Abstract
Consumer genomics databases have reached the scale of millions of individuals. Recently, law enforcement authorities have exploited some of these databases to identify suspects via distant familial relatives. Using genomic data of 1.28 million individuals tested with consumer genomics, we investigated the power of this technique. We project that about 60% of the searches for individuals of European descent will result in a third-cousin or closer match, which theoretically allows their identification using demographic identifiers. Moreover, the technique could implicate nearly any U.S. individual of European descent in the near future. We demonstrate that the technique can also identify research participants of a public sequencing project. On the basis of these results, we propose a potential mitigation strategy and policy implications for human subject research.
Consumer genomics has gained popularity (1). As of April 2018, more than 15 million people have undergone direct-to-consumer (DTC) autosomal genetic tests, with about 7 million kits sold in 2017 alone (2). Nearly all major DTC providers use dense genotyping arrays that probe ~700,000 genomic variants and let participants download their raw genotype files in a plain-text format. This has led to the advent of third-party services, such as DNA.Land and GEDmatch, which allow participants to upload their raw genotype files for further analysis (table S1) (3). Nearly all of these services offer the option to find genetic relatives by locating identity-by-descent (IBD) segments that can indicate a shared ancestor. Finding genetic relatives can accurately link even distant relatives, such as second or third cousins (4–6) (fig. S1), and has led to multiple “success stories” within the genetic genealogy community, such as reunions of adoptees with their biological families (7).
In the past few months, law enforcement agencies have started exploiting third-party consumer genomics services to trace suspects by finding their distant genetic relatives. This route to identify individuals, dubbed long-range familial search, has been predicted before (8). It offers a powerful alternative to familial searches in forensic databases, which can only identify close (first to second degree) relatives (9, 10), and is highly regulated (11). In one notable case, law enforcement used a long-range familial search to trace the Golden State Killer (12, 13). Investigators generated a genome-wide profile of the perpetrator from a crime scene sample and uploaded the profile to GEDmatch, a database that contains ~1 million DNA profiles. The GEDmatch search identified a third-degree cousin (12). Extensive genealogical data traced the identity of the perpetrator, which was confirmed by a standard DNA test. Between April and August 2018, at least 13 cases were reportedly solved by long-range familial searches (Table 1 and table S2). Most of these investigations focused on cold cases, for which decades of investigation failed to identify the offender. Nonetheless, one case involved a crime from April 2018, suggesting that some law enforcement agencies have incorporated long-range familial DNA searches into active investigations. Parabon NanoLabs, a forensic DNA company, has announced that it set up a division that will use long-range familial searches and has already uploaded 100 cold cases to third-party DTC services (14). All of these lines of evidence suggest that long-range familial searches may become a standard investigative tool.
Table 1. Public cases of long-range familial cases.
A “−” indicates data not available.
| Case | Announcement | Solved by | Closest match | Comments |
|---|---|---|---|---|
| Buckskin Girl | 9 April 2018 | DNA Doe Project | First cousin once removed | |
| Golden State Killer | 24 April 2018 | Barbara Rae-Venter | Third cousin | |
| Lyle Stevik | 8 May 2018 | DNA Doe Project | Second cousin | Inbreeding complicated the estimation of the match. |
| William Earl Talbott II | 21 May 2018 | Parabon | Half-first cousin once removed | Second cousins were identified as well. |
| Joseph Newton Chandler III | 21 June 2018 | DNA Doe Project | Second cousin once removed | |
| Gary Hartman | 22 June 2018 | Parabon | Half-first cousin | Genealogists were able to overcome a nonpaternity event in the family tree of the suspect. |
| Raymond “DJ Freez” Rowe | 25 June 2018 | Parabon | – | |
| James Otto Earhart | 26 June 2018 | Parabon | Second cousin | |
| John D. Miller | 15 July 2018 | Parabon | – | |
| Matthew Dusseault and Tyler Grenon | 28 July 2018 | Parabon | – | |
| Spencer Glen Monnett | 29 July 2018 | Parabon | – | This was an active case for a crime that occurred in April 2018. |
| Darold Wayne Bowden | 23 August 2018 | Parabon | – | |
| Michael F. Henslick | 29 August 2018 | Parabon | – |
We took an empirical approach to investigate the probability that a long-range familial search will identify an individual. To this end, we analyzed a dataset of 1.28 million individuals who were tested with a DTC provider (15). We retained relatives with at least two IBD segments of >6 centimorgans (cM) each to increase the chance of correctly inferring genealogical relationships. Next, we removed pairs with IBD segments greater than 700 cM (i.e., first cousin and closer relationships) to circumvent ascertainment biases owing to the tendency of close relatives to undergo genetic testing together. Finally, considering each individual in turn as our “target,” we counted the number of individuals with a total IBD sharing of between 30 and 600 cM with the target (15). The low end of our range corresponds to approximately fourth cousins and the high end to second cousins, on the basis of a crowdsourcing project (16).
Our results show that nearly 60% of long-range familial searches return a relative with IBD segments with a total length of 100 cM or more (Fig. 1A). This level of IBD sharing usually corresponds to a third cousin or closer relative, similar to the case of the Golden State Killer. Interestingly, these success rates are higher than with surname inference from the Y chromosome, which is another genetic reidentification tactic (17). In 15% of the searches with our data, the top match had IBD segments of a total length of at least 300 cM, which corresponds to a second cousin or closer relative.
Fig. 1. The performance of long-range familial searches for various database sizes.
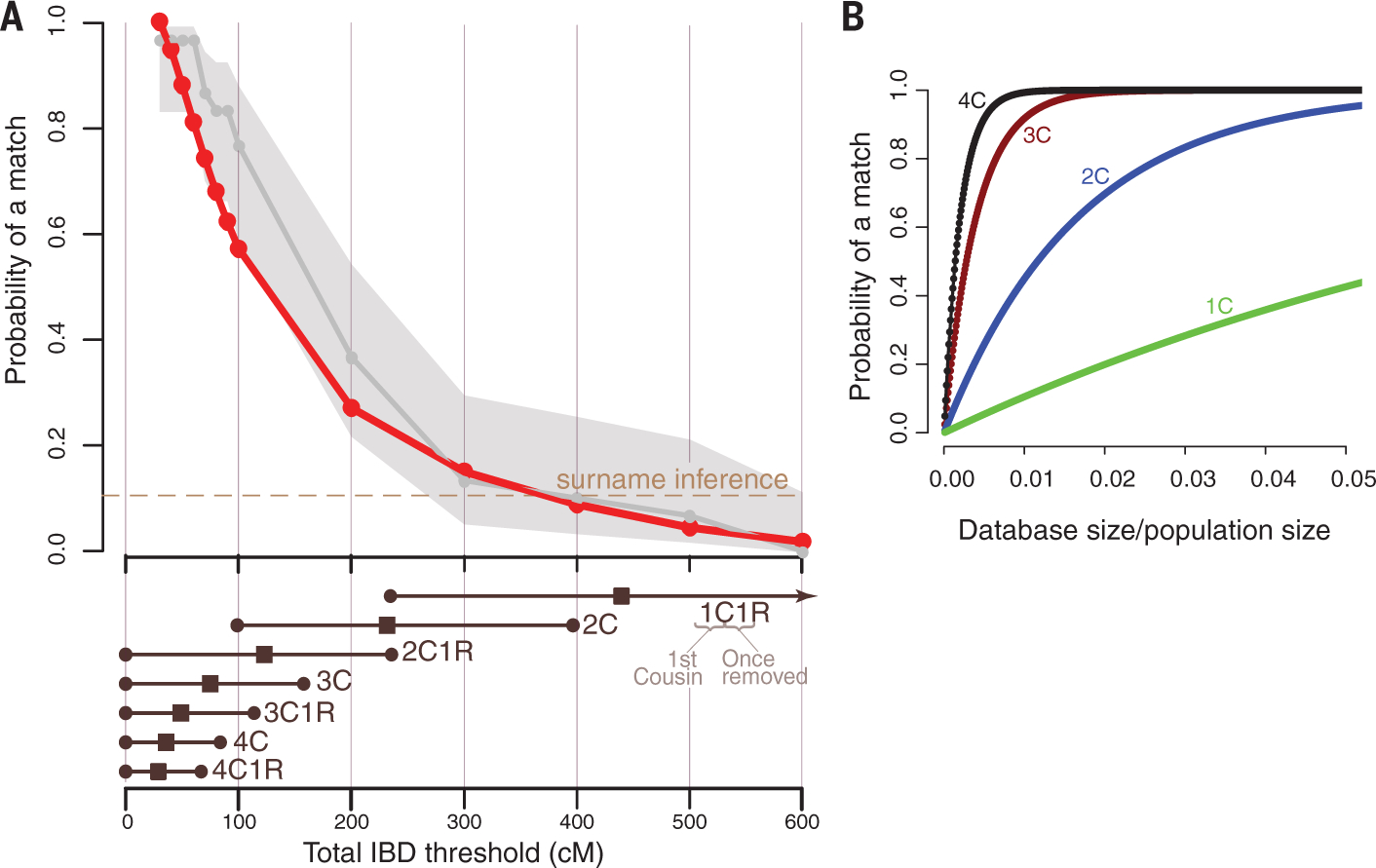
(A) The probability of finding at least one relative for various IBD thresholds (top) with 1.28 million searches of DTC-tested individuals (red) and 30 random GEDmatch searches (gray). Light gray shading indicates the 95% CI for the GEDmatch estimates. The dashed line indicates the probability of a surname inference from Y chromosome data (17). The bottom panel shows the 95% CIs (circles) and average total IBD length (squares) for a first cousin once removed (1C1R) to a fourth cousin once removed (4C1R) (20). (B) A population-genetic theoretical model for the probability of finding relatives up to a certain type of cousinship as a function of the database coverage of the population. 1C to 4C indicate first to fourth cousins.
We validated our results by performing 30 random long-range familial searches in GEDmatch. The results were similar: The top match in GEDmatch shared >100 cM in 76% of the cases [confidence interval (CI) of 59 to 88%] and >300 cM in 10% of the searches (CI of 3 to 25%), similar to the results with our 1.28 million individuals (Fig. 1A).
Long-range familial searches create racial disparity that is the opposite of disparities documented in traditional forensic databases (11). About 75% of the 1.28 million individuals were primarily of Northern European genetic background (fig. S2 and table S3), similar to previous reports of DTC genomics data (18). Individuals of primarily Northern European background were 30% more likely to have a >100-cM match than individuals whose genetic background was primarily from sub-Saharan Africa (fig. S3).
More broadly, a genetic database needs to cover only 2% of the target population to provide a third-cousin match to nearly any person (Fig. 1B). This assertion relies on a population genetics model that takes into account the probability of sharing at least two IBD segments of length >6 cM and assumes that the population grows at similar rates to the observed growth rates in the Western world during the past 200 years. (15) (fig. S4). This model has multiple simplifying assumptions, such as no population structure, no inbreeding, and random sampling of participants, and thus should be interpreted only as a rough guideline. Nevertheless, the model showed consistency between our empirical results and the IBD sharing profile of Northern Europeans in the United States (fig. S5). Using this model, we predict that with a database size of ~3 million U.S. individuals of European descent (2% of the adults of this population), more than 99% of the people of this ethnicity would have at least a single third-cousin match and more than 65% are expected to have at least one second-cousin match. With the exponential growth of consumer genomics (1), we posit that such a database scale is foreseeable for some third-party websites in the near future.
Next, we examined the theoretical ability to find the person of interest after finding a relative in a long-range familial search. We focused on reducing the search space using basic demographic information, such as geography, age, and sex. Using genealogical records of population-scale family trees (19), we computed the number of relatives of a third-cousin match after filtering them on the basis of place of residence, age, and sex. A study of serial criminals indicates that the place of crime is nearly always within 40 km (25 miles) of the criminal’s place of residence (20). To be conservative, we thus assumed that the location of the target can be estimated within a radius of 160 km (100 miles). We also assumed that the age of the target can be estimated within a ±5-year interval based on eyewitnesses or camera footage, as previously estimated (21). Finally, we assumed that the biological sex is known from the DNA sample.
We found that the suspect list can be pruned from basic demographic information. On the basis of counting relevant relatives of the match, the initial list of candidates contains an average of ~850 individuals (Fig. 2A). Our simulations indicate that localizing the target to within 160 km (100 miles) will exclude 57% of the candidates on average (Fig. 2B and table S4). Next, availability of the target’s age to within ±5 years will exclude 91% of the remaining candidates (Fig. 2C). Finally, inference of the biological sex of the target will halve the list to just around 16 to 17 individuals, a search space that is small enough for manual inspection. We also considered a scenario of reidentification of anonymized clinical genetic data. The safe-harbor provisions of the Health Insurance Portability and Accountability Act of 1996 (HIPAA) privacy law permit the release of the year of birth. An age specified at a single-year resolution is, as expected, a more powerful identifier compared to a 10-year interval (Fig. 2D). Together with geography (<160 km (100 miles)) and sex, it is expected to reduce the search space to just one to two individuals (Fig. 2E). To conclude, the main barrier is not finding a match or theoretical power to prune the search space. Rather, successfully tracing an individual depends mainly on the accessibility of genealogical data for the matched relative, their accuracy, and the time invested in organizing the genealogical data.
Fig. 2. Tracing a person of interest from a distant match using demographic identifiers.
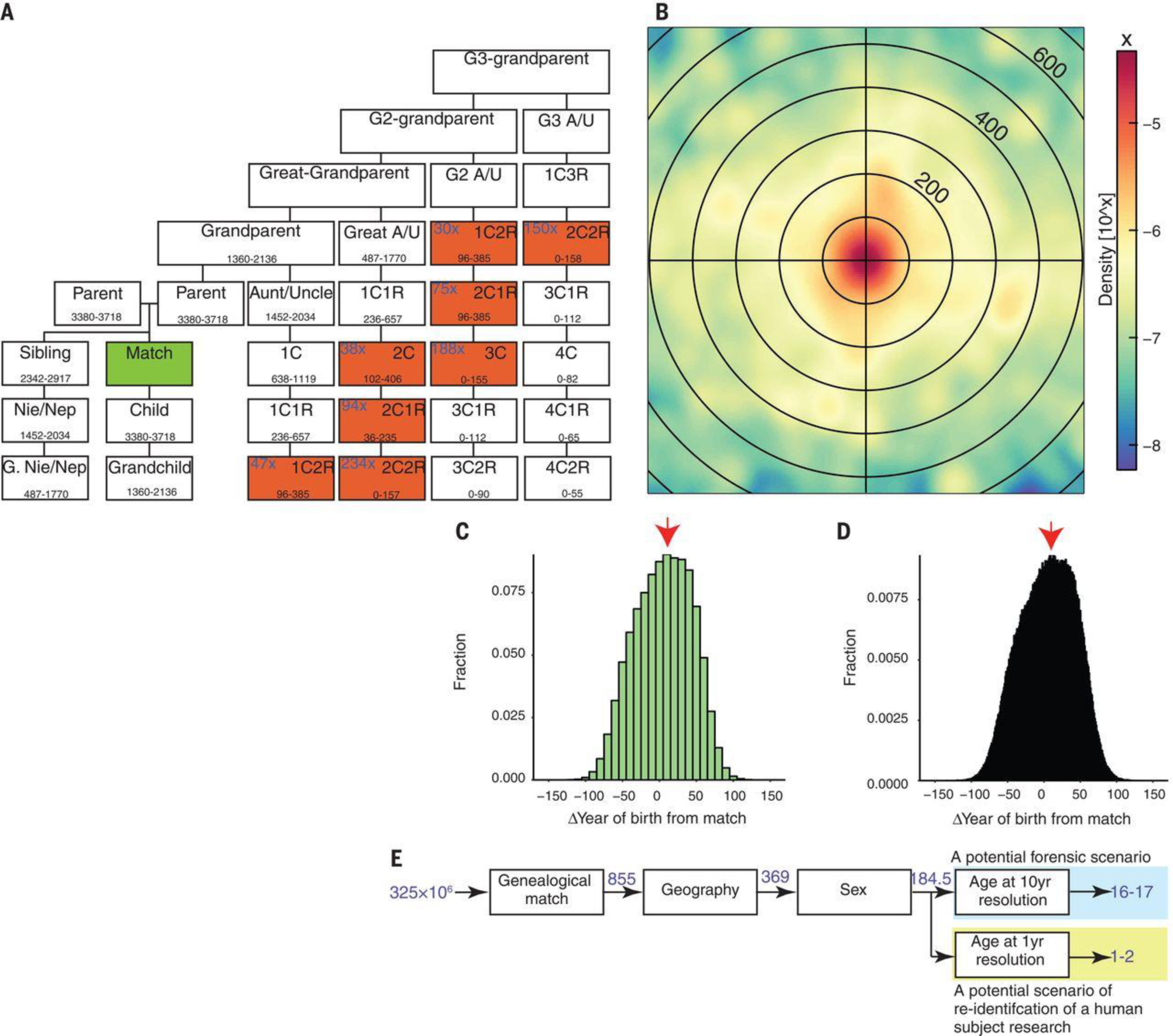
(A) The possible relatives of a match (green) in a database. Each square represents a potential degree of relatedness. The range corresponds to the 5th to 95th percentile of shared IBD in centimorgans from (16). Red indicates relatives that could fit a bona fide 3C match (~100 cM).The average number of relatives is indicated in the top-left corner of each square on the basis of a fertility rate of 2.5 children per couple. Only genealogical relationships that are within 100-cM range include the average number of relatives. Nie/Nep, Niece/Nephew; G, Great; G2, Great-great; G3,Great-great-great; A/U, Aunt/Uncle. (B) An example of the geographical dispersion of third cousins or second cousins once removed around the matched relative. Every circle indicates 100 km. (C and D) The distribution of the expected age differences between matches and their potential relatives with a genetic distance of third cousins.The main text reports a conservative scenario, in which the age estimator of the target is in the highest bin of each histogram (red arrow).The age distribution is shown at a 10-year resolution (C) and at a 1-year resolution (D). (E) The entire pipeline of using demographic identifiers along with a long-range familial match to identify a U.S. person (blue type indicates the average number of people after incorporating each piece of information.).
To better understand the risk of reidentifying human subjects, we conducted a long-range familial search on a specific 1000 Genomes Project individual. We selected a female from the CEU (Utah residents with Northern and Western European ancestry) cohort, whose husband has been identified using surname inference (17). We extracted her genome from the (publicly available) 1000 Genomes data repository, reformatted her genotype to resemble a file released by DTC providers, and uploaded the genotype to GEDmatch. Searching GEDmatch returned two relatives, one from North Dakota and one from Wyoming, with sufficient genetic and genealogical details (Fig. 3). Both relatives shared about 170 to 180 cM with the 1000Genomes sample, which corresponds to six to seven degrees of separation. They also shared 62 cM between each other, indicating that they were distantly related via an ancestral couple who lived four to six generations ago. In about 1 hour of work, we identified the ancestral couple from publicly available genealogical records. Next, we searched for descendants of the ancestral couple that matched the publicly available demographic data of the 1000Genomes sample, such as her expected year of birth and pedigree structure. This step, performed manually, was time consuming and not trivial, because the ancestral couple had more than 10 children and hundreds of descendants. After a full day of work, we eventually excluded all other candidates and traced the identity of our target, which was the same person we had previously reidentified based on surname inference of her husband.
Fig. 3. Tracing a 1000Genomes sample using a long-range familial search.
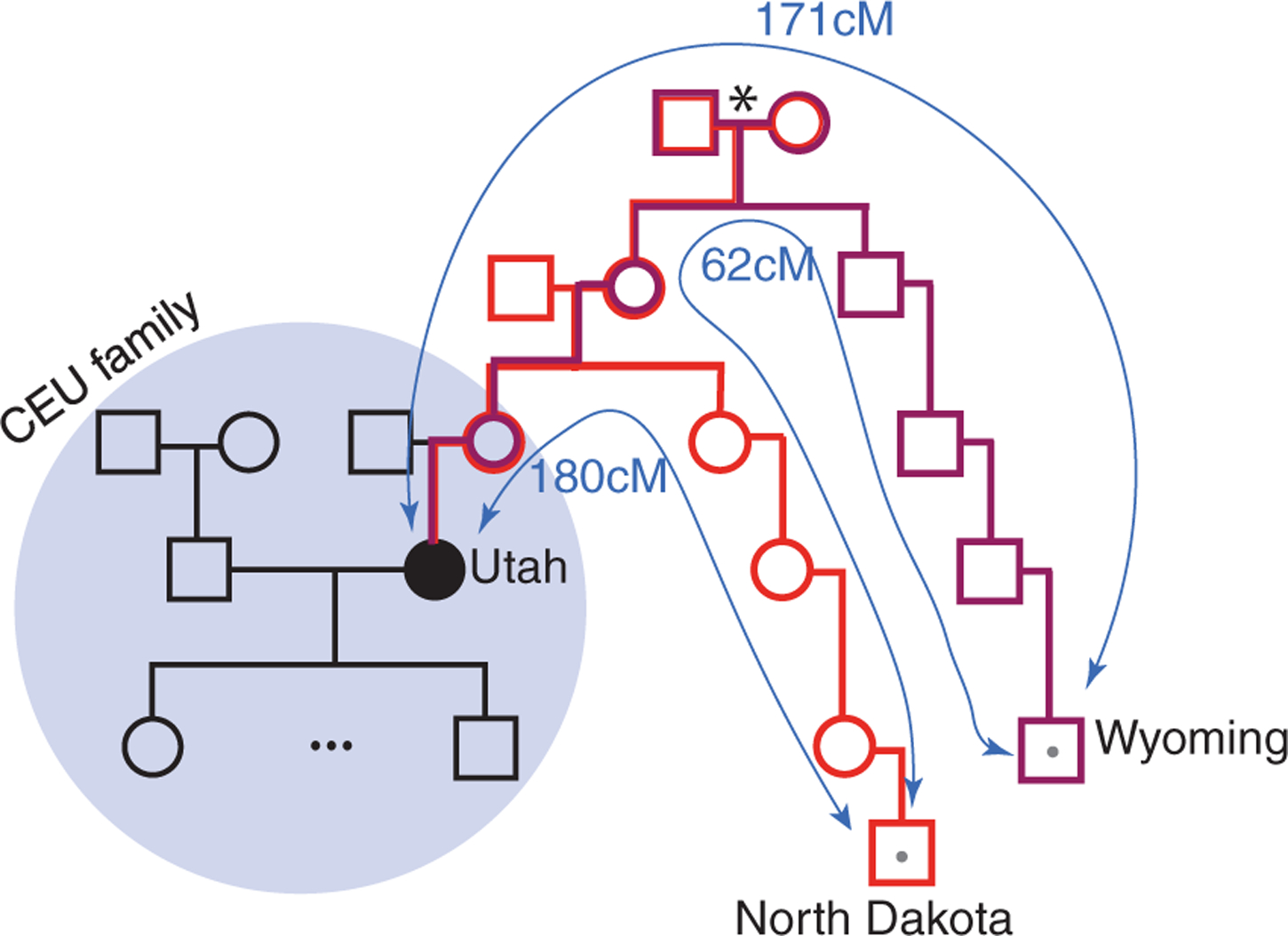
The CEU pedigree is shown in black. To respect the privacy of the family, we omitted the sample identifiers and the exact pedigree structure. A GEDmatch search of the person of interest (black circle) returned two males (squares with gray dots) with a total IBD sharing of 180 and 171 cM to the target, respectively, and 62 cM between themselves. Using public genealogical records, we identified the ancestral couple (asterisk) of the matches and the person of interest.
Taken together, we posit that our results warrant a reevaluation of the status quo regarding the identifiability of DNA data, especially of U.S. individuals. Although policy-makers and the general public may be in favor of such enhanced forensic capabilities for solving crimes, it relies on databases and services that are open to everyone. Thus, the same technique could also be exploited for harmful purposes, such as reidentification of research subjects from their genetic data. The Revised Common Rule, which will regulate federally funded human subject research starting in January 2019, does not de fine genome-wide genetic datasets as identifiable information (22). However, the rule permits the U.S. Department of Health and Human Services (HHS) to revise the scope of identifiable private information on the basis of technological developments. In light of our results, we encourage HHS to consider genome-wide information as identifiable.
Finally, we propose a measure to mitigate some of the risks and restore control to data custodians. In our proposal, DTC providers should cryptographically sign the text file containing the raw data available to customers (fig. S6). Third-party services will be able to authenticate that a raw genotyping file was created by a valid DTC provider and not further modified. If adopted, our approach has the potential to prevent the exploitation of long-range familial searches to identify research subjects from genomic data. Moreover, it will complicate the ability to conduct unilaterally long-range familial searches from DNA evidence (15). As such, it can complement previous proposals regarding the regulation of long-range familial searches by law enforcement (23) and offers better protection in cases in which the law cannot deter misuse. To facilitate consideration of our approach by the community, we provide a demo source code on GitHub that can sign and verify the raw genotype files using a previously published digital signature scheme (24). Overall, we believe that technical measures, clear policies for law enforcement in using long-range familial searches, and respecting the autonomy of participants in genetic studies are necessary components for long-term sustainability of the genomics ecosystem.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Japhet and A. Gordon for their contributions to the cryptographic signature scheme and Y. Naveh, Y. Ben-David, C. Moore, and the DNA Doe Project for valuable comments.
Funding: Y.E. holds a Burroughs Wellcome Fund Career Award at the Scientific Interface. S.C. thanks the Israel Science Foundation, grant no. 407/17.
Footnotes
Competing interests: Y.E. and T.S. adapted the code for the cryptographic signatures. Y.E. and T.S are MyHeritage employees. Y.E. is also a consultant of ArcBio. I.P. holds equity in 23andMe. S.C. is a paid consultant of MyHeritage. When multiple companies are mentioned in this manuscript, we listed them in a lexicographic order.
Data and materials availability: The code for the cryptographic signatures is available on https://github.com/erlichya/signature with an MIT license. The millions of genealogical records for the demographic analysis data are available on http://familinx.org/ under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported license. The code for the population genetics simulation and 1000Genomes extraction is available in the supplementary materials under an MIT license. Following the MyHeritage terms, we cannot share the individual-level genomic data. We will share the anonymized IBD network topology on request and subject to the MyHeritage Terms and Conditions and Privacy Policy under the following terms: (i) researchers will need an IRB approval for their study, (ii) the data can only be processed in a MyHeritage facility and cannot be used to reidentify individuals, (iii) the results can only be used for noncommercial purposes, and (iv) MyHeritage does not ask authorship in new publications that use the anonymized IBD network. Researchers who are interested in the data or in pursuing research collaboration opportunities can contact dnaresearch@myheritage.com.
REFERENCES AND NOTES
- 1.Khan R, Mittelman D, Genome Biol. 19, 120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larkin L, Autosomal DNA testing comparison chart, The DNA Geek; http://thednageek.com/dna-tests/.
- 3.Nelson SC, Fullerton SM, J. Genet. Couns 27, 770–781 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Gusev A et al. , Genome Res. 19, 318–326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huff CD et al. , Genome Res. 21, 768–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henn BM et al. , PLOS ONE 7, e34267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Society of Genetic Genealogy Wiki, Success stories (2018); https://isogg.org/wiki/Success_stories.
- 8.Erlich Y, Narayanan A, Nat. Rev. Genet 15, 409–421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge J, Chakraborty R, Eisenberg A, Budowle B, J. Forensic Sci 56, 1448–1456 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Garrison NA, Rohlfs RV, Fullerton SM, Nat. Rev. Genet 14, 445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Mammo D, Siegel MB, Katsanis SH, Investig. Genet 2, 22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gafni M, “Here’s the ‘open-source’ genealogy DNA website that helped crack the Golden State Killer case,” Mercury News, 26 April 2018; www.mercurynews.com/2018/04/26/ancestry-23andme-deny-assisting-law-enforcement-in-east-area-rapist-case/.
- 13.Jouvenal J, “To find alleged Golden State Killer, investigators first found his great-great-great-grandparents,” Washington Post, 30 April 2018; www.washingtonpost.com/local/public-safety/to-find-alleged-golden-state-killer-investigators-first-found-his-great-great-great-grandparents/2018/04/30/3c865fe7-dfcc-4a0e-b6b2-0bec548d501f_story.html?utm_term=.6ff5cff1630e.
- 14.Aldhous P, “DNA data from 100 crime scenes has been uploaded to a genealogy website—just like the Golden State Killer,” BuzzFeed, 17 May 2018; www.buzzfeed.com/peteraldhous/parabon-genetic-genealogy-cold-cases?utm_term=.tkKXDVOWq#.yyz8oGQWd.
- 15. See supplementary materials.
- 16.Bettinger BT, The Shared cM Project – Version 3.0 (2017); https://thegeneticgenealogist.com/wp-content/uploads/2017/08/Shared_cM_Project_2017.pdf.
- 17.Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y, Science 339, 321–324 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Yuan J et al. , Nat. Genet 50, 160–165 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Kaplanis J et al. , Science 360, 171–175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren J et al. , J. Quant. Criminol 14, 35–59 (1998). [Google Scholar]
- 21.Han H, Otto C, Jain AK, “Age estimation from face images: Human vs. machine performance,” paper presented at the 6th International Association for Pattern Recognition (IAPR) International Conference on Biometrics (ICB), Madrid, Spain, 4 to 7 June 2013. [Google Scholar]
- 22.Department of Health and Human Services, Fed. Regist 82, 7149–7274 (2017). [PubMed] [Google Scholar]
- 23.Ram N, Guerrini CJ, McGuire AL, Science 360, 1078–1079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein DJ, Duif N, Lange T, Schwabe P, Yang B-Y, J. Cryptogr. Eng 2, 77–89 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


