ABSTRACT
Originally considered futile degradation products, tRNA-derived RNA fragments (tdRs) have been shown over the recent past to be crucial players in orchestrating various cellular functions. Unlike other small non-coding RNA (ncRNA) classes, tdRs possess a multifaceted functional repertoire ranging from regulating transcription, apoptosis, RNA interference, ribosome biogenesis to controlling translation efficiency. A subset of the latter tdRs has been shown to directly target the ribosome, the central molecular machine of protein biosynthesis. Here we describe the function of the mammalian tRNAPro 5ʹ half, a 35 residue long ncRNA associated with ribosomes and polysomes in several mammalian cell lines. Addition of tRNAPro halves to mammalian in vitro translation systems results in global translation inhibition and concomitantly causes the upregulation of a specific low molecular weight translational product. This tRNAPro 5ʹ half-dependent translation product consists of both RNA and amino acids. Transfection of the tRNAPro half into HeLa cells leads to the formation of the same product in vivo. The migration of this product in acidic gels, the insensitivity to copper sulphate treatment, the resistance to 3ʹ polyadenylation, and the association with 80S monosomes indicate that the accumulated product is peptidyl-tRNA. Our data thus suggest that binding of the tRNAPro 5ʹ half to the ribosome leads to ribosome stalling and to the formation of peptidyl-tRNA. Our findings revealed a so far unknown functional role of a tdR thus further enlarging the functional heterogeneity of this emerging class of ribo-regulators.
KEYWORDS: tRNA-derived fragments, tRNA halves, ncRNA, rancRNA, translation regulation, ribosome
Introduction
In early high-throughput studies, tRNA-derived fragments (tdRs) were identified in a wide variety of model systems yet they were considered as degradation products devoid of physiological relevance [1]. Nowadays, with the improved deep-sequencing protocols, tailor-made bioinformatics tools, and accumulating functional studies it became clear that tdRs represent a so far unknown class of regulatory ncRNAs. It could be demonstrated that many tdRs, which include tRNA half molecules as well as even shorter tRNA fragments (tRFs), are specifically processed by endonucleases to participate in and regulate diverse biological process [2]. tRNA halves and tRNA fragments are involved in controlling cell proliferation, cellular homoeostasis, priming of viral reverse transcriptases, regulating gene expression, trans-generational epigenetic inheritance, RNA processing, modulation of the DNA damage response, tumour suppression, regulation of transposition, neurodegeneration, ribosome biogenesis, and translation regulation [2–5]. Accumulating evidence further link misregulated tRF production to certain human diseases, while other studies highlight tRFs as potential biomarkers for disease states [6–8].
Recently, ribosome-associated non-coding RNAs (rancRNAs) were recognized as a new class of regulatory ncRNAs. It was shown that small rancRNAs influence protein synthesis by directly targeting the ribosome in organisms representing all three domains of life [9]. A major fraction of these rancRNAs is processed from longer functional precursors including mRNAs, snoRNAs, and tRNAs [10,11]. Functional analyses of selected ribosome-bound tRFs revealed an unexpected functional heterogeneity. For example, an alkaline stress-induced archaeal tRNAVal fragment associates with the small ribosomal subunit and globally inhibits translation initiation by competing with mRNA binding [12,13]. Another study suggests that yeast ribosome-bound tRFs have the potential of inhibiting tRNA aminoacylation [14]. On the other hand, in the human parasite T. brucei a tRNAThr 3ʹ half associates with ribosomes under nutrient deprivation conditions and stimulates global translation during stress recovery [15]. Ribosome-targeted tdRs have recently also been linked to cell proliferation in mammalian cells and might promote cancer progression in a mouse model [16]. These findings reveal ribosome-bound tdRs as functionally versatile regulators capable of fine-tuning protein biosynthesis in a stress-dependent manner.
Despite recent studies highlighting regulatory roles during gene expression in different model organisms, rancRNA has not yet been studied in detail in human cells or any other mammalian species so far. In this study, we performed screening for ribosome-bound tRFs in different mammalian cell lines and uncovered the highly conserved tRNAPro 5ʹ half. We demonstrate that upon ribosome-binding the tRNAPro 5ʹ half inhibits global protein biosynthesis with the concomitant appearance of a low molecular weight translation product, dubbed ProTiP. Cumulative experimental evidence suggests ProTiP to be peptidyl-tRNA likely accumulating as a by-product of tRNAPro half-mediated translation inhibition.
Results
Expression and ribosome-association of the tRNAPro half in mammalian cell lines
Previous deep sequencing of the small RNome in CHO-K1 cells identified differentially expressed miRNAs [17,18] and piRNAs [19]. Here we re-analysed the data for other small ncRNAs and indeed found indications for high expression of tRNA halves and shorter tdRs (Fig. S1). The expression and function of tRFs and halves were shown before to be associated with stress in several studies, therefore, CHO-K1 cells from different stress conditions (heat shock, cold shock, stationary phase growth, oxidative stress, and oxidative stress followed by recovery) were used for screening for tdRs via northern blot analysis. Since there are characterized examples of tRFs and tRNA halves directly binding to ribosomes and in this way regulating translation [13,15], northern blot screening was performed on total RNA and on RNA isolated from the crude ribosome pellets in parallel. As a result of this screen, the tRNAPro 5ʹ half was detected predominately in the ribosome-containing pellet, but under stationary growth conditions also in the total RNA fraction (Fig. 1A). The tRNAPro 5ʹ half was also apparent in other mammalian cell lines such as in HeLa, HEK, BON, NCI, and Hep3B cells (Fig. 1B). As observed in CHO cells, the tRNAPro half was constitutively expressed and did not show a clear change upon stress induction neither in HeLa nor in HEK cells (Fig. S2). Polysome profiling and consequent northern blot analysis confirmed the ribosome association of the tRNAPro half in CHO, HEK, and HeLa cells. In all three cell lines the tRNAPro half is present mainly in the 80S monosome fraction (HeLa, HEK) and in CHO and HEK cells also in the 60S subunit peak (Fig. 1C-E). The northern blot signal obtained in the small ribosomal subunit fractions (e.g. Fig. 1C) most certainly results from a spill-over from the light gradient fractions.
Figure 1.

Expression and ribosome association of the tRNAPro half in different mammalian cell lines. (A) Northern blot analysis of the tRNAPro 5ʹ half of 20 µg of total RNA or 20 µg of RNA isolated from the crude ribosome pellet (P100) from CHO-K1 at several stress conditions: heat shock (heat), oxidative stress (oxid), oxidative stress with recovery, stationary (stat) growth phase and as a control exponential (exp) growth phase. Lower panel shows the same northern blot in the region of the tRNA half with increased contrast. (B) Northern blot analysis of the tRNAPro 5ʹ half of 30 µg total RNA isolated from HeLa, HEK, BON, NCI, and Hep3B cells. Ethidium bromide-stained 5 S rRNA serves as a loading control. (C-E) Polysome profiling of exponentially grown CHO-K1, HeLa, and HEK cells, respectively (top panels). RNA was isolated from the indicated fractions (free RNA, 40 S, 60 S, 80 S, and polysomes), its integrity, and identity monitored by agarose gel electrophoresis (middle panel) and used for northern blot analysis (lower panels). Full length (FL) tRNAPro and the tRNAPro 5ʹ half are indicated.
Binding of tRNAPro 5ʹ half to ribosomes and ribosomal subunits was confirmed by in vitro filter binding assays. P32-labelled tRNAPro half was shown to bind the crude ribosomal pellets (P100) isolated from CHO or HeLa cells (Fig. 2A). Filter binding assay using human or yeast ribosomal subunits showed binding to both 60 S and 40 S subunits, with slightly varying binding preferences (Fig. 2A). An unrelated short archaeal ncRNA was used as binding specificity control [20] which resulted in no or very diminished binding to mammalian ribosomes. The obtained binding data with the tRNAPro half are compatible with a binding site at or near the subunit interface. To test this assumption, UV crosslinking experiments employing a 5ʹ-32P labelled and 4-Thio-U-containing tRNAPro half was performed. The tRNAPro half crosslinked exclusively to 18S rRNA of HeLa 80S ribosomes (Fig. 2B). Using a similarly 5ʹ-32P labelled and 4-Thio-U-containing tRNA half from T. brucei [15] did not result in any detectable crosslink to rRNAs (Fig. S3) therefore highlighting the specificity of the observed effects. Employing a 3ʹ-biotinylated and 4-Thio-U-containing tRNAPro half in crosslinking and streptavidin pull-down experiments did not identify any ribosomal protein (Table S1), thus suggesting the ribosome binding site being primarily composed of rRNA. RNaseH-mediated cleavage guided by anti-sense DNA oligos complementary to different parts the 18S rRNA allowed us to narrow down the cross-linking site(s) to the last 306 nucleotides of the 3′ end of 18S rRNA (Fig. 2C, D). Highlighting this region in the context of the human 80S cryo-EM structure [21] showed its location at the subunit interface (Fig. S4). These data are therefore in line with the assumption that both subunits contribute to the binding site as suggested by the filter binding results (Fig. 2A).
Figure 2.

Binding of the tRNAPro half to ribosomes and ribosomal subunits in vitro. (A) Binding of the P32-labelled tRNAPro 5ʹ half to the crude ribosomal pellet (P100) and gradient-purified ribosomal subunits (40 S and 60 S) isolated from CHO cells, HeLa, and yeast cells was monitored by filter binding. Negative control (-) shows background signals in the absence of ribosomal particles. An unrelated small ncRNA (control RNA) from H. volcanii showed no (HeLa, yeast) or severely reduced (CHO) binding to ribosomal particles and serves as a binding specificity control. (B-C) Photocrosslinking of the tRNAPro half to human ribosomes. 5ʹ P32 – labelled tRNAPro half-carrying 4-thio-uracils was crosslinked to crude HeLa ribosomes at 366 nm followed by protease K treatment and RNA isolation. (B) After photo-crosslinking, RNAs were resolved on a 1% agarose gel. The ethidium bromide-stained gel and the autoradiogram thereof are shown. 18S rRNA and 28 S rRNA are labelled. (C) After photo-crosslinking, purified RNAs were subjected to RNase H treatment guided by DNA oligos complementary to different regions of 18S rRNA and the cleavage products were separated on 4% polyacrylamide denaturing gels. Ethidium bromide staining and autoradiogram are shown. Expected sizes of the produced rRNA fragments are indicated above each lane.
tRNAPro 5ʹ half inhibits translation and triggers a low molecular weight product
To test if the ribosome-associated tRNAPro half can influence protein synthesis, in vitro translation was performed based on a CHO cell lysate in the presence of the synthetic 35 nt long tRNAPro half. Addition of tRNAPro half reproducibly led to an inhibition of global protein biosynthesis to a varying extent (Figs. 3A, B). However, when using a rabbit reticulocyte-based translation system with a single mRNA reporter, the picture became clearer and very low levels of protein synthesis were reproducibly observed upon addition of the tRNAPro 5ʹ half (Fig. 3C). Addition of other tRNA 5ʹ halves had only comparably mild effects on translation (Figs. 3C, D). Addition of a DNA version of the tRNAPro half on the other hand showed no effect on protein biosynthesis (Fig. S5). Based on these experiments we decided to focus our subsequent research on the tRNAPro 5ʹ half.
Figure 3.

Effect of tRNAPro 5ʹ half on translation. (A) Autoradiogram of an SDS polyacrylamide gel after in vitro translation in the presence of the synthetic tRNAPro half using a CHO cell lysate. (B) Quantification of six in vitro translation experiments performed with CHO cell lysates in the absence (-) or presence (+) of the tRNAPro half. Values were normalized to the in vitro translation reaction with no tRNAPro half added. Box shows ± 25% of the mean, error bars show standard deviation. (C) Autoradiogram of an SDS polyacrylamide gel after in vitro translation of one reporter in the absence (-) presence of various synthetic tRNA 5ʹ halves (5 or 8 µM) using a rabbit reticulocyte cell lysate. In all cases, the tRNA 5ʹ halves are abbreviated by their respective amino acid three letter code. The asterisk marks the position of the full-length reporter protein. (D) Quantification of three in vitro translation experiments performed with rabbit reticulocyte cell lysate and one mRNA reporter in the presence of different tRNA 5ʹ halves (5 or 8 µM). Values were normalized to the in vitro translation reaction in the absence of the tRNAPro half. The horizontal lines indicate the mean and the error bars show standard deviations. (E) CHO in vitro translation in the presence of 8 µM synthetic tRNA halves. The apparent tRNAPro half-induced product (ProTiP) is indicated with a black arrowhead (here and in all panels). (F) CHO in vitro translation with increasing amounts of tRNAPro half added. (G) Quantification of ProTiP formation of three in vitro translation reactions as a function of increasing tRNAPro 5ʹ half concentration. The horizontal lines indicate the mean and the error bars show standard deviations. In (A), (C), (E), and (F) the coomassie stained protein bands of the lysate serve as a loading control. In (B) and (D) significant differences were determined using the 2-tailed paired Student’s t-test (***p < 0.001, **p < 0.01, *p < 0.05).
During these experiments, we noticed that besides inhibition of translation, addition of the synthetic 5ʹ tRNAPro half into in vitro translation reactions led to the formation of a specific band that migrates like a ~ 17 kDa product on SDS-containing polyacrylamide gels (Figs. 3A, C). This proline tRNA half-induced product (to which we refer to as ProTiP) is specific to the tRNAPro 5ʹ half and is not produced in the presence of any other tested tRNA half (Fig. 3C, E). Formation of ProTiP was found to be dose-dependent as addition of increasing amounts of the tRNAPro 5ʹ half stimulates its production (Fig. 3F, G).
Alignment of different tRNAPro genes shows high sequence identities in its 5ʹ region among several species (Fig. S6) suggesting a possible conservation of the tRNAPro 5ʹ half function. To test the effect of this tRNA half on translation in other biological systems, the same tRNAPro half was added to in vitro translation systems based on HeLa, HEK, or yeast cell extracts. Similar to the CHO and rabbit reticulocytes systems, addition of the tRNAPro 5ʹ half led to global translation inhibition and accumulation of ProTiP in all tested systems (Fig. 4A-C). We noticed that the addition of the tRNAPro half in the presence of low concentrations of the translation elongation inhibitor cycloheximide did not inhibit but even enhanced ProTiP formation. This increased ProTiP abundance, however, does not result from a putative stimulatory effect of low cycloheximide concentrations on tRNAPro half ribosome binding (Fig. S7). In cycloheximide titration experiments it turned out that only under higher concentrations of the inhibitor also ProTiP formation was reduced (Fig. 4D). The fact that tRNAPro half-mediated translation inhibition and ProTiP formation were observed in mammalian and yeast cell extracts suggests that the ribosome-binding site and/or the mode of action is conserved between different eukaryal translation systems. To test if ProTiP formation represents a peculiarity of in vitro translation systems or is also produced in living cells, the tRNAPro half was transfected into HeLa cells. Metabolic labelling in the presence of 35S methionine after transfection led to the production of ProTiP in vivo (Fig. 4E). These data show that tRNAPro half-mediated ProTiP production is not solely an in vitro translation phenomenon but occurs in living mammalian cells as well.
Figure 4.
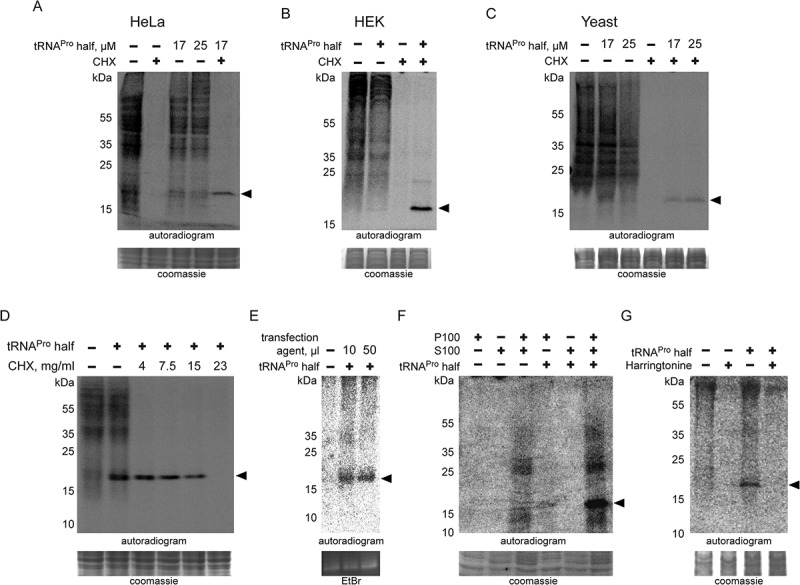
Effect of the tRNAPro half on translation in different eukaryotic systems. Autoradiograms of in vitro translation reactions performed using (A) HeLa, (B) HEK and (C) yeast extracts in the absence or presence of the tRNAPro 5ʹ half. Cycloheximide (CHX) with an f.c. of 7.5 mg/ml was used to inhibit global translation. ProTiP is indicated with a black arrowhead. (D) In vitro translation using CHO extracts and ProTiP formation (arrow head) was monitored in the presence of increasing amounts of cycloheximide (CHX). (E) Formation of ProTiP in vivo. 300 pmol of synthetic tRNAPro half was transfected into HeLa cells and the cells were subsequently incubated with S35 methionine. RNA isolated after metabolic labelling performed with (+) or without (-) transfected tRNAPro half was separated on a denaturing gel and visualized via autoradiography. The ethidium brome (EtBr) strained gel serves as a loading control. (F) Autoradiogram of in vitro translation reactions with HeLa ribosome-containing pellets (P100) or the corresponding ribosome-free supernatants (S100). Newly synthesized proteins, as well as ProTiP (black arrowhead), were produced only once P100 and S100 were combined. 100 pmol of tRNAPro half was used. (G) Inhibition of global translation in CHO extracts and ProTiP formation in the absence (-) or presence (+) of harringtonine (f.c. 2.5 mg/ml). In (A), (B), (C), (D), (F), and (G) the coomassie stained protein gels serve as loading controls.
ProTiP formation requires genuine protein biosynthesis
Several of the above-shown experiments suggest that ProTiP formation is a translation-dependent event. To substantiate these observations, HeLa cell extracts were divided via 100,000 xg centrifugation into the crude ribosomal pellet (P100) and the post-ribosomal supernatant (S100). In vitro translation activity only occurred once P100 and S100 fractions were combined (Fig. 4F). Addition of the tRNAPro 5ʹ half to P100 or S100 separately did neither result in translation activity nor in ProTiP formation. However, upon combination of the P100 and S100 fractions protein synthesis occurred and caused the appearance of ProTiP in the presence of the tRNAPro half (Fig. 4F). This experiment demonstrates that ProTiP formation indeed requires genuine translation. Support for this conclusion came from the fact that ProTiP synthesis could be influenced by the translational inhibitor harringtonine, which immobilizes eukaryotic ribosomes immediately after the initiation of translation. Harringtonine completely abolished global translation and also the production of ProTiP in the presence of the tRNAPro half (Fig. 4G). As mentioned above, similar effects were observed upon addition of elevated concentration of the tRNA translocation inhibitor cycloheximide (Fig. 4D).
The chemical nature of ProTiP
ProTiP requires translation and it migrates as a ~ 17 kDa band on SDS gels thus indicating ProTiP of being a polypeptide. Indeed, in vitro translation followed by protease K digestion completely eliminated the ProTiP band on the gel (Fig. 5A). These findings are compatible with the view that addition of tRNAPro 5ʹ half into in vitro or in vivo translation reactions stimulates the production of one or several proteins that run as ~17 kDa products on denaturing gels. Ethidium bromide staining of the SDS gel showed that small RNAs, in particular tRNAs, also migrated in the same region of the gel as ProTiP (Fig. 5B). To test if ProTiP also contains RNA, in vitro translation reactions were subjected to an RNase I treatment. ProTiP was shown to be RNase sensitive since the band disappeared after nuclease digestion (Fig. 5A). The observation that ProTiP could be extracted into the water phase during phenol/chloroform extraction further indicates that ProTiP contains not only a peptidyl or protein part but also an RNA component (Fig. 5C).
Figure 5.

Chemical nature of ProTiP. (A) The products of in vitro translation using CHO cell lysates were incubated with proteinase K or RNase I. Protease K digests all newly translated and cell lysate proteins, as well as ProTiP (arrowhead). Also, RNAse I digestion removes ProTiP. The coomassie or ethidium bromide-stained gels serve as controls for efficient protein and RNA digestions, respectively. (B) Spike-in of yeast bulk tRNA (lane 3) after CHO in vitro translation in the absence (lane 1) or presence (lane 2) of 8 µM tRNAPro 5ʹ halves shows that ProTiP (arrowhead) migrates in the size range of tRNAs in the SDS polyacrylamide gel used. The gel was stained with coomassie (loading control) or with ethidium bromide (for visualization of the spiked-in tRNAs). (C) In vitro translation reactions (with or without tRNAPro halves) were loaded before (-) or after (+) phenol/chloroform/isoamyl alcohol (PCI) extraction on a denaturing SDS gel. ProTiP (black arrowhead) can be extracted into the water phase during PCI treatment. (D) Autoradiogram of an SDS polyacrylamide gel after in vitro translation in the presence of the internally 32P-labelled tRNAPro half (open arrowhead). Migration of the labelled tRNAPro half did not change after reactions in the presence of the cell lysate and the translational mix, thus demonstrating that ProTiP does not contain the tRNAPro 5ʹ half. Expected size of ProTiP is indicated with the black arrowhead.
To better reveal the chemical identity of ProTiP, we first focused on the characterization of its RNA part. By utilizing a 5ʹ-32P labelled (Fig. 5D) or a 3ʹ biotinylated tRNAPro half (Fig. S8) during in vitro translation we could clearly demonstrate that the tRNAPro 5ʹ half itself is not part of ProTiP. To better estimate the size of ProTiP, samples extracted via phenol/chloroform treatment after in vitro translation were loaded on an acidic gel. On acidic gels, ProTiP migrated in the range of the tRNA pool, slightly above methionyl-tRNAs (Fig. 6A). To characterize the 3ʹend of ProTiP, a polyadenylation experiment was designed. If the RNA moiety of ProTiP has a free 3ʹ end, it will be susceptible to polyadenylation and consequently would migrate more slowly on polyacrylamide gels. In contrast, if the RNA 3ʹ end is protected it would be insensitive to polyadenylation and, thus, would not change its electrophoretic mobility. Polyadenylation performed on RNA extracted after in vitro translation in the presence of tRNAPro half lead to an upshift of the majority of RNA on ethidium bromide-stained gels, while the autoradiogram of the same gel showed that ProTiP migration remained unchanged upon polyadenylation (Fig. 6B). As a control, northern blot analysis was performed on the same gel using a probe against the 5.8S rRNA. After polyadenylation, the 5.8 S rRNA signal was quantitatively upshifted on the gel demonstrating efficient poly(A) addition (Fig. 6B). Therefore, the insensitivity of ProTiP to polyadenylation demonstrates that it does not contain a free 3ʹ OH end. Electrophoretic mobility of ProTiP within the size range of tRNAs on acidic gels and the insensitivity to polyadenylation suggests that ProTiP could be a tRNA with one or several amino acids attached to its 3ʹ end thus resembling aminoacyl – or peptidyl-tRNAs, respectively.
Figure 6.

ProTiP is likely a peptidyl-tRNA. (A) Autoradiogram and ethidium bromide staining of an acidic gel after in vitro translation performed in the absence (-) or presence (+) of the tRNAPro 5ʹ half. Charged tRNAiMet and tRNAMet are labelled by asterisks. ProTiP is indicated with the black arrowhead. (B) Autoradiogram of an 8% denaturing polyacrylamide gel showing the position of ProTiP (arrowhead) before (-) and after (+) polyadenylation. Middle and left panels: Ethidium bromide staining and northern blot against 5.8S rRNA demonstrates the efficiency of polyadenylation. (C) Copper sulphate treatment of RNA isolated after in vitro translation performed in the presence (+) or absence (-) the tRNAPro half. The location of ProTiP, which was insensitive to copper sulphate treatment, is indicated in the autoradiogram with the black arrowhead. The Met-tRNAMet molecules are labelled with asterisks. (D) Fractions of the polysome profile after in vitro translation in the presence of the tRNAPro half were subjected to isopropanol precipitation. Precipitated RNA was separated on a denaturing gel and visualized via phosphorimaging. ProTiP is indicated with the black arrowhead. (E) Binding of ProTiP to purified HeLa 80 S in vitro with and without pre-incubated bulk tRNAs. Ribosome-bound 35S-ProTiP was determined by filter binding. (F) Quantification of three individual binding experiments. Values were normalized to the binding of ProTiP to ribosomes without the addition of bulk tRNAs. The error bars show standard deviations (*p < 0.05).
To distinguish if ProTiP is an aminoacyl-tRNA or a peptidyl-tRNA, copper sulphate treatment was performed. Copper sulphate selectively deacylates aminoacyl-tRNAs with a free N-terminus but leaves peptidyl-tRNA intact [22]. This method is based on the ability of heavy metal ions to form stable complexes with α-amino acid esters and to promote hydrolysis of the ester bond, while any substituent on the α-carbon of the amino acid ester decreases the rate of hydrolysis [23]. Copper sulphate treatment on RNA extracted after in vitro translation showed that ProTiP stays intact while the methionyl-tRNAs were effectively deacylated (Fig. 6C). Polysome profiling performed after in vitro translation reactions in the absence or presence of the tRNAPro 5ʹ half showed that addition of tRNAPro half leads to a slight but reproducible increase of the 80S monosome peak on polysome gradients (Fig. S9). To trace ProTiP within the density gradient, fractions of the polysome gradient were subjected to RNA precipitation and the samples subsequently loaded on a denaturing SDS gel. The autoradiogram of the dried gel reproducibly showed the association of ProTiP with the 80S monosome fraction (Fig. 6D). Cumulatively, the presented evidence suggests that ProTiP is a tRNAPro 5ʹ half-dependent translation product composed of both an RNA and a peptidyl part and is likely a peptidyl-tRNA. To substantiate this conclusion, two control experiments were performed. First, we subjected ProTiP to low concentrations of RNase T1 or RNase I and expected the radioactive signal to down shift in denaturing gels. Indeed, the full-length ProTiP band diminished with the concomitant appearance of specific shorter radioactive products as one would expect if ProTiP is a peptidyl-tRNA (Fig. S10). Second, we designed ribosome-binding competition experiments of 35S-ProTiP with deacylated tRNAs under P-site binding conditions. Indeed, the signal of ribosome-bound ProTiP reproducibly decreased upon tRNA addition indicating that they share an overlapping binding site on the 80S ribosome (Fig. 6E, F).
Discussion
In this study, we screened for tRNA halves and tRNA fragments (tRFs) in various mammalian cell lines and uncovered a ribosome-associated tRNAPro 5ʹ half (Fig. 1C-E and Fig. S2). Ribosome association was confirmed in vitro via filter binding assays and photo-crosslinking experiments (Fig. 2). Addition of synthetic tRNAPro 5ʹ half to in vitro translation reactions showed unique effects on protein biosynthesis. First, the tRNAPro 5ʹ half inhibits global translation (Fig. 3A-D). Previously, it has been described that several stress-induced tRNA 5ʹ halves (5ʹ tiRNAs) inhibit translation in human cells by titrating away crucial translation initiation factors [24] and promote stress granule formation [25]. In contrast to the previously described tiRNAs, the tRNAPro 5ʹ half identified here does not contain the 5ʹ terminal oligoguanosine motif uncovered before on tiRNAs of being central for its inhibitory function and it also does not show stress-dependent expression (Fig. 1A). Moreover, while a DNA analogue of a 5ʹ tiRNA was sufficient to inhibit translation [26], a DNA version of the tRNAPro half failed to inhibit protein biosynthesis (Fig. S5). These findings also suggest that the interaction of the tRNA half with the rRNA target is not entirely depending on simple base pairing. Altogether, our data suggest that the tRNAPro 5ʹ half possesses a so far unknown mechanism of translation inhibition in mammalian cells. The second effect of the 5ʹ tRNAPro half on translation was the formation of a low molecular weight product that is bound to 80S monosomes (Fig. 6D) and migrates like a ~ 17 kDa product in SDS gels (Fig. 3A,C,E,F). The tRNAPro half–induced product (named ProTiP) was not only observed in vitro but also after the transfection of tRNAPro halves into HeLa cells (Fig. 4E). By performing streptavidin pull-down experiments employing a 3ʹ biotinylated tRNAPro half after in vitro translation (Fig. S8), or by utilizing a 32P-labelled tRNAPro half during the translation reaction (Fig. 5D), we could exclude the possibility that the tRNAPro half itself is part of ProTiP. Both, translation inhibition and ProTiP formation were observed in several eukaryotic translation systems therefore indicating a conserved function and ribosome-binding site of the tRNAPro 5ʹ half (Fig. 4A-C). Cumulative experimental evidence indicates that ProTiP is a hybrid molecule containing both an RNA and a peptidyl part (Fig. 5A, C and Fig. S10). Furthermore, ProTiP migrates in denaturing gels in the size range of tRNAs (Figs. 5B and 6A-C), associates with 80S monosomes in polysome profiles (Fig. 6D) and can be chased off the ribosome by tRNA addition (Fig. 6E,F). Taken together the obtained data are in line with the conclusion that ProTiP is a peptidyl-tRNA.
Previous studies have shown that under certain conditions distinct aminoacyl-tRNAs are modified at the N-terminus resulting in the block of translation [27,28]. In both cases, the produced modified aminoacyl-tRNAs were detected as a sharp clean band migrating on polyacrylamide gels in the region of the tRNA pool, in a similar fashion like ProTiP. However, if ProTiP was an N-terminally modified aminoacyl-tRNA, its production would not be translation dependent and ProTiP would migrate together with free tRNAs on polysome profiles after in vitro translation. The fact that after in vitro translation ProTiP stays associated with 80S monosomes, but was absent in the light density gradient fractions containing unbound RNAs (Fig. 6D) suggests that the accumulated product is a peptidyl-tRNA rather than N-modified aminoacyl-tRNA. Thus, our data are compatible with the scenario that binding of the tRNAPro 5ʹ half to ribosomes might lead to ribosome stalling, and consequently to translation inhibition followed by the accumulation of peptidyl-tRNA(s) (ProTiP) (Fig. 7). Despite the fact that we were unable to unequivocally identify the molecular nature of ProTiP by a variety of complementary methods, several conclusions can be drawn based on our findings. Since ProTiP always migrates as a sharp band on denaturing gels suggests that it contains one specific tRNA body but could still be heterogeneous in the length and composition of the peptidyl part. As ProTiP is produced in different translation systems on different mRNAs, it is likely that the formed peptidyl-tRNAs have heterogeneous amino acid sequences with a varying number of residues. Variation in the amino acid sequence and slight differences of the peptidyl length would not dramatically change the bulk charge of the peptidyl-tRNA, and consequently its migration on denaturing polyacrylamide gels. Therefore, it is reasonable to assume that ProTiP represents a group of peptidyl-tRNAs with one particular tRNA body and different short peptidyl chains. The ability to extract ProTiP during a phenol/chloroform treatment into the water phase indicates that the peptidyl part is probably shorter than 80 amino acids [29]. Since cycloheximide allows only one round of elongation after ribosome binding [30], the production of ProTiP in the presence of cycloheximide pre-incubated with ribosomes (see Material and Methods) suggests that the peptidyl part contains only a few amino acids.
Figure 7.
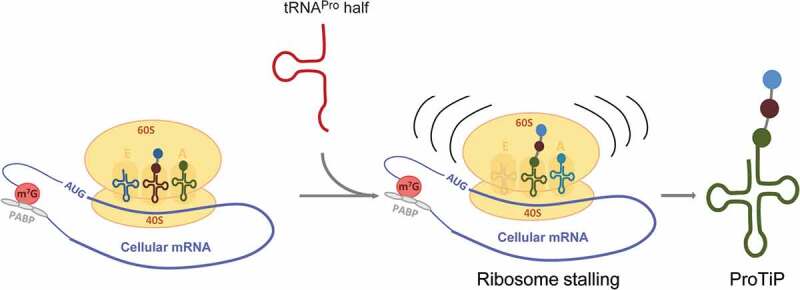
Model of the effect of the tRNAPro 5ʹ half on translation. The tRNAPro is processed into the tRNAPro 5ʹ half through cleavage at the anticodon region. The tRNAPro 5ʹ half binds to the ribosome and likely causes ribosome stalling. Consequently, translation is inhibited globally resulting in the accumulation of the peptidyl-tRNA inside the stalled ribosomes that we identified as ProTiP.
Some of the previously identified tdRs, including short tRFs as well as tRNA half molecules, have been appreciated in the recent past of possessing biological roles in various model organisms and are therefore more than degradation intermediates of tRNA homoeostasis. Accumulating evidence suggests that tdRs are a functionally very heterogonous class of ribo-regulators [2,6,31]. A subclass of these tRNA fragments effectively associates with the ribosome and are therefore included into the growing class of ribosome-associated ncRNAs (rancRNAs) [9]. However, even those tdRs that target the ribosome turned out to influence protein production in a markedly different manner. While some affect translation globally in an inhibitory [12,24] or stimulatory [15] manner, the tRNAPro 5ʹ half reported here, reveals yet another aspect of tdR biology. While it is functionally analogous to previously identified tRNA 5ʹ halves [24] in the sense that it globally down regulates protein production, it is unique in triggering the formation of ProTiP, a small molecular weight product. Our attempts to characterize the molecular nature of ProTiP suggest that it is peptidyl-tRNA likely produced as a consequence of tRNAPro half-mediated ribosome stalling. Future work will have to reveal whether ProTiP is a mere byproduct of the ribosome arrest or if it possesses a physiological role by itself. While previously reported tRNA halves capable of inhibiting translation were generated by angiogenin specifically during certain stress conditions, called tiRNAs [24], the tRNAPro 5ʹ half reporter herein is produced in a stress-independent manner. Therefore, it is safe to suspect that the tRNAPro 5ʹ half is generated via a different biogenesis pathway and it might fulfil a cellular role distinct from tiRNAs in mammalian cells.
Materials and methods
Cell cultivation and stress application
CHO-K1 cells were cultivated in CD CHO medium (Gibco) supplemented with 8 mM glutamine (Gibco), 1x PenStrep (Gibco), and 2 ml/l anticlumping agent (Gibco) at 37ºC, 5% CO2. CHO cells were harvested during exponential growth at approximately 48 h after seeding with a cell density of 0.2 × 106 cells/ml. HEK cells were cultivated in DMEM medium (Gibco) supplemented with 10% FCS (Thermo Scientific), 4 mM Glutamine (Gibco), and 1 x PenStrep (Gibco). For HeLa cells RPMI (Gibco) medium supplemented with 10% FCS, 4 mM glutamine (Gibco) and 1 x PenStrep (Gibco) was used. For northern blot analysis, the following stress conditions were applied: (I) stationary growth phase: for CHO-K1 cells 7 days after seeding with a cell density of 0.2 × 106 cells/ml, for HeLa and HEK cells growth to the overconfluent stage, (II) heat shock treatment: 42°C for 30 min, (III) oxidative stress: incubation with 2 mM H2O2 for 2 h, (IV) recovery from oxidative stress: 2 mM H2O2 for 2 h followed by 2 h recovery in fresh media, (V) nutritional deprivation by incubation the cells in PBS for 2 h.
Northern blot analysis
For northern blotting, 10–30 μg of RNA was separated by denaturating polyacrylamide gels (7 M urea) and subsequently electro-blotted onto nylon membranes (Amersham Hybond N+, GE Healthcare) as described [13]. All membranes were hybridized to a 32P-5′-end-labelled DNA probe complementary to the 5ʹ part of tRNAPro AATCATACCCCTAGACCAACGAGCC.
Bulk ribosome pelleting
To generate crude ribosomal pellets, cell lysates were loaded on 0.5 ml 1.1 M sucrose cushion in 30 mM Hepes pH 7.6, 150 mM KOAc, 3.9 mM Mg(OAc)2, 4 mM DTT, 1 mM PMSF and centrifuged for 2.5 h at 200,000 xg in a Sorval M120+ miniultracentrifuge equipped with an S140AT rotor (Thermo Scientific). Obtained ribosomal pellets were resuspended in 1 x resuspension buffer (30 mM HEPES-KOH, pH 7.6; 150 mM KOAc, 3.9 mM Mg(OAc)2; 4 mM DTT; 1 x Tm complete (Roche); RNase inhibitor (Thermo Fisher or Promega). To prepare the more concentrated S100 extracts used in HeLa in vitro translation experiments, the cell lysate was directly ultracentrifuged without a sucrose cushion.
Polysome profiling
For polysome profiling, exponentially growing cells were treated with freshly prepared cycloheximide to an f. c. of 100 µg/ml for 10 min. Cells were washed with PBS containing 100 µg/ml cycloheximide and opened in 1 x resuspension buffer containing 100 µg/ml cycloheximide by passing through a 25 G needle 30 times. Cell lysates were layered onto a 10–50% (w/v) linear sucrose gradient prepared in 30 mM HEPES-KOH, pH 7.6; 150 mM KOAc; 5 mM Mg(OAc)2, 4 mM DTT; 1 mM PMSF; 100 µg/ml cycloheximide. Samples were centrifuged at 39 krpm for 2 h 45 min at 4°C using an SW-41 swing-out rotor (Beckman). The gradient was pumped out from the top to the bottom using a Density Gradient Fractionation System (Brandel). Polysome profiles were generated by continuous measurement of the absorbance at 254 nm.
Ribosome filter binding
In order to investigate the binding of the tRNAPro 5ʹ half to ribosomes and ribosomal subunits, the tRNAPro half was radioactively labelled at the 5ʹend with P32 – ATP or at the 3ʹ end with P32–pCp. For the binding reaction, 5 pmol of 80S/60S/40S was incubated with P32 tRNAPro half in 1 x binding buffer (30 mM HEPES-KOH, pH 7.6, 150 mM KOAc, 3.9 mM Mg(OAc)2) for 30 min at 35°C for CHO ribosomes and at 30°C for yeast ribosomes/ribosomal subunits. The reaction was diluted by the addition of 175 μl 1 x binding buffer. Then, samples were filtrated as described before [12]. As negative binding control an archaeal ncRNA was used (5ʹ-CAGAGUAACCCCUCGGACGCAUCCACAUGACCGAACUCAUCCCCCCUUCCCCCCUUCCCCAUUAGGUUCGGUCACCCCUUUA-3) (ref. 20).
In order to characterize the binding site of ProTiP on HeLa ribosomes by binding competition experiments with deacylated tRNA, ProTiP was first produced in in vitro translation reaction in the presence of the tRNAPro half in 20 replicates (see details below). To deplete endogenous tRNAs from the ribosome pool and to remove unincorporated S35 methionine, all reactions were pooled and subjected to ultracentrifugation for 1 h at 200,000 xg in a Sorval M120+ mini-ultracentrifuge equipped with an S140AT rotor (Thermo Scientific). To deacylate tRNAMet, the obtained pellet was resuspended in 2 x Laemmli buffer and boiled for 5 min at 95°C. Then, RNA was extracted using phenol/chloroform and the RNA pellet was resuspended in 70 μl water. For the binding competition reaction, 5 pmol of sucrose gradient – purified HeLa 80 S ribosomes in 1 x resuspension buffer in a volume of 18 μl was pre-incubated with 1 μl water or 1 μl of 40 μM yeast bulk tRNAs for 10 min at 35°C and 450 rpm. After pre-incubation, 5 μl of ProTiP-containing RNA was added to each sample and the reactions were incubated for another 10 min at 35°C and 450 rpm. Subsequently, the reactions were diluted by the addition of 175 μl 1 x binding buffer. Finally, the samples were filtered as described before [12].
Crosslinking of 4–Thio-U-tRNAPro half to HeLa ribosomes
For the crosslinking experiments, 5–10 pmol of 5ʹ P32 labelled synthetic tRNAPro half carrying three photo-reactive 4-thiouridine residues at positions 4, 13, and 27 was incubated with 10 pmol of HeLa ribosomal pellet in 1 x resuspension buffer in a final volume of 20 μl for 15 min at 35°C and 450 rpm. Crosslinking was performed on ice at 366 nm for 10 min as described before [12]. After crosslinking, samples were subjected first to protease K treatment (f.c. 0.5 mg/ml of protease K, incubation for 30 min at 37°C), and then to phenol/chloroform extraction RNA extraction. Obtained RNA was resuspended in 20 μl of water. To test if the 32P-labled tRNAPro half was crosslinked to the rRNAs, 3 μl of RNA was separated on 1% agarose gel. After staining with ethidium bromide, the agarose gel was vacuum-dried at 70°C for 2 h and exposed to a phosphor imaging screen overnight. To narrow down the crosslinking site(s) of tRNAPro half on the 18 S rRNA, RNase H cleavage experiments were performed with DNA oligos annealing to different regions of 18S rRNA (18 S_563-582: AAAGGATTTAAAGTGGACTC, 18S_653-672: TTTTTAACTGCAGCAACTTT, 18 S_1247-1266: GTGAGGTTTCCCGTGTTGAG, 18S_1375-1394: CCAGAGTCTCGTTCGTTATC, 18S_1544-1563: CGTAGGGTAGGCACACGCTG). Prior to RNase H treatment, the extracted crosslinked RNAs were denatured at 70°C for 2 min. Each reaction contained 3 μl of denatured RNA, 10 pmol DNA oligo, 1x RNase H buffer (New England Biolabs), RNase inhibitor RNasine (Promega), and 3 U of RNase H. Cleavage was performed at 37°C for 1 hour. Cleavage products were analysed on 4% polyacrylamide gels containing 7 M urea. After staining with ethidium bromide, gels were vacuum-dried at 70°C for 2 h and exposed to phosphorimager screens.
In vitro translation
Harvested cells were resuspended in equal volume of 1 x resuspension buffer. Cells were opened by passing through a 25 G needle 30 times. Cell debris was removed by centrifugation at 20,000 xg for 15 min at 4°C. To deplete endogenous methionine, the cell lysate was passed through a G25 sephadex spin column prepared in 30 mM HEPES pH 7.6, 150 mM KOAc, 3.9 mM Mg(OAc)2. Every in vitro translation reaction at the pre-incubation step had a total volume of 9.9 μl and contained 6–7 μl of cell lysate and 2.9–3.9 μl of water. For translation inhibition by antibiotics, cycloheximide, or harringtonine was dissolved in abs. ethanol and added first to the tubes and the solvent was evaporated. To test the effect of tRNA 5ʹ halves on translation 100–200 pmol of synthetic RNAs were added to the reactions (tRNAPro 5ʹ half GGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUAGG, tRNAAsp 5ʹ half UCCUCGUUAGUAUAGUGGUGAGUAUCCCCGCCUG, tRNAHis 5ʹ half GCCGUGAUCGUAUAGUGGUUAGUACUCUGCGUUG, tRNAAsn 5ʹ half GUCUCUGUGGCGCAAUCGGUUAGCGCGUUCGGCUG). Reactions were pre-incubated at 35°C at 450 rpm for 10 min. After pre-incubation, 2.1 μl of freshly prepared translational mix consisting of 1.2 µl 10 x translation cocktail (150 mM HEPES-KOH, pH 7.6, 750 mM KOAc, 19.5 mM Mg(OAc)2, 4 mM GTP, 17.5 mM ATP and 500 µM each of all 19 amino acids except methionine), 0.17 µl water, 0.08 µl 3 M creatine phosphate, 0.06 µl 20 mg/ml creatine phosphokinase and 0.625 µl S35-Methionine was added to every pre-incubated mixture increasing a total volume of reaction to 12 μl. In vitro translation was performed at 35°C at 450 rpm for 30 min. Reactions were stopped by the addition of 4 µl of 4 x Laemmli buffer and boiling at 95°C for 5 min.
For the translation of one reporter mRNA, Xef1 mRNA (Thermo Scientific) and rabbit reticulocyte cell lysate (Thermo Scientific or Promega) were used according to the manufactory’s protocol. To test the effect of tRNA halves on translation 50–150 pmol of synthetic RNAs were used. In vitro translation was stopped by the addition of Laemmli buffer and heating at 95°C for 5 min.
Samples were separated on 10-14% SDS-PAGE, stained with Coomassie Brilliant Blue and destained (several hours to overnight). For RNA visualization, gels were stained by soaking in a 1 x TBE solution containing 0.5 µg/ml ethidium bromide for 5–10 min. Afterwards, the gel was vacuum-dried at 70°C for 1 h and exposed to phosphor imaging screen overnight.
Metabolic labelling
For metabolic labelling experiments, Hela cells were grown in 48-well plates to around 90% confluence. To transfect RNA into HeLa cells, 100 µl of FBS free Opti-MEM medium (Gibco) were combined with 300 pmol of the tRNAPro half, 10 µl or 50 µl of Lipofectamine RNAi MAX (Invitrogen) and incubated for 5 min at room temperature. Then, the mixture was added to one well and the cells were kept at 37°C and 5% CO2 for 5 min. To start the metabolic labelling 1 µl of S35 methionine (10 μCi/μl, Hartmann Analytic) was added to each well and the cells were incubated for another 30 min. Afterwards, the RNA was isolated with TRI reagent and separated on 12% SDS PAGE. To check the efficiency of RNA isolation and equal loading, the gel was stained with 1 x TBE containing 0.5 µg/ml ethidium bromide for 5–10 min. The gel was vacuum-dried at 70°C and exposed for 1 week to a phosphor imager screen to visualize ProTiP.
Acidic gel electrophoresis
To separate charged tRNAs, RNA were separated on freshly prepared 15% denaturing urea acidic polyacrylamide gels (8 M Urea, 50 mM CH3 COONa pH 5.5). For the loading one volume of the RNA sample was combined with one volume of loading dye (93% v/v formamide, 150 mM NaOAc, pH 5.5; 10 mM EDTA, bromphenol blue, xylene cyanol). Subsequently, the gel was stained with 1 x TBE containing 0.5 µg/ml ethidium bromide for 10–15 min. The gel was vacuum-dried at 70°C and exposed overnight to a phosphorimaging screen.
Polyadenylation
To characterize the 3ʹ end of ProTiP, total RNA was extracted from in vitro translation reactions performed in the presence of tRNAPro 5ʹ half and subjected to polyadenylation. Each reaction (total volume of 20 µl) contained 2 µl of 10 x Poly(A)Pol buffer (New England Biolabs), 1 µl RNA template (corresponding to 0.2 x of one in vitro translation reaction), 4 µl of 10 mM ATP, 1 µl RNasine (Promega), and 2 µl of Poly(A) Polymerase (New England Biolabs). Polyadenylation was carried out for 30 min at 37⁰C in a water bath. The reaction was stopped by addition of 22 µl of 2 x RNA loading dye. Afterwards, samples were separated on denaturing 8% polyacrylamide 7 M Urea 1 x TBE gel. After RNA separation, the gel was stained with 1 x TBE containing 0.5 µg/ml ethidium bromide for 10–15 min and then dried for 3 h at 70 ⁰C under vacuum. To detect the S35 signal of ProTiP, the dried gel was exposed to a phosphor imaging screen overnight. After the screen was scanned, the dried gel was rehydrated by incubation in 1 x TBE containing 0.5 µg/ml ethidium bromide. Once the gel was rehydrated, the quality of the RNA in the gel was checked under the UV light. To control for correct polyadenylation, the gel was blotted onto a nylon membrane and subsequently probed against 5.8S rRNA TCCTGCAATTCACATTAATTCTCGAGCTAGC by northern blot analysis.
Copper sulphate treatment
Copper sulphate treatment was performed using a protocol developed previously [22] on RNA isolated after in vitro translation with CHO cell lysate. Each reaction in a total volume of 30 µl contained 40 mM CuSO4 x 5 H2O, 40 mM Tris, pH 7.5, or pH 8.9 and RNA isolated from 3 in vitro translation reactions. Reactions were incubated at 37°C for 1 h and afterwards subjected to RNA extraction. After PCI extraction, 1/10 of the obtained RNA was loaded on a 15% acidic gel.
Supplementary Material
Acknowledgments
We thank Marina Cristodero, Julia Reuther, and Robert Rauscher for valuable suggestions and discussions as well as Roman Ott for experimental help during his internship. Our thanks are extended to Fabian Nagelreiter for his much appreciated help with mammalian cell culture.
Funding Statement
This study was supported by the D-A-CH grant 310030E-162559/1 (to N.P.), and in part by the grants 31003A_166527 and 310030-188969 (to N.P.) all funded by the Swiss National Science Foundation. Additional funding came from the D-A-CH grant I2514 (to J.G.) by the Austrian Science Fund FWF.
Disclosure Statement
No potential conflict of interest was reported by the authors.
Supplementary Material
The supplementary data for this article can be accessed here.
References
- [1].Diebel KW, Zhou K, Clarke AB, et al. Beyond the ribosome: extra-translational functions of tRNA Fragments. Biomark Insights. 2016;11(Suppl 1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cristodero M, Polacek N.. The multifaceted regulatory potential of tRNA-derived fragments. Non-cod RNA Investgat. 2017;1:7. [Google Scholar]
- [3].Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci. 2016;41(8):679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Megel C, Morelle G, Lalande S, et al. Surveillance and cleavage of eukaryotic tRNAs. Int J Mol Sci. 2015;16(1):1873–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kim HK, Fuchs G, Wang S, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552(7683):57–62. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oberbauer V, Schaefer MR. tRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes (Basel). 2018;9(12):607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Soares AR, Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA. 2017;8(5):e1423. [DOI] [PubMed] [Google Scholar]
- [8].Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588(23):4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pircher A, Gebetsberger J, Polacek N. Ribosome-associated ncRNAs: an emerging class of translation regulators. RNA Biol. 2014;11(11):1335–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pircher A, Bakowska-Zywicka K, Schneider L, et al. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell. 2014;54(1):147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res. 2012;40(9):4013–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gebetsberger J, Wyss L, Mleczko AM, et al. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017;14(10):1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gebetsberger J, Zywicki M, Kunzi A, et al. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mleczko AM, Celichowski P, Bakowska-Zywicka K. Transfer RNA-derived fragments target and regulate ribosome-associated aminoacyl-transfer RNA synthetases. Biochim Biophys Acta. 2018;1861(7):647–656. [DOI] [PubMed] [Google Scholar]
- [15].Fricker R, Brogli R, Luidalepp H, et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun. 2019;10(1):118. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen Z, Qi M, Shen B, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47(5):2533–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hackl M, Jadhav V, Jakobi T, et al. Computational identification of microRNA gene loci and precursor microRNA sequences in CHO cell lines. J Biotechnol. 2012;158(3):151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hackl M, Jakobi T, Blom J, et al. Next-generation sequencing of the Chinese hamster ovary microRNA transcriptome: identification, annotation and profiling of microRNAs as targets for cellular engineering. J Biotechnol. 2011;153:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gerstl MP, Hackl M, Graf AB, et al. Prediction of transcribed PIWI-interacting RNAs from CHO RNAseq data. J Biotechnol. 2013;166(1–2):51–57. [DOI] [PubMed] [Google Scholar]
- [20].Wyss L, Waser M, Gebetsberger J, et al. mRNA-specific translation regulation by a ribosome-associated ncRNA in Haloferax volcanii. Sci Rep. 2018;8(1):12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Behrmann E, Loerke J, Budkevich TV, et al. Structural snapshots of actively translating human ribosomes. Cell. 2015;161(4):845–857. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schofield P, Zamecnik PC. Cupric ion catalysis in hydrolysis of aminoacyl-tRNA. Biochim Biophys Acta. 1968;155(2):410–416. [DOI] [PubMed] [Google Scholar]
- [23].Kroll H. The participation of heavy metal ions in the hydrolysis of amino acid esters. J Am Chem Soc. 1952;74(8):2036–2039. [Google Scholar]
- [24].Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lyons SM, Achorn C, Kedersha NL, et al. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016;44(14):6949–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ivanov P, O’Day E, Emara MM, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111(51):18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cheverton AM, Gollan B, Przydacz M, et al. A salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell. 2016;63(1):86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jurenas D, Chatterjee S, Konijnenberg A, et al. AtaT blocks translation initiation by N-acetylation of the initiator tRNA(fMet). Nat Chem Biol. 2017;13(6):640–646. [DOI] [PubMed] [Google Scholar]
- [29].Lintner NG, McClure KF, Petersen D, et al. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol. 2017;15(3):e2001882. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schneider-Poetsch T, Ju J, Eyler DE, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6(3):209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10(12):1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


