Magnetotactic bacteria are aquatic or sediment-dwelling microorganisms able to take advantage of the Earth’s magnetic field for directed motility. The source of this amazing trait is magnetosomes, unique organelles used to synthesize single nanometer-sized crystals of magnetic iron minerals that are queued up to build an intracellular compass. Most of these microorganisms cannot be cultivated under controlled conditions, much less genetically engineered, with only few exceptions. However, two of the genetically amenable Magnetospirillum species have emerged as tractable model organisms to study magnetosome formation and magnetotaxis.
KEYWORDS: magnetotaxis, magnetosome, magnetospirillum, magnetoskeleton, cytoskeleton, cell shape, flagella
ABSTRACT
Magnetotactic bacteria are aquatic or sediment-dwelling microorganisms able to take advantage of the Earth’s magnetic field for directed motility. The source of this amazing trait is magnetosomes, unique organelles used to synthesize single nanometer-sized crystals of magnetic iron minerals that are queued up to build an intracellular compass. Most of these microorganisms cannot be cultivated under controlled conditions, much less genetically engineered, with only few exceptions. However, two of the genetically amenable Magnetospirillum species have emerged as tractable model organisms to study magnetosome formation and magnetotaxis. Recently, much has been revealed about the process of magnetosome biogenesis and dedicated structures for magnetosome dynamics and positioning, which suggest an unexpected cellular intricacy of these organisms. In this minireview, we summarize new insights and place the molecular mechanisms of magnetosome formation in the context of the complex cell biology of Magnetospirillum spp. First, we provide an overview on magnetosome vesicle synthesis and magnetite biomineralization, followed by a discussion of the perceptions of dynamic organelle positioning and its biological implications, which highlight that magnetotactic bacteria have evolved sophisticated mechanisms to construct, incorporate, and inherit a unique navigational device. Finally, we discuss the impact of magnetotaxis on motility and its interconnection with chemotaxis, showing that magnetotactic bacteria are outstandingly adapted to lifestyle and habitat.
INTRODUCTION
In 1975, Richard Blakemore rediscovered what had been observed by Salvatore Bellini in 1963 as “a unique behavior of freshwater bacteria”: some of them seemed able to sense and swim along magnetic fields, apparently by use of magnetic iron minerals (1, 2). Today, 45 and 57 years later, respectively, much has been learned about this exceptional sense in what are now known as “magnetotactic bacteria” (MTB), which actually represent a phylogenetically diverse collection of single and multicellular bacteria that are abundant and widespread in almost any aquatic habitat (3, 4). These bacteria all share the ability to form magnetosomes, which are organelles consisting of dedicated vesicles, each synthesizing a perfect crystal of a magnetic iron mineral that operates as part of a sensor for the Earth’s magnetic field. This sensor is assumed to direct the swimming of MTB along magnetic field lines and vertical redox gradients within sediments or oxic-anoxic transition zones of natural waters (5–8). The number, size, shape, subcellular arrangement, and chemical composition of the magnetic crystals vary considerably between phylogenetic groups (4, 9, 10), reflecting a high diversity of biomineralization processes and adaptation to lifestyle, biotope, and cell morphology. This suggests that magnetosomes represent one of the most complex structures found in prokaryotic cells (11, 12). The precise control that is exerted during all stages of biomineralization yields magnetic nanoparticles with exceptionally well-defined characteristics such as high crystallinity, strong magnetization, and a uniform size distribution, mostly outcompeting technically produced magnetic nanocrystals (13, 14). Moreover, the particular magnetosome envelope provides an excellent target to add artificial functions by genetic and biochemical coupling of diverse bioactive moieties such as fluorophores, enzymes, antibody fragments, or other ligands (15, 16). Since the organelles can readily be isolated from disrupted cells, they can also be modified in vitro, and much of the interest in their biosynthesis has been motivated by their potential use in several biotechnical and biomedical settings. For example, the use of bacterial magnetosomes has been successfully tested in pilot applications such as magnetic imaging techniques like magnet resonance imaging and magnetic particle imaging (17) or magnetic hyperthermia (18, 19). Because of the precisely controlled, unique properties of magnetosomes, MTB have attracted interdisciplinary scientific attention for decades not only from microbiologists and biomedical researchers but also from physicists, materials scientists, geologists, paleontologists, and others. Therefore, much progress has been made in applied research on MTB and the use of magnetosomes, which has been reviewed, for example, by Vargas et al. (16) and others.
Inevitably, MTB have also emerged as an attractive model to study formation of prokaryotic organelles and bacterial cell biology. The increasing and diversifying scientific community and the approach of new techniques in recent years have led to surprising discoveries about the genuine biological function of magnetosomes, the molecular details of their synthesis, and bacterial magnetotaxis in general. On the other hand, the ability of eukaryotic organisms to sense magnetic fields remains a mystery.
Most of our knowledge about magnetosome structure and biosynthesis comes from studies of the two closely related Alphaproteobacteria, Magnetospirillum magneticum and Magnetospirillum gryphiswaldense, which, compared to most other MTB, can be cultivated and genetically manipulated reasonably well. Both species are microaerophiles that can also grow anaerobically by denitrification; they share many similarities with respect to their magnetosome biosynthesis and cell morphology, and yet they also exhibit several notable differences in magnetosome structure and intracellular organization. While increasing knowledge is accumulating also from other cultured and uncultured MTB, we will mostly focus here on magnetosome structure, biosynthesis, and biological function in magnetospirilla.
ARCHITECTURE AND STEPWISE BIOSYNTHESIS OF MAGNETOSOMES
In both M. magneticum and M. gryphiswaldense, magnetosomes consist of single cuboctahedral crystals of magnetite (Fe3O4), which in their mature state are about 45 nm in size (Fig. 1C). Early studies in the related M. magnetotacticum indicated that each magnetite particle is enveloped by a membrane containing phospholipids and proteins, and the complete entity comprising the mineral core plus the surrounding membrane was termed a “magnetosome” (20, 21). The presence of a similar magnetosome membrane was later confirmed in M. magneticum, M. gryphiswaldense, and apparently all other MTB.
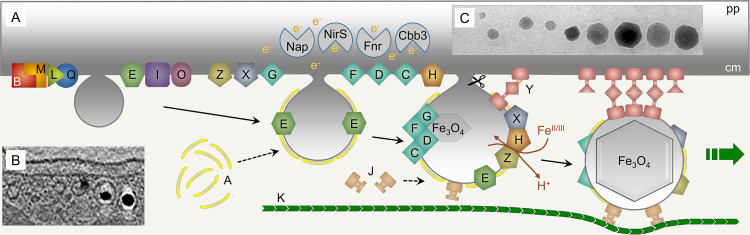
FIG 1.
Steps and some key proteins of magnetosome biogenesis. (A) Essential membrane-bound Mam proteins (labeled with respective letters) (see also Table 1) are thought to tightly interact at the cytoplasmic membrane and eventually facilitate formation and growth of vesicles. Later, more proteins that function in iron transport, redox control, and magnetite precipitation are recruited. Soluble proteins are associated with the periphery of the vesicles and either play a role in magnetosome membrane assembly, e.g., MamA, or are involved in the positioning and mobility of the vesicles, e.g., MamJ and MamK. Magnetite precipitation and crystal growth require not only magnetosome vesicles equipped with Mam and Mms proteins as a “nanoreactor” but also appropriate cellular redox conditions, depending on nitrate and oxygen respiration controlled by Nap, NirS, Fnr, and Cbb3 that act in the periplasmic (pp) space or the cytoplasmic membrane (cm). In M. gryphiswaldense, vesicles eventually become pinched off the cytoplasmic membrane by an unknown mechanism. Note that only a subset of the essential and accessory factors is shown. For a synopsis of magnetosome-associated proteins in magnetospirilla, refer to Table 1. (B) Cryo-electron tomography image of nascent magnetosomes in an M. gryphiswaldense cell. (Adapted from PLoS Genetics [22].) (C) Transmission electron microscopy (TEM) image of magnetite crystals in a wild-type cell close to the end of a chain highlighting their even cuboctahedral shape.
The biosynthesis of magnetosomes was subsequently revealed to be a complex, stepwise process that can be genetically dissected (11, 12, 22, 23). First, the magnetosome membrane is invaginated from the cytoplasmic membrane. Second, a set of specific magnetosome proteins is sorted to the magnetosome membrane. Third, iron is transported into the membrane vesicle and mineralized as magnetite crystal. Fourth, a magnetosome chain is assembled, positioned, and partitioned during cell division.
Proteomic and comparative genomic studies revealed that all steps are highly controlled by a distinct set of genes that are harbored within a genomic island in M. gryphiswaldense (24, 25) but also in the related M. magneticum and, apparently, in all MTB (9, 26–30) (Table 1). A core set of these so-called mam, mms, mad, and man genes seems conserved and proved useful as genomic signature for the potential to build magnetosomes (31). Transfer of 32 essential and accessory mam and mms genes from M. gryphiswaldense into the photosynthetic alphaproteobacterium Rhodospirillum rubrum resulted in its “magnetization,” that is, it caused the biosynthesis of magnetosomes resembling those of the donor M. gryphiswaldense and demonstrated that these genes are basically sufficient to confer magnetosome biosynthesis to this hitherto-nonmagnetic microbe (32). This also highlights that the genetic equipment to form magnetosomes can be transmitted horizontally (33, 34), although magnetotaxis capabilities of the recent MTB mostly seem to descend from a common ancestor (31, 35, 36).
TABLE 1.
Synopsis of genes known to control magnetosome biosynthesis in magnetotactic spirillaa
| Genes | Affected function upon deletion |
|---|---|
| mamA (“mms24”), mamB, mamQ/mamQ-like | Vesicle formation |
| mamC (“mms13”), mamD (“mms7”)/mamD-like, mamE/limE/mamE-like, mamF/mamF-like, mamG, mamH, mamI, mamL/mamL-like, mamM, mamN, mamO/limO, mamP, mamR, mamS, mamT, mamX, mamZ, mms5, mms6, mms36, mms48, mmsF, mmxF, feoB1, feoB2, ftsZm, feR5, feR6, amb411 | Iron transport/magnetite biomineralization and crystal size control |
| mamJ/limJ/mamJ-like, mamK/mamK-like, mamY | Magnetosome chain formation, localization and dynamics |
| mamU, mamV, mamW, mms5 | Uncertain |
| nap, nirS, nirN, norC, norB, fnr, cbb3, fur (not magnetosome island encoded) | Redox balance or iron homeostasis |
All essential and most of the accessory genes cluster in a genomic island (“magnetosome island”). A few genes have been identified and named twice, such as mamA/mms24. The “-like” genes are restricted to M. magneticum and have been identified as paralogs of the respective mam genes in a genomic “islet” of this organism. This redundancy is partially responsible for some differences in deletion mutant phenotypes of M. gryphiswaldense and M. magneticum. In other MTB, homologs for most of the mam and mms genes have been detected as well. However, with increasing phylogenetic distance to magnetospirilla, distinctive genes occur such as mad genes in greigite mineralizing Deltaproteobacteria or man genes in Nitrospirae. Moreover, some gene functions are redundant and some deletion phenotypes are pleiotropic so that unique functions cannot yet be assigned to all of the listed genes (for example, MamB plays a crucial role in vesicle formation but also in iron transport [47]). Biomineralization phenotypes owing to nap, nir, nor, fnr, cbb3, and fur deletion are likely an indirect effect of perturbed cellular redox balance or iron homeostasis. Products of further genes (not listed) have been found associated with magnetosomes (43), but their significance is not yet clear.
FORMATION OF MAGNETOSOME MEMBRANE VESICLES
Magnetosome vesicles are formed by invagination of the cytoplasmic membrane and apparently become pinched off at later stages of biogenesis in M. gryphiswaldense (Fig. 1A and B) but seemingly remain connected to it in M. magneticum, raising the possibility of a continuum between magnetosome interior and periplasm at least temporarily during vesicle growth. However, plug-like structures that separate the magnetosome lumen from the periplasm are conceivable and probable.
The formation of the magnetosome membrane is independent of magnetite biomineralization, as shown by the presence of empty vesicles in iron-starved cells or biomineralization-defective mutants. Likewise, empty vesicles are formed when cells are cultivated under aerobic conditions which suppress magnetite biomineralization, likely due to inappropriate redox conditions (37–39). In both Magnetospirillum strains, these invaginations originate simultaneously from several nonspecific cellular locations, as was revealed by tracking de novo magnetosome biogenesis by time-lapse fluorescence microscopy and cryo-electron tomography (22, 23, 40).
The magnetosome membrane has a lipid composition similar to that of the cytoplasmic membrane, but it contains a distinct set of proteins encoded in the genomic magnetosome island and with functions in magnetite biomineralization (21, 41). Magnetosome proteins are present in different quantities from one or few (e.g., MamZ) up to 120 copies (MamC) per particle (41–43). The high protein content of the magnetosome membrane suggests a crowded composition and a tight packing with transmembrane domains of integral proteins. It has been proposed that a lipid raft-like association of magnetosome-membrane proteins takes place prior to the magnetosome invagination (22). How proteins are specifically targeted to the magnetosome membrane is not known, and no conserved motifs that encode sorting signals to the magnetosome membrane have been identified.
The four proteins MamB, MamI, MamL, and MamQ were recognized as key factors in the early biogenesis of the magnetosome membrane (22, 26, 29). Among them, elimination of MamB completely abolished formation of regular magnetosome membranes, while mutants of MamI, MamL, or MamQ still contained fewer immature vesicles in M. gryphiswaldense (22). The crucial role of the cation diffusion facilitator (CDF) protein MamB in membrane biogenesis is independent from its function in iron transport but involves interactions with other magnetosome proteins, including the paralogous CDF transporter MamM and the protease MamE, which acts in processing and protein sorting to magnetosomes (44–47). From available genetic and biochemical data, a model for magnetosome membrane formation has been proposed in which MamB serves as landmark protein that interacts with a subset of proteins at the inner cell membrane (Fig. 1A). This initial protein complex is then thought to recruit further interaction partners that by protein crowding eventually induce lateral pressure to generate membrane curvature and finally membrane vesicles, possibly by a “blebbing” mechanism (22).
BIOMINERALIZATION OF MAGNETITE CRYSTALS
Compartmentalization of biomineralization by the magnetosome membrane provides a specialized “nanoreactor” in which the iron redox species and pH environments of biomineralization can be strictly regulated for the formation of single crystals of magnetite. Most available evidence supports a model in which extracellular iron is first imported into the cytoplasm by generic transporters. Subsequently, iron is transported from the cytoplasm into membrane vesicles by the magnetosome-specific transporters MamB and MamM (for ferrous iron), which are members of the Fe/Zn-transporting subfamily of divalent metal CDF proteins (47), as well as MamH and MamZ (for ferric iron), which are members of the major facilitator superfamily (48). The proper Fe2+/Fe3+ ratio for production of the mixed-valence iron mineral magnetite inside magnetosome membrane vesicles is thought to be regulated by MamE, MamP, MamT, and MamX, which are also constituents of the magnetosome membrane (41). These proteins each contain two or three conserved MTB-specific CXXCH c-type cytochrome heme-binding motifs denoted the “magnetochrome” domain that may oxidize magnetosomal Fe2+ for biomineralization (49, 50). In addition, the redox balance for magnetite biomineralization is also affected by the activity of cellular electron transport chains (Fig. 1). For instance, magnetite formation is linked to dissimilatory nitrate reduction, and M. gryphiswaldense cells are impaired in magnetosome biomineralization upon deletion of genes encoding the periplasmic nitrate and nitrite reductases Nap and NirS, as well in cells lacking the fumarate and nitrate reduction regulator protein Fnr. In a similar manner, inactivation of the terminal oxidase Cbb3 involved in aerobic respiration caused pleiotropic effects on magnetosomes under microaerobic conditions, probably by disturbing the redox balance required for proper magnetite biomineralization (38, 39, 51).
After nucleation of the magnetite crystal, several magnetosome proteins regulate their maturation into particles of defined size and shape in a positive (MamG, MamF, MamD, MamC, MamS, MamR, MamN, Mms6, and MmsF) or negative (Mms36 and Mms48) manner (Fig. 1 and Table 1). However, an understanding of how they exactly interact with the surface of the nascent magnetite crystals is just emerging (27, 52–54). Mature magnetosomes finally contain a single-domain magnetite (or greigite) crystal of species-specific shape and size, mostly between 30 and 100 nm. Under optimal growth conditions, an M. gryphiswaldense cell contains up to 100 cuboctahedral magnetosome particles of about 40 nm in size. However, the molecular mechanisms which ensure an optimal number of magnetosomes are not well understood. Apparently, the key biosynthetic genes are constitutively expressed (25), as well as poorly regulated, and simultaneous overexpression of almost all mam and mms gene clusters substantially increases the numbers and sizes of magnetosomes (55).
However, synthesis of magnetosomes alone is not sufficient for magnetotaxis because mature but disorganized magnetosomes would be still unsuitable to guide the cell within magnetic fields.
COMPASS CONSTRUCTION AND MAGNETOTAXIS
Magnetotaxis differs from conventional chemotaxis paradigms known from, e.g., Escherichia coli. Here, chemotactic swimming resembles a three-dimensional (3D) trial-and-error walk where periods of straight movement are interrupted by tumbling pauses where cells turn randomly before they resume straight swimming. These so-called run-and-tumble sequences are biased by chemosensory signal cascades, which control the frequency of runs and tumbling by interaction with flagellar motor proteins in response to detected gradients of nutrients, repellents or electron acceptors (reviewed, for example, in reference 56). Magnetospirilla and all other characterized MTB thus far also use flagella for motility, and aerotaxis or even phototaxis have been described (57, 58), but in contrast to E. coli, magnetospirilla were observed to swim in long runs interrupted by short reversals, resulting in an immediate change of direction of almost 180° (10). So, what is the benefit of detecting the Earth’s magnetic field in addition to chemical gradients, and how does this sense feed into motility of MTB?
Magnetotaxis could, for example, be beneficial for a bacterium if it combined the sense of direction provided by magnetosomes with decisions on locomotion, i.e., on run-and-tumble/reversal frequencies which could be conveyed by interaction of MamK with the chemosensory system (as suggested previously [59–61]). However, until now, neither deviant chemotaxis patterns in a mamK mutant nor direct evidence for a biochemical signal transduction between magnetosome chain and flagellar motor have been found. This and the lack of any canonical signal transduction motif (as, for example, in kinases or methylases) in the magnetosome gene clusters suggest that magnetotaxis functions in a different way and exploits magnetic forces directly. In recent years, the evidence for the latter idea has increased, but some biophysical considerations are necessary to comprehend this paradigm.
It has been shown that the force generated by a single magnetosome in the geomagnetic field is too weak to align a cell effectively to the field vector (62, 63). On the other hand, multiple free magnetosomes within a cell agglomerate by their own magnetic pull, which again minimizes their net magnetic moment close to zero (64–66). Hence, to serve as efficient magnetic field receptor, single magnetosomes must become stacked into a linear structure that adds the single magnetic moment of each unit to form a much stronger magnetic dipole. In fact, almost all MTB regardless of their phylogenetic affiliation, cell morphology, or lifestyle seem to carefully organize their magnetosomes into chains, most likely by conserved actin-like proteins. This also suggests that the number of magnetosomes per cell is controlled so that the assembled magnet becomes strong enough to align whole cells to the rather weak geomagnetic field akin to a compass needle (62, 67–69). Indeed, the cell size and number of magnetosomes appear to be correlated. Whereas ∼45 particles on average within a single chain (70) are sufficient to align a cell of the model organism M. gryphiswaldense (66), MTB with larger cells, such as members of the Nitrospirae phylum or multicellular Deltaproteobacteria, tend to possess a multitude of magnetosomes and chains (9, 71), which increase the magnetic force for their alignment. These findings suggest both that cell alignment to the magnetic field is a passive, physical effect and that MTB possess a dedicated cytoskeleton allowing them to form, stabilize and (re)position a modular magnetic field sensor suitable for navigation.
With such systems in place, the need for cell tumbling motion in MTB is likely obviated. The spatial orientation of their cells is controlled by the magnetosome chain, and their motility axis is preset by the position of flagella (incidentally even in fast swimming magnetotactic cocci [72]). The alignment to the geomagnetic field then reduces three-dimensional trial-and-error-swimming to a linear (or quasilinear for cocci and spherical multicellular MTB [73]) movement along vertical inclines of the Earth’s magnetic field. In this way, together with integration of chemotactic responses such as aerotaxis, MTB are efficiently guided to their preferred oxygen concentration in stratified environments (74). However, it turned out recently that spirilla, in particular, have evolved sophisticated adaptations to optimize and inherit their magnetic navigation skills, which will be highlighted in the following section.
DYNAMICS AND PARTITIONING OF MAGNETOSOME CHAINS
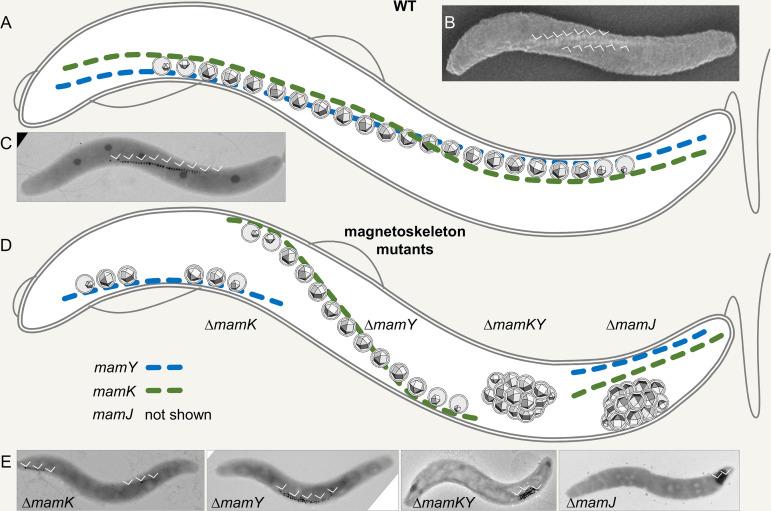
During maturation, nascent magnetosomes become organized and assembled into chains. Essential active parts of this assembly and positioning machinery consist of dedicated cytoskeletal proteins first identified years ago in the two model organisms M. gryphiswaldense and M. magneticum. In both bacteria, the magnetosomes become concatenated by the joint action of the actin-like MamK, which polymerizes into cell-spanning dynamic filaments (75–78) (Fig. 2A to C), and MamJ, an adaptor protein with less understood function that seems to be weakly structured, and central parts of it are dispensable (79, 80). In M. magneticum, both proteins exist as two paralogs with overlapping but also slightly different functions causing less pronounced phenotypes upon gene deletion compared to M. gryphiswaldense (81, 82). mamK mutants fail to assemble wild-type-like continuous magnetosome chains but contain either disordered chains (in M. magneticum [75]) or short and fragmented chains in M. gryphiswaldense (77) (Fig. 2D and E). The mamJ deletion mutant phenotype in M. gryphiswaldense is striking since magnetosomes are completely detached from MamK filaments and cluster irregularly, or they form magnetic flux-closed rings due to their unconstrained magnetic interaction (79, 83, 84) (Fig. 2D and E). In M. magneticum, which contains two MamJ paralogs, the phenotype is less pronounced, but magnetosome chain formation is also perturbed (78). These phenotypes and suggested MamK-MamJ interactions (79) led to the early model of an only two-part cytoskeletal structure, consisting of a presumably static backbone made of MamK filaments and MamJ, which attaches magnetosomes to that scaffold. However, this model recently proved incomplete in at least two aspects: first, the magnetosome chain was revealed to be highly dynamic, and second, the localization of the chain in spirilla is much more controlled than initially assumed.
FIG 2.
Magnetosome organization in wild-type and phenotypes of magnetoskeleton mutants in M. gryphiswaldense. (A to C) Wild type. (A) Magnetosomes in wild-type cells are organized in extended straight chains along the magnetoskeleton formed by membrane-bound MamY, the cytoplasmic actin-like MamK, and MamJ proteins (MamJ is not shown). Due to the two-dimensional (2D) nature of the scheme, the chain seemingly detaches from the cell envelope in its central part but actually stays in close proximity to it. (B) Scanning electron microscopy image of an M. gryphiswaldense wild-type cell harboring two magnetosome chains (arrowheads, iron detected by energy dispersive X-ray spectroscopy) and indicating the twisted cell morphology. (C) TEM (2D) image of an M. gryphiswaldense wild-type cell. Arrowheads indicate the magnetosome chain. (D and E) Phenotypes of magnetoskeleton mutants in M. gryphiswaldense. (D) Schematic view of magnetosome organization in mamK, mamY, mamKY, and mamJ mutants. mamK, short, fragmented, off-center chains attached to positive membrane curvature; mamY, chain shifted to the negative inner cell curvature; mamKY, agglomerated magnetosomes or magnetic flux-closed rings; mamJ, similar to mamKY. (E) Representative TEM images are shown in the lower panel. The position of the magnetosomes is indicated by arrowheads.
MamK has been analyzed extensively in vitro, and the pure protein was found to polymerize into filaments when ATP or GTP are present (85–87). The crystal structure of the protein and molecular details of its polymeric form have been solved (88) and underpinned its structural relatedness to eukaryotic actin proteins. In vivo studies analyzed the dynamics of polymeric MamK filaments in their cellular context. By fluorescence recovery after photobleaching experiments it was found that MamK treadmilling speed in M. gryphiswaldense is about 300 nm/min (89) and depends on its ATP hydrolysis capacity. Similar observations were made in M. magneticum (78, 82, 90). Interestingly, the dynamics of the MamK filaments was found to also depend on MamJ, which therefore seems not only necessary to tether magnetosomes to MamK filaments but also necessary for tuning MamK turnover rates.
The dynamics of MamK filaments in M. gryphiswaldense already suggested that a magnetosome chain may be not a static structure and that individual magnetosomes are moved along the track. In fact, active movement of the organelles seems a crucial prerequisite to assemble a chain: magnetosome vesicles emerge more or less randomly at the cytoplasmic membrane (22). Mature magnetosomes, however, are regularly found in chains, suggesting an active collecting and positioning mechanism, although spontaneous incorporation of maturing magnetosomes based on their increasing magnetism may also play a role (91). The function of MamK in M. magneticum seems somewhat different, but MamK is still needed to restrict movement and to position magnetosomes (23, 92). There is, however, other evidence for active translocation of magnetosomes: when M. gryphiswaldense cells divide, each daughter cell receives exactly half the number of magnetosomes from the mother cell indicating that cells distribute their magnetosomes with highest possible precision to their offspring (89, 93). This is achieved by strict positioning of the chain center at the cell division site. After cytokinesis, the daughter chains undergo repositioning from the new cell poles to midcell, and they are maintained at this position during growth by an unknown mechanism. Photobleaching experiments suggest that MamK filaments in M. gryphiswaldense nucleate close to the cell poles and grow toward midcell, i.e., into the direction where magnetosomes do migrate. In catalytic site mutants with severely impaired dynamics of MamK filaments, chain dynamics are also perturbed (i.e., chains are not efficiently relocalized to midcell), leading to unequal distribution of the organelles, a phenotype that is also seen in mamK and mamJ deletion mutants (89, 94). Despite these data, the molecular mechanism by which magnetosomes move along MamK filaments is still unknown. On one hand, effective repositioning of magnetosomes seems to depend on intact MamK turnover rates. On the other hand, the treadmilling speed of MamK filaments seems much higher than the speed of magnetosomes (89). Since MamK monomers are immobile once incorporated into the filament, and homologs of myosin-like cargo proteins that might walk along the actin-like MamK filaments have not been found, the mechanism of magnetosome dynamics cannot yet be fully explained. However, a candidate for further studies might be the so far poorly characterized MamJ protein.
Potential factors that possibly control nucleation of MamK filaments and their polarity are also unknown. It is also not clear what happens if the filaments of opposite polarity converge at midcell and if MamK filaments stop growing at all. For example, it has been observed that filaments that reach the opposite cell pole bend and turn, sometimes even at physiological expression levels (Fig. 3Aii) (75, 89, 94), which has never been observed for magnetosome chains. The reason for the different phenotypes of the mamK and the mamJ mutants (fragmented chains versus clustered magnetosomes) in M. gryphiswaldense has been also enigmatic for many years but this mystery became elucidated recently.
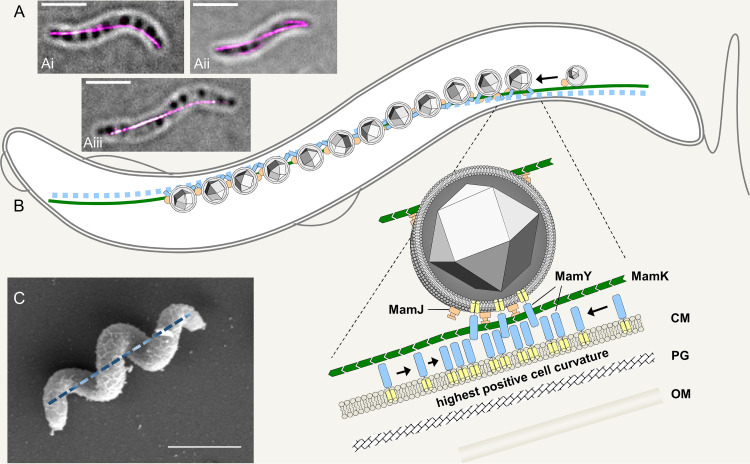
FIG 3.
Localization and function of magnetoskeleton constituents. (A) Localization of mCherry-MamK (i and ii) and mCherry-MamY (iii) in live M. gryphiswaldense cells imaged by 3D structured illumination microscopy (3D-SIM; maximum-intensity projection overlaid with a bright-field image; scale bar, 2 μm). (B) 2D model of the current view of the “magnetoskeleton” in M. gryphiswaldense, as suggested by Toro-Nahuelpan et al. (99). The actin-like MamK (green) polymerizes into cell-spanning dynamic cytoplasmic filaments that nucleate at the cell poles and “treadmill” toward midcell. Maturating magnetosomes become attached to this filament via MamJ (orange). MamY is a protein of the cytoplasmic and magnetosome membranes (membrane helices, yellow; cytoplasmic domain, blue) with high potential to self-interact. MamY assemblies are suggested to become curvature sensitive and localize to sites of highest positive inner membrane curvature coinciding with the geodetic cell axis, where they recruit magnetosome chains. CM, cytoplasmic membrane; PG, peptidoglycan; OM, outer membrane. The model is not drawn to scale. (C) SEM image of an M. gryphiswaldense cell illustrating a corkscrew-like cell morphology. The geodetic cell axis is indicated as a dashed line. Scale bar, 2 μm.
SOPHISTICATED POSITIONING OF MAGNETOSOME CHAINS IN HELICAL CELLS
On top of the amazingly organized assembly, dynamics, and partitioning of magnetosome chains, magnetospirilla have evolved even more sophisticated means to reconcile helical cell morphology with a straight magnetoreceptor and to optimize magnetotaxis. Again, we will discuss some biophysical considerations that can help to appreciate these remarkable adaptations.
For efficient magnetotaxis, the magnetosome chain must adopt and maintain a very distinct position in the cell for several reasons. First, a mechanically fixed position is important because a flexible, “floating” magnetosome chain would move within the cell rather than aligning it. However, a physical connection of magnetosome chains to dynamic cytoplasmic content such as DNA (as described for other organelles such as carboxysomes [95] or carbonosomes, i.e., polyhydroxybutyrate granules [96, 97]) seems not sufficient. Tethering the chain to a rigid and more static structure such as the cell envelope would meet this requirement much better. Second, for efficient magnetotaxis, the magnetic moment of the magnetosome chain must perfectly match the swimming direction of the cell, which is predefined by the position of the flagella on the cell surface (62). Spirilla are propelled by polar flagella and hence move along the longitudinal cell axis, accompanied by fast rotations of the cell body. If swimming direction and magnetosome chain were misaligned, cells would tumble when they swim because two forces pull in slightly different directions. Third, to maximize the net magnetic moment similar to a corresponding bar magnet, the magnetosome chain must be maintained as straight as possible and stabilized against its inherent tendency to bend or to even form energetically favored, magnetic flux-closed rings (98). This is intriguing because unlike rod-shaped bacteria, spirilla lack any straight cell surface to support a rod-like magnetoreceptor. Nevertheless, a three-dimensional analysis of magnetosome chain positioning in M. gryphiswaldense wild-type cells revealed that the chains indeed meet all these criteria: they tightly follow a path along the inner cell envelope with the highest positive curvature. This path coincides with the shortest connection between the cell poles and hence represents the geodetic axis of the helix, i.e., the track which comes closest to a bar (Fig. 2A to C). Importantly, this path also coincides with the cellular motility axis (99). The question of how spirilla could achieve this has remained unaddressed for a long time, and mechanisms for magnetosome chain positioning in curved cells have been unknown until recently, when an in-depth analysis of the magnetosome protein MamY and the mamY mutant in M. gryphiswaldense have shed light on this phenomenon.
MamY is found among the magnetosome-associated proteins and conserved in magnetotactic spirilla and vibrios, i.e., in MTB of curved cell shape. Initially proposed to be involved in vesicle formation in M. magneticum (100), it represents a protein of the inner of the cytoplasmic and associated with the magnetosome membranes in M. gryphiswaldense. The protein self-interacts and forms higher-ordered structures at the cytoplasmic membrane. Upon achieving a certain size, these polymers are supposed to become curvature sensitive and further enrich along the membrane with highest positive curvature (99), eventually resulting in an extended assembly reaching from pole to pole and designating the geodetic cell axis. With its cytoplasmic domain, MamY recruits the magnetosome chain made by MamK and MamJ and forces it to a path that is parallel to the cellular motility axis (Fig. 3) (99).
Upon deletion of mamY, magnetosome chains lose their straight appearance, detach from the inner (convex) curvature of the helical cell, and shift to the outer (concave) curvature (Fig. 2D and E). Consequently, the ability of the cells to align to the magnetic field drops similar to the mamK mutant (66). Additional evidence that MamY functions as a scaffold for the chain comes from a mamKY double deletion mutant, which phenocopies a mamJ mutant where all magnetosomes agglomerate due to the missing tether to MamK filaments (Fig. 2D and E). This also suggests that in the mamK mutant, the magnetosomes are likely still attached to MamY structures but not concatenated into coherent chains because the cytomotive MamK filaments are missing. Correspondingly, short, fragmented chains of the mamK mutant are still observed at sites of inner positive cell curvature (Fig. 2D and E). The formation of short chains may be explained by the linear localization of MamY and magnetic attraction of the particles (91).
Taking these results together, MamY seems to be the key to (i) connect the magnetosome chain to the cell envelope ensuring efficient force transmission and cell alignment, (ii) keep the magnetosome chain straight to maximize its magnetic moment, and (iii) bring the chain into alignment with the cellular motility axis. Finally, its discovery reconciles the previously observed different phenotypes of the mamK and mamJ mutants.
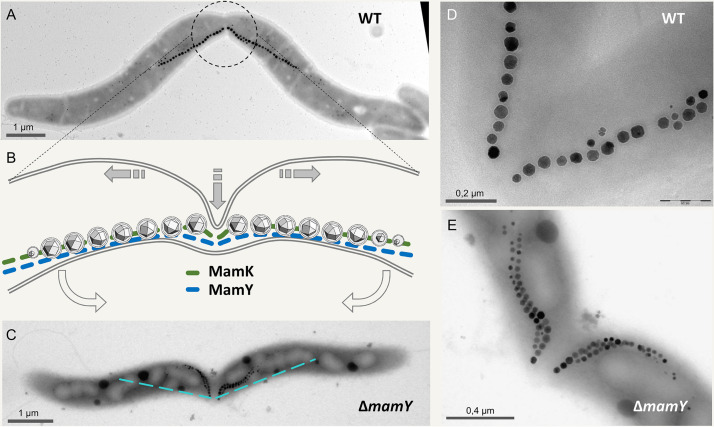
However, the tightly fixed “compass” poses a challenge during cytokinesis when not only the magnetoskeleton but also the magnetic forces that act between magnetosomes or even chains must be overcome. Experimental data and biophysical calculations suggest that the magnetoskeleton of M. gryphiswaldense withstands forces up to 25 pN (84, 101, 102). The magnetic force that attracts magnetosomes was calculated to be at least 40 pN (94). These resistances must be mastered in addition to the turgor and cell wall tension, which may add up to 400 pN (103). Two mechanisms can be inferred to facilitate chain splitting and hence cell division. First, the magnetoskeleton may reverse its polarity, i.e., the direction of MamK treadmilling may become inverted at the nascent septum, weakening the overall rigidity of the scaffold. This idea is supported by nonseparating, chained cells that occur in a mutant where MamK dynamics is perturbed (89). Second, a mechanical trick could help to “crack” the pulling magnetosome chains. Within the division plane of a magnetospirillum cell, the septum grows asymmetrically starting from the side of highest negative inner curvature toward positive curvature, resulting in a buckling-like deformation of the dividing cell (93) (Fig. 4A). Placing the chain at the positive curvature, i.e., opposite to the indentation results in a leverage-like mechanism, which has been interpreted as specific adaptation to overcome the magnetostatic interactions between separating daughter chains (94) (Fig. 4B and D). Obviously, this peculiarity is lost in the mamY mutant (Fig. 4C and E) and yet seems not to compromise cell division under laboratory conditions. However, it might matter in natural, energy-limiting environments.
FIG 4.
Cytokinesis in the presence of a magnetosome chain, as suggested by Katzmann et al. (94). (A) TEM image of a dividing M. gryphiswaldense wild-type cell. The center of the magnetosome chain is precisely localized at the division plane, ensuring that each daughter cell inherits exactly half of the magnetosomes (89, 93). Note the “buckling” cell center reflecting a distortion of the otherwise preserved helical cell morphology. (B) 2D scheme of the center from a dividing wild-type cell. Shaded arrows indicate direction of main cell wall growth at the division plane. Open arrows indicate the direction of cell bending. (In three dimensions, some twisting may occur as well.) Within the division plane, the septum grows by asymmetric indentation starting unilaterally from the site of negative inner cell curvature, resulting in a fracture-like appearance of the magnetosome chain. (C) In the mamY mutant, the magnetosome chain seemingly becomes disrupted by the wedge-like growing division septum. However, the buckling cell shape upon division is still apparent. The dashed line indicates the magnetosome chain position in wild-type cells. (D and E) Magnification of magnetosome chains immediately after splitting in the wild type (D) and the mamY mutant (E).
CELL SHAPE, MOTILITY, AND MAGNETOCHEMOTAXIS
In contrast to the magnetoskeleton, knowledge of the generic cytoskeleton in MTB is very limited, and yet some recent insights again from magnetospirilla allow a first glimpse of its organization. Genomic data suggest that magnetospirilla possess the common cytoskeletal elements and cytokinesis key organizers known from other Alphaproteobacteria such as FtsZ, MipZ, PopZ, MreB, and others (104–108). Interestingly, however, some of the factors, such as MipZ or FtsZ, have paralogous or even xenologous counterparts. For example, the essential tubulin-like cell division organizer FtsZ possesses a C-terminally truncated duplicate called FtsZm (or FtsZ-like) intriguingly encoded in the magnetosome island and seemingly colocalizing with its canonical equivalent at the future division site readily suggesting an interplay of cyto- and magnetoskeleton. Yet, the function of the paralogous genes is not fully understood. Deletion of ftsZm does not cause a cell or magnetosome chain division phenotype under standard growth conditions. Instead, the mutant seems to have a biomineralization defect in the absence of the anaerobic electron acceptor nitrate (51, 109), suggesting that the tubulin-like FtsZm is not a core constituent of the magnetoskeleton in contrast to the actin-like MamK, although its polymerization properties seem conserved (51, 110). The second MipZ (midcell positioning of FtsZ) (108) protein (encoded outside the magnetosome island) was found to localize to the division plane as well, which stands in stark contrast to its canonical counterpart that forms pole-to-midcell concentration gradients that inhibit FtsZ polymerization (111). Again, deletion of mipZ2 does not elicit an obvious phenotype under standard conditions.
The polar organizing protein Z PopZ was found to have slightly different properties than in the related Caulobacter crescentus and Agrobacterium tumefaciens, since it localizes not predominantly to one but to both cell poles throughout the cell cycle. Its essential role as landmark for accurate cell division and chromosome segregation still seems conserved in M. gryphiswaldense. Upon deletion or overexpression severe defects in magnetosome segregation occur that are, however, likely an indirect effect of perturbed cell division (112).
The obvious corkscrew-like cell morphology of magnetospirilla (Fig. 3C) suggests the presence of a particular cell shape-determining cytoskeleton. Several factors for curved cell shape are known from non-MTB with vibroid or helical cell shape. In these examples, commonly, intracellular scaffolding structures were found acting in concert with the actin-like MreB and periplasmic cell wall biosynthetic enzymes to introduce curvature and helical twist via spatial patterning of peptidoglycan synthesis (113). Whereas MreB promotes elongated rod cell shape by organizing centers of predominant cell wall synthesis (114, 115), nucleotide-independent elements such as coiled-coil rich proteins or bactofilins (which are characterized by a right-handed β-helix core domain structure [116]) introduce curvature by functioning as a mechanical support with intrinsic curvature preference and/or scaffold for cell wall synthesis-related enzymes in a rather static manner (113). Knowledge of cell shape determinants in magnetospirilla is still very limited and hardly based on experimental evidence, but the presence of mostly uncharacterized genes in M. gryphiswaldense and M. magneticum coding for MreB (90), a putative bactofilin, a structural coiled-coil rich protein, as well as multiple enzymes involved in peptidoglycan remodeling, suggest that their helical cell morphology is likely controlled by a multifactorial cell shaping system. Time-lapse microscopy of M. gryphiswaldense further indicates that cells elongate by an unequal predominant polar mode of cell growth (94, 112), although it has not been analyzed quantitatively whether new cell wall material is incorporated more toward the old or new cell pole. Future studies related to the function of a shape-determining cytoskeleton in magnetospirilla might reveal novel insights into the control of helical cell morphology, e.g., by use of fluorogenic cell wall labels, and also focus on the intriguing question whether underlying structures are interconnected with the magnetoskeleton.
The morphology of magnetospirilla is part of the motility system because in helical cells, the cell body rotates in the opposite direction of flagellar rotation (117). This agrees with live-cell observations on M. magneticum with fluorescently labeled cell poles which revealed right-handed helical trajectories, indicating that the cell body of magnetospirilla rotates clockwise during swimming (118). Fluorescent labeling of flagella in bipolarly flagellated M. magneticum revealed that the leading flagellum forms a parachute-like structure, likely caused by wrapping around the cell body, with the lagging counterclockwise-rotating flagellum forming a “tuft” oriented away from the cell body (118). The rotational direction of the leading flagellum could not be clearly resolved due to its association with the cell body but was suggested as clockwise (if observed from the tip of the flagellum toward the cell pole), i.e., suggesting that the two flagellar motors rotated in opposite directions. Bipolarly flagellated M. gryphiswaldense and M. magneticum were described to reach swimming speeds up to ∼70 μm s−1 (58, 66, 119) and ∼100 μm s−1 (120), respectively, in liquid, but the corkscrew-like cell morphology of magnetospirilla might be of particular advantage for motility in complex structured habitats such as aquatic sediments (117, 121) since the helical cell shape might prevent slipping of the rotating cell body under such conditions and hence facilitates to achieve higher swimming speeds (113). An increase in average swimming speeds of soft agar-grown M. gryphiswaldense (66) suggests that cells might undergo specific ultrastructural alterations to increase flagellar torque in more viscous environments (e.g., changes in flagellum [motor] architecture or number [122, 123]) or even increased helicity of the cell body (Fig. 5A).
FIG 5.
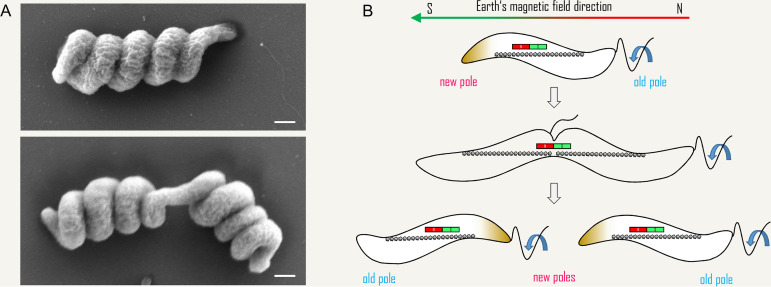
Helicity of M. gryphiswaldense cells grown on solid media and a hypothetical mechanism for preservation of cell polarity upon cytokinesis in MTB. (A) SEM images of M. gryphiswaldense cells that were recovered from a colony growing on agar-solidified medium. Note the highly spiralized cell morphology in comparison to cells grown in liquid medium (e.g., Fig. 2B). Cell division (lower image) seems to require (or to cause) some “unwinding” of the tight helix at the division plane, and yet the “buckling” appearance of the dividing cell (Fig. 4) is still visible. Scale bars, 500 nm. (B) Maintenance of polarity during cytokinesis in a monopolarly flagellated MTB. Before cytokinesis, swimming polarity is defined by the internal magnetic dipole (indicated by the bar magnet) and a gradient in redox potential (data not shown), leading to preferred swimming toward one pole of the external magnetic field. However, cell division generates daughter cells of opposite magnetic polarity with respect to the new cell pole. To ensure that both cells have the same magnetic polarity with respect to their flagellated pole, the daughter cell that does not inherit the flagellum must synthesize a new flagellum at the new cell pole (127).
Magnetotaxis, aerotaxis, and flagellum-mediated swimming motility are likely integrated with complex sensory systems, since multiples of putative chemotaxis-related proteins (i.e., Che proteins and methyl-accepting chemotaxis transducers) are encoded within magnetospirillum genomes, and the first experimental evidence (58) underpins this notion. Chemoreceptor arrays are located near flagellar motors at both cell poles in M. gryphiswaldense, as revealed by cryo-electron tomography and fluorescent labeling of the chemoreceptor-associated adaptor protein CheW1 (77, 124). Since the flagellar polymorphic structure and conformational changes during clockwise/counterclockwise rotation are likely physically imprinted (i.e., determined by the ultrastructure of the flagellum), as well as motor switching by CheY response regulators, it is currently unknown how motor rotation in opposite senses (as proposed earlier [118]) might be integrated with chemotactic signaling and switched in a coordinated manner. Given the vast number of putative motility- and chemotaxis-related proteins in magnetospirilla, it was suggested that distinct proteins might govern some sort of motor asymmetry (58, 118). Other models suggest simultaneous counterclockwise rotation of both flagellar motors in a nonmagnetotactic bipolarly flagellated spirillum (125) or pausing of one flagellum in M. gryphiswaldense (119). Whereas rotation of both motors in the same direction during straight swimming is conclusive in terms of chemotactic signaling, pausing of one motor appears to be unlikely due to the observation of tethered magnetospirilla, which change rotational direction without extended pauses, as well as “tumbling-like” motions observed in M. magneticum cells with both flagella spinning at the same time in an uncoordinated manner (58, 118).
NORTH- AND SOUTH-SEEKING SWIMMING POLARITY
An intriguing phenomenon was observed in early studies when populations of MTB enriched from environmental samples were observed under the light microscope. When exposed to a magnetic field, the cells not only aligned to it and moved along the field lines, but they also seemed to prefer one direction over the other, i.e., they ran either toward the magnetic north or south pole (5, 8, 126). This behavior was termed “swimming polarity” (to be distinguished from the magnetic polarity which is a physical property set by the magnetosome chain). Even more exciting was the observation that in the Earth’s northern hemisphere predominantly north-seeking cells and in the southern hemisphere predominantly south-seeking cells were isolated. Because the magnetic south pole currently coincides with the geographic north pole and vice versa, and because the Earth’s magnetic field is inclined, the north-seeking cells of the northern hemisphere swim actually down toward a magnetic south pole. This swimming polarity is thought to assist magnetoaerotaxis to efficiently guide cells from oxic zones down into regions of preferred low oxygen concentration toward the bottom of the water column (6). Interestingly, swimming polarity was lost in lab strains of magnetospirilla, but could be restored within only few generations by cultivation in an oxygen gradient with superimposed magnetic field, suggesting that this characteristic is an individually acquired adaptation rather than a “hardwired” physical or genomic trait (10, 58). This means that swimming polarity in magnetospirilla is set by the direction of the oxygen gradient with respect to the magnetic field orientation during growth but is lost in the absence of an oxygen gradient (for example, if cell cultures are stirred) and is reversibly triggered and switched by changes in oxygen concentration (58). These observations also suggest that swimming polarity is closely linked to aerotaxis at the molecular level, which was confirmed by the finding that deletion of the chemosensory pathway cheOp1 in M. gryphiswaldense results in loss of aerotaxis and swimming polarity (58). Currently, it is not understood how magnetotactic swimming polarity is exactly controlled and perhaps inherited at the cellular and molecular levels. Faithful segregation of magnetosome chains by the magnetoskeleton might be important. As stated above, the magnetosome chain is divided into half during cell division. For a polarized cell, this results in daughter cells with opposing magnetic polarities with respect to their new cell poles but similar directional preferences toward one of the magnetic poles (58, 112, 118, 127) (Fig. 5B). It was hypothesized that asymmetric activity or localization of motility-related structures such as flagella might determine a cellular asymmetry with respect to the physically imprinted magnetic polarity of the magnetosome chain leading to equal cellular and magnetic polarity of the offspring. In other words, direction of polarized swimming could be maintained by a monopolar flagellation pattern for both cells when one of them synthesized a new flagellum at the division site (127), so that the flagellated cell pole of both daughter cells is oriented toward the same pole of the magnetic field (Fig. 5B). Interestingly, in magnetospirilla, synthesis of new flagella at the division site has been observed to occur before cytokinesis has completed (89, 94), raising the question about their rotational sense (as discussed above) and how swimming polarity is determined in cells with two flagella.
CONCLUSIONS
Altogether, past and present studies emphasize that MTB are a unique and rewarding model to study a plethora of bacterial adaptations. Apart from micro- and anaerobic physiology, iron metabolism and biomineralization, organelle formation, applied research, and ecology/phylogeny, which were only briefly touched upon in this minireview, recent work highlighted that magnetospirilla possess distinctive cellular adaptations to take advantage of their magnetic sense and to link it to life style and motility. A sophisticated “magnetoskeleton” can now be defined, which in M. gryphiswaldense to date consists of three cytoskeletal factors dedicated to magnetotaxis. The actin-like MamK and its adaptor MamJ represent the dynamic part of the magnetoskeleton as they actively concatenate magnetosomes and position the chain at midcell, thereby also ensuring precise equipartitioning of the magnetoreceptor upon cell division. The static MamY structure by contrast is responsible to identify and to tether the dynamic assembly to the geodetic cell axis, i.e., to fix it along the shortest path from cell pole to cell pole, where the flagella are inserted. This straightens the “compass needle” within the helix and aligns it with the motility axis, thereby perfectly superimposing the Earth’s magnetic field vector on swimming movements. These recent results highlight that magnetosomes are not only one of the most complex structures found in bacteria, but they require dedicated cytoskeletal elements to form a dynamic and distinctly localized higher ordered structure and to function in a cooperative manner. Moreover, magneto-aerotaxis turned out not only to comprise magnetosome chains but also to require dynamic integration of chemosensory signals, coordination with motility systems, and even cell shape to perfect magnetic navigation.
ACKNOWLEDGMENTS
We are grateful to the Deutsche Forschungsgemeinschaft for grant Schu1080/9-2 and for European Research Council Advanced Grant ERC-ADG-2015 Syntomagx.
REFERENCES
- 1.Blakemore R. 1975. Magnetotactic bacteria. Science 190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 2.Bellini S. 2009. On a unique behavior of freshwater bacteria. Chin J Ocean Limnol 27:3–5. doi: 10.1007/s00343-009-0003-5. [DOI] [Google Scholar]
- 3.Lefèvre CT, Bazylinski DA. 2013. Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol Mol Biol Rev 77:497–526. doi: 10.1128/MMBR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin W, Pan Y, Bazylinski DA. 2017. Diversity and ecology of and biomineralization by magnetotactic bacteria. Environ Microbiol Rep 9:345–356. doi: 10.1111/1758-2229.12550. [DOI] [PubMed] [Google Scholar]
- 5.Blakemore RP, Frankel RB, Kalmijn AJ. 1980. South-seeking magnetotactic bacteria in the southern hemisphere. Nature 286:384–385. doi: 10.1038/286384a0. [DOI] [Google Scholar]
- 6.Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat Rev Microbiol 2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 7.Simmons SL, Bazylinski DA, Edwards KJ. 2006. South-seeking magnetotactic bacteria in the Northern Hemisphere. Science 311:371–374. doi: 10.1126/science.1122843. [DOI] [PubMed] [Google Scholar]
- 8.Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys J 73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jogler C, Wanner G, Kolinko S, Niebler M, Amann R, Petersen N, Kube M, Reinhardt R, Schüler D. 2011. Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep-branching Nitrospira phylum. Proc Natl Acad Sci U S A 108:1134–1139. doi: 10.1073/pnas.1012694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefèvre CT, Bennet M, Landau L, Vach P, Pignol D, Bazylinski DA, Frankel RB, Klumpp S, Faivre D. 2014. Diversity of magneto-aerotactic behaviors and oxygen sensing mechanisms in cultured magnetotactic bacteria. Biophys J 107:527–538. doi: 10.1016/j.bpj.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uebe R, Schüler D. 2016. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol 14:621–637. doi: 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- 12.McCausland HC, Komeili A. 2020. Magnetic genes: studying the genetics of biomineralization in magnetotactic bacteria. PLoS Genet 16:e1008499. doi: 10.1371/journal.pgen.1008499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem Rev 108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- 14.Staniland SS, Rawlings AE. 2016. Crystallizing the function of the magnetosome membrane mineralization protein Mms6. Biochem Soc Trans 44:883–890. doi: 10.1042/BST20160057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mickoleit F, Schüler D. 2019. Generation of nanomagnetic biocomposites by genetic engineering of bacterial magnetosomes. Bioinspired Biomim Nanobiomaterials 8:86–98. doi: 10.1680/jbibn.18.00005. [DOI] [Google Scholar]
- 16.Vargas G, Cypriano J, Correa T, Leão P, Bazylinski DA, Abreu F. 2018. Applications of magnetotactic bacteria, magnetosomes and magnetosome crystals in biotechnology and nanotechnology: mini-review. Molecules 23:2348. doi: 10.3390/molecules23102438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraupner A, Eberbeck D, Heinke D, Uebe R, Schüler D, Briel A. 2017. Bacterial magnetosomes: nature’s powerful contribution to MPI tracer research. Nanoscale 9:5793. doi: 10.1039/c7nr01530e. [DOI] [PubMed] [Google Scholar]
- 18.Hergt R, Dutz S, Röder M. 2008. Effects of size distribution on hysteresis losses of magnetic nanoparticles for hyperthermia. J Phys Condens Matter 20:385214. doi: 10.1088/0953-8984/20/38/385214. [DOI] [PubMed] [Google Scholar]
- 19.Le Fèvre R, Durand-Dubief M, Chebbi I, Mandawala C, Lagroix F, Valet J-P, Idbaih A, Adam C, Delattre J-Y, Schmitt C, Maake C, Guyot F, Alphandéry E. 2017. Enhanced antitumor efficacy of biocompatible magnetosomes for the magnetic hyperthermia treatment of glioblastoma. Theranostics 7:4618–4631. doi: 10.7150/thno.18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balkwill DL, Maratea D, Blakemore RP. 1980. Ultrastructure of a magnetotactic spirillum. J Bacteriol 141:1399–1408. doi: 10.1128/JB.141.3.1399-1408.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorby YA, Beveridge TJ, Blakemore RP. 1988. Characterization of the bacterial magnetosome membrane. J Bacteriol 170:834–841. doi: 10.1128/jb.170.2.834-841.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raschdorf O, Forstner Y, Kolinko I, Uebe R, Plitzko JM, Schüler D. 2016. Genetic and ultrastructural analysis reveals the key players and initial steps of bacterial magnetosome membrane biogenesis. PLoS Genet 12:e1006101. doi: 10.1371/journal.pgen.1006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornejo E, Subramanian P, Li Z, Jensen GJ, Komeili A. 2016. Dynamic remodeling of the magnetosome membrane is triggered by the initiation of biomineralization. mBio 7:e01898-15. doi: 10.1128/mBio.01898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol 187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schübbe S, Würdemann C, Peplies J, Heyen U, Wawer C, Glöckner FO, Schüler D. 2006. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 72:5757–5765. doi: 10.1128/AEM.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murat D, Quinlan A, Vali H, Komeili A. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the stepwise assembly of a prokaryotic organelle. Proc Natl Acad Sci U S A 107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohsse A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. 2011. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One 6:e25561. doi: 10.1371/journal.pone.0025561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefèvre CT, Menguy N, Abreu F, Lins U, Pósfai M, Prozorov T, Pignol D, Frankel RB, Bazylinski DA. 2011. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science 334:1720–1723. doi: 10.1126/science.1212596. [DOI] [PubMed] [Google Scholar]
- 29.Lohsse A, Borg S, Raschdorf O, Kolinko I, Tompa E, Pósfai M, Faivre D, Baumgartner J, Schüler D. 2014. Genetic dissection of the mamAB and mms6 operons reveals a gene set essential for magnetosome biogenesis in Magnetospirillum gryphiswaldense. J Bacteriol 196:2658–2669. doi: 10.1128/JB.01716-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin W, Deng A, Wang Z, Li Y, Wen T, Wu L-F, Wu M, Pan Y. 2014. Genomic insights into the uncultured genus “Candidatus Magnetobacterium” in the phylum Nitrospirae. ISME J 8:2463–2477. doi: 10.1038/ismej.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monteil CL, Perrière G, Menguy N, Ginet N, Alonso B, Waisbord N, Cruveiller S, Pignol D, Lefèvre CT. 2018. Genomic study of a novel magnetotactic Alphaproteobacteria uncovers the multiple ancestry of magnetotaxis. Environ Microbiol 20:4415–4430. doi: 10.1111/1462-2920.14364. [DOI] [PubMed] [Google Scholar]
- 32.Kolinko I, Lohsse A, Borg S, Raschdorf O, Jogler C, Tu Q, Pósfai M, Tompa E, Plitzko JM, Brachmann A, Wanner G, Müller R, Zhang Y, Schüler D. 2014. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat Nanotechnol 9:193–197. doi: 10.1038/nnano.2014.13. [DOI] [PubMed] [Google Scholar]
- 33.Dziuba MV, Zwiener T, Uebe R, Schüler D. 2020. Single-step transfer of biosynthetic operons endows a non-magnetotactic Magnetospirillum strain from wetland with magnetosome biosynthesis. Environ Microbiol 22:1603–1618. doi: 10.1111/1462-2920.14950. [DOI] [PubMed] [Google Scholar]
- 34.Monteil CL, Grouzdev DS, Perrière G, Alonso B, Rouy Z, Cruveiller S, Ginet N, Pignol D, Lefevre CT. 2020. Repeated horizontal gene transfers triggered parallel evolution of magnetotaxis in two evolutionary divergent lineages of magnetotactic bacteria. ISME J 14:1783–1794. doi: 10.1038/s41396-020-0647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin W, Paterson GA, Zhu Q, Wang Y, Kopylova E, Li Y, Knight R, Bazylinski DA, Zhu R, Kirschvink JL, Pan Y. 2017. Origin of microbial biomineralization and magnetotaxis during the Archean. Proc Natl Acad Sci U S A 114:2171–2176. doi: 10.1073/pnas.1614654114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin W, Zhang W, Zhao X, Roberts AP, Paterson GA, Bazylinski DA, Pan Y. 2018. Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. ISME J 12:1508–1519. doi: 10.1038/s41396-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heyen U, Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl Microbiol Biotechnol 61:536–544. doi: 10.1007/s00253-002-1219-x. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Katzmann E, Borg S, Schüler D. 2012. The periplasmic nitrate reductase Nap is required for anaerobic growth and involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. J Bacteriol 194:4847–4856. doi: 10.1128/JB.00903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Bali S, Borg S, Katzmann E, Ferguson SJ, Schüler D. 2013. Cytochrome cd1 nitrite reductase NirS is involved in anaerobic magnetite biomineralization in Magnetospirillum gryphiswaldense and requires NirN for proper d1 heme assembly. J Bacteriol 195:4297–4309. doi: 10.1128/JB.00686-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komeili A, Vali H, Beveridge TJ, Newman DK. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci U S A 101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grünberg K, Müller E-C, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 70:1040–1050. doi: 10.1128/aem.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mickoleit F, Schüler D. 2018. Generation of multifunctional magnetic nanoparticles with amplified catalytic activities by genetic expression of enzyme arrays on bacterial magnetosomes. Adv Biosys 2:1700109. doi: 10.1002/adbi.201700109. [DOI] [Google Scholar]
- 43.Raschdorf O, Bonn F, Zeytuni N, Zarivach R, Becher D, Schüler D. 2018. A quantitative assessment of the membrane-integral subproteome of a bacterial magnetic organelle. J Proteomics 172:89–99. doi: 10.1016/j.jprot.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, Zarivach R, Kasama T, Wanner G, Pósfai M, Böttger L, Matzanke B, Schüler D. 2011. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol Microbiol 82:818–835. doi: 10.1111/j.1365-2958.2011.07863.x. [DOI] [PubMed] [Google Scholar]
- 45.Hershey DM, Browne PJ, Iavarone AT, Teyra J, Lee EH, Sidhu SS, Komeili A. 2016. Magnetite biomineralization in Magnetospirillum magneticum is regulated by a switch-like behavior in the HtrA protease MamE. J Biol Chem 291:17941–17952. doi: 10.1074/jbc.M116.731000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hershey DM, Ren X, Melnyk RA, Browne PJ, Ozyamak E, Jones SR, Chang MCY, Hurley JH, Komeili A. 2016. MamO is a repurposed serine protease that promotes magnetite biomineralization through direct transition metal binding in magnetotactic bacteria. PLoS Biol 14:e1002402. doi: 10.1371/journal.pbio.1002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uebe R, Keren-Khadmy N, Zeytuni N, Katzmann E, Navon Y, Davidov G, Bitton R, Plitzko JM, Schüler D, Zarivach R. 2018. The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization. Mol Microbiol 107:542–557. doi: 10.1111/mmi.13899. [DOI] [PubMed] [Google Scholar]
- 48.Raschdorf O, Müller FD, Pósfai M, Plitzko JM, Schüler D. 2013. The magnetosome proteins MamX, MamZ, and MamH are involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. Mol Microbiol 89:872–886. doi: 10.1111/mmi.12317. [DOI] [PubMed] [Google Scholar]
- 49.Siponen MI, Legrand P, Widdrat M, Jones SR, Zhang W-J, Chang MCY, Faivre D, Arnoux P, Pignol D. 2013. Structural insight into magnetochrome-mediated magnetite biomineralization. Nature 502:681–684. doi: 10.1038/nature12573. [DOI] [PubMed] [Google Scholar]
- 50.Jones SR, Wilson TD, Brown ME, Rahn-Lee L, Yu Y, Fredriksen LL, Ozyamak E, Komeili A, Chang MCY. 2015. Genetic and biochemical investigations of the role of MamP in redox control of iron biomineralization in Magnetospirillum magneticum. Proc Natl Acad Sci U S A 112:3904–3909. doi: 10.1073/pnas.1417614112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller FD, Raschdorf O, Nudelman H, Messerer M, Katzmann E, Plitzko JM, Zarivach R, Schüler D. 2014. The FtsZ-like protein FtsZm of Magnetospirillum gryphiswaldense likely interacts with its generic homolog and is required for biomineralization under nitrate deprivation. J Bacteriol 196:650–659. doi: 10.1128/JB.00804-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheffel A, Gärdes A, Grünberg K, Wanner G, Schüler D. 2008. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol 190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rong C, Zhang C, Zhang Y, Qi L, Yang J, Guan G, Li Y, Li J. 2012. FeoB2 functions in magnetosome formation and oxidative stress protection in Magnetospirillum gryphiswaldense strain MSR-1. J Bacteriol 194:3972–3976. doi: 10.1128/JB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nudelman H, Lee Y-Z, Hung Y-L, Kolusheva S, Upcher A, Chen Y-C, Chen J-Y, Sue S-C, Zarivach R. 2018. Understanding the biomineralization role of magnetite-interacting components (MICs) from magnetotactic bacteria. Front Microbiol 9:2480. doi: 10.3389/fmicb.2018.02480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohsse A, Kolinko I, Raschdorf O, Uebe R, Borg S, Brachmann A, Plitzko JM, Müller R, Zhang Y, Schüler D. 2016. Overproduction of magnetosomes by genomic amplification of biosynthesis-related gene clusters in a magnetotactic bacterium. Appl Environ Microbiol 82:3032–3041. doi: 10.1128/AEM.03860-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bi S, Sourjik V. 2018. Stimulus sensing and signal processing in bacterial chemotaxis. Curr Opin Microbiol 45:22–29. doi: 10.1016/j.mib.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, Ma Q, Jiang W, Song T. 2011. Phototaxis in the magnetotactic bacterium Magnetospirillum magneticum strain AMB-1 is independent of magnetic fields. Appl Microbiol Biotechnol 90:269–275. doi: 10.1007/s00253-010-3017-1. [DOI] [PubMed] [Google Scholar]
- 58.Popp F, Armitage JP, Schüler D. 2014. Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun 5:5398. doi: 10.1038/ncomms6398. [DOI] [PubMed] [Google Scholar]
- 59.Philippe N, Wu L-F. 2010. An MCP-like protein interacts with the MamK cytoskeleton and is involved in magnetotaxis in Magnetospirillum magneticum AMB-1. J Mol Biol 400:309–322. doi: 10.1016/j.jmb.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 60.Zhu X, Ge X, Li N, Wu L-F, Luo C, Ouyang Q, Tu Y, Chen G. 2014. Angle sensing in magnetotaxis of Magnetospirillum magneticum AMB-1. Integr Biol (Camb) 6:706–713. doi: 10.1039/c3ib40259b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.González LM, Ruder WC, Mitchell AP, Messner WC, LeDuc PR. 2015. Sudden motility reversal indicates sensing of magnetic field gradients in Magnetospirillum magneticum AMB-1 strain. ISME J 9:1399–1409. doi: 10.1038/ismej.2014.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frankel RB, Blakemore RP. 1980. Navigational compass in magnetic bacteria. J Magn Magn Mater 15–18:1562–1564. doi: 10.1016/0304-8853(80)90409-6. [DOI] [Google Scholar]
- 63.Frankel RB, Blakemore RP. 1981. Magnetic navigation in bacteria. Sci Am 245:58–49. doi: 10.1038/scientificamerican1281-58. [DOI] [Google Scholar]
- 64.Kirschvink JL. 1982. Paleomagnetic evidence for fossil biogenic magnetite in western Crete. Earth Planet Sci Lett 59:388–392. doi: 10.1016/0012-821X(82)90140-6. [DOI] [Google Scholar]
- 65.Kobayashi A, Kirschvink JL, Nash CZ, Kopp RE, Sauer DA, Bertani LE, Voorhout WF, Taguchi T. 2006. Experimental observation of magnetosome chain collapse in magnetotactic bacteria: sedimentological, paleomagnetic, and evolutionary implications. Earth Planet Sci Lett 245:538–550. doi: 10.1016/j.epsl.2006.03.041. [DOI] [Google Scholar]
- 66.Pfeiffer D, Schüler D. 2019. Quantifying the benefit of a dedicated “magnetoskeleton” in bacterial magnetotaxis by live-cell motility tracking and soft agar swimming assay. Appl Environ Microbiol 86:e01976-19. doi: 10.1128/AEM.01976-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosenblatt C, de Araujo FFT, Frankel RB. 1982. Light scattering determination of magnetic moments of magnetotactic bacteria (invited). J Appl Phys 53:2727–2729. doi: 10.1063/1.330948. [DOI] [Google Scholar]
- 68.Rosenblatt C, de Araujo FFT, Frankel RB. 1982. Birefringence determination of magnetic moments of magnetotactic bacteria. Biophys J 40:83–85. doi: 10.1016/S0006-3495(82)84461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moskowitz BM, Frankel RB, Flanders PJ, Blakemore RP, Schwartz BB. 1988. Magnetic properties of magnetotactic bacteria. J Magn Magn Mater 73:273–288. doi: 10.1016/0304-8853(88)90093-5. [DOI] [Google Scholar]
- 70.Zahn C, Keller S, Toro-Nahuelpan M, Dorscht P, Gross W, Laumann M, Gekle S, Zimmermann W, Schüler D, Kress H. 2017. Measurement of the magnetic moment of single Magnetospirillum gryphiswaldense cells by magnetic tweezers. Sci Rep 7:3558. doi: 10.1038/s41598-017-03756-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leão P, Chen Y-R, Abreu F, Wang M, Zhang W-J, Zhou K, Xiao T, Wu L-F, Lins U. 2017. Ultrastructure of ellipsoidal magnetotactic multicellular prokaryotes depicts their complex assemblage and cellular polarity in the context of magnetotaxis. Environ Microbiol 19:2151–2163. doi: 10.1111/1462-2920.13677. [DOI] [PubMed] [Google Scholar]
- 72.Bente K, Mohammadinejad S, Charsooghi MA, Bachmann F, Codutti A, Lefèvre CT, Klumpp S, Faivre D. 2020. High-speed motility originates from cooperatively pushing and pulling flagella bundles in bilophotrichous bacteria. Elife 9:e47551. doi: 10.7554/eLife.47551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keim CN, Melo RD, de Almeida FP, de Barros HGPL, Farina M, Acosta-Avalos D. 2018. Effect of applied magnetic fields on motility and magnetotaxis in the uncultured magnetotactic multicellular prokaryote “Candidatus Magnetoglobus multicellularis.” Environ Microbiol Rep 10:465–474. doi: 10.1111/1758-2229.12640. [DOI] [PubMed] [Google Scholar]
- 74.Bennet M, McCarthy A, Fix D, Edwards MR, Repp F, Vach P, Dunlop JWC, Sitti M, Buller GS, Klumpp S, Faivre D. 2014. Influence of magnetic fields on magneto-aerotaxis. PLoS One 9:e101150. doi: 10.1371/journal.pone.0101150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 76.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 77.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. 2010. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol 77:208–224. doi: 10.1111/j.1365-2958.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 78.Draper O, Byrne ME, Li Z, Keyhani S, Barrozo JC, Jensen G, Komeili A. 2011. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol 82:342–354. doi: 10.1111/j.1365-2958.2011.07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scheffel A, Schüler D. 2007. The acidic repetitive domain of the Magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J Bacteriol 189:6437–6446. doi: 10.1128/JB.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nudelman H, Zarivach R. 2014. Structure prediction of magnetosome-associated proteins. Front Microbiol 5:9. doi: 10.3389/fmicb.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rioux J-B, Philippe N, Pereira S, Pignol D, Wu L-F, Ginet N. 2010. A second actin-like MamK protein in Magnetospirillum magneticum AMB-1 encoded outside the genomic magnetosome island. PLoS One 5:e9151. doi: 10.1371/journal.pone.0009151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abreu N, Mannoubi S, Ozyamak E, Pignol D, Ginet N, Komeili A. 2014. Interplay between two bacterial actin homologs, MamK and MamK-Like, is required for the alignment of magnetosome organelles in Magnetospirillum magneticum AMB-1. J Bacteriol 196:3111–3121. doi: 10.1128/JB.01674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bennet M, Bertinetti L, Neely RK, Schertel A, Körnig A, Flors C, Müller FD, Schüler D, Klumpp S, Faivre D. 2015. Biologically controlled synthesis and assembly of magnetite nanoparticles. Faraday Discuss 181:71–83. doi: 10.1039/c4fd00240g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kiani B, Faivre D, Klumpp S. 2018. Self-organization and stability of magnetosome chains: a simulation study. PLoS One 13:e0190265. doi: 10.1371/journal.pone.0190265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sonkaria S, Fuentes G, Verma C, Narang R, Khare V, Fischer A, Faivre D. 2012. Insight into the assembly properties and functional organization of the magnetotactic bacterial actin-like homolog, MamK. PLoS One 7:e34189. doi: 10.1371/journal.pone.0034189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozyamak E, Kollman J, Agard DA, Komeili A. 2013. The bacterial actin MamK: in vitro assembly behavior and filament architecture. J Biol Chem 288:4265–4277. doi: 10.1074/jbc.M112.417030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bergeron JRC, Hutto R, Ozyamak E, Hom N, Hansen J, Draper O, Byrne ME, Keyhani S, Komeili A, Kollman JM. 2017. Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Protein Sci 26:93–102. doi: 10.1002/pro.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Löwe J, He S, Scheres SHW, Savva CG. 2016. X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization. Proc Natl Acad Sci U S A 113:13396–13401. doi: 10.1073/pnas.1612034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toro-Nahuelpan M, Müller FD, Klumpp S, Plitzko JM, Bramkamp M, Schüler D. 2016. Segregation of prokaryotic magnetosomes organelles is driven by treadmilling of a dynamic actin-like MamK filament. BMC Biol 14:88. doi: 10.1186/s12915-016-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pradel N, Santini C-L, Bernadac A, Fukumori Y, Wu L-F. 2006. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc Natl Acad Sci U S A 103:17485–17489. doi: 10.1073/pnas.0603760103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klumpp S, Faivre D. 2012. Interplay of magnetic interactions and active movements in the formation of magnetosome chains. PLoS One 7:e33562. doi: 10.1371/journal.pone.0033562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grant CR, Wan J, Komeili A. 2018. Organelle formation in Bacteria and Archaea. Annu Rev Cell Dev Biol 34:217–238. doi: 10.1146/annurev-cellbio-100616-060908. [DOI] [PubMed] [Google Scholar]
- 93.Staniland SS, Moisescu C, Benning LG. 2010. Cell division in magnetotactic bacteria splits magnetosome chain in half. J Basic Microbiol 50:392–396. doi: 10.1002/jobm.200900408. [DOI] [PubMed] [Google Scholar]
- 94.Katzmann E, Müller FD, Lang C, Messerer M, Winklhofer M, Plitzko JM, Schüler D. 2011. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol 82:1316–1329. doi: 10.1111/j.1365-2958.2011.07874.x. [DOI] [PubMed] [Google Scholar]
- 95.MacCready JS, Basalla JL, Vecchiarelli AG. 2020. Origin and evolution of carboxysome positioning systems in cyanobacteria. Mol Biol Evol 37:1434–1451. doi: 10.1093/molbev/msz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
- 97.Wahl A, Schuth N, Pfeiffer D, Nussberger S, Jendrossek D. 2012. PHB granules are attached to the nucleoid via PhaM in Ralstonia eutropha. BMC Microbiol 12:262. doi: 10.1186/1471-2180-12-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Philipse AP, Maas DGBM. 2002. Magnetic colloids from magnetotactic bacteria: chain formation and colloidal stability. Langmuir 18:9977–9984. doi: 10.1021/la0205811. [DOI] [Google Scholar]
- 99.Toro-Nahuelpan M, Giacomelli G, Raschdorf O, Borg S, Plitzko JM, Bramkamp M, Schüler D, Müller F-D. 2019. MamY is a membrane-bound protein that aligns magnetosomes and the motility axis of helical magnetotactic bacteria. Nat Microbiol 4:1978–1912. doi: 10.1038/s41564-019-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanaka M, Arakaki A, Matsunaga T. 2010. Identification and functional characterization of liposome tubulation protein from magnetotactic bacteria. Mol Microbiol 76:480–488. doi: 10.1111/j.1365-2958.2010.07117.x. [DOI] [PubMed] [Google Scholar]
- 101.Körnig A, Dong J, Bennet M, Widdrat M, Andert J, Müller FD, Schüler D, Klumpp S, Faivre D. 2014. Probing the mechanical properties of magnetosome chains in living magnetotactic bacteria. Nano Lett 14:4653–4659. doi: 10.1021/nl5017267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Günther E, Klauss A, Toro-Nahuelpan M, Schüler D, Hille C, Faivre D. 2019. The in vivo mechanics of the magnetotactic backbone as revealed by correlative FLIM-FRET and STED microscopy. Sci Rep 9:19615. doi: 10.1038/s41598-019-55804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lan G, Wolgemuth CW, Sun SX. 2007. Z-ring force and cell shape during division in rod-like bacteria. Proc Natl Acad Sci U S A 104:16110–16115. doi: 10.1073/pnas.0702925104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Busiek KK, Margolin W. 2015. Bacterial actin and tubulin homologs in cell growth and division. Curr Biol 25:R243–R254. doi: 10.1016/j.cub.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van den Ent F, Amos LA, Löwe J. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 106.Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 107.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 109.Ding Y, Li J, Liu J, Yang J, Jiang W, Tian J, Li Y, Pan Y, Li J. 2010. Deletion of the ftsZ-like gene results in the production of superparamagnetic magnetite magnetosomes in Magnetospirillum gryphiswaldense. J Bacteriol 192:1097–1105. doi: 10.1128/JB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Q, Wu S, Li X, Zhang T, Yang J, Wang X, Li F, Li Y, Peng Y, Li J. 2018. Work patterns of MamXY proteins during magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. Appl Environ Microbiol 85:e02394-18. doi: 10.1128/AEM.02394-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Toro-Nahuelpan M, Corrales-Guerrero L, Zwiener T, Osorio-Valeriano M, Müller F-D, Plitzko JM, Bramkamp M, Thanbichler M, Schüler D. 2019. A gradient-forming MipZ protein mediating the control of cell division in the magnetotactic bacterium Magnetospirillum gryphiswaldense. Mol Microbiol 112:1423–1439. doi: 10.1111/mmi.14369. [DOI] [PubMed] [Google Scholar]
- 112.Pfeiffer D, Toro-Nahuelpan M, Bramkamp M, Plitzko JM, Schüler D. 2019. The polar organizing protein PopZ is fundamental for proper cell division and segregation of cellular content in Magnetospirillum gryphiswaldense. mBio 10:e02716-18. doi: 10.1128/mBio.02716-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taylor JA, Sichel SR, Salama NR. 2019. Bent bacteria: a comparison of cell shape mechanisms in proteobacteria. Annu Rev Microbiol 73:457–480. doi: 10.1146/annurev-micro-020518-115919. [DOI] [PubMed] [Google Scholar]
- 114.Hussain S, Wivagg CN, Szwedziak P, Wong F, Schaefer K, Izoré T, Renner LD, Holmes MJ, Sun Y, Bisson-Filho AW, Walker S, Amir A, Löwe J, Garner EC. 2018. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. Elife 7:e32471. doi: 10.7554/eLife.32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi H, Bratton BP, Gitai Z, Huang KC. 2018. How to build a bacterial cell: MreB as the foreman of Escherichia coli construction. Cell 172:1294–1305. doi: 10.1016/j.cell.2018.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi C, Fricke P, Lin L, Chevelkov V, Wegstroth M, Giller K, Becker S, Thanbichler M, Lange A. 2015. Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR. Sci Adv 1:e1501087. doi: 10.1126/sciadv.1501087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Berg HC. 1975. Chemotaxis in bacteria. Annu Rev Biophys Bioeng 4:119–136. doi: 10.1146/annurev.bb.04.060175.001003. [DOI] [PubMed] [Google Scholar]
- 118.Murat D, Hérisse M, Espinosa L, Bossa A, Alberto F, Wu L-F. 2015. Opposite and coordinated rotation of amphitrichous flagella governs oriented swimming and reversals in a magnetotactic spirillum. J Bacteriol 197:3275–3282. doi: 10.1128/JB.00172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reufer M, Besseling R, Schwarz-Linek J, Martinez VA, Morozov AN, Arlt J, Trubitsyn D, Ward FB, Poon WCK. 2014. Switching of swimming modes in Magnetospirillum gryphiswaldense. Biophys J 106:37–46. doi: 10.1016/j.bpj.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seong S, Park TH. 2001. Swimming characteristics of magnetic bacterium, Magnetospirillum sp. AMB-1, and implications as toxicity measurement. Biotechnol Bioeng 76:11–16. doi: 10.1002/bit.1021. [DOI] [PubMed] [Google Scholar]
- 121.Rismani Yazi S, Nosrati R, Stevens CA, Vogel D, Escobedo C. 2018. Migration of magnetotactic bacteria in porous media. Biomicrofluidics 12:011101. doi: 10.1063/1.5024508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Subramanian S, Kearns DB. 2019. Functional regulators of bacterial flagella. Annu Rev Microbiol 73:225–246. doi: 10.1146/annurev-micro-020518-115725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dufrêne YF, Persat A. 2020. Mechanomicrobiology: how bacteria sense and respond to forces. 4. Nat Rev Microbiol 18:227–240. doi: 10.1038/s41579-019-0314-2. [DOI] [PubMed] [Google Scholar]
- 124.Borg S, Popp F, Hofmann J, Leonhardt H, Rothbauer U, Schüler D. 2015. An intracellular nanotrap redirects proteins and organelles in live bacteria. mBio 6:e02117-14. doi: 10.1128/mBio.02117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cohen EJ, Nakane D, Kabata Y, Hendrixson DR, Nishizaka T, Beeby M. 2020. Campylobacter jejuni motility integrates specialized cell shape, flagellar filament, and motor, to coordinate action of its opposed flagella. PLoS Pathog 16:e1008620. doi: 10.1371/journal.ppat.1008620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.de Araujo FF, Germano FA, Gonçalves LL, Pires MA, Frankel RB. 1990. Magnetic polarity fractions in magnetotactic bacterial populations near the geomagnetic equator. Biophys J 58:549–555. doi: 10.1016/S0006-3495(90)82398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lefèvre CT, Bennet M, Klumpp S, Faivre D. 2015. Positioning the flagellum at the center of a dividing cell to combine bacterial division with magnetic polarity. mBio 6:e02286-14. doi: 10.1128/mBio.02286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]