ABSTRACT
We examined sequence conservation and signatures of selection in Rab7 proteins across 11 Paramecium aurelia species, and determined the localization patterns of two P. tetraurelia Rab7 paralogs when expressed as GFP fusions in live cells. We found that, while there is a variable number of Rab7 paralogs per genome, Rab7 genes are highly conserved in sequence and appear to be under strong purifying selection across aurelias. Additionally, and surprisingly based on earlier studies, we found that two P. tetraurelia Rab7 proteins have virtually identical localization patterns. Consistent with this, when we examined the gene family of a highly conserved Rab binding partner across aurelias (Rab-Interacting Lysosomal Protein, or RILP), we found that residues in key binding sites in RILPs were absolutely conserved in 13 of 21 proteins, representing genes from 9 of the 11 species examined. Of note, RILP gene number appears to be even more constrained than Rab7 gene number per genome.
Abbreviation: WGD: Whole genome duplication
KEYWORDS: Rab7, effector, RILP, Paramecium, phagosome, lysosome, protein evolution, paralog, ortholog
Rab7 genes are both essential and highly conserved in the majority of eukaryotic genomes [1]. Additionally, the Rab7 subfamily does not appear to have undergone the large expansions that other subfamilies of Rab GTPases have. Why some genes or proteins follow certain evolutionary trajectories with regard to duplicate retention, sequence constraint, and/or functional innovation is still little-understood for most gene families. We wanted to better understand the evolutionary constraints that these proteins are under, particularly at the subcellular level.
Members of the Rab family of small GTPases control fusion steps in eukaryotic membrane trafficking; subfamilies of Rabs are further associated with specific subcellular trafficking pathways. Proteins in the Rab7 subfamily are key regulators of endosomal maturation, as well as lysosome biogenesis [2]. Concomitant with this integral role in endosome-to-lysosome maturation, at least one Rab7 is highly conserved in virtually every eukaryote examined thus far [1,3–5]. This trafficking step is also one of the most-well-characterized at the molecular level: the cascade of activation and deactivation that leads to the handoff from active Rab5 (and early endosome identity) to active Rab7 (to the acquisition of late endosome identity) through the SAND effector homolog in C. elegans is particularly elegantly characterized [6]. Interestingly, this interaction is conserved in plants, but has been co-opted for vacuolar transport [7]. In mammalian cells, Rab7 also has direct roles in: controlling acidification of lysosomes by organizing the assembly and function of V-ATPases on lysosomal membranes [8,9]; both plus and minus-end-directed transport of late endosomes and phagosomes [10]; late endosomal and phagosomal positioning near to the microtubule organizing center (MTOC) [11,12]; and in mediating traffic between lysosomes and various degradative compartments, as they mature, including those involved in autophagy, mitophagy and lipophagy (extensively reviewed in [3]). For most of these functions, the extent of functional conservation beyond mammalian cells remains to be determined.
The ancestor of all eukaryotes possessed a single Rab7 gene [1]. While Rab7 is one of the most retained and essential members of this gene family, study of both recent and ancient Rab7 duplications has indicated that duplication of Rab7 genes has led, in several cases, to functional diversification over time. One of the oldest examples was the duplication of an ancestral Rab7 gene that led to the creation and diversification of Rab9 in the opisthokont lineage, and involved in lysosome-to-trans golgi network traffic [13]. Other well-studied Rab7 duplicates, or paralogs include human Rab7b [14], the Entamoeba Rab7 paralogs [4], fission yeast Ypt7 and Ypt71 [5], the eight Arabidopsis Rab7 genes [15], and Paramecium octaurelia Rab7a and Rab7b [16,17]. However, only the latter study focuses on a functional comparison of relatively closely-related duplicates, that have high sequence similarity. The existence of multiple Rab7 paralogs in a genome also appears to correlate with recent whole genome or large-scale duplications. This indicates that, over time, most Rab7 duplicates are lost. For example, in a recent survey of kinetoplastid trafficking proteins, it was found that most kinetoplastids have only one Rab7 gene, while Bodo saltans and Trypanosoma borreli have two and three paralogs respectively, with the B. saltans duplicates correlating with a possible whole genome duplication event [18].
The ancestral lineage that led to the Paramecium aurelia species complex underwent at least two whole genome duplications (WGDs) in its evolutionary history [19]. After the most recent WGD, WGD1, the species radiated into the 16 characterized species that we observe today. The duplicates have experienced ongoing loss, but on average each species has retained ~50% of the duplicates in sum from both recent WGDs, with differential loss experienced among specific duplicates [20]. While three Paramecium aurelia species and one pre-duplication outgroup species, P. caudatum, have been previously sequenced and published [20,21]., other P. aurelia genome sequencing efforts (both macronuclear and micronuclear) are in progress. We obtained Rab7 and RILP sequences from 11 genomes across the aurelia complex, as well as from two outgroups (the published P. caudatum and the as-yet-unpublished P. multimicronucleatum) by requesting that the authors search the unpublished genomes for us (Olivier Arnaiz and Jean-François Gout, personal communication). With this larger set of duplicates from across the species complex, as well as the two outgroups, we were able to observe larger patterns of retention and loss, and examine the extent of sequence conservation within this subfamily of essential proteins. Additionally, functional diversification was recently reported between Rab7 duplicates for one species, P. octaurelia [16,17]. To test whether this functional diversification extended to other sets of Rab7 paralogs in the P. aurelia complex, we localized the corresponding Rab7s in P. tetraurelia.
Results
Rab7 paralog sequences were retrieved from each genome through blastp analysis of the predicted protein sequences. Each genome contains between one and four Rab7 genes, for a total of 28 genes, representing 11 of the 16 recorded P. aurelia species (Table 1). In the 11 P. aurelia genomes analyzed, 6 contain three Rab7 paralogs. While it is possible that a paralog has been missed due to an incomplete genome assembly or missed genes in genome annotation, this is unlikely to significantly affect this study. Indeed, genome assembly sizes are highly conserved across the aurelia species studied here, suggesting that most of the genomes were captured and only a very small fraction of genes might be missing from each assembly. P. jenningsi, with four Rab7 paralogs, has the highest number among the 11 genomes; interestingly, it is likely that this species harbors an additional whole or partial genome duplication, which may explain the additional one or two Rab7 duplicates in its genome (Jean-François Gout, personal communication).
Table 1.
Paramecium aurelia genomes contain a variable number of Rab7 genes. Number of Rab7 and RILP genes in each genome; number of RILP paralogs with invariant conserved amino acid residues, out of a possible 17 sites.
| Organism | Rab7 paralogs in genome |
RILP paralogs in genome |
# of RILP paralogs with invariant conserved residues |
|---|---|---|---|
| Paramecium primaurelia | 2 | 2 | 1 |
| Paramecium biaurelia | 3 | 2 | 1 |
| Paramecium tetraurelia | 3 | 2 | 2 |
| Paramecium sexaurelia | 2 | 1 | 0 |
| Paramecium septaurelia | 1 | 2 | 1 |
| Paramecium octaurelia | 3 | 2 | 1 |
| Paramecium novaurelia | 3 | 2 | 2 |
| Paramecium decaurelia | 3 | 2 | 2 |
| Paramecium dodecaurelia | 3 | 2 | 2 |
| Paramecium quadecaurelia | 1 | 2 | 1 |
| Paramecium jenningsi | 4 | 2 | 0 |
| Outgroup species | |||
| Paramecium caudatum | 1 | 1 | 0 |
| Paramecium multimicronucleatum | 2 | N/A | N/A |
| Tetrahymena thermophila | 1 | N/A | N/A |
To better understand the evolutionary history of Rab7s in P. aurelia species, we constructed both maximum likelihood (ML) and neighbor joining (NJ) phylogenies by aligning the nucleotide coding sequences each Rab7 gene (Figure 1 and S1; alignment files figure S2 and S3). In both trees, many branching relationships have only low bootstrap support; this reflects that these genes are very closely related and have only a few nucleotide changes between them, making strong branching relationships difficult to decipher. Overall, the gene sequences group into roughly two major clades, which we have named the Rab7a clade and the Rab7b clade, consistent with previous naming [22]. The clade with the strongest bootstrap support in both phylogenies is the Rab7b clade, as well a subset of the 7a clade, with clear outgroups of often-paired Rab7a paralogs from different species. The most closely related pairs or triplets of P. aurelia species [23] share the same numbers of Rab7 paralogs: octaurelia and tetraurelia (with 3 Rab7 genes); and novaurelia, dodecaurelia and decaurelia (with 3). Otherwise, there is not a strong trend in Rab7 expansion or loss across the species complex. Two sets of P. jenningsi Rab7 paralogs group together, consistent with more recent WGDs in this lineage. Generally, since the whole genome duplication events predate the speciation events in these species, orthologs should be more closely related by descent than paralogs. However, multiple pairs of intraspecies paralogs group together, including the Rab7a1 and a2 pairs in P. biaurelia, tetraurelia, sexaurelia, octaurelia, novaurelia, decaurelia, dodecaurelia; this is consistent with previously detected gene conversion in these species [20]. In general, relationships between orthologs track species relationships, with the exception of the P. biaurelia Rab7 genes. Two outgroup species, which speciated before the two recent WGDs, were also analyzed. Outgroup species P. caudatum harbors just one Rab7, and P. multimicronucleatum has an additional more divergent gene that, when reciprocally blasted, yields Rab7 genes as the closest hits. The long branch of P. mm Rab7b appears to be affecting the branching relationships in the trees, in different ways depending on the method used.
Figure 1.
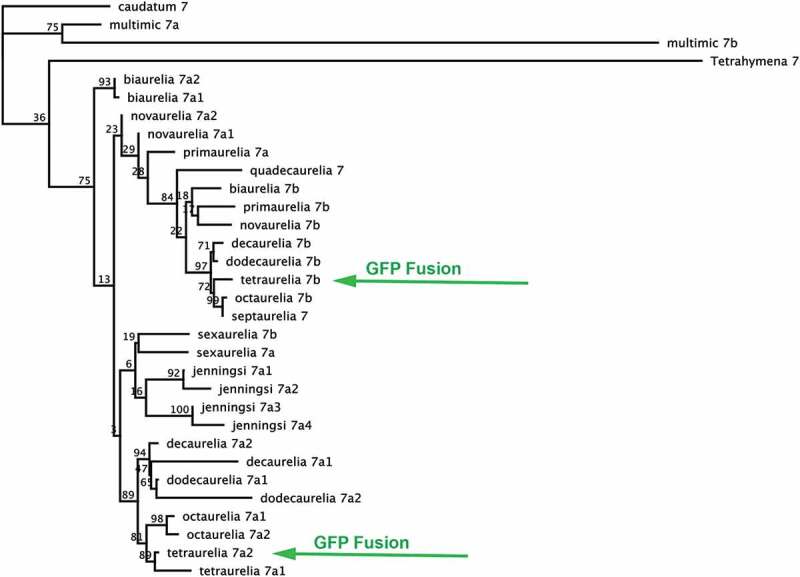
Maximum likelihood phylogeny showing evolutionary relationships between 28 P. aurelia Rab7 coding sequences. Rab7s appear to form two major clades, and roughly follow species relationships, with several exceptions. Genes localized in this study are marked with green arrows.
The Rab7 subfamily is highly conserved across the aurelias, both at the nucleotide and protein level. When the 28 genes are aligned, 60.9% (380 sites) of the 624 nucleotide sites are absolutely conserved, with an average pairwise identity of 91.6%. The protein sequences are similarly conserved, with 59.7% (123 sites) of the 208 sites absolutely conserved across all 34 proteins, and an average pairwise identity of 96.9%. When compared to previous whole-genome analysis of conservation between paralogs in P. tetraurelia, this amount of conservation is higher than the majority of both WGD1 and 2 paris in the genome [19]. In many cases, there are only one, two or three amino acid differences across the entire protein between a pair of paralogs or orthologs. This is remarkable for genes that have been diverging for millions, often tens of millions, of years [20]. This indicates high purifying selection across the entire protein. To test this further, we estimated the dN/dS for these genes among orthologs using the Mega7 program [24]. We used a Z-test in Mega7 to test the hypotheses that first, the sequences are evolving neutrally (dN = dS, or non-synonymous changes are equal to synonymous changes) and second, that the sequences are under positive selection (dN > dS, or that there have been significantly more non-synonymous changes). We were able to reject both of these alternative hypotheses to purifying selection, with P-values of 4.58 x 10−5 and 1.9 x 10−5, respectively, while the hypothesis of purifying selection could not be rejected.
Rab-Interacting Lysosomal Protein, or RILP, is a canonical effector of Rab7, linking Rab7 to dynein motors, which direct minus-end-directed transport on microtubules [10]. RILP genes are conserved across eukaryotes [25]. We asked if the number of RILP paralogs in a genome correlated with the number of Rab7 genes. We used the BLAT algorithm to search the P. aurelia and P. caudatum genomes for RILP homologs, retrieving 21 RILP genes in the 11 P. aurelia genomes surveyed. We found that, somewhat surprisingly, the number of RILP genes in a given P. aurelia genome did not correlate strongly with the number of Rab7s (Table 1). Indeed, the number of RILP genes seems to be more constrained than the number of Rab7 genes: every aurelia genome examined has two RILP paralogs, with the exception of P. sexaurelia, which has one. We also found that the RILP protein sequences overall were less conserved than the Rab7 protein sequences; when aligned, among the 21 genes, 56.6% of the sites (289 sites out of a total of 517) were absolutely conserved across the aurelia complex, with an average pairwise identity of 88.7% between the protein sequences. We also examined specific residues in the RILP proteins known to be involved in lysosome biogenesis [26] and interaction with dynein [27,28]; across the 17 sites posited to be important for canonical RILP function [25], the vast majority of the sites were conserved; indeed, in the majority (9 out of 11 examined) of aurelia genomes, at least one paralog contains absolutely conserved residues at these 17 sites (Table 1, specific sites in the proteins marked on figure S4). On the other side of the binding interface, RILP-interacting residues in Rab7 proteins are absolutely conserved across aurelias [28], even in the species that have divergent RILP genes, indicating that there have not been compensatory, or even equivalent, divergent changes in the Rab7 proteins with regard to RILP function.
Because the Wyroba group [16,17] found localization changes between P. octaurelia Rab7a1 and Rab7b proteins, and correlated the localization changes with specific amino acid changes, we predicted that the corresponding (and closely-related, see Figure 1) P. tetraurelia orthologs of each of those genes would also have divergent localization patterns when expressed as full-gene GFP fusions in live cells. However, we found that the P. tetraurelia Rab7a2 and Rab7b paralogs had virtually identical localization patterns when expressed in live cells as N-terminally tagged GFP fusions (Figure 2). This identical localization pattern appears to be consistent with the canonical Rab7 role in controlling transport between late endosome-to-lysosomal trafficking, and between different types of lysosome-related compartments. For both paralogs, the Rab-GFP signal targets small vesicles in the cytoplasm, as well as the outer membrane of maturing phagosomes. To identify acidified compartments that Rab7 proteins might target to, we also treated the cells with LysoTracker Red (LT Red) dye, which labels acidified lysosomes, phagosomes, and other acidic compartments in the cell. The GFP signal on small cytoplasmic vesicles overlaps with LT Red signal sometimes, but not always, indicating partial or dynamic overlap with acidified lysosomes. The GFP signal on the outer membrane of phagosomes (Figure 2, arrows and details) is consistent with vesicles that are docking onto phagosomes and mediating interactions with the lysosomal pathway as well. Importantly, the phagosomes ringed with GFP-labeled vesicles are either not LT Red-stained or only slightly red, indicating targeting of Rab7 to the membrane or membrane-associated vesicles of either pre-acidified or post-acidified phagosomes. Essentially, both fusions had patterns consistent with a conserved canonical function, including overlap with lysosomes and phagolysosomes. Lastly, GFP-Rab7 protein localizes to and overlaps with LT Red in irregularly-shaped (possibly tubulated) compartments (Figure 2) that appear to be shaped like the structures in which Rab11s proteins overlap with LT Red [29]. There is not enough evidence to determine for certain what these structures represent; an identity as sorting or late endosomes would be consistent with the canonical Rab7 function in late endosome-to-lysosome trafficking. Rab7-GFP and LT Red also label what appear to be undocked trichocysts (Figure 2).
Figure 2.
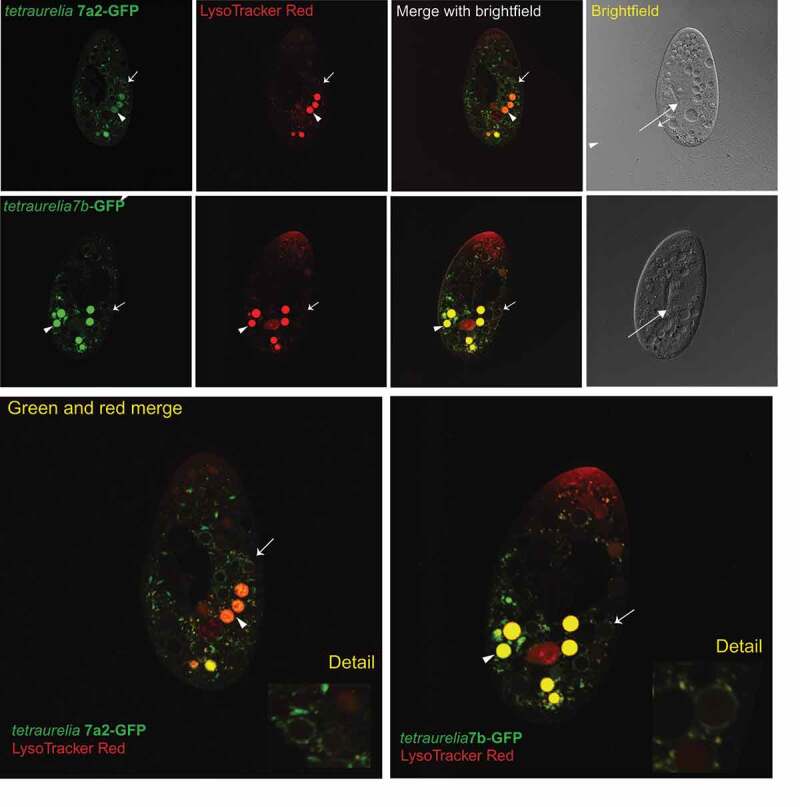
Representative localization patterns for P. tetraurelia Rab7a2 and Rab7b, respectively. Green signal is that of the protein-GFP fusion (green) as expressed in P. tetraurelia cells. Red signal is LysoTracker Red; areas of overlap are yellow. In each fluorescing cell image: arrows mark representative non-acidified food vacuoles ringed by GFP signal, and arrowheads mark representative acidified food vacuoles. In brightfield images: arrows mark the base of the oral apparatus. Each cell image represents >10 identical localization patterns recorded for each fusion.
The Wyroba group proposed that specific amino acids, and in particular specific post-translational modifications (PTMs) of those residues, drive localization differences between the paralogs [16]. We examined these sites across the aurelia Rab7 phylogeny and could not find a clear phylogenetic association, within either the Rab7a or Rab7b clade, between the Thr200 residue that Wyroba et al. suggested was responsible for the localization change, and either clade (Figure S4B). This does not mean that it does not drive a functional difference between the two proteins; however, in our live cell experiments, we were unable to detect a difference in localization between two paralogs carrying the two different residues, Thr200 in P. tetraurelia Rab7b and Ala200 in Pt Rab7a2 (marked in Figure S4B).
Discussion
Recent studies have shown that gene family and subfamily size is a product of sculpting, or loss of, duplicates, as much as it is an expansion of specific clades of genes [30]. In line with this finding, we wanted to better understand how a smaller clade of essential genes might be constrained, especially when compared with more expanded, apparently dynamic protein clades in the same gene family. In other recent work, we explored the evolution of duplicates within the expanded Rab11 clade in P. aurelias [29].We next chose to focus on a smaller, potentially more constrained subfamily, the Rab7 genes, for which there were also indications that there might be functional diversification occurring between some paralogs.
One question regarding small, constrained clades of proteins is whether it is the possession of a particular function in the cell that constrains their evolution, or whether it is some character of the molecule itself that limits its potential evolutionary paths. Further evolutionary analysis combined with structural analysis at the level of individual residues is required to understand how evolutionarily ‘rigid’ the Rab7 proteins themselves are; however, it is clear that Rab7 duplicates diversified or specialized in function at least several times over eukaryotic evolution. One clue from the plant lineage is that even while Rab7 protein function has changed, some of its key binding partners have not [7]. Consistent with this observation, we also found that, while the number of RILP effectors in the genome has stayed relatively constant, and that the majority of RILP effectors studied contained absolutely conserved residues in key binding sites, many genomes contained at least on paralog with divergent binding site residues. In the future, it will be interesting to see whether the RILP paralogs with divergent binding site residues have also diverged in binding or other functions. Comparisons within and between gene families of other binding partners that are involved in the same processes in the cell, but may be differentially constrained, can give further insight into these questions as well.
The extent of diversification between paralogs that have diverged for longer times – as compared to shorter timescales – has not been fully explored, but recent studies of two closely related Rab7 duplicates in P. octaurelia [16,17] extensively tracked functional diversification at the protein and post-translation modification (PTM) level. This group found that while one of the P. octaurelia Rab7 paralogs localizes as expected to phagosomes and lysosomes, the other differs in both localization pattern and post-translational modifications (PTMs), and that one of these PTMs, glycosylation of a single residue, Thr200, is sufficient to drive a localization change [16]. In our localization studies of the orthologs of these protein in P. tetraurelia, we found virtually identical patterns between the two paralogs, in contrast to the earlier study. The two studies were conducted in different species, so they are not directly comparable. Additionally, the other study was conducted in situ on fixed cells, while in our study, we injected fusion genes rather than proteins into P. tetraurelia individual cells, creating multiple independent lines, and then imaging the resulting cell lines expressing the GFP-Rab fusion proteins in live cells.
Overall, much of this data is complementary; it is quite interesting that the different Rab7 paralogs have different PTMs, particularly in their C-terminal HVDs. However, whether these varying PTMs lead to functional changes between the proteins in live cells remains to be shown. The different localization results between the studies highlight issues with defining function and the large amount of work necessary to say something definitive about the function of a protein, rather than its biochemical activity in vitro, or its localization pattern in live cells. Also, based on our evolutionary analysis of the phylogenetic pattern of these residue differences, the residue(s) that Wyroba et al. posit as the source of changes in PTMs leading to changes in localization do not correlate with either the Rab7a or Rab7b clade in the phylogeny, which might be predicted if their evolutionary history included functional changes occurring as a result of different PTMs. Instead, this suggests that overall sequence evolution is not correlated with changes at this residue. Based on our localization results, it is unlikely that each of these amino acid changes corresponds to a localization change. In any case, further functional work of other orthoparalogs across the P. aurelia species complex is needed, in order to better understand the patterns of functional conservation and innovation.
Methods
Sequence and evolutionary analysis
Blastp analysis was performed using the predicted protein sequences in Wyroba et al., 2007 as queries to search the predicted protein sequences from each Paramecium genome; the BLAT program was used to search for the RILP homologs; the default parameters of the algorithm were used, except that to retrieve all putative proteins, we reduced the cutoff to return hits that cover ≥30% of the length of the protein. Unpublished P. aurelia genomes were assembled and annotated using the Eugene pipeline [31], and were graciously shared by the researchers assembling and analyzing the genomes (personal communication, Jean-François Gout and Olivier Arnaiz). Rab7 and RILP alignments and neighbor joining and maximum likelihood phylogenies were made using the Muscle algorithm in the Geneious software program (http://www.geneious.com). Full nucleotide alignments can be found in Figures S2 and S3. The neighbor-joining (NJ) tree was made using the Jukes-Cantor substitution model and 100 bootstrapped replicates to construct the consensus tree in Figure S1. P. tetraurelia Rab32 was the the outgroup used for this tree. The maximum likelihood tree was constructed using the PhyML program [32]; the TN93 substitution matrix was used, and the Subtree Pruning and Regrafting (SPR) topological moves algorithm [33] was used to search the tree space and determine the best topology. Rabs were named according to phylogenetic grouping with existing published P. octaurelia Rabs [22]. RILP analysis was conducted using a protein sequence alignment (Figure S5), due to lower overall sequence homology between the the proteins.
For the codon-based tests for selection using the Mega7 program, the following parameters were used: 500 bootstrap replications, the Kumar model/method for substitutions, and the complete deletion treatment of gaps/missing data.
Rab-GFP expression in live cells
Paramecium tetraurelia cells were grown in wheat grass medium infused with Klebsiella bacteria [34]. Two N-terminally tagged gene fusions to green fluorescent protein (GFP) were constructed by molecular cloning: P. tetraurelia Rab7a2- GFP and P. tetraurelia Rab7b-GFP. Rab-GFP gene fusions were cloned using the Gateway system (ThermoFisher). PCR-amplified Rab genes were TOPO cloned into the pENTR-D-TOPO entry vector. CACC was added to each forward primer to enable directional cloning into pENTR-D. The expression vector for constitutive expression under the calmodulin promoter was obtained from Jean Cohen as pPXV [35] and converted into a Gateway destination vector using the Gateway conversion kit (ThermoFisher). The genes were recombined into this final destination expression vector using the Clonase reaction (ThermoFisher), and screened by diagnostic digest and Sanger sequencing. Linearized Rab-GFP expression constructs containing Paramecium telomeres on each end were injected into the macronucleus of the relevant Paramecium species using a Nikon Diaphot inverted microscope and a Narishige IM300 microinjector. Single cells were immobilized under silicon oil using an Eppendorf CellTram Vario manual microinjector, injected, re-mobilized and recovered into single cell culture. Needles for injection and immobilization were pulled using a Sutter Model P-87 micropipette puller. Each resulting line was screened for either rescue, by injection of an Nd7 gene-containing construct, of a trichocyst release phenotype in the P. tetraurelia in an Nd7- mutant [36] and GFP expression, or solely for GFP expression (all other Paramecium species) on a Zeiss Axioplan widefield epifluorescent microscope. Several GFP-positive lines for each construct were screened to check for consistency of expression pattern at different expression levels. Cellular organelles, including contractile vacuoles and the oral apparatus, were identified by correlating the brightfield channels of each Z series of sections taken through the cell with the green fluorescent channel. Localization patterns were virtually all consistent, although different levels of expression, or brightness of GFP signal, were observed. GFP-expressing cell lines were grown up without allowing starvation, as triggering autogamy and the ensuing remaking of the macronucleus would discard the exogenous expression vector. For confocal imaging, cells were dyed with a 5 µM final concentration of LysoTracker red acidic dye (LysoTracker Red DND-99 from ThermoFisher) for 5 minutes and immobilized by centrifuging a 500 µl culture into 30 µl of a 2% methyl cellulose solution. The supernatant was then removed, and the live cells embedded in the methyl cellulose were transferred onto a Fisher SuperFrost slide under a 22 × 22 mm no. 1 coverslip, and immediately imaged on a Leica SP5 laser scanning confocal microscope. Post-processing of images was performed using the public domain program ImageJ (http://imagej.nih.gov/ij/).
Supplementary Material
Acknowledgments
We thank Jean-François Gout for the blastp and blat results, as well as Olivier Arnaiz and Linda Sperling for sharing the unpublished genomic data from the recently-sequenced P. aurelias. The GFP-Rab7 plasmids were constructed by undergraduates from Indiana University, particularly Ugne Zekonyte, Samantha Freijie, and Jordan Crowe. Microinjection was conducted in the Indiana Drosophila Genomics Initiative (INGEN) facility. Microscopy was conducted in the Light Microscopy Imaging Facility at Indiana University, with assistance from Jim Powers. We thank Elzbieta Wyroba for informative discussion of her group’s results, Tom Doak for invaluable experimental discussions and editing, and Parul Johri for helpful evolutionary discussions.
Funding Statement
This research was supported by a National Institute of Health Ruth L. Kirschstein National Research Service Award (NRSA) NIH-NIGMS,1F32GM101891-02 to L.J.B., and National Science Foundation award MCB-1050161 to M.L.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Elias M, Brighouse A, Gabernet-Castello C, et al. Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J Cell Sci. 2012;125Pt 10:2500–2508. PubMed PMID: 22366452; PubMed Central PMCID: PMCPMC3383260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bucci C, Thomsen P, Nicoziani P, et al. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11(2): 467–480. PubMed PMID: 10679007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Guerra F, Bucci C.. Multiple roles of the small GTPase Rab7. Cells. 2016;5(3). PubMed PMID: 27548222; PubMed Central PMCID: PMCPMC5040976. DOI: 10.3390/cells5030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Saito-Nakano Y, Mitra BN, Nakada-Tsukui K, et al. Two Rab7 isotypes, EhRab7A and EhRab7B, play distinct roles in biogenesis of lysosomes and phagosomes in the enteric protozoan parasite Entamoeba histolytica. Cell Microbiol. 2007;97:1796–1808. PubMed PMID: 17359234. [DOI] [PubMed] [Google Scholar]
- [5].Kashiwazaki J, Iwaki T, Takegawa K, et al. Two fission yeast rab7 homologs, ypt7 and ypt71, play antagonistic roles in the regulation of vacuolar morphology. Traffic (Copenhagen, Denmark). 2009;107:912–924. PubMed PMID: 19453973. [DOI] [PubMed] [Google Scholar]
- [6].Poteryaev D, Datta S, Ackema K, et al. Identification of the switch in early-to-late endosome transition. Cell. 2010;1413:497–508. PubMed PMID: 20434987. [DOI] [PubMed] [Google Scholar]
- [7].Cui Y, Zhao Q, Gao C, et al. Activation of the Rab7 GTPase by the MON1-CCZ1 Complex Is Essential for PVC-to-Vacuole trafficking and plant growth in arabidopsis. Plant Cell. 2014;26(5):2080–2097. PubMed PMID: 24824487; PubMed Central PMCID: PMCPMC4079370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].De Luca M, Cogli L, Progida C, et al. RILP regulates vacuolar ATPase through interaction with the V1G1 subunit. J Cell Sci. 2014;127(Pt 12):2697–2708. PubMed PMID: 24762812. [DOI] [PubMed] [Google Scholar]
- [9].De Luca M, Bucci C.. A new V-ATPase regulatory mechanism mediated by the Rab interacting lysosomal protein (RILP). Commun Integr Biol. 2014;7(5). PubMed PMID: 26843904; PubMed Central PMCID: PMCPMC4594554. DOI: 10.4161/cib.29616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Johansson M, Rocha N, Zwart W, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176(4):459–471. PubMed PMID: 17283181; PubMed Central PMCID: PMCPMC2063981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Harrison RE, Bucci C, Vieira OV, et al. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol. 2003;23(18): 6494–6506. PubMed PMID: 12944476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Johansson M, Lehto M, Tanhuanpaa K, et al. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005;1612:5480–5492. PubMed PMID: 16176980; PubMed Central PMCID: PMCPMC1289395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Klöpper TH, Kienle N, Fasshauer D, et al. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biology. 2012;10:71. PubMed PMID: 22873208; PubMed Central PMCID: PMCPMC3425129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yao M, Liu X, Li D, et al. Late endosome/lysosome-localized Rab7b suppresses TLR9-initiated proinflammatory cytokine and type I IFN production in macrophages. J Immunol. 2009;1833:1751–1758. PubMed PMID: 19587007. [DOI] [PubMed] [Google Scholar]
- [15].Ebine K, Inoue T, Ito J, et al. Plant vacuolar trafficking occurs through distinctly regulated pathways. Current Biology: CB. 2014;24(12):1375–1382. PubMed PMID: 24881878. [DOI] [PubMed] [Google Scholar]
- [16].Wyroba E, Kwasniak P, Miller K, et al. Site-directed mutagenesis, in vivo electroporation and mass spectrometry in search for determinants of the subcellular targeting of Rab7b paralogue in the model eukaryote Paramecium octaurelia. Eur J Histochem. 2016;602:2612. PubMed PMID: 27349314; PubMed Central PMCID: PMCPMC4933825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Osińska M, Wiejak J, Wypych E, et al. Distinct expression, localization and function of two Rab7 proteins encoded by paralogous genes in a free-living model eukaryote. Acta Biochimica Polonica. 2011;58(4): 597–607. PubMed PMID: 22030555. [PubMed] [Google Scholar]
- [18].Venkatesh D, Boehm C, Barlow LD, et al. Evolution of the endomembrane systems of trypanosomatids - conservation and specialisation. J Cell Sci. 2017;130(8):1421–1434. PubMed PMID: 28386020; PubMed Central PMCID: PMCPMC5399786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aury J-M, Jaillon O, Duret L, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444(7116):171–178. PubMed PMID: 17086204. [DOI] [PubMed] [Google Scholar]
- [20].McGrath CL, Gout JF, Johri P, et al. Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 2014. PubMed PMID: 25085612; PubMed Central PMCID: PMCPMC4199370. DOI: 10.1101/gr.173740.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McGrath CL, Gout J-F, Doak TG, et al. Insights into three whole-genome duplications gleaned from the Paramecium caudatum genome sequence. Genetics. 2014;1974:1417–1428. PubMed PMID: 24840360; PubMed Central PMCID: PMCPMC4125410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mackiewicz P, Wyroba E. Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evol Biol. 2009;9:101. PubMed PMID: 19442299; PubMed Central PMCID: PMCPMC2693434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Catania F, Wurmser F, Potekhin AA, et al. Genetic diversity in the Paramecium aurelia species complex. Mol Biol Evol. 2009;262:421–431. PubMed PMID: 19023087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;337:1870–1874. PubMed PMID: 27004904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wyroba E, Surmacz L, Osinska M, et al. Phagosome maturation in unicellular eukaryote Paramecium: the presence of RILP, Rab7 and LAMP-2 homologues. Eur J Histochem. 2007;51(3): 163–172. PubMed PMID: 17921111. [PubMed] [Google Scholar]
- [26].Wang T, Wong KK, Hong W. A unique region of RILP distinguishes it from its related proteins in its regulation of lysosomal morphology and interaction with Rab7 and Rab34. Mol Biol Cell. 2004;152:815–826. PubMed PMID: 14668488; PubMed Central PMCID: PMCPMC329395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Marsman M, Jordens I, Rocha N, et al. A splice variant of RILP induces lysosomal clustering independent of dynein recruitment. Biochem Biophys Res Commun. 2006;3443:747–756. PubMed PMID: 16631113. [DOI] [PubMed] [Google Scholar]
- [28].Wu M, Wang T, Loh E, et al. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J. 2005;248:1491–1501. PubMed PMID: 15933719; PubMed Central PMCID: PMCPMC1142575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bright LJ, Gout JF, Lynch M. Early stages of functional diversification in the Rab GTPase gene family revealed by genomic and localization studies in Paramecium species. Mol Biol Cell. 2017;288:1101–1110. PubMed PMID: 28251922; PubMed Central PMCID: PMCPMC5391186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schlacht A, Herman EK, Klute MJ, et al. Missing pieces of an ancient puzzle: evolution of the eukaryotic membrane-trafficking system. Cold Spring Harb Perspect Biol. 2014;610:a016048. PubMed PMID: 25274701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Arnaiz O, Van Dijk E, Betermier M, et al. Improved methods and resources for paramecium genomics: transcription units, gene annotation and gene expression. BMC Genomics. 2017;18(1):483. 10.1186/s12864-017-3887-z. PubMed PMID: 28651633; PubMed Central PMCID: PMCPMC5485702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;593:307–321. PubMed PMID: 20525638. [DOI] [PubMed] [Google Scholar]
- [33].Hordijk W, Gascuel O. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics. 2005;2124:4338–4347. PubMed PMID: 16234323. [DOI] [PubMed] [Google Scholar]
- [34].Beisson J, Bétermier M, Bré M-H, et al. Mass culture of Paramecium tetraurelia. Cold Spring Harb Protoc. 2010;2010(1):pdb.prot5362. PubMed PMID: 20150121. [DOI] [PubMed] [Google Scholar]
- [35].Hauser K, Haynes W, Kung C, et al. Expression of the green fluorescent protein in Paramecium tetraurelia. Eur J Cell Biol. 2000;79(2): 144–149. PubMed PMID: 4841347477311106025related:6atcS-HrL0MJ. [DOI] [PubMed] [Google Scholar]
- [36].Skouri F, Cohen J. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol Biol Cell. 1997;8(6): 1063–1071. PubMed PMID: 9201716; PubMed Central PMCID: PMCPMC305714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


