Abstract
Background
Protein kinase membrane-associated tyrosine/threonine (PKMYT1) has been found in many tumors, but its association with clear cell renal cell carcinoma (ccRCC) remains unclear.
Material/Methods
PKMYT1 expression in ccRCC was examined in the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), and Tumor Immune Estimation Resource databases. The correlation between PKMYT1 expression and clinicopathological parameters was explored via the chi-square test. Receiver operating characteristic curves were used to estimate the diagnostic performance of PKMYT1. Kaplan-Meier curves, a Cox model, nomogram, time-dependent receiver operating characteristic curves, and decision curve analysis (DCA) were used to evaluate the prognostic value and clinical utility of PKMYT1. Genes coexpressed with PKMYT1 in ccRCC were identified based on TCGA, the gene expression profiling interactive, and cBioPortal. Gene Set Enrichment Analysis revealed biological pathways associated with PKMYT1 in ccRCC.
Results
Weighted gene coexpression network analysis identified PKMYT1 as one of the genes most significantly correlated with progression of histological grade. PKMYT1 was significantly upregulated in ccRCC compared with normal tissue (P<0.001), with a trend toward differentiating between individuals with ccRCC and those who were healthy (area under the curve=0.942). High PKMYT1 expression was correlated with unsatisfactory survival (hazard ratio=1.67, P=0.001), indicating that it is a risk factor for ccRCC. A nomogram incorporating PKMYT1 level was created and showed a clinical net benefit. PKMYT1 was strongly positively correlated with the anti-silencing function of 1B histone chaperone (ASF1B) gene in ccRCC.
Conclusions
PKMYT1 is upregulated in ccRCC and its presence indicates poor prognosis, making it a potential therapeutic target for ccRCC.
MeSH Keywords: Biological Processes; Carcinoma, Renal Cell; Database
Background
Kidney cancer is the 12th most common solid neoplasm, with approximately 400 000 new diagnoses and 175 000 tumor-associated deaths globally in 2018 [1]. Renal cell carcinoma (RCC) accounts for approximately 90% of all renal tumors and is derived from epithelial cells in renal tubules [2,3]. There are 3 primary histologic subtypes of RCC: clear cell RCC (ccRCC) (about 75–80% of RCC), papillary RCC (pRCC) (about 10%), and chromophobe RCC (chRCC) (about 5%) [4,5]. Rapid development in the molecular characterization of ccRCC has facilitated advances in targeted therapy, thus improving median survival of patients with advanced-stage RCC stage from less than 10 months before 2004 to 30 months by 2011 [6]. Specifically, ccRCC is usually characterized by few specific symptoms and/or laboratory abnormalities. About 20% to 30% of individuals with RCC present with advanced-stage disease at initial diagnosis. In addition, approximately 30% of patients who have localized ccRCC experience recurrence or metastasis after tumor-targeted surgery [7,8]. Thus, it is essential to further investigate reliable ccRCC-associated molecular biomarkers, thereby facilitating early diagnosis, monitoring of tumor development, and discovery of novel therapeutic targets [9].
PKMYT1 (also known as MYT1), a member of the WEE1 family, exerts a crucial effect on Golgi and endoplasmic reticulum assembly in mammalian cells. It is a kinase that efficiently phosphorylates cell division cycle 2 (cDC2) in both threonine-14 (Thr14) and tyrosine-15 (Thr15) in the African clawed frog, Xenopus [10–12]. Throughout the cell cycle, DNA-damage checkpoints can help preserve genomic stability in normal cells, a monitoring mechanism that is commonly deregulated in tumors. Because of the action of tumor-suppressor genes such as P53, mutations in which result in deactivation of the G1 checkpoint, most tumor cells are heavily dependent on the G2/M checkpoint, thus ensuring its genomic stability and survival advantage [13,14]. PKMYT1 is involved in G2 arrest in oocytes and its activity is modulated through Akt phosphorylation [12]. PKMYT1 serves as a negative modulator of the cell cycle, preventing cells from transforming from G2 to the mitosis phase through 2 pathways [15]. One is binding of PKMYT1 localized to the cytoplasm to the cDC2/cyclin B complex, which restrains the complex from entering the nucleus [16]. In addition, PKMYT1 suppresses cDC2 activity by phosphorylating the Thr14/Thr15 residue on cDC2 [17–20]. Considering that PKMYT1 secures the G2/M phase transition, inhibitors that target PKMYT1 could potentially effectively diminish the survival ability of tumor cells, which would endowed them with potential in clinical therapy [13]. Currently, an increasing number of studies have demonstrated that overexpression of PKMYT1 facilitates proliferation, migration, invasion, and colony-forming ability, as well as epithelial-mesenchymal transition, (EMT) in multiple tumors. Specifically, a study has demonstrated that PKMYT1 is positively associated with polo-like kinase 1 (PLK1) in breast cancer. In breast cancer, the 2 genes may act synergistically to modulate tumor growth by maintaining precise control of the cell cycle and genome stability [13]. PKMYT1 facilitates the progression of prostate cancer by targeting cyclin B1 and cyclin E1 expression [10]. PKMYT1 also interacts with microspherule protein 1 (MCRS1) – an oncogene known to play a role in several tumors – to promote the malignant phenotype of gastric cancer (GC) [21]. PKMYT1 can bind and inactivate glycogen synthase kinase 3beta and further activate beta-catenin/T-cell factor signaling, thus facilitating the development of hepatocellular carcinoma (HCC) [22]. PKMYT1 facilitates development of ovarian cancer (OC) by negatively modulating SIRT3 [23]. Diminished expression of PKMYT1 is also accompanied by decreased levels of Notch1, p21, and Hes1, indicating that PKMYT1 has potential to strengthen the activity of the Notch signal pathway in non-small cell lung cancer (NSCLC) [24]. Silencing PKMYT1 expression eliminates irradiation-induced G2/M phase arrest and enhances the sensitivity of cancer cells to radiation [15].
Nevertheless, an understanding of the correlation between PKMYT1 and ccRCC and the effect of PKMYT1 on ccRCC progression remains elusive, and further investigation in this area is warranted. In the present study, multiple integrated bioinformatics approaches were used to assess the expression, prognostic effect, and potential biological function of PKMYT1 in ccRCC.
Material and Methods
Extraction of differentially expressed gene
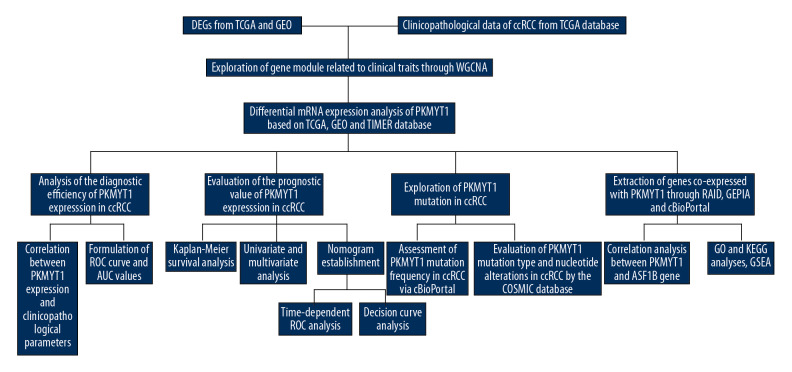
A flow diagram of our research approach is shown in Figure 1. The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/) was used to acquire publicly available messenger RNA (mRNA) profiles and corresponding clinicopathological data from 539 ccRCC tissues and 72 adjacent normal samples, Information recorded included: sex; race; laterality; tumor, node classification, and metastasis classification; tumor, node metastasis (TNM) stage; histologic grade; tumor status; relapse; and vital status. Differentially expressed genes (DEGs) were identified from TCGA database and GEO datasets (http://www.ncbi.nlm.nih.gov/geo/) (including GSE15641 [25], GSE36895 [26] and GSE40435 [27], as well as GSE105261 [28]) through the “DEseq2” package and via the “Limma” package, respectively. The thresholds were |log2-fold change (FC)| >2.0 and false discovery rate (FDR) <0.01 [29].
Figure 1.
Flowchart of the study design.
Weighted gene coexpression network analysis was applied for screening PKMYT1
Coexpression methodology with weighted gene coexpression network analysis was transformed into connection weights or topology overlap measurements to assess the correlations among the identified DEGs [30,31]. The R package of weighted gene coexpression network analysis software was used to formulate a coexpression network for 2369 DEGs in 519 ccRCC tissues with specific clinicopathological data. We considered module significance (MS) as the average gene significance (GS) for all genes in a module [32]. We estimated the association between the module eigengene (ME) and clinical traits to investigate relevant modules. Commonly, the module with the greatest absolute value of MS was considered to be the one the most associated with a clinical trait, which then was selected for further analysis. The module membership (MM) was used to describe the significance of genes in the module. Thus, we identified the PKMYT1 gene in a module via MM and GS [33]. The levels of expression of PKMYT1 in different tumors were identified through the Tumor Immune Estimation Resource (TIMER) database (http://timer.cistrome.org/).
Survival analysis
Pearson’s test was used to assess the correlation between PKMYT1 expression and clinicopathological variables in ccRCC. Kaplan-Meier plots with log-rank tests were applied to evaluate the association between overall survival (OS) of patients with ccRCC and PKMYT1 expression. A receiver operating characteristic curve (ROC) with areas under the curve (AUC) was formulated to assess the diagnostic efficiency of PKMYT1 in ccRCC [34,35]. Cox regression analysis was used to calculate the hazard ratio (HR) with 95% confidence interval (CI) to estimate the prognostic effect of PKMYT1 in ccRCC.
Nomogram formulation and validation
A nomogram was developed with the “rms” R package to estimate the probability of 3-, 5-, and 10-year OS in patients with ccRCC. A concordance index (C-index) and calibration plot were applied to evaluate the nomogram discrimination and the consistence between nomogram prediction and practical observation, respectively [34,35]. The time-dependent ROC curves were applied to evaluate the predictive capability of the nomogram, PKMYT1 expression level, and additional clinicopathologic variables for 3-, 5-, and 10-year OS. DCA is a novel statistical method for evaluating the utility of a predictive model in clinical decision-making and to help select the best model for use in clinical practice [36]. DCA can estimate the clinical net benefit of every model in contrast to all or no strategies. The best model is the one with the greatest net benefit, as calculated [35,37]. We conducted DCA to compare the potential predictive benefit of the nomogram, PKMYT1 expression, and traditional clinical parameters.
PKMYT1 mutations analysis
The cBioPortal database (https://www.cbioportal.org/) (last accessed May 20, 2020) was used to analyze the frequency of PKMYT1 mutations in ccRCC. The Catalogue of Somatic Mutations in Cancer (COSMIC) (https://cancer.sanger.ac.uk/cosmic/) is a high-resolution database used to investigate the impact of somatic mutations on multiple human cancers, which was used to explore mutations in PKMYT1 in ccRCC [38,39].
Gene coexpression analysis
We identified long noncoding RNAs (lncRNAs), micro-RNAs (miRNAs), mRNA, and RNA-binding protein (RBP), as well as transcription factors (TFs), that interact with PKMYT1 using the RNA InterActome (RAID) database (RAID v2.0, www.rna-society.org/raid/) [40]. The Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) is a database-based resource which incorporates 9736 tumors and 8587 normal tissues from TCGA and the Genotype-Tissue Expression datasets, thus analyzing RNA sequencing expression [41]. The correlation coefficient was generated via the Spearman method, which was used to estimate the degree of gene expression correlation. PKMYT1 and the genes positively coexpressed with PKMYT1 expression in ccRCC are shown on the x-axis and the y-axis, respectively.
Function enrichment analyses
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were formulated by using the clusterProfiler package (version 3.14.3). Thus, the biological attributes of the genes that were positively correlated with PKMYT1 were investigated [42]. Gene Set Enrichment Analysis (GSEA) (https://www.broadinstitute.org/gsea/) was used to determine the significance of the potential biological mechanisms in the high and low PKMYT1 expression groups. Gene sets with a normalized (NOM) P<0.05 and FDR <0.25 were considered significantly enriched.
Statistical analysis
R language software (R 3.6.3 version) was used for statistical analysis. A Wilcoxon rank sum test was conducted to compare PKMYT1 expression in ccRCC tissues and adjacent non-tumor samples using TCGA and GEO datasets. All ccRCC cases were stratified into the high- or low-expression group according to the median value of PKMYT1 expression. The correlation between clinicopathological parameters and PKMYT1 mRNA level was estimated via the chi-square test and the Fisher exact test. The Kaplan-Meier method was used to compare OS. Univariate and multivariate Cox analyses were formulated to determine the correlations between PKMYT1 mRNA level and OS and with additional clinicopathological parameters. Pearson correlation analysis and Spearman correlation analysis were used to assess the relevance of PKMYT1 and ASF1B gene expression values. P<0.05 was deemed to be statistically significant (* P<0.05; ** P<0.01; *** P<0.001).
Results
PKMYT1 is a differentially expressed gene significantly associated with histological grade in ccRCC
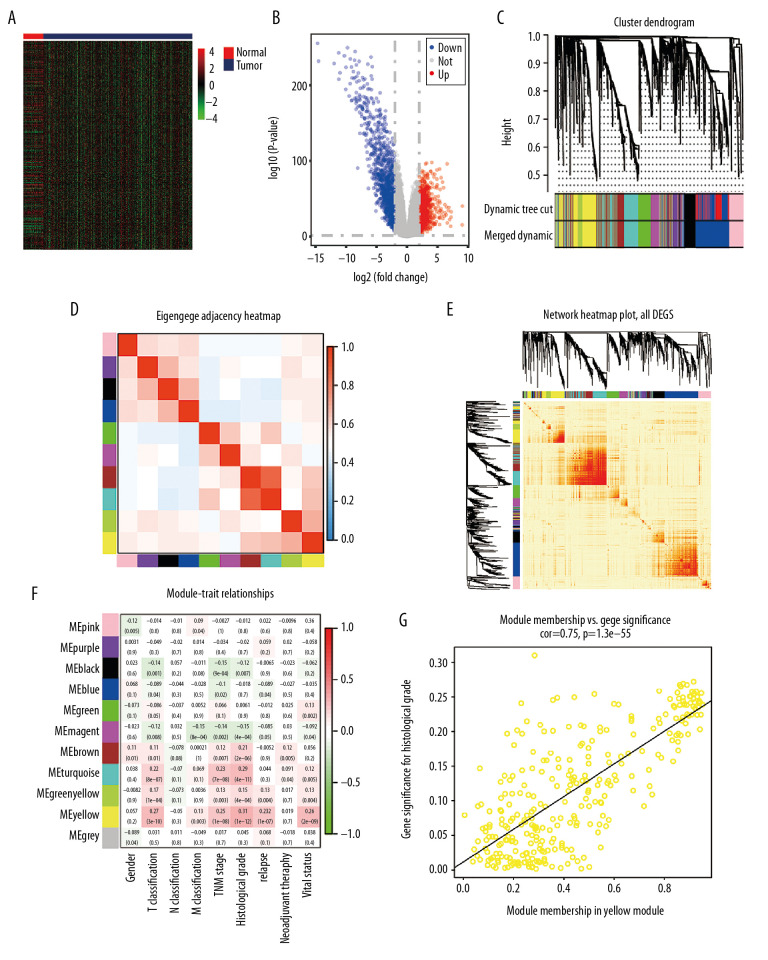
Based on the cutoff value of FDR <0.05 and |FC|>2, there were 2369 DEGs between ccRCC and normal kidney samples, including 1115 upregulated and 1254 downregulated DEGs. The heat map and volcano plot for mRNA profiles are shown in Figure 2A and 2B, respectively. A total of 2369 DEGs were clustered to formulate the coexpression network through weighted correlation network analysis [43]. The soft threshold power value of 3 defined the adjacency matrix, and MEs up to 0.75 were merged (Supplementary Figure 1). Ten different gene coexpression modules were identified in ccRCC after the insignificant gray module was excluded (Figure 2C). The results of an eigengene connectivity analysis of those modules are shown in Figure 2D and 2E. The yellow module, which was most significantly correlated with histological grade of ccRCC, was pivotal for predicting development of ccRCC (r=0.31; P=1e–12) (Figure 2F). PKMYT1 was one of the most significant genes in the yellow module (GS=0.951435, MM=0.935643) (Figure 2G), which indicates that PKMYT1 is one of the DEGs that is the most significantly correlated with histological grade in ccRCC and it potentially predicts the prognosis of ccRCC based on histological grade.
Figure 2.
Bioinformatic analysis of ccRCC and normal kidney samples in the TCGA database.(A, B) The heatmap and volcano plot of the DEGs between 539 ccRCC cases and 72 normal samples. (C) Cluster dendrogram of DEGs and module screening based on gene expression profiles of 539 ccRCC samples. (D, E) Heatmap of the correlation coefficient expressed between modules. (F) Relationships between consensus module eigengenes and various clinical traits. (G) Analogous scatter plots for the yellow module. ccRCC – clear cell renal cell carcinoma; TCGA – The Cancer Genome Atlas; DEGs – differently expressed genes.
PKMYT1 is significantly upregulated in ccRCC
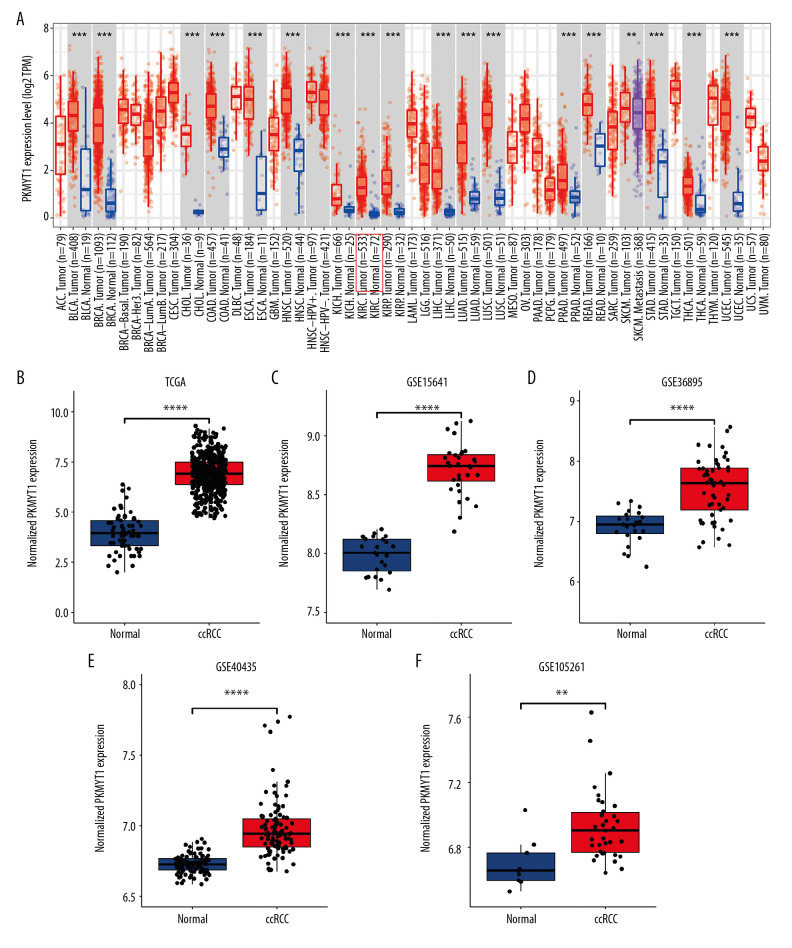
As shown in Figure 3A, PKMYT1 expression was significantly increased in multiple solid tumors, especially in bladder urothelial carcinoma and esophageal carcinoma. Mining TCGA and multiple GEO datasets (including GSE15641, GSE36895, GSE40435, and GSE105261) further demonstrated that PKMYT1 was markedly upregulated in ccRCC samples compared with normal kidney tissues (Figure 3B–3F).
Figure 3.
PKMYT1 is prominently overexpressed in ccRCC. (A) PKMYT1 mRNA expression in multiple neoplasms and adjacent normal samples from the TIMER database. PKMYT1 mRNA level in ccRCC and normal kidney tissues from (B) the TCGA database, (C) GSE15641, (D) GSE36895, (E) GSE40435, and (F) GSE105261. ccRCC – clear cell renal cell carcinoma; TIMER – Tumor Immune Estimation Resource.
PKMYT1 expression is associated with clinicopathological parameters in ccRCC
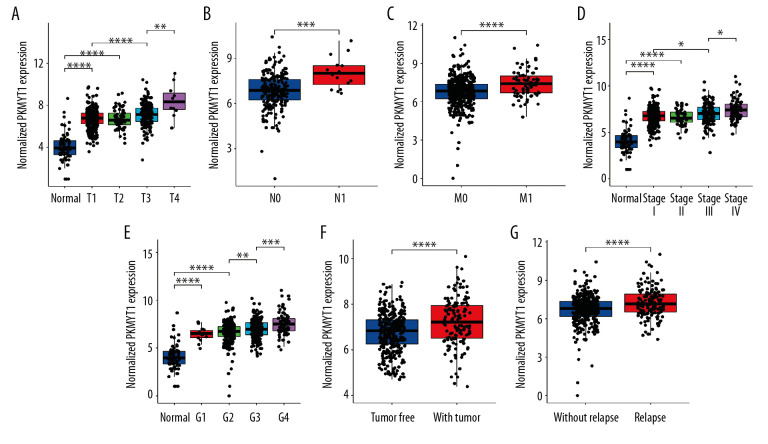
The association between PKMYT1 expression and clinicopathological variables in 530 cases of ccRCC from the TCGA database was further analyzed through Pearson’s χ2 test. As shown in Table 1, PKMYT1 was significantly correlated with T classification (P<0.0001), N classification (P=0.027), M classification (P=0.0058), TNM stage (P=0.0021), histologic grade (P=0.0002), tumor status (P=0.0037), status of relapse (P=0.0065), and vital status (P=0.0009). More specifically, PKMYT expression was enhanced in patients whose tumors had more advanced T, N, M classification, TNM stage, and histological grade, and those in whom ccRCC had relapsed (Figure 4A–4G). There was no significant association between PKMYT1 mRNA levels and additional clinicopathological factors, such as age, sex, and laterality of the neoplasm (Supplementary Figure 2A–2C).
Table 1.
Correlation between PKMYT1 mRNA levels and clinicopathological parameters in 530 patients with ccRCC from the TCGA database.
| Variable | Groups | N | Low | High | χ2 | P value |
|---|---|---|---|---|---|---|
| Age (years) | ≤60 | 264 | 129 | 135 | 0.27136 | 0.6024 |
| >60 | 266 | 137 | 129 | |||
| NA | ||||||
| Sex | Male | 344 | 164 | 180 | 2.2003 | 0.138 |
| Female | 186 | 102 | 84 | |||
| NA | ||||||
| Race | White | 459 | 226 | 233 | 0.4298* | |
| Asian | 8 | 4 | 4 | |||
| Black | 56 | 33 | 23 | |||
| NA | 7 | |||||
| Laterality | Right | 280 | 148 | 132 | 1.5888 | 0.2075 |
| Left | 249 | 117 | 132 | |||
| Bilateral | 1 | |||||
| NA | ||||||
| T classification | T1 | 271 | 149 | 122 | 22.122 | P<0.0001 |
| T2 | 69 | 44 | 25 | |||
| T3 | 179 | 72 | 107 | |||
| T4 | 11 | 1 | 10 | |||
| TX | ||||||
| NA | ||||||
| N classification | N0 | 239 | 121 | 118 | 4.8908 | 0.027 |
| N1 | 16 | 3 | 13 | |||
| NX | 275 | |||||
| NA | ||||||
| M classification | M0 | 440 | 231 | 209 | 7.6072 | 0.0058 |
| M1 | 80 | 28 | 52 | |||
| MX | 10 | |||||
| NA | ||||||
| TNM stage | I | 265 | 145 | 120 | 14.688 | 0.0021 |
| II | 57 | 36 | 21 | |||
| III | 123 | 55 | 68 | |||
| IV | 82 | 29 | 53 | |||
| NA | 3 | |||||
| Histologic grade | G1 | 14 | 11 | 3 | 19.3 | 0.0002 |
| G2 | 227 | 127 | 100 | |||
| G3 | 206 | 99 | 107 | |||
| G4 | 75 | 23 | 52 | |||
| NA | 8 | |||||
| Tumor status | With tumor | 138 | 54 | 84 | 8.4434 | 0.0037 |
| Tumor-free | 358 | 194 | 164 | |||
| NA | 34 | |||||
| Relapse | No | 373 | 202 | 171 | 7.3991 | 0.0065 |
| Yes | 157 | 64 | 93 | |||
| NA | ||||||
| Vital status | Dead | 172 | 68 | 104 | 10.939 | 0.0009 |
| Alive | 358 | 198 | 160 | |||
| NA |
P value of Fisher’s exact test conducted on a small sample.
mRNA – messenger RNA; ccRCC – clear cell renal cell carcinoma; TCGA – The Cancer Genome Atlas.
Figure 4.
PKMYT1 mRNA expression is significantly correlated with diversiform clinical variables in ccRCC cases from the TCGA database. PKMYT1 in individuals with ccRCC stratified by (A) T classification, (B) N classification, (C) M classification, (D) TNM stage, (E) histologic grade, (F) tumor status, and (G) the status of relapse. ccRCC – clear cell renal cell carcinoma; TCGA – The Cancer Genome Atlas; TNM – tumor, node, metastasis.
PKMYT1 for diagnosing and predicting prognosis of ccRCC
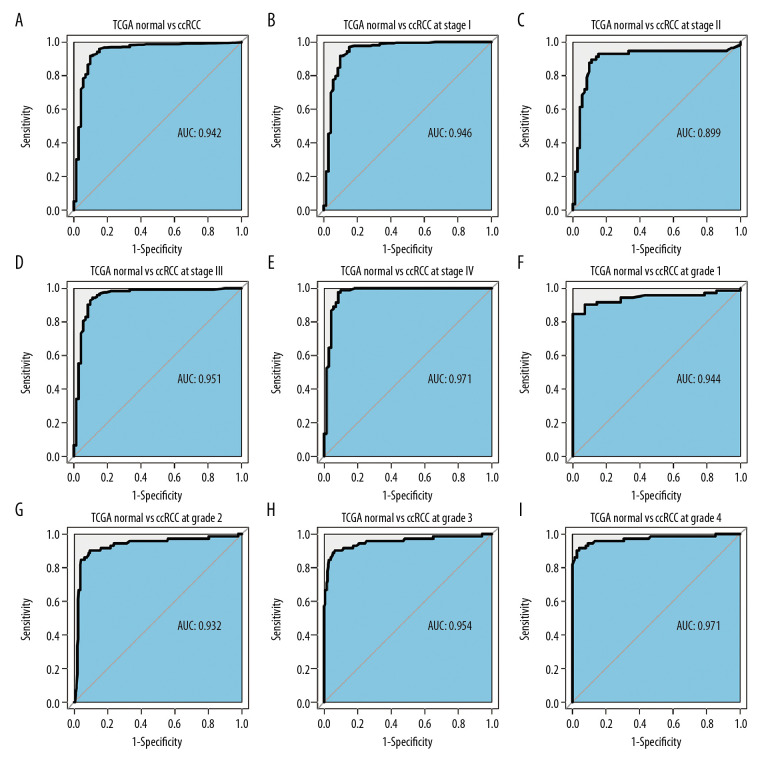
An ROC curve was used to further clarify the diagnostic efficiency of PKMYT1 expression in ccRCC cases for which information was extracted from the TCGA dataset. It showed that expression of the gene was satisfactory for differentiating between ccRCC and tissue from healthy individuals, achieving an AUC of 0.942 (95% CI: 0.906–0.978) (Figure 5A). In addition, the AUC values for the ability of PKMYT1 to distinguish between normal renal tissue and ccRCC at TNM stages I, II, III, and IV were 0.946 (95% CI: 0.908–0.985), 0.899 (95% CI: 0.833–0.965), 0.951 (95% CI: 0.914–0.988), and 0.971 (95% CI: 0.942–0.999), respectively (Figure 5B–5E). Similarly, PKMYT1 also demonstrated significant diagnostic value in distinguishing normal tissue from ccRCC of histological grades G1, G2, G3, and G4, with AUCs of 0.944 (95% CI: 0.897–0.991), 0.932 (95% CI: 0.890–0.973), 0.954 (95% CI: 0.919–0.989), and 0.971 (95% CI: 0.943–0.999), respectively (Figure 5F–5I). These findings demonstrate that PKMYT1 is a potentially reliable and promising biomarker for diagnosis of even early-stage ccRCC.
Figure 5.
PKMYT1 can effectively diagnose ccRCC. ROC curves reveal that PKMYT1 expression can differentiate adjacent normal tissues from (A) ccRCC and ccRCC at (B) TNM I stage, (C) TNM II stage, (D) TNM III stage, (E) TNM IV stage, (F) histological stage 1, (G) histological stage 2, (H) histological stage 3, and (I) histological stage 4. ccRCC – clear cell renal cell carcinoma; ROC: receiver operating characteristic; TNM – tumor, node, metastasis.
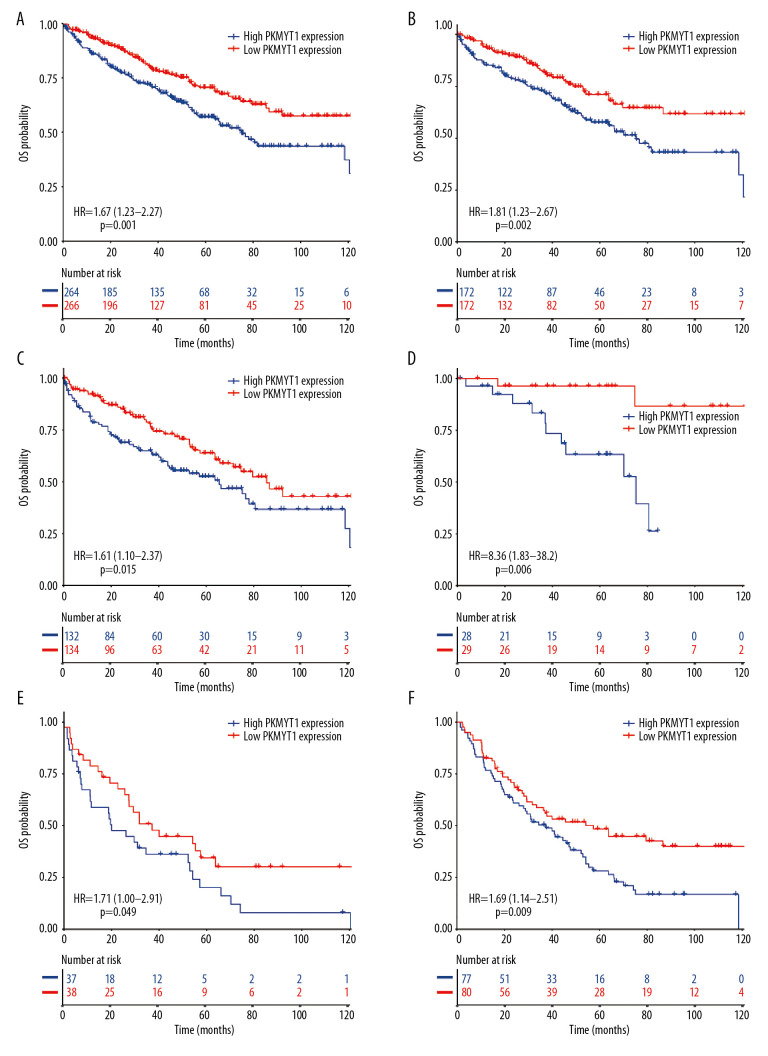
To determine whether PKMYT1 exerted a significant effect on the prognosis of ccRCC, all ccRCC cases for which information was extracted from the TCGA database were stratified into high- and low-expression groups, based on the median value for PKMYT1 expression. The Kaplan-Meier curve with log-rank test showed that prognosis of ccRCC was worse in those with high expression of PKMYT1 than in those with low expression of the gene (HR=1.67, 95% CI: 1.23–2.27, P=0.001) (Figure 6A). High PKMYT1 expression indicated shorter OS than low PKMYT1 expression in men (HR=1.81, 95% CI: 1.23–2.67, P=0.002) and older patients (HR=1.61, 95% CI: 1.10–2.37, P=0.015) with TNM stage II ccRCC (HR=8.36, 95% CI: 1.83–38.2, P=0.006), histological grade G4 disease (HR=1.71, 95% CI: 1.00–2.91, P=0.049), and relapse (HR=1.69, 95% CI: 1.14–2.51, P=0.009) (Figure 6B–6F).
Figure 6.
Overexpressed PKMYT1 exerts negative effects on ccRCC prognosis. Survival analysis reveals OS between high PKMYT1 and low PKMYT1 mRNA levels in (A) all ccRCC cases from the TCGA database, (B) male patients, (C) cases with age >60 years, (D) TNM stage II, (E) cases at histological grade 4, and (F) ccRCC that has relapsed. ccRCC – clear cell renal cell carcinoma; OS – overall survival; TCGA – The Cancer Genome Atlas; TNM, tumor, node, metastasis.
Cox regression analysis was conducted to assess the prognostic performance of PKMYT1 mRNA level in ccRCC. Univariate analysis showed that high PKMYT1 expression (HR=1.82, 95% CI: 1.32–2.52, P<0.001) was significantly associated with inferior OS (Table 2). The multivariate analysis also found that a high PKMYT1 mRNA level (HR=1.79, 95% CI: 1.29–2.49, p=0.001) had the ability to predict unsatisfactory prognosis. Thus, high PKMYT1 expression can be considered an independent risk factor for OS in ccRCC patients.
Table 2.
Univariate and multivariate analyses of OS in patients with ccRCC in the TCGA database.
| Variable | Mean survival (Mo) | 95% CI | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| Total | 44.9 | 42.0–47.9 | ||||
| Age (years) | ||||||
| ≤60 | 46.9 | 42.6–51.2 | Ref | |||
| >60 | 43 | 39–47.1 | 1.61 (1.17–2.21) | 0.004 | 1.25 (0.9–1.75) | 0.188 |
| Sex | ||||||
| Female | 45 | 39.6–50.3 | Ref | |||
| Male | 44.9 | 41.4–48.5 | 0.96 (0.69–1.34) | 0.811 | ||
| Race | ||||||
| Asian | 30.6 | 17.1–44.2 | Ref | |||
| Black | 26.2 | 19.2–33.1 | 2.01 (0.26–15.7) | 0.507 | ||
| White | 47.3 | 44.1–50.4 | 1.89 (0.26–13.52) | 0.527 | ||
| T classification | ||||||
| T1+T2 | 47.6 | 44–51.3 | Ref | |||
| T3+T4 | 41 | 36–46.1 | 2.88 (2.09–3.97) | <0.001 | 0.64 (0.34–1.2) | 0.164 |
| N classification | ||||||
| N0 | 45.2 | 40.8–49.5 | Ref | |||
| N1 | 27.2 | 11.7–42.6 | 2.81 (1.36–5.83) | 0.005 | ||
| M classification | ||||||
| M0 | 47.1 | 43.9–50.3 | Ref | |||
| M1 | 35.2 | 27.4–43.1 | 4.57 (3.3–6.32) | <0.001 | 1.31 (0.84–2.05) | 0.236 |
| TNM stage | ||||||
| I+II | 48 | 44.4–51.7 | Ref | |||
| III+IV | 40.2 | 35.3–45 | 3.61 (2.59–5.03) | <0.001 | 2.54 (1.19–5.41) | 0.016 |
| Histologic grade | ||||||
| G1+G2 | 46.6 | 42.4–50.9 | Ref | |||
| G3+G4 | 43.5 | 39.4–47.6 | 2.63 (1.83–3.76) | <0.001 | 1.57 (1.06–2.32) | 0.024 |
| Tumor status | ||||||
| Tumor-free | 46.5 | 43.1–50 | Ref | |||
| With tumor | 40.9 | 35.3–46.4 | 5.41 (3.89–7.53) | <0.001 | 4.75 (2.32–9.75) | <0.001 |
| Laterality | ||||||
| Left | 43.5 | 39.4–47.7 | Ref | |||
| Right | 46.2 | 42–50.3 | 0.66 (0.48–0.91) | 0.010 | 0.73 (0.53–1.01) | 0.057 |
| Relapse | ||||||
| No | 45.3 | 41.8–48.8 | Ref | |||
| Yes | 44.2 | 38.7–49.8 | 4.17 (3.0–5.8) | <0.001 | 0.64 (0.31–1.32) | 0.225 |
| Neoadjuvant treatment history | ||||||
| No | 45.1 | 42.1–48.1 | Ref | |||
| Yes | 39.7 | 24–55.3 | 2.19 (1.15–4.16) | 0.017 | 1.22 (0.62–2.39) | 0.561 |
| PKMYT1 expression | ||||||
| Low | 47.7 | 43.7–51.7 | Ref | |||
| High | 42.1 | 37.8–46.4 | 1.82 (1.32–2.52) | <0.001 | 1.79 (1.29–2.49) | 0.001 |
OS – overall survival; ccRCC – clear cell renal cell carcinoma; TCGA – The Cancer Genome Atlas; HR – hazard ratio; 95% CI – 95% confidence interval.
A novel nomogram model was established and validated using ROC and DCA
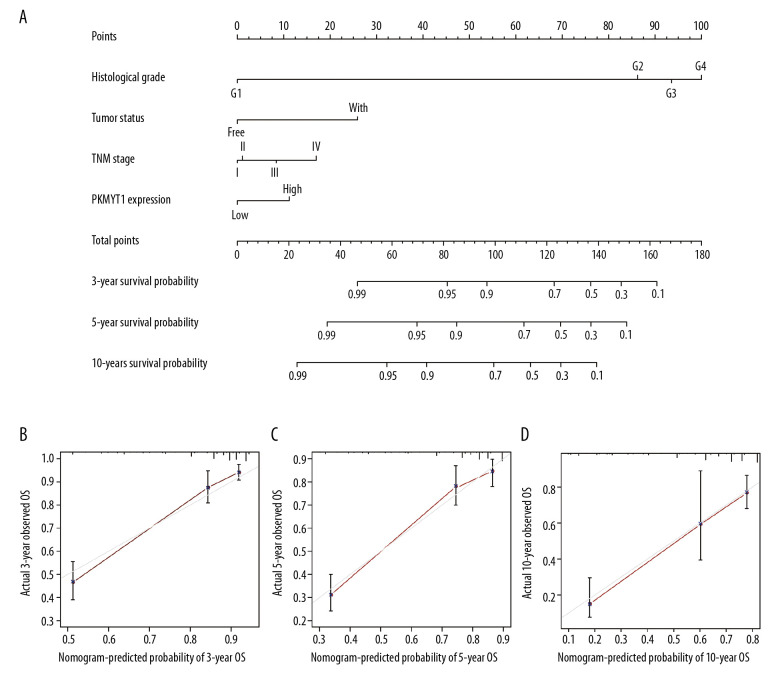
We next formulated a nomogram that incorporated statistically significant clinical parameters identified by multivariate analysis, thus predicting 3-, 5-, and 10-year OS for patients with ccRCC. Histologic grade made the greatest contribution to prognosis, followed by tumor status, TNM stage, and PKMYT1 expression (Figure 7A). The nomogram performed favorably for prediction, with a C-index of 0.784. The calibration plot for the probability of survival at 3, 5, and 10 years revealed great concordance between the prediction of the nomogram and actual observations (Figure 7B–7D).
Figure 7.
Nomogram model incorporating PKMYT1 mRNA expression and significant clinical variables to predict OS of ccRCC. (A) Nomogram for predicting the 3-, 5-, and 10-year OS of patients with ccRCC from the TCGA database. The calibration curves of OS at (B) 3, (C) 5, and (D) 10 years for ccRCC. OS – overall survival; ccRCC – clear cell renal cell carcinoma; TCGA – The Cancer Genome Atlas.
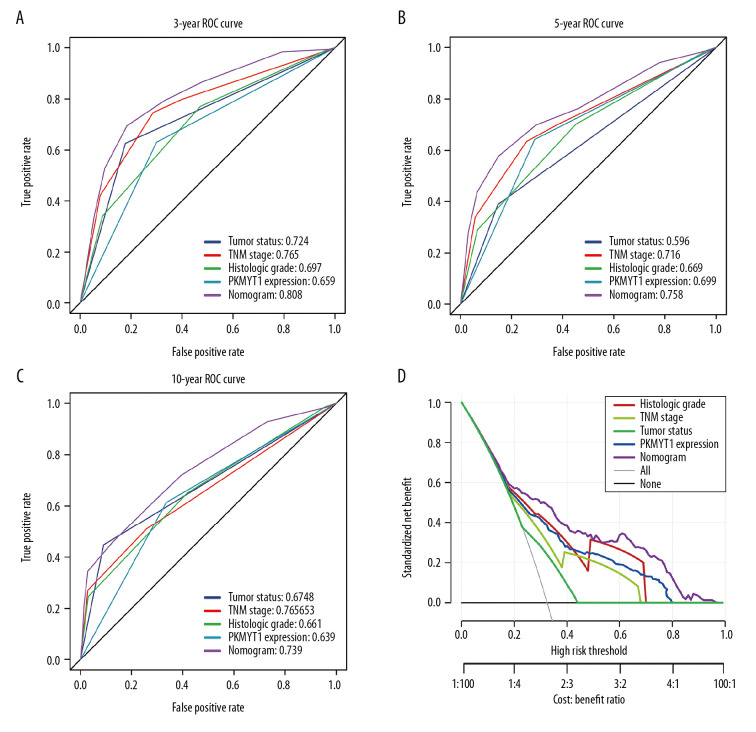
We further performed ROC analysis to compare the predictive efficiency of our nomogram with that for PKMYT1 expression, histological grade, TNM stage, and tumor status. In the ROC curves of 3-year OS in the TCGA dataset, our nomogram displayed the highest AUC value of 0.808 (95% CI: 0.779–0.867), followed by that for TNM stage (0.765, 95% CI: 0.659–0.797), tumor status (0.724, 95% CI: 0.687–0.767), histological grade (0.697, 95% CI: 0.662–0.717), and PKMYT1 expression (0.659, 95% CI: 0.599–0.707) (Figure 8A). Similarly, the AUC for the nomogram in predicting 5-year OS achieved a value of 0.758 (95% CI: 0.693–0.801), which was greater than those for TNM stage (0.716, 95% CI: 0.654–0.736), PKMYT1 expression (0.699, 95% CI: 0.628–0.746), histological grade (0.669, 95% CI: 0.592–0.698), and tumor status (0.596, 95% CI: 0.518–0.622) (Figure 8B). The AUC values for the nomogram, tumor status, histological grade, TNM stage, and PKMYT1 expression for 10-year OS were 0.739 (95% CI: 0.652–0.863), 0.678 (95% CI: 0.592–0.723), 0.661 (95% CI: 0.604–0.738), 0.653 (95% CI: 0.588–0.768), and 0.639 (95% CI: 0.572–0.743) (Figure 8C). These results underscore that our nomogram, which comprised PKMYT1 expression level and other clinical parameters, is an optimal model for predicting long-term prognosis of ccRCC. PKMYT1 expression had a slightly lower or similar predictive performance for predicting prognosis of ccRCC compared with that of the additional clinical indicators (including histologic grade, TNM stage, and tumor status).
Figure 8.
The predictive value and clinical utility of PKMYT1 expression level was assessed through time-dependent ROC and DCA. ROC curves of nomogram, PKMYT1 expression level, and additional clinical variables for (A) 3-, (B) 5- and (C) 10-year OS of ccRCC. (D) DCA curves of above parameters for prediction of OS in ccRCC. ROC – receiver operating characteristic; DCA – decision curve analysis; OS – overall survival; ccRCC – clear cell renal cell carcinoma.
In addition, we developed DCA curves to assess the clinical effect of our nomogram and other indicators, thus helping us to visually estimate their utility. As shown in Figure 8D, the DCA of our nomogram showed the greatest net benefits, followed by histological grade, PKMYT1 expression, TNM stage, and tumor status, which suggests that PKMYT1 expression level has relatively satisfactory clinical utility for predicting the prognosis of ccRCC.
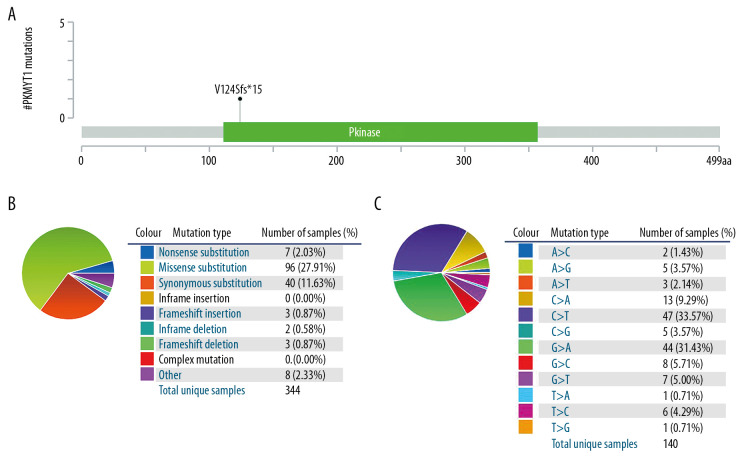
PKMYT1 mutations are rare in ccRCC
The mutation frequency of PKMYT1 in ccRCC was assessed through cBioPortal, which accounted for 0.1% (Figure 9A). Specifically, the mutant types of PKMYT1 in ccRCC were investigated through the COSMIC database. As shown in Figure 9B, missense mutation was the most frequent type of PKMYT1 mutation in ccRCC (27.91%), followed by synonymous mutation (11.63%), nonsense mutation (2.03%), and frameshift mutation (0.87%). Nucleotide alterations primarily consisted of C>T, G>A, C>A and G>C mutations, the greatest proportion of which (33.57%) were C>T (Figure 9C).
Figure 9.
PKMYT1 mutations are infrequent in ccRCC. (A) Schematic diagram describing PKMYT1 mutations in ccRCC based on the cBioportal. (B, C) Different PKMYT1 mutation types and the corresponding mutation proportions in ccRCC. ccRCC – clear cell renal cell carcinoma.
PKMYT1 positively modulates cell proliferation in ccRCC
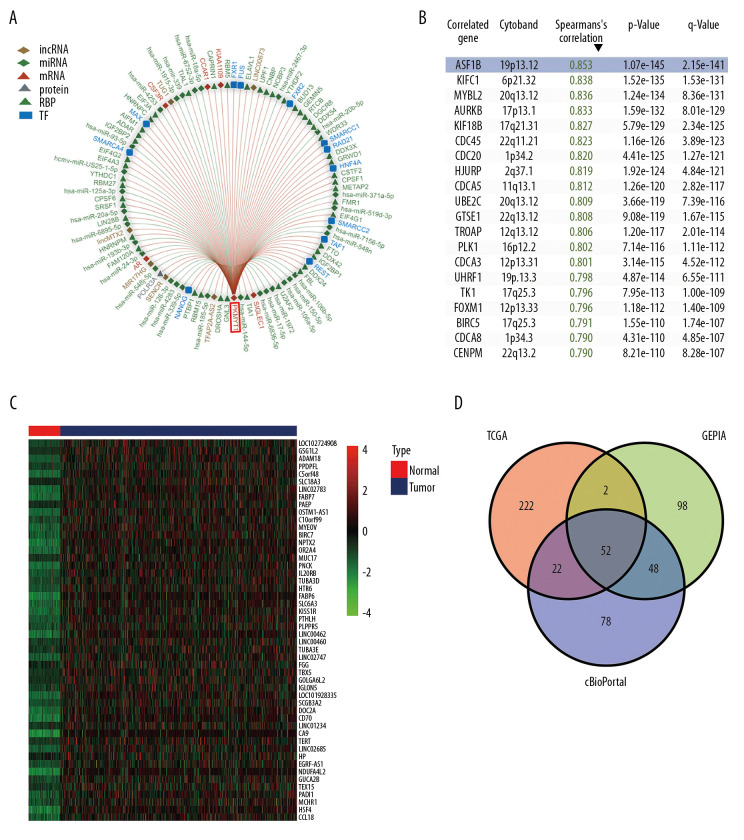
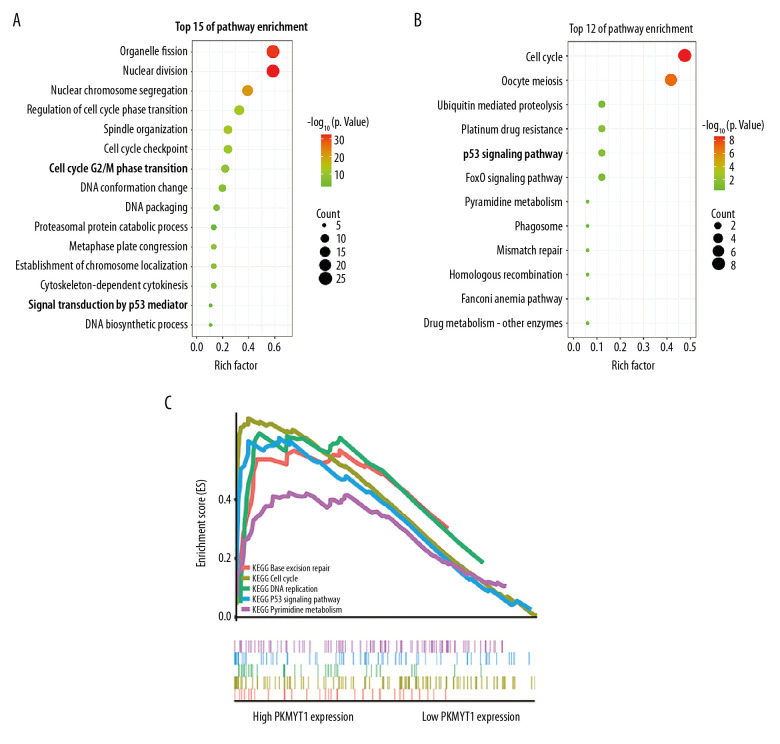
Using the RAID database, we investigated molecules that have potential to interact with PKMYT1. Specifically, PKMYT1 displayed crosstalk with 3 lncRNAs (lncMTX2, TUG1, and linc00673), multiple miRNAs (such as has-miR-20b-5p, has-miR-371b-5p, has-miR-519d-3p, and has-miR-106b-5p), mRNAs (such as AR, CSF3R, CCAR1, KIAA1109, and SIGLEC1), RBPs (such as WDR33, GRWD1, CPSF1, METAP2, and FMR1), and TFs (such as HNF4A, SWARCC2, TAF1, REST, and FXR1) (Figure 10A). Furthermore, TCGA database (Figure 10B), the cBioPortal dataset (Figure 10C), and the GEPIA dataset (Table 3) were overlapped to select 52 common genes that are positively coexpressed with PKMYT1 in ccRCC (Figure 10D). The functional and pathway enrichment analysis was conducted to explore the biological significance of these coexpressed genes. GO analysis demonstrated that pathway enrichment with the top 15 genes was associated with organelle fission, nuclear division, cell cycle G2/M phase transition, and signal transduction by p53 class mediator (Figure 11A), which was roughly consistent with the findings of the KEGG analysis (Figure 11B). As revealed in the results from GSEA, high expression of PKMYT1 was positively correlated with the cell cycle (NES=2.389, P<0.0001), p53 signaling pathway (NES=2.164, P<0.0001), DNA replication (NES=1.908, P=0.009), and base excision repair (NES=1.854, P=0.007) (Figure 11C). Therefore, PKMYT1 plays an essential role in facilitating cell proliferation in ccRCC.
Figure 10.
A series of genes coexpressed with PKMYT1 are revealed based on multi-database mining methods. (A) RAID map concerning miRNA, lncRNA, RBP, and TF interacting with PKMYT1. (B) The top 20 genes positively correlated with PKMYT1 expression in ccRCC based on the cBioPortal. (C) Heatmap of the top 50 DEGs that positively are coexpressed with PKMYT1 in ccRCC from the TCGA. (D) Venn diagram of the 52 common genes positively coexpressed with PKMYT1 in ccRCC through overlapping the TCGA, cBioPortal and GEPIA database. Abbreviations: RAID – RNA InterActome Database; miRNA – microRNA; lncRNA – long noncoding RNA; RBP – RNA-binding protein; TF – transcription factor; ccRCC– clear cell renal cell carcinoma; TCGA – The Cancer Genome Atlas; GEPIA – Gene Expression Profiling Interactive Analysis.
Table 3.
The top 20 genes positively correlated with PKMYT1 mRNA expression in ccRCC from the GEPIA database.
| Gene symbol | Gene ID | PCC |
|---|---|---|
| SPC24 | ENSG00000161888.11 | 0.85 |
| CENPM | ENSG00000100162.14 | 0.77 |
| TOP2A | ENSG00000131747.14 | 0.76 |
| RECQL4 | ENSG00000160957.12 | 0.74 |
| C16orf59 | ENSG00000162062.14 | 0.72 |
| PTTG1 | ENSG00000164611.12 | 0.72 |
| RP11-360L9.7 | ENSG00000253174.2 | 0.71 |
| POLD1 | ENSG00000062822.12 | 0.71 |
| TK1 | ENSG00000167900.11 | 0.71 |
| CDC20 | ENSG00000117399.13 | 0.69 |
| CDC45 | ENSG00000093009.9 | 0.69 |
| TROAP | ENSG00000135451.12 | 0.69 |
| B3GNTL1 | ENSG00000175711.8 | 0.67 |
| RNASEH2A | ENSG00000104889.4 | 0.67 |
| ATP5G1P4 | ENSG00000227440.1 | 0.66 |
| DDX39A | ENSG00000123136.14 | 0.66 |
| BIRC5 | ENSG00000089685.14 | 0.66 |
| PAQR4 | ENSG00000162073.13 | 0.66 |
| CDCA3 | ENSG00000111665.11 | 0.66 |
| ASF1B | ENSG00000105011.8 | 0.65 |
mRNA – messenger RNA; ccRCC – clear cell renal cell carcinoma; GEPIA – Gene Expression Profiling Interactive Analysis; PCC – Pearson correlation coefficient.
Figure 11.
PKMYT1 accelerates ccRCC progression by modulating the tumor cell cycle. The most significant signaling pathways in ccRCC found through (A) GO, (B) KEGG analysis, and (C) GSEA. Abbreviations: ccRCC – clear cell renal cell carcinoma; GO – gene ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; GSEA – gene set enrichment analysis.
Expressions of PKMYT1 and ASF1B in ccRCC are significantly correlated
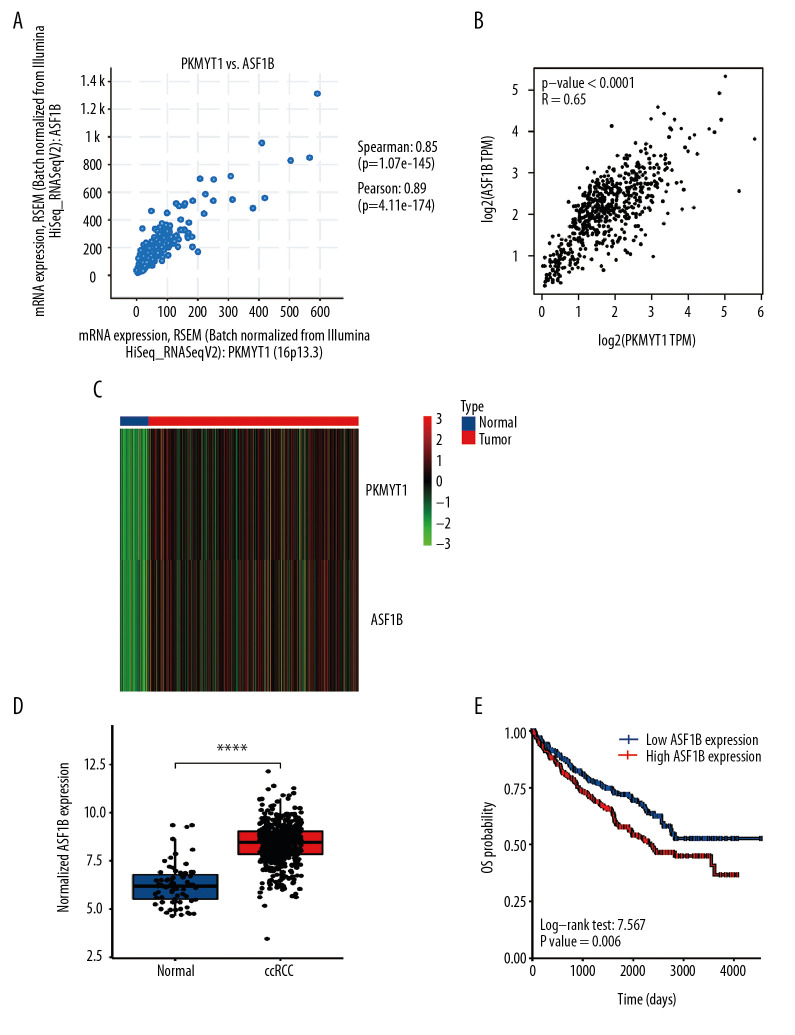
To further explore the underlying modulation of PKMYT1 in ccRCC, data extraction was conducted in a ccRCC cohort via cBioPortal. The regression analysis revealed that PKMYT1 and the anti-silencing function 1B (ASF1B) gene displayed the greatest correlation coefficient (Spearman’s correlation=0.85, P=1.07e–145; Pearson’s correlation=0.89, P=4.11e–174) (Figure 12A). Similarly, data based on GEPIA also demonstrated that there was a strongly positive correlation between PKMYT1 and ASF1B transcription (R=0.65, P<0.0001) (Figure 12B). A heat map in accordance with the TCGA database further confirmed the previously described positive correlation (Figure 12C). These findings suggest that PKMYT1 may be involved in ASF1B signaling pathways, and may be a functional partner of ASF1B in ccRCC. An analysis of RNA sequencing and clinical information extracted from TCGA dataset demonstrated that levels of ASF1B expression were significantly enhanced in ccRCC compared with normal tissue (Figure 12D). Furthermore, based on the Human Protein Atlas (HPA) database, a high ASF1B level was significantly correlated with diminished OS in ccRCC (P=0.006) (Figure 12E).
Figure 12.
PKMYT1 is significantly correlated with ASF1B gene expression in ccRCC. There was a positive correlation between PKMYT1 and ASF1B based on (A) cBioPortal regression analysis, (B) GEPIA, and (C) TCGA dataset. (D) ASF1B was significantly increased in ccRCC compared with normal tissue. (E) High expression of ASF1B was associated with poor prognosis for ccRCC. ccRCC – clear cell renal cell carcinoma; GEPIA – Gene Expression Profiling Interactive Analysis; TCGA – The Cancer Genome Atlas.
Discussion
Globally, RCC is the 9th most common tumor in men and the 10th most common in women, resulting in over 140 000 cancer-associated deaths annually [44–46]. ccRCC is the most frequent histological subtype of RCC, with high likelihood of invasion, metastasis, and resistance to chemotherapy, which makes eliminating this neoplasm more challenging [47,48]. Thus, a growing number of studies are focusing on the pathogenesis and diagnosis of and therapy for ccRCC.
In our study, we showed the level of expression and clinical significance of PKMYT1 in ccRCC for the first time. Initially, PKMYT1 was found to be one of the genes significantly associated with histologic progression of ccRCC via the WGCNA method. PKMYT1 mRNA levels were prominently increased in ccRCC compared with adjacent normal kidney samples in the TIMER, TCGA, and multiple GEO datasets. Notably, PKMYT1 mRNA levels were positively correlated with TNM stage and histological grade of ccRCC. ROC analysis demonstrated that PKMYT1 could distinguish healthy individuals from patients with ccRCC. In addition, enhanced PKMYT1 levels also corresponded to unsatisfactory prognosis in patients who were male, older, had TNM stage II or histological grade G4 disease, or disease that had relapsed. PKMYT1 expression was further identified as an independent adverse factor for prognosis of ccRCC through a survival analysis that integrated a Cox analysis and a nomogram. ROC curves and DCA both demonstrated that PKMYT1 expression levels were slightly less useful for predicting prognosis of ccRCC prognosis than traditional clinical indicators such as histological grade and TNM stage. Consistent with the present results, PKMYT1 has been shown to be upregulated in oncogenesis and progression of multiple human tumors, including colorectal cancer [49], esophageal squamous cell carcinoma (ESCC) [11], hepatocellular carcinoma (HCC) [22], non-small-cell lung cancer (NSCLC) [24], gastric cancer (GC) [21], and breast cancer [13], as well as OC [23]. These reports have also revealed a significant correlation between increased PKMYT1 expression and advanced clinical stage and progressive histological grade, as well as unsatisfactory prognosis of diversiform tumors [11,13,21–24,49], which coincides with the results from our study. One report revealed that mutation in the C-terminal domain of PKMYT1 influenced the prognosis of neuroblastoma by altering its catalytic activity [50]. A previous study also demonstrated that PKMYT1 was a potential ccRCC metastasis-related novel gene, based on expression profiles in cultures and tumor tissues [51]. Thus, high PKMYT1 expression potentially indicates tumor progression and it may be a promising diagnostic and prognostic biomarker for ccRCC.
In the present study, we highlighted that PKMYT1, as a pivotal regulator of G2/M transition, exerted a strong effect on DNA-damage recovery and maintenance of genomic stability during rapid ccRCC cell proliferation. Given the challenge of developing effective therapies for ccRCC and the significance of the G2/M checkpoint for tumor cell survival, we speculate that PKMYT1 may be an attractive molecular target for ccRCC treatment. Another report revealed that depletion of PKMYT1 suppressed the β-catenin/TCF pathway, which is considered a driver of the EMT in multiple tumors and that the gene can interact with primary EMT-related biomarkers, such as Twist, Snail, and Slug [22,52,53]. Similarly, downregulation of PKMYT1 impeded Twist expression by hindering the Akt/mTOR pathway, thus suppressing migration and metastasis as well the EMT process in ESCC cells [11]. Considering that EMT is a crucial determinant for tumor metastasis and progression [52,53], targeting the β-catenin/TCF and Akt/mTOR pathway through PKMYT1 may be a promising strategy for tumor treatment. Some PKMYT1 inhibitors have been developed, such as the tyrosine kinase inhibitors dasatinib and bosutinib. Studies of the pyridopyrimidine derivatives PD-0166285, PD-173952, and PD-173955 also are under way [11]. No potential PKMYT1 inhibitors have yet been studied in clinical trials [14].
ASF1, a histone H3–H4 chaperone that comprises 2 ASF1 isoforms (ASF1A and ASF1B), has a critical effect on modulation of S-phase progression in multiple organisms. Depletion of both ASF1 isoforms triggers primary defects in S-phase progression. ASF1A and ASF1B are crucial to handle histones at replication forks through interaction with the MCM2-7 complex, which is a putative helicase with the ability to unwind DNA ahead of the replication fork [54,55]. Importantly, high ASF1B expression correlates with faster disease development and metastasis in various tumors, including small-cell breast cancer [54], prostate cancer [56], and ccRCC [57]. Notably, in prostate cancer, downregulation of ASF1B suppressed expression of cyclin D1 and resulted in cell cycle arrest [56]. ASF1B overexpression accelerated tumor cell proliferation and migration in an Akt/P70 S6K1-dependent pathway in ccRCC [57]. Thus, the positive association of ASF1B and PKMYT1 in ccRCC suggests an underlying cell cycle checkpoint mechanism related to rapid tumor cell proliferation, based on maintenance of genomic stability. Overexpression of PKMYT1 may result in genomic stability through modulation of G2/M in cancer cells. The ASF1B regulatory pathway simultaneously participates in controlling the S-phase duration for prompt cell proliferation. Co-targeting PKMYT1 and ASF1B may be a promising approach to preventing ccRCC progression.
Our study deepens our comprehension of the correlation between PKMYT1 and ccRCC, but several limitations need to be discussed. Initially, our report was performed on the basis of data extracted from multiple public biological databases. Although we identified PKMYT1 expression at mRNA and protein levels and performed a survival analysis of ccRCC cases using information from TCGA, GEO, and HPA databases, validation of our results in further experiments is required to unravel the molecular mechanisms related to PKMYT1 in ccRCC development and prognosis. It is also essential to incorporate more prognostic factors into the nomogram to enhance the precision of survival analysis. Ultimately, the effects of PKMYT1 on drug resistance associated with ccRCC require validation in future studies.
Conclusions
Our study is the first to assess the upregulation and negative prognostic role of PKMYT1 in development of ccRCC, and our results suggest that the gene may be a promising therapeutic target for the management of ccRCC.
Supplementary Data
Analysis of network topology for multiple soft-thresholding powers.
PKMYT1 mRNA expression in ccRCC is stratified by (A) age, (B) sex, and (C) laterality. mRNA – messenger RNA; ccRCC – clear cell renal cell carcinoma.
Footnotes
Conflicts of interest
None.
Source of support: This work was supported by the National Natural Science Foundation of China [81770693 (Xiang Chen)]
References
- 1.Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–53. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Meskawi M, Sun M, Trinh QD, et al. A review of integrated staging systems for renal cell carcinoma. Eur Urology. 2012;62(2):303–14. doi: 10.1016/j.eururo.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 3.Carril-Ajuria L, Santos M, Roldán-Romero JM, et al. Prognostic and predictive value of PBRM1 in clear cell renal cell carcinoma. Cancers. 2019;12(1):16. doi: 10.3390/cancers12010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf MM, Kimryn Rathmell W, Beckermann KE. Modeling clear cell renal cell carcinoma and therapeutic implications. Oncogene. 2020;39(17):3413–26. doi: 10.1038/s41388-020-1234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linehan WM, Ricketts CJ. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat Rev Urol. 2019;16(9):539–52. doi: 10.1038/s41585-019-0211-5. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moch H, Cubilla AL, Humphrey PA, et al. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs – Part A: Renal, penile, and testicular tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 8.Srigley JR, Delahunt B, Eble JN, et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of renal neoplasia. Am J Surg Pathol. 2013;37(10):1469–89. doi: 10.1097/PAS.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- 9.López-Fernández E, López JI. The Impact of tumor eco-evolution in renal cell carcinoma sampling. Cancers. 2018;10(12):485. doi: 10.3390/cancers10120485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Wang L, Chen S, et al. PKMYT1 is associated with prostate cancer malignancy and may serve as a therapeutic target. Gene. 2020;744:144608. doi: 10.1016/j.gene.2020.144608. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Zhao X, Zhang C, et al. Overexpressed PKMYT1 promotes tumor progression and associates with poor survival in esophageal squamous cell carcinoma. Cancer Manage Res. 2019;11:7813–24. doi: 10.2147/CMAR.S214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: A membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270(5233):86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Qi J, Dou Z, et al. Systematic expression analysis of WEE family kinases reveals the importance of PKMYT1 in breast carcinogenesis. Cell Prolif. 2020;53(2):e12741. doi: 10.1111/cpr.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt M, Rohe A, Platzer C, et al. Regulation of G2/M transition by inhibition of WEE1 and PKMYT1 kinases. Molecules. 2017;22(12):2045. doi: 10.3390/molecules22122045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long HP, Liu JQ, Yu YY, et al. PKMYT1 as a potential target to improve the radiosensitivity of lung adenocarcinoma. Front Genet. 2020;11:376. doi: 10.3389/fgene.2020.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lescarbeau RS, Lei L, Bakken KK, et al. Quantitative phosphoproteomics reveals Wee1 kinase as a therapeutic target in a model of proneural glioblastoma. Mol Cancer Ther. 2016;15(6):1332–43. doi: 10.1158/1535-7163.MCT-15-0692-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurvitz SA, Martin M, Symmans WF, et al. Neoadjuvant trastuzumab, pertuzumab, and chemotherapy versus trastuzumab emtansine plus pertuzumab in patients with HER2-positive breast cancer (KRISTINE): A randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2018;19(1):115–26. doi: 10.1016/S1470-2045(17)30716-7. [DOI] [PubMed] [Google Scholar]
- 18.Suganuma M, Kawabe T, Hori H, et al. Sensitization of cancer cells to DNA damage-induced cell death by specific cell cycle G2 checkpoint abrogation. Cancer Res. 1999;59(23):5887–91. [PubMed] [Google Scholar]
- 19.Wells NJ, Watanabe N, Tokusumi T, et al. The C-terminal domain of the Cdc2 inhibitory kinase Myt1 interacts with Cdc2 complexes and is required for inhibition of G(2)/M progression. J Cell Sci. 1999;112(Pt 19):3361–71. doi: 10.1242/jcs.112.19.3361. [DOI] [PubMed] [Google Scholar]
- 20.Passer BJ, Nancy-Portebois V, Amzallag N, et al. The p53-inducible TSAP6 gene product regulates apoptosis and the cell cycle and interacts with Nix and the Myt1 kinase. Proc Natl Acad Sci USA. 2003;100(5):2284–89. doi: 10.1073/pnas.0530298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XM, Li QY, Ren LL, et al. Effects of MCRS1 on proliferation, migration, invasion, and epithelial mesenchymal transition of gastric cancer cells by interacting with PKMYT1 protein kinase. Cell Signal. 2019;59:171–81. doi: 10.1016/j.cellsig.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Wu J, Wang S, et al. PKMYT1 promoted the growth and motility of hepatocellular carcinoma cells by activating beta-catenin/TCF signaling. Exp Cell Res. 2017;358(2):209–16. doi: 10.1016/j.yexcr.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Xuan ZH, Wang HP, Zhang XN, et al. PKMYT1 aggravates the progression of ovarian cancer by targeting SIRT3. Eur Rev Med Pharmacol Sci. 2020;24(10):5259–66. doi: 10.26355/eurrev_202005_21308. [DOI] [PubMed] [Google Scholar]
- 24.Sun QS, Luo M, Zhao HM, Sun H. Overexpression of PKMYT1 indicates the poor prognosis and enhances proliferation and tumorigenesis in non-small cell lung cancer via activation of Notch signal pathway. Euro Rev Med Pharmacol Sci. 2019;23(10):4210–19. doi: 10.26355/eurrev_201905_17925. [DOI] [PubMed] [Google Scholar]
- 25.Yang JF, Shi SN, Xu WH, et al. Screening, identification and validation of CCND1 and PECAM1/CD31 for predicting prognosis in renal cell carcinoma patients. Aging. 2019;11(24):12057–79. doi: 10.18632/aging.102540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–59. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Wang Y, Liu F. A 44-gene set constructed for predicting the prognosis of clear cell renal cell carcinoma. Int J Mol Med. 2018;42(6):3105–14. doi: 10.3892/ijmm.2018.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi SN, Qin X, Wang S, et al. Identification of potential novel differentially-expressed genes and their role in invasion and migration in renal cell carcinoma. Aging. 2020;12:9205–23. doi: 10.18632/aging.103192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu D, Huber W, Vitek O. Shrinkage estimation of dispersion in Negative Binomial models for RNA-seq experiments with small sample size. Bioinformatics. 2013;29(10):1275–82. doi: 10.1093/bioinformatics/btt143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4(8):e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan L, Qian G, Chen L, et al. Coexpression network analysis of biomarkers for adrenocortical carcinoma. Front Genet. 2018;9:328. doi: 10.3389/fgene.2018.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui H, Shan H, Miao MZ, et al. Identification of the key genes and pathways involved in the tumorigenesis and prognosis of kidney renal clear cell carcinoma. Sci Rep. 2020;10(1):4271. doi: 10.1038/s41598-020-61162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Chen P, Xie H, Cao P. Using circulating tumor DNA as a novel biomarker to screen and diagnose hepatocellular carcinoma: A systematic review and meta-analysis. Cancer Med. 2020;9(4):1349–64. doi: 10.1002/cam4.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Xie H, Chen P, Cao P. Development and identification of a nomogram prognostic model for patients with primary clear cell carcinoma of the liver. Med Sci Monit. 2020;26:e919789. doi: 10.12659/MSM.919789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Calster B, Wynants L, Verbeek JFM, et al. Reporting and interpreting decision curve analysis: a guide for investigators. Eur Urol. 2018;74(6):796–804. doi: 10.1016/j.eururo.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int. 2019;19:138. doi: 10.1186/s12935-019-0858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–83. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forbes SA, Beare D, Bindal N, et al. COSMIC: High-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr Prot Human Genet. 2016;91:10.11–11.37. doi: 10.1002/cphg.21. [DOI] [PubMed] [Google Scholar]
- 40.Yi Y, Zhao Y, Li C, et al. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017;45(D1):D115–18. doi: 10.1093/nar/gkw1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–87. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, Lai W, Shi Y, et al. Annotation and cluster analysis of long noncoding RNA linked to male sex and estrogen in cancers. NPJ Precis Oncol. 2020;4:5. doi: 10.1038/s41698-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang F, Yi X, Guo J, et al. Association of plasma and urine metals levels with kidney function: A population-based cross-sectional study in China. Chemosphere. 2019;226:321–28. doi: 10.1016/j.chemosphere.2019.03.171. [DOI] [PubMed] [Google Scholar]
- 45.Zhang B, Liu P, Zhou Y, et al. Dihydroartemisinin attenuates renal fibrosis through regulation of fibroblast proliferation and differentiation. Life Sci. 2019;223:29–37. doi: 10.1016/j.lfs.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X, Cui Y, Chen J, et al. UCA1 promotes cell viability, proliferation and migration potential through UCA1/miR-204/CCND2 pathway in primary cystitis glandularis cells. Biomed Pharmacother. 2019;114:108872. doi: 10.1016/j.biopha.2019.108872. [DOI] [PubMed] [Google Scholar]
- 47.Du Y, Liu P, Chen Z, et al. PTEN improve renal fibrosis in vitro and in vivo through inhibiting FAK/AKT signaling pathway. J Cell Biochem. 2019;120(10):17887–97. doi: 10.1002/jcb.29057. [DOI] [PubMed] [Google Scholar]
- 48.Huang W, Cen S, Kang XL, et al. TGF-β1-induced Fascin1 promotes cell invasion and metastasis of human 786-0 renal carcinoma cells. Acta Histochemica. 2016;118(2):144–51. doi: 10.1016/j.acthis.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Jeong D, Kim H, Kim D, et al. Protein kinase, membrane-associated tyrosine/threonine 1 is associated with the progression of colorectal cancer. Oncol Rep. 2018;39(6):2829–36. doi: 10.3892/or.2018.6371. [DOI] [PubMed] [Google Scholar]
- 50.Novak EM, Halley NS, Gimenez TM, et al. BLM germline and somatic PKMYT1 and AHCY mutations: Genetic variations beyond MYCN and prognosis in neuroblastoma. Med Hypotheses. 2016;97:22–25. doi: 10.1016/j.mehy.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 51.Tan X, Zhai Y, Chang W, et al. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008;123(5):1080–88. doi: 10.1002/ijc.23637. [DOI] [PubMed] [Google Scholar]
- 52.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Ann Rev Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 53.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–26. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Corpet A, De Koning L, Toedling J, et al. ASF1B, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30(3):480–93. doi: 10.1038/emboj.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klimovskaia IM, Young C, Strømme CB, et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat Commun. 2014;5:3394. doi: 10.1038/ncomms4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han G, Zhang X, Liu P, et al. Knockdown of anti-silencing function 1B histone chaperone induces cell apoptosis via repressing PI3K/Akt pathway in prostate cancer. Int J Oncol. 2018;53(5):2056–66. doi: 10.3892/ijo.2018.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiangqiao Z, Tao Q, Zhongbao C, et al. Anti-silencing function 1B histone chaperone promotes cell proliferation and migration via activation of the AKT pathway in clear cell renal cell carcinoma. Biochem Biophys Res Commun. 2019;511(1):165–72. doi: 10.1016/j.bbrc.2019.02.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of network topology for multiple soft-thresholding powers.
PKMYT1 mRNA expression in ccRCC is stratified by (A) age, (B) sex, and (C) laterality. mRNA – messenger RNA; ccRCC – clear cell renal cell carcinoma.