ABSTRACT
Fragmentation of tRNAs generates a family of small RNAs collectively known as tRNA-derived fragments. These fragments vary in sequence and size but have been shown to regulate many processes involved in cell homoeostasis and adaptations to stress. Additionally, the field of extracellular RNAs (exRNAs) is rapidly growing because exRNAs are a promising source of biomarkers in liquid biopsies, and because exRNAs seem to play key roles in intercellular and interspecies communication. Herein, we review recent descriptions of tRNA-derived fragments in the extracellular space in all domains of life, both in biofluids and in cell culture. The purpose of this review is to find consensus on which tRNA-derived fragments are more prominent in each extracellular fraction (including extracellular vesicles, lipoproteins and ribonucleoprotein complexes). We highlight what is becoming clear and what is still controversial in this field, in order to stimulate future hypothesis-driven studies which could clarify the role of full-length tRNAs and tRNA-derived fragments in the extracellular space.
KEYWORDS: Exosomes, tRFs, tRNA halves, intercellular communication
1. Introduction
The presence of metabolites derived from tRNAs in blood or urine has been noticed in the past [1]. However, it was not until the discovery of small regulatory RNAs and the coming of age of high-throughput sequencing that tRNA-derived small RNAs started to be identified in a variety of cells, tissues and organisms [2–6], and later in biofluids such as blood serum [7,8].
It was clear from almost the beginning that these tRNA-derived fragments were involved in the regulation of cellular stress, as their generation by endonucleolytic cleavage at the tRNA anticodon loop was upregulated in cells exposed to different types of stress stimuli [6,9,10]. Later, molecular mechanisms influenced or directly regulated by tRNA-derived fragments started to emerge. The regulation of translation initiation by the so called stress-induced tRNA halves [10,11] is particularly interesting because it shows that tRNAs are a hub of adaptive responses of the cell and not simply static adaptors between codons and amino acids. To date, tRNA-derived fragments have been functionally linked to a variety of molecular processes including translational regulation [10–14], proliferation [15], apoptosis [16], stress-granule formation [17,18], mRNA stabilization [19], tRNA aminoacylation [20], transposon expression [21], ribosome biogenesis [22] and the inheritance of acquired traits [23,24].
In the extracellular space, tRNA-derived fragments have been shown to exist in extracellular vesicles (EVs) derived from the epididymis, from where they are transferred to sperm and later to oocytes during fertilization in mice [23–25]. In embryos, they modulate the stability and expression of ncRNAs and histones, ultimately regulating global chromatin organization [26] and the expression of key developmental genes [24]. Importantly, many of these functions depend on 28 nt-long tRNAGlyGCC 5ʹ halves [24,26]. These fragments are only two nucleotides shorter than the most abundant tRNA-derived sequences that we have identified in the extracellular space [27] and later showed to possess dimerization capacity and high extracellular stability [28]. Considering EVs can mediate the transfer of these specific tRNA halves between human epithelial cells, we have hypothesized they could play a role in intercellular communication, especially during the stress response [29]. Alternatively, the release of tRNA-derived fragments to the extracellular space has been suggested as a way to derepress mRNA targets of these fragments, allowing T cells to transit towards an activated phenotype [30]. Whatever their function could be, tRNA-derived fragments are abundant in the extracellular space and have the potential to serve as useful disease biomarkers in liquid biopsies.
The scope of this review is to survey the different descriptions of extracellular tRNAs and tRNA-derived fragments, in different types of samples and associated to different extracellular RNA (exRNA) carriers. We expect to find consensus on which species are more representative of different extracellular fractions (EVs, lipoproteins, or ribonucleoprotein complexes) or highlight situations where this is still controversial.
The review is structured in different chapters which have specific aims and motivations. We will initially summarize descriptions of tRNAs and tRNA-derived fragments in blood serum or plasma or other biofluids (chapters two and three). The aims of these chapters are: (i) to identify which exRNA carriers and which subtypes of tRNA-derived fragments are more representative in human or mammalian biofluids and to (ii) stablish the biomarker potential of extracellular tRNA-derived fragments. Chapter four describes what is known about extracellular tRNAs and tRNA-derived fragments from studies performed in cell culture. The main advantage of these studies is that they tend to be insightful about the mechanisms used by cells to release different RNAs to the extracellular space. Chapter five addresses the question of the evolutionary conservation of extracellular tRNAs or tRNA-derived fragments. Finally, chapter six proposes a model to explain the presence of tRNAs and tRNA halves in different extracellular fractions based on studies discussed in the previous chapters and recent data from our group. We highlight several questions that we hope will stimulate hypothesis-driven research in this field and ultimately lead to a better understanding of the functionality of extracellular tRNAs and their fragments.
1.1. Nomenclature and working definitions
There is currently no universal consensus on the nomenclature of tRNA-derived fragments. In addition, efforts to standardize nomenclature are only recent in the exRNA field [31]. Therefore, we will define the basic concepts which will be used throughout this review.
There are two main types of EVs. Those which have an endosomal origin are termed ‘exosomes’ while vesicles generated by outward budding of the plasma membrane have received a variety of names including ‘ectosomes’ and ‘microvesicles’ (Fig. 1A). However, once released into the extracellular space we will refer to them collectively as ‘EVs’ because there is still no reliable method to infer their subcellular origin. Other relevant exRNA carriers include soluble ribonucleoprotein complexes (RNPs) and lipoprotein particles (LPPs). The latter being highly relevant in certain biofluids such as blood plasma or serum, where they can outnumber EVs (Fig. 1B).
Figure 1.
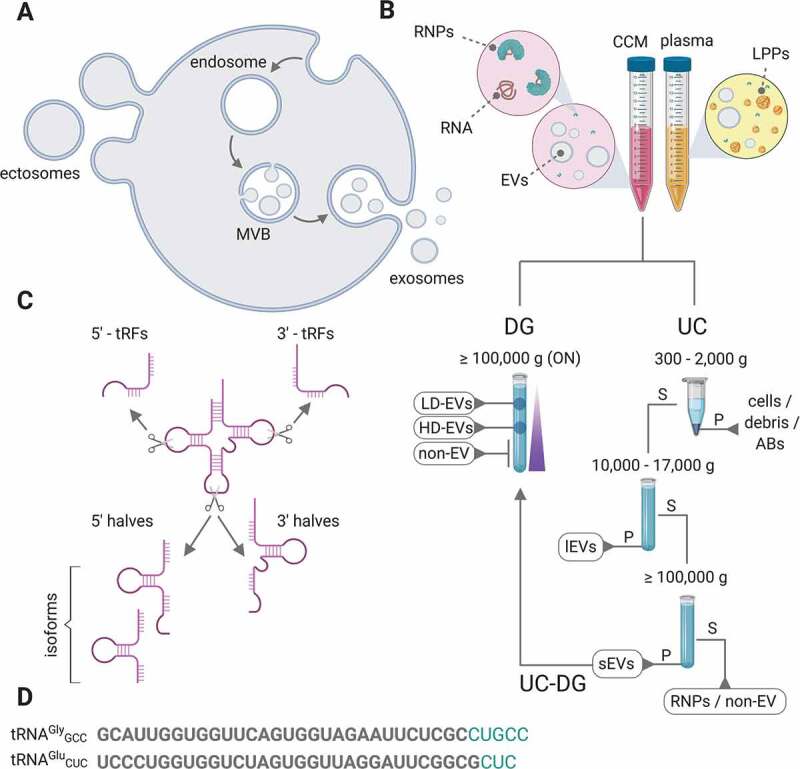
Nomenclature and purification strategies of extracellular RNA carriers and tRNA-derived fragments. (A) Diagram of a cell releasing ‘ectosomes’ (also called microvesicles or membrane-shed vesicles) to the extracellular space by outward budding of its plasma membrane. Alternatively, late endosomes can transform into multivesicular bodies (MVBs) by invagination of their membrane. If MVBs then fuse with the plasma membrane of the cell, they can release their intraluminal vesicles (ILVs) into the extracellular space. Once outside the cells, these ILVs are called ‘exosomes’. (B) Cell-conditioned medium (CCM) contains a collection of ectosomes and exosomes. Because their subcellular origin cannot be precisely inferred, they are called collectively ‘extracellular vesicles’ (EVs). Beyond EVs, CCM also contains extravesicular RNAs, mainly in the form of extracellular ribonucleoprotein particles (RNPs) although the presence of naked RNA cannot be discarded. Biofluids such as blood plasma or serum also contain lipoprotein particles (LPPs) of different densities and size. These LPPs have been shown to contain RNA. Purification of different exRNA carriers can be achieved by a variety of methods. Those most common are sequential ultracentrifugation (UC) and density gradient separation (DG), or a combination of both (UC-DG). Vesicular fractions obtained by UC are called ‘large EVs’ (lEVs, collected at medium speed) and ‘small EVs’ (sEVs, collected at high speed). Ultracentrifugation supernatants are considered as the extravesicular (non-EV) or RNP fraction. In DG, different populations of vesicles can be obtained depending on their buoyant densities (low density EVs, LD-EVs; high density EVs, HD-EVs). RNPs are usually present in the deeper (i.e., denser) fractions of the gradient. S: supernatant. P: pellet. (C) Nomenclature of tRNA-derived fragments. Short fragments (<26 nt) are called ‘tRFs’, while fragments generated by endonucleolytic cleavage at the anticodon loop are called 5ʹ and 3ʹ ‘tRNA halves’. We will extend this name to slightly shorter fragments, and consider sequences of 30 or 31 nucleotides as tRNA halves as long as they contain the ends of their parental tRNAs. (D) The two most abundant tRNA-derived sequences in the non-EV fraction of MCF-7 CCM [27] are shown in black. Extensions in green show the most abundant 5ʹ tRNA halves in the intracellular space of the same cells, sequenced in parallel.
The most frequently-used method to isolate EVs is differential or sequential ultracentrifugation (UC; Fig. 1B). When EVs are isolated based on this methodology, we will use the term ‘large EVs’ (lEVs) for those vesicles pelleted at intermediate speeds (10,000–20,000 × g). Conversely, vesicles obtained by high-speed (≥100,000 × g) centrifugation of samples previously devoid of lEVs will be considered ‘small EVs’ (sEVs), as suggested by Clotilde Théry and co-workers [32] and implemented in the most recent guidelines of the International Society for Extracellular Vesicles [31]. The concentrated supernatant of this high-speed centrifugation will be defined as the ‘non-EV’ or the RNP fraction. If EVs are purified based on their flotation in linear sucrose or iodixanol density gradients (DG), different subpopulations of EVs will be termed ‘low-density EVs’ (LD-EVs) or ‘high-density EVs’ (HD-EVs) (Fig. 1B). Other frequent purification techniques include size-exclusion chromatography, polymer-based precipitation and immunoaffinity capture.
Nomenclature of tRNA-derived fragments will be based in the following criteria: short fragments (<26 nt) starting or ending at the 5ʹ or the 3ʹ end of their parental tRNAs will be termed as 5ʹ-tRFs and 3ʹtRFs, respectively [33] (Fig. 1C). When these fragments are equal or longer than 30 nt but do not extend beyond the anticodon loop they will receive the name of 5ʹ and 3ʹ tRNA halves. Fragments starting and ending at internal positions of their parental tRNA will be considered internal tRFs or i-tRFs. There are other classes of tRNA-derived fragments which have been described in the literature [34], but this classification covers those sequences most frequently detected in the extracellular space and is therefore useful for the purpose of this work.
2. tRNAs and tRNA-derived fragments circulating in the bloodstream
Extracellular RNAs (exRNAs) need to be protected from degradation catalysed by extracellular RNases. This is achieved by their encapsulation or association with different exRNA carriers such as EVs, LPPs and RNPs. The RNP fraction remains to be the least studied exRNA carrier. Paradoxically, most extracellular tRNA-derived fragments circulating in blood serum seem to be associated with this fraction.
2.1. The extra-vesicular or ribonucleoprotein (RNP) fraction
Autoantibodies against transfer RNAs have been identified long ago in the sera of autoimmune patients [35,36]. This suggests that, at least under pathological conditions, tRNAs are capable of leaving the cells and interact with the immune system. Modified nucleotides derived from tRNAs have also been identified in biofluids, particularly in urine from normal and cancer-bearing monkeys [1]. However, the first report specifically aimed to the analysis of tRNA-derived fragments circulating in the bloodstream was published in 2013 by Joseph Dhahbi and co-workers [7]. The authors were looking for changes in the profiles of cell-free RNAs in mouse serum that could be associated to ageing and/or calorie restriction. In addition to microRNAs, they found an unexpected peak of sequences in the range of 30–33 nt where the vast majority (67%) mapped to tRNAs. Virtually all of them corresponded to the category of 5ʹ tRNA halves and were derived from tRNAGly (46%), tRNAVal (44%), tRNAGlu (8%) and tRNAHis (1%). The presence of some of these fragments was confirmed by Northern blot. In contrast, full-length tRNAs or 3ʹ tRNA halves were undetectable. Interestingly, the Northern blot signal remained in the supernatant after ultracentrifugation of the samples, suggesting that most 5ʹ tRNA halves in mouse serum are not associated to EVs. By sequential filtration, the authors estimated the MW of the macromolecular complexes containing these tRNA halves to be in the range 100–300 kDa. Similar results were reported shortly after in serum samples from healthy human volunteers [37].
The authors also observed that the Northern blot signals were weaker when treating serum with EDTA for 15 minutes before RNA extraction or when using EDTA-treated plasma. Conversely, when heparin was used as an anticoagulant the Northern blot signal was unmodified. Overall, these results suggest that 5ʹ tRNA halves are transported in the bloodstream of at least mice and humans as a complex which protects them from degradation, and this hypothetical macromolecular complex is dependent on the concentration of divalent cations.
To infer the origin of the tRNA halves detected in sera, the authors analysed different mouse organs and found robustly higher levels of glycine and valine 5ʹ tRNA halves in haematopoietic and lymphoid tissues [7]. However, it is still unknown whether serum tRNA halves are actually derived from haematopoietic cells.
An independent study later confirmed that glycine and glutamic acid 5ʹ tRNA halves are predominantly not associated to EVs in mouse serum [8]. There are two additional aspects of this study which deserve further attention. In first place, the authors showed that endogenous 5ʹ tRNA halves were stable in mouse serum when incubated at 37°C for at least two hours, while synthetic mimics were lost in as little as ten minutes. A second assay which deserves consideration is the observation that both tRNAGly and tRNAGlu 5ʹ halves were highly upregulated (20 and 40-fold, respectively) two days after injection of lipopolysaccharides (LPS) to mice, with levels returning to baseline at day four. This demonstrates that acute inflammation alters the levels of tRNA-derived fragments in the circulation. Consequently, biomarker discovery based on the profiling of circulating tRNA halves can be affected by disease- or treatment-associated inflammation. These aspects will be discussed in chapter 3.
Studies performed in serum from cattle [38], rats and monkeys [8] also confirmed that 5ʹ halves from tRNAGly, tRNAGlu, tRNAVal and tRNAHis are the most abundant circulating tRNA-derived sequences. In human serum, circulating tRNA-derived fragments are even more biased towards 5ʹ halves from tRNAGly and tRNAGlu [39].
The effect of EDTA in circulating tRNA halves was confirmed in humans by Thomas Tuschl’s group [40]. A profiling of circulating Trizol-purified small RNAs in heart failure showed that tRNA-derived fragments comprised 28% of sequencing reads in serum, while they were only 0.6% (47-fold decrease) in EDTA-treated plasma. Similar results were later obtained in healthy subjects [41,42]
Other studies still reported a substantial amount of reads mapping to tRNAs (23%) in EDTA-treated plasma obtained from human subjects [43]. However, the size distribution of the sequences aligning to tRNAs was not reported. Therefore, we do not know whether these sequences represent tRNA halves or shorter tRFs.
2.2. The extracellular vesicle (EV)-associated fraction
In an early report, tRNA-derived fragments represented only a small proportion (1.24%) of the reads in EVs purified from human plasma by a precipitation-based kit [44]. Similar results were obtained in a more recent study performed in a large cohort of healthy individuals and cancer patients [45]. This strongly contrasts with the high proportion of tRNA-derived fragments which other authors have found in total serum. Although a direct quantitative comparison is not possible across studies, this suggests that most tRNA-derived fragments circulating in the bloodstream are not found inside EVs. This is further supported by the low percentage of tRNA-derived fragments found in ultracentrifugation pellets of pooled plasma from healthy human subjects (< 0.5% of mapped reads) [46].
Recently, 5ʹ tRNA halves have been identified in preparations of syncytiotrophoblast-derived extracellular vesicles purified by UC from placental perfusions [47]. Strikingly, >95% of small RNA sequences corresponded to tRNA-derived species in this study. In a different study where EVs were obtained from blood plasma also by UC [48], rRNA-derived sequences accounted for the majority of sequencing reads (73%). However, tRNA-derived fragments were well represented in the remaining set of sequences. A nuance of this study is that the purified RNA was fragmented with RNase III (dsRNA endonuclease) before library preparation. Consequently, the high representation of rRNA and tRNA-derived fragments in this study could correspond to lab-derived fragmentation of full-length rRNAs and tRNAs present in the EVs (see chapter 4.4).
In summary, EVs do not seem to carry relevant amounts of tRNA-derived fragments in human blood plasma or serum as opposed to the RNP fraction which is highly enriched in 5ʹ tRNA halves.
2.3. The plasma lipoprotein (LPP) fraction
High density lipoproteins (HDL) were reported as carriers of microRNAs in human plasma [49]. To address whether LPPs could also carry additional exRNAs families, the authors later purified HDL and Apolipoprotein B-containing lipoproteins (low and very low density LPPs) from EDTA-collected mouse plasma by size-exclusion chromatography [50]. Small RNA-seq on total RNA was performed on LPPs, unfractionated biofluids (bile and urine) and liver tissue. Surprisingly, less than 5% of quality sequencing reads in LPPs were microRNAs. A high proportion of small RNAs mapped to the murine genome in both HDL and low density LPPs were 5ʹ tRNA halves. The most prevalent fragments were derived from tRNAGluCUC, tRNAGlyGCC, tRNAAspGUC and tRNAValCAC.
2.4. Are exogenous RNAs transported in lipoproteins?
One of the most surprising observations of the work which has just been described [50] is the high proportion of sequencing reads which the authors could not annotate as endogenous transcripts in LPP samples (approximately 75%). For instance, host microRNAs represented approximately 5% and 2% of total reads in HDL and APOB-LPPs, respectively. In contrast, exogenous tRNA-derived fragments could represent up to 5% and 7% of total reads in certain LPP samples. More strikingly, LPP samples had exogenous rRNA-derived fragments representing above 25% of total reads. These non-host sequences could be perfectly aligned to bacterial genomes in the majority of cases. Based predominantly on these results, the authors concluded that lipoproteins transport exogenous RNAs in the mammalian bloodstream.
The presence of exogenous RNAs in mammalian biofluids is an open controversy, especially in the case of exogenous RNAs that are presumably acquired from the diet [51–53]. In 2012, a widely-commented paper reported the presence of several plant microRNAs in human plasma [54], but the identification of the same plant-specific microRNAs in small RNA datasets of benthic filter feeding chordates made us somehow sceptical of this finding [55]. Other authors have shown that human plasma contains a variety of exogenous RNAs from bacterial and fungal origin (including many tRNA-derived sequences) which could represent a supply of exogenous RNAs from the gut microbiota [56].
It is important to mention that the authors of the LPP-RNA study performed a series of measurements directed to assess the extent of microbial contamination in their plasma samples [50]. However, it is known that the impact of environmental contaminants is inversely proportional to the amount of RNA in a sample [57,58], and the quantities of extracellular RNAs purified from biofluids are usually low. The fact that the percentage of reads not mapping to the host genome followed the trend: tissue < biofluids ⋘ purified LPPs [50] can be considered a consequence of this principle. In the most extreme scenario, sequencing of a purified particle known to lack any associated RNAs will only give a breadth of the environmental contaminants present in the sample and the labware used for its purification. By saying this we are not denying the possibility that LPPs could transport exogenous small RNAs in the bloodstream. However, the study of exogenous sequences in situations where minute amounts of endogenous nucleic acids are obtained demands cautionary interpretation.
2.5. The overlap between circulating piRNAs and ncRNA fragments
Small RNA sequencing in a large cohort of citrate-treated plasma identified 144 small RNAs that mapped to Piwi–interacting RNAs (piRNAs) [59], a small RNA family important for transposon silencing in the germline. Furthermore, the authors evaluated these sequences by RT-qPCR and showed that they were not affected by sodium periodate treatment, suggesting that they were 2ʹ-o-methylated at their 3ʹ end as is characteristic of piRNAs. Other authors also found piRNAs in saliva [60], in human serum [61], and in EVs purified from cells [62] or plasma [45], where they accounted for 40% of mapped reads compared to 2.1% of sequences aligning to tRNAs. This was a striking discovery because piRNA expression in mammals is usually restricted to the germline or germline-associated cells [63], although arthropods [64] and molluscs [65] are known to possess somatic piRNA-based regulatory networks.
When piRNAs were first described, fragments cloned with a median size of 29–30 nt in mouse testes were annotated as potential piRNAs as long as they did not match to other known ncRNAs [63]. However, some ncRNA fragments were not filtered out and they were later included in piRNA databases. We have shown that, while the number of ncRNA fragments in piRNA databases is rather small (< 1%), the most abundant piRNAs identified in plasma belong to this group and are therefore probably not bona fide piRNAs [66]. By filtering out sequences which also mapped to ncRNAs, other authors have shown that the number of putative piRNA sequences in human adult plasma dropped from 13% of total reads to < 0.1% [67]. Some tRNA-derived fragments have been shown to co-IP with PIWI proteins in arthropod cells, and were therefore termed tRNA-derived piRNAs [68]. However, it is still not clear whether putative plasma or serum piRNAs overlapping ncRNA fragments are transported in association to a PIWI-clade protein, as has been shown in Tetrahymena [69]. Until this evidence is available, we suggest referring to these sequences as ncRNA fragments rather than somatic piRNAs.
2.6. The ratio between tRNAs and their fragments depends on sequencing techniques
Alan Lambowitz’s group has developed sequencing library preparation methods which rely on the reverse transcriptase encoded by mobile group II introns (bacterial retrotransposons) instead of retroviruses [70]. This method has been useful for tRNA sequencing in human plasma [71] and cell-culture purified EVs [72]. The RTs from group II introns encoded in the genomes of thermophilic bacteria (termed TGIRTs) have a higher thermostability, processivity and fidelity than retroviral RTs, and can attach sequencing adapters and primers by a template-switching reaction which is independent of RNA ligation, a step which introduces profound biases in small RNA-seq data [73,74].
The capacity of TGIRT enzymes of performing the RT reaction at higher temperatures (60°C) and their tendency to read-through modified bases [71] which would otherwise induce the abortion of conventional retroviral RTs [75,76] are desirable features when sequencing highly-structured and hypermodified tRNAs. Additionally, the authors in [71] compared TGIRT sequencing libraries performed in untreated and T4 PNK-treated plasma. The T4 PNK enzyme removes the 3ʹ phosphate or terminal 2ʹ,3ʹ cyclic phosphates which results from the cleavage by Angiogenin or other members of the RNase A family, enhancing detection of 5ʹ tRNA halves [15]. Consistent with the development of an optimized method for tRNA detection, 83 – 93% of small RNA sequencing reads in human plasma aligned to tRNAs [71]. More importantly, most of these reads corresponded to full-length tRNAs, and sequences included the post-transcriptionally added 3ʹ CCA and 5ʹ G (in tRNAHis). Removal of the 3ʹ (cyclic) phosphate increased the representation of 5ʹ tRNA halves from 0.4% of the reads mapped to tRNAs to 7.1%. However, since the authors used EDTA-treated plasma, it is likely that many 5ʹ tRNA halves were lost [7,37]. Thus, a quantitative estimation of the actual ratio between full-length tRNAs and their fragments circulating in the bloodstream is still not possible. Previous work using Northern blot did not show any evident band for full-length tRNAs in mouse serum or in heparin-treated plasma, despite 5ʹ tRNA halves were highly abundant based on the same technique [7]. Thus, there are still some inconsistencies in the literature that future work will need to solve.
2.7. tRNA-derived fragments in other biofluids
Purification of extracellular vesicles from seminal plasma (by UC-DG) showed that semen EVs contain tRNA-derived fragments including 5ʹ tRNA halves and 5ʹ tRFs derived from tRNAGly and tRNAAla, respectively [77]. More recently, EVs released from cells in the epididymis (epididymosomes) were shown to contain glycine tRNA halves, and these fragments were transferred first to sperm and then to the oocytes during fertilization in mice, where they were linked to epigenetic inheritance of a paternally-acquired metabolic disorder [23–25].
Several recent reports have analysed a variety of human biofluids. In a first study published in 2017, healthy male college athletes provided multiple plasma, urine and saliva samples [78]. One obvious difference between biofluids was the high percentage of reads in saliva which mapped to bacterial species instead of the human genome. This was further confirmed in another study performed in 145 human samples spanning 12 biofluid types [67]. Urine and bile were the biofluids with the highest proportion of tRNA-derived fragments. After removing rRNA reads and aligning the rest to the human transcriptome, tRNA-derived fragments accounted for > 90% of aligned reads in these samples. They were followed by seminal plasma and amniotic fluid. Cerebrospinal fluid (CSF), saliva, serum and ovarian follicle fluid contained comparable amounts of tRNA-derived fragments, in the range of 25–50% of transcriptome-aligned reads. Cord blood and adult blood plasma contained negligible amounts of tRNA-derived fragments, consistent with the demonstrated loss of tRNA halves in the presence of chelating agents [7]. It is still unclear whether urine and bile contain higher amounts of tRNA-derived fragments per unit of volume, or whether their higher proportion is a consequence of very low levels of microRNAs and other RNA biotypes in these biofluids. Sequencing does not inform about absolute concentrations.
A recent study was aimed to identify optimal RNA purification methods for the enrichment of specific exRNA carriers in biofluids [39]. Interestingly, the authors compared methods like ultracentrifugation or affinity purification of EVs with total RNA extraction without pre-purification steps. There was no obvious effect between methods aimed to pre-enrich EVs or LPPs and those intended for total RNA isolation in their capacity to capture tRNA-derived fragments. However, based on clustering analysis, the authors inferred that some biofluids such as serum could contain tRNA-derived fragments in more than one carrier (e.g., EVs, LPP or soluble RNPs). This study also showed that most tRNA-derived sequences were 30–34 nt 5ʹ tRNA halves derived from tRNAGly and tRNAGlu, irrespectively of the biofluid type. Based on the nomenclature and definitions which were presented in Chapter 1, we can state that tRFs are far less represented than tRNA halves in human biofluids.
An analysis of rat mesenteric lymph showed very similar results [79]. There, >90% of reads were tRNA-derived fragments, most of them being 5ʹ tRNA halves derived from tRNAGlyGCC. It is remarkable that whatever biofluid seems to be chosen, tRNA-derived fragments are a relevant population among small RNAs, and glycine and glutamic acid 5ʹ tRNA halves constitute the majority of all tRNA-derived sequences. However, it is still not clear how these RNAs are internally distributed between EVs, LPPs and RNPs in each bio fluid type.
3. Biomarker potential of circulating tRNA halves and tRFs
Early attempts to establish the biomarker potential of circulating tRNA-derived fragments were performed by the Dhahbi’s group [80]. After finding some differences in the levels of circulating tRNA-derived fragments in a cohort of five heterogeneous breast cancer patients and an equal number of control subjects, the authors reanalysed small RNA sequencing data present in public repositories. The reanalysed dataset [81] is compelling because it contains serum samples from 42 female stage II–III locally advanced and inflammatory breast cancer patients before they received treatment. Because it does not provide information on healthy subjects, reanalysis performed in [80] looked for differences between groups of breast cancer patients. For some fragments including those from tRNAValCAC, there was a fold-change of roughly 20 in those patients which later relapsed (n = 10) in comparison with those who did not (n = 31). However, most of the variation was ascribable to only two patients in the relapsed group where the levels of circulating 5ʹ tRNA halves were abnormally high.
A later study performed in the sera of seven male patients with oral cancer and seven healthy subjects also showed variations in certain tRNA halves between the control and the study groups [82]. Although some of the differences between groups were highly significant, inter-individual variation was not shown. Additionally, cancer and control patients were enrolled in two different institutions from two different continents, so it is not clear whether these differences are consequential to the cancer status or other confounding variables.
A study performed in clear cell renal cell carcinoma (ccRCC) identified 32 differentially expressed tRNA-derived fragments [83]. Among them, tRNAValACC 5ʹ halves were highly and significantly reduced in cancer tissue, and this was further validated by using an independent cohort. When their levels were compared in normal and patient derived sera they were highly similar and the difference in the median of both groups was not significant. This negative result place a cautionary note in expecting that differences observed between biopsies of solid tumours and normal tissue will be necessarily reflected in the profiles of circulating small RNAs. However, a later report from the same group analysed the levels of other four 5ʹ tRNA halves downregulated in ccRCC tissue and reported that their levels in serum were also significantly lower than in control subjects [84].
How should these results be interpreted? If a renal tumour overexpresses an RNA which is normally not found in blood serum, then it is feasible to identify higher levels of this hypothetical RNA circulating in the bloodstream. However, downregulated RNAs are certainly not expected to induce a significant change in circulating RNA profiles, unless assumed that kidney cells are major contributors to the circulating small RNAome under physiological conditions.
The same group has recently reported that tRNA halves are prognostic biomarkers for patients with prostate cancer [85]. By segregating patients above and below a cut-off determined for each tRNA half, the authors showed that high levels of some tRNA halves in cancer tissue biopsies were associated with a shorter recurrence-free survival period. The authors then looked for the levels of these tRNA halves in the sera (obtained pre-operative) of some of these patients, but they did not find a significant correlation between levels in serum and in cancer tissue. However, when comparing with additional serum samples obtained from patients with metastatic, castration-resistant prostate cancer, they found that tRNAGluCUC 5ʹ halves were significantly increased in metastatic patients. For an adequate interpretation of these results, it should be considered that sera from primary prostate cancer patients was obtained pre-operative and probably before pharmacological interventions, while metastatic patients had received various therapeutic interventions at the time of blood collection.
The assumption that RNA profiles in the circulation would mirror their expression in solid tumours is not infrequent. Several reports have performed differential expression analysis between relevant cells lines and then looked for some of the differentially expressed tRNA-derived fragments in the sera of cancer patients [86,87]. The differences observed between cell lines and between cancer patients and control subjects showed poor correlation [86].
Perhaps more promising, circulating tRNA-derived fragments could be used for monitoring the advancement of a disease rather than for diagnostic purposes. For instance, tRNALeuCAG 5ʹ halves were significantly upregulated in non-small cell lung cancer in comparison with normal tissue [88]. The authors then looked for the expression of this specific fragment in the sera of normal and classified stage I–IV patients. Their RT-qPCR analysis showed that stage III and IV patients had significantly higher levels of this tRNA half in their sera.
Circulating tRNA-derived fragments could also be used to predict the benefit of targeted oncological therapies such as those using monoclonal antibodies. In a recent study, blood was collected from women with HER-2 positive breast cancer before treatment with trastuzumab [89]. Levels of selected tRNA-derived fragments based on previous results were then assayed in EDTA-treated plasma and compared between patients which later responded to the therapy or showed primary resistance. Levels of two fragments classified as i-tRF and derived from tRNACysGCA were significantly different between sensitive and resistant patients. While EDTA impairs detection of circulating non-vesicular tRNA halves-containing complexes [7], its effects on shorter tRFs such as i-tRFs is unclear.
Beyond applications in oncology, circulating tRNA-derived fragments might be informative in patients with mental illness. In a recent study, small RNA-seq was performed on pooled plasma from control subjects and pre-seizure or post-seizure samples from patients with drug-refractory focal epilepsy [90]. Most of the tRNA-derived reads corresponded to 24–26 nt 5ʹ-tRFs. We attribute the low representation of tRNA halves to the fact that plasma was used instead of serum. The authors then selected three tRFs based on their fold-change between pre-seizure and control individuals and measured them by RT-qPCR in an expanded cohort. Consistent with sequencing results, the levels of the three 5ʹ-tRFs were higher in pre-seizure than in control samples and were also significantly lower in post-seizure than in matched pre-seizure sera. Thus, it is tempting to speculate that a rise in the circulating levels of these tRFs precedes seizure events in epileptic patients. However, the authors point out at an important cofounder effect in the interpretation of their data. Because patient’s medication was withdrawn in most cases before arrival at the epilepsy monitoring unit, post-seizure samples were taken with predictably lower levels of the medication in the patient’s bloodstream in comparison with the pre-seizure samples. Methodological details like this are very important for an adequate interpretation of results but not always disclosed.
Profiling of tRNA-derived fragments in sera is also being used with the aim of applying it to biomarker analysis in veterinary medicine [91,92].
In the biomarker field, most studies typically compare total unfractionated plasma or serum between patients and control subjects irrespectively of the exRNA carrier. However, some reports have tried EV purification before comparing tRNA-derived fragment levels between cancer patients and healthy controls [93].
As has been attempted multiple times in serum, tRNA halves are also being studied as potential biomarkers in other biofluids such as seminal plasma [94]. Urine is also an attractive biofluid because its collection is not invasive. A comparison of RNA contents in EVs obtained by ultracentrifugation of urine from chronic kidney disease patients and healthy subjects showed a significant reduction of 5ʹ halves from valine and leucine tRNAs in the patients [95].
In summary, tRNA-derived fragments can be found in the bloodstream and their levels can change in response to stimuli. For this reason, it is not surprising that many studies are trying to position extracellular tRNA-derived fragments as promising biomarkers for the liquid biopsy era that is under development. A recent report has suggested that many ncRNAs, including mitochondrial tRNA-derived fragments, can be sequenced from a single droplet of human serum and their profiling enables almost perfect separation between breast cancer patients and normal donors [96]. However, knowledge on the basic variables influencing the extracellular levels of tRNA-derived fragments is still limited. For instance, it has been shown that serological levels of some tRNA halves peak during acute inflammation [8]. Thus, it is not clear whether changes in the concentration of specific tRNA-derived fragments are going to be necessarily specific for a single pathology. Additionally, differences between patients and normal donors can be a consequence of the treatment rather than the disease, especially when the patient group has been exposed to cytotoxic drugs such as those used in oncology. The same concerns apply for microRNA-based biomarker discovery [97], but tRNAs have the additional drawback that they usually do not have tissue-specific expression. This being said, circulating levels of tRNA-derived fragments may be useful as a surrogate measurement of disease progression even if they do not have enough specificity to work as diagnostic molecules.
4. Cell-conditioned medium
Extracellular RNA profiling in biofluids is the best alternative for biomarker discovery. However, exRNA analysis in cell culture media has the advantage that RNA profiling can be done simultaneously in the extracellular and intracellular spaces. Biofluids contain a mixed population of RNAs released from different cell types at different rates, and it is still not clear which cell types contribute more to reference extracellular profiles. In contrast, the origin of exRNAs is more controlled when working with isolated cells and therefore exRNA analysis in cell-conditioned medium is usually the choice when interrogating RNA secretion mechanisms and exRNA dynamics. However, it remains to be determined how representative are the mechanisms derived from cell culture of the situation occurring in vivo.
4.1. Serum and culture media-derived exRNA contaminants
Foetal bovine serum (FBS) is a growth factor-rich, widely used additive in mammalian cell culture. Since FBS is known to contain EVs, the extracellular RNA field has long relied on the use of the so-called ‘vesicle-depleted serum’ for sustaining growth of mammalian cell lines. Vesicle-depleted serum is typically a term used to call the supernatant obtained after performing ultracentrifugation of FBS under sterile conditions and for long periods of time. However, the way this method is performed impacts in the amount of EVs that are left in the supernatant and even prolonged centrifugation does not deplete all bovine RNAs, which are also present outside EVs as soluble RNPs [98,99]. Thus, ‘EV-depleted serum’ contains lower levels of EV-associated RNAs than FBS but it is still a source of contaminating sequences that can greatly affect the interpretation of results in exRNA studies [99,100].
This can be even more problematic when studying tRNA-derived fragments. The reason is simple: (a) tRNAs tend to be highly conserved between humans and cows and (b) most tRNA-derived fragments in bovine sera do not pellet at 100,000 × g [7] and are therefore expected to remain in ‘vesicle-depleted serum’. Attempts to further reduce the amounts of EVs by methods alternative to ultracentrifugation have reduced total microRNAs 100-fold but were not efficient in reducing sequencing reads which aligned to bovine tRNAs [101].
Moving towards serum-free media does not completely circumvent the problem because some growth factors used in the formulation of ‘chemically defined’ media also contain RNAs which can strongly affect results [102]. Correcting for background levels of exRNAs obtained from non-cell-conditioned-media is an important control that should be performed routinely.
4.2. EV-associated tRNA-derived fragments
The first description of tRNA-derived fragments (and pieces of other ncRNAs) in EVs obtained from mammalian cell culture was published by Esther Nolte-t’ Hoen and co-workers in 2012 [103]. Sequencing libraries were size-selected in order to exclude full-length tRNAs. Small RNA analysis was performed in parallel in the EVs and in the cells, allowing the authors to look for small RNAs which were preferentially packed in the vesicles. They found that, while both 5ʹ and 3ʹ fragments were abundant at the cellular level, only 5ʹ tRNA-derived fragments were prominent in EVs. Also, a subpopulation of 5ʹ tRNA-derived fragments starting at position 5–10 of the tRNA and extending beyond the anticodon and up to position 50 was only observed in the vesicles.
In 2015, the group of Michiel Pegtel isolated EVs from bone marrow- and adipose tissue-derived human mesenchymal stem cells (BM-MSC and A-MSC, respectively) and analysed their small RNA content as well as in their parental cells [104]. The authors pretreated their vesicular preparations with RNase A in order to degrade unprotected RNA, before RNA purification with the TRIzol reagent. Interestingly, while tRNA-derived fragments represented only 5–11% of the entire cellular small RNA pool, they represented > 50% of total small RNAs in A-MSC-EVs and 23–35% in BM-MSC-EVs. Adipose MSC EVs contained mostly tRNAGlyGCC 5ʹ halves, which were only 5% of all tRNA reads in the cells. The 5ʹ halves from tRNAGluCUC were also frequently detected in both cells and EVs, but one subpopulation of BM-MSC-EVs also contained a substantial fraction of 70–75 nt reads which corresponded to the full-length glutamic acid tRNA. To the best of our knowledge, this is the first report describing the simultaneous detection of tRNAs and their fragments in extracellular vesicles.
Another comparison of cells (MCF-7 and MCF-10A) and different vesicular populations was performed by our group in 2015 [27]. Two nuances in the design of this study were: (a) the use of serum-free media to avoid serum-derived contaminants, as was described in chapter 4.1 and (b) the inclusion and analysis of the extracellular RNP fraction (i.e., 100,000 × g supernatants; Fig. 1B) alongside EV preparations. Even though tRNA-derived sequences were not uncommon in our EVs and were biased towards 5ʹ tRNA halves, their relative abundance was much lower than in the extra-vesicular fraction. This observation will be addressed in chapter 4.3. Nevertheless, it is important to mention that the 100,000 × g supernatants were highly enriched in 5ʹ halves from tRNAGlyGCC and tRNAGluCUC. Furthermore, the total RNA content of this non-EV or RNP fraction was higher than the RNA content in EVs purified from the same media. Therefore, we can conclude that the non-EV fraction contains higher amounts of tRNA-derived fragments than the EVs (in absolute numbers, not just in terms of relative representation).
Another compositional classification at the level of small RNAs between cells, large and small EVs and the non-EV fraction was performed by the group of Anna Krichevsky in primary cultures of glioblastoma [105]. A sequential filtration-based protocol was used to obtain the different extracellular fractions. Interestingly, the non-EV fraction showed the highest enrichment in tRNA-derived fragments, in agreement with our previous report [27]. Again, these sequences mostly corresponded to 5ʹ halves from tRNAGlyGCC and tRNAGluCUC [105]. In contrast to cells, the relative abundance of these RNAs was much lower in the non-EV fraction than in EVs when cerebrospinal fluid was used as a source of exRNAs.
Are extracellular tRNA-derived fragments responsive to changes in their intracellular levels induced by stimuli? This question was addressed in a recent report by Nolte-‘t Hoen’s group [106]. In this work, immune activating (LPS) or suppressing stimuli (1α,25-dihydroxyvitamin D3) was imposed on primary dendritic cells, and the RNA content in EVs was compared between conditions. Interestingly, stimulation of DC with any of the compounds did not lead to major changes in the distribution of sequencing reads across different RNA classes in cells and EVs. The more abundant tRNA-derived fragments in both LPS-EVs and Vit D3-EVs did not change either.
Another interesting study was performed in a model of T cell activation [30]. Here, small EVs were obtained from the media of activated T cells by UC-DG (Fig. 1B). By studying the sensitivities of the small RNAs to RNase alone or to combined detergent plus RNase treatment, the authors concluded that the RNAs in the low density (LD) fractions were protected inside EVs, while the high density (HD) fraction probably corresponded to RNPs. Interestingly, the LD fraction was enriched in tRNA-derived fragments when compared to cells, while the HD fraction was not. However, these tRNA-derived fragments were mainly short 5ʹ tRFs (18–21 nt, Fig. 1C) rather than the 5ʹ tRNA halves (30–31 nt) predominantly found in the non-EV fraction in other studies [27,105]. Interestingly, the extracellular enrichment in 5ʹ tRFs was less pronounced in EVs from resting T cells than in EVs from activated T cells. Based on this and other findings, the authors proposed a model where activated T cells selectively release those 5ʹ tRFs which would inhibit their activation if they remained inside the cells.
Methodological details can explain apparent disparities between different reports. We [27] as well as others [105] have found that, while all extracellular fractions are enriched in tRNA-derived fragments compared to parental cells, this enrichment is much higher in the non-EV or RNP fraction. In contrast, LD-EVs from activated T cells contain higher relative levels of tRNA-derived fragments than HD aggregates [30]. However, this high-density aggregates were pelleted at 100,000 × g before sucrose gradient separation, meaning that they correspond to large, high-density particles which cannot be compared to the rather small particles which do not pellet at 100,000 × g [27] or that cross 0.02 µm filters [105].
In summary, many reports have shown that EVs are enriched in sequences aligning to tRNAs compared to their parental cells, while microRNAs usually show the opposite trend [27,103–105,107–110]. The universe of EV-enriched RNAs also extends to fragments of tRNAs encoded in the mitochondrial genome [111]. However, tRNA-derived fragments are enriched to much higher levels in the non-EV or RNP fraction, where tRNA halves comprise the majority of the RNAs amenable to standard small RNA sequencing procedures [27,105]. This being comparable to the situation in sera [7,8,37].
4.3. EV-mediated transfer of tRNA-derived fragments between cells
Two papers showing that sperm cells from mice can transmit an acquired metabolic disorder to their offspring through tRNA-derived fragments were published in 2016 [23,24]. Zygotic injection of a mixture of purified tRNA-derived fragments [23] or synthetic tRNAGlyGCC 5ʹ halves [24] induced profound effects on zygotic gene expression, demonstrating the bioactivity of tRNA-derived fragments. Interestingly, these small RNAs were acquired by sperm from EVs released from the epididymis (epididymosomes) during post-testicular sperm maturation [24,25]. The model proposed in this work is extremely interesting and provocative and highlights a biologically relevant function for extracellular tRNA halves and/or tRFs. In brief, paternal diet would alter tRNA fragmentation in the epididymal epithelium, resulting in epididymosomes enriched in some tRNA-derived fragments which could then be transferred to sperm and subsequently to embryos.
Published in this issue of RNA Biology, we have shown that cells transfected with synthetic tRNAGlyGCC 5ʹ halves can release these RNAs in EVs and, subsequently, deliver them to other cells [29]. Implications in intercellular communication will be discussed in chapter six.
4.4. EVs contain more full-length tRNAs than tRNA-derived fragments
The group of Jan Lötvall, who helped develop interest into the exRNA field by describing the functional delivery of microRNAs and mRNAs to recipient cells via EVs [112], performed an interesting study in which cells and different types of EVs (small, large and very large EVs purified by UC, Fig. 1B) were directly compared [113]. The presence of full-length 18 S and 28 S rRNAs and full-length tRNAs could be inferred from electrophoretic profiles in cells, very large EVs and large EVs, but not in small EVs. However, later work from the same group showed the presence of full-length tRNAs also in small EVs [114]. A more robust purification protocol (UC-DG) was used to separate two different exRNA carriers according to their density, using 120,000 × g pellets of cell-conditioned media as input before sucrose gradient-based separation [115]. In contrast to their previous studies [113,114] full-length tRNAs and rRNAs were now clearly evident in low density, small EVs (LD-sEVs) [115], but not in the high density fraction. Interestingly, there was a shift in the main tRNA-derived fragments between both exRNA subpopulations. While the HD fraction contained mostly tRNAGlyGCC 5ʹ halves, the LD-EVs were more enriched in 5ʹ halves from glutamic acid, as in cells. When compared to cells, both exRNA fractions contained less sequences assigned to the 3ʹ region of their parental tRNAs.
Regarding the HD aggregates purified by UC-DG in [115], they can correspond to a mixture of high density EVs and large aggregates of ribonucleoprotein particles that are co-isolated during ultracentrifugation. By now, it is well stablished that subpopulations of EVs with different densities can be isolated from cell culture media [32,116]. However, the density of the HD fraction in [115] is more consistent with RNPs [117]
In summary, full-length tRNAs as well as 5ʹ tRNA halves can be identified in low density, small EVs, but only 5ʹ tRNA halves (mainly Gly) are found in high-density RNP aggregates, in agreement with our analysis based solely on UC [27]. The presence of full-length tRNAs and rRNAs in LD-sEVs purified by UC-DG was confirmed by other groups [30,117]. What these reports cannot tell is the relative proportion between full-length tRNAs and their fragments in purified EVs.
We have previously discussed how the use of a thermostable group II intron reverse transcriptase changed the extracellular RNA profile of human plasma, greatly increasing the proportion of reads represented by full-length tRNAs [71]. In a collaboration with the group of Randy Schekman, this enzyme was later applied to the characterization of EVs derived from cell culture [72]. Similar to plasma, full-length tRNAs were abundantly detected in EVs purified by DG. There was also a strong correlation between intracellular and EV-associated tRNAs, meaning that EVs are preferentially loaded with those tRNAs which are highly abundant inside cells. Importantly, these RNAs were resistant to RNase A treatment but they were sensitive to combined detergent and RNase A treatment, suggesting that they are present inside EVs and not simply co-purifying with them. Some additional findings of this work are remarkable. First, the authors treated RNA with T4 polynucleotide kinase (PNK) in order to remove 3ʹ phosphates and increase the probability of sequencing tRNA halves, which bare this modification [15]. Because no differences were found with or without T4 PNK, the authors concluded that HEK293-derived EVs contain full-length tRNAs, while their fragments were not abundant in EVs [72]. Second, the authors measured a significant reduction of tRNA levels in EVs derived from Y-box-binding protein 1 (YBX1)-null cells, with a concomitant upregulation inside the cells. Thus, they suggested that YBX1 is responsible for sorting ncRNAs to EVs, as they previously showed for microRNAs [118]. Third, the authors identified an RT stop present in the D-loop of certain tRNAs, and this RT stop (presumably corresponding to a post-transcriptional tRNA modification incompatible with group II intron RTs) was more abundant in EVs than in cells [72,116].
Others have shown that cell culture-derived EVs contain more full-length tRNAs than tRNA halves by Northern blot (a technique which is expected to give a better picture of tRNA-to-tRF ratios when compared to sequencing) [95]. However, the authors of this study purified EVs by ultracentrifugation, which results in less pure preparations compared to the density gradient method used in [72]. We have combined density gradient separation with non-radioactive digoxigenin-based Northern blot, and found that, indeed, EVs from non-stressed cells contain mostly full-length tRNAs instead of tRNA-derived fragments [119]. We will resume this discussion in chapter six.
It remains unknown whether tRNAs are forming complexes with other proteins inside EVs. Proteomic studies have identified high levels of rRNA-, poly(A)- and tRNA-binding proteins in EVs [120,121], but for the moment this information indicates just presence and not direct physical association.
4.5. Stability as a key factor influencing extra-vesicular abundance of tRNA halves
There is evidence that most tRNA-derived fragments circulating in blood serum are not present inside EVs because they do not pellet at 100,000 × g [7,8,80] and are not retained by 20 nm filters [105]. If they are not encapsulated in EVs, how are they stabilized and protected from the action of extracellular RNases?
Our work with breast epithelial cell lines [27,28] can help to answer this question. In MCF-7 cell lysates, tRNA-derived fragments were only a small proportion of reads mapped to the human genome (2.2%), without any evident accumulation of specific tRNA-derived sequences [27]. In contrast, they accounted for roughly 60% of mapped reads in 100,000 × g supernatants of cell-conditioned medium. Close to 90% of these sequences were 5ʹ tRNA halves derived from tRNAGlyGCC and tRNAGluCUC. Their size distribution was restricted to 30 or 31 nt, while longer and shorter fragments could be sequenced inside cells. To confirm that extracellular 5ʹ tRNA halves were EV-free, we injected concentrated 100,000 × g supernatants of MCF-7 cell-conditioned media in a size exclusion chromatography column and observed the elution of two RNA-containing peaks. We initially thought that the larger of them corresponded to a complex between tRNA halves and an associated RNA-binding protein. Surprisingly, we later found that it consisted of RNAs eluting as dimers, explaining the unexpected elution profile of these tRNA halves [28].
The capacity of forming homodimers seems to be a property unique to glycine 5ʹ tRNA halves of 30 or 31 nt [28]. We also predicted that these dimeric structures should be highly resistant to both endo- and 3ʹ-5ʹ exonucleases. Structural predictions were then validated with synthetic RNAs, in which we introduced mutations to destabilize the predicted dimer interface. Biotinylated RNAs capable of forming these hybrids had a longer half-life when transfected in MCF-7 cells in comparison with RNAs where the dimerization interface was disrupted or when using scrambled oligonucleotides. Finally, we analysed cell lysates by size exclusion chromatography and found that endogenous tRNA halves could also form supramolecular aggregates inside cells. This is remarkable because other 5ʹ tRNA halves, such as those derived from alanine and cysteine, have been shown by Pavel Ivanov’s group to form very stable tetramers stabilized by a G-quadruplex [122].
It is possible that tRNA-derived fragments have an intrinsic tendency to self-associate, and this can partially explain the high extracellular stability of certain 5ʹ tRNA halves.
4.6. Biogenesis of 5ʹ tRNA halves in the extracellular space
Considering that glycine 5ʹ tRNA halves seem to be more resistant to enzymatic degradation than other single-stranded RNAs [28], we hypothesized that their accumulation in the non-EV extracellular fraction could be a consequence of their differential extracellular stability rather than their preferential or selective secretion. In other words, cells could release a representative sample of their intracellular transcriptome, but most RNAs could be later degraded or fragmented by extracellular RNases. Those RNAs being more resistant to degradation would then accumulate.
To test this hypothesis, we added recombinant ribonuclease inhibitor (RI) directly to the cell-conditioned medium of different mammalian cell lines. To our surprise, addition of RI dramatically changed extracellular RNA profiles [119]. Without RI, most exRNAs outside EVs were 30–31 nt 5ʹ tRNA halves derived from acceptors of glycine and glutamic acid, as has been described in previous sections of this review. In the presence of RI, most extracellular RNAs were now full-length tRNAs and ribosomal RNAs. Furthermore, these rRNAs were actually present in the form of extracellular ribosomes. We could also detect intact, non-fragmented mRNAs in cell-conditioned medium when RI was added, consistent with a recent report in human serum [41].
The origin of these extracellular tRNAs and ribosomes is not completely clear, but it is reasonable to speculate that they are at least partially derived from damaged or dead cells. Whatever their source could be, we have provided clear evidence that full-length tRNAs are released to the extracellular space outside EVs and in high amounts. We then observed that extracellular RNases, especially those present in serum, can rapidly cleave these tRNAs and generate tRNA halves and other smaller fragments. We also discovered that different tRNAs and different tRNA-derived fragments have very different sensitivities to extracellular RNases. For instance, in situations of low RNase activity, tRNALysUUU is more resistant than tRNAGlyGCC, which is fragmented very rapidly once released from cells. At low RNase activity, the predominant 5ʹ fragments of tRNAGlyGCC are 33–35 nt, and correspond to endonucleolytic cleavage products at the level of the anticodon. However, with higher RNase activity these fragments are lost while those of 30–31 nt accumulate, presumably because of their capacity to form dimers. When serum-containing cell-conditioned medium was analysed using equilibrium density gradient centrifugation, we found that full-length tRNAs were only present in the fractions corresponding to EVs, while the non-EV fraction was enriched in tRNA halves. These tRNA halves were originated in the extracellular space, by enzymatic fragmentation of full-length tRNAs. Thus, in samples with high RNase activity such as serum, full-length tRNAs are only found inside EVs, while the most stable tRNA halves can accumulate in the outside [119].
5. Extracellular tRNA-derived fragments in the three domains of life
Despite the fact that most research on exRNAs has been performed in humans and some mammals, several reports have focused on their potential as mediators of intercellular communication and host-pathogen interactions.
Recent experimental evidence revealed that most bacterial and eukaryotic microorganisms release significant amounts of RNAs packed in EVs, raising the idea that EV-mediated secretion of RNAs could represent a widespread pathway of intercellular communication used by organisms in all domains of life (reviewed in [123]). Although most studies in archaea, bacteria and protists are related to exRNA packed into EVs, the release of naked extravesicular RNAs upon cell death [124] or by active secretion pathways [125] is also documented.
5.1. Extracellular RNAs in prokaryotes
One of the first descriptions of bacterial extracellular nucleic acids was reported in membrane blebs of Neisseria gonorrheae, which served as an intermicrobial mechanism of genetic exchange [126]. However, the bacterial extracellular RNA complement has been only recently characterized by deep sequencing of EVs from the marine cyanobacteria Prochlorococcus [127]. In total, 89% of the genome of Prochlorococcus was represented at least once in sequencing libraries from the EV fraction.
Subsequently, the release of RNA packaged in EVs was reported in archaea [128], Gram-negative [129] and Gram-positive bacteria [130]. Much of our understanding of bacterial EV composition and biogenesis derives from studies in Gram-negative bacteria. These bacteria release EVs which are commonly known as outer membrane vesicles (OMVs). OMVs are actively formed through the budding and release of the outer membrane and contain numerous virulence-associated and immunomodulatory factors [131]. They have been implicated in many aspects of bacterial pathogenesis, including delivery of toxins, induction of inflammatory responses, transfer of DNA and RNA and protection against phages and antimicrobial peptides. On the other hand, the release of EVs from Gram-positive bacteria was cautiously considered because EVs would need to cross the cell wall on their way to the extracellular space, but substantial evidence supporting their existence and importance has accumulated with time [132].
Most RNAs reported in EVs form bacteria and archaea are mainly below the length of 250 nt and include mRNAs, rRNAs, tRNAs and small RNAs derived from intra- and intergenic regions [123]. The mechanisms underlying sRNA sorting into bacterial EVs and the relative bacterial cell and EV sRNA profiles remain to be clarified.
Additionally, the function of these exported small RNAs is still unknown, but it is important to note that OMVs can be taken up by human cells. Thus, they may contribute to pathogenesis, as demonstrated for OMVs derived from Helicobacter pylori, which are taken up by gastric epithelial cells [133]. In this context, OMVs have been found to enhance the carcinogenic potential of this specific bacterium [134].
A complete characterization of the extracellular RNA complement of both OMV-associated and extravesicular RNAs by high-throughput sequencing was reported in E. coli [135]. The authors reported that the majority of the RNA obtained from the OMV and the extravesicular fractions have a length below 60 nt with an enrichment of RNAs ranging between 15 and 40 nt. In both fractions, more than 80% of reads were represented by 5ʹ and 3ʹ tRNA fragments. Full-length tRNAs were only observed inside cells, but not in the extracellular fractions. Moreover, the identities and abundances of the secreted RNAs differed between extravesicular and vesicular RNAs. In contrast, similar experimental approaches showed that in OMVs from Vibrio cholerae the most abundant RNAs were from intergenic regions [129]. Similarly, in OMVs from Salmonella typhimurium, all RNA classes could be observed, but most RNAs were represented by rRNAs followed by several full-length and fragmented mRNAs and other ncRNAs [136]. These reports suggest that OMV-associated RNAs vary significantly among different bacterial species, although subtle differences in methodological details can have profound effects on experimental outcomes.
5.2. Relevance of exRNAs in bacterial-host interactions
Recent years have seen increased interest in the role of bacterial EV-associated RNAs in intercellular and interspecies communication [125,137]. For instance, an OMV-associated small RNA has been demonstrated to participate in Pseudomonas pathogenicity by reducing the host immune response [138]. This small RNA was derived from the P. aeruginosa methionine tRNA, comprised its first 24 nucleotides (i.e., a 5ʹ tRF), was abundant in OMVs and reduced LPS-induced as well as OMV–induced IL-8 secretion by cultured primary human airway epithelial cells.
5.3. Extracellular RNAs in protozoan parasites and in host-parasite interactions
Early reports searching for small RNAs in unicellular eukaryotes reported the generation of stress-induced small RNAs derived from tRNAs, rRNAs and other non-coding RNAs. In Tetrahymena thermophila, the response to amino acid deprivation triggers the generation of tRNA halves through the cleavage of mature tRNA [4]. Similarly, generation of tRNA halves and small RNAs derived from rRNAs, snoRNAs and CDS was also reported in the protozoan parasite Trypanosoma cruzi as a stress response [139,140] and in the encystation process of Giardia lamblia [5].
Shortly after, in T. cruzi, 5ʹ tRNA halves of 30–33 nt in length packed in EVs were reported as one of the most abundant intravesicular RNAs along with rRNA-derived fragments [141–143]. Similar results were obtained in Leishmania [144], demonstrating that the release of tRNA-derived fragments to the extracellular space is conserved in trypanosomatids. However, the overlap at the level of tRNA-derived fragments between EVs from T. cruzi [142] and Leishmania [144] was rather small. Although Leishmania EVs were competent to deliver their small RNA cargo to macrophages [144], their eventual functional influence in recipient macrophages was not analysed and remains to be elucidated.
Our lab also reported that tRNA-derived fragments induced in T. cruzi by nutrient starvation [140], colocalized with an argonaute protein distinctive of trypanosomatids [145], and were secreted inside extracellular vesicles [143]. We showed that the vesicular cargo including tRNA-derived fragments was transferred between parasites and from parasites to mammalian cells. This transference promoted life cycle transition of epimastigotes to infective trypomastigotes and also increased the susceptibility to infection in mammalian cells [143] by inducing gene expression changes in these cells [146]. Indeed, T. cruzi-derived EVs elicited profound transcriptomic changes in HeLa cells involving host cell cytoskeleton, extracellular matrix, and immune responses pathways. Interestingly, a significant proportion of these responses were reproduced by transfecting cells with specific 5ʹ tRNA halves from RNAThr and RNALeu which were among the most abundant tRNA-derived fragments in the EVs secreted by T. cruzi.
A heterogeneous population of small RNAs ranging between 25 and 200 nt in size were also reported in EVs secreted by Trichomonas vaginalis [147] but the identity of these small RNAs was not analysed. In cultures of red blood cells infected with the malaria parasite Plasmodium falciparum, tRNA-derived fragments from the host and the parasite were identified [148]. It remains to be determined if the profile of host tRNA-derived fragments in EVs from infected cells differs from that obtained in control, noninfected red blood cells.
5.4. Extracellular RNAs in helminths, plants and fungi
Although several recent reports have demonstrated that EVs secreted from several helminths carry a variety of small RNAs that may be taken up by host cells, only few reports analysed the presence of tRNA-derived small RNAs in these extracellular vesicles [149].
In Heligmosomoides polygyrus [150] tRNA-derived small RNAs were minor components of vesicular cargo. On the contrary, it was shown that Schistosoma mansoni secrete EVs during in vitro development and that these vesicles contain many 5ʹ and 3ʹ tRNA fragments derived from most tRNAs [151]. Interestingly, the authors analysed both EV-enriched (120,000 × g pellet) and extravesicular (120,000 × g supernatant) fractions from the culture media, and found that some tRNA-derived fragments were equally distributed between both fractions, while others displayed contrasting patterns. Even though sequencing parameters were not optimized to capture full-length tRNAs, a lysine tRNA was read with a 93% of coverage. This implies that full-length tRNAs are probably present in the extracellular space of S. mansoni. Transfer RNA sequences mapping to the S. mansoni genome were also identified in the sera of infected mice, but the high sequence conservation and post-transcriptional modifications of tRNAs did not permit to unambiguously identify these sequences as helminth-derived [152]. Most if not all of them probably corresponded to murine sequences because they were identified at similar levels in both infected and naïve mice. This is one of the challenges involved when working with tRNAs and their fragments. Sequences aligning to tRNAs were also described in EVs derived from the eggs of S. japonicum, but their relative representation was rather low [153].
In plants, it is known that 21–24 nt small silencing RNAs can move within the plant, presumably through plasmodesmata or the vasculature and from the plant to invading pathogens and parasites [154]. Although there are several documented cases of mobile sRNAs in plants, the mechanisms by which these RNAs move are only partially understood [155]. An early study performed in pumpkin analysed 30–90 nt RNAs present in phloem sap, the fluid transported in phloem sieve tube elements in plants [156]. Here, the authors identified a high proportion (27%) of tRNA-derived fragments among sequenced small RNAs. These tRNA-derived sequences were variable in size and matched both the 5ʹ and 3ʹ halves of their precursors, with predicted cleavage sites located at the anticodon and the D loop of their parental tRNAs. By Northern blot, full-length tRNAs and tRNA-derived fragments were simultaneously detected in the phloem sap. However, in contrast to leaf and stem protein extracts, phloem sap protein extracts were unable to cleave an in vitro-produced tRNA. This result suggests that circulating tRNA-derived fragments in phloem sap were not generated in the extracellular space by enzymatic cleavage of extracellular full-length tRNAs. A possible function of these extracellular tRNAs could be related to the transport of mRNAs in the phloem, since it was shown that tRNA-like structures in mRNAs are important for their transport in Arabidopsis [157].
Extracellular RNAs have been shown to play important roles in plant-pathogen interactions. At least in Arabidopsis, EVs can function as exRNA carriers of small RNAs [158] and mediate a process known as host-induced gene silencing where small RNAs produced in plant cells can regulate the expression of genes in invading pathogens or parasites. Interestingly, the cross-kingdom trafficking of small RNAs between plants and invading fungal parasites is bi-directional [159]. How exactly this bidirectional fungi-plant interaction works and whether tRNA-derived fragments are part of this phenomenon is currently unknown. Nevertheless, analysis of EVs purified from A. thaliana has shown a minor population of small RNAs ranging 18–34 nt in length that included reads mapping to tRNAs [160]. Because 5ʹ tRFs are known to be upregulated in phosphate-starved Arabidopsis roots [161], it would be interesting to study how stress affects the levels of extracellular tRNA-derived fragments in plants.
In fungi, the presence of exRNA was formerly suggested in EVs from the human pathogen Cryptococcus neoformans [162]. More recently, the RNA content of EVs from Saccharomyces cerevisiae and the human pathogens Cryptococcus neoformans, Paracoccidioides brasiliensis and Candida albicans was analysed by deep sequencing [163]. The most abundant small RNA species were derived from snoRNAs and tRNAs. Depending on the fungal species considered, tRNA-derived sequences accounted for 22 to 75% of total small RNAs. Interestingly, fungal EVs were enriched in 3ʹ halves of nuclear and mitochondrial tRNAs, which were not among the most abundant intracellular fragments in these organisms.
6. Conclusion: towards an integrated model of extracellular tRNAs and tRNA-derived fragments
Transfer RNAs are ancient molecules already present in the Last Universal Common Ancestor (LUCA) and key players in defining the genetic code and the flux of genetic information. Despite their central role in molecular biology, interest in tRNAs started to decline after the 70’s, presumably because their role as adapter molecules between the worlds of nucleic acids and proteins seemed understood and rather static. With the passing of time it became clear that, precisely because of their pivotal role as genetic decoders, tRNAs participate in many gene regulatory networks including adaptive translation during stress [164,165]. Because tRNA fragmentation was described in all domains of life and their generation was induced by acute cellular stress [2–6,9,10,13,33,140,166], it became clear that at least some tRNA-derived fragments are small regulatory RNAs rather than functionless byproducts of tRNA turnover.
These tRNA-derived fragments were then detected in extracellular vesicles released from murine immune cells in culture [103], from whole unicellular organisms [143] and circulated in mammalian blood serum as soluble complexes which did not pellet with EVs [7,8]. By a combination of ultracentrifugation and size-exclusion chromatography we demonstrated that glycine and glutamic acid 5ʹ tRNA halves were indeed present as vesicle-free extracellular complexes in the conditioned media of human cell lines [27]. We then realized that their high extracellular abundance could be explained by their intrinsic resistance to degradation, which was observed before [8], but that we linked to their capacity of forming RNA dimers [28]. Interestingly, higher-order RNA ‘quaternary’ (i.e., intermolecular) interactions were described for other tRNA halves such as those derived from tRNACys and tRNAAla [11,17,167]. Although these tRNA-derived fragments are not among the most frequently detected in the extracellular space, as our hypothesis of ‘stability-defines-abundance’ would predict, it is possible that the G-quadruplex-stabilized tetrameric structures they form [167] are refractory to standard sequencing techniques. Additionally, post-transcriptionally modified bases which are widely present in tRNAs are probably ‘inherited’ by tRNA halves and will have an impact in their extracellular (and intracellular) stabilities. With the advent of new sequencing techniques which are sensitive to base modifications [168,169] we expect that a more complete picture will emerge in the following years.
Divalent ions seem to play an important role in stabilizing tRNA halves in the extracellular, non-EV fraction. Independent groups have shown that tRNA halves are usually lost when EDTA-treated plasma is used instead of heparinized plasma [7,40,67]. Based on this evidence, it is tempting to speculate that tRNA halves are protected outside EVs by their complexation with RNA-binding proteins in a divalent ion-depending manner. However, it has been shown that the extracellular stability of tRNA halves is still high even when purified from proteins and added back to sera [8]. Alternatively, divalent ions can be important to preserve intermolecular RNA interactions. If proteins are involved, their identity still needs to be revealed.
Based on the use of a non-retroviral thermostable reverse transcriptase and sequencing [70,72], electrophoretic profiles [114,115] or Northern blot [95,119] it seems clear that EVs contain more full-length tRNAs than tRNA-derived fragments. However, the mechanisms used by cells to encapsulate tRNAs in EVs are still not completely understood. It has been suggested that the RNA-binding protein YBX1 could be involved in the process [72], but we have also observed EVs containing full-length tRNAs in YBX1-null cells [119]. An alternative possibility is that YBX1 defines tRNA sorting into EVs indirectly, by impacting the pool of tRNAs not engaged in translation and, therefore, in the probability of non-selective tRNA encapsulation. The high correlation between intracellular and EV-associated tRNA abundances observed by the authors [72] is indicative of non-selective RNA sorting at the tRNA level. There might still be additional constrains related to size, as was shown for mRNA encapsulation inside EVs [105]. This can explain why we see higher levels of tRNAs than rRNAs in density gradient-purified EVs [119], and why large EVs seem to contain more full-length 18 S and 28 S rRNAs than small EVs [113].
How dynamic is the RNA content of EVs? Even though full-length tRNAs seem to be in excess compared to tRNA-derived fragments inside EVs, this can change in situations of stress. Under stress, Angiogenin [9,10] or another ribonuclease [170] cleaves some tRNAs at their anticodon loop to generate tRNA halves which receive the name of tiRNAs, for tRNA-derived, stressed-induced small RNAs. A similar mechanisms might be responsible of generating tRNA halves in sex-hormone responsive cells exposed to oestrogens or androgens [15]. Published alongside this review, we have shown that the overexpression of tRNA halves resistant to degradation is sufficient to trigger their encapsulation inside EVs [29]. Although this needs to be demonstrated with endogenous tRNA halves, it is reasonable to speculate that stress-derived EVs will contain higher levels of tRNA halves when compared to EVs released from non-stressed cells. Additionally, cells detecting higher payloads of certain tRNA halves in EVs could decipher this as a sign of stress in nearby cells.
An attempt to connect the different pieces of information described in the past three paragraphs into a unified and coherent model that could explain the extracellular profiles of tRNAs and their fragments is depicted in Fig. 2 and Table 1. Even if incorrect, this model highlights relevant questions which can increase our understanding once addressed. The most intriguing aspects are related to how cells sense and respond to extracellular tRNAs and their fragments, including those present inside and outside EVs. At least for alanine and cysteine 5ʹ tRNA halves, spontaneous uptake from the extracellular space was demonstrated [17]. Although this applies to exRNAs in general, stoichiometric considerations need to be addressed rigorously, because the uptake of only a few copies of a given small RNA are likely not to be functional [171]. However, tRNA halves are highly upregulated upon stress, and if the background levels of tRNA halves in neighbouring non-stressed cells are low, uptake of moderate amounts of molecules capable of inducing translational arrest [11] could be impactful.
Figure 2.
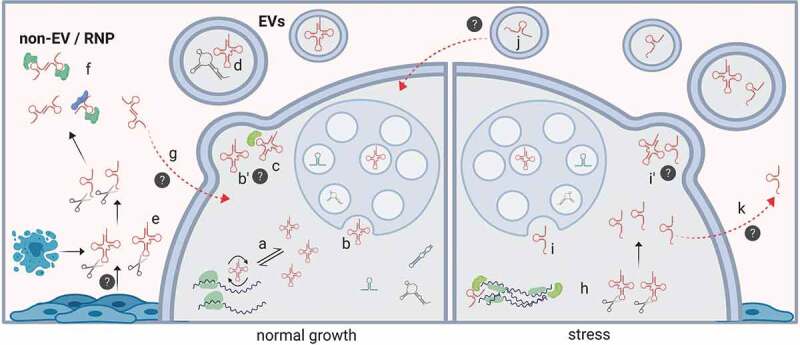
A model for the release of tRNAs and tRNA-derived fragments to the extracellular space, their extracellular dynamics, and unexplored avenues in intercellular communication. (A) During normal growth, tRNAs are engaged in translation. However, it is possible that at least two different pools of tRNAs exist in a cell. Those tRNAs being recycled by translating ribosomes and aminoacyl tRNA synthetases might not be available for encapsulation at the sites of EV biogenesis. We hypothesize that different factors can affect the equilibrium between both pools. Among them: any factors affecting translation, including YBX1, or dynamic post-transcriptional modifications. The encapsulation of tRNAs can simply depend on their relative concentration at the sites of EV encapsulation (non-selective secretion, B and B’) or can be facilitated by proteins such as YBX1 (C). Whatever the case, EVs seem to contain high levels of full-length tRNAs and other ncRNAs (D) compared to their respective fragments. Full-length tRNAs are also released to the extracellular, non-EV fraction (also called RNP or extravesicular fraction), either by an unknown tRNA pumping pathway or passively by dead cells (E). These nonvesicular tRNAs are usually undetectable in the absence of added ribonuclease inhibitors because they are rapidly fragmented by extracellular RNases. Some degradation products seem to be more resistant to RNases, either because they form supramolecular structures or extracellular RNP complexes (F). Comparisons between plasma and serum suggest that divalent ions are involved in the process. Collectively, resistant ncRNA fragments define the extracellular nonvesicular RNAome. It is still unknown whether these extravesicular ncRNA fragments can be internalized or sensed by other cells, and the downstream signalling pathways involved (G). In stressed cells, Angiogenin and/or other RNase A family members cleave at least some tRNAs at their anticodon loop and generate tRNA-derived, stress-induced tRNA halves (H). Some of these tRNA halves and associated proteins are recruited to stress-granules, but it is reasonable to speculate that others would be encapsulated inside EVs and released to the extracellular space (I and I’). We hypothesize that the recognition of EV-associated stress-induced tRNA halves by neighbouring cells could prepare them to upcoming advert growth conditions (J). Effects on immune cells are expected to be different. Finally, whether stressed but fully viable cells can release tRNA-derived fragments to the extracellular, non-vesicular space remains to be elucidated (K).
Table 1.
Studies supporting claims or hypothesis presented in Fig. 2.
| # | Claim/hypothesis | Support |
|---|---|---|
| a | Subcellular localization of tRNAs and their association to the translation apparatus is dynamic | Reviewed in Wilusz [165] |
| b, b’ | tRNAs are incorporated into EVs in a non-selective, concentration-driven manner | Extrapolation from Gámbaro et al. [29] |
| c | The RNA-binding protein YBX1 mediates active incorporation of tRNAs into EVs | [72] |
| d | EVs contain more full-length tRNAs than tRNA-derived fragments | [72,119] |
| e | Passive release of tRNAs to the non-EV fraction by damaged or dead cells | [119] |
| f | Some ncRNA fragments are resistant to degradation and accumulate in the non-EV fraction | [8,28,119] |
| g | Cells can sense extracellular, nonvesicular tRNA-derived fragments | Ivanov et al. [17] (spontaneous uptake of Ala/Cys 5ʹ tRNA halves) |
| h | Stressed cells cleave tRNAs and generate stress-induced tRNA halves | [9,6,10] |
| i, i’ | Stress-induced tRNA halves are then encapsulated in EVs and released to the extracellular space | Extrapolation from Gámbaro et al. 2020 |
| j | Recognition of EV-associated stress-induced tRNA halves prepare neighbouring cells to imminent stress | Speculation inspired by interferon response during viral infections |
| k | Active release of tRNA-derived fragments to the non-EV extracellular fraction [in stressed cells] | Extrapolation from Jeppesen et al. [117] |
A recent report has shown that T cells need to release specific tRNA-derived fragments because they inhibit intracellular pathways related to T cell activation [30]. However, we would like to highlight that excretion and intercellular communication are not mutually exclusive. What one cell needs to release might be informative to other cells in its surroundings. Studies in biofluids are promising for biomarker discovery, but we still do not know where are exRNAs coming from and what is their final fate. Even if they are extracellularly-generated byproducts of tRNAs released by damaged or dead cells, it is by now well stablished that glycine and glutamic acid 5ʹ tRNA halves accumulate in biofluids [39]. As we also tried to highlight in this review, evolution has had enough time to reassign a purpose to these very ancient molecules, which can be detected in the extracellular space of prokaryotes, protozoa, fungi and animals. Moreover, glycine 5ʹ tRNA halves are transferred from somatic cells to germ cells to embryos, where they are involved in widespread gene-expression reprogramming [24–26]. It took some time to accept that ncRNA fragments are functionally relevant molecules. Now we need to fully understand what they are doing at the other side of the plasma membrane.
Acknowledgments
The authors are members of the Sistema Nacional de Investigadores (SNI, ANII). Partial funding for this work was received from Agencia Nacional de Investigación e Innovación, (ANII, Uruguay) [FCE_3_2018_1_148745, FSS_X_2018_1_149070] and from the National Institutes of Health, USA [UG3CA241694, supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director]. The authors want to thank Marco Li Calzi for proofreading this manuscript.
Funding Statement
This work was supported by the Agencia Nacional de Investigación e Innovación (ANII, Uruguay) [FCE_3_2018_1_148745, FSS_X_2018_1_149070]; National Institutes of Health, USA [UG3CA241694, supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Lakings DB, Waalkes TP, Borek E, et al. Composition, associated tissue methyltransferase activity, and catabolic end products of transfer RNA from carcinogen-induced hepatoma and normal monkey livers. Cancer Res. 1977;37:285–292. [PubMed] [Google Scholar]
- [2].Haiser HJ, Karginov FV, Hannon GJ, et al. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jöchl C, Rederstorff M, Hertel J, et al. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lee SR, Collins K.. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744–42749. [DOI] [PubMed] [Google Scholar]
- [5].Li Y, Luo J, Zhou H, et al. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thompson DM, Lu C, Green PJ, et al. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dhahbi JM, Spindler SR, Atamna H, et al. 5ʹ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang Y, Zhang Y, Shi J, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol. 2014;6:172–174. [DOI] [PubMed] [Google Scholar]
- [9].Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. [DOI] [PubMed] [Google Scholar]
- [10].Yamasaki S, Ivanov P, Hu G-F, et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fricker R, Brogli R, Luidalepp H, et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun. 2019;10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gebetsberger J, Zywicki M, Künzi A, et al. TRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gebetsberger J, Wyss L, Mleczko AM, et al. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017;14:1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Saikia M, Jobava R, Parisien M, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34:2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ivanov P, O’Day E, Emara MM, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lyons SM, Achorn C, Kedersha NL, et al. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Goodarzi H, Liu X, Nguyen HCB, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mleczko AM, Celichowski P, Bąkowska-Żywicka K. Transfer RNA-derived fragments target and regulate ribosome-associated aminoacyl-transfer RNA synthetases. Biochim Biophys Acta - Genet Regul Mech. 2018;1861:647–656. [DOI] [PubMed] [Google Scholar]
- [21].Schorn AJ, Gutbrod MJ, LeBlanc C, et al. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim HK, Fuchs G, Wang S, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. [DOI] [PubMed] [Google Scholar]
- [24].Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sharma U, Sun F, Conine CC, et al. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell. 2018;46:481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Boskovic A, Bing XY, Kaymak E, et al. Control of noncoding RNA production and histone levels by a 5′ tRNA fragment. Genes Dev. 2020;34:118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tosar JP, Gámbaro F, Sanguinetti J, et al. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015;43:5601–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tosar JP, Gambaro F, Darre L, et al. Dimerization confers increased stability to nucleases in 5ʹ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 2018;46:9081–9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gámbaro F, Li Calzi M, Fagúndez P, et al. Stable tRNA halves can be sorted into extracellular vesicles and delivered to recipient cells in a concentration-dependent manner. RNA Biol. 2019;1–15. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chiou N-T, Kageyama R, Ansel KM. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 2018;25:3356–3370.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Matsumura M, Ohosone Y, Miyachi K, et al. Novel autoantibodies directed against the common tertiary configuration of transfer RNA in a patient with interstitial lung disease. Arthritis Rheum. 1996;39:1308–1312. [DOI] [PubMed] [Google Scholar]
- [36].Wilusz J, Keene JD. Autoantibodies specific for U1 RNA and initiator methionine tRNA. J Biol Chem. 1986;261:5467–5472. [PubMed] [Google Scholar]
- [37].Dhahbi JM, Spindler SR, Atamna H, et al. 5-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics. 2013;45:990–998. [DOI] [PubMed] [Google Scholar]
- [38].Casas E, Cai G, Neill JD. Characterization of circulating transfer RNA-derived RNA fragments in cattle. Front Genet. 2015;6:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Srinivasan S, Yeri A, Cheah PS, et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177:446–462.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Akat KM, Moore-McGriff D, Morozov P, et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci U S A. 2014;111:11151–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Akat KM, Lee YA, Hurley A, et al. Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. JCI Insight. 2019;4:127317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Max KEA, Bertram K, Akat KM, et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc Natl Acad Sci U S A. 2018;115:E5334–E5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Danielson KM, Rubio R, Abderazzaq F, et al. High throughput sequencing of extracellular RNA from human plasma. PLoS One. 2017;12:e0164644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yuan T, Huang X, Woodcock M, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Savelyeva AV, Baryakin DN, Chikova ED, et al. Vesicular and extra-vesicular RNAs of human blood plasma. Adv Exp Med Biol. 2016;924:117–119. [DOI] [PubMed] [Google Scholar]
- [47].Cooke WR, Cribbs A, Zhang W, et al. Maternal circulating syncytiotrophoblast-derived extracellular vesicles contain biologically active 5ʹ-tRNA halves. Biochem Biophys Res Commun. 2019;518:107–113. [DOI] [PubMed] [Google Scholar]
- [48].Amorim MG, Valieris R, Drummond RD, et al. A total transcriptome profiling method for plasma-derived extracellular vesicles: applications for liquid biopsies. Sci Rep. 2017;7:14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Allen RM, Zhao S, Ramirez Solano MA, et al. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J Extracell Vesicles. 2018;7:1506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fromm B, Kang W, Rovira C, et al. Plant microRNAs in human sera are likely contaminants. J Nutr Biochem. 2019;65:139–140. [DOI] [PubMed] [Google Scholar]
- [52].Hou D, Zhou Z, Chen X, et al. Reply to Fromm et al. J Nutr Biochem. 2019;65:140–141. [DOI] [PubMed] [Google Scholar]
- [53].Witwer KW, Zhang CY. Diet-derived microRNAs: unicorn or silver bullet? Genes Nutr. 2017;12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tosar JP, Rovira C, Naya H, et al. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA. 2014;20:754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang K, Li H, Yuan Y, et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One. 2012;7:e51009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Heintz-Buschart A, Yusuf D, Kaysen A, et al. Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biol. 2018;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Freedman JE, Gerstein M, Mick E, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. 2016;7:11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Umu SU, Langseth H, Bucher-Johannessen C, et al. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2018;15:242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chahar HS, Corsello T, Kudlicki AS, et al. Respiratory syncytial virus infection changes cargo composition of exosome released from airway epithelial cells. Sci Rep. 2018;8:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Girard A, Sachidanandam R, Hannon GJ, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. [DOI] [PubMed] [Google Scholar]
- [64].Lewis SH, Quarles KA, Yang Y, et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol. 2018;2:174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jehn J, Gebert D, Pipilescu F, et al. PIWI genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun Biol. 2018;1:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tosar JP, Rovira C, Cayota A. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun Biol. 2018;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Godoy PM, Bhakta NR, Barczak AJ, et al. Large differences in small RNA composition between human biofluids. Cell Rep. 2018;25:1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Honda S, Kawamura T, Loher P, et al. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 2017;45:9108–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24:2742–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mohr S, Ghanem E, Smith W, et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Qin Y, Yao J, Wu DC, et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016;22:111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Shurtleff MJ, Yao J, Qin Y, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci U S A. 2017;114:E8987–E8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Alon S, Vigneault F, Eminaga S, et al. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011;21:1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Giraldez MD, Spengler RM, Etheridge A, et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat Biotechnol. 2018;36:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cozen AE, Quartley E, Holmes AD, et al. ARM-seq: alkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zheng G, Qin Y, Clark WC, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vojtech L, Woo S, Hughes S, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yeri A, Courtright A, Reiman R, et al. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci Rep. 2017;7:44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hong J, Blenkiron C, Tsai P, et al. Extracellular RNA profile in mesenteric lymph from exemplar rat models of acute and critical illness. Lymphat Res Biol. 2019;17:512–517. [DOI] [PubMed] [Google Scholar]
- [80].Dhahbi JM, Spindler SR, Atamna H, et al. Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer. 2014;6:BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wu X, Somlo G, Yu Y, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012;10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Martinez BV, Dhahbi JM, Nunez Lopez YO, et al. Circulating small non coding RNA signature in head and neck squamous cell carcinoma. Oncotarget. 2015;6:19246–19263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Nientiedt M, Deng M, Schmidt D, et al. Identification of aberrant tRNA-halves expression patterns in clear cell renal cell carcinoma. Sci Rep. 2016;6:37158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zhao C, Tolkach Y, Schmidt D, et al. 5′-tRNA halves are dysregulated in clear cell renal cell carcinoma. J Urol. 2018;199:378–383. [DOI] [PubMed] [Google Scholar]
- [85].Zhao C, Tolkach Y, Schmidt D, et al. tRNA-halves are prognostic biomarkers for patients with prostate cancer. Urol Oncol Semin Orig Investig. 2018;36:503.e1–503.e7. [DOI] [PubMed] [Google Scholar]
- [86].Feng W, Li Y, Chu J, et al. Identification of tRNA-derived small noncoding RNAs as potential biomarkers for prediction of recurrence in triple-negative breast cancer. Cancer Med. 2018;7:5130–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Huang Y, Ge H, Zheng M, et al. Serum tRNA‐derived fragments (tRFs) as potential candidates for diagnosis of nontriple negative breast cancer. J Cell Physiol. 2019. DOI: 10.1002/jcp.29185 [DOI] [PubMed] [Google Scholar]
- [88].Shao Y, Sun Q, Liu X, et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sun C, Yang F, Zhang Y, et al. TRNA-derived fragments as novel predictive biomarkers for trastuzumab-resistant breast cancer. Cell Physiol Biochem. 2018;49:419–431. [DOI] [PubMed] [Google Scholar]
- [90].Hogg MC, Raoof R, El Naggar H, et al. Elevation of plasma tRNA fragments precedes seizures in human epilepsy. J Clin Invest. 2019;129:2946–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Casas E, Cai G, Kuehn LA, et al. Association of circulating transfer RNA fragments with antibody response to mycoplasma bovis in beef cattle. BMC Vet Res. 2018;14:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Taxis TM, Bauermann FV, Ridpath JF, et al. Analysis of tRNA halves (tsRNAs) in serum from cattle challenged with bovine viral diarrhea virus. Genet Mol Biol. 2019;42:374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Li C, Qin F, Hu F, et al. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018;8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Dhahbi JM, Atamna H, Selth LA. Data mining of small RNA-seq suggests an association between prostate cancer and altered abundance of 5′ transfer RNA halves in seminal fluid and prostatic tissues. Biomark Cancer. 2018;10:1179299X1875954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Khurana R, Ranches G, Schafferer S, et al. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA. 2017;23:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Zhou Z, Wu Q, Yan Z, et al. Extracellular RNA in a single droplet of human serum reflects physiologic and disease states. Proc Natl Acad Sci. 2019;116:19200–19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61:56–63. [DOI] [PubMed] [Google Scholar]
- [98].Driedonks TAP, Nijen Twilhaar MK, Nolte-‘t Hoen ENM. Technical approaches to reduce interference of fetal calf serum derived RNA in the analysis of extracellular vesicle RNA from cultured cells. J Extracell Vesicles. 2019;8:1552059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wei Z, Batagov AO, Carter DRF, et al. Fetal bovine serum RNA interferes with the cell culture derived extracellular RNA. Sci Rep. 2016;6:31175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Tosar JP, Cayota A, Eitan E, et al. Ribonucleic artefacts: are some extracellular RNA discoveries driven by cell culture medium components? J Extracell Vesicles. 2017;6:1272832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Mannerström B, Paananen RO, Abu-Shahba AG, et al. Extracellular small non-coding RNA contaminants in fetal bovine serum and serum-free media. Sci Rep. 2019;9:5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Auber M, Fröhlich D, Drechsel O, et al. Serum-free media supplements carry miRNAs that co-purify with extracellular vesicles. J Extracell Vesicles. 2019;8:1656042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Nolte’T Hoen ENM, Buermans HPJ, Waasdorp M, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Baglio SR, Rooijers K, Koppers-Lalic D, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wei Z, Batagov AO, Schinelli S, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Driedonks TAP, van der Grein SG, Ariyurek Y, et al. Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cell Mol Life Sci. 2018;75:3857–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Cha DJ, Franklin JL, Dou Y, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Li CCY, Eaton SA, Young PE, et al. Glioma microvesicles carry selectively packaged coding and noncoding RNAs which alter gene expression in recipient cells. RNA Biol. 2013;10:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Santangelo L, Giurato G, Cicchini C, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17:799–808. [DOI] [PubMed] [Google Scholar]
- [110].Stik G, Crequit S, Petit L, et al. Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells. J Cell Biol. 2017;216:2217–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Van Balkom BWM, Eisele AS, Michiel Pegtel D, et al. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J Extracell Vesicles. 2015;4:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- [113].Lunavat TR, Cheng L, Kim DK, et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells – evidence of unique microRNA cargos. RNA Biol. 2015;12:810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lunavat TR, Cheng L, Einarsdottir BO, et al. BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc Natl Acad Sci U S A. 2017;114:E5930–E5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Lässer C, Shelke GV, Yeri A, et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017;14:58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, et al. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife. 2019;8:e47544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, et al. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Tosar JP, Segovia M, Gámbaro F, et al. Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. BioRxiv. 2020. DOI: 10.1101/2020.01.29.923714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sork H, Corso G, Krjutskov K, et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci Rep. 2018;8:10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Statello L, Maugeri M, Garre E, et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS One. 2018;13:e0195969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lyons SM, Fay MM, Akiyama Y, et al. RNA biology of angiogenin: current state and perspectives. RNA Biol. 2017;14:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tsatsaronis JA, Franch-Arroyo S, Resch U, et al. Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol. 2018;26:401–410. [DOI] [PubMed] [Google Scholar]
- [124].Rodriguez-Rodrigues N, Castillo LA, Landoni VI, et al. Prokaryotic RNA associated to bacterial viability induces Polymorphonuclear neutrophil activation. Front Cell Infect Microbiol. 2017;7:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Abdullah Z, Schlee M, Roth S, et al. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. Embo J. 2012;31:4153–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:2499–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Biller SJ, Schubotz F, Roggensack SE, et al. Bacterial vesicles in marine ecosystems. Science. 2014;343:183–186. [DOI] [PubMed] [Google Scholar]
- [128].Choi DH, Kwon YM, Chiura HX, et al. Extracellular vesicles of the hyperthermophilic archaeon “Thermococcus onnurineus” NA1T. Appl Environ Microbiol. 2015;81:4591–4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Sjöström AE, Sandblad L, Uhlin BE, et al. Membrane vesicle-mediated release of bacterial RNA. Sci Rep. 2015;5:15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Resch U, Tsatsaronis JA, Le Rhun A, et al. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group a streptococcus. MBio. 2016;7:e00207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Coelho C, Casadevall A. Answers to naysayers regarding microbial extracellular vesicles. Biochem Soc Trans. 2019;47:1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Parker H, Chitcholtan K, Hampton MB, et al. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun. 2010;78:5054–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Chitcholtan K, Hampton MB, Keenan JI. Outer membrane vesicles enhance the carcinogenic potential of Helicobacter pylori. Carcinogenesis. 2008;29:2400–2405. [DOI] [PubMed] [Google Scholar]
- [135].Ghosal A, Upadhyaya BB, Fritz JV, et al. The extracellular RNA complement of Escherichia coli. Microbiologyopen. 2015;4:252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Malabirade A, Habier J, Heintz-Buschart A, et al. The RNA complement of outer membrane vesicles from Salmonella enterica Serovar Typhimurium under distinct culture conditions. Front Microbiol. 2018;9:2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Obregón-Henao A, Duque-Correa MA, Rojas M, et al. Stable extracellular RNA fragments of mycobacterium tuberculosis induce early apoptosis in human monocytes via a caspase-8 dependent mechanism. PLoS One. 2012;7:e29970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Koeppen K, Hampton TH, Jarek M, et al. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLOS Pathog. 2016;12:e1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Franzén O, Arner E, Ferella M, et al. The short non-coding transcriptome of the protozoan parasite Trypanosoma cruzi. PLoS Negl Trop Dis. 2011;5:e1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Garcia-Silva MR, Frugier M, Tosar JP, et al. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol. 2010;171:64–73. [DOI] [PubMed] [Google Scholar]
- [141].Bayer-Santos E, Lima FM, Ruiz JC, et al. Characterization of the small RNA content of Trypanosoma cruzi extracellular vesicles. Mol Biochem Parasitol. 2014;193:71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Fernandez-Calero T, Garcia-Silva R, Pena A, et al. Profiling of small RNA cargo of extracellular vesicles shed by Trypanosoma cruzi reveals a specific extracellular signature. Mol Biochem Parasitol. 2015;199:19–28. [DOI] [PubMed] [Google Scholar]
- [143].Garcia-Silva MR, Cura Das Neves RF, Cabrera-Cabrera F, et al. Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol Res. 2014;113:285–304. [DOI] [PubMed] [Google Scholar]
- [144].Lambertz U, Oviedo Ovando ME, Vasconcelos E, et al. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA packaging. BMC Genomics. 2015;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Garcia Silva MR, Tosar JP, Frugier M, et al. Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene. 2010;466:26–35. [DOI] [PubMed] [Google Scholar]
- [146].Garcia-Silva MR, Cabrera-Cabrera F, Cura Das Neves RF, et al. Gene expression changes induced by Trypanosoma cruzi shed Microvesicles in mammalian host cells: relevance of tRNA-derived halves. Biomed Res Int. 2014;2014:305239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Twu O, de Miguel N, Lustig G, et al. Trichomonas vaginalis exosomes deliver cargo to host cells and mediate host: parasite interactions. PLoS Pathog. 2013;9:e1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Babatunde KA, Mbagwu S, Hernández-Castañeda MA, et al. Malaria infected red blood cells release small regulatory RNAs through extracellular vesicles. Sci Rep. 2018;8:884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Claycomb J, Abreu-Goodger C, Buck AH. RNA-mediated communication between helminths and their hosts: the missing links. RNA Biol. 2017;14:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Buck AH, Coakley G, Simbari F, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Nowacki FC, Swain MT, Klychnikov OI, et al. Protein and small non-coding RNA-enriched extracellular vesicles are released by the pathogenic blood fluke Schistosoma mansoni. J Extracell Vesicles. 2015;4:28665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Hoy AM, Lundie RJ, Ivens A, et al. Parasite-derived microRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl Trop Dis. 2014;8:e2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Zhu S, Wang S, Lin Y, et al. Release of extracellular vesicles containing small RNAs from the eggs of Schistosoma japonicum. Parasit Vectors. 2016;9:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Molnar A, Melnyk CW, Bassett A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. [DOI] [PubMed] [Google Scholar]
- [155].Liu L, Chen X. Intercellular and systemic trafficking of RNAs in plants. Nat Plants. 2018;4:869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Zhang W, Thieme CJ, Kollwig G, et al. TRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell. 2016;28:1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Cai Q, Qiao L, Wang M, et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360:1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Weiberg A, Wang M, Lin FM, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Baldrich P, Rutter BD, Karimi HZ, et al. Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-nucleotide “Tiny” RNAs. Plant Cell. 2019;31:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Hsieh LC, Lin SI, Shih ACC, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Nicola AM, Frases S, Casadevall A. Lipophilic dye staining of cryptococcus neoformans extracellular vesicles and capsule. Eukaryot Cell. 2009;8:1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Da Silva RP, Puccia R, Rodrigues ML, et al. Extracellular vesicle-mediated export of fungal RNA. Sci Rep. 2015;5:7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16:98–112. [DOI] [PubMed] [Google Scholar]
- [165].Wilusz JE. Controlling translation via modulation of tRNA levels. Wiley Interdiscip Rev RNA. 2015;6:453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Levitz R, Chapman D, Amitsur M, et al. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. Embo J. 1990;9:1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Lyons SM, Gudanis D, Coyne SM, et al. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017;8:1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kimura S, Dedon PC, Waldor MK. Surveying the landscape of tRNA modifications by combining tRNA sequencing and RNA mass spectrometry. BioRxiv. 2019. DOI:0.1101/723049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Liu H, Begik O, Lucas MC, et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun. 2019;10:4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Su Z, Kuscu C, Malik A, et al. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J Biol Chem. 2019;294:16930–16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]


