ABSTRACT
Transfer RNA-derived fragments (tRFs) exist in all branches of life. They are involved in RNA degradation, regulation of gene expression, ribosome biogenesis. In archaebacteria, kinetoplastid, yeast, and human cells, they were also shown to regulate translation. In Arabidopsis, the tRFs population fluctuates under developmental or environmental conditions but their functions are yet poorly understood. Here, we show that populations of long (30–35 nt) or short (19–25 nt) tRFs produced from Arabidopsis tRNAs can inhibit in vitro translation of a reporter gene. Analysing a series of oligoribonucleotides mimicking natural tRFs, we demonstrate that only a limited set of tRFs possess the ability to affect protein synthesis. Out of a dozen of tRFs, only two deriving from tRNAAla(AGC) and tRNAAsn(GUU) strongly attenuate translation in vitro. Contrary to human tRF(Ala), the 4 Gs present at the 5ʹ extremity of Arabidopsis tRF(Ala) are not implicated in this inhibition while the G18 and G19 residues are essential. Protein synthesis inhibition by tRFs does not require complementarity with the translated mRNA but, having the capability to be associated with polyribosomes, tRFs likely act as general modulation factors of the translation process in plants.
KEYWORDS: Protein synthesis, polyribosomes, small non-coding RNAs, tRNAs, plant
Introduction
Beyond their primordial role in protein synthesis, transfer RNAs (tRNAs) appear to have many other multiple functions [1,2]. Among them, the production of regulatory small non-coding RNAs (sncRNAs) called tRFs (tRNA-derived Fragments) has been put forward in the last few years. These tRFs have various origins within tRNA transcripts. They can originate from tRNA precursor molecules, but most of them derive from mature tRNAs (for reviews see e.g. [3,4]). Mature tRNAs generate two main classes of tRFs depending on the cleavage site: long tRFs (l-tRFs, 30–35 nt in size) when cleavage occurs in the anticodon region and short ones (s-tRFs, 19–25 nt) if the cleavage is located in the D or T regions of tRNA molecules. Several nomenclatures exist and we will use here the nomenclature we have proposed in [3] where l-tRFs mainly correspond to tRF-5A or tRF-3A and s-tRFs to tRF-5D or tRF-3T whether the RNA fragments derive from the 5ʹ or 3ʹ extremities of mature tRNAs.
The cleavage of tRNAs likely represents an essential process to recycle molecules such as phosphate or nitrogen upon nutrient starvation. Nevertheless, in all evolutionary divergent organisms from bacteria to human cells, a growing list of other important functions have been assigned to tRFs (for a review, see [2]). For instance, numerous l-tRFs were often shown to be upregulated upon stress ([5–7] and references therein). Others are involved in tumorigenesis [8] and haematopoiesis [9]. The implication of tRFs to promote RNA degradation [10], to prime viral reverse transcription [11] or to regulate gene expression via the RNA silencing pathway (e.g. [12,13]) has also been described. Recent work also showed that tRFs can regulate epigenetic inheritance [14], the expression of retro-elements [15], or plant nodulation [16].
Regulation of translation appears to be the most documented process where tRFs are involved. Upon abiotic stress, l-tRFs produced in human cells promote stress granule assembly and translation repression [17]. Reduction of translation was also observed when some human stress-induced l-tRFs, including tRF-5A (Ala) and tRF-5A (Cys), can displace the eIF4F factor of the initiation elongation complex [18]. Also in human, Sobala and Hutvagner [19] showed that some human s-tRFs (e.g. a tRF-5D (Gln) of 19 nt), can inhibit protein synthesis. More recently, a proteomic approach allowed us to demonstrate that this tRF can interact with the multi-synthetase complex (MSC), a complex interacting with ribosomes [20]. Furthermore, in the yeast Saccharomyces cerevisiae [21] and the archaea, Haloferax volcanii [22], stress-induced tRFs can bind ribosomes and inhibit translation in vitro. By contrast, it was recently demonstrated that in the kinetoplastid Trypanosoma brucei, a tRF-3A (Thr) produced during nutrient deprivation can associate with ribosomes and subsequently stimulate translation [23].
The existence of tRFs has also been proven in various plants (e.g. [7,13,24–27]). In Arabidopsis thaliana, the tRFs population varies upon stress and plant development and some are specifically enriched in Argonaute (AGO) immunoprecipitates [28,29]. This suggests that tRFs likely also play key roles in photosynthetic cells. Currently, except for the study reporting that Arabidopsis AGO1-associated tRFs can target and cleave transposable elements, little is known about their functions [30].
Some work on pumpkin provided evidence that a phloem small non-coding RNA (sncRNA) population triggers translation inhibition in vitro [24]. This sncRNA population comprises many tRFs, and here we have addressed the question of whether plant tRFs could act as a regulator of protein synthesis. We observed that both tRFs purified from A. thaliana leaves and specific synthetic tRFs deriving from the 5ʹ extremity of Arabidopsis tRNAAla and tRNAAsn efficiently inhibit protein synthesis in vitro. This inhibition is cap-independent and does not require a sequence complementarity with the mRNA to be translated. Furthermore, analysis of Arabidopsis ribosomal fractions shows that tRFs can bind and be associated with heavy polysomes, suggesting their involvement in global regulation of the translation process in the land plant.
Results
Arabidopsis short and long tRFs inhibit protein synthesis in vitro
We previously showed that Arabidopsis tissues (roots, stems, leaves, flowers, siliques) contain complex and variable pools of tRFs [28]. Furthermore, several RNases T2, called RNS1, 2 and 3, expressed in these tissues are involved in the biogenesis of both l-tRFs and s-tRFs [31]. Indeed, incubation of in vitro synthesized cytosolic tRNAAla transcript or Arabidopsis total tRNAs with a crude enzymatic extract from Arabidopsis leaves produces l-tRF (Ala) and s-tRF (Ala), (Fig. 1A and B). As a first step to knowing whether plant tRFs can modulate translation, l- and s-tRFs pools were produced by incubating total Arabidopsis tRNAs in the presence of a crude enzymatic extract from Arabidopsis leaves (Fig. 1C). According to our previous results, this enzymatic extract contains the RNS involved in tRFs production in Arabidopsis [31], and the tRFs population must correspond to the previously identified one when analysing sncRNA libraries [28]. Then, an in vitro coupled transcription/translation wheat germ system was used to test the effect of the tRFs pools on the translation of an mRNA coding for the pGFP (i.e. GFP fused at its N-terminal extremity to a mitochondrial targeting sequence) in the presence of 35S methionine. No difference in protein synthesis efficiency, as compared to the mock sample, was observed when purified Arabidopsis tRNA fractions were added to the system (Fig. 1D). By contrast, both types of tRFs efficiently inhibit the production of radiolabeled pGFP when added to the wheat germ system. Only two concentrations of tRFs (4 and 8 µM) were used, but the data obtained suggest that the inhibition displays dose-dependency. At a concentration of 8 µM, l- or s-tRFs can inhibit up to 95% of pGFP expression (Fig. 1E). Globally, each set of Arabidopsis tRFs (long or short) inhibits translation. However, both the Arabidopsis l- and s-tRFs populations are generated from a vast collection of Arabidopsis tRNAs [32], and the question of whether all of them can modulate protein-coding gene expression needed to be addressed.
Figure 1.
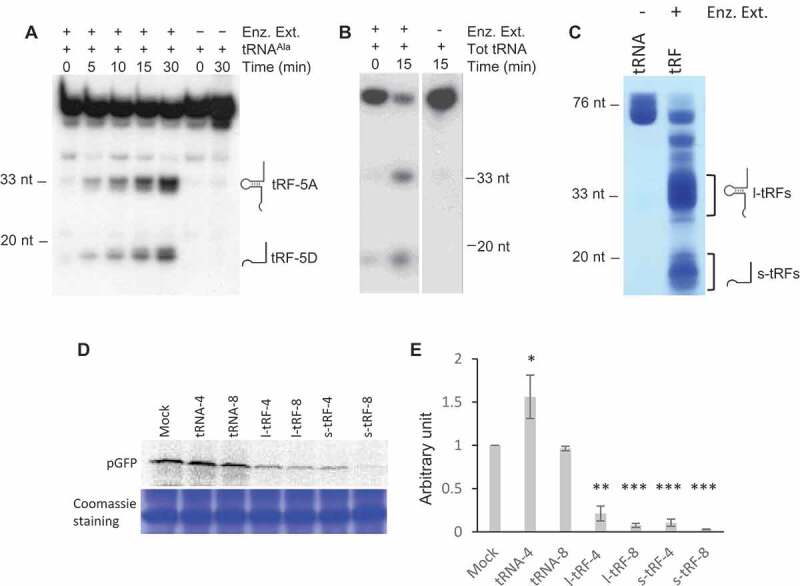
A. thaliana l- or s-tRFs can inhibit translation in vitro. (A) Kinetics of cleavage of Arabidopsis in vitro synthesized tRNAAla(UGC) transcript upon incubation in the presence (+) or absence (-) of a crude enzymatic extract from Arabidopsis leaves (Enz. Ex.). After incubation, RNAs were phenol extracted and fractionated on 15% polyacrylamide gel, followed by northern blot analysis with a radiolabeled oligonucleotide specific for the 5ʹ extremity of cytosolic Arabidopsis tRNAAla. (B) Cleavage of Arabidopsis total tRNA (Tot RNA) upon incubation in the presence (+) or absence (-) of a crude enzymatic extract from Arabidopsis leaves (Enz. Ex.). After incubation, RNAs were phenol extracted and fractionated on a 15% polyacrylamide gel, followed by northern blot analysis with a radiolabeled oligonucleotide specific for the 5ʹ extremity of cytosolic Arabidopsis tRNAAla. (C) Production of long (l-tRF) and short (s-tRF) tRFs by in vitro cleavage of total Arabidopsis tRNAs in the presence (+) of a crude enzymatic extract from Arabidopsis leaves (Enz. Ex.). After incubation, RNAs were phenol extracted, fractionated on 15% polyacrylamide gel and all l-tRFs and s-tRFs were purified separately from the gel. (D) Effect of the addition of total Arabidopsis tRNAs or tRFs purified in (B) on the synthesis of GFP in a wheat germ coupled transcription/translation system in the presence of 35S methionine. Radiolabeled GFP was detected by autoradiography or phosphor imaging after fractionation on 15% acrylamide gel. Two concentrations of tRNAs or tRFs were used: 4 µM (tRNA-4 and tRF-4) and 8 µM (tRNA-8 and tRF-8). Mock = control experiment without added tRNAs or tRFs. Coomassie blue staining of the gel is shown as a loading control. (E) Relative quantification of in vitro synthesized pGFP in experiments similar to that described in (D). Error bars show standard deviations (mean of three biological replicates). One-way ANOVA tests were used to calculate p-values. Asterisks indicate statistically significant differences between Mock and each treatment (***P< 0.001; **P< 0.01; *P< 0.1). A value of 1 has been given to the Mock sample.
Only a limited set of arabidopsis tRFs affects translation in vitro
The Arabidopsis tRFs are highly variable in origin, sequence, and length, with a majority deriving from the 5ʹ or 3ʹ extremities of mature tRNAs [28]. On the one hand, we previously identified a specific population of tRFs associated with AGO1, thus suggesting their implication at least in the regulation of gene expression via the RNA silencing pathway. On the other hand, abundant tRFs that were not found in AGO precipitates could be involved in other molecular processes, including regulation of translation as shown in other organisms. As a first attempt to know whether translation inhibition is a general function of tRFs, we synthesized 10 sequences (Fig. 2A; Supplemental Table S1) representative of the Arabidopsis tRFs population that we previously analysed in [28]. These sequences correspond to three tRFs that were shown to be among the most abundant ones associated with AGO1. The two tRFs-3T reflect two different abundances: Thr-AGU is rather abundant whereas Gly-GCC is present in low amount in sncRNA libraries. Finally, among the five selected tRF-5, Ala-AGC, Gly-GCC, and Glu-CUC belong to the most abundant tRFs and by contrast, Asn-GUU and Cys-GCA are present in a low amount. We tested them at two concentrations (0.2 and 1 µM) for their ability to inhibit the translation of pGFP using the in vitro coupled transcription/translation wheat germ system (Fig. 2B and C). As control experiments, two random oligoribonucleotides of 15 and 21 nt corresponding to an internal sequence of GFP have no significant effect on expression (Supplementary Figure 1). The three tRFs previously found associated with AGO1 in leaves have no effect on protein expression (rather a slight positive effect of pGFP synthesis was noted). For five others [e.g. tRF-5D (Gly-GCC), tRF-5A (Cys-GCA) or tRF-3T (Thr-AGU)], inhibition of expression is also not observed. Only two tRFs of 20 nt, namely tRF-5D (Ala-AGC) and tRF-5D (Asn-GUU) strongly inhibit GFP expression already at a concentration of 0.2 µM, inhibition reaching 75–90% at 1 µM. It is important to note that several isomers exist for each of these tRFs in vivo. Indeed, from the 33 tRNAAla and the 15 tRNAAsn Arabidopsis nuclear genes, various tRFs of 20 nt can be generated (Supplementary Figure 2) and are found in sncRNA libraries [28]. For simplification, these two tRFs are now called Ala20 and Asn20 and correspond to one specific isomer (see Supplementary Figure 2). To have an idea of the amount of Ala20 versus tRNAAla in vivo, we quantified them by northern blot in an Arabidopsis total tRNA extract and we observed that roughly there are 104 more tRNAs than tRFs in the extract (Supplementary Figure 3). It is to note that tRFs-5D (Ala) are the most abundant tRFs found in sncRNA libraries, whereas tRF-5D (Asn) is 15 times less present than Ala20 [28]. In the wheat germ extract, the tRNA concentration is about 20 µM (Supplementary Figure 3), thus to keep physiological conditions, we should have a 2 nM tRF concentration. However, in our assays, we used 0.2 to 1 µM. There is an excess of tRFs but we also cannot exclude that, in vivo, there is a higher localized concentration within the cell. Though this analysis has been performed at a small scale, our observations indicate that Arabidopsis tRFs behave differently (from null to strong effect) in their capacity to modulate expression. Interestingly, none of the major tRFs associated with AGO1 affects mRNA expression and more, the tRF capacity to modulate expression is not correlated with their abundance in the total Arabidopsis tRFs population. Thus, likely only a limited set of tRFs are efficient inhibitors, among them, Asn20 and Ala20.
Figure 2.
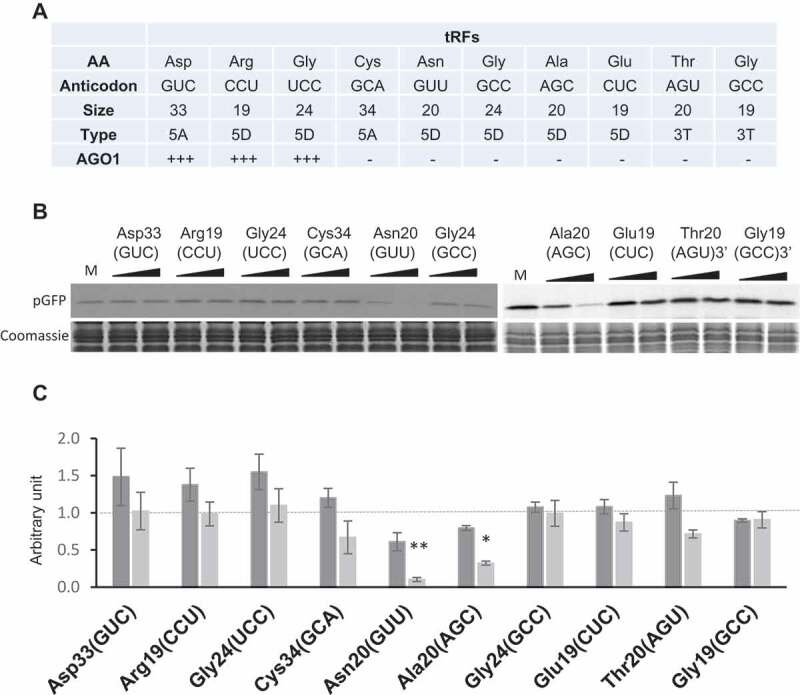
A subset of Arabidopsis tRFs can inhibit efficiently translation in vitro. (A) Features of the 10 tRFs analysed in this work. Their sequences are presented in Supplemental Table 1 and the data derive from [28]. Besides, tRF-5A (Asp) was also found in AGO1 immunoprecipitates (L. Drouard, personal communication). (B) The synthetic tRFs presented in (A) were tested for their ability to inhibit the in vitro protein synthesis of pGFP. Experiments were performed essentially as described in Fig. 1C. Two concentrations of oligoribonucleotides (0.2 and 1 µM) were used. Quantifications of the results are presented in (C). An arbitrary value of 1 (dotted vertical line) has been given to in vitro synthesized pGFP in the absence of added tRF (Mock experiment: M). Error bars represent standard errors of the mean of independent biological experiments (n = 3 to 13). One-way ANOVA tests were used to calculate p-values. Asterisks indicate statistically significant differences between Mock and each treatment (**P< 0.01; *P< 0.1). Dark grey: 0.2 µM, pale grey: 1 µM.
tRF Ala20 inhibits translation by a cap- and sequence-independent mechanism
Interestingly, a stress-induced tRF of 29 nt deriving from human tRNAAla and called 5′-tiRNAAla, was shown to strongly inhibit translation [17,18]. We thus wondered whether inhibition by tRF (Ala) is an evolutionary conserved molecular process and focused our work on Ala20. In Arabidopsis, under normal growth conditions, tRF-5 of 16, 20 and 33 nt deriving from tRNAAla are abundant [28]. Thus, in addition to Ala20, we further tested two other synthetic oligoribonucleotides corresponding to tRF-5D of 16 nt (Ala16) and 33 nt (Ala33) deriving from tRNAAla (Fig. 3B–D). In contrast to Ala20, no significant inhibition of pGFP translation is observed when Ala16 is added in the transcription/translation system at a concentration of 1 µM, and a 40% decrease of expression is observed at a 4 µM concentration. When Ala33 is added in the transcription/translation system, an intermediate inhibitory effect is observed with a 40% decrease of expression at 1 µM. Altogether, this suggests that the size and/or the sequence is important for determining the efficiency of inhibition. Yet, no tRF was shown to be implicated in the inhibition of both transcription and translation. The coupled transcription/translation system does not allow us to discriminate at which step the tRF is implicated. We thus performed in vitro translation experiments with a wheat germ extract translation kit where no transcription is possible. In this case, in the presence of in vitro synthesized capped or uncapped pGFP mRNA reporter, a similar important decrease in pGFP synthesis is observed when Ala20 or Asn20 is present (Fig. 3E). This further demonstrates that the decrease in protein synthesis is not due to inhibition of transcription but rather that the translation process is indeed affected by the addition of the tRF. Finally, to exclude the possibility that the Ala20 is inhibiting translation by directly interfering with the sequence of the pGFP mRNA (although no sequence similarity has been bioinformatically detected), an in vitro transcription/translation assay was performed in the presence of another recombinant plasmid coding for pDHFR (the mammalian DiHydroFolateReductase fused to a mitochondrial targeting sequence) [33]. As for pGFP translation, pDHFR synthesis is not affected by GFP15, GFP21 (Supplementary Figure 1) and Ala16 but is efficiently repressed by the addition of Ala20 (Fig. 3F). This shows that plant tRF-5D Ala20 inhibits translation by a transcript sequence-independent mechanism.
Figure 3.
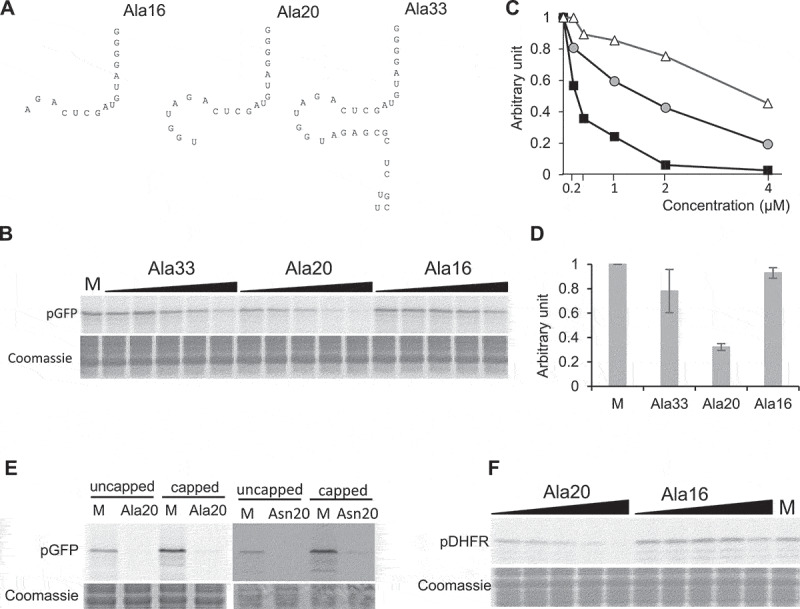
The tRF-5D (Ala) of 20 nt efficiently inhibits translation in vitro. (A) Sequences of the three tRF-5D of 16, 20, and 33 nt (Ala16, Ala20, and Ala33) deriving from Arabidopsis tRNAAla(AGC). (B) Effect of the addition of Ala16, Ala20, and Ala33 on the in vitro synthesis of pGFP protein. The experiment was performed as described in Fig. 1C. Increasing concentrations of synthetic tRFs were used: 0.2, 0.4, 1, 2 and 4 µM. M = Mock, control experiment without tRF. Coomassie blue staining of the gel is shown as a loading control. (C) Relative quantification of in vitro synthesized GFP in the experiment presented in (B). Ala16: white triangles, Ala20: black squares, Ala33: Grey circles. (D) Relative quantification of in vitro synthesized pGFP in experiments similar to that described in (A) and for a tRF concentration of 1 µM. Error bars show standard deviations (mean of three biological replicates). A value of 1 has been given to M. (E) In vitro protein synthesis in a wheat germ translation system of synthesized pGFP transcript in the presence of 35S methionine. Capped or uncapped transcript was used in the absence or presence of Ala20 or Asn20 oligoribonucleotide at 1 µM concentration. M = Mock, control experiment without tRF. Coomassie blue staining of the gel is shown as a loading control. (F) Effect of the addition of Ala16 and Ala20 oligoribonucleotides on the in vitro synthesis of DHFR protein. The experiment was performed as described in Fig. 2B with the same concentration range.
Conserved G18 and G19, and A16 are important for tRF-mediated translation inhibition in arabidopsis
Several residues of mammalian tRFs (also called tiRNA) were identified as important for translation inhibition. On the one hand, the four G residues present at the 5ʹ-end of 5′-tiRNAAla and 5′-tiRNACys can form a G-quadruplex that binds the YB1 protein. This complex then displaces eIF4F complex from the cap and inhibits translation initiation [18]. Arabidopsis Ala20 also starts with four Gs (Fig. 3A). To determine if these residues are also involved in translation inhibition in plants, we replaced them by Us (Ala20-5U, Fig. 4A). As shown in Fig. 4B and C, the inhibition of translation is not affected when a synthetic Ala20-5U is added to the wheat germ system, thus demonstrating that in Arabidopsis, contrary to human tRF-5 (Ala), the G1 to G4 residues are not essential for tRF-mediated translation inhibition. This result is in agreement with the work of Nowacka and collaborators [34] showing that human tRF-5 Ala does not inhibit translation in a wheat germ extract. On the other hand, Sobala and Hutvagner showed that in several human tRFs-5D, the conserved two Gs present in the D loop at positions 18 and 19 of tRNAs are important for translation inhibition [19]. Therefore, we also replaced G18 and G19 by Us in Ala20 (Ala20-3U). As compared to the wild type sequence, the synthetic tRFAla20-3U has lost its capacity to promote translation inhibition (Fig. 4B and C), suggesting that these two residues are important for tRF(Ala)-mediated translation inhibition. To confirm these data, when the two Gs located at the same position in Asn20 oligoribonucleotide (Fig. 4A) are replaced by Us, the same loss in translation inhibition is observed (Fig. 4D). Furthermore, sequence comparison between Ala20 and Asn20 (the two tRFs that efficiently inhibit translation) and the sequence of Arg19 (unable to reduce translation), reveals a high degree of conservation (Fig. 4E). The major difference is that G18 and G19 are at positions 17 and 18 in Arg19 due to the absence of an additional A at position 16 in Ala20 and Asn20. To know whether A16 residue is also important for translation inhibition, other oligoribonucleotides were designed. When the A16 of Ala20 is deleted (Ala20-16A) or replaced by a U (Ala20-A16U), translation inhibition is strongly affected. Conversely, when an A is introduced at position 16 in Arg19 sequence (Arg19+ A16), this tRF acquires the ability to inhibit translation as efficiently as Ala20 or Asn20 (Fig. 4E and F). The introduction of a U rather than an A (Arg19+ U16), although less efficient, still leads to a 50% inhibition. To sum up, among tRF sequences, G18 and G19 appear to be crucial to get the capacity of inhibiting translation; nucleotides such as A16 may have a synergistic effect, and the size of the tRF and/or the position of the nucleotides within the sequence may also be crucial.
Figure 4.
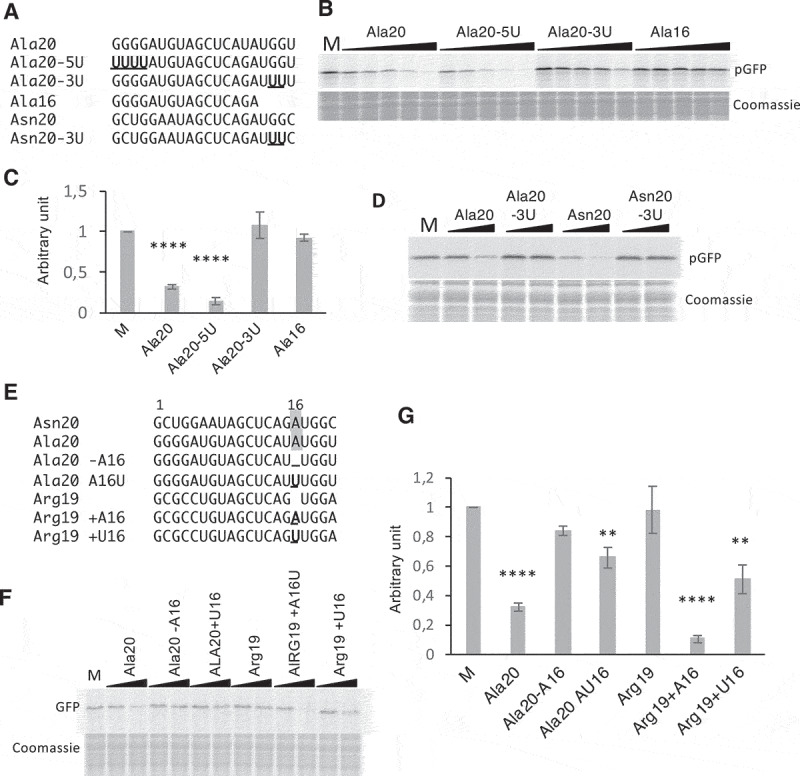
Importance of A16, G18, and G19 of tRF-5D (Ala) for the inhibition of translation. (A) Sequence alignment of Ala20, Ala16, Asn20 with the mutated versions Ala20-5U, Ala20-3U, and Asn20-3U. Us replacing Gs are in bold and underlined (B) Effect of the addition of the Ala20-5U and Ala20-3U presented in (A) on the in vitro synthesis of pGFP. The experiment was performed as described in Fig. 3B with the same concentration range. (C) Quantification of in vitro synthesized pGFP in experiments similar to that described in (B). Error bars represent standard errors of the mean of independent biological experiments (n = 3 to 13). One-way ANOVA tests were used to calculate p-values. Asterisks indicate statistically significant differences between Mock and each treatment (****P< 0.0001). A value of 1 has been given to M (Mock, control experiment without tRF). (D) Effect of the addition of Ala20-3U or Asn20-3U on the in vitro synthesis of pGFP. Experiment was performed as in (B). Two concentrations of oligoribonucleotides (0.2 and 1 µM) were used. (E) Sequence alignment of Ala20 and Arg19 with the mutated versions Ala20-A16, Ala20A16U, Arg19+ A16, Arg19+ U16. Mutated or added nucleotides are in bold and underlined. The A residue at position 16 in Asn20 and Ala20 is under grey background. In vitro synthesis of pGFP in the presence of these oligoribonucleotides and quantifications of the results are presented in (F) and (G) respectively. The experiments were performed as in Fig. 3B, except that only two concentrations of oligoribonucleotides (0.2 and 1 µM) were used. Error bars represent standard errors of the mean of independent biological experiments (n = 3 to 13). A value of 1 has been given to M (Mock, control experiment without tRF). One-way ANOVA tests were used to calculate p-values. Asterisks indicate statistically significant differences between Mock and each treatment (****P< 0.0001; **P< 0.01).
tRF-5d (Ala) associates with polyribosomes
A subset of Arabidopsis s-tRFs appears to efficiently repress translation, raising the question at which level this regulation is achieved. In organisms such as Archaea [35] or human cells [19], it has been shown that regulation of translation by short tRFs can be due to their association with ribosomal subunits and we hypothesize a similar mechanism in plants.
To determine if Arabidopsis tRF Ala20 can act similarly, in vitro binding studies were performed. For that purpose, crude lysates from Arabidopsis seedlings, pre-incubated in the presence of the synthetic oligoribonucleotide Ala20, were fractionated on sucrose gradient to separate free RNAs, ribosomal subunits and polyribosomes. The quality of the fractionation is attested by the ribosome profile (Fig. 5A) and northern blots (Fig. 5B). An enrichment of 5S rRNA was observed in the polyribosomes, ribosomes, and free ribosomal subunits fractions, while few 5S rRNA are present in the free RNA fractions. As expected, most of the tRNAs (as attested by tRNAAla) is found in the low-density fractions and in the polyribosomes where active translation occurs. When polyribosomes are disrupted in the presence of puromycin, an antibiotic causing premature chain termination of translation, the distribution of 5S rRNA and tRNAAla is shifted towards the light fractions of the sucrose gradient (right panels of Fig. 5A). The profile of the binding of tRF Ala20 follows the profile of tRNAAla (Fig. 5C). As expected, more than 98% of Ala20 is recovered in the low-density fractions. It corresponds to the excess of tRF Ala20 added to the Arabidopsis extract before fractionation on the sucrose gradient. The remaining 2% of Ala20 is mainly found in the heavy polyribosomal fractions compared to the 80S and ribosomal subunit fractions (Fig. 5B and C). By contrast, when a puromycin treatment is performed, Ala20 is not detected anymore in the polyribosomal fractions. Altogether this indicates that Arabidopsis tRF Ala20 does not efficiently associate with 80S ribosomes or with free ribosomal subunits but rather binds active polyribosomes. Similar binding experiments were performed with Ala16 and Ala33, which were previously shown to not efficiently inhibit translation in vitro. Both tRFs can associate with polyribosomes as Ala20 does (Supplemental Figure 4). In the same way, the Arg19 that does not affect mRNA expression in vitro has the same binding profile than Ala20 (Supplemental Figure 5). Thus, the capacity of tRFs to bind polyribosomes in vitro cannot explain their differential translation inhibition observed in wheat germ extract.
Figure 5.
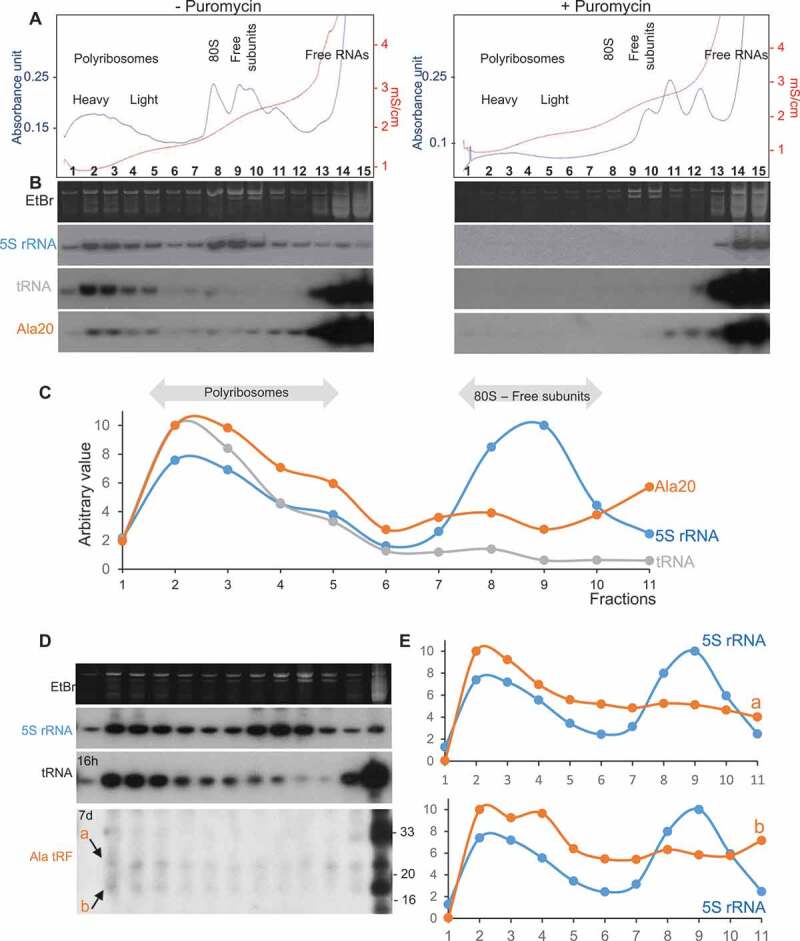
tRF-5D (Ala) associates with polyribosomes in vitro and in vivo. (A) Polyribosomes profile of Arabidopsis seedlings pre-incubated in vitro with Ala20 was determined using sucrose gradient sedimentation and OD254nm measurement (blue line) and conductivity (red line). Samples were treated or not with Puromycin. The positions of the heavy and light polyribosomes, 80S and ribosomal subunits and free RNAs are shown on the graphs. (B) RNAs extracted from each fraction shown in (A) were fractionated on 15% polyacrylamide gels and analysed by northern blots using probes specific for 5S rRNA, tRNAAla and tRF-5D (Ala). Ethidium bromide (EtBr) profiles are also shown. (C) Profiles obtained after quantification of the signals from the northern blots (5S rRNA in blue, tRNA in grey and tRF Ala20 in orange) performed with samples not treated with puromycin. Due to saturation of the free RNA fractions with the alanine probe, quantification was done only for fractions 1 to 11. An arbitrary value of 10 was given to the highest value obtained for each curve. Note that the intensity of the signals is not comparable from one curve to the other. In (D) a similar experiment was performed without adding any tRF to determine in vivo association of tRF-5D (Ala) with polyribosomes. The RNA fragments corresponding to the signals a and b are indicated by arrows. In (E) the profiles obtained after northern blots quantification with 5S rRNA probes (in blue) are compared to those obtained for a or b (in orange).
In Arabidopsis seedlings and leaves, under normal condition of growth, the amount of tRF Ala20 is very low as compared to the amount of the corresponding tRNA. Nevertheless, we raised the question of whether endogenous tRFs can be detected in polyribosomal fractions in a crude lysate from Arabidopsis seedlings under unstressed conditions. As shown in Fig. 5D, although Ala33 is visible in the light fractions, it is not detected in the polyribosomal ones, thus suggesting that in vivo this l-tRF is weakly or not associated with active ribosomes. Two weak signals (called a and b; Fig. 5D) of around 18–19 and 24–26 nt in size, abundant in the free fractions but also observed all along the gradient, are slightly enriched in the heavy fractions (Fig. 5D and E). They represent less than 0.5% of the total tRF Ala of the same size found in the free fractions. As different tRF-5D (Ala) in the size range of 19–26 nt were previously identified in sncRNA libraries from Arabidopsis leaves [28], it is presently impossible to precisely identify which ones are detected in polyribosomes. In vitro, tRF-5D Ala20 strongly inhibits translation and can bind polyribosomes; however, in vivo, the amount of s-tRF-5D (Ala) associated with polyribosomes is very low. Thus, taking into account these two facets of Ala20, we propose that tRFs may rather act as modulators rather than strong inhibitors of translation.
Discussion
Among the functions attributed to tRFs, regulation of translation has been observed in various organisms such as Archaea, protozoan, yeast, and human. In plants, previous work on sncRNAs found in the phloem sap of pumpkin and on Arabidopsis tRNA degradants suggested that plant tRFs could also have an inhibitory effect on protein biosynthesis [24,34]. Here, our work brings further evidence that indeed, in Arabidopsis, some specific tRFs possess the ability to modulate protein synthesis.
Out of a dozen of tested tRFs, only two tRF-5D, derived from tRNAAla and tRNAAsn, were shown to strongly inhibit translation. These two tRFs are 20 nt long, but we also showed that a total fraction of Arabidopsis l-tRFs can efficiently inhibit translation in vitro. Thus, the population of Arabidopsis tRFs able to negatively affect translation is likely more widespread. However, considering the high complexity of the Arabidopsis tRFs population [28], no rule can be established yet to predict which tRFs will affect the translation. Starting from total s-tRFs or l-tRFs populations of Arabidopsis, a SELEX approach may be a valuable tool to identify them. Besides, the in vitro synthesized tRFs lack modifications that can influence the affinity of nucleotide-binding, thus the use of natural post-transcriptionally modified tRFs as those generated from the Arabidopsis tRNA fraction are likely useful.
Intriguingly, while the function of tRFs as a translation inhibitor appears as a rather widespread process, the modes of action seem to differ depending on the organism and the type of tRF. Yet, only a few data on the mode of actions have been reported, they are summarized in Fig. 6. In the human alanine l-tRF, the four G residues of the 5ʹ extremity enable the formation of a G-quadruplex structure essential for translation repression by interacting with the translational silencer YB-1 and displacing the eukaryotic initiation factor eIF4G/A from mRNAs, thus inhibiting the binding of the small ribosomal subunit [18,36]. By contrast, we show here that the four Gs also present in Arabidopsis tRF (Ala) are not essential to affect protein synthesis. Interestingly, the G18 and 19 residues of tRF-5D (Ala) and tRF-5D (Asn) appear necessary. However, they belong to the conserved nucleotides of the D-loop present on mostly all tRNA molecules and are thus not sufficient to explain the specificity of inhibition. Nevertheless, similar observations were obtained in human cells with a global protein synthesis inhibition by s-tRFs, in particular, tRF-Gln19 [19]. Both in Arabidopsis and human cells, these tRFs are not or are weakly associated with AGO proteins and repression of translation is independent of the mRNA sequence, thus suggesting that their mode of action is independent of the silencing pathway. Rather, such tRFs can associate with actively elongating polyribosomes, but not efficiently with ribosomal subunits. This may suggest the existence of a still unknown but conserved process of global translation regulation by tRFs between animals and plants, and future work is required to verify this hypothesis. This is different to what has been described in the archaea Haloferax volcanii where s-tRFs inhibit translation by binding the small ribosomal subunit and competing with mRNA association [22] or in Saccharomyces cerevisiae where l-tRFs negatively affect protein synthesis by interacting with aminoacyl-tRNA synthetases and the small ribosomal subunit [37]. Finally, in Trypanosoma brucei, an l-tRF deriving from a tRNAThr induced under nutrient starvation has an opposite effect, which is to stimulate translation by binding to ribosomes and polysomes [23].
Figure 6.
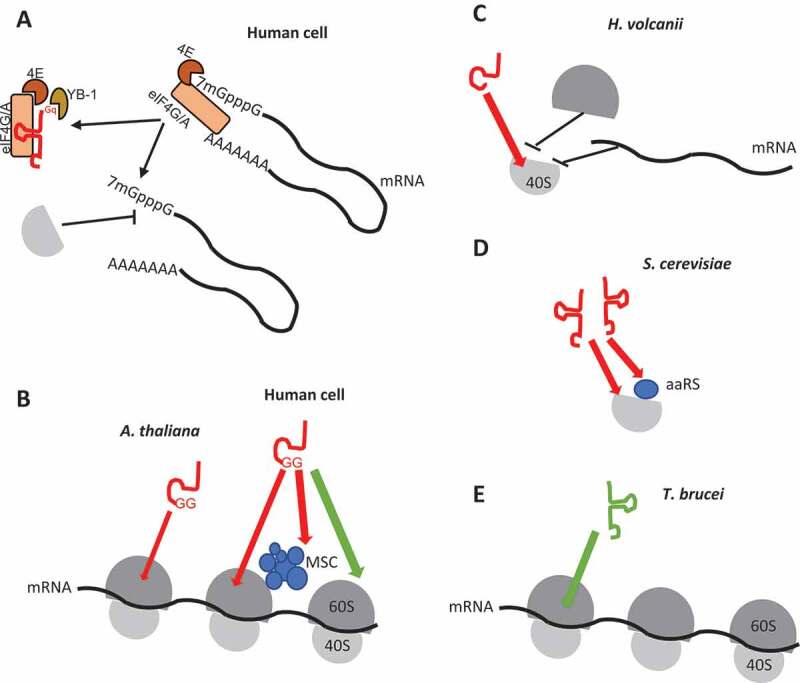
Representative examples of modulation of translation by tRFs. (A) Inhibition of translation by human l-tRF (Ala), via a G-quadruplex structure (Gq), by displacement of eIF4G/A and interaction with YB-1. (B) In human cells and this work in Arabidopsis shows that s-tRFs can associate with active polyribosomes. The two residues G18 and G19 (GG) are essential. Also, an interaction between the MultiSynthetases Complex (MSC) and tRFs have been characterized in humans. Note that in human cells, while global repression of translation has been observed [19], the protein synthesis of a specific set of mRNAs (coding for ribosomal and RNA-binding proteins) is, by contrast, stimulated [19]. (C) In the archaea H. volcanii, s-tRFs bind to the small ribosomal subunit and compete with mRNA for ribosome binding. (D) In the yeast S. cerevisiae, l-tRFs directly interact with the small ribosomal subunit and with aminoacyl-tRNA synthetases associated with ribosomes, thus impairing tRNA aminoacylation. (E) The tRNAThr 3ʹ half (depicted in green) stimulates protein synthesis in the protozoan T. brucei, by interacting with ribosomes.
How Arabidopsis tRFs associate with active polyribosomes is presently unknown. Interestingly, by providing the first tRF protein interactome, Keam et al. demonstrated recently that the human tRF (Gln)19 which globally represses translation can associate with the Multisynthetase Complex MSC, a complex known to be associated with ribosomes, and paradoxically increase the translation of ribosomal and RNA-binding proteins [20]. Such an MSC complex that was shown to interact with polyribosomes in mammals or archaea also exists in protozoans and yeast [38] and is likely present in Arabidopsis. Deciphering the Arabidopsis tRF interactome will be required to see whether a similar process may be relevant in plants or whether other partners are identified. In human, archaea and yeast, many tRFs are induced upon various stress conditions, from oxidative stress to nutrient deprivation or pH change. In these organisms, the regulation of translation by tRFs seems to be linked to the stress response. Here, in Arabidopsis, a small proportion of tRF (Ala) was detected in active polyribosomal fractions from young leaves of plants grown under normal conditions. This low amount of tRFs may represent a way to slightly and regularly modulate the global translation process under normal conditions and to play an important biological role by allowing a rapid change in translation efficiency upon demand, i.e. during plant development or under adverse growth conditions. Indeed, as already shown, the population of plant tRFs dynamically fluctuates upon various stresses or in different tissues or organs. For instance, tRFs deriving from Arabidopsis Alanine tRNAs are among the most abundant in leaves [28] and their population may even be much higher in senescing tissues such as old leaves or senescing seeds where the RNases T2 responsible for tRFs production are upregulated [31]. Unfortunately, polyribosomes fractionation from such tissues has not been successful yet to allow tRFs analysis.
Materials and methods
Plant material and growth conditions
A. thaliana is of Columbia 0 (Col-0) ecotype. Eight-week-old leaves were harvested from plants grown on the soil at 22°C under a 16 h light photoperiod. For in vitro grown seedlings, surface-sterilized seeds stratified 2 days at 4°C in the dark were sown on agar plates containing MSP01 (Caisson Labs UT, USA) supplemented with 3% (wt/vol) sucrose and grown 8 days at 22°C under a 16 h light photoperiod.
Crude leaf enzymatic preparation
The protocol used is from [31]. Briefly, 200 mg of leaves powder was resuspended in 1 ml of enzymatic extraction buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 10% glycerol, 1 mM EDTA, 1 mM β-mercaptoethanol, 50 mM Phenylmethanesulfonyl fluoride, 1X proteases inhibitor cocktail Complete® (Roche)) and shaked at 4°C for 20 min. After centrifugation at 10 000 x g for 15 min, the supernatant was used for in vitro cleavage assays.
In vitro synthesized tRNA and GFP transcript
Plasmid containing A. thaliana cytosolic tRNAAla(UGC) gene sequence was obtained previously [39]. The tRNA gene was fused to the T7 RNA polymerase promoter at its 5ʹ terminus and included a BstNI site at its 3ʹ terminus. The BstNI linearized constructs were used as a substrate to synthesize an in vitro transcript with T7 RNA polymerase using RibomaxTM transcription kit (Promega, Madison WI) under conditions previously described in [40]. Capped pGFP transcript was obtained using the same kit in the presence of a pSu9GFP recombinant plasmid [33]. Reaction was performed in a final volume of 10 µl following manufacturer’s protocol, with slight differences: rGTP was added to a 0.9 mM final concentration, the Cap analog used was from New England Biolabs, and 50 µM BSA and 50 µM RNase OUTTM (Invitrogen) were added. Reaction was run 25 min at 37°C, then 0.75 µl of rGTP (100 mM) was added, and the reaction was pursued during 2h30. To synthesize uncapped GFP transcript, no cap analog was added and rGTP was supplied to a final concentration of 7.5 mM. Plasmid DNA was eliminated by a 40 min incubation at 37°C after addition of 80 µl of water, 10 µl of DNase buffer (400 mM Tris-HCl pH 8.0, 100 mM MgSO4, 10 mM CaCl2) and 1 µl of DNase RQ1 (1U/µl, Promega). RNAs were then extracted with phenol/chloroform (1/1) and ethanol precipitated.
In vitro tRNA cleavage assay
Assays [31] were performed at 20°C in a 175 µl reaction volume containing 1 µg of synthesized tRNAAla transcript, 12.5 µl of leaf enzymatic extract and completed with water. At time points 0, 5, 10, 15, and 30 min, 30 µl aliquots were rapidly transferred into a microtube containing 200 µl of water-saturated phenol and 170 µl of tRNA extraction solution (10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1% SDS) and vortexed until all samples were collected. After centrifugation (2300 x g, 10 min), the aqueous phase was ethanol precipitated, and RNAs were analysed by northern blot as described in [28] using as a probe an oligonucleotide (Supplemental Table S1) complementary to the 5ʹ extremity of most Arabidopsis tRNAsAla (Supplementary Figure 2). As a negative control, tRNAAla transcript was incubated in the absence of leaf enzymatic extract.
Arabidopsis tRNAs and tRFs purification
Total tRNA was prepared from A. thaliana Col-0 leaves as described in [41]. This protocol includes a LiCl precipitation step that allows enrichment, in the supernatant, of RNAs of a size smaller than 150 nt (mainly 5S rRNA, tRNAs and sncRNAs). The tRNA fraction was further purified on 15% polyacrylamide gel by cutting out the piece of gel containing tRNAs after staining by methylene blue. tRNAs were eluted from the gel by a 16-h incubation at 20°C in the presence of extraction solution (500 mM ammonium acetate, 10 mM magnesium acetate, 100 mM EDTA, 0.1% SDS). Eluted tRNAs were submitted to a phenol extraction, followed by ethanol precipitation. The tRNA pellet was dissolved in water. To produce l-tRFs and s-tRFs, 5 µg of purified tRNAs were incubated 30 min at 20°C in the presence of 15 µl of leaf enzymatic extract in a total volume of 90 µl, essentially as described above for in vitro tRNA cleavage assay. After phenol extraction and ethanol purification, the RNA sample was fractionated on 15% polyacrylamide gel. Purification of l-tRFs and s-tRFs was performed as described above for total tRNA sample preparation. As a negative control, tRNAs incubated in the absence of Arabidopsis leaf enzymatic extract were purified following the same protocol.
In vitro protein synthesis assays
In vitro radiolabeled protein synthesis assays were performed using the TnT-T7 Coupled Wheat Germ Extract System kit (Promega) following manufacturer instructions. Recombinant plasmids expressing pSu9GFP (pGFP) or pSu9DHFR (pDHFR) constructs under the control of T7 promoter [33] were used and synthetic oligoribonucleotides were added at different concentrations. After fractionation of the protein samples on 15% polyacrylamide gels, gels were further stained with Coomassie blue, dried on a Whatmann paper and analysed by autoradiography and Phosphor imaging. For translation assays only, the TnT Wheat Germ Extract kit (Promega) was used following manufacturer instructions, in the presence of 1.5 µg of capped or uncapped GFP transcripts and synthetic oligoribonucleotides.
Polyribosomes extraction and RNA analysis
Polyribosomes were extracted from A. thaliana seedlings and analysed as described previously [42], with few modifications. Crude polysomal extracts were obtained from 200 mg of seedlings powder (in 600 µl), and 300 µl were resolved on 15–60% sucrose gradient (9 ml) centrifuged for 3 h at 178 000 x g (rotor SW41-Ti, Beckman Coulter). Analysis of polysome profiles was performed with an absorbance detector at 254 nm and sucrose gradients collected with a BioLogic Duoflow fractions collector (Biorad) into 13 to 16 fractions of 600 µl each. When mentioned, 1.2 µl of a synthetic oligoribonucleotide (100 µM) was added to the lysis buffer. For puromycin treatment, 200 mg of seedlings powder was homogenized in the presence of 600 µl of lysis buffer (200 mM Tris-HCl pH 9.0, 200 mM KCl, 35 mM MgCl2, 1% Brij-35, 1% Triton X-100, 1% NP40, 1% Tween 20). After 10 min of incubation on ice and 10 min centrifugation at 16 000 x g at 4°C, supernatant (340 µl) was supplied with 85 µl of puromycin solution (3 mg/ml puromycin, 1.5 M KCl) and 75 µl of 2 M KCl. After 30 min of incubation at 37°C, 27.5 µl of 10% sodium deoxycholate and 22.5 µl of water were added. After centrifugation (5 min, 16 000 x g, 4°C), 485 µl of supernatant were loaded on a 15–60% sucrose gradient.
RNAs were extracted from sucrose gradient fractions or from the crude polysomal extract by water-saturated phenol/chloroform (1/1), chloroform extracted, and ethanol precipitated. Then, they were analysed by northern blot as previously described [28].
Miscellaneous
The oligonucleotide sequences used in this study are listed in Supplementary Table S1. The tRFs were chemically synthesized and HPLC purified by the company Integrated DNA Technologies IDT.
Supplementary Material
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS) in association with the University of Strasbourg. This study is set within the framework of the “Laboratoires d’Excellences (LABEX)” TULIP (ANR-10-LABX-41) and MITOCROSS (ANR-AA-LABX-0057). We wish to thank Cyrille Megel and Elodie Ubrig for technical support and Anne-Marie Duchêne for advices.
Funding Statement
This work benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program [MITOCROSS ANR-11-LABX-0057].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
References
- [1].Raina M, Ibba M.. tRNAs as regulators of biological processes. Front Genet. 2014;5:171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2018;19:45–58. [DOI] [PubMed] [Google Scholar]
- [3].Megel C, Morelle G, Lalande S, et al. Surveillance and cleavage of eukaryotic tRNAs. Int J Mol Sci. 2015;16:1873–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shen Y, Yu X, Zhu L, et al. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med. 2018;96:1167–1176. [DOI] [PubMed] [Google Scholar]
- [5].Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2019;138:215–219. [DOI] [PubMed] [Google Scholar]
- [6].Park EJ, Kim T-H. Fine-tuning of gene expression by tRNA-derived fragments during abiotic stress signal transduction. Int J Mol Sci. 2018;19:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hsieh LC, Lin SI, Kuo HF, et al. Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots. Plant Signal Behav. 2010;5:537–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang S-Q, Sun B, Xiong Z-P, et al. The dysregulation of tRNAs and tRNA derivatives in cancer. J Exp Clin Cancer Res. 2018;37:101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dhahbi JM. 5ʹ tRNA halves: the next generation of immune signaling molecules. Front Immunol. 2015;6:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Couvillion MT, Bounova G, Purdom E, et al. A Tetrahymena Piwi bound to mature tRNA 3ʹ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell. 2012;48:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ruggero K, Guffanti A, Corradin A, et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol. 2014;88:3612–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Maute RL, Schneider C, Sumazin P, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang H, Zhang X, Liu J, et al. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 2011;67:292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schorn AJ, Gutbrod MJ, LeBlanc C, et al. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ren B, Wang X, Duan J, et al. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science. 2019;365:919–922. [DOI] [PubMed] [Google Scholar]
- [17].Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sobala A, Hutvagner G. Small RNAs derived from the 5ʹ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Keam SP, Sobala A, Ten Have S, et al. tRNA-derived RNA fragments associate with human Multisynthetase Complex (MSC) and modulate ribosomal protein translation. J Proteome Res. 2017;16:413–420. [DOI] [PubMed] [Google Scholar]
- [21].Bąkowska-Żywicka K, Kasprzyk M, Twardowski T. tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS Yeast Res. 2016;16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gebetsberger J, Zywicki M, Künzi A, et al. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909–260919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fricker R, Brogli R, Luidalepp H, et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun. 2019;10:118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang Y, Li H, Sun Q, et al. Characterization of small RNAs derived from tRNAs, rRNAs and snoRNAs and their response to heat stress in wheat seedlings. PLoS One. 2016;11:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Thompson A, Zielezinski A, Plewka P, et al. tRex: A web portal for exploration of tRNA-derived fragments in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:e1–e8. [DOI] [PubMed] [Google Scholar]
- [27].Alves CS, Vicentini R, Duarte GT, et al. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol Biol. 2017;93:35–48. [DOI] [PubMed] [Google Scholar]
- [28].Cognat V, Morelle G, Megel C, et al. The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res. 2017;45:3460–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Loss-Morais G, Waterhouse PM, Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol Direct. 2013;8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Martinez G, Choudury SG, Slotkin RK. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017;45:5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Megel C, Hummel G, Lalande S, et al. Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res. 2019;47:941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cognat V, Pawlak G, Duchêne AM, et al. PlantRNA, a database for tRNAs of photosynthetic eukaryotes. Nucleic Acids Res. 2013;41:D273–D279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sieber F, Placido A, El Farouk-Ameqrane S, et al. A protein shuttle system to target RNA into mitochondria. Nucleic Acids Res. 2011;39:e96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nowacka M, Strozycki PM, Jackowiak P, et al. Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. Plant Mol Biol. 2013;83:191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gebetsberger J, Wyss L, Mleczko AM, et al. tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017;14:1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ivanov P, O’Day E, Emara MM, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci USA. 2014;111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mleczko AM, Celichowski P, Bąkowska-Żywicka K. Transfer RNA-derived fragments target and regulate ribosome-associated aminoacyl-transfer RNA synthetases. Biochim Biophys Acta - Gene Regul Mech. 2018;1861:647–656. [DOI] [PubMed] [Google Scholar]
- [38].Raina M, Elgamal S, Santangelo TJ, et al. Association of a multi-synthetase complex with translating ribosomes in the archaeon Thermococcus kodakarensis. FEBS Lett. 2012;586:2232–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Carneiro VT, Dietrich A, Maréchal-Drouard L, et al. Characterization of some major identity elements in plant alanine and phenylalanine transfer RNAs. Plant Mol Biol. 1994;26:1843–1853. [DOI] [PubMed] [Google Scholar]
- [40].Salinas T, El Farouk-Ameqrane S, Ubrig E, et al. Molecular basis for the differential interaction of plant mitochondrial VDAC proteins with tRNAs. Nucleic Acids Res. 2014;42:9937–9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maréchal-Drouard L, Small I, Weil JH, et al. Transfer RNA import into plant mitochondria. Methods Enzymol. 1995;260:310–327. [DOI] [PubMed] [Google Scholar]
- [42].Merret R, Nagarajan VK, Carpentier M-C, et al. Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Res. 2015;43:4121–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


