ABSTRACT
Translation initiation is a critical facet of gene expression with important impacts that underlie cellular responses to stresses and environmental cues. Its dysregulation in many diseases position this process as an important area for the development of new therapeutics. The gateway translation factor eIF4E is typically considered responsible for ‘global’ or ‘canonical’ m7G cap-dependent translation. However, eIF4E impacts translation of specific transcripts rather than the entire translatome. There are many alternative cap-dependent translation mechanisms that also contribute to the translation capacity of the cell. We review the diversity of these, juxtaposing more recently identified mechanisms with eIF4E-dependent modalities. We also explore the multiplicity of functions played by translation factors, both within and outside protein synthesis, and discuss how these differentially contribute to their ultimate physiological impacts. For comparison, we discuss some modalities for cap-independent translation. In all, this review highlights the diverse mechanisms that engage and control translation in eukaryotes.
KEYWORDS: Translation, RNA processing, RNA regulons, therapeutics, eIF4E, cancer
Introduction
A plethora of studies into epigenetics, genomics and transcriptional regulation have yielded important insights into the pathogenesis of human disease. However, proteomic studies revealed that the transcriptome does not always predict the proteome [1]. This prompts the question: why not? This dissociation is due, at least in part, to dysregulation at the post-transcriptional level, including processes such as nuclear RNA export and translation. As a consequence, it can elevate the production or alter the form of proteins that function in survival, proliferation, migration, invasion and metastases. Often, networks of RNAs encoding proteins involved in the same biological process, known as RNA regulons, are disrupted simultaneously, leading to a wide array of biological changes supporting disease phenotypes [2,3]. Several studies demonstrated that RNA processing, including dysregulated RNA export and translation, can contribute to aberrant control of RNA regulons thereby driving human diseases such as cancer and neurodegenerative conditions [4–6]. Thus, it is critical to understand the molecular bases driving and controlling these processes.
Recent studies have highlighted what has been known for some time: there are multiple means to initiate translation in eukaryotes and that this diversity underpins responses to stress and other environmental cues [5,7–12]. Furthermore, key factors in translation also play direct roles in other aspects of RNA processing and thereby impact on multiple levels of post-transcriptional control simultaneously [12,13]. For instance, some proteins recognize the methyl-7-guanosine (m7G) cap on the 5ʹend of RNAs, and these proteins can act in both the recruitment of RNAs to the ribosome via their cap-binding activity as well as function in other levels of RNA processing or independent roles in translation [11] (Table 1). Examples of this include the eukaryotic translation initiation factor eIF4E and the cap-binding complex CBC [12,13]. This pliability and modularity of translation is not limited to the cap-binding components of the initiation machinery, but also include diversity in the platform proteins required for assembly of the initiation complex including eIF4G, DAP5 (Death Associated protein 5) also known as p97, the CBC-dependent translation initiation factor (CTIF), and the threonyl-tRNA synthetase (TRS) [5,7–12]. Further, there is also cap-independent translation which implements Internal Ribosome Entry Site (IRES), Cap-Independent Translation Elements (CITEs) or even small potyviral proteins that can substitute for the m7G cap in vitro [14–16]. This modularity of translation initiation and its integration into other forms of RNA processing highlight the plasticity of post-transcriptional control. Their relative contributions to steady state and stress-responsive translation differ. In this review, we explore these different strategies and compare these with regard to the machinery used, the RNAs targeted and the relevant contexts. We also discuss the impact of dysregulation of these processes and the relevance to their therapeutic development including results of early phase clinical studies targeting eIF4E as a model. Unfortunately, due to space restrictions, we were not able to describe all the outstanding papers that merited discussion.
Table 1.
Multiplicity in function of translation initiation factors.
| Factor | Translation Function | Other Functions |
|---|---|---|
| eIF4E | m7G cap engagement | RNA export, 3ʹend processing, effects on eIF4A helicase activity |
| CBC | m7G cap engagement | General processing (splicing, export etc.) |
| eIF3d | m7G cap engagement | Recruitment of 40S ribosome subunit to eIF4F by eIF4G interactions and roles in IRES translation |
| PARN | m7G cap engagement | Shortening of poly(A) tails of mRNAs |
| eIF4G | Assembly platform for initiation binding eIF4E and eIF3d | |
| CTIF | Assembly platform for initiation binding CBC and eIF3g | |
| DAP5/p97 | Assembly platform for eIF3d or PARN dependent translation | Acts in some forms of IRES dependent translation and others |
| TRS | Assembly platform for eIF4E2 cap-dependent translation | tRNA synthetase |
eIF4E-dependent, cap-dependent translation: a reference for translation initiation
Translation initiation via eIF4E is often considered ‘canonical translation’ [17]. As we discuss in this review, there are a multiplicity of translation initiation mechanisms and thus, it may be more relevant to describe eIF4E-mediated translation as ‘reference translation’. eIF4E binds the m7G cap structure on the 5ʹend of mRNAs and associates with the eIF4F complex (Fig. 1A) which in turn associates with the 43 S pre-initiation complex (PIC) [17]. eIF4F is comprised of the assembly platform protein eIF4G and the DEAD box helicase eIF4A. The 43S PIC is comprised of the 40S ribosomal subunit and several other eIFs e.g. eIF1, eIF1A, eIF3, eIF5 and the initiator tRNAMeti-eIF2-GTP complex [17]. eIF4E and eIF4G play central roles in this process. eIF4G utilizes a YXXXXLΦ (where X is any residue and Φ is any hydrophobic) consensus binding motif and a separate auxiliary motif to directly bind the dorsal surface of eIF4E [18–20] (Fig. 1A, 2A–B). eIF4E directly interacts with the RNAs via the m7G cap in its cap-binding site, which is located on the opposite face to its dorsal surface [18,19] (Figs. 1A, 2A–C). In this way, eIF4E can bind both m7G-capped RNAs and eIF4G simultaneously. The poly(A) tail of mRNAs also stimulates translation [17]. This is usually considered to occur because the poly(A) binding protein (PABP) binds to eIF4G, circularizing the RNA and increases the affinity of eIF4E for the cap via its interactions with eIF4G [17] (Fig. 1A). However, recent live-cell imaging studies also demonstrate that the RNAs may not be circularized during translation leading to potential questions as to how the poly(A) tail and PABP stimulate translation [21,22]. eIF4G serves as the platform for assembly of 43S PIC and eIF4F, including eIF4E bound to m7G-capped RNA [17]. Following assembly, the 43S PIC scans the 5ʹUTR until it finds a start codon, and then engages the 60S ribosome subunit to form the 80S ribosome, which can then engage translation. Cryo-EM studies have revealed the first snapshots of this complex [23].
Figure 1.
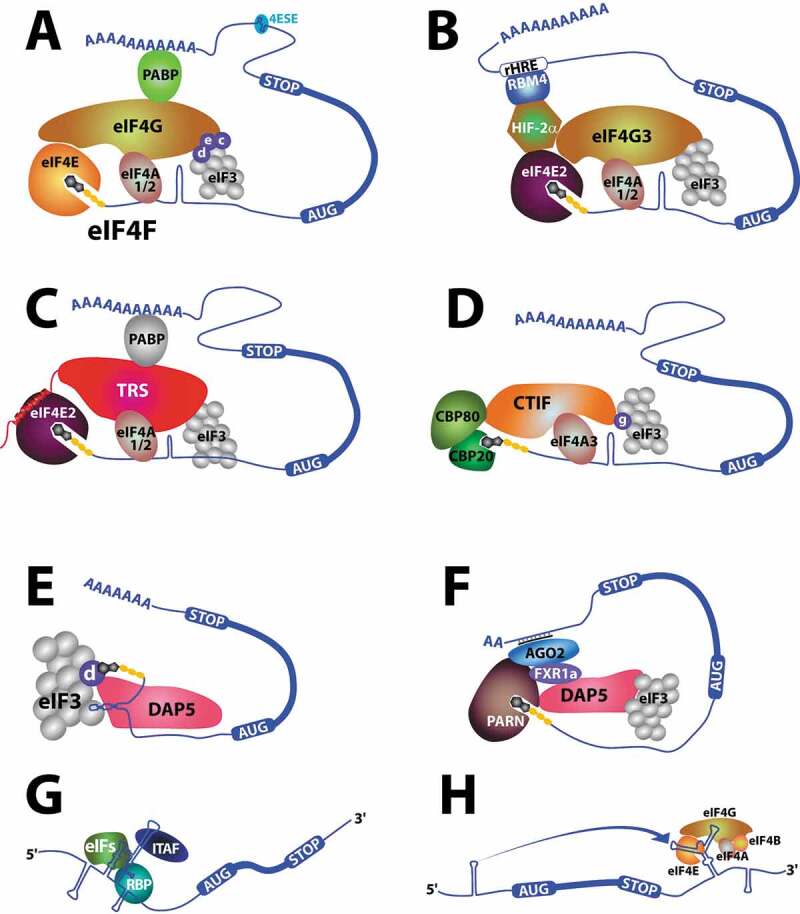
Models of different forms of cap-dependent translation and for comparison, two models of cap-independent translation. Not all factors are shown for clarity and positioning of factors in the complexes is approximate. The m7G cap is depicted as a grey ring system on the 5ʹ end of the RNAs. Circularization of RNAs is traditionally considered to be accomplished via the 3ʹ poly(A) tail, but this is controversial (see text). For panels G & H, there are multiple forms of IRES (G) and CITE (H) translation, only one modality of each is shown here. In panel G, RBP (RNA binding protein) represents a multitude of factors that can be co-opted for translation.
Figure 2.
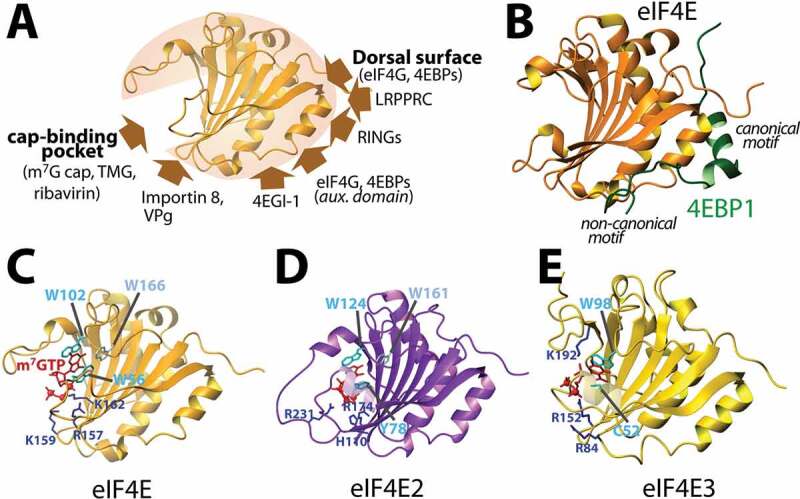
Structural insights into the eIF4E family. Panel A. Structure of apo-eIF4E demonstrating the relative location for the binding of described factors which engage and/or regulate this protein. Panel B. eIF4E/4E-BP1 demonstrates the arrangement of 4E-BP1 on the dorsal surface. Panels C, D and E. Structures of all three members of the eIF4E family demonstrating the similar means by which they bind the m7G cap. This same pocket is used to bind TMG cap for eIF4E. PDB codes are as follow: 4UED (B); 3AM7 (C); 2JGB (D); 4B6U (E).
There are substantial biochemical and structural studies focussed on the engagement and regulation of eIF4E [18,19]. The dorsal surface of eIF4E is used as an important control point for eIF4E’s translation (via eIF4G) and other biochemical activities (see below) (Figs. 1A, 2A). The eIF4E binding proteins (4E-BPs) bind to eIF4E, compete for eIF4G binding by using the same structural strategies to interact with the dorsal surface of eIF4E and thereby impair translation [17,19](Fig. 2B). The phosphorylation status of 4E-BPs impacts on their affinity for eIF4E by causing a structural change whereby hypo-phosphorylated 4E-BP binds to eIF4E and competes for eIF4G-binding, disrupting this form of translation [24]. Phosphorylated 4E-BPs have substantially reduced affinity for eIF4E [17]. Interestingly, structural studies indicate that hypo-phosphorylated 4E-BP1 is an intrinsically disordered protein whereby phosphorylation reduces binding by inducing 4E-BP1 to adopt a tertiary fold and this form of the protein no longer binds eIF4E [24]. Structural and biochemical studies revealed that eIF4E also binds to both the m7G cap and 4E-BPs simultaneously suggesting that 4E-BP1 could suppress translation by sequestering both eIF4E and the target RNA [25,26]. There are several other factors that compete for eIF4G-binding using a similar dorsal-surface competition strategy including a series of homeodomain proteins, but in other cases factors use distinct structural modalities [5,18,19,27–32] (Fig. 2A). For example, the RING domains from the promyelocytic leukaemia protein (PML) or the arenaviral protein Z bind an overlapping but distinct region of the dorsal surface of eIF4E, as compared to eIF4G, and this leads to a wide-scale conformational changes in the cap-binding site thereby reducing m7G cap affinity [27,33–35]. The RING domain from HHARI similarly binds the dorsal surface of the eIF4E family member eIF4E2 [27,33,34,36] (Fig. 2A). eIF4E also has cap-independent effects on translation. Notably, eIF4E can alleviate eIF4G-mediated repression of eIF4A helicase activity through protein-protein interactions [37]. eIF4E does not need its cap-binding activity to modulate translation in this manner [37]. This finding provides an interesting model as to the preferential effects of eIF4E on mRNAs with highly structured 5ʹUTRs.
Importantly, many findings from several groups support the notion that eIF4E is likely NOT a global mediator of translation. For example, studies using antisense oligonucleotides (ASOs) to eIF4E demonstrated that while the ASOs substantially reduced eIF4E RNA and protein levels, the global impact on protein synthesis, as measured by 35S Met incorporation, was very modest. Indeed, there was ~60-80% of translation relative to controls [38]. Thus, the reduction in global translation upon eIF4E reduction was only between 20–40%. At the same time, the ASO treatments clearly impacted on the cancer-associated activities of eIF4E, just not global translation. This is consistent with eIF4E impacting on the production of specific proteins involved in these activities. This is supported by observations that genetic reduction of eIF4E in mouse models did not reduce total translation, and indeed could be reduced by ~50% with little impact on the mice [39]. These findings agree with the observation that eIF4E can be targeted with inhibitors (e.g. ribavirin) and in all of these cases, global translation is not targeted but rather only the translation of specific subgroups of RNAs is impaired [38,40]. For instance, ACTIN and GAPDH RNAs are often used for controls in translation experiments because housekeeping transcripts are well known to be unaffected by eIF4E expression [41]. In agreement with these findings, genetic reduction in eIF4G also had a modest impact on global protein synthesis as assessed by 35S Met incorporation [42]. Reduction in eIFGI or eIF4GI/II only reduced global protein synthesis by ~20% i.e. translation was ~80% of controls. Finally, others have shown that different mechanisms are used for about 20–30% of translation e.g. via eIF3d [9]. In summary, there is ~60-80% of translation that is independent of eIF4E and/or eIF4G. Consistently, eIF4E preferentially engages translation of a large subgroup of transcripts with highly complex 5ʹ untranslated regions (UTRs) but not housekeeping transcripts [17–19,43]. In this way, eIF4E overexpression leads to increased numbers of ribosomes per transcript on a select group of RNAs thereby elevating translation efficacy. Many of these transcripts encode proteins involved in oncogenic RNA regulons [2]. Indeed, eIF4E is elevated in a wide variety of cancers and eIF4E has been targeted in multiple cancer clinical trials [43–47] (see cancer section).
eIF4E: cap-dependent functions beyond translation
While most focus on eIF4E has related to its role in the cytoplasm, it is also localizes to the nucleus [43,44,48–54] (Table 1). eIF4E forms nuclear bodies in many organisms e.g. yeast, Drosophila, Xenopus, mouse and human [27,48,55,56]. Depending on the cell type, between 33% to 70% of eIF4E is found within the nucleus [48,52,55]. Its best characterized nuclear role is in the export of a subset of RNAs, thereby increasing their cytoplasmic concentrations and thus, providing better availability to the translation apparatus; and in some cases, increases their translational efficiency (number of ribosomes per mRNA) in the cytoplasm [1,10,11,27,57]. At the biochemical level, eIF4E requires target RNAs to have the m7G cap; and additionally, translation targets need a complex 5ʹUTR while RNA export targets require a ~50-nucleotide element in their 3ʹUTR denoted an eIF4E sensitivity element (4ESE) to select RNA targets [1,10,11,27,57]. Genome-wide analyses revealed that ~3000 RNAs are nuclear targets of eIF4E, many acting in pathways driving proliferation, survival, invasion and malignant transformation [40,58,59]. By contrast, housekeeping transcripts e.g. GAPDH are neither mRNA export nor translation targets of eIF4E [27,57].
Nuclear eIF4E plays a direct role in the RNA export of these transcripts. eIF4E forms a 4ESE-RNA export complex comprised of LRPPRC (leucine rich pentatricopeptide repeat C-terminus protein), the 4ESE RNA and the CRM1/XPO1 export receptor [40,43] (Fig. 2A). Indeed, LRPPRC directly interacts with all of these components acting as an assembly platform binding the dorsal surface of eIF4E, using similar structural features to eIF4G and 4E-BPs [43,60]. eIF4E-dependent export is distinct from bulk mRNA export on many levels e.g. it uses CRM1 rather than the bulk mRNA export receptor NXF1 to transit through the nuclear pore [49,50,60,61]. Indeed, knockdown of NXF1/TAP1 does not impair this RNA export activity while Leptomycin B treatment does [50]. Moreover, eIF4E remodels the nuclear pore complex to promote 4ESE mRNA export [62]. In all, eIF4E drives protein expression by increasing the cytoplasmic levels of specific transcripts (via mRNA export) and in some cases, additionally enhancing their translation efficiency in the cytoplasm. Other RNAs are only translation targets of eIF4E providing further layers of regulation. Additionally, nuclear eIF4E plays newly described roles in 3ʹend processing [63] and m7G RNA capping [64] and these too could play important roles with regard to its oncogenic activities. In all, eIF4E appears to be a cap-chaperone protein, escorting transcripts through various types of RNA processing defined by cis-acting elements in RNAs and context specific trans-acting factors [13]. Indeed, eIF4E appears to bind other elements than the m7G cap, suggesting it may have a broader chaperone activity (see below).
A central question relates to eIF4E trafficking given it has no nuclear localization signal (NLS). One factor involved in its nuclear import, likely of many, is Importin 8 [54,61]. NMR and biochemical studies revealed that Importin 8 binds the cap-binding site of eIF4E (Fig. 2A); thereby revealing a new RNA surveillance mechanism which protects eIF4E engaged in translation from nuclear import [54,61]. Further, this interaction would prevent the re-import of eIF4E-RNA export complexes and thus, reduce futile re-import cycles of these transcripts. eIF4E nuclear import and export are important in several human diseases including Acute Myeloid Leukaemia (AML). The PML and Z proteins prevent eIF4E-dependent RNA export through binding the dorsal surface (Fig. 2A) and reducing the affinity of eIF4E for m7G-capped RNAs by ~50-100 fold by altering motions in the cap-binding site [27,33–35]. These also provide examples of different structural modalities for eIF4E regulation beyond the strategies used by eIF4G and 4E-BPs.
eIF4E is also found in cytoplasmic foci including in P-bodies and stress granules [65–68]. In P-bodies, eIF4E likely plays a role in sequestration of transcripts from the translation machinery, as these bodies do not typically have ribosomes and could be involved in decay. RNAs have been shown to move between active translation and these P-bodies, consistent with the idea that they can re-engage in translation once released from sequestration [68]. eIF4E may also play a role in RNA stability in this case by protecting the 5ʹend of the transcripts from decapping enzymes and subsequent degradation in these foci [65–68].
eIF4E can interact with transcripts independently of the m7G cap
The basic principle of engagement of mRNAs by this translation initiation complex is that eIF4E binds their m7G cap on the 5ʹend of RNAs [18,19]. High-resolution crystal structures and NMR solution structures indicate that eIF4E uses two tryptophan residues (W56 and W102 in humans) to intercalate the methyl-7-guanosine moiety. The partial positive charge on the m7G cap is thought to be required for its interactions with the pi-electron clouds of the tryptophan, or in some cases, tyrosine or phenylalanine residues. It also uses a positively charged patch constituted of R157, K159, K162 which associates with the phosphate moieties of the m7G cap [18,19] (Fig. 2C). eIF4E also binds other types of caps [18,19]. Crystal structures of nematode and human eIF4E with 2,2,7-trimethyl guanosine (TMG) indicate that the TMG is bound in a similar manner to m7G cap albeit with lower affinity (100 fold less in humans; 6 fold in nematodes) [69]. The TMG cap is used to actively engage eIF4E’s translation activity in nematodes [69]. However, given the high concentrations of UsnRNAs in cells, it may be possible that eIF4E-TMG interactions occur in mammals. This remains to be tested. Also of relevance, eIF4E does not require m7G cap-binding for folding, but cap-binding does cause local conformational changes around this site [70].
The viral protein genome linked (VPg) from plant potyviruses provides insights into other ways that eIF4E can be engaged to act in translation (Fig. 3A). Potyviruses are small RNA viruses which present a major threat to a wide array of agricultural crops. After infection, VPg protein is covalently attached via a hydroxyl group of a tyrosine residue to the 5ʹ end of the viral genomic RNA [71,72]. Importantly, potyvirus VPg has no similarity at the sequence or structure level with VPgs from other viruses [15,73]. Botanists and biochemists had identified a strong genetic link between VPg and eIF4E [74–78]. Indeed, plants evolve resistance to infection through mutation of eIF4E. Studies into the structure of potyvirus VPg and the VPg-eIF4E complex were conducted using NMR and crosslink-mass spectrometry [15]. VPg is a beta-sheet protein with adjacent alpha helices which is used to directly bind the cap-binding site of eIF4E [15]. Indeed, NMR and biochemical studies demonstrated that VPg competed for m7G cap-binding, with similar affinities for eIF4E. Free VPg (in the absence of conjugated RNA) successfully competed for all the cap-dependent activities of eIF4E in the cell suppressing translation, RNA export and oncogenic transformation [15]. VPg-RNA conjugates were translated with similar efficiency to m7G-capped RNAs indicating that VPg could both bind eIF4E and engage the translation machinery, as expected for a ‘cap’ [15]. Thus, VPg functionally substitutes for the m7G cap in vitro, but this remains to be tested during viral infection. Furthermore, VPg exhibited structural homology to the human kinesin EG5 (but with no sequence similarity), suggesting that human proteins may bind to eIF4E in a similar manner [15]. In this case, one would predict that EG5 would only bind RNA-free eIF4E.
Figure 3.
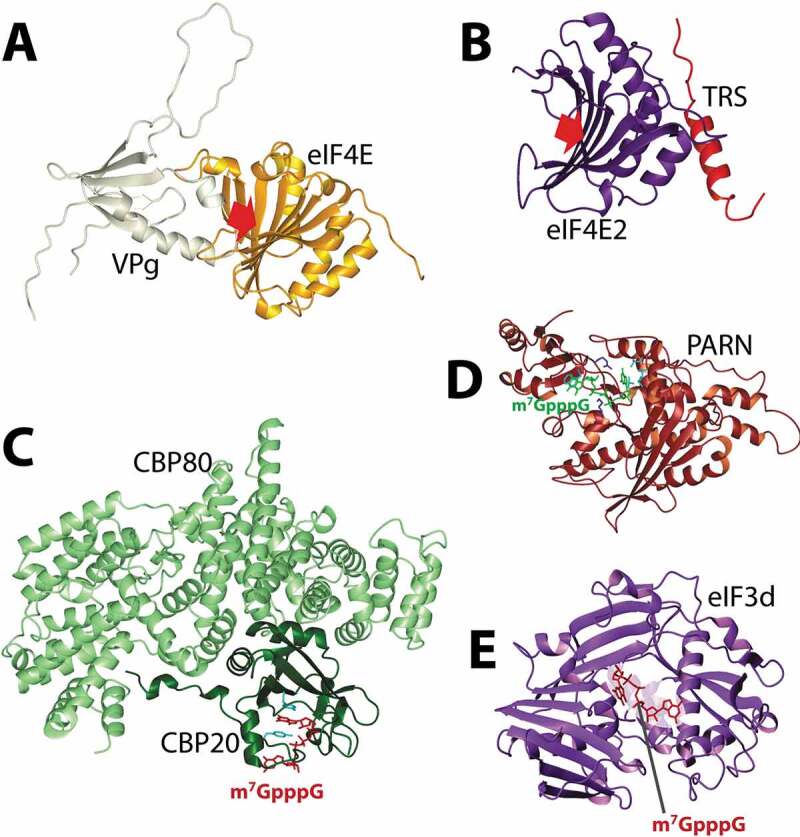
Structural insights into cap substitutions and other cap-binding proteins. Panel A. NMR based model of eIF4E/VPg complex highlighting the competition with the m7G cap in the cap-binding pocket (red arrow). Panel B. eIF4E2 bound to TRS, using a similar surface used by eIF4E to bind eIF4G or 4E-BP1. The crystal structure was not solved in the presence of a cap analogue, but the cap-binding pocket is shown with the red arrow. Panels C and D. Structures of factors identified demonstrating the similar means by which they interact with the m7G cap. This same pocket is use to bind TMG cap for CBC. Panel E. eIF3d cap-binding domain modelled with the cap analogue placed according to its position in DXO. PDB codes are as follow: 5XLN (B); 1H2U (C); 3D45 (D); 5K4B (E).
Interestingly, eIF4E also binds RNA elements. eIF4E binds the 22-nucleotide trans-spliced leader sequence in nematodes. Here, association of this element induces conformation changes in the cap-binding pocket of eIF4E which increases affinity for TMG [69]. In an example of cap-independent translation mediated by eIF4E, eIF4E binds the PTE (Panicum mosaic virus-like translational enhancer) element of Pea enation mosaic virus [79]. eIF4E also binds a paired stem loop element in the 5ʹUTR of Histone H4 mRNA, structurally similar to the 4ESE export element in the 3ʹUTRs according to RNase mapping, to promote translation [80]. This element binds independently of the m7G cap. Conversely, eIF4E can interact with the m7G cap of this RNA independently of this other element. These studies provide strong evidence that eIF4E can engage a wide variety of modalities to interact with RNAs, and by inference recruit these to the translation and/or RNA export machineries.
eIF4E family homologues can be activators and repressors: context is everything
There are three eIF4E family members: eIF4E1 referred to here as eIF4E, eIF4E2 (also known as 4EHP, homologous protein) and eIF4E3 [81]. These three proteins have highly similar structures. Like eIF4E, eIF4E2 binds the m7G cap by intercalating it between two aromatic residues [82] whereas eIF4E3 binds the m7G cap using a novel arrangement of one conserved tryptophan and a series of hydrophobic interactions including a cysteine at the position of the other tryptophan [81] (Fig. 2C–E). eIF4E2 both activates and inhibits translation [11,83,84]. Structural studies demonstrated that eIF4E2 can form complexes with the Grb10-interacting GYF protein 2 (GIGYF2) and the zinc finger protein 598 to repress translation during embryonic development [84]. During hypoxia, eIF4E2 replaces eIF4E in translation initiation [83]. Here it assembles with hypoxia induced factor 2α (HIF-2α) and the RNA binding protein RBM4 to regulate global protein synthesis during hypoxia [84] (Fig. 1B). Recent structural and biochemical studies revealed that eIF4E2 can form a completely novel translation initiation complex with the tRNAThr synthetase complex (TRS) and eIF4A [11] (Fig. 1C). Here, the TRS substitutes for eIF4G. TRS specifically bound eIF4E2, not eIF4E or eIF4E3 and conversely, TRS was the only aminoacyl-tRNA synthetase examined that bound eIF4E2 [11]. A fragment of TRS uses a consensus eIF4E-binding site motif to associate with the dorsal surface in a manner similar to eIF4G (Fig. 3B). Consistent with this role, TRS also physically interacts with eIF3 (b, d, f, l) and PABP [11]. TRS-mediated translation is positioned to modulate many transcripts with interactions reported for over 2000 RNAs [11]. These RNAs play important biological roles and indeed, eIF4E2 and TRS impact on translation of transcripts that ultimately impact on endothelial migration and tubal formation [11].
In parallel, eIF4E3 can act both as a repressor of, or initiate, translation [81,85]. eIF4E3 overexpression correlated with reduction in eIF4E target proteins (but not housekeeping factors), and reduced oncogenic foci formation [81]. Mutation of the tryptophan 98, (equivalent to W102 in eIF4E) which is important for cap recognition, reversed these effects [81]. Under these conditions, eIF4E3 did not immunoprecipitate with eIF4G [81]. However, during MNK inhibition which reduces the levels of phosphorylated eIF4E and thus reduces eIF4E activity in both RNA export and translation [86], eIF4E3 bound eIF4G and activated translation [85]. In all, the biochemical roles played by eIF4E family members are diverse and context dependent.
CBC: pioneer round, translation under stress and viral hijacking
The cap binding complex (CBC), comprised of a heterodimer of CPB20 (NCBP2) and CBP80 (NCPB1), plays roles in nuclear RNA processing binding transcripts shortly after they are capped, and escorts these through multiple maturation steps [12] (Fig. 1D, 3C, Table 1). CBC also binds TMG and plays important roles in the processing of UsnRNAs including exporting them to the cytoplasm for their maturation [12]. One of the best-characterized roles for CBC-dependent translation is in nonsense mediated decay (NMD), which is used to detect premature termination codons (PTCs) [87,88]. The pioneer round of translation occurs during or just after nuclear export of transcripts. RNAs with PTCs would be less stable and thus degraded after detection of PTCs via this mechanism. Interestingly, NMD can also occur on eIF4E-bound RNAs [89,90]. In this way, CBC-dependent translation is considered to be a facet of RNA quality control which precedes eIF4E-dependent, and perhaps other forms, of translation [12].
However, recent studies suggest that the line between CBC and eIF4E may be more blurred [12]. There are many commonalities and some important distinctions relating to the translation initiation complexes formed. For CBC-dependent translation, CBC binds to CTIF which plays a parallel role to eIF4G (Fig. 1A versus 1D). Both CTIF and eIF4G bind to eIF3 through a common evolutionarily conserved domain known as MIF4G (middle of 4G domain) [12]. However, this MIF4G domain binds to different constituents of eIF3 to recruit the 40S ribosome. For example, CTIF associates with eIF3g while eIF4G interacts with eIF3c, eIF3d and eIF3e. For the case of CBC-dependent translation, eIF3g is thought to act as a platform to access the start codon. In terms of the helicase activity associated with unwinding during scanning, these two types of translation again exhibit substantial differences. CBC-mediated translation utilizes eIF4A3 while eIF4E uses eIF4A1/A2 with co-factors eIF4B and eIF4H [91–95]. Indeed, genetic targeting of eIF4A3 only impairs CBC-dependent translation [91]. In eIF4E-dependent translation, the poly(A) tail plays an important role, particularly through interactions between PABP and eIF4G presumably by increasing the affinity of eIF4E for the m7G cap [17,26,96] (Fig. 1A). The role of the poly(A) tail in CBC-dependent translation is not yet well understood [12] (Fig. 1D). Aside from PTCs and NMD, some transcripts and cellular contexts favour CBC-dependent translation. For instance, replication-dependent histones do not contain a poly(A) tail but undergo alternative 3ʹend processing generating a stem loop structure. It has been proposed that the stem-loop binding protein (SLBP) which binds the histone stem-loop structure could bind eIF4G and thereby substitute for a poly(A) tail [12]. Recent studies suggest that these transcripts undergo CBC-dependent translation at steady state [12]. Stress conditions such as nutrient deprivation or hypoxia also influence the type of translation, as can development or differentiation. For instance, during stress, eIF2α is phosphorylated while 4E-BP1 is dephosphorylated [5]. Phosphorylation of eIF2α leads to reduced amounts of available eIF2-GTP, thus reducing amounts of eIF2-GTP-tRNAMeti initiator complexes which decreases both CBC and eIF4E-dependent translation [12]. By contrast, 4E-BP1 hypo-phosphorylation increases its affinity for eIF4E, reducing binding of eIF4E to eIF4G and thereby selectively repressing eIF4E-dependent translation [17]. In this way, CBC-dependent translation may be more resilient under stress conditions that do not affect eIF2-GTP availability. An example of this is hypoxia. In acute hypoxia, reduction in eIF2-GTP availability represses both types of translation while for prolonged hypoxia, 4E-BP1 is hypo-phosphorylated and eIF4E is sequestered into cytoplasmic foci thereby selectively repressing eIF4E-dependent translation [97–99]. Under these conditions, CBC-dependent translation increases [99]. Another example of this comes from hyperosmotic stress in yeast. Here, CBC actively engages polysomes during hyperosmotic stress, where it mediates translation of around 600 transcripts, about 10% of the transcriptome [100]. These same conditions inhibit eIF4E-mediated translation. Indeed, deletion of eIF4E during osmotic stress leads to increased growth while simultaneously targeting eIF4E and CBC causes synthetic sickness at both restrictive and permissive temperatures [100].
CBC-dependent translation is also co-opted during HIV infection. In lymphocytes, eIF4E mediated translation is inhibited during infection because of reduced levels of phospho-eIF4E and decreased phospho-4E-BP1. In this case, the virus uses CBC-mediated translation of unspliced HIV RNAs in order to bypass loss of eIF4E activity [101]. CBC binds Rev which in turn associates with eIF4A1 [102]. Some HIV RNAs are TMG capped [103]. The CBC link here is interesting since CPB20 has a ~70-fold increased affinity for the TMG (for the dinucleotide form) while eIF4E has a ~100 fold higher affinity for m7G cap [69,104].
eIF3: roles in eIF4E-dependent and eIF4E-independent translation
eIF3 is a multiprotein complex which is typified by its role in binding the 40S ribosome during translation [17]. Mammalian eIF3 is comprised of 13 subunits [17]. In the context of the 43S PIC, the cryo-EM structure of eIF3 revealed that eIF3d is positioned near the RNA exit, far from the m7G cap [23]. However, recent studies demonstrated that two components of this system, eIF3d and eIF3l, can directly bind the m7G cap [9,10]. Indeed, in the case of eIF3d, a high-resolution crystal structure reveals what was an unexpected cap-binding site which is highly similar to the one observed in the DXO cap-cleaving enzyme [10] (Figs. 1E, 3E, Table 1). Initial studies revealed that eIF3d regulated the translation of c-Jun suggesting that eIF3d cap-binding may be a rare form of translation to overcome stress [10]. However, global studies in cancer cells showed that ~20-30% of translation occurred via the cap-binding activity of eIF3d [9]. Here, eIF3d binds the eIF4G homologue DAP5/p97, which importantly lacks the eIF4E binding domains of eIF4G (Fig. 1E). Biochemical analysis demonstrated a direct interaction between eIF3d and DAP5 [9]. Further the eIF4F factors (eIF4E, eIF4GI, and eIF4A1) were not present in the eIF3d-DAP5 complexes [9]. Indeed, inhibition of eIF4E-dependent translation did not alter translation by eIF3d-DAP5 [9,10]. These studies also demonstrated that about 9% of poorly translated and 13% of well-translated RNAs were DAP5-dependent highlighting the global relevance of DAP5 to translation [9]. The RNA determinants for eIF3d-DAP5 versus eIF4E-eIF4G translation are not yet known, and it seems that some RNAs may be substrates of both pathways. Along the same theme, eIF3l has also been shown to directly bind both the m7G methylated and unmethylated forms of the cap positioning it as another factor that might play a role in eIF4E-independent, cap-dependent translation [105]. Finally, eIF3-mediated, eIF4E-independent translation was shown to be important for the translation initiation of specific transcripts during Drosophila development [106]. During pruning of sensory neurons, eIF4E-dependent translation is turned off. Here, eIF3 and eIF4A are required for translation initiation of Mical mRNA. This depends on the 5ʹUTR of Mical RNA and the helicase activity of eIF4A [106].
MicroRNA-dependent translation using PARN as the cap-binding protein
There is typically a decline in eIF4E-dependent translation upon mTOR inhibition due to reductions in phosphorylation of eIF4E and 4E-BPs. This is observed during G0 in the AML cell line THP1 [7]. Studies into the molecular basis of this revealed a novel translation initiation complex associated with Fragile-X-mental-retardation protein 1a (FXA1a) [7] (Figs. 1F, 3D, Table 1). Here, FXR1a and Argonaut family member 2 (AGO2) are part of a complex that activates translation initiation of transcripts including TNFα and MYT1 [7]. Here, RNAs with short or no-poly(A) tails undergo increased translation (Fig. 1F). FXR1a-AGO2 complexes directly bind microRNA-binding sites in the 3ʹUTR of target transcripts, thereby selecting the RNA targets. FXR1a-microRNP also physically associates with poly(A) specific ribonuclease (PARN) and DAP5/p97, the eIF4G homologue [7]. Importantly, there is no eIF4E or eIF4G present in this translation complex. In this process, PARN plays a dual role: 1. ensures that the 3ʹend remains with short or no A tails and 2. directly binds the m7G cap of the target RNAs. Indeed, there are several biochemical and structural studies that demonstrate PARN binds the m7G cap, and this interaction is independent of its de-adenylase activity [107–109] (Fig. 3D). Interestingly when mTOR activity is low during G0, PARN binding to the m7G cap increases ~2.5 fold relative to controls [7]. Under these same conditions, eIF4E still binds the cap but this activity is reduced by ~25%. Thus, both forms of translation likely occur side-by-side. DAP5 also acts as the assembly platform for PARN and the FXR1a-microRNP. Like eIF4G, DAP5 recruits eIF3 and the 40S ribosome subunits to mediate translation. DAP5 has also been implicated in IRES driven translation [110–112] (Table 1, see below) and additionally interacts with eIF3d to engage translation of different subsets of RNAs indicating it is a widely utilized translation platform [68,69]. The number of transcripts associated with FXR1a-PARN-DAP5 translation is not yet known, but provides an elegant model for the initiation of translation of RNAs without A tails or with short A tails as well as a model of microRNA engagement.
Cap-independent, IRES-mediated translation
Internal ribosome entry site (IRES) mediated translation is a strategy used by many positive-sense single-stranded RNA viruses which do not have capped RNAs and lack the capacity to steal caps. IRES’s are cis-acting RNA elements that either directly interact with the ribosome or interact with adaptor proteins (IRES trans-acting factor, ITAFs) which mediate the association with the translation machinery [14,113]. ITAFs can act as adaptors and/or remodel the IRES RNA in order to activate it [14,113]. Viruses often reduce host cell translation initiation in order to compete for the machinery. These include such tactics as cleavage of factors such as PABP, eIF4G, eIF4A or eIF3, or inactivation of eIF2 which compromises host RNA translation [114–117]. In other cases, viruses must co-opt some of the translation machinery in order to translate their RNAs (see below) (Fig. 1G). While viral IRES are well characterized both biochemically and structurally, the existence of IRES elements in host RNAs is somewhat controversial [14,113]. A genome-wide study of IRESs in human and virus cells, suggested that up to ~10% of host-cell translation could be IRES mediated [118]. Interestingly in this study, IRESs were nearly equally distributed between 5ʹ and 3ʹUTRs and could be short, unstructured elements or complex structures [118]. However, these elements were not confirmed to act in translation initiation. Mechanisms for putative IRES activity in human transcripts are not delineated but suggested to run along similar lines to viral IRESs [118]. One of the roadblocks in developing a fuller understanding of IRESs in human transcripts is the complexity of the assays required to definitively prove IRES-mediated translation; indeed there are many pitfalls with regard to commonly used assays which have been summarized elsewhere and thus are not repeated here [14,119,120]. Here, we discuss a few well-described mechanisms for viral IRESs. There are many excellent reviews highlighting in particular the structural aspects of IRES-ribosome interaction which we do not have space to cover here [14,113].
Picornaviruses were the first to be identified as IRES elements and provide an example of the diversity of these mechanisms [121,122]. There is substantial diversity even within this genus with 5 different classes of IRES elements identified, all characterized by substantial structural diversity [14,113]. These co-opt different facets of translation. For class I and class II IRESs, eIF4A, eIF2, eIF3 and the C-terminal domain of eIF4G are required to form the 48S initiation complex in vitro, and do not require eIF4E [123] (Fig. 1G). Type III IRESs depend on eIF4G and eIF4E, and increase the affinity of this eIF4F complex to the IRES as well as unwinding of the IRES by eIF4A [124]. Type IV IRES elements require eIF2 and eIF3 but not eIF4G. DAP5 is also proposed to be utilized also for some human IRES mediated activity [125].
Studies into Hepacivirus reveal a structural basis for the recruitment of eIF3 and provide an excellent example of how RNA structure can substitute for protein factors. The IRES is divided into three parts: domains II, III and IV. The L-shaped Domain II is involved in eIF5 induced GTP hydrolysis of eIF2 and other regions of Domain II bind the small ribosomal subunit [126,127]. Domain III uses different subdomains to contact eIF3 and the 40S ribosomal subunit. This includes the use of a GGG motif to bind the CCC triplet in 18S rRNA causing a conformational change [128–131]. Importantly, there is no one universal IRES structure. These elements can adopt a variety of structures which can either act directly through RNA-RNA interactions or use adaptor proteins to recruit the ribosome. These elements do not appear conserved at the sequence level and in some cases can be short unstructured elements. However, in viruses, specific virus families do retain some common structural features.
Cap-independent translation elements (CITEs)
CITEs are 3ʹ RNA elements that can facilitate cap-independent translation, usually via increasing the affinity of RNAs for translation initiation factors [16,132–135] (Fig. 1H). These elements are found in many plant positive-stranded RNA viruses which do not have transcripts with a m7G cap or 5ʹ VPg [16]. CITEs are characterized by different classes of structural elements. CITEs typically contain elements that bind a part of the eIF4F complex and additionally an RNA-RNA kissing-loop interaction forms between the CITEs and the 5ʹ end of the RNA to promote circularization. It remains unknown if these elements are found in, and/or contribute to, translation in animal cells.
eIF4E and cancer
eIF4E governs RNA regulons that underpin its oncogenic activities and is dysregulated in ~30% of cancers [4,40,57]. In cell lines, eIF4E overexpression promotes foci formation, growth in soft agar, and apoptotic rescue from a variety of stimuli [5,57,136]. In xenograft mouse models, elevated eIF4E correlates with increased tumour numbers, invasion and metastases [137]. In transgenic models of eIF4E overexpression, mice develop a variety of cancers [138]. eIF4E-mediated transformation was thought to rely only on increased translation of oncogenic mRNAs [58]. However, eIF4E’s nuclear functions are also critical for its oncogenic activities. For example, the W73A eIF4E mutant is deficient in stimulating translation but exports RNAs and transforms cells as well as wildtype eIF4E [27,49,50,139], while the S53A mutant is active in translation but deficient in RNA export and oncogenic transformation [62,140]. eIF4E’s nuclear import and ability to modify the nuclear pore also substantially contribute to its oncogenic activity [54,62]. eIF4E levels are increased in many cancers where it generally correlates with poor prognosis [57]. In a subset of AMLs, eIF4E is substantially elevated and forms abnormally large nuclear bodies relative to CD34+ or bone marrow mononuclear cells from healthy volunteers [43,44,52,141]. This subset includes all examined French American British (FAB) M4/M5 AML subtypes as well as ~20-30% of M1 and M2 AML subtypes (>150 specimens examined to date) [43,44,52,141]. While FAB subtypes are no longer used, these provide evidence that a substantial number of AML patients have elevated, nuclear eIF4E relative to healthy volunteers. The nuclear enrichment of eIF4E in these AML specimens correlated with elevated eIF4E-dependent RNA export relative to normal cells [43,44,52,141]. Indeed, this can even lead to restructuring the extracellular surface architecture of cells which is related to its invasion and migration activities [136].
Ribavirin, an old antiviral drug, acts as a m7G-cap competitor directly binding eIF4E as shown by NMR and other biophysical techniques [51,142,143]. 3H ribavirin binds eIF4E demonstrating interactions in cells. Ribavirin inhibits eIF4E’s activities in mRNA export, translation, and oncogenic transformation [51,141,142,144,145]. In cell lines and primary specimens, ribavirin alone or in combination with other drugs targeted eIF4E in glioblastoma, multiple myeloma, diffuse large B-cell lymphoma (DLBCL), AML, infant acute lymphoblastic leukaemia and others [40,43,44,54,146]. RNAi knockdown of eIF4E reduces ribavirin activity supporting it acts via eIF4E [147,148]. These findings prompted multiple clinical trials targeting eIF4E in cancer. In AML patients, ribavirin treatment resulted in objective clinical responses including remissions [43,44] (ClinialTrials.gov NCT02073838). In patients, ribavirin blocks eIF4E’s association with Importin 8 leading to cytoplasmic retention of eIF4E, impaired eIF4E-dependent mRNA export and clinical responses [43,44,54]. Relapse correlated with nuclear re-entry of eIF4E and increased mRNA export due to chemical deactivation of ribavirin [3,43,54,142]. Other groups have also completed early-stage clinical trials targeting eIF4E with ribavirin e.g. castration-resistant prostate cancer and head and neck cancers and observed objective clinical responses [45,46]. There are >15 ongoing trials targeting eIF4E with ribavirin in cancer, e.g. Follicular Lymphoma (ClinicalTrials.gov NCT03585725).
eIF4E has also been targeted using antisense oligonucleotides (ASO) in the clinic. This strategy was very promising in early mouse models of prostate cancer in which ASOs substantially reduced eIF4E levels [38]. Importantly, ASOs would be expected to impact on the nuclear and cytoplasmic activities of eIF4E. The efficacy of eIF4E ASOs was investigated in the setting of advanced solid tumours in a phase I trial [47]. Unfortunately, the ASO treatment was not successful in humans. No patients achieved remissions, with 7 stable diseases and 15 progressive diseases out of 22 patients with only 2 patients on the study for more than 3 months [47]. The reduction in eIF4E levels in the mouse models was substantial, but in the patients, much less so. This suggests that there is a delivery issue with the ASO that needs to be overcome to improve efficacy.
Another translation initiation inhibitor was developed to interfere with the eIF4E-eIF4G interaction [149]. This compound, known as 4EGI-1 (Fig. 2A), strongly inhibits eIF4E-dependent, cap-dependent translation in cell lines without targeting eIF4F levels, but has not been used in patients to date [150,151]. Also, it appears to have multiple activities including increasing the interaction of 5ʹ cap RNAs with ribosomal complexes associated with inactive eIF2, suggesting that it is not only targeting eIF4E-eIF4G-mediated translation events [152].
There have also been substantial efforts targeting phosphorylation of eIF4E. Inhibition of eIF4E phosphorylation is linked to a strong reduction in its oncogenic activities and is linked to repression of both its RNA export and translation initiation activities, and perhaps its other functions [86]. Phosphorylation of eIF4E is carried out by MNK1 and MNK2 [153,154]. It should be noted that MNKs also phosphorylates a major RNA processing protein hnRNPA1 and thus the impact of MNK inhibition is not limited to its effects on eIF4E alone [153,154]. Ongoing studies seek to determine the efficacy of combining the MNK1/2 inhibitor eFT508 with anti-PD1/anti-PD-L in patients with solid tumours who could not achieve a response with anti-PD therapy alone (ClinicalTrials.gov NCT03616834). In summary, it is clear that targeting eIF4E activity is safe and well tolerated. This is consistent with the overall plasticity of translation initiation described in this review. Inhibition of other translation modalities might give more targeted effects. To date other translation initiation elements have not been targeted in clinical trials, but this is an exciting new direction.
Conclusions
The biological relevance of translation is clear. RNA processing including RNA translation enables well-poised adaptive responses to a wide array of environmental cues and contexts. Thus, it is not surprising that there are a variety of intricate molecular mechanisms in place to both engage and control protein synthesis. While eIF4E is often considered the gateway to translation, given its role in ~40% of translation at ‘steady state’, there are several other translation initiation mechanisms and these can have biological impacts independently of eIF4E. For example, the eIF3d-DAP5 pathway is responsible for ~20-30% of translation and likely also contributes to cancer. It will be exciting to quantify the translation capacity of the other pathways outlined here and their relative impact in different environments and conditions. The biochemical diversity highlighted here provides an understanding as to why targeting eIF4E with ribavirin or ASOs does not lead to overt toxicity in patients. Indeed, this multiplicity has important implications for developing therapeutic interventions for translation. Further, these translation modalities likely allow specific subgroups of RNAs to be translated (or to have their translation inhibited). It will be critical to determine which RNAs are controlled by what pathways and when. Likely the RNAs are selected via specific cis-acting elements known as USER codes to enable coordinated translation control as in the RNA regulon model [3]. Furthermore, it is clear that many of the factors involved in translation initiation are actors in other aspects of RNA processing (e.g. eIF4E, CBC) or other facets of translation (e.g. eIF3d, TRS) (Table 1). Given this multiplicity of function, dissection of the biological impacts of these differing roles must be considered. This is an exciting time to study the diversity of translation initiation and its impact on human disease.
Acknowledgments
We are grateful for critical comments from Drs Osborne and Culjkovic-Kraljacic. KLBB acknowledges support from CIHR, NIH, LLS USA and holds a Senior Canada Research Chair in Molecular Biology of the Cell Nucleus.
Funding Statement
This work was supported by the Leukemia and Lymphoma Society (R6513); National Institutes of Health [80728]; National Institutes of Health [98471]; Canadian Institutes for Health Research [PJT159785].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].de Sousa Abreu R, Penalva LO, Marcotte EM, et al. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5(12):1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Culjkovic-Kraljacic B, Borden KLB.. The impact of post-transcriptional control: better living through RNA regulons. Front Genet. 2018;9:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Keene JD, Lager PJ. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13(3):327–337. [DOI] [PubMed] [Google Scholar]
- [4].Culjkovic-Kraljacic B, Borden KL. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 2013;23(7):328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Carroll M, Borden KL. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res. 2013;33(5):227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Amorim IS, Lach G, Gkogkas CG. the role of the eukaryotic translation initiation factor 4E (eIF4E) in neuropsychiatric disorders. Front Genet. 2018;9:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bukhari SIA, Truesdell SS, Lee S, et al. A specialized mechanism of translation mediated by FXR1a-associated MicroRNP in cellular quiescence. Mol Cell. 2016;61(5):760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee S, Micalizzi D, Truesdell SS, et al. A post-transcriptional program of chemoresistance by AU-rich elements and TTP in quiescent leukemic cells. Genome Biol. 2020;21(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].de la Parra C, Ernlund A, Alard A, et al. A widespread alternate form of cap-dependent mRNA translation initiation. Nat Commun. 2018;9(1):3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lee AS, Kranzusch PJ, Doudna JA, et al. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016;536(7614):96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jeong SJ, Park S, Nguyen LT, et al. A threonyl-tRNA synthetase-mediated translation initiation machinery. Nat Commun. 2019;10(1):1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ryu I, Kim YK. Translation initiation mediated by nuclear cap-binding protein complex. BMB Rep. 2017;50(4):186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Borden KL. The eukaryotic translation initiation factor eIF4E wears a “cap” for many occasions. Translation (Austin). 2016;4:e1220899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yamamoto H, Unbehaun A, Spahn CMT. Ribosomal chamber music: toward an understanding of IRES mechanisms. Trends Biochem Sci. 2017;42(8):655–668. [DOI] [PubMed] [Google Scholar]
- [15].Coutinho de Oliveira L, Volpon L, Rahardjo AK, et al. Structural studies of the eIF4E-VPg complex reveal a direct competition for capped RNA: implications for translation. Proc Natl Acad Sci U S A. 2019;116(48):24056–24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Simon AE, Miller WA. 3′ Cap-independent translation enhancers of plant viruses. Annu Rev Microbiol. 2013;67(1):21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83(1):779–812. [DOI] [PubMed] [Google Scholar]
- [18].Osborne MJ, Borden KL. The eukaryotic translation initiation factor eIF4E in the nucleus: taking the road less traveled. Immunol Rev. 2015;263(1):210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Volpon L, Osborne MJ, Borden KLB. Biochemical and structural insights into the eukaryotic translation initiation factor eIF4E. Curr Protein Pept Sci. 2019;20(6):525–535. [DOI] [PubMed] [Google Scholar]
- [20].Gruner S, Peter D, Weber R, et al. The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol Cell. 2016;64(3):467–479. [DOI] [PubMed] [Google Scholar]
- [21].Adivarahan S, Livingston N, Nicholson B, et al. Spatial organization of single mrnps at different stages of the gene expression pathway. Mol Cell. 2018;72(4):727–738 e725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vicens Q, Kieft JS, Rissland OS. Revisiting the closed-loop model and the nature of mRNA 5ʹ-3ʹ communication. Mol Cell. 2018;72(5):805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Des Georges A, Dhote V, Kuhn L, et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525(7570):491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bah A, Vernon RM, Siddiqui Z, et al. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519(7541):106–109. [DOI] [PubMed] [Google Scholar]
- [25].Siddiqui N, Tempel W, Nedyalkova L, et al. Structural insights into the allosteric effects of 4EBP1 on the eukaryotic translation initiation factor eIF4E. J Mol Biol. 2012;415(5):781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ptushkina M, von der Haar T, Vasilescu S, et al. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5’ cap in yeast involves a site partially shared by p20. Embo J. 1998;17(16):4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cohen N, Sharma M, Kentsis A, et al. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. Embo J. 2001;20(16):4547–4559. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Topisirovic I, Culjkovic B, Cohen N, et al. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. Embo J. 2003;22(3):689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Topisirovic I, Kentsis A, Perez JM, et al. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol Cell Biol. 2005;25(3):1100–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Topisirovic I, Borden KL. Homeodomain proteins and eukaryotic translation initiation factor 4E (eIF4E): an unexpected relationship. Histol Histopathol. 2005;20(4):1275–1284. [DOI] [PubMed] [Google Scholar]
- [31].Brunet I, Weinl C, Piper M, et al. The transcription factor engrailed-2 guides retinal axons. Nature. 2005;438(7064):94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Prochiantz A, Joliot A. Can transcription factors function as cell-cell signalling molecules? Nat Rev Mol Cell Biol. 2003;4(10):814–819. [DOI] [PubMed] [Google Scholar]
- [33].Volpon L, Osborne MJ, Capul AA, et al. Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc Natl Acad Sci U S A. 2010;107(12):5441–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kentsis A, Dwyer EC, Perez JM, et al. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J Mol Biol. 2001;312(4):609–623. [DOI] [PubMed] [Google Scholar]
- [35].Kentsis A, Gordon RE, Borden KL. Self-assembly properties of a model RING domain. Proc Natl Acad Sci U S A. 2002;99(2):667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tan NGS, Ardley HC, Scott GB, et al. Human homologue of ariadne promotes the ubiquitylation of translation initiation factor 4E homologous protein, 4EHP. FEBS Lett. 2003;554(3):501–504. [DOI] [PubMed] [Google Scholar]
- [37].Feoktistova K, Tuvshintogs E, Do A, et al. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A. 2013;110(33):13339–13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Graff JR, Konicek BW, Vincent TM, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117(9):2638–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Truitt ML, Conn C, Shi Z, et al. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162(1):59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Culjkovic-Kraljacic B, Fernando TM, Marullo R, et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood. 2016;127(7):858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rousseau D, Kaspar R, Rosenwald I, et al. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci U S A. 1996;93(3):1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ramirez-Valle F, Braunstein S, Zavadil J, et al. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol. 2008;181(2):293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Assouline S, Culjkovic B, Cocolakis E, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114(2):257–260. [DOI] [PubMed] [Google Scholar]
- [44].Assouline S, Culjkovic-Kraljacic B, Bergeron J, et al. A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haematologica. 2015;100(1):e7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dunn LA, Fury MG, Sherman EJ, et al. Phase I study of induction chemotherapy with afatinib, ribavirin, and weekly carboplatin and paclitaxel for stage IVA/IVB human papillomavirus-associated oropharyngeal squamous cell cancer. Head Neck. 2017;40(2):233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kosaka T, Maeda T, Shinojima T, et al. a clinical study to evaluate the efficacy and safety of docetaxal with ribavirin in patients with progressive castration resistant prostate cancer who have previously received docetaxol alone. J clin oncol. 2017;35(15_suppl):e14010-e14010. [Google Scholar]
- [47].Hong DS, Kurzrock R, Oh Y, et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin Cancer Res. 2011;17(20):6582–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lejbkowicz F, Goyer C, Darveau A, et al. A fraction of the mRNA 5ʹ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci U S A. 1992;89(20):9612–9616. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Culjkovic B, Topisirovic I, Skrabanek L, et al. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3ʹUTR. J Cell Biol. 2005;169(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Culjkovic B, Topisirovic I, Skrabanek L, et al. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175(3):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kentsis A, Topisirovic I, Culjkovic B, et al. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101(52):18105–18110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Topisirovic I, Guzman ML, McConnell MJ, et al. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol. 2003;23(24):8992–9002. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Topisirovic I, Siddiqui N, Borden KL. The eukaryotic translation initiation factor 4E (eIF4E) and HuR RNA operons collaboratively regulate the expression of survival and proliferative genes. Cell Cycle. 2009;8(7):960–961. [PubMed] [Google Scholar]
- [54].Volpon L, Culjkovic-Kraljacic B, Osborne MJ, et al. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc Natl Acad Sci U S A. 2016;113(19):5263–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Iborra FJ, Jackson DA, Cook PR. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293(5532):1139–1142. [DOI] [PubMed] [Google Scholar]
- [56].Lang V, Zanchin NI, Lunsdorf H, et al. Initiation factor eIF-4E of saccharomyces cerevisiae. Distribution within the cell, binding to mRNA, and consequences of its overproduction. J Biol Chem. 1994;269(8):6117–6123. [PubMed] [Google Scholar]
- [57].Culjkovic B, Borden KL. Understanding and targeting the eukaryotic translation initiation factor eIF4E in head and neck cancer. J Oncol. 2009;2009:981679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sonenberg N, Gingras AC. The mRNA 5ʹ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10(2):268–275. [DOI] [PubMed] [Google Scholar]
- [59].Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle. 2007;6(1):65–69. [DOI] [PubMed] [Google Scholar]
- [60].Topisirovic I, Siddiqui N, Lapointe VL, et al. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. Embo J. 2009;28(8):1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Volpon L, Culjkovic-Kraljacic B, Sohn HS, et al. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA. 2017;23(6):927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Culjkovic-Kraljacic B, Baguet A, Volpon L, et al. The oncogene eIF4E reprograms the nuclear pore complext to promote mRNA export and oncogenic transformation. Cell Rep. 2012;2(2):207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Davis MR, Delaleau M, Borden KLB. Nuclear eIF4E stimulates 3ʹ-end cleavage of target RNAs. Cell Rep. 2019;27(5):1397–1408 e1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Culjkovic-Kraljacic B, Skrabanek LA, Revuelta MV, et al. The eukaryotic translation initiation factor eIF4E elevates m7G capping efficiency of selected coding and non-coding transcripts. PNAS. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Andrei MA, Ingelfinger D, Heintzmann R, et al. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11(5):717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8(1):9–22. [DOI] [PubMed] [Google Scholar]
- [67].Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25(5):635–646. [DOI] [PubMed] [Google Scholar]
- [68].Buchan JR. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11(8):1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Liu W, Jankowska-Anyszka M, Piecyk K, et al. Structural basis for nematode eIF4E binding an m 2,2,7 G-Cap and its implications for translation initiation. Nucleic Acids Res. 2011;39(20):8820–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Volpon L, Osborne MJ, Topisirovic I, et al. Cap-free structure of eIF4E suggests a basis for conformational regulation by its ligands. Embo J. 2006;25(21):5138–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Murphy JF, Klein PG, Hunt AG, et al. Replacement of the tyrosine residue that links a potyviral VPg to the viral RNA is lethal. Virology. 1996;220(2):535–538. [DOI] [PubMed] [Google Scholar]
- [72].Murphy JF, Rychlik W, Rhoads RE, et al. A tyrosine residue in the small nuclear inclusion protein of tobacco vein mottling virus links the VPg to the viral RNA. J Virol. 1991;65(1):511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Coutinho de Oliveira L, Volpon L, Osborne MJ, et al. Chemical shift assignment of the viral protein genome-linked (VPg) from potato virus Y. Biomol NMR Assign. 2019;13(1):9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Léonard S, Plante D, Wittmann S, et al. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J Virol. 2000;74(17):7730–7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bastet A, Robaglia C, Gallois JL. eIF4E resistance: natural variation should guide gene editing. Trends Plant Sci. 2017;22(5):411–419. [DOI] [PubMed] [Google Scholar]
- [76].German-Retana S, Walter J, Doublet B, et al. Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J Virol. 2008;82(15):7601–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Robaglia C, Caranta C. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 2006;11(1):40–45. [DOI] [PubMed] [Google Scholar]
- [78].Grzela R, Strokovska L, Andrieu J-P, et al. Potyvirus terminal protein VPg, effector of host eukaryotic initiation factor eIF4E. Biochimie. 2006;88(7):887–896. [DOI] [PubMed] [Google Scholar]
- [79].Wang Z, Parisien M, Scheets K, et al. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure. 2011;19(6):868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Martin F, Barends S, Jaeger S, et al. Cap-assisted internal initiation of translation of histone H4. Mol Cell. 2011;41(2):197–209. [DOI] [PubMed] [Google Scholar]
- [81].Osborne MJ, Volpon L, Kornblatt JA, et al. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc Natl Acad Sci U S A. 2013;110(10):3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Rosettani P, Knapp S, Vismara MG, et al. Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J Mol Biol. 2007;368(3):691–705. [DOI] [PubMed] [Google Scholar]
- [83].Uniacke J, Perera JK, Lachance G, et al. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res. 2014;74(5):1379–1389. [DOI] [PubMed] [Google Scholar]
- [84].Morita M, Ler LW, Fabian MR, et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol. 2012;32(17):3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Landon AL, Muniandy PA, Shetty AC, et al. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat Commun. 2014;5(1):5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64(23):8639–8642. [DOI] [PubMed] [Google Scholar]
- [87].Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142(3):368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fatscher T, Boehm V, Gehring NH. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell Mol Life Sci. 2015;72(23):4523–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Rufener SC, Muhlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2013;20(6):710–717. [DOI] [PubMed] [Google Scholar]
- [90].Durand S, Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol. 2013;20(6):702–709. [DOI] [PubMed] [Google Scholar]
- [91].Choe J, Ryu I, Park OH, et al. eIF4AIII enhances translation of nuclear cap-binding complex-bound mRNAs by promoting disruption of secondary structures in 5ʹUTR. Proc Natl Acad Sci U S A. 2014;111(43):E4577–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19(18):2212–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Parsyan A, Svitkin Y, Shahbazian D, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12(4):235–245. [DOI] [PubMed] [Google Scholar]
- [94].Lu WT, Wilczynska A, Smith E, et al. The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc Trans. 2014;42(1):166–172. [DOI] [PubMed] [Google Scholar]
- [95].Marintchev A, Edmonds KA, Marintcheva B, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136(3):447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].von Der Haar T, Ball PD, McCarthy JE. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5ʹ-Cap by domains of eIF4G. J Biol Chem. 2000;275(39):30551–30555. [DOI] [PubMed] [Google Scholar]
- [97].Koritzinsky M, Magagnin MG, van den Beucken T, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. Embo J. 2006;25(5):1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Koumenis C, Naczki C, Koritzinsky M, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22(21):7405–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Oh N, Kim KM, Choe J, et al. Pioneer round of translation mediated by nuclear cap-binding proteins CBP80/20 occurs during prolonged hypoxia. FEBS Lett. 2007;581(26):5158–5164. [DOI] [PubMed] [Google Scholar]
- [100].Garre E, Romero-Santacreu L, De Clercq N, et al. Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol Biol Cell. 2012;23(1):137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sharma A, Yilmaz A, Marsh K, et al. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8(3):e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Toro-Ascuy D, Rojas-Araya B, García-de-Gracia F, et al. A Rev-CBP80-eIF4AI complex drives Gag synthesis from the HIV-1 unspliced mRNA. Nucleic Acids Res. 2018;46(21):11539–11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yedavalli VS, Jeang KT. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci U S A. 2010;107(33):14787–14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Worch R, Niedzwiecka A, Stepinski J, et al. Specificity of recognition of mRNA 5’ cap by human nuclear cap-binding complex. RNA. 2005;11(9):1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kumar P, Hellen CU, Pestova TV. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016;30(13):1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Rode S, Ohm H, Anhäuser L, et al. Differential requirement for translation initiation factor pathways during ecdysone-dependent neuronal remodeling in drosophila. Cell Rep. 2018;24(9):2287–2299 e2284. [DOI] [PubMed] [Google Scholar]
- [107].Monecke T, Schell S, Dickmanns A, et al. Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m(7)G-cap-binding mode. J Mol Biol. 2008;382(4):827–834. [DOI] [PubMed] [Google Scholar]
- [108].Nilsson P, Henriksson N, Niedzwiecka A, et al. A multifunctional RNA recognition motif in poly(A)-specific ribonuclease with cap and poly(A) binding properties. J Biol Chem. 2007;282(45):32902–32911. [DOI] [PubMed] [Google Scholar]
- [109].Nagata T, Suzuki S, Endo R, et al. The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition. Nucleic Acids Res. 2008;36(14):4754–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci U S A. 2005;102(38):13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Henis-Korenblit S, Shani G, Sines T, et al. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci U S A. 2002;99(8):5400–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lee SH, McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. Embo J. 2006;25(17):4008–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, et al. Insights into structural and mechanistic features of viral IRES elements. Front Microbiol. 2017;8:2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Garrey JL, Lee YY, Au HH, et al. Host and viral translational mechanisms during cricket paralysis virus infection. J Virol. 2010;84(2):1124–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Rodriguez Pulido M, Serrano P, Saiz M, et al. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology. 2007;364(2):466–474. [DOI] [PubMed] [Google Scholar]
- [116].Kobayashi M, Arias C, Garabedian A, et al. Site-specific cleavage of the host poly(A) binding protein by the encephalomyocarditis virus 3C proteinase stimulates viral replication. J Virol. 2012;86(19):10686–10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6(6):513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Weingarten-Gabbay S, Elias-Kirma S, Nir R, et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351(6270):aad4939. [DOI] [PubMed] [Google Scholar]
- [119].Jackson RJ. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol. 2013;5(2):a011569-a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285(38):29033–29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Pelletier J, Kaplan G, Racaniello VR, et al. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5ʹ noncoding region. Mol Cell Biol. 1988;8(3):1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Jang SK, Kräusslich HG, Nicklin MJ, et al. A segment of the 5ʹ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62(8):2636–2643. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].de Breyne S, Yu Y, Unbehaun A, et al. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A. 2009;106(23):9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Avanzino BC, Fuchs G, Fraser CS. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc Natl Acad Sci U S A. 2017;114(36):9611–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Marash L, Liberman N, Henis-Korenblit S, et al. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell. 2008;30(4):447–459. [DOI] [PubMed] [Google Scholar]
- [126].Fuchs G, Petrov AN, Marceau CD, et al. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc Natl Acad Sci U S A. 2015;112(2):319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Quade N, Boehringer D, Leibundgut M, et al. Cryo-EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat Commun. 2015;6(1):7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Malygin AA, Shatsky IN, Karpova GG. Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry (Mosc). 2013;78(1):53–59. [DOI] [PubMed] [Google Scholar]
- [129].Angulo J, Ulryck N, Deforges J, et al. LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 2016;44(3):1309–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Filbin ME, Vollmar BS, Shi D, et al. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat Struct Mol Biol. 2013;20(2):150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Matsuda D, Mauro VP. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc Natl Acad Sci U S A. 2014;111(43):15385–15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Treder K, Pettit Kneller EL, Allen EM, et al. The 3ʹ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14(1):134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Wang Z, Treder K, Miller WA. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem. 2009;284(21):14189–14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Timmer RT, Benkowski LA, Schodin D, et al. The 5ʹ and 3ʹ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5ʹ cap structure. J Biol Chem. 1993;268(13):9504–9510. [PubMed] [Google Scholar]
- [135].Danthinne X, Seurinck J, Meulewaeter F, et al. The 3ʹ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol Cell Biol. 1993;13(6):3340–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Zahreddine HA, Culjkovic-Kraljacic B, Emond A, et al. The eukaryotic translation initiation factor eIF4E harnesses hyaluronan production to drive its malignant activity. Elife. 2017;6. doi: 10.7554/eLife.29830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].De Benedetti A, Harris AL. eIF4E expression in tumors: its possible role in progression and malignacies. Int J Biochem Cell Biol. 1999;31(1):59–72. [DOI] [PubMed] [Google Scholar]
- [138].Ruggero D, Montanaro L, Ma L, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10(5):484–486. . [DOI] [PubMed] [Google Scholar]
- [139].Culjkovic B, Tan K, Orolicki S, et al. The eIF4E RNA regulon promotes the Akt signaling pathway. J Cell Biol. 2008;181(1):51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Zhang Y, Klein HL, Schneider RJ. Role of Ser-53 phosphorylation in the activity of human translation initiation factor eIF-4E in mammalian and yeast cells. Gene. 1995;163(2):283–288. [DOI] [PubMed] [Google Scholar]
- [141].Kraljacic BC, Arguello M, Amri A, et al. Inhibition of eIF4E with ribavirin cooperates with common chemotherapies in primary acute myeloid leukemia specimens. Leukemia. 2011;25(7):1197–1200. [DOI] [PubMed] [Google Scholar]
- [142].Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511(7507):90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Volpon L, Osborne MJ, Zahreddine H, et al. Conformational changes induced in the eukaryotic translation initiation factor eIF4E by a clinically relevant inhibitor, ribavirin triphosphate. Biochem Biophys Res Commun. 2013;434(3):614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Kentsis A, Volpon L, Topisirovic I, et al. Further evidence that ribavirin interacts with eIF4E. Rna. 2005;11(12):1762–1766. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Bollmann F, Fechir K, Nowag S, et al. Human inducible nitric oxide synthase (iNOS) expression depends on chromosome region maintenance 1 (CRM1)- and eukaryotic translation initiation factor 4E (elF4E)-mediated nucleocytoplasmic mRNA transport. Nitric Oxide. 2013;30:49–59. [DOI] [PubMed] [Google Scholar]
- [146].Urtishak KA, Wang L-S, Culjkovic-Kraljacic B, et al. Targeting EIF4E signaling with ribavirin in infant acute lymphoblastic leukemia. Oncogene. 2019;38(13):2241–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Pettersson F, Yau C, Dobocan MC, et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin Cancer Res. 2011;17(9):2874–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Pettersson F, Del Rincon SV, Emond A, et al. Genetic and pharmacolgic inhibition of eIF4E reduces breast cancer cell migration, invasion and metastasis. Cancer Res. 2015;75(6):1102–1112. [DOI] [PubMed] [Google Scholar]
- [149].Moerke NJ, Aktas H, Chen H, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128(2):257–267. [DOI] [PubMed] [Google Scholar]
- [150].Chen L, Aktas BH, wang Y, et al. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget. 2012;3(8):869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Willimott S, Beck D, Ahearne MJ, et al. Cap-translation inhibitor, 4EGI-1, restores sensitivity to ABT-737 apoptosis through cap-dependent and -independent mechanisms in chronic lymphocytic leukemia. Clin Cancer Res. 2013;19(12):3212–3223. [DOI] [PubMed] [Google Scholar]
- [152].McMahon R, Zaborowska I, Walsh D. Noncytotoxic inhibition of viral infection through eIF4F-independent suppression of translation by 4EGi-1. J Virol. 2011;85(2):853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Joshi S, Platanias LC. Mnk kinases in cytokine signaling and regulation of cytokine responses. Biomol Concepts. 2015;6(1):85. [DOI] [PubMed] [Google Scholar]
- [154].Joshi S, Platanias LC. Mnk kinase pathway: cellular functions and biological outcomes. World J Biol Chem. 2014;5(3):321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]


