 575:217–223. 10.1038/s41586-019-1694-1 [DOI] [PubMed] [Google Scholar]
- Chen Y.N., LaMarche M.J., Chan H.M., Fekkes P., Garcia-Fortanet J., Acker M.G., Antonakos B., Chen C.H., Chen Z., Cooke V.G., et al. 2016. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 535:148–152. 10.1038/nature18621 [DOI] [PubMed] [Google Scholar]
- Chow M.T., and Luster A.D.. 2014. Chemokines in cancer. Cancer Immunol. Res. 2:1125–1131. 10.1158/2326-6066.CIR-14-0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G., Miller M.L., Aksoy B.A., Senbabaoglu Y., Schultz N., and Sander C.. 2013. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45:1127–1133. 10.1038/ng.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C.E., Hingorani S.R., Mick R., Combs C., Tuveson D.A., and Vonderheide R.H.. 2007. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 67:9518–9527. 10.1158/0008-5472.CAN-07-0175 [DOI] [PubMed] [Google Scholar]
- Duncan J.S., Whittle M.C., Nakamura K., Abell A.N., Midland A.A., Zawistowski J.S., Johnson N.L., Granger D.A., Jordan N.V., Darr D.B., et al. 2012. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 149:307–321. 10.1016/j.cell.2012.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele C., Ran H., Diskin B., Wei W., Jen J., Geer M.J., Araki K., Ozerdem U., Simeone D.M., Miller G., et al. 2018. SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer Discov. 8:1237–1249. 10.1158/2159-8290.CD-18-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellouse F.A., Esaki K., Birtalan S., Raptis D., Cancasci V.J., Koide A., Jhurani P., Vasser M., Wiesmann C., Kossiakoff A.A., et al. 2007. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373:924–940. 10.1016/j.jmb.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Garcia Fortanet J., Chen C.H., Chen Y.N., Chen Z., Deng Z., Firestone B., Fekkes P., Fodor M., Fortin P.D., Fridrich C., et al. 2016. Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor. J. Med. Chem. 59:7773–7782. 10.1021/acs.jmedchem.6b00680 [DOI] [PubMed] [Google Scholar]
- Govindan R., Fakih M.G., Price T.J., Falchook G.S., Desai J., Kuo J.C., Strickler J.H., Krauss J.C., Li B.T., Denlinger C.S., et al. 2019. Phase I study of AMG 510, a novel molecule targeting KRAS G12C mutant solid tumours. Ann. Oncol. •••:30. [Google Scholar]
- Gremer L., Merbitz-Zahradnik T., Dvorsky R., Cirstea I.C., Kratz C.P., Zenker M., Wittinghofer A., and Ahmadian M.R.. 2011. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 32:33–43. 10.1002/humu.21377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán C., Bagga M., Kaur A., Westermarck J., and Abankwa D.. 2014. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 9 e92444 10.1371/journal.pone.0092444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin J., Engstrom L.D., Hargis L., Calinisan A., Aranda R., Briere D.M., Sudhakar N., Bowcut V., Baer B.R., Ballard J.A., et al. 2020. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 10:54–71. 10.1158/2159-8290.CD-19-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R., Peters U., Babbar A., Chen Y., Feng J., Janes M.R., Li L.S., Ren P., Liu Y., and Zarrinkar P.P.. 2018. The reactivity-driven biochemical mechanism of covalent KRASG12C inhibitors. Nat. Struct. Mol. Biol. 25:454–462. 10.1038/s41594-018-0061-5 [DOI] [PubMed] [Google Scholar]
- Hao H.X., Wang H., Liu C., Kovats S., Velazquez R., Lu H., Pant B., Shirley M., Meyer M.J., Pu M., et al. 2019. Tumor Intrinsic Efficacy by SHP2 and RTK Inhibitors in KRAS-Mutant Cancers. Mol. Cancer Ther. 18:2368–2380. 10.1158/1535-7163.MCT-19-0170 [DOI] [PubMed] [Google Scholar]
- Hattori T., Lai D., Dementieva I.S., Montaño S.P., Kurosawa K., Zheng Y., Akin L.R., Świst-Rosowska K.M., Grzybowski A.T., Koide A., et al. 2016. Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation. Proc. Natl. Acad. Sci. USA. 113:2092–2097. 10.1073/pnas.1522691113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., and Tuveson D.A.. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 7:469–483. 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Hobbs G.A., Der C.J., and Rossman K.L.. 2016. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129:1287–1292. 10.1242/jcs.182873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof P., Pluskey S., Dhe-Paganon S., Eck M.J., and Shoelson S.E.. 1998. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 92:441–450. 10.1016/S0092-8674(00)80938-1 [DOI] [PubMed] [Google Scholar]
- Hui E., Cheung J., Zhu J., Su X., Taylor M.J., Wallweber H.A., Sasmal D.K., Huang J., Kim J.M., Mellman I., et al. 2017. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 355:1428–1433. 10.1126/science.aaf1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J.C., Manandhar A., Carrasco M.A., Gurbani D., Gondi S., and Westover K.D.. 2015. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 13:1325–1335. 10.1158/1541-7786.MCR-15-0203 [DOI] [PubMed] [Google Scholar]
- Hymowitz S.G., and Malek S.. 2018. Targeting the MAPK Pathway in RAS Mutant Cancers. Cold Spring Harb. Perspect. Med. 8 a031492 10.1101/cshperspect.a031492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambusaria A., Hong Z., Zhang L., Srivastava S., Jana A., Toth P.T., Dai Y., Malik A.B., and Rehman J.. 2020. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. eLife. 9 e51413 10.7554/eLife.51413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes M.R., Zhang J., Li L.S., Hansen R., Peters U., Guo X., Chen Y., Babbar A., Firdaus S.J., Darjania L., et al. 2018. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell. 172:578–589.e17. 10.1016/j.cell.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Reid D., Parker J.A., Salter S., Knihtila R., Kuzmic P., and Mattos C.. 2017. The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects. J. Biol. Chem. 292:12981–12993. 10.1074/jbc.M117.778886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda N., and Watanabe S.. 2007. Prolactin enhances interferon-gamma-induced production of CXC ligand 9 (CXCL9), CXCL10, and CXCL11 in human keratinocytes. Endocrinology. 148:2317–2325. 10.1210/en.2006-1639 [DOI] [PubMed] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A., et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature. 502:333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczkowski D.J., Johannessen C.M., and Garraway L.A.. 2018. A Convergence-Based Framework for Cancer Drug Resistance. Cancer Cell. 33:801–815. 10.1016/j.ccell.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlever R.M., Sodir N.M., Wilson C.H., Burkhart D.L., Pellegrinet L., Brown Swigart L., Littlewood T.D., and Evan G.I.. 2017. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 171:1301–1315.e14. 10.1016/j.cell.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRochelle J.R., Fodor M., Ellegast J.M., Liu X., Vemulapalli V., Mohseni M., Stams T., Buhrlage S.J., Stegmaier K., LaMarche M.J., et al. 2017. Identification of an allosteric benzothiazolopyrimidone inhibitor of the oncogenic protein tyrosine phosphatase SHP2. Bioorg. Med. Chem. 25:6479–6485. 10.1016/j.bmc.2017.10.025 [DOI] [PubMed] [Google Scholar]
- LaRochelle J.R., Fodor M., Vemulapalli V., Mohseni M., Wang P., Stams T., LaMarche M.J., Chopra R., Acker M.G., and Blacklow S.C.. 2018. Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition. Nat. Commun. 9:4508 10.1038/s41467-018-06823-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne E., Combadière C., Iga M., Boissonnas A., Bonduelle O., Maho M., Debré P., and Combadiere B.. 2004. Intratumoral CC chemokine ligand 5 overexpression delays tumor growth and increases tumor cell infiltration. J. Immunol. 173:3755–3762. 10.4049/jimmunol.173.6.3755 [DOI] [PubMed] [Google Scholar]
- Lazear H.M., Lancaster A., Wilkins C., Suthar M.S., Huang A., Vick S.C., Clepper L., Thackray L., Brassil M.M., Virgin H.W., et al. 2013. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 9 e1003118 10.1371/journal.ppat.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu S., Deng J., Akbay E.A., Hai J., Ambrogio C., Zhang L., Zhou F., Jenkins R.W., Adeegbe D.O., et al. 2018. Assessing Therapeutic Efficacy of MEK Inhibition in a KRASG12C-Driven Mouse Model of Lung Cancer. Clin. Cancer Res. 24:4854–4864. 10.1158/1078-0432.CCR-17-3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Overman M.J., Boutin A.T., Shang X., Zhao D., Dey P., Li J., Wang G., Lan Z., Li J., et al. 2019. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell. 35:559–572.e7. 10.1016/j.ccell.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Sabnis A.J., Chan E., Olivas V., Cade L., Pazarentzos E., Asthana S., Neel D., Yan J.J., Lu X., et al. 2015. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47:250–256. 10.1038/ng.3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay C.R., and Blackhall F.H.. 2019. Direct Ras G12C inhibitors: crossing the rubicon. Br. J. Cancer. 121:197–198. 10.1038/s41416-019-0499-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P., Solomon M., Li L.S., Hansen R., and Rosen N.. 2016. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 351:604–608. 10.1126/science.aad6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou K., Steri V., Ge A.Y., Hwang Y.C., Yogodzinski C.H., Shkedi A.R., Choi A.L.M., Mitchell D.C., Swaney D.L., Hann B., et al. 2019. KRASG12C inhibition produces a driver-limited state revealing collateral dependencies. Sci. Signal. 12 eaaw9450 10.1126/scisignal.aaw9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi S., Mulero-Sánchez A., Prahallad A., Germano G., Bosma A., Krimpenfort P., Lieftink C., Steinberg J.D., de Wit N., Gonçalves-Ribeiro S., et al. 2018. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat. Med. 24:961–967. 10.1038/s41591-018-0023-9 [DOI] [PubMed] [Google Scholar]
- Manchado E., Weissmueller S., Morris J.P. IV, Chen C.C., Wullenkord R., Lujambio A., de Stanchina E., Poirier J.T., Gainor J.F., Corcoran R.B., et al. 2016. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 534:647–651. 10.1038/nature18600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhab-Jafari M.T., Marshall C.B., Smith M.J., Gasmi-Seabrook G.M., Stathopulos P.B., Inagaki F., Kay L.E., Neel B.G., and Ikura M.. 2015. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA. 112:6625–6630. 10.1073/pnas.1419895112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S., Fatherree J.P., Cortez E., Li C., Bilton S., Timonina D., Myers D.T., Lee D., Gomez-Caraballo M., Greenberg M., et al. 2019. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin. Cancer Res. 25:796–807. 10.1158/1078-0432.CCR-18-0368 [DOI] [PubMed] [Google Scholar]
- Muthalagu N., Monteverde T., Raffo-Iraolagoitia X., Wiesheu R., Whyte D., Hedley A., Laing S., Kruspig B., Upstill-Goddard R., Shaw R., et al. 2020. Repression of the Type I Interferon pathway underlies MYC & KRAS-dependent evasion of NK & B cells in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 10:872–887. 10.1158/2159-8290.CD-19-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarsheth N., Wicha M.S., and Zou W.. 2017. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17:559–572. 10.1038/nri.2017.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H., Chen Z., Lee M.K., Attar N., Sazegar H., et al. 2010. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 468:973–977. 10.1038/nature09626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R.J., Haderk F., Stahlhut C., Schulze C.J., Hemmati G., Wildes D., Tzitzilonis C., Mordec K., Marquez A., Romero J., et al. 2018. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 20:1064–1073. 10.1038/s41556-018-0169-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly A.M., and Neel B.G.. 1998. Structural determinants of SHP-2 function and specificity in Xenopus mesoderm induction. Mol. Cell. Biol. 18:161–177. 10.1128/MCB.18.1.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem J.M., and Shokat K.M.. 2016. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15:771–785. 10.1038/nrd.2016.139 [DOI] [PubMed] [Google Scholar]
- Ou S.I., Koczywas M., Ulahannan S., Janne P., Pacheco J., Burris H., McCoach C., Wang J.S., Gordon M., Haura E., et al. 2020. A12 The SHP2 Inhibitor RMC-4630 in Patients with KRAS-Mutant Non-Small Cell Lung Cancer: Preliminary Evaluation of a First-in-Man Phase 1 Clinical Trial. J. Thorac. Oncol. 15:S15–S16. 10.1016/j.jtho.2019.12.041 [DOI] [Google Scholar]
- Pádua R.A.P., Sun Y., Marko I., Pitsawong W., Stiller J.B., Otten R., and Kern D.. 2018. Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2. Nat. Commun. 9:4507 10.1038/s41467-018-06814-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli M.P., Janes M.R., Li L.S., Hansen R., Peters U., Kessler L.V., Chen Y., Kucharski J.M., Feng J., Ely T., et al. 2016. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 6:316–329. 10.1158/2159-8290.CD-15-1105 [DOI] [PubMed] [Google Scholar]
- Peled M., Tocheva A.S., Sandigursky S., Nayak S., Philips E.A., Nichols K.E., Strazza M., Azoulay-Alfaguter I., Askenazi M., Neel B.G., et al. 2018. Affinity purification mass spectrometry analysis of PD-1 uncovers SAP as a new checkpoint inhibitor. Proc. Natl. Acad. Sci. USA. 115:E468–E477. 10.1073/pnas.1710437115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas C.A., Taylor B.S., Ye Q., Viale A., Sander C., Solit D.B., and Rosen N.. 2009. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. USA. 106:4519–4524. 10.1073/pnas.0900780106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I.A., Hood F.E., and Hartley J.L.. 2020. The frequency of Ras mutations in cancer. Cancer Res. 80:2969–2974. 10.1158/0008-5472.CAN-19-3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., and Joyce J.A.. 2013. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19:1423–1437. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E., Schulze C.J., Myers D.R., Choy T.J., Mordec K., Wildes D., Shifrin N.T., Belwafa A., Koltun E.S., Gill A.L., et al. 2020. Allosteric inhibition of SHP2 stimulates anti-tumor immunity by transforming the immunosuppressive environment. Cancer Res. 80:2889–2902. 10.1158/0008-5472.CAN-19-3038 [DOI] [PubMed] [Google Scholar]
- Ran H., Tsutsumi R., Araki T., and Neel B.G.. 2016. Sticking It to Cancer with Molecular Glue for SHP2. Cancer Cell. 30:194–196. 10.1016/j.ccell.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruess D.A., Heynen G.J., Ciecielski K.J., Ai J., Berninger A., Kabacaoglu D., Görgülü K., Dantes Z., Wörmann S.M., Diakopoulos K.N., et al. 2018. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med. 24:954–960. 10.1038/s41591-018-0024-8 [DOI] [PubMed] [Google Scholar]
- Ryan M.B., and Corcoran R.B.. 2018. Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 15:709–720. 10.1038/s41571-018-0105-0 [DOI] [PubMed] [Google Scholar]
- Ryan M.B., Fece de la Cruz F., Phat S., Myers D.T., Wong E., Shahzade H.A., Hong C.B., and Corcoran R.B.. 2020. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRASG12C Inhibition. Clin. Cancer Res. 26:1633–1643. 10.1158/1078-0432.CCR-19-3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. 2020. Grail of RAS cancer drugs within reach. Nat. Biotechnol. 38:6–8. 10.1038/s41587-019-0382-x [DOI] [PubMed] [Google Scholar]
- Simanshu D.K., Nissley D.V., and McCormick F.. 2017. RAS Proteins and Their Regulators in Human Disease. Cell. 170:17–33. 10.1016/j.cell.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.J., and Ikura M.. 2014. Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat. Chem. Biol. 10:223–230. 10.1038/nchembio.1435 [DOI] [PubMed] [Google Scholar]
- Stephen A.G., Esposito D., Bagni R.K., and McCormick F.. 2014. Dragging ras back in the ring. Cancer Cell. 25:272–281. 10.1016/j.ccr.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Sun C., Hobor S., Bertotti A., Zecchin D., Huang S., Galimi F., Cottino F., Prahallad A., Grernrum W., Tzani A., et al. 2014. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 7:86–93. 10.1016/j.celrep.2014.02.045 [DOI] [PubMed] [Google Scholar]
- Tokunaga R., Zhang W., Naseem M., Puccini A., Berger M.D., Soni S., McSkane M., Baba H., and Lenz H.J.. 2018. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat. Rev. 63:40–47. 10.1016/j.ctrv.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheide R.H., and Bayne L.J.. 2013. Inflammatory networks and immune surveillance of pancreatic carcinoma. Curr. Opin. Immunol. 25:200–205. 10.1016/j.coi.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G.S., Zhou J., Liu J.B., Wu Z., Xu X., Li T., Xu D., Schumacher S.E., Puschhof J., McFarland J., et al. 2018. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat. Med. 24:968–977. 10.1038/s41591-018-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Xu G., Li X., Xu W., Li Q., Liu W., Kirby K.A., Loh M.L., Li J., Sarafianos S.G., et al. 2019. A small molecule inhibitor that stabilizes the autoinhibited conformation of the oncogenic tyrosine phosphatase SHP2. J. Med. Chem. 62:1125–1137. 10.1021/acs.jmedchem.8b00513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J.Y., Zhao Y., Aronowitz J., Mai T.T., Vides A., Qeriqi B., Kim D., Li C., de Stanchina E., Mazutis L., et al. 2020. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 577:421–425. 10.1038/s41586-019-1884-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi K., Milarski K.L., Saltiel A.R., and Pessin J.E.. 1995. Protein-tyrosine-phosphatase SHPTP2 is a required positive effector for insulin downstream signaling. Proc. Natl. Acad. Sci. USA. 92:664–668. 10.1073/pnas.92.3.664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T., Takamatsu M., Kobayashi-Imanishi W., Hashimoto-Tane A., Azuma M., and Saito T.. 2012. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 209:1201–1217. 10.1084/jem.20112741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L., Sung J., Stacey G., and Masters J.R.. 2010. Detection of Mycoplasma in cell cultures. Nat. Protoc. 5:929–934. 10.1038/nprot.2010.43 [DOI] [PubMed] [Google Scholar]
- Zawistowski J.S., Bevill S.M., Goulet D.R., Stuhlmiller T.J., Beltran A.S., Olivares-Quintero J.F., Singh D., Sciaky N., Parker J.S., Rashid N.U., et al. 2017. Enhancer Remodeling during Adaptive Bypass to MEK Inhibition Is Attenuated by Pharmacologic Targeting of the P-TEFb Complex. Cancer Discov. 7:302–321. 10.1158/2159-8290.CD-16-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., Srinivasan P., Gao J., Chakravarty D., Devlin S.M., et al. 2017. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23:703–713. 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Sachsenmeier K., Zhang L., Sult E., Hollingsworth R.E., and Yang H.. 2014. A New Bliss Independence Model to Analyze Drug Combination Data. J. Biomol. Screen. 19:817–821. 10.1177/1087057114521867 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data have been deposited in the Gene Expression Omnibus database under the accession code GSE149815. All other data supporting the findings of this study are available within the article, in the supplemental information files, or from the corresponding author upon request.