ABSTRACT
RNA- and DNA-binding domains are essential building blocks for specific regulation of gene expression. While a number of canonical nucleic acid binding domains share sequence and structural conservation, others are less obviously linked by evolutionary traits. In this review, we describe a protein fold of about 150 aa in length, bearing a conserved β-β-β-β-α-linker-β-β-β-β-α topology and similar nucleic acid binding properties but no apparent sequence conservation. The same overall fold can also be achieved by dimerization of two proteins, each bearing a β-β-β-β-α topology. These proteins include but are not limited to the transcription factors PC4 and P24 from humans and plants, respectively, the human RNA-transport factor Pur-α (also termed PURA), as well as the ssDNA-binding SP_0782 protein from Streptococcus pneumonia and the bacteriophage coat proteins PP7 and MS2. Besides their common overall topology, these proteins share common nucleic acids binding surfaces and thus functional similarity. We conclude that these PC4-like domains include proteins from all kingdoms of life and are much more abundant than previously known.
KEYWORDS: RNA/DNA binding, PC4-like, protein domain, protein fold, structural biology, topology, PURA, RRM, KH-domain
Introduction
To date a considerable number of globular RNA- and DNA-binding domains have been described. The RNA binders include domains such as the RNA Recognition Motif (RRM), the double-stranded RNA-Binding Domain (dsRBD), the K-Homology (KH) Domain, the Pumilio- and FBP-homology domain (PUF domain) and the zinc finger [1]. Well-described DNA-binding domains include the helix-turn-helix motif, homeodomain, leucine zipper, zinc finger, winged helix, and the HMG-box [2,3].
Amongst them, a number of domains are known to bind to both, DNA and RNA [4]. Nucleic acid binding domains that are found in these non-discriminating proteins include RRMs, KH domains, dsRBDs, zinc-finger domains, and so-called PUR domains [4–6]. Even classical DNA-binding domains such as the homeodomain have been shown to specifically recognize RNA targets [7,8]. Thus, it appears that for several of the known nucleic-acid binding domains their fold allows for the recognition of both targets. Steric limitations such as a narrower major groove in RNAs might be overcome by specific sequence features that create for instance bulges.
For many proteins, sequence homology and similarities in domain folds go hand in hand, suggesting a common evolutionary origin. However, similar three-dimensional structures can also originate from completely unrelated protein sequences [9]. Well-known examples for related domain folds without sequence homologies are the α-β barrel and β-barrel proteins.
The Purine-rich binding protein α (Pur-α, also termed PURA) is a DNA- and RNA-binding protein that has reported functions in DNA replication, transcriptional regulation, as well as the temporal and spatial regulation of mRNAs [5]. Mutation in the PURA gene that encodes Pur-α results in a neuro-developmental disorder, termed PURA syndrome [10–12]. While studying the structural properties of Pur-α [13–15] and its similarities (using DALI server) [16], we noted that a number of sequence-unrelated nucleic-acid binding proteins adopt the same domain fold with identical connectivity of their secondary structures. Here we describe this family of proteins and highlight similarities as well as differences.
Material and methods
The structure of D. melanogaster Pur-α (fragment 41–185, chain A, PDB ID: 3k44) [14] has been used as a model for the similarities search using Dali server (http://ekhidna2.biocenter.helsinki.fi/dali/) [16]. Sequence comparison as well as a phylogenetic tree have been calculated using Clustal Omega software (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Results
Members of the PC4-like superfamily are structurally and functionally related
According to the Structural Classification of Proteins SCOP [17] proteins adopting a similar fold as Pur-α have been classified as members of the ssDNA-binding transcriptional regulator domain superfamily (SCOPID: 3000513) with four families including the transcriptional coactivator PC4 C-terminal domain family (SCOP identifier: 4001029). The name is derived from the human replication and transcription cofactor PC4 (PDB ID: 1pcf), which represents the first published high-resolution structure of this fold [18]. The protein structure classification database CATH [19] describes the transcriptional co-activator PC4 as 2.30.31 family.
Besides human PC4 [18] and Pur-α from pro- and eukaryotes [13–15], representative proteins of this domain family include the Whirly-family of single-stranded DNA-binding proteins from mitochondria and chloroplasts, like the P24 subunit of the plant transcription factor PBF-2 [20] and the mitochondrial transcriptional regulators MRP1/2 from Trypanosoma brucei [21]. Furthermore, the coat proteins of the bacteriophages PP7 and MS2 [22,23] show structural similarities and similar nucleic acids binding properties to the PC4 family. Below we describe in more detail these and other structural entities that adopt closely related three-dimensional arrangements with identical connectivity of their secondary-structure elements and provide a structure-guided suggestion for their sub-classification. In addition, a number of functionally uncharacterized protein domains also adopt this fold, albeit it is unclear whether they also bind nucleic acids (for details, see Supplementary text and Supplementary Fig. S1).
The overall domain arrangement of PC4-like domains
Despite high structural homology, most PC4-like domains lack sequence similarity and show a wide occurrence in all three kingdoms of life (Supplementary Table S1 and S2, Supplementary Fig. S2). This may suggest that the common overall domain fold and nucleic-acid binding function appeared through convergent evolution. Such domains can either be built from a single peptide chain with a β-β-β-β-α-linker-β-β-β-β-α topology (type I; Fig. 1A) or from two identical peptides with β-β-β-β-α fold, forming an inter-molecular homodimer (type II; Fig. 1B). In both cases the α-helices swap between both β-β-β-β-α repeats, so that the helix from one repeat interacts with the β-sheet of a second repeat and vice versa. This swapping of secondary structure elements gives extra stability to this fold. By observing this effect, we assume that the existence of just one repeat as folded entity is highly unlikely, unless the α-helix adopts a different orientation, much closer to the β-sheet of the same repeat. With just one exception known so far, there are no proteins containing both types of domains (Fig. 1C).
Figure 1.
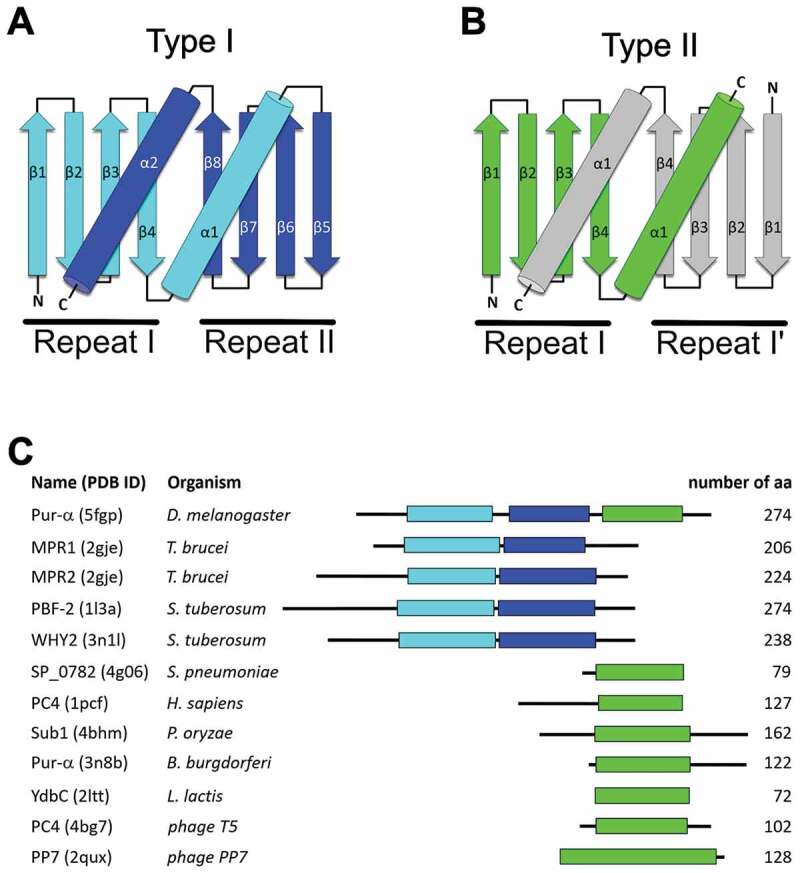
Topology diagram of type I (A) and type II (B) PC4-like fold. Domain organization for the selected proteins containing PC4-like domains (C).
Type I domains fold as intra-molecular dimers
P24 subunit of the chloroplast transcription factor PBF-2. The first described nucleic-acid binding protein of this type was the P24 subunit of the potato chloroplast transcription factor PBF-2 (WHY1, PDB ID: 1l3a) [20] (Fig. 2A and Supplementary Fig. S3A). The respective domain of this protein folds from a single peptide chain. The four β-strands of each repeat form a concave β-sheet that is additionally stabilized by the α-helix at the C-terminus of this repeat. The two β-β-β-β-α repeats interact with each other by undergoing coiled-coil-like contacts between their two swapped α-helices of the repeats. In addition, the domain is stabilized by hydrophobic interactions of residues from the helices and the sheets. The two β-sheets adopt a dome-like shape, each side binding to ssDNA in a sequence-specific manner [20]. Compared to the other representatives of type I domain proteins outlined here, the P24 nucleic acid binding domain is the least symmetric and structurally most divergent example (Fig. 2A). PBF-2 forms a homotetramer, which adopts a whirligig appearance [20,24] (Supplementary Fig. S4A). Characteristic for this quaternary complex of P24 is its central pore, with β-strands radiating outwards. P24 shares high structural similarity to other Whirly proteins and to the mitochondrial MRP1/2 protein complex.
Figure 2.
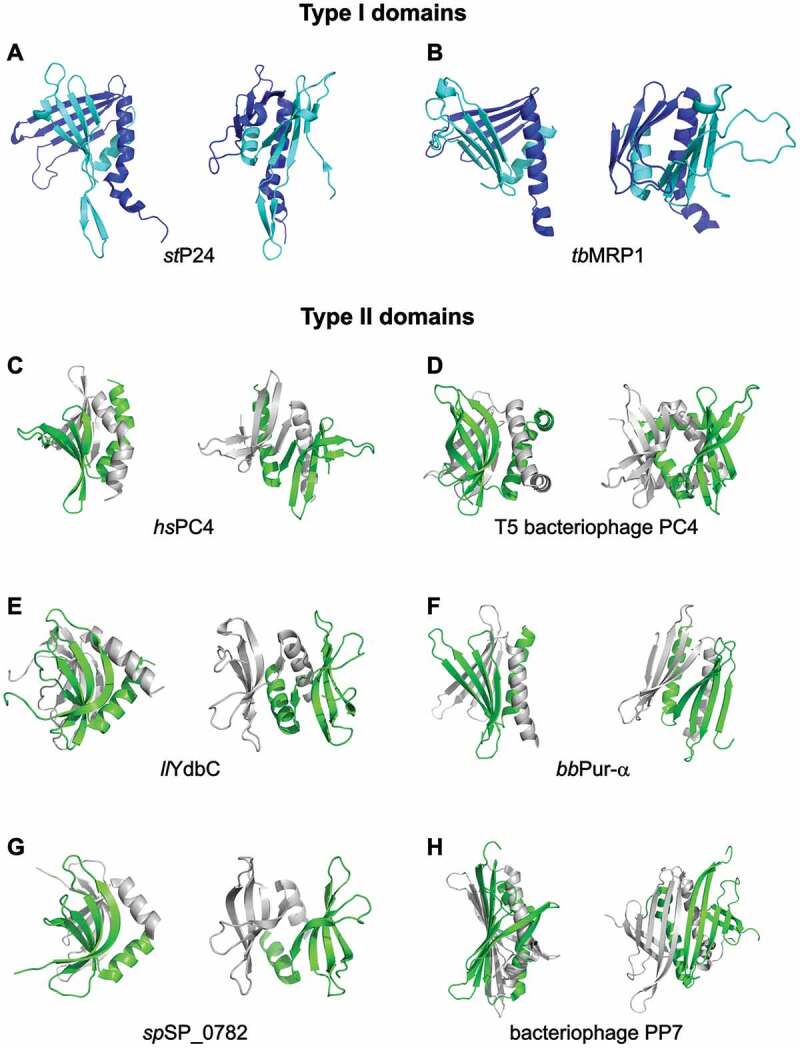
Comparison of different protein structures exhibiting type I and II PC4-like fold. Each structure is depicted in two different orientations. The coordinates are taken from: (A) Solanum tuberosum P24 subunit of PBF-2 (PDB ID: 1l3a) [20], (B) Trypanosoma brucei MRP1 (PDB ID: 2gje) [21], (C) The human replication and transcription cofactor PC4 (PDB ID: 1pcf) [18], (D) Bacteriophage T5 homologue of PC4 (PDB ID: 4bg7) [36], (E) Lactococcus lactis DUF2128 family member YdbC (PDB ID: 2ltt) [37], (F) Pur-α from Borrelia burgdorferi (PDB ID: 3n8b) [13], (G) Streptococcus pneumonia SP_0782 protein (PDB ID: 5zkl) [39], (H) The bacteriophage coat protein PP7 (PDB ID: 2qux) [22]. The structures of type I PC4-like fold have been represented as cyan (repeat I) and navy blue (repeat II), whereas in the structures of type II PC4-like fold individual chains have been shown in green and grey. The figure has been prepared in PyMol v 1.3 (pymol.org).
The mitochondrial RNA-binding proteins MRP1 and MRP2. In the parasite Trypanosoma brucei mitochondrial editing of kinetoplastid RNA (kRNA) is carried out by the Ligase-containing supramolecular complex [25–28]. This multiprotein complex consists of critical core enzymes [29–33] as well as substoichiometric amounts of RNA-associated proteins, including the so-called Mitochondrial RNA binding Protein (MRP) complex with its two subunits MRP1 and MRP2 [33–35]. MRP1/MRP2 serve as a matchmaker by binding to guide RNAs (gRNAs) and facilitating their hybridization with cognate pre-edited mRNAs [21]. These two RNA binding proteins adopt a hetero-tetrameric arrangement (PDB ID: 2gia, 2gje, and 2gid) [21] similar to P24 (r.m.s.d. of 3.17 Å for 447 superimposed residues). Like the other type I domains, MRP1 and MRP2 share an overall conserved β-β-β-β-α-linker-β-β-β-β-α topology, where each of the four β-strands within a given β-β-β-β-α repeat contributes to the formation of a curved antiparallel β-sheet that packs perpendicularly against the β-sheet from the other repeat (Fig. 2B and Supplementary Fig. S3B). The folds of MRP1 and MRP2 are remarkably similar, despite the fact that their protein sequences display only 18% identity. Both proteins interact with RNA via one of its two β-sheets, mostly by recognizing its phosphate backbone.
Type II domains fold as inter-molecular dimers
The human replication and transcription cofactor PC4 dimerizes and binds ssDNA through its C-terminal domain (CTD). The crystal structure also revealed that each subunit is formed by a curved four-stranded antiparallel β-sheet followed by a 45° kinked α-helix (PDB ID: 1pcf) [18] (Fig. 2C and Supplementary Fig. S3C). ssDNA binding cavities are oriented in opposite directions to each other on the surface of both four-stranded β-sheets. Although the two α-helices of the PC4 CTD dimer can be superimposed onto those of the WHY proteins, the corresponding β-sheet surfaces, which harbour their DNA binding regions, vary considerably in length, curvature and orientation. In addition, the WHY proteins do not possess the high symmetry of their monomers observed in the PC4 dimer [18]. This high symmetry as well as the curvature of its β-sheets allows the PC4 homodimer to form an unusually steep β-ridge, which is rarely present in such a pronounced form in other PC4-like domains. As discussed below, such β-ridges may be functionally important by contributing to dsDNA unwinding [18].
In this context it is also worth mentioning the bacteriophage recombination-dependent DNA replication factor T5 [36]. Besides its function that is similar to the eukaryotic transcription coactivator PC4, both proteins share a sequence identity of 25% (Supplementary Table S2 and Supplementary Fig. S2). Its dimeric structure is extended by two helices in addition to the two conserved helices of the PC4-like fold, yielding the topology of the single repeat to be β-β-β-β-α-α. The additional helix gives extra stability to the already swapped dimer (PDB ID: 4bg7; Fig. 2D and Supplementary Fig. S3D).
YdbC is an ssDNA- and RNA-binding protein from the bacterium Lactococcus lactis and a member of the DUF2128 family, which also folds into a type II domain structure (PDB ID: 2ltd and 2ltt) [37] (Fig. 2E and Supplementary Fig. S3E). Its ability to bind single-stranded nucleic acids indicates that this class of proteins is not only structurally but also functionally related to the other proteins described here.
The same conclusion can be made after analysing the structure of Pur-α from gram-negative bacteria Borrelia burgdorferi (PDB ID: 3n8b) [13] (Fig. 2F and Supplementary Fig. S3F). Apart of its high structural similarity to our search model Pur-α from D. melanogaster [14], the ability to bind DNA oligomers with (GGN)n sequences has been shown for B. burgdorferi Pur-α indicating that this protein is likely also involved in cell cycle control and transcription.
SP_0782 protein (fragment 7–79) from Streptococcus pneumoniae yielded a high score in our structural similarity search (Z = 5.1). It is potentially involved in maintenance of genome stability and natural transformation [38]. Its structure has been determined in complex with ssDNA (Northeast Structural Genomics Consortium; PDB ID: 5zkl [39]; Fig. 2G and Supplementary Fig. S3G). Binding of ssDNA to SP_0782 suggests its potential involvement in gene transcription, recombination, DNA repair or replication.
Bacteriophage coat protein PP7. This protein serves as the structurally most dissimilar example for the type II domain fold. The structure of a truncated version of PP7 that is deficient in capsid assembly (PP7ΔFG) in complex with a 25-nt RNA hairpin has been solved to 2.4 Å resolution (PDB ID: 2qux) [22]. While the canonical arrangement of secondary structure elements is maintained in this domain, in contrast to all other mentioned examples its two β-sheets form a continuous, flat surface to which dsRNA can bind. Another unique feature of the PP7ΔFG protein is that the β-sheet of each repeat is composed of five β-strands and it contains an additional β-loop (Fig. 2H and Supplementary Fig. S3H). The topology of each PP7ΔFG monomer is characteristic also for other ssRNA bacteriophage proteins such as the MS2 coat protein [40], the GA coat protein [41], the Q-beta capsid protein [42], and the bacteriophage FR capsid [43].
Proteins containing both, type I and type II domains
An interesting addition to this theme is offered by the eukaryotic Pur-α protein. This protein is on one hand a transcription factor that binds to ssDNA [5,44–46]. On the other hand, Pur-α has been described as core factor of cytoplasmic mRNA-transport complexes, for which it binds to ssRNA [47–50]. To our knowledge, this DNA- and RNA-binding protein is the only reported factor containing type I as well as type II domains (Fig 1C).
Pur-α contains three repeating sequence elements in its peptide chain [14,15]. Its N-terminal type I domain is formed by the so-called PUR repeats I and II, whereas its C-terminal type II domain folds via the interaction of two PUR repeats III from two distinct Pur-α molecules [14]. The PUR repeats I–II interact with each other to form a stable domain, in which about 1/3 of the surfaces of each β-β-β-β-α repeat is buried (PDB ID: 3k44 and 5fgp; Fig. 3A and Supplementary Fig. S5A) [14,15]. The overall fold of the type I domain is very similar to its type II domain (PDB ID: 5fgo; Fig. 3B and Supplementary Fig. S5B), with a root mean square deviation (r.m.s.d.) of only 1.5 Å [15]. DNA- and RNA-binding studies combined with biophysical assessment of the oligomeric states of this protein suggest a division of functions between these domains. Although nucleic acid binding of the C-terminal PC4-like domain (type II; consisting of two PUR repeats III) has been demonstrated, the major interaction with nucleic acids seems to be mediated by its N-terminal PC4-like domain (type I; consisting of PUR repeats I–II) [13–15].
Figure 3.
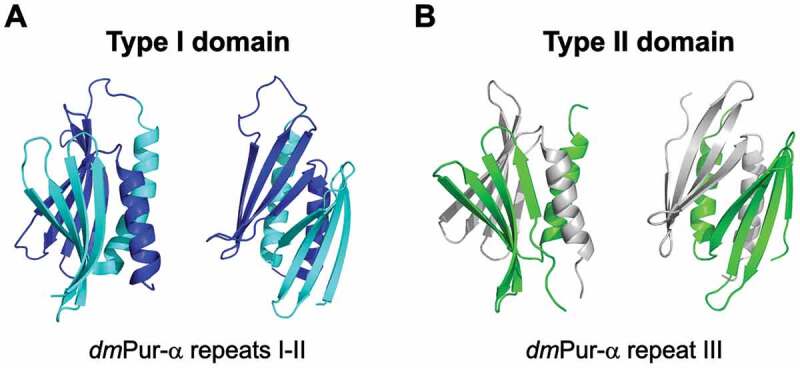
Structure of Drosophila melanogaster Pur-α protein exhibiting type I (A) (PUR repeat I-II; PDB ID: 5fgp) [15] and type II (B) (PUR repeat III; PDB ID: 5fgo) [15] PC4-like fold. Each structure is shown in two different orientations. The structure of type I is represented in cyan (repeat I) and navy blue (repeat II), whereas in the structure of type II PC4-like fold individual chains are shown in green and grey. The figure has been prepared in PyMol v 1.3 (pymol.org).
In humans, two paralogs of Pur-α have been reported, termed Pur-β and Pur-γ [51,52]. Based on co-immunoprecipitation experiments with cellular extracts it has been suggested that Pur-α forms heterodimers with Pur-β [53] to modulate their respective functions. Based on our knowledge on the homodimerization of Pur-α, such heterodimerization could be formally possible via repeat III.
Mutations have been reported in humans that result in a deletion of the C-terminal PC4-like dimerization domain of Pur-α while leaving the more N-terminal DNA/RNA-binding domain intact. Patients with such mutations develop the neurodevelopmental disorder PURA syndrome [12], indicating that in addition to the N-terminal domain oligomerization by its type II domain is important for the function of Pur-α.
Nucleic-acid binding by type I domains
Dedicated DNA-repair mechanisms are crucial to maintain the integrity of the genetic information. Plant chloroplasts and mitochondria express proteins belonging to the Whirly family, which contribute to DNA repair by binding single-stranded DNA. P24 and other Whirly proteins show high sequence conservation and are present in Arabidopsis, tomato and more distantly related species, such as wheat, rice, maize and loblolly pine [24]. The structural studies on the Whirly protein WHY2 from Solanum tuberosum in complex with ssDNA (PDB ID: 3n1l, 3n1 k, 3n1 j, 3n1i, and 3ra0) [54,55] revealed details of its sequence-unspecific ssDNA binding mechanism. In WHY2 proteins, binding of ssDNA is observed primarily in a circular arrangement on the outer edges and in between the β-sheets of adjacent protomers of the Whirly-like tetramer (Fig. 4A and Supplementary Fig. S6A). While most single-stranded nucleic acid binding proteins use the core of their β-sheets as a primary binding platform [28], the WHY2-ssDNA interaction relies mainly on binding of the DNA between properly positioned domains. Binding to ssDNA exploits and is stabilized by the four-fold symmetry of the Whirly tetramer. The mode of ssDNA binding is dominated by stacking and hydrophobic interactions between adjacent nucleobases and between nucleobases and aromatic/hydrophobic protein residues (Supplementary Fig. S6B). Most of the nucleobases have their sequence-specific binding moieties exposed to the solvent, whereas the faces of the nucleobases make intimate contacts with residues of the protein surface [54]. Cappadocia and colleagues proved that the Whirly protein WHY2 can not only bind to melted DNA but actively melts dsDNA [54]. Thus, WHY2 and most likely other plant Whirly family members can destabilize DNA duplexes, probably by binding with a higher affinity to the single-stranded form of DNA and hence shifting the equilibrium in favour of ssDNA.
Figure 4.
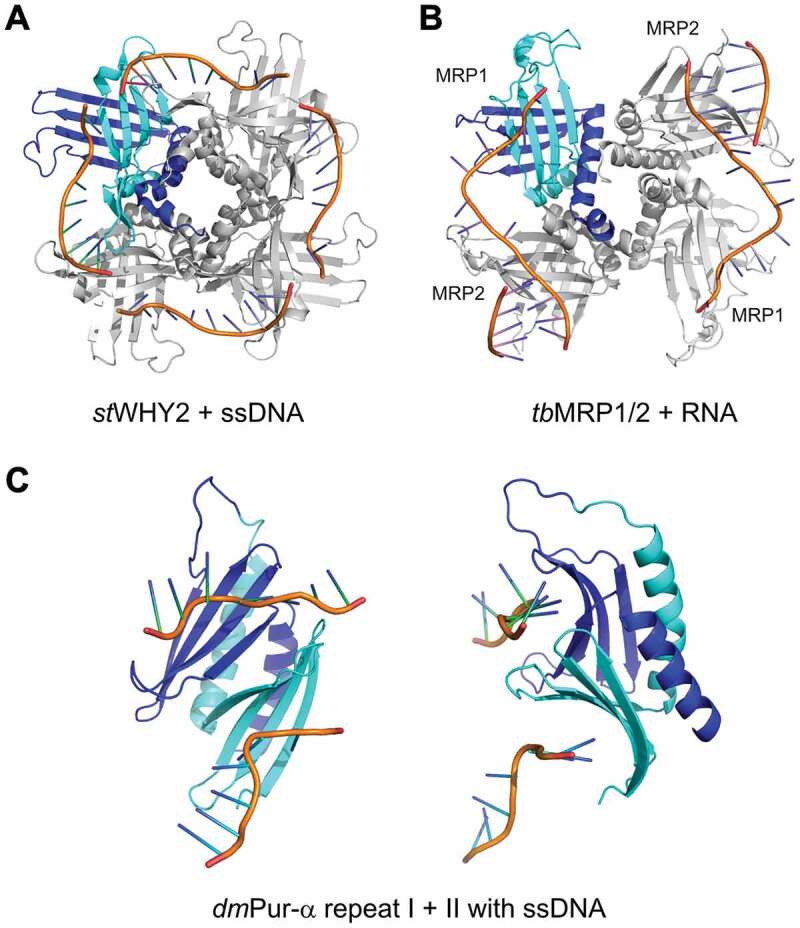
DNA/RNA binding by type I PC4-like domains. The coordinates taken from: (A) Solanum tuberosum WHY2 in complex with ssDNA (PDB ID: 3n1l) [54], (B) Trypanosoma brucei MRP1/MRP2 complex with RNA (PDB ID: 2gje) [21], (C) Drosophila melanogaster Pur-α repeat I–II in complex with ssDNA (PDB ID: 5fgp) [15]. The figure has been prepared in PyMol v 1.3 (pymol.org).
To assess the versatility of nucleic acid binding by the Whirly domain containing proteins, we also analysed the structure of the MRP1/MRP2 complex involved in kinetoplastid RNA editing (PDB ID: 2gje) [21]. This complex also exhibits a tetrameric whirligig appearance, but in contrast to the previously described structures, it forms a heterotetramer with affinity to RNA (Fig. 4B and Supplementary Fig. S6C). Despite the lack of sequence homology between MRP1/MRP2 and plant WHY proteins, their similarity in tertiary structure is obvious (Fig. 4A,B). However, there are differences between these two complexes with respect to their binding to nucleic acids. The MRP1/MRP2 complex interacts with the phosphate backbone of the RNA via its β-sheet, while the RNA bases are exposed to the solvent (Supplementary Fig. S6D). As a consequence, there are no base stacking or base-specific interactions observed between MRP1, MRP2 and RNA. Surprisingly, the MRP domains bind RNAs with nanomolar affinity despite their lack of sequence specificity [21,56–58]. These binding features are different from the WHY2-DNA complex, where ssDNA is observed primarily on the edges and in between the β-sheets of adjacent protomers (compare Fig. 4A,B) [54,55].
Whereas MRP1/MRP2 binds ssRNA, WHY2 has been shown to bind ssDNA as well as to unwind dsDNA. Eukaryotic Pur-α combines the functions of both protein classes. Pur-α repeats I and II fold into the N-terminal, PC4-like nucleic acid binding domain that mediates sequence-specific binding to ssDNA, dsDNA, and ssRNA, with a reported preference for GGN repeats [48,51,59]. NMR titration experiments of Drosophila Pur-α repeat I–II with CGG repeats revealed almost indistinguishable chemical shift perturbations for DNA and RNA. This finding and very similar KD values for CGG DNA and RNA indicate the same mode of binding for both types of nucleic acids [15]. Furthermore, the crystal structure of Drosophila Pur-α repeat I–II in complex with the GCGGCGG ssDNA (Fig. 4C and Supplementary Fig. S6E) reveals that one molecule of Pur-α repeat I–II can bind two molecules of ssDNA via base-specific contacts to guanines [15] (Supplementary Fig. S6F). Both binding events appear at overlapping but non-identical surface regions and therefore might prefer different nucleic-acid motifs as has been previously suggested [48].
Besides its ability to bind RNA and DNA, eukaryotic Pur-α possesses dsDNA-destabilizing activity in an ATP-independent fashion [60]. It was suggested that this feature enables Pur-α to open up dsDNA at the replication fork, thus explaining its requirement in cell division. The crystal structure of Drosophila Pur-α in complex with DNA offered an explanation for its unwindase activity [15]. Most likely spontaneous breathing of odd dsDNA helices allows Pur-α to intercalate between both strands and to stabilize ssDNA. Since Pur-α forms a dimer, two domains with unwindase activity (i.e. repeats I–II) would be able to act in close vicinity on the DNA, allowing for unwinding of a larger DNA region (Supplementary Fig. S4B). Such locally melted DNA regions could then be further unwound by ATP-dependent DNA helicases. Since in NMR and binding experiments no difference in binding to DNA and RNA was observed for Pur-α, this mechanism could in principle also apply to dsRNA unwinding. Consistent with such a function is the report that Drosophila Pur-α interacts with the RNA helicase Rm62 [61].
Nucleic-acid binding bytype II domains
The human replication and transcription cofactor PC4 dimerizes, adopting a type II domain fold, and binds ssDNA through its C-terminal domain (CTD). A crystal structure of the human PC4 CTD in complex with a single-stranded 20-mer DNA was solved by Werten and Moras [62]. It revealed how symmetry-related β-sheets of the PC4 CTD homodimer interact with juxtaposed five nucleotide-long DNA strands running in opposite directions (Fig. 5A and Supplementary Fig. S7A). Such melted DNA is bound with higher affinity than regular ssDNA in a sequence-independent manner [62,63]. Similar to the observed DNA binding by Pur-α, several DNA bases are involved in stacking interactions with the aromatic side chains of PC4 CTD (Supplementary Fig. S7B). For binding by two PC4 monomers of the same dimer ssDNA adopts a U-like shape (U mode) (Fig. 5A and Supplementary Fig. S7A) [64,65].
Figure 5.
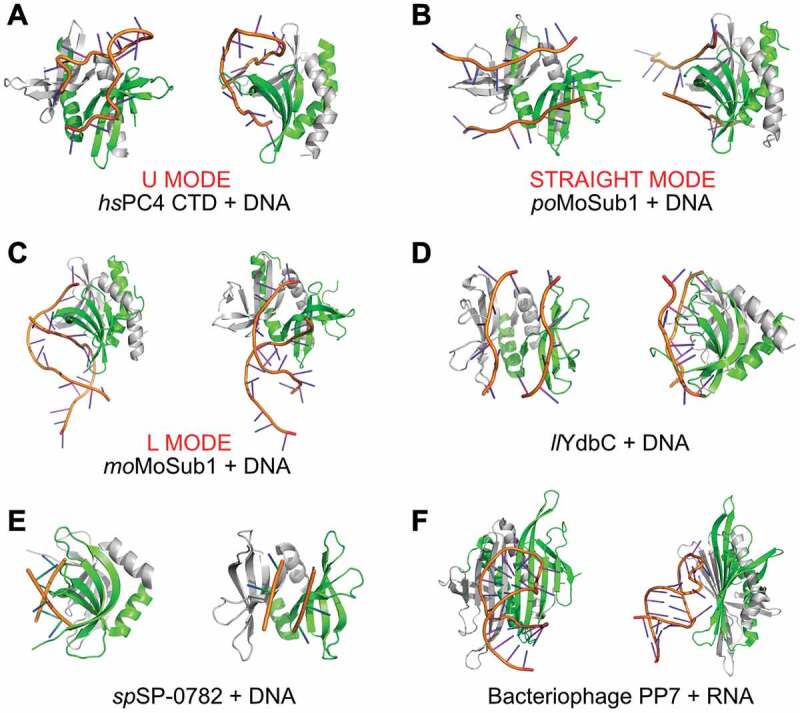
DNA/RNA binding by type II PC4-like fold proteins. The coordinates are taken from: (A) hsPC4 CTD in complex with ssDNA (U mode; PDB ID: 2c62) [64], (B) Pyricularia oryzae MoSub1 in complex with ssDNA (Straight mode; PDB ID: 4bhm) [66], (C) Magnaporthe oryzae MoSub1 in complex with ssDNA (L mode; PDB ID: 5zg9) [65], (D) Lactococcus lactis Ydbc in complex with ssDNA (PDB ID: 2ltt) [37], (E) Streptococcus pneumonia SP_0782 protein in complex with ssDNA dT12 (PDB ID: 5zkl) [39], (F) Pseudomonas phage protein PP7 in complex with RNA (PDB ID: 2qux) [22]. The figure has been prepared in PyMol v 1.3 (pymol.org).
Another interesting example of versatile ssDNA binding is MoSub1, whose co-structure with ssDNA (PDB ID: 4bhm) exhibits an extended, straight conformation of two individual DNA strands (Fig. 5B and Supplementary Fig. S7C and S7D) [66]. In the crystal packing two neighbouring MoSub1 dimers are linked to another by their bound ssDNA. This observation suggests that complementary strands of longer unwound regions interact with multimers of MoSub1 by forming a continuous left-handed double helix around a central protein filament. Such a filament could grow as the DNA unwinds further and additional MoSub1 homodimers join in (Supplementary Fig. S4C). DNA-dependent multimerization of MoSub1 homodimers is likely to be the driving force for the experimentally observed ATP-independent unwinding of duplex DNA by PC4 domains in vitro [62]. Similar modes of multimerization-driven unwinding have also been proposed for other ssDNA-binding proteins [67]. The MoSub1 ortholog from rice blast fungus shows another mode of nucleic acid binding (PDB ID: 5zg9) [65]. Here, the dT19G ssDNA is bound between two MoSub1 dimers, thereby connecting them (Fig. 5C, Supplementary Figs. S7E, S7F and S4D). The bound ssDNA adopts an L‐like conformation, termed here L mode.
Also YdbC from Lactococcus lactis, which is a multifunctional nucleic acid-binding protein of the DUF2128 family, binds ssDNA in a straight mode (PDB ID: 2ltt) [37] (Fig. 5D and Supplementary Fig. S8A and S8B). With its type II fold, YdbC has remarkable structural similarity to the PC4 CTD and the Pur-α domains (Supplementary Table S1). The ability of YdbC dimers to bind to ssDNA with nanomolar affinities and thus the potential to disrupt DNA duplexes indicates that these proteins also share strong functional similarities to the other proteins described here.
Very recently, DNA binding has been shown also for SP_0782 protein (fragment 7–79) from S. pneumonia. The complex of this protein with single stranded DNA dT12 (Northeast Structural Genomics Consortium; PDB ID: 5zkl [39]) shows a straight mode of DNA binding (Fig. 5E and Supplementary Fig. S8C and S8D).
The bacteriophage PP7ΔFG homodimer forms a type II domain, whose co-structure with a 25-nt translational operator RNA hairpin has been solved at 2.4 Å resolution (PDB ID: 2qux) [22] (Fig. 5F and Supplementary Fig. S8E and S8F). The antiparallel association of two PP7ΔFG protomers form a ten-stranded, flat β-sheet that is not suited to separate two ssRNA strands. Instead, this protein interacts with a double-stranded RNA stem-loop. Although also other bacteriophage coat proteins such as MS2 share similar protein scaffolds, their RNA-binding surfaces have evolved to recognize distinct asymmetric RNA hairpins in a sequence-specific mode [22,23,68,69].
PP7 and MS2 were not classified in SCOP and CATH as members of the PC4-like family, however their folds as well as functional features suggest that these proteins belong to the PC4-like family. In addition, a critical argument for including them in the PC4-like family is their helix swapping feature. It is present in all members of the PC4-like family and stabilizes their domain fold.
In addition to the cases discussed above, we found other examples that did not pass our criteria to be classified as a member of the PC4-like family, but constitute interesting cases with considerable similarities to this family of proteins. For an example, see Supplementary Text and Supplementary Fig. S4E.
Discussion
From the examples given above a picture emerges in which PC4-like domains show a conserved fold and a function in nucleic acids binding. The presence or absence of the β-ridge and the additional secondary structure elements, the length of the β-strands, curvature of the β-sheets and their relative orientation affect the binding preferences, stoichiometry of the formed complexes with the nucleic acids as well as an oligomeric state of the PC4-like domains. Since no significant sequence similarity has been found between most members of this superfamily, it seems likely that the PC4-like fold with its RNA/DNA binding properties evolved independently in all kingdoms of life.
A common feature of many nucleic acid binding proteins, especially RNA binding proteins, is a curved β-sheet surface, which serves as binding site for extended RNAs [70]. For PC4-like domains with nucleic acids binding properties, we found a great variation in their shapes of β–sheets. Symmetric β-sheets with rather shallow curvatures are observed in the P24 subunit of PBF-2 (Fig. 2A). Both, the type I and II domains of Pur-α show β-sheets with a modest curvature, while keeping the overall two-fold symmetry of the molecule (Figs. 2F, 3A,B & 4C). In the human replication and transcription cofactor PC4 (Figs. 2C & 5A), DUF2128 family member YdbC (Figs. 2E & 5D) as well as in SP_0782 protein (Figs. 2G & 5E) the domain still shows high symmetry but its β-sheets show a significantly stronger curvature. In other cases, the β-sheets differ in their orientation towards each other, resulting in asymmetric domains. For instance, in stWHY2 (Fig. 4A), as well as in tbMRP1/MRP2 (Fig. 4B) one β-sheet is rather planar while the second one tends to wrap around it. This lack of symmetry limits the domain to bind only one ssDNA/RNA chain. Based on the cases reported here, such asymmetric features appear to be constrained to type I domains, as to our knowledge only symmetric homodimers of type II domains have been reported yet. Since many of the examples described in the manuscript are proteins involved in DNA unwinding, we assume that high symmetry of the PC4-like fold would be favourable. As observed from all the examples, this high symmetry can be achieved by assembling homodimer from two identical repeats.
In cases of WHY2 and MRP1/MRP2 their symmetries are increased by forming higher order oligomers and thus allowing for unwinding of double-stranded nucleic acids. Even for Pur-α, whose N-terminal type I domain already has a symmetric fold (Fig. 3A), dimerization via its C-terminal PUR repeat III might help to render unwinding more efficient [14,15]. Upon dimerization, two N-terminal domains are brought into close vicinity, potentially allowing for cooperativity of their unwinding activity (Supplementary Fig. S4B).
β-sheets of nucleic acids binding proteins are common interaction surfaces for RNAs and to a much lesser extent also for DNAs [70]. Similar to PC4-like domains, several of these domains stabilize the folding of their β-sheets via interactions with α-helices. For instance, the RNA recognition motif (RRM) is a protein domain consisting of a four-stranded β-sheet, which accommodates nucleic acids on its curved surface [71–77]. Despite different topology (β-α-β-β-α-β), the three-dimensional arrangement of the four β-strands and one or more α-helices in an RRM domain resembles a single repeat of the PC4-like scaffold (Fig. 6A,B). Similar to the PC4-like scaffold, RRM shows high versatility in RNA sequence and shape recognition [78]. Considering these structural and functional similarities, one may speculate that the RRM and PC4-like folds co-evolved. In contrast to these examples, KH-domains with β-α-(α)-β-β-α topology do not interact with the nucleic acids via their β-sheet surfaces. However, it is interesting to note that also this domain, with its three-stranded β-sheet and two or three associated helices, has a related three-dimensional orientation of secondary structure elements (Fig. 6C). It seems obvious that during evolution domains with a β-sheet and associated α-helices have proven particularly useful for RNA interactions.
Figure 6.
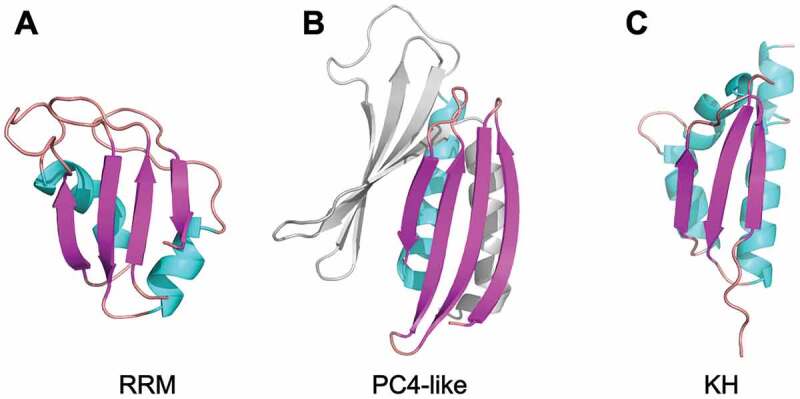
Structural comparison of the scaffold of the RNA binding domains: (A) RRM (PDB ID: 2up1) [79], (B) PC4-like domain (PDB ID: 3k44) [14], and (C) KH domain (PDB ID: 6gqe) [80]. The figure has been prepared in PyMol v 1.3 (pymol.org).
Supplementary Material
Acknowledgments
We would like to thank Dr. Thomas Monecke for his valuable comments on the manuscript.
Funding Statement
This work was supported by the Care-for-Rare Foundation [2018]; Deutsche Forschungsgemeinschaft [FOR2333].
Availability
Dali server used for the analysis describerd in this manuscript is available under this link: http://ekhidna2.biocenter.helsinki.fi/dali/(16)
Clustal Omega software for sequence comparison as well as a phylogenetic tree calculation is available here: https://www.ebi.ac.uk/Tools/msa/clustalo/
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
References
- [1].Lunde BM, Moore C, Varani G.. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Luscombe NM, Austin SE, Berman HM, et al. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1:REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rohs R, Jin X, West SM, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol. 2014;15:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].White MK, Johnson EM, Khalili K. Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle. 2009;8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cassiday LA, Maher LJ 3rd.. Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. [DOI] [PubMed] [Google Scholar]
- [8].Rivera-Pomar R, Niessing D, Schmidt-Ott U, et al. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. [DOI] [PubMed] [Google Scholar]
- [9].Doolittle RF. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994;19:15–18. [DOI] [PubMed] [Google Scholar]
- [10].Hunt D, Leventer RJ, Simons C, et al. Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet. 2014;51:806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lalani SR, Zhang J, Schaaf CP, et al. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet. 2014;95:579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Reijnders MRF, Janowski R, Alvi M, et al. PURA syndrome: clinical delineation and genotype-phenotype study in 32 individuals with review of published literature. J Med Genet. 2018;55:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Graebsch A, Roche S, Kostrewa D, et al. Of bits and bugs–on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure. PLoS One. 2010;5:e13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Graebsch A, Roche S, Niessing D. X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc Natl Acad Sci USA. 2009;106:18521–18526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Weber J, Bao H, Hartlmuller C, et al. Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha. Elife. 2016;5. DOI: 10.7554/eLife.11297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Holm L, Kaariainen S, Rosenstrom P, et al. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Murzin AG, Brenner SE, Hubbard T, et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. [DOI] [PubMed] [Google Scholar]
- [18].Brandsen J, Werten S, van der Vliet PC, et al. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat Struct Biol. 1997;4:900–903. [DOI] [PubMed] [Google Scholar]
- [19].Dawson NL, Lewis TE, Das S, et al. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017;45:D289–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Desveaux D, Allard J, Brisson N, et al. A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat Struct Biol. 2002;9:512–517. [DOI] [PubMed] [Google Scholar]
- [21].Schumacher MA, Karamooz E, Zikova A, et al. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. [DOI] [PubMed] [Google Scholar]
- [22].Chao JA, Patskovsky Y, Almo SC, et al. Structural basis for the coevolution of a viral RNA-protein complex. Nat Struct Mol Biol. 2008;15:103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Valegard K, Murray JB, Stockley PG, et al. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371:623–626. [DOI] [PubMed] [Google Scholar]
- [24].Desveaux D, Despres C, Joyeux A, et al. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell. 2000;12:1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Benne R, Van den Burg J, Brakenhoff JP, et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. [DOI] [PubMed] [Google Scholar]
- [26].Feagin JE, Shaw JM, Simpson L, et al. Creation of AUG initiation codons by addition of uridines within cytochrome b transcripts of kinetoplastids. Proc Natl Acad Sci U S A. 1988;85:539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shaw JM, Feagin JE, Stuart K, et al. Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988;53:401–411. [DOI] [PubMed] [Google Scholar]
- [28].Horvath A, Berry EA, Maslov DA. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 2000;287:1639–1640. [DOI] [PubMed] [Google Scholar]
- [29].Corell RA, Read LK, Riley GR, et al. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol Cell Biol. 1996;16:1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Panigrahi AK, Schnaufer A, Carmean N, et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol Cell Biol. 2001;21:6833–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Panigrahi AK, Schnaufer A, Ernst NL, et al. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Aphasizhev R, Aphasizheva I, Nelson RE, et al. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. Embo J. 2003;22:913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Simpson L, Aphasizhev R, Gao G, et al. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aphasizhev R, Aphasizheva I, Nelson RE, et al. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vondruskova E, van den Burg J, Zikova A, et al. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem. 2005;280:2429–2438. [DOI] [PubMed] [Google Scholar]
- [36].Steigemann B, Schulz A, Werten S. Bacteriophage T5 encodes a homolog of the eukaryotic transcription coactivator PC4 implicated in recombination-dependent DNA replication. J Mol Biol. 2013;425:4125–4133. [DOI] [PubMed] [Google Scholar]
- [37].Rossi P, Barbieri CM, Aramini JM, et al. Structures of apo- and ssDNA-bound YdbC from Lactococcus lactis uncover the function of protein domain family DUF2128 and expand the single-stranded DNA-binding domain proteome. Nucleic Acids Res. 2013;41:2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gong Y, Li S, Li Y, et al. Mutation of leucine 20 causes a change of local conformation indirectly impairing the DNA binding of SP_0782 from Streptococcus pneumoniae. Biochem Biophys Res Commun. 2020. DOI: 10.1016/j.bbrc.2020.01.057 [DOI] [PubMed] [Google Scholar]
- [39].Li S, Lu G, Fang X, et al. Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins. Nucleic Acids Res. 2020;48:432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Valegard K, Liljas L, Fridborg K, et al. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. [DOI] [PubMed] [Google Scholar]
- [41].Ni CZ, White CA, Mitchell RS, et al. Crystal structure of the coat protein from the GA bacteriophage: model of the unassembled dimer. Protein Sci. 1996;5:2485–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Golmohammadi R, Fridborg K, Bundule M, et al. The crystal structure of bacteriophage Q beta at 3.5 A resolution. Structure. 1996;4:543–554. [DOI] [PubMed] [Google Scholar]
- [43].Liljas L, Fridborg K, Valegard K, et al. Crystal structure of bacteriophage fr capsids at 3.5 A resolution. J Mol Biol. 1994;244:279–290. [DOI] [PubMed] [Google Scholar]
- [44].Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chepenik LG, Tretiakova AP, Krachmarov CP, et al. The single-stranded DNA binding protein, Pur-alpha, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene. 1998;210:37–44. [DOI] [PubMed] [Google Scholar]
- [46].Gupta M, Sueblinvong V, Raman J, et al. Single-stranded DNA-binding proteins PURalpha and PURbeta bind to a purine-rich negative regulatory element of the alpha-myosin heavy chain gene and control transcriptional and translational regulation of the gene expression. Implications in the repression of alpha-myosin heavy chain during heart failure. J Biol Chem. 2003;278:44935–44948. [DOI] [PubMed] [Google Scholar]
- [47].Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. [DOI] [PubMed] [Google Scholar]
- [48].Aumiller V, Graebsch A, Kremmer E, et al. Drosophila Pur-alpha binds to trinucleotide-repeat containing cellular RNAs and translocates to the early oocyte. RNA Biol. 2012;9:633–643. [DOI] [PubMed] [Google Scholar]
- [49].Ohashi S, Kobayashi S, Omori A, et al. The single-stranded DNA- and RNA-binding proteins pur alpha and pur beta link BC1 RNA to microtubules through binding to the dendrite-targeting RNA motifs. J Neurochem. 2000;75:1781–1790. [DOI] [PubMed] [Google Scholar]
- [50].Johnson EM, Kinoshita Y, Weinreb DB, et al. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res. 2006;83:929–943. [DOI] [PubMed] [Google Scholar]
- [51].Bergemann AD, Ma ZW, Johnson EM. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol Cell Biol. 1992;12:5673–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Liu H, Johnson EM. Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res. 2002;30:2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kelm RJ Jr., Cogan JG, Elder PK, et al. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter. J Biol Chem. 1999;274:14238–14245. [DOI] [PubMed] [Google Scholar]
- [54].Cappadocia L, Marechal A, Parent JS, et al. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell. 2010;22:1849–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cappadocia L, Parent JS, Zampini E, et al. A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res. 2012;40:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Koller J, Muller UF, Schmid B, et al. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J Biol Chem. 1997;272:3749–3757. [DOI] [PubMed] [Google Scholar]
- [57].Lambert L, Muller UF, Souza AE, et al. The involvement of gRNA-binding protein gBP21 in RNA editing-an in vitro and in vivo analysis. Nucleic Acids Res. 1999;27:1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Muller UF, Lambert L, Goringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. Embo J. 2001;20:1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wortman MJ, Johnson EM, Bergemann AD. Mechanism of DNA binding and localized strand separation by Pur alpha and comparison with Pur family member, Pur beta. Biochim Biophys Acta. 2005;1743:64–78. [DOI] [PubMed] [Google Scholar]
- [60].Darbinian N, Gallia GL, Khalili K. Helix-destabilizing properties of the human single-stranded DNA- and RNA-binding protein Puralpha. J Cell Biochem. 2001;80:589–595. [PubMed] [Google Scholar]
- [61].Qurashi A, Li W, Zhou JY, et al. Nuclear accumulation of stress response mRNAs contributes to the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2011;7:e1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Werten S, Langen FW, van Schaik R, et al. High-affinity DNA binding by the C-terminal domain of the transcriptional coactivator PC4 requires simultaneous interaction with two opposing unpaired strands and results in helix destabilization. J Mol Biol. 1998;276:367–377. [DOI] [PubMed] [Google Scholar]
- [63].Ballard DW, Philbrick WM, Bothwell AL. Identification of a novel 9-kDa polypeptide from nuclear extracts. DNA binding properties, primary structure, and in vitro expression. J Biol Chem. 1988;263:8450–8457. [PubMed] [Google Scholar]
- [64].Werten S, Moras D. A global transcription cofactor bound to juxtaposed strands of unwound DNA. Nat Struct Mol Biol. 2006;13:181–182. [DOI] [PubMed] [Google Scholar]
- [65].Zhao Y, Zhang Y, Huang J, et al. The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus. Proteins. 2019;87:257–264. [DOI] [PubMed] [Google Scholar]
- [66].Huang J, Zhao Y, Liu H, et al. Substitution of tryptophan 89 with tyrosine switches the DNA binding mode of PC4. Sci Rep. 2015;5:8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dekker J, Kanellopoulos PN, Loonstra AK, et al. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. Embo J. 1997;16:1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lim F, Peabody DS. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 2002;30:4138–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wu HN, Uhlenbeck OC. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987;26:8221–8227. [DOI] [PubMed] [Google Scholar]
- [70].Nagai K. RNA-protein complexes. Curr Opin Struct Biol. 1996;6:53–61. [DOI] [PubMed] [Google Scholar]
- [71].Hardin JW, Hu YX, McKay DB. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J Mol Biol. 2010;402:412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hargous Y, Hautbergue GM, Tintaru AM, et al. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. Embo J. 2006;25:5126–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Oberstrass FC, Auweter SD, Erat M, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. [DOI] [PubMed] [Google Scholar]
- [74].Auweter SD, Fasan R, Reymond L, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. Embo J. 2006;25:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Skrisovska L, Bourgeois CF, Stefl R, et al. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007;8:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Clery A, Jayne S, Benderska N, et al. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nat Struct Mol Biol. 2011;18:443–450. [DOI] [PubMed] [Google Scholar]
- [77].Dominguez C, Fisette JF, Chabot B, et al. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol. 2010;17:853–861. [DOI] [PubMed] [Google Scholar]
- [78].Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–2131. [DOI] [PubMed] [Google Scholar]
- [79].Ding J, Hayashi MK, Zhang Y, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Schneider T, Hung LH, Aziz M, et al. Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. Nat Commun. 2019;10:2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


