Veillonella parvula is an anaerobic commensal and opportunistic pathogen whose ability to adhere to surfaces or other bacteria and form biofilms is critical for it to inhabit complex human microbial communities such as the gut and oral microbiota. Although the adhesive capacity of V. parvula has been previously described, very little is known about the underlying molecular mechanisms due to a lack of genetically amenable Veillonella strains. In this study, we took advantage of a naturally transformable V. parvula isolate and newly adapted genetic tools to identify surface-exposed adhesins called autotransporters as the main molecular determinants of adhesion in this bacterium. This work therefore provides new insights on an important aspect of the V. parvula lifestyle, opening new possibilities for mechanistic studies of the contribution of biofilm formation to the biology of this major commensal of the oral-digestive tract.
KEYWORDS: Veillonella, adhesins, autotransporter proteins, biofilms, diderm firmicutes
ABSTRACT
The Negativicutes are a clade of the Firmicutes that have retained the ancestral diderm character and possess an outer membrane. One of the best studied Negativicutes, Veillonella parvula, is an anaerobic commensal and opportunistic pathogen inhabiting complex human microbial communities, including the gut and the dental plaque microbiota. Whereas the adhesion and biofilm capacities of V. parvula are expected to be crucial for its maintenance and development in these environments, studies of V. parvula adhesion have been hindered by the lack of efficient genetic tools to perform functional analyses in this bacterium. Here, we took advantage of a recently described naturally transformable V. parvula isolate, SKV38, and adapted tools developed for the closely related Clostridia spp. to perform random transposon and targeted mutagenesis to identify V. parvula genes involved in biofilm formation. We show that type V secreted autotransporters, typically found in diderm bacteria, are the main determinants of V. parvula autoaggregation and biofilm formation and compete with each other for binding either to cells or to surfaces, with strong consequences for V. parvula biofilm formation capacity. The identified trimeric autotransporters have an original structure compared to classical autotransporters identified in Proteobacteria, with an additional C-terminal domain. We also show that inactivation of the gene coding for a poorly characterized metal-dependent phosphohydrolase HD domain protein conserved in the Firmicutes and their closely related diderm phyla inhibits autotransporter-mediated biofilm formation. This study paves the way for further molecular characterization of V. parvula interactions with other bacteria and the host within complex microbiota environments.
IMPORTANCE Veillonella parvula is an anaerobic commensal and opportunistic pathogen whose ability to adhere to surfaces or other bacteria and form biofilms is critical for it to inhabit complex human microbial communities such as the gut and oral microbiota. Although the adhesive capacity of V. parvula has been previously described, very little is known about the underlying molecular mechanisms due to a lack of genetically amenable Veillonella strains. In this study, we took advantage of a naturally transformable V. parvula isolate and newly adapted genetic tools to identify surface-exposed adhesins called autotransporters as the main molecular determinants of adhesion in this bacterium. This work therefore provides new insights on an important aspect of the V. parvula lifestyle, opening new possibilities for mechanistic studies of the contribution of biofilm formation to the biology of this major commensal of the oral-digestive tract.
INTRODUCTION
Negativicutes are atypical and poorly studied lineages of the Firmicutes displaying an outer envelope with lipopolysaccharide (1). Among the Negativicutes, Veillonella spp. are anaerobic diderm cocci that commonly inhabit the human and animal microbiota. One of their best-studied species, Veillonella parvula (2), is a natural inhabitant of multiple different microbiota, including the human gut (3, 4). V. parvula is considered a commensal organism and is proposed to play a role in the development of immunity through its capacity to colonize the infant gut (5, 6). It is a key early colonizer of the dental plaque during the establishment of sessile microbial communities called biofilms (7), promoting multispecies growth and playing a central role in the metabolism of community members through lactic acid consumption (8). However, V. parvula is also described as an opportunistic pathogen and has been associated with diverse infections, including osteomyelitis, endocarditis, spondylodiscitis, and abscesses as well as systemic infections (9–13).
The importance of V. parvula in the development of the microbial community spurred our interest in identifying the determinants of its adhesion and biofilm formation capacities. Moreover, considering the presence of an outer membrane (OM) in this atypical firmicute, we wondered whether V. parvula uses known diderm or monoderm biofilm determinants or currently undescribed adhesion factors. We recently studied V. parvula DSM2008 as a model diderm firmicute strain (14) to investigate its OM protein composition and detected 78 OM proteins, 13 of which are potential adhesins belonging to the type V family of secreted autotransporter (AT) proteins (T5SS) (15). Autotransporter proteins are specifically found in diderms, and all share common structural and functional features: a Sec-dependent signal peptide, a passenger domain providing the protein function, and an outer membrane β-barrel domain that allows secretion of the passenger domain (16). However, the challenge of genetic manipulation in V. parvula DSM2008 severely limited the study of these adhesins in this strain.
Here, we have sequenced and annotated the genome of V. parvula SKV38, a recently isolated, naturally transformable, and genetically amenable strain (17). We adapted and developed genetic tools for this organism, permitting random and site-directed mutagenesis, plasmid complementation, and controlled expression using an inducible promoter. This enabled us to identify and characterize factors involved in V. parvula biofilm formation. We find that the main V. parvula biofilm-modulating determinants are T5SS adhesins, i.e., typical diderm determinants. Interestingly, the identified adhesins possess an additional C-terminal domain compared to the known domain architecture of classical autotransporters. We also show that a locus encoding a metal-dependent phosphohydrolase HD domain protein is involved in biofilm formation, similarly to what was shown in the prototypical monoderm Bacillus subtilis (18). Therefore, our results demonstrate that diderm firmicutes use a mixture of diderm and monoderm factors to modulate their ability to engage in a biofilm lifestyle, supporting the idea that monoderm and diderm molecular systems could have coevolved in these atypical firmicutes.
RESULTS
Random transposon mutagenesis reveals two V. parvula SKV38 genes involved in biofilm formation.
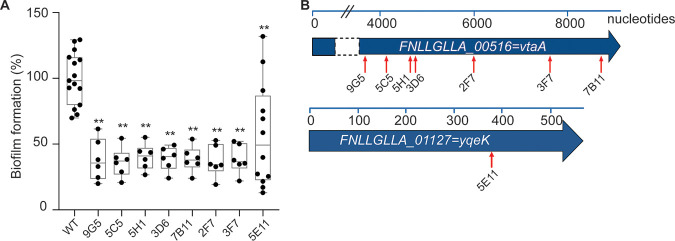
In order to obtain a framework for genetic work in the recently described naturally competent V. parvula SKV38 isolate, we sequenced it using PacBio technology. We obtained a completely assembled genome of 2.146 Mbp, carrying 1,912 predicted protein-encoding open reading frames (ORFs), 12 rRNAs, 49 tRNAs, and one transfer-messenger RNA (tmRNA) (see Materials and Methods). We performed random transposon mutagenesis in V. parvula SKV38 using the pRPF215 plasmid carrying an inducible transposase and a mariner-based transposon, previously used to mutagenize Clostridioides difficile (19), a close relative of the Negativicutes. We screened 940 individual transposon mutants for biofilm formation using crystal violet (CV) staining static biofilm assay in 96-well microtiter plates and identified eight independent mutants with significant reduction in biofilm formation (Fig. 1A). Whole-genome sequencing localized the transposons in two loci putatively implicated in biofilm formation (Fig. 1B). The most affected mutants correspond to insertions in FNLLGLLA_00516 (seven mutants), encoding a T5SS type Vc trimeric autotransporter. One transposon mutant corresponded to an insertion in FNLLGLLA_01127, encoding a putative HD phosphatase (Fig. 1B).
FIG 1.
Random transposon mutagenesis in Veillonella parvula SKV38 led to identification of mutants with reduced biofilm formation. (A) Biofilm assay in 96-well polystyrene plates after CV staining of nine transposon mutants identified by random mutagenesis grown for 24 h in BHILC. The mean for the WT is adjusted to 100%. Min-max box plots for 6 or 5 biological replicates for each strain are represented; each replicate is the mean of two technical replicates. *, P < 0.05; **, P < 0.005 (Mann-Whitney test). (B) Schematic representation of the identified transposon insertion point (red arrow) for the 8 transposon mutants. The blue bar represents the size of the gene in nucleotides.
FNLLGLLA_00516 encodes a trimeric autotransporter involved in autoaggregation.
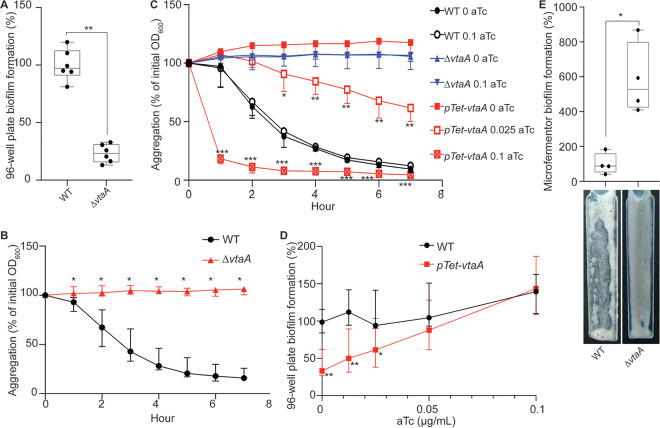
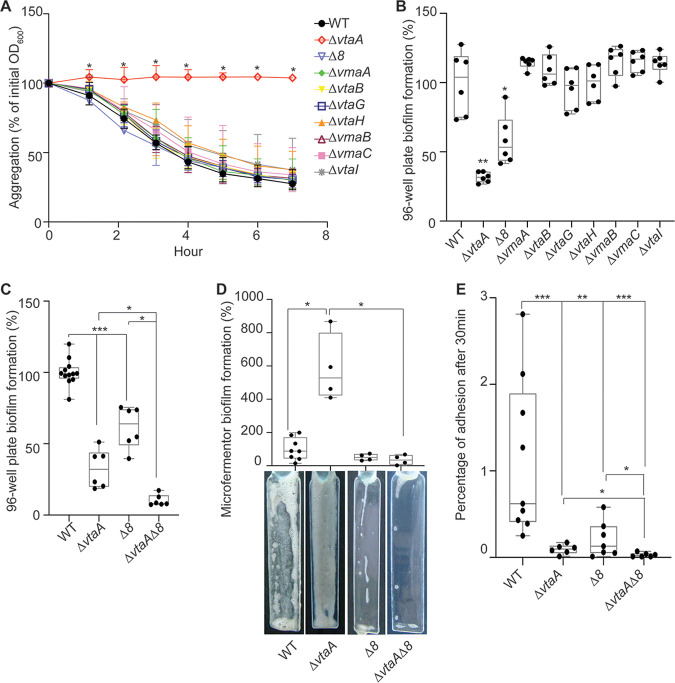
FNLLGLLA_00516 encodes a protein containing several domains usually identified in the T5SS type Vc trimeric autotransporters. Trimeric autotransporters are OM proteins specific to diderm bacteria that have been widely studied for their ability to bind to different surfaces or to other bacteria (20). FNLLGLLA_00516 is a homolog of V. parvula DSM2008 vpar_0464, which encodes a protein that was detected in the OM (15). FNLLGLLA_00516 was annotated by PROKKA as BtaF, a trimeric autotransporter identified in Brucella suis involved in adhesion to extracellular matrix and abiotic surfaces (21). Here, we renamed it Veillonella trimeric autotransporter A (VtaA), as the first trimeric autotransporter involved in biofilm formation identified in V. parvula SKV38. We deleted the vtaA coding sequence and showed that the ΔvtaA strain had no growth defect (see Fig. S1A in the supplemental material) but displayed a marked reduction of biofilm formation in 96-well polystyrene microtiter plates (Fig. 2A). Moreover, while V. parvula SKV38 cultures strongly aggregated, the ΔvtaA mutant did not (Fig. 2B; see Fig. S2 in the supplemental material). We constructed the Ptet-vtaA strain, where the chromosomal vtaA gene is placed under the control of a functional tetracycline/anhydrotetracycline (aTc)-inducible promoter (see Fig. S3 in the supplemental material), and showed that its aggregation capacity and biofilm formation in 96-well polystyrene microtiter plates directly correlated with the aTc concentration (Fig. 2C and D), demonstrating that VtaA-mediated cell-to-cell interactions are critical for biofilm formation under these conditions. Whereas the microtiter plate assay corresponds to a static biofilm assay, we also used continuous-flow glass microfermentors to investigate the contribution of VtaA to biofilm formation under dynamic conditions. Surprisingly, the ΔvtaA strain formed almost six times more biofilm than the wild-type (WT) strain under these conditions (Fig. 2E). Accordingly, scanning electronic microscopy (SEM) images of mature biofilms on microscopic plastic slides in a microfermentor showed that the ΔvtaA strain formed a much thicker biofilm than the WT (see Fig. S4 in the supplemental material). Altogether, these results suggest that autoaggregation differentially contributes to biofilm formation under static conditions on hydrophobic surfaces versus continuous-flow conditions on hydrophilic surfaces.
FIG 2.
VtaA is an adhesin involved in autoaggregation and biofilm formation. (A) Results of 96-well plate biofilm assay after 24 h growth in BHILC. The mean for the WT is adjusted to 100%. Min-max box plots of 6 biological replicates for each strain are shown. *, P < 0.05; **, P < 0.005 (Mann-Whitney test between strains). (B and C) Aggregation curves in spectrophotometry cuvette of the WT and ΔvtaA strains (B) and of an inducible vtaA strain with 0, 0.025, or 0.1 μg/ml of the inducer aTc (C). A value of 100% represent lack of aggregation, and 0% represents complete sedimentation of the culture. Medians from 6 biological replicates are shown, and error bars represent 95% confidence interval. At each time point we performed the Mann-Whitney test between conditions. We applied the Bonferroni correction for multiple testing. Significant P values are as follows: *, P < 0.004; **, P < 0.0004; ***, P < 0.00004. The indicated P values were calculated by comparing the WT and ΔvtaA strains (B) or the Ptet-vtaA strain without aTc and with different aTc concentrations (C). (D) Results of 96-well plate biofilm assay after 24 h of growth of an inducible vtaA mutant in BHILC with different concentrations of aTc. The value for the WT without aTc is adjusted to 100%. Medians of 6 biological replicates are shown; each replicate corresponds to the mean of two technical replicates, and error bars represent 95% confidence interval. *, P < 0.05; **, P < 0.005 (Mann-Whitney test). (E) Biofilm formation in a continuous-flow microfermentor on a glass spatula during 48 h in BHILC. The value for the WT was adjusted to 100%. Min-max box plots of 4 biological replicates for each strain are shown. A picture of the spatula before resuspension is shown below each box plot bar. *, P < 0.05 (Mann-Whitney test).
V. parvula SKV38 encodes 16 putative autotransporters in addition to VtaA.
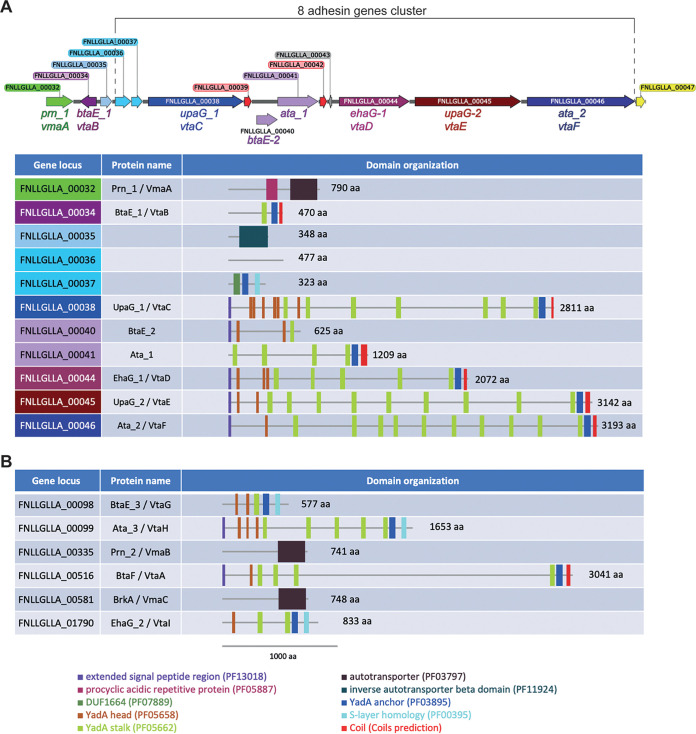
The strong biofilm phenotype displayed by the ΔvtaA mutant in a microfermentor led us to suspect that additional adhesins could modulate V. parvula biofilm formation capacity. Indeed, searching the V. parvula SKV38 genome revealed multiple genes encoding autotransporters (Table 1): three Va classical monomeric autotransporters with a characteristic PFAM_PF03797 autotransporter β domain (renamed Veillonella monomeric autotransporters A to C [VmaA to -C]) and eight other putative Vc trimeric autotransporters with a characteristic PFAM_PF03895 YadA anchor domain (renamed Veillonella trimeric autotransporters B to I [VtaB to -I]). We also identified several partial autotransporters: FNLLGLLA_00035, which contains only a PFAM_PF11924 Ve inverse autotransporter β domain but no putative α domain that normally carries the function of the protein, and FNLLGLLA_00036-37 and FNLLGLLA_00040-41, which are homologs of V. parvula DSM2008 Vpar_0041 and Vpar_0048, respectively, and appear to be split in SKV38 (Table 1). Interestingly, domain analysis of all trimeric ATs of V. parvula SKV38 showed that they possess an extra C-terminal domain (S-layer homology [SLH] or coiled-coil domain) after the YadA anchor domain that is not found in classical trimeric ATs. Among those, six autotransporter genes plus FNLLGLLA_00035, FNLLGLLA_00036-37, and FNLLGLLA_00040-41 form a potential genomic cluster coding for adhesins (Fig. 3A), whereas the six others are located in different areas of the genome (Fig. 3B).
TABLE 1.
V. parvula SKV38 autotransporters
| Locus tag | PROKKA gene name | Genome position |
Gene size (kb) | Strand | Description | DSM2008 homolog | Name | Class | |
|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||
| FNLLGLLA_00032 | prn 1 | 39354 | 41723 | 2.370 | Forward | Autotransporter | Fusion Vpar_0036-0037 | VmaA | Va |
| FNLLGLLA_00034 | btaE 1 | 42345 | 43754 | 1.410 | Reverse | Trimeric autotransporter, YadA like | Vpar_0039 | VtaB | Vc |
| FNLLGLLA_00035 | Hypothetical protein | 44146 | 45189 | 1.040 | Forward | Autotransporter (partial) | Vpar_0040 | Ve | |
| FNLLGLLA_00036 | Hypothetical protein | 45453 | 46883 | 1.431 | Forward | None | Split Vpar_0041 | ? | |
| FNLLGLLA_00037 | omp-alpha | 46910 | 47878 | 0.969 | Forward | Trimeric autotransporter/S-layer homology domain | Split Vpar_0041 | Vc? | |
| FNLLGLLA_00038 | upaG 1 | 48397 | 56829 | 8.433 | Forward | Trimeric autotransporter, YadA like | Vpar_0042 | VtaC | Vc |
| FNLLGLLA_00040 | btaE 2 | 57966 | 59840 | 1.875 | Forward | Trimeric autotransporter, YadA like (partial) | Split Vpar_0048 | ? | |
| FNLLGLLA_00041 | ata 1 | 59837 | 63463 | 3.627 | Forward | Trimeric autotransporter, YadA like | Split Vpar_0048 | Vc? | |
| FNLLGLLA_00044 | ehaG 1 | 65300 | 71515 | 6.216 | Forward | Trimeric autotransporter, YadA like | Vpar_0051 | VtaD | Vc |
| FNLLGLLA_00045 | upaG 2 | 71995 | 81420 | 9.426 | Forward | Trimeric autotransporter, YadA like | Vpar_0052 | VtaE | Vc |
| FNLLGLLA_00046 | ata 2 | 81941 | 91519 | 9.579 | Forward | Trimeric autotransporter, YadA like | Vpar_0053 | VtaF | Vc |
| FNLLGLLA_00098 | btaE 3 | 151792 | 153522 | 1.731 | Forward | Trimeric autotransporter/S-layer homology domain | Vpar_0100 | VtaG | Vc |
| FNLLGLLA_00099 | ata 3 | 154024 | 158982 | 4.959 | Forward | Trimeric autotransporter/S-layer homology domain | Absent | VtaH | Vc |
| FNLLGLLA_00335 | prn 2 | 414666 | 416888 | 2.223 | Forward | Autotransporter | Vpar_0330 | VmaB | Va |
| FNLLGLLA_00516 | btaF | 581236 | 590358 | 9.123 | Forward | Trimeric autotransporter, YadA like | Vpar_0464 | VtaA | Vc |
| FNLLGLLA_00581 | brkA | 668340 | 670583 | 2.244 | Forward | Autotransporter | Vpar_1322 | VmaC | Va |
| FNLLGLLA_01790 | ehaG 2 | 1943661 | 1946159 | 2.499 | Reverse | Trimeric autotransporter/S-layer homology domain | Vpar_1664 | VtaI | Vc |
FIG 3.
Veillonella parvula autotransporter domain organization. (A) Genetic organization of the V. parvula SKV38 autotransporter adhesin gene cluster and the corresponding adhesin domain organization. (B) Domain organization of the six remaining V. parvula SKV38 autotransporter adhesins encoded by genes located outside the cluster. Domains were detected with the HMMER package (59); only the domains with E values lower than 10−3 are shown. The presence of a C-terminal coil structure was determined using the COILS program (https://embnet.vital-it.ch/software/COILS_form.html). All V. parvula trimeric ATs display an additional C-terminal domain (an SLH or a coiled-coil domain) following the YadA anchor domain compared to classical trimeric autotransporters. aa, amino acids.
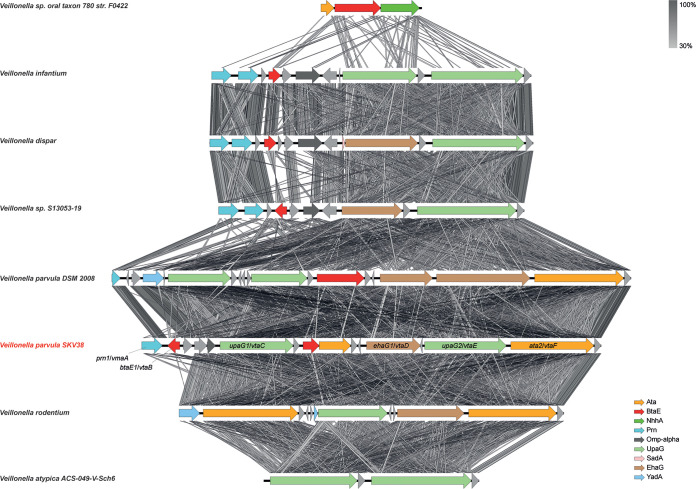
We selected eight Veillonella strains, including SKV38 and DSM2008, to study more precisely the evolution of the adhesin cluster. The trimeric autotransporter adhesins seem to evolve dynamically with numerous domain swaps, duplications, and reductions of gene copies, likely through homologous recombination, suggesting rapid evolutionary changes in the repertoire of Veillonella adhesins (Fig. 4). Duplications and deletions could be eased by the presence of short ORFs annotated as hypothetical proteins presenting a high degree of sequence identity. The most basal strain in the Veillonella phylogeny has a minimal cluster of only three adhesin genes. Throughout the Veillonella genus, the size of the cluster is very variable, with a minimal form in V. atypica, with only two adhesins. This specific adhesin locus, immediately upstream of rRNA-coding genes, is to our knowledge a peculiar genomic character of the Veillonella genus and is not found in other genera of the Veillonellaceae, suggesting that it originated in the common ancestor of all Veillonella species.
FIG 4.
Synteny of the adhesin gene cluster in a selection of Veillonella species. The synteny of the proteins of the cluster between the closest relatives was assessed using EasyFig (66). Oblique lines between genes represent tblastx identities (program parameters: maximum E value of 1012, minimum length of 30, minimum identity of 30). The V. parvula SKV38 strain used in this study is presented in red. The annotation of the genes of the cluster is indicated on the right.
The cluster of trimeric autotransporters is involved in surface binding and not aggregation.
To assess the function of the potential adhesins identified in the V. parvula SKV38 genome, we constructed, within the cluster of adhesin genes, independent deletion mutants for the two first autotransporters (vmaA and vtaB) and a large deletion for the eight adjacent genes encoding trimeric autotransporters or partial trimeric autotransporters, here called Δ8 (Δ[FNLLGLLA_00036 to vtaF]). We also generated independent individual mutants for each of the six additional autotransporters located outside the cluster. These mutants were all tested for biofilm formation in 96-well polystyrene plates and for aggregation capacities. None of the mutants, with the exception of the previously mentioned ΔvtaA strain, was affected for aggregation capacities (Fig. 5A). The Δ8 mutant was the sole mutant, in addition to the ΔvtaA mutant, to display lower biofilm formation in 96-well polystyrene microtiter plates (Fig. 5B and C), suggesting that the adhesins of this cluster could be involved in biofilm formation independently of cell-to-cell interactions. When tested in a microfermentor, the Δ8 mutant displayed a slightly reduced ability to form mature biofilm, but this was not statistically different from that of the WT (Fig. 5D). This reduced ability to form mature biofilms was actually more visible when observing SEM images, since the Δ8 mutant only poorly covered the coverslip, with sporadic aggregates of cells producing extracellular matrix (Fig. S4). An initial assay of adhesion to a glass spatula showed that both the ΔvtaA and Δ8 strains displayed a lower percentage of initial adhesion than the WT, suggesting that VtaA-mediated autoaggregation contributed to initial adhesion of the WT strain while the adhesin cluster is probably directly involved in surface binding (Fig. 5E). This also indicates that the ΔvtaA strain does not adhere to glass better than the WT, and so the increased biofilm formation of the Δvta strain in a microfermentor arises during the continuous-flow culture step. The effect of deleting vtaA and the 8 adhesin genes on initial adhesion was additive, since a ΔvtaA Δ8 double mutant showed a reduced initial adhesion on the microfermentor spatula compared to that of either the WT, ΔvtaA, or Δ8 strain (Fig. 5E). In addition, the ΔvtaA Δ8 mutant formed 17 times less biofilm than the ΔvtaA mutant in the microfermentor, indicating that in the absence of VtaA, the adhesins encoded by some of these eight genes strongly promote mature biofilm formation in the microfermentor (Fig. 5D).
FIG 5.
A cluster of eight trimeric autotransporters is involved in surface binding. (A) Aggregation curve in a spectrophotometry cuvette. A value of 100% represents lack of aggregation, and 0% represents complete sedimentation of the culture. Medians of 6 biological replicates are shown, and error bars represent 95% confidence interval. *, significant by Mann-Whitney test, corrected for multiple testing with Bonferroni correction; significance is achieved if the P value is <0.007. (B and C) Results of 96-well plate biofilm assay after 24 h of growth in BHILC. The mean for the WT is adjusted to 100%. Min-max box plots of 6 biological replicates for each strain are shown; each replicate is the mean of two technical replicates. For panel B, we applied a Mann-Whitney test: *, P < 0.05; **, P < 0.005. For panel C, we applied the Bonferroni correction for multiple testing, and tests were deemed significant only if the P value was <0.01: *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (D) Biofilm formation in a continuous-flow microfermentor on a glass spatula during 48 h in BHILC. The value for the WT was adjusted to 100%. Min-max boxplots of 4 biological replicates for each strain are shown. *, P < 0.05 (Mann-Whitney test). A picture of the spatula before resuspension is shown for each mutant below the box plot. (E) Initial adhesion on a glass spatula. The percentage of CFU that adhered to the spatula, controlled by the number of CFU of the inoculation solution, is shown. Min-max box plots of 6 to 9 replicates per strain are represented. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (Mann-Whitney test).
Taken together, these results demonstrate the differential contribution of VtaA and part of the adhesin cluster to V. parvula SKV38 adhesion and highlight the existence of potential interference mechanisms between them.
FNLLGLLA_01127 encodes an HD phosphatase that inhibits biofilm formation.
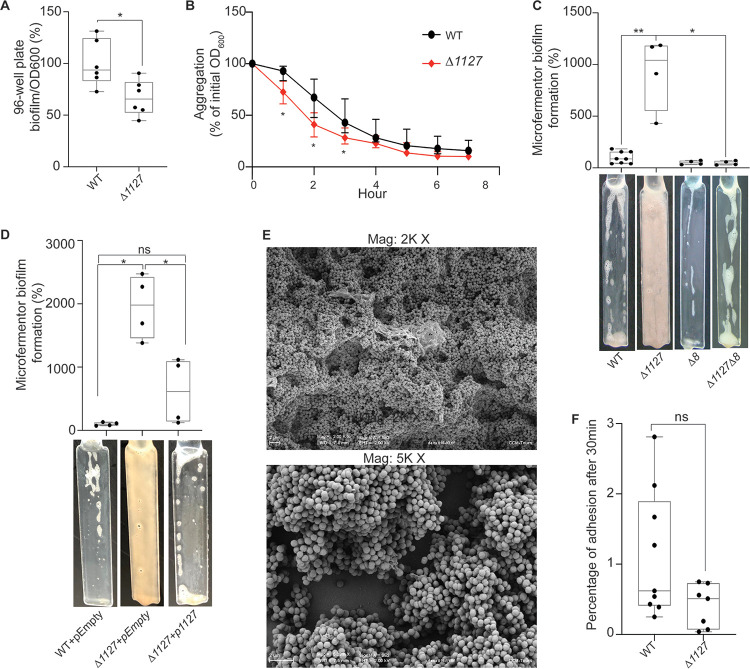
In addition to genes encoding potential T5SS proteins, we also identified a transposon mutant in FNLLGLLA_01127, encoding a protein of the HD phosphatase superfamily (Fig. 1B). The FNLLGLLA_01127 gene product is homologous to YqeK, a putative phosphatase required for pellicle formation and the development of biofilm in B. subtilis (18). FNLLGLLA_01127/yqeK is found in a cluster of genes (obg, yhbY, proB, proA, nadD, yqeK, lytR, and rsfS) whose synteny is very well conserved among Negativicutes. This cluster, or part of it, is also well conserved in almost all Firmicutes genomes we analyzed, both monoderm and diderm, as well as in members of other diderm phyla phylogenetically close to the Firmicutes, notably Deinococcus-Thermus (Fig. 6; see Fig. S5 and Data Set S2 in the supplemental material). A FNLLGLLA_01127 deletion mutant (Δ1127 mutant) had a lower carrying capacity than the WT, perhaps due to higher mortality during the stationary phase (Fig. S1), and a moderate 1.5-fold decrease in biofilm formation in microtiter plates after correcting for the growth defect (Fig. 7A). This mutant also displayed a slightly higher aggregation rate than the WT at early time points (Fig. 7B). The strongest phenotype of this mutant was detected in the microfermentor, with a 9-fold increase in biofilm formation compared to that of the WT (Fig. 7C). Expression of the FNLLGLLA_01127 gene in trans (plasmid p1127) did not complement the observed growth defect (Fig. S1B), but it did complement the increased biofilm formation in the microfermentor (Fig. 7D), showing that deletion of FNLLGLLA_01127 might have had polar effects on downstream genes of the operon, causing a growth defect, but that FNLLGLLA_01127 alone was responsible for the observed inhibition of biofilm formation. Scanning electronic microscopy showed that the Δ1127 mutant, similarly to the ΔvtaA mutant, formed a thick layered biofilm, although with fewer filaments and protein deposits than the WT (Fig. 7E). However, in contrast to the ΔvtaA or Δ8 mutant, the Δ1127 mutant showed no defect in initial adhesion to a glass spatula (Fig. 7F). Interestingly, a Δ1127 Δ8 double mutant formed almost 20 times less biofilm than the Δ1127 mutant in the microfermentor (Fig. 7C), suggesting that at least some of the autotransporters of the cluster were necessary for the observed strong biofilm formation by the Δ1127 mutant in the microfermentor.
FIG 6.
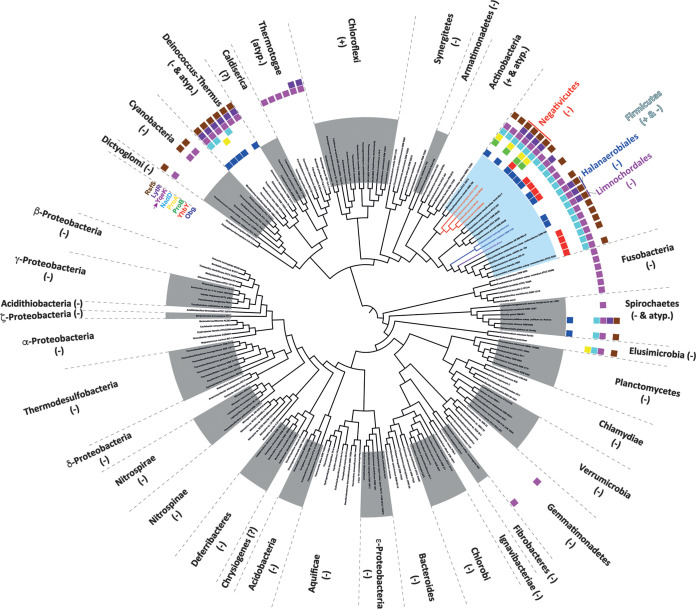
Occurrence and synteny of HD phosphatase (YqeK) in diderm and monoderm bacteria. The presence of the cluster was investigated using MacSyFinder (61), and the results were plotted onto a schematic reference tree of 187 cultivable bacteria among the 390 of the analyzed data bank. The cell wall status of each phylum is indicated as follows: −, diderm with LPS, +. monoderm; atyp., diderm without LPS; ?, unclear. For the Firmicutes, the diderm lineages are indicated in red (Negativicutes), blue (Halanaerobiales), and purple (Limnochordales).
FIG 7.
FNLLGLLA_01127 represses biofilm formation in a microfermentor. (A) Results of 96-well plate biofilm assay after 24 h of growth in BHLC corrected by optical density at 600 nm (0D600) after 24 h growth in plate. The mean value for the WT is adjusted to 100%. Min-max box plots of 6 biological replicates for each strain are shown, and each replicate is the mean of two technical replicates. *, P < 0.05 (Mann-Whitney test). (B) Aggregation curve in a spectrophotometry cuvette. A value of 100% represents lack of aggregation, and 0% represents complete sedimentation of the culture. Medians of 6 biological replicates are shown, and error bars represent 95% confidence interval. *, significant by Mann-Whitney test, corrected for multiple testing with Bonferroni correction; significance is achieved if the P value is <0.007. (C) Biofilm formation in a continuous-flow microfermentor on a glass spatula during 48 h in BHILC. The mean value for the WT is adjusted to 100%. Min-max box plots of 4 biological replicates for each strain are shown. *, P < 0.05; **, P < 0.005 (Mann-Whitney test). A picture of a spatula before resuspension is shown for each strain below the histogram. (D) Biofilm formation in a continuous-flow microfermentor on a glass spatula during 48 h in BHILC plus chloramphenicol. The mean value for WT+pEmpty is adjusted to 100%. Min-max box plots of 4 biological replicates for each strain are shown. *, P < 0.05 (Mann-Whitney test). A picture of a spatula before resuspension is shown for each strain below the boxplot. (E) Scanning electronic microscopy of Δ1127 biofilm grown under continuous flow of BHILC in a microfermentor on a plastic microscopy slide. Magnifications, ×2,000 and ×5,000. (F) Initial adhesion on glass spatula. The percentage of CFU that adhered to the spatula in 30 min, controlled by the number of CFU of the inoculation solution, is shown. Min-max box plots of 6 to 9 replicates per strain are shown. *, P < 0.05 (Mann-Whitney test); ns, not significant.
DISCUSSION
Originally described as a social organism mostly living in biofilm communities (8), Veillonella is a known bacterial member of multiple human microbiota. Biofilm formation and adhesion are important in these niches, but their study in Veillonella has been hindered by the lack of efficient genetic tools. Here, we used genetic tools adapted from Clostridia to characterize factors promoting biofilm formation in a naturally competent Veillonella parvula isolate.
We identified a T5SS type Vc trimeric autotransporter, FNLLGLLA_0516 (VtaA), as an important biofilm factor promoting V. parvula SKV38 autoaggregation. In addition to Hag1, a YadA-like autotransporter identified from the related species V. atypica involved in interspecies interactions (22), VtaA represents the second Veillonella protein described which is involved in adhesion and the first involved in abiotic surface adhesion and autoaggregation in diderm firmicutes. Beyond the potential impact on Veillonella niche colonization, aggregation capacity is known to contribute to bacterial protection from environmental stresses or host responses (23), promotion of host colonization (24), or pathogenesis (25) in various bacterial species. VtaA is homologous to the Brucella suis trimeric autotransporter BtaF. However, while B. suis BtaF promotes biofilm formation in vitro, it was not shown to promote aggregation (21), suggesting that these two proteins have different functions.
In diderm bacteria such as Escherichia coli, self-associating autotransporters (SAATs) from the type Va family and type Vc trimeric autotransporters were shown to contribute to biofilm formation through their self-recognition properties (26–32). However, in V. parvula, VtaA-mediated autoaggregation either promoted (on plastic surfaces and under static conditions) or strongly impaired (on glass surfaces and under continuous-flow conditions) biofilm formation, depending on the model used. The ΔvtaA mutant initially adhered less to the glass spatula than the WT, even though later it formed much more biofilm; thus, we suspect that the material (glass versus plastic) is not responsible for the observed difference between our two systems. We hypothesize instead that in the WT, VtaA-mediated aggregates are more sensitive to flow than individual cells and are thus washed out of the microfermentor faster and that adhesion to surfaces or to the biofilm extracellular matrix is more important than cell-to-cell interactions when the culture is performed under continuous flow.
Interference between cell surface structures is a well-described mechanism by which bacteria modulate their adhesion properties. In E. coli, multiple structures, such as chaperone-usher fimbriae, lipopolysaccharide (LPS) O antigen, or capsules, interfere with the self-recognizing autotransporter Ag43 though unknown mechanisms (33–36). Therefore, it is possible that in V. parvula, VtaA could compete with other adhesins through steric hindrance or competition for membrane export and thus limit biofilm formation under continuous-flow conditions. Consistently, the enhanced biofilm formation of the ΔvtaA mutant in the microfermentor was dependent on the presence of eight genes of the cluster of trimeric autotransporters, suggesting a competition between VtaA and an adhesin(s) of this cluster. Moreover, we noticed that both VtaA and the 8-gene cluster are necessary for full initial adhesion to a glass spatula in an independent manner, suggesting that any competition between them arises only later on, during continuous-flow cultures. Understanding the exact contributions of these different trimeric autotransporters to biofilm formation and their interplay with VtaA will require further characterization.
Analysis of the V. parvula SKV38 genome revealed the presence of seven other potential full-length autotransporters but no other types of classical diderm adhesins. None of them appeared to be involved in cell-to-cell interactions or biofilm formation on abiotic surfaces, and their function remains to be fully elucidated. As V. parvula is present in different microbiota, it is expected that a large arsenal of adhesion factors is necessary to adhere under different mechanical constraints and on different surfaces, such as tooth enamel or various epithelia. Moreover, Veillonella is known to coaggregate with streptococci (37–39), which produce the favored Veillonella carbon source, lactate (8), and it was shown to specifically coaggregate with Streptococcus and Actinomyces strains from the same microbiota, showing that coaggregation could have strong implications for niche colonization of these bacteria (40). V. parvula and other Veillonella spp. are also associated with different opportunistic infections, and the contribution of their adhesins to pathogenicity remains to be addressed. Finally, some autotransporters have been shown to carry nonadhesive functions, including protease activity (41), but we detected no classical protease domain in the Veillonella autotransporters.
Trimeric autotransporters possess a characteristic YadA anchor domain (PF03895) that is found mainly in Proteobacteria but also in Cyanobacteria, Verrumicrobia, Planctomycetes, Kiritimatiellaeota, Chlorobi, Synergistetes, Fusobacteria, and Negativicutes (https://pfam.xfam.org/family/PF03895 [December 2019]) (42). Interestingly, the YadA anchor of V. parvula SKV38 and all Veillonella trimeric autotransporters is not at the very end of the C terminus, where it is usually found in Proteobacteria, but is before the C terminus, followed by either a coiled domain or a S-layer homology (SLH) domain (Fig. 3; see Data Set S1 in the supplemental material). While the function of the coiled domain is unknown, in some bacteria the periplasmic SLH domain binds to peptidoglycan (43), suggesting that Veillonella trimeric autotransporters could be noncovalently attached to the peptidoglycan. These extra domains after the YadA anchor are also found in other Negativicutes (notably the extra SLH domain) and in some other diderm phyla phylogenetically related to the Firmicutes, such as Synergistetes and Fusobacteria (Data Set S1). In addition to possessing trimeric autotransporters with an extra coiled C-terminal domain, the fusobacterium Streptobacillus moniliformis ATCC 14647 carries eight genes encoding unique trimeric autotransporters with an extra OmpA family domain (PF00691) at their extreme C termini, a domain known to display affinity to peptidoglycan (44) (Data Set S1). These data suggest that a subset of phylogenetically close diderm bacteria have evolved trimeric autotransporters integrating different peptidoglycan binding domains. Whether these domains have an impact on trimeric autotransporter function or exposure to the surface, or more generally on outer membrane stabilization, is presently unknown.
Our screening also led to the identification of FNLLGLLA_01127, the homolog of B. subtilis YqeK, a putative phosphatase required for pellicle formation and the development of biofilm (18). Staphylococcus aureus YqeK was recently shown to be a nucleosidase hydrolyzing diadenosine tetraphosphate (Ap4A) into ADP (45). In Pseudomonas fluorescens, an increased level of Ap4A increases the cyclic di-GMP (c-di-GMP) concentration and enhances cell surface exposure of the large adhesin LapA, thus inducing biofilm formation (46). c-di-GMP regulates biofilm formation by modulating production of a variety of cell surface appendages or exopolysaccharides in both monoderm and diderm bacteria (50, 51). Interestingly, B. subtilis YqeK induces the epsA-O operon, which is involved in the production of biofilm matrix-forming polysaccharides (52). Deletion of V. parvula FNLLGLLA_01127 led to only a minor decrease in biofilm formation in 96-well plates but to a strong increase in continuous-flow biofilm formation that was dependent on the presence of the cluster of trimeric autotransporters. Further work is needed to determine whether FNLLGLLA_01127 directly impacts production of the adhesins of the cluster or participates to the production/regulation of an unknown exopolysaccharide, which, in contrast to the case for B. subtilis, would interfere with the function or exposure of the adhesins of the cluster rather than favor community development.
In this study, we have shown that classical diderm trimeric autotransporters and a potential nucleotidase, conserved in both monoderms and diderms, are crucial for adhesion between cells and/or to surfaces in the diderm firmicute V. parvula. Our work also underscores the rapid evolution of a diverse arsenal of trimeric autotransporters in the Veillonella genus, both in numbers and size, probably by efficient recombination favored by gene clustering, allowing rapid adaptation to changing environments. Taken together, our results suggest a complex interplay at the surface of V. parvula between different cell surface structures that may have coevolved for a long time in these atypical firmicutes. Much remains to be discovered about the regulatory circuits controlling these adhesion factors and their role in diderm firmicute biology.
MATERIALS AND METHODS
Genome preparation and sequencing.
V. parvula SKV38 genomic DNA was extracted using the Qiagen genomic tip 20G kit. It was sequenced to 1,500× coverage using PacBio sequencing of one single-molecule real-time (SMRT) cell with no multiplexing using the V2.1 chemistry. Only one SMRT cell was used but with no multiplexing, leading to an unusually large amount of subreads: 3 Gbp, meaning about 1,500× coverage assuming a 2.1-Mbp genome. This yielded 338,310 reads with a mean subread length of 9,080 bp and N50 read length of 13,500 bp. The longest subread length is above 70 kbp. We randomly subsampled the data to avoid misassemblies, keeping only 100,000 subreads, which resulted in a 430× coverage. The genome was then assembled using Canu version 1.8 (53) with the default parameters. In particular, subreads below 1,000 bp were dropped. The error correction steps of the Canu algorithm were not tuned, keeping the parameters that control alignment seed length, read length, overlap length, and error rates to their default values. We obtained one contig of 2.146 Mbp and an additional contig of only 1,972 bp that was abandoned due to lack of supporting data and was removed by the circularization process. The obtained assembled genome closely matched the genome size (2.1422 Mbp) and GC content (38.7%; expected, 38.6%) of the reference V. parvula DSM2008 strain. The resulting assembled genome was polished using Pilon (54), but no correction was required. No gaps or drops of coverage was detected based on sequana_coverage output (55, 56). The completeness of the candidate assembly was assessed to be 98% using the bacterial mode and the bacteria_db9 lineage-specific profile library of BUSCO software (57), while the number of complete duplicated or fragmented BUSCOs remained at 0, indicative of complete assembly. Alignment of all reads showed that only 4% (13,028) remained unmapped, and 80% of their length was below 2 kbp. The remaining reads (2,000 reads) mapped on various species and could not be further assembled. Overall, these analyses indicate that the final genome assembly is complete and of good quality.
Bioinformatic analyses.
The V. parvula SKV38 genome was annotated using PROKKA (58).
For protein domain visualization, PFAM domains (pfam.xfam.org, Pfam 32.0. [42]) were detected using HMMER (59). Domains with an E value lower than 10−3 were kept, and in the case of overlapping domains, the domain having the best E value was kept. The presence of C-terminal coil structures was determined using the COILS program (https://embnet.vital-it.ch/software/COILS_form.html) (60).
The search for HD phosphatase (YqeK) cluster homologs was conducted as follows. A local data bank containing 390 genomes representative of bacterial diversity was mined for the presence of a phosphatase containing the HD domain (PF01966) using HMMSEARCH and the –cut_ga option. Protein sequences were then filtered using alignment, functional annotation, protein domain presence, and phylogeny. Synteny was investigated in the locus around yqeK by looking, using MacSyFinder (61), for the presence of at least one of the 7 genes surrounding yqeK in V. parvula SKV38, namely, obg (containing the GTP1_OBG domain, PF01018), yhbY (containing the CRS1_YhbY domain, PF01985), proB (containing the AA_kinase domain, PF00696), proA (containing the Aldedh domain, PF00171), nadD (containing the CTP_transf_like domain, PF01467), lytR (containing the LytR_cpsA_psr domain, PF03816), and rsfS (containing the RsfS domain, PF02410), with no more than eight other genes separating them. All HMM profiles were downloaded from the PFAM site (pfam.xfam.org). As YqeK homologs are widespread in the Firmicutes, another local data bank containing 230 representative Firmicutes genomes was queried by the MacSyFinder approach as described above. All trees were visualized with ITOL (62). Details of the results are presented in Data Set S2 in the supplemental material.
Strains and growth conditions.
Bacterial strains and plasmids are listed in Table 2. V. parvula was grown in either brain heart infusion (BHI) medium (Bacto brain heart infusion; Difco) supplemented with 0.1% l-cysteine and 0.6% sodium dl-lactate (BHILC) or SK medium (10 g liter−1 tryptone [Difco], 10 g liter−1 yeast extract [Difco], 0.4 g liter−1 disodium phosphate, 2 g liter−1 sodium chloride, and 10 ml liter−1 60% [wt/vol] sodium dl-lactate; described in reference 17) and incubated at 37°C under anaerobic conditions in anaerobic bags (GENbag anaero; bioMérieux no. 45534) or in a C400M Ruskinn anaerobic-microaerophilic station. Escherichia coli was grown in lysogeny broth (LB) (Corning) medium under aerobic conditions at 37°C. When needed, 20 mg liter−1 chloramphenicol (Cm), 200 mg liter−1 erythromycin (Ery), or 2.5 mg liter−1 tetracycline (Tc) was added to V. parvula cultures, and 100 mg liter−1 carbenicillin (Cb) or 5 mg liter−1 Tc was added to E. coli cultures. Unless stated otherwise, 100 μg liter−1 anhydrotetracycline (aTc) was added to induce the Ptet promoter. All chemicals were purchased from Sigma-Aldrich unless stated otherwise.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Veillonella parvula strains | ||
| SKV38 | WT | 17 |
| 9G5 | SKV38 FNLLGLLA_00516::transposon | This study |
| 5C5 | SKV38 FNLLGLLA_00516::transposon | This study |
| 5H1 | SKV38 FNLLGLLA_00516::transposon | This study |
| 3D6 | SKV38 FNLLGLLA_00516::transposon | This study |
| 7B11 | SKV38 FNLLGLLA_00516::transposon | This study |
| 2F7 | SKV38 FNLLGLLA_00516::transposon | This study |
| 3F7 | SKV38 FNLLGLLA_00516::transposon | This study |
| 5E11 | SKV38 FNLLGLLA_01127::transposon | This study |
| ΔvtaA mutant | SKV38 ΔFNLLGLLA_00516::tetM | This study |
| Ptet-vtaA mutant | SKV38 catP-Term(fdx)-Ptet-FNLLGLLA_00516 | This study |
| Δ8 mutant | SKV38 ΔFNLLGLLA_00036-46::tetM | This study |
| ΔvmaA mutant | SKV38 ΔFNLLGLLA_00032::tetM | This study |
| ΔvtaB mutant | SKV38 ΔFNLLGLLA_00034::tetM | This study |
| ΔvtaG mutant | SKV38 ΔFNLLGLLA_00098::tetM | This study |
| ΔvtaH mutant | SKV38 ΔFNLLGLLA_00099::tetM | This study |
| ΔvmaB mutant | SKV38 ΔFNLLGLLA_00335::tetM | This study |
| ΔvmaC mutant | SKV38 ΔFNLLGLLA_00581::tetM | This study |
| ΔvtaI mutant | SKV38 ΔFNLLGLLA_01790::tetM | This study |
| ΔvtaA Δ8 mutant | SKV38 ΔFNLLGLLA_ 00516::catP ΔFNLLGLLA_00036-46::tetM | This study |
| Δ1127 mutant | SKV38 ΔFNLLGLLA_01127::tetM | This study |
| Δ1127 Δ8 mutant | SKV38 ΔFNLLGLLA_01127::tetM ΔFNLLGLLA_00036-46::catP | This study |
| WT+pEmpty | SKV38-pBSJL2-catP-Pmdh | This study |
| Δ1127+pEmpty mutant | SKV38 ΔFNLLGLLA_01127::tetM-pBSJL2-catP-Pmdh | This study |
| Δ1127+p1127 mutant | SKV38 ΔFNLLGLLA_01127::tetM-pBSJL2-catP-Pmdh-FNLLGLLA_01127 | This study |
| Ptet-ϕ mutant | SKV38-pRPF185ΔgusA | This study |
| Ptet-gusA mutant | SKV38-pRPF185 | This study |
| PCwp2 -gusA mutant | SKV38-pRPF144 | This study |
| Plasmids | ||
| pRPF215 | mariner Tn delivery plasmid, Ptet::Himar1 ITR-ermB-ITR catP tetR | 19 |
| pRPF185 | Tetracycline-inducible expression system fused with β-glucuronidase gusA Term(fdx)-Ptet-gusA-Term(slpA), catP | 63 |
| pRPF185ΔgusA | pDIA6103, tetracycline-inducible expression system Term(fdx)-Ptet-Term(slpA), catP | 67 |
| pRPF144 | Carries a Clostridium constitutive promoter fused with gusA PCwp2-gusA | 63 |
| pBSJL2 | E. coli-Veillonella shuttle plasmid, PgyrA::tetM | 68 |
| pBSJL2-cat | E. coli-Veillonella shuttle plasmid, Pcat::catP Pmdh promoter | This study |
| p1127 | pBSJL2-catP-Pmdh-FNLLGLLA_01127 | This study |
Natural transformation.
Cells were resuspended in 1 ml SK medium adjusted to an optical density at 600 nm (OD600) of 0.4 to 0.8, and 10 μl was dotted on SK agar petri dishes. On each drop, 0.5 to 1 μg plasmid or 75 to 200 ng μl−1 linear double-stranded DNA (dsDNA) PCR product was added, or water for the negative control. The plates were incubated for 48 h. The biomass was resuspended in 500 μl SK medium, plated on SK agar supplemented with the corresponding antibiotic, and incubated for another 48 h. Colonies were streaked on fresh selective plates, and the correct integration of the construct was confirmed by PCR and sequencing.
Random mariner transposon mutagenesis.
Plasmid pRPF215, described for Clostridium mutagenesis (Addgene 106377) (19), was transformed into V. parvula SKV38 by natural transformation and selected on Cm-supplemented SK agar medium. An overnight culture of V. parvula SKV38-pRPF215 in BHILC was then diluted to an OD600 of 0.1 in the same medium, supplemented with aTc, and grown for 5 h to induce the transposase. After induction, the culture was diluted and plated on BHILC supplemented with Ery and aTc for selection and incubated for 48 h. From the resulting colonies, 940 were inoculated in Greiner Bio-one polystyrene flat-bottom 96-well plates (655101), grown in BHILC supplemented with either Ery and aTc or Cm to confirm both the presence of the transposon and the loss of pRPF215, and then kept in 15% glycerol at –80°C. Selected transposon mutants were grown overnight, and the genomic DNA was harvested using the DNeasy blood and tissue kit (Qiagen). The genomic DNA was then sent for whole-genome sequencing at the Mutualized Platform for Microbiology of the Institut Pasteur.
Cloning-independent allelic exchange mutagenesis.
Site-directed mutagenesis of V. parvula strain SK38 was performed as described by Knapp and colleagues (17). Briefly, 1-kb regions upstream and downstream of the target sequence and the V. atypica tetracycline resistance cassette (tetM in pBSJL2) or catP resistance cassette from C. difficile (catP in pRPF185; Addgene 106367 [63]) were PCR amplified with overlapping primers using Phusion Flash high-fidelity PCR master mix (Thermo Scientific, F548). PCR products were used as templates in a second PCR round using only the external primers, which generated a linear dsDNA with the tetracycline resistance cassette flanked by the upstream and downstream sequences. This construct was transformed into V. parvula by natural transformation, and its integration into the genome was selected by plating on Tc- or Cm-supplemented medium. Positive candidates were further confirmed by a set of PCRs and sequencing around the site. Primers used in this study are listed in Table S1 in the supplemental material.
Complementation.
We replaced the tetracycline resistance gene and its gyrA promoter of the shuttle vector pBSJL2 by a chloramphenicol resistance gene, Pcat::cat from pRPF185, by Gibson assembly. Briefly, the inserts and the plasmids were PCR amplified and then mixed with 2× Gibson master mix (100 μl 5× ISO buffer, 0.2 μl 10,000-U/ml T5 exonuclease [NEB number M0363S], 6.25 μl 2,000-U/ml Phusion HF polymerase [NEB number M0530S], 50 μl 40,000-U/ml Taq DNA ligase [NEB number M0208S], 87 μl distilled water [dH2O]) for 24 reactions and incubated at 50°C for 30 to 60 min.
The resulting plasmid, pBSJL2-cat, was digested by Fastdigest BamHI (Thermo Scientific), and the band obtained was purified from the agarose gel using the QIAquick gel extraction kit (Qiagen) to be used as a linear plasmid in a second Gibson assembly. The genes and the Pmdh promoter of V. parvula SKV38 were amplified by PCR using PhusionFlash master mix and cloned in pBSJL2-cat using Gibson assembly. The mix was then transformed in E. coli DH5α and plated on LB with carbenicillin. The plasmid was harvested using the QIAprep spin miniprep kit (Qiagen) and transformed in V. parvula as described above.
Alternatively, the anhydrotetracycline-inducible expression cassette of pRPF185, here referred to as Ptet (Addgene 106367) (63), was inserted along with a chloramphenicol marker right before the ATG of the target gene, following the procedure described above for cloning-independent allelic exchange mutagenesis. The functionality of Ptet in V. parvula was previously verified using measurement of the aTc-dependent β-glucuronidase activity generated by the presence of pRPF185 transformed in V. parvula SKV38 (see Fig. S3 in the supplemental material).
Biofilm formation in 96-well microtiter plates.
Overnight cultures in BHILC medium were diluted to an OD600 of 0.05 and transferred to three Greiner Bio-one polystyrene flat-bottom 96-well plates, adding 150 μl per well. After 24 h of static incubation, one of the three plates was resuspended by pipetting to measure OD600 using a Tecan Infinite-M200-Pro spectrophotometer. The two other plates were used for coloration, as follows. Cultures were removed by carefully pipetting the supernatant out and biofilms fixed with 150 μl Bouin solution (HT10132; Sigma-Aldrich) for 15 min. Bouin solution was removed by inversion, and the biofilms were washed once in water. The biofilms were stained with 150 μl of 1% crystal violet (V5265; Sigma-Aldrich) for 15 min without shaking and then washed in water twice and left to dry. All washes were made by flicking the plate. After drying the plate, crystal violet was dissolved with 200 μl absolute ethanol and transferred to a clean 96-well plate for OD620 measurement (Tecan Infinite-M200-Pro spectrophotometer).
Biofilm formation in microfermentor.
Continuous-flow nonbubbled microfermentors containing a removable spatula were used as described previously (64, 65; https://research.pasteur.fr/en/tool/biofilm-microfermenters/). Briefly, a glass spatula was dipped in an overnight culture diluted to an OD600 of 0.5 in 15 ml BHILC for 15 min and returned to the fermentor. Biofilm was grown on the spatula for 48 h at 37°C. BHILC was constantly supplied through a peristaltic pump at 4 rpm. During the last hour, the speed was increased to 10 rpm to remove planktonic bacteria. A mix of filtered 90% nitrogen–5% hydrogen–5% carbon dioxide was also constantly supplied to maintain anaerobic conditions. After 48 h of growth, the spatula was removed, and the biofilm was resuspended by vortexing in 15 ml BHILC. We measured the OD600 of the resuspended biofilms with a Smart Spec Plus spectrophotometer (Bio-Rad).
Aggregation curve.
Overnight cultures were diluted to an OD600 of 0.8 in BHI medium in a semimicrospectrophotometry cuvette (Fisherbrand) and left to sediment on the bench in the presence of oxygen, so no growth should occur. The OD600 was measured every hour in a single point of the cuvette using a SmartSpec spectrophotometer (Bio-Rad).
Initial adhesion on glass.
Glass spatulas from microfermentors (described above) were dipped in overnight cultures diluted to an OD600 of 0.5 in 15 ml BHI medium for 30 min to let bacteria adhere. The spatulas were washed once in 15 ml BHI by submersion, and the adhering bacteria were resuspended in 15 ml clean BHI by vortexing. The culture used for inoculation, as well as the resuspended bacteria, was serially diluted and plated on an SK agar plate for CFU counting.
Statistical analysis.
Statistical analysis was performed using either R and Rstudio software or Prism8 (GraphPad Software, Inc.). We used only the nonparametric test and, when applicable, corrected for multiple testing. For microfermentor experiments, 4 replicates of each condition were used. For all the other experiments, at least 6 biological replicates in at least 2 independent experiment were used. A cutoff P value of 5% was used for all tests (*, P < 0.05; **, P < 0.05; ***, P < 0.005).
For growth curve analyses, we computed the growth rate and carrying capacity of each biological replicate using the Growthcurver 0.3.0 package in R, and we performed a Mann-Whitney test comparing both parameters for each mutant to those for the corresponding WT.
Data availability.
The SKV38 annotated genome sequence was deposited in the National Center for Biotechnology Information (NCBI) database under accession number NZ_LR778174.1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Justin Merritt for providing the V. parvula SKV38 strain, Bruno Dupuy and Robert P. Fagan for providing the different Clostridium plasmids, Pierre Simon Garcia for help with preparation of Fig. 3, Daniela Megrian Nuñez and Panagiotis Adam for genome data bank preparation, and the platforms France Génomique and IBISA.
We acknowledge funding from the French National Research Agency (ANR) (Fir-OM ANR-16-CE12-0010), from the Institut Pasteur “Programmes Transversaux de Recherche” (PTR 39-16), from the French government’s Investissement d'Avenir Program, Laboratoire d'Excellence “Integrative Biology of Emerging Infectious Diseases” (grant ANR-10-LABX-62-IBEID), and from the Fondation pour la Recherche Médicale (grant DEQ20180339185). N.B. was supported by a MENESR (Ministère Français de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche) fellowship. A.J.-F. was supported by a PRESTIGE program from Campus France.
N.B., A.J.-F., and C.B. designed the experiments. N.B., A.J.-F., E.B., and L.M. performed the experiments. J.W., N.T., and T.C. carried out all genomics and phylogeny analyses under the supervision of S.G. N.B., A.J.-F., and C.B. wrote the paper, with contributions from J.W., T.C., J.-M.G., and S.G. All authors read and approved the manuscript.
We declare no competing financial interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Megrian D, Taib N, Witwinowski J, Beloin C, Gribaldo S. 2020. One or two membranes? Diderm Firmicutes challenge the Gram-positive/Gram-negative divide. Mol Microbiol 113:659–671. doi: 10.1111/mmi.14469. [DOI] [PubMed] [Google Scholar]
- 2.Veillon A, Zuber A. 1898. Recherches sur quelques microbes strictement anaérobies et leur rôle en pathologie humaine. Arch Med Exp Anat Pathol 10. [Google Scholar]
- 3.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Bogert B, Erkus O, Boekhorst J, de Goffau M, Smid EJ, Zoetendal EG, Kleerebezem M. 2013. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol 85:376–388. doi: 10.1111/1574-6941.12127. [DOI] [PubMed] [Google Scholar]
- 5.Aujoulat F, Roudière L, Picaud J-C, Jacquot A, Filleron A, Neveu D, Baum T-P, Marchandin H, Jumas-Bilak E. 2014. Temporal dynamics of the very premature infant gut dominant microbiota. BMC Microbiol 14:325. doi: 10.1186/s12866-014-0325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrieta M-C, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, CHILD Study Investigators, Mohn WW, Turvey SE, Finlay BB. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 7.Kolenbrander PE. 2011. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci 3:49–54. doi: 10.4248/IJOS11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Periasamy S, Kolenbrander PE. 2010. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol 192:2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Chen P, Li J, Gao X, Chen X, Chen J. 2017. A new treatment of sepsis caused by Veillonella parvula: a case report and literature review. J Clin Pharm Ther 42:649–652. doi: 10.1111/jcpt.12559. [DOI] [PubMed] [Google Scholar]
- 10.Hirai J, Yamagishi Y, Kinjo T, Hagihara M, Sakanashi D, Suematsu H, Fujita J, Mikamo H. 2016. Osteomyelitis caused by Veillonella species: case report and review of the literature. J Infect Chemother 22:417–420. doi: 10.1016/j.jiac.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Gouze H, Noussair L, Padovano I, Salomon E, de Laroche M, Duran C, Felter A, Carlier R, Breban M, Dinh A. 2019. Veillonella parvula spondylodiscitis. Med Mal Infect 49:54–58. doi: 10.1016/j.medmal.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Hyo Y, Fukushima H, Harada T, Hara H. 2019. Nasal septal abscess caused by anaerobic bacteria of oral flora. Auris Nasus Larynx 46:147–150. doi: 10.1016/j.anl.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Wellens L, Casteels I, Huygens M. 2019. Veillonella parvula periorbital cellulitis: an unusual pathogen causing a common clinical sign. GMS Ophthalmol Cases 9:Doc17. doi: 10.3205/oc000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gronow S, Welnitz S, Lapidus A, Nolan M, Ivanova N, del Rio TG, Copeland A, Chen F, Tice H, Pitluck S, Cheng JF, Saunders E, Brettin T, Han C, Detter JC, Bruce D, Goodwin L, Land M, Hauser L, Chang YJ, Jeffries CD, Pati A, Mavromatis K, Mikhailova N, Chen A, Palaniappan K, Chain P, Rohde M, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lucas S. 2010. Complete genome sequence of Veillonella parvula type strain (Te3 T). Stand Genomic Sci 2:57–65. doi: 10.4056/sigs.521107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poppleton DI, Duchateau M, Hourdel V, Matondo M, Flechsler J, Klingl A, Beloin C, Gribaldo S. 2017. Outer membrane proteome of Veillonella parvula: a diderm firmicute of the human microbiome. Front Microbiol 8:1215. doi: 10.3389/fmicb.2017.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berne C, Ducret A, Hardy GG, Brun YV. 2015. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knapp S, Brodal C, Peterson J, Qi F, Kreth J, Merritt J. 2017. Natural competence is common among clinical isolates of Veillonella parvula and is useful for genetic manipulation of this key member of the oral microbiome. Front Cell Infect Microbiol 7:139. doi: 10.3389/fcimb.2017.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF, Fagan RP. 2015. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio 6:e02383. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Łyskowski A, Leo JC, Goldman A. 2011. Structure and biology of trimeric autotransporter adhesins. Adv Exp Med Biol 715:143–158. doi: 10.1007/978-94-007-0940-9_9. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Ranwez V, Posadas DM, Estein SM, Abdian PL, Martin FA, Zorreguieta A. 2013. The BtaF trimeric autotransporter of Brucella suis is involved in attachment to various surfaces, resistance to serum and virulence. PLoS One 8:e79770. doi: 10.1371/journal.pone.0079770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou P, Liu J, Merritt J, Qi F. 2015. A YadA-like autotransporter, Hag1 in Veillonella atypica is a multivalent hemagglutinin involved in adherence to oral streptococci, Porphyromonas gingivalis, and human oral buccal cells. Mol Oral Microbiol 30:269–279. doi: 10.1111/omi.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trunk T, Khalil HS, Leo JC. 2018. Bacterial autoaggregation. AIMS Microbiol 4:140–164. doi: 10.3934/microbiol.2018.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bongrand C, Ruby EG. 2019. The impact of Vibrio fischeri strain variation on host colonization. Curr Opin Microbiol 50:15–19. doi: 10.1016/j.mib.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonazzi D, Lo Schiavo V, Machata S, Djafer-Cherif I, Nivoit P, Manriquez V, Tanimoto H, Husson J, Henry N, Chaté H, Voituriez R, Duménil G. 2018. Intermittent pili-mediated forces fluidize Neisseria meningitidis aggregates promoting vascular colonization. Cell 174:143–155. doi: 10.1016/j.cell.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Klemm P, Vejborg RM, Sherlock O. 2006. Self-associating autotransporters, SAATs: functional and structural similarities. Int J Med Microbiol 296:187–195. doi: 10.1016/j.ijmm.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Ageorges V, Schiavone M, Jubelin G, Caccia N, Ruiz P, Chafsey I, Bailly X, Dague E, Leroy S, Paxman J, Heras B, Chaucheyras-Durand F, Rossiter AE, Henderson IR, Desvaux M. 2019. Differential homotypic and heterotypic interactions of antigen 43 (Ag43) variants in autotransporter-mediated bacterial autoaggregation. Sci Rep 9:11100. doi: 10.1038/s41598-019-47608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol 10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 29.Totsika M, Wells TJ, Beloin C, Valle J, Allsopp LP, King NP, Ghigo J-M, Schembri MA. 2012. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl Environ Microbiol 78:2179–2189. doi: 10.1128/AEM.06680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo J-M, Beloin C. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol 190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paton AW, Srimanote P, Woodrow MC, Paton JC. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun 69:6999–7009. doi: 10.1128/IAI.69.11.6999-7009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leo JC, Lyskowski A, Hattula K, Hartmann MD, Schwarz H, Butcher SJ, Linke D, Lupas AN, Goldman A. 2011. The structure of E. coli IgG-binding protein D suggests a general model for bending and binding in trimeric autotransporter adhesins. Structure 19:1021–1030. doi: 10.1016/j.str.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 33.Beloin C, Michaelis K, Lindner K, Landini P, Hacker J, Ghigo J-M, Dobrindt U. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J Bacteriol 188:1316–1331. doi: 10.1128/JB.188.4.1316-1331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korea C-G, Badouraly R, Prevost M-C, Ghigo J-M, Beloin C. 2010. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ Microbiol 12:1957–1977. doi: 10.1111/j.1462-2920.2010.02202.x. [DOI] [PubMed] [Google Scholar]
- 35.Hasman H, Chakraborty T, Klemm P. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol 181:4834–4841. doi: 10.1128/JB.181.16.4834-4841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schembri MA, Dalsgaard D, Klemm P. 2004. Capsule shields the function of short bacterial adhesins. J Bacteriol 186:1249–1257. doi: 10.1128/jb.186.5.1249-1257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes CV, Andersen RN, Kolenbrander PE. 1992. Characterization of Veillonella atypica PK1910 adhesin-mediated coaggregation with oral Streptococcus spp. Infect Immun 60:1178–1186. doi: 10.1128/IAI.60.3.1178-1186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mashima I, Nakazawa F. 2015. Interaction between Streptococcus spp. and Veillonella tobetsuensis in the early stages of oral biofilm formation. J Bacteriol 197:2104–2111. doi: 10.1128/JB.02512-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mashima I, Nakazawa F. 2014. The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe 28:54–61. doi: 10.1016/j.anaerobe.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Hughes CV, Kolenbrander PE, Andersen RN, Moore LV. 1988. Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology. Appl Environ Microbiol 54:1957–1963. doi: 10.1128/AEM.54.8.1957-1963.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells TJ, Totsika M, Schembri MA. 2010. Autotransporters of Escherichia coli: a sequence-based characterization. Microbiology 156:2459–2469. doi: 10.1099/mic.0.039024-0. [DOI] [PubMed] [Google Scholar]
- 42.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J 19:4473–4484. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park JS, Lee WC, Yeo KJ, Ryu K-S, Kumarasiri M, Hesek D, Lee M, Mobashery S, Song JH, Kim S Il, Lee JC, Cheong C, Jeon YH, Kim H-Y. 2012. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J 26:219–228. doi: 10.1096/fj.11-188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minazzato G, Gasparrini M, Amici A, Cianci M, Mazzola F, Orsomando G, Sorci L, Raffaelli N. 2020. Functional characterization of COG1713 (YqeK) as a novel diadenosine tetraphosphate hydrolase family. J Bacteriol 202:e00053-20. doi: 10.1128/JB.00053-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monds RD, Newell PD, Wagner JC, Schwartzman JA, Lu W, Rabinowitz JD, O'Toole GA. 2010. Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways. J Bacteriol 192:3011–3023. doi: 10.1128/JB.01571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Reference deleted.
- 49.Reference deleted.
- 50.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 51.Krasteva PV, Sondermann H. 2017. Versatile modes of cellular regulation via cyclic dinucleotides. Nat Chem Biol 13:350–359. doi: 10.1038/nchembio.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan F, Yu Y, Wang L, Luo Y, Guo J-H, Chai Y. 2016. The comER gene plays an important role in biofilm formation and sporulation in both Bacillus subtilis and Bacillus cereus. Front Microbiol 7:1025. doi: 10.3389/fmicb.2016.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cokelaer T, Desvillechabrol D, Legendre R, Cardon M, Cokelaer T, Desvillechabrol D, Legendre R, Cardon M. 2017. “Sequana”: a set of Snakemake NGS pipelines. JOSS 2:352. doi: 10.21105/joss.00352. [DOI] [Google Scholar]
- 56.Desvillechabrol D, Bouchier C, Kennedy S, Cokelaer T. 2018. Sequana coverage: detection and characterization of genomic variations using running median and mixture models. Gigascience 7:giy110. doi: 10.1093/gigascience/giy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 58.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 59.Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 61.Abby SS, Néron B, Ménager H, Touchon M, Rocha EPC. 2014. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One 9:e110726. doi: 10.1371/journal.pone.0110726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghigo J-M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 65.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JAJ, Molin S, Prensier G, Arbeille B, Ghigo J-M. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- 66.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J, Xie Z, Merritt J, Qi F. 2012. Establishment of a tractable genetic transformation system in Veillonella spp. Appl Environ Microbiol 78:3488–3491. doi: 10.1128/AEM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SKV38 annotated genome sequence was deposited in the National Center for Biotechnology Information (NCBI) database under accession number NZ_LR778174.1.