ABSTRACT
Each gene typically has multiple alternatively spliced transcripts. Different transcripts are assumed to play a similar biological role; however, some transcripts may simply lose their function due to loss of important functional domains. Here, we show that two different transcripts of lncRNA gene ANRIL associated with coronary artery disease (CAD) play antagonizing roles against each other. We previously reported that DQ485454, the short transcript, is downregulated in coronary arteries from CAD patients, and reduces monocyte adhesion to endothelial cells (ECs) and transendothelial monocyte migration (TEM). Interestingly, the longest transcript NR_003529 is significantly upregulated in coronary arteries from CAD patients. Overexpression of ANRIL transcript NR_003529 increases monocyte adhesion to ECs and TEM, whereas knockdown of NR_003529 expression reduces monocyte adhesion to ECs and TEM. Much more dramatic effects were observed for the combination of overexpression of NR_003529 and knockdown of DQ485454 or the combination of knockdown of NR_003529 and overexpression of DQ485454. The antagonizing effects of ANRIL transcripts NR_003529 and DQ485454 were associated with their opposite effects on expression of downstream target genes EZR, CXCL11 or TMEM106B. Our results demonstrate that different transcripts of lncRNA can exert antagonizing effects on biological functions, thereby providing important insights into the biology of lncRNA. The data further support the hypothesis that ANRIL is the causative gene at the 9p21 CAD susceptibility locus.
KEYWORDS: lncRNA, ANRIL (CDKN2B-AS1), EZR, TMEM106B, coronary artery disease (CAD) and myocardial infarction (MI), monocyte adhesion to endothelial cells, transendothelial migration of monocytes
Introduction
Coronary artery disease (CAD) and its complication myocardial infarction (MI) are the most common heart disease that causes 400,000 deaths each year in the US alone [1]. CAD is caused by accumulation or build-up of cholesterol and fatty lipids, leading to the development of plaques in the coronary arteries, a process referred to as atherosclerosis [2–4]. Atherosclerosis is initiated by monocyte adhesion to the endothelium of coronary arteries upon inflammatory stimulation or endothelial injury, followed by the transmigration of monocytes across the endothelial layer into the intima [2–4]. The monocytes then differentiate into macrophages, which further develop into lipid-laden foam cells known as a fatty streak, the hallmark of atherosclerosis [2–4]. For largely unknown reasons, some arterial beds are more prone to atherosclerosis than others, and coronary arteries are most often involved [5]. Moreover, genetic factors play a critical role in the development of atherosclerotic CAD. Genome-wide association studies (GWAS) have identified genomic variants at more than 150 loci that confer risk of CAD, including the chromosome 9p21.3 locus that contain a long noncoding RNA gene ANRIL [6].
Noncoding RNAs (ncRNAs) are a class of short and long transcripts that act as genetic, epigenetic and translational regulators for multiple biological processes and the development of some human diseases [7]. Examples of ncRNAs include microRNAs (miRNAs), circular RNAs, and long noncoding RNAs (lncRNAs) that are more than 200 base pairs in length [8]. Although the function of most lncRNAs remains unknown, some of these molecules are also implicated in various biological processes [9–11]. LncRNAs can regulate gene expression through a variety of mechanisms, such as epigenetic modifications of DNA, alternative splicing, posttranscriptional regulation, mRNA stability and translation [9–11]. Some lncRNAs are involved in cis-inactivation of larger genomic regions by epigenetic mechanisms [9–11]. Notably, many lncRNAs are known to regulate the expression of genes by a trans mechanism [11]. LncRNAs can act as scaffolds to bringing together multiple proteins to form ribonucleoprotein complexes [12]. In some cases, lncRNAs can function as molecular ‘decoys’ for transcription factors, other regulatory proteins, and miRNAs [11]. Interestingly, lncRNAs can positively or negatively control the expression of their target genes. For example, we recently showed that lncRNA ANRIL can positively regulate the expression of ANHAK1, CLIP1, CXCL11, ENC1, EZR, LYVE1 and WASL, but negatively regulates expression of TMEM100 and TMEM106B [13], although the underlying molecular mechanism(s) are unknown.
Recent studies suggest that lncRNAs are involved in development of atherosclerosis and CAD. Knockout (KO) of lncRNA MeXis gene impaired cholesterol metabolism and accelerated atherosclerosis in mice via an LXR-MeXis-Abca1 axis [14]. For lncRNA MALAT1, heterozygous Malat1± KO mice showed increased atherosclerosis with an ApoE−/- KO background due to massive immune system dysregulation [15]. The lncRNA ANRIL gene is located within the most robust GWAS locus for CAD on chromosome 9p21.3 locus [16,17]. Many variants in ANRIL show significant association with risk of CAD [18]. Previously, we identified the most abundant transcript of ANRIL in ECs as DQ485454, and showed that DQ485454 was significantly downregulated in coronary arteries from CAD patients compared with normal arteries [13]. DQ485454 overexpression significantly reduced monocyte adhesion to endothelial cells (ECs) and transendothelial migration of monocytes (TEM), two critical cellular processes involved in the initiation of CAD [13]. The siRNA-mediated knockdown of ANRIL expression showed the opposite effects [13]. Mechanistic studies showed that overexpression of ANRIL downstream target genes CLIP1, EZR, and LYVE1 reversed the effects of ANRIL KD on monocyte adhesion to ECs and TEM. These data suggest that ANRIL regulates EC functions directly related to CAD, and support the hypothesis that ANRIL is the causal gene for CAD at the 9p21 genetic locus. Moreover, in addition to ECs, ANRIL was also shown to regulate proliferation of vascular smooth muscle cells, a property also relevant to the development of atherosclerosis and CAD [18].
Despite the fact that accumulating evidence implicates ANRIL in development of CAD, much more work is needed to make a stronger case. Moreover, ANRIL shows extensive alternative splicing, which results in more than 20 different transcripts. The function of the longest ANRIL transcript, NR_003529, remains to be further defined. Therefore, in this study, we characterized the expression levels of NR_003529 in CAD vs. non-CAD coronary arteries, and examined the functional effects of overexpression or knockdown of NR_003529 on EC functions involved in the pathogenesis of CAD. Moreover, we investigated the interaction of NR_003529 with DQ485454 in EC functions. Identifying functions of different transcripts of ANRIL may be a key to identify new lncRNA-based molecular mechanisms contributing to the development of CAD.
Results
Upregulation of the longest transcript of ANRIL, NR_003529, in coronary arteries from CAD patients
We recently reported that ANRIL transcript DQ485454 was significantly downregulated in CAD coronary artery tissue samples [13]. However, it is unknown whether the expression level of the full-length or the longest transcript NR_003529 is also changed in CAD coronary artery tissue. We investigated the transcript-specific expression of the reported major transcripts of ANRIL, NR_003529, DQ485454, and circANRIL7-5, with the tissue samples from CAD patients vs. non-CAD controls (Fig. 1A). Using quantitative RT-PCR (qRT-PCR) analysis, we found that in contrast to DQ485454, the expression level of ANRIL transcript NR_003529 (detected using primers in exons 12 and 13b) was significantly upregulated by 2.5-fold in CAD patients compared to that of the non-CAD controls (Fig. 1B). ANRIL has more than 20 different transcripts, and it was technically challenging to design unique primers for individual transcripts and examine their expression levels using qRT-PCR analysis. However, we found that many transcripts share exons 5–7 and our qRT-PCR analysis revealed that the overall expression level across all transcripts of ANRIL sharing exons 5–7 was significantly increased in CAD coronary artery tissue compared to non-CAD tissue (Fig. 1C). Interestingly, we also found that the expression level of circANRIL containing exons 7–5 was significantly downregulated in CAD coronary artery tissue compared to the healthy controls (Fig. 1D). Similarly, the expression level of DQ485454 (detected using primers in exons 11–13) was significantly downregulated in CAD coronary arteries as previously reported [13] (Fig. 1E). Quantitative RT-PCR analysis also revealed that the expression level of transcripts containing exon 19 (Fig. 1F) was significantly increased in CAD coronary artery tissue compared to non-CAD tissue. These data suggest that ANRIL transcripts show differential expression patterns in CAD coronary arteries. Specifically, ANRIL transcripts NR_003529 and DQ485454 show significant upregulation and downregulation in coronary artery tissue samples from CAD patients, respectively.
Figure 1.
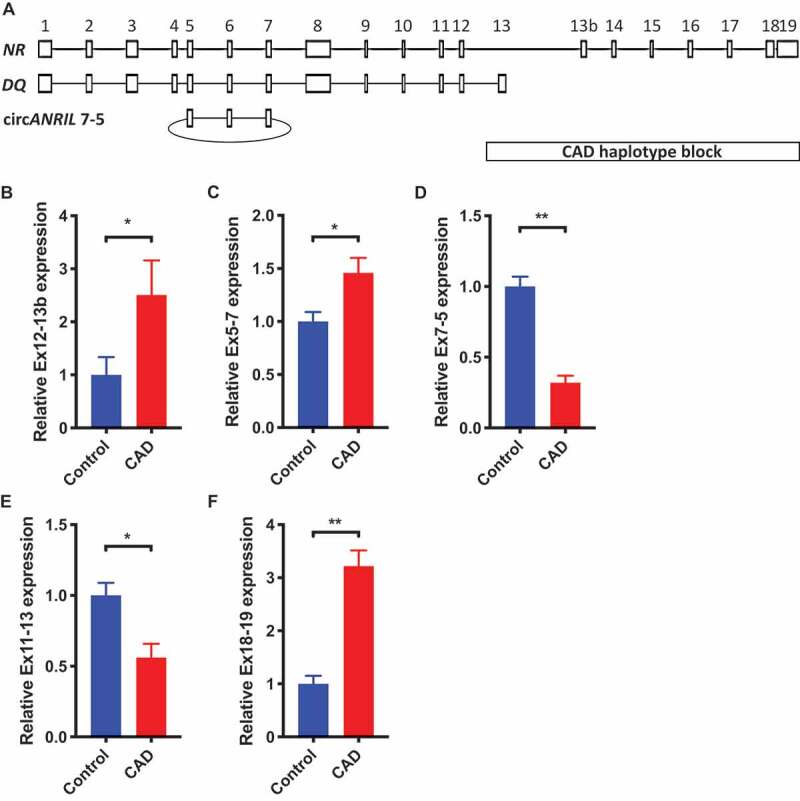
Increased expression of the longest ANRIL transcript NR_003529 (NR) in coronary arteries from CAD patients vs. non-CAD controls. Coronary arteries from 6 CAD patients and 6 age-matched non-CAD individuals were studied with real-time RT-PCR analysis. (A) Schematic diagram for the structure of ANRIL transcripts NR_003529 and DQ485454, and circANRIL with exons 5–7. The location of the CAD haplotype block is shown. (B-E) The relative abundance of different transcripts of ANRIL in CAD patients vs. non-CAD controls as represented by expression of (B) exons 12–13b for the detection of NR_003529, (C) exons 5–7 for the detection of both NR_003529 and DQ485454, (D) exons 7–5 for the detection of circANRIL7-5, and (E) exons 11–13 for the detection of DQ485454. (F) exons 18–19 for the detection of 3ʹ end of NR_003529 *p < 0.05 and **p < 0.01 vs. vector, N = 6.
Overexpression of ANRIL transcript NR_003529 in endothelial cells increases monocyte adhesion and transmigration
Based on the finding of significant upregulation of NR_003529 transcript in CAD tissue, we hypothesized that NR_003529 is involved in the development of CAD. To test the hypothesis, we overexpressed NR_003529 transcript in different ECs, including human coronary artery ECs (HCAECs), human umbilical vein ECs (HUVECs), and an EC cell line, EA.hy926 cells, by transient transfection with a NR_003529 expression plasmid. Our qRT-PCR analysis showed that the expression level of NR_003529 was increased by 13–17 fold depending on the type of ECs (Fig. 2A). Moreover, our qRT-PCR analysis showed that the overexpression was specific to NR_003529 and did not affect the expression of transcript DQ485454 (Fig. 2B). The transfected ECs were then used for assays for monocyte adhesion to ECs and transendothelial migration of monocytes, two key processes involved in the initiation of atherosclerosis during the pathogenesis of CAD [5,19,20]. We used THP-1 cells as monocytes in these assays. As shown in Fig. 2C, specific overexpression of NR_003529 in HCAECs and EA.hy926 cells, but not HUVECs, significantly increased monocyte adhesion to ECs (Fig. 2C) and transendothelial migration of monocytes (Fig. 2D). The data suggest that overexpression of ANRIL transcript NR_003529 confers a risk role in some ECs in development of CAD.
Figure 2.
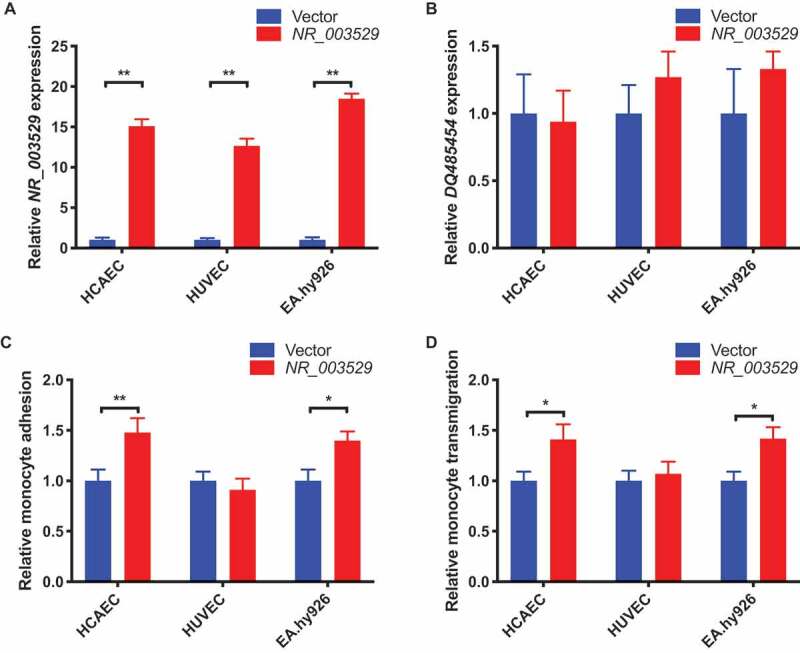
Overexpression of NR_003529 in HCAEC and EA.hy926 cells increases monocyte adhesion and transendothelial migration. (A) Expression of the NR_003529 transcript of ANRIL in HCAECs, HUVECs, and EA.hy926 cells transiently transfected with an expression plasmid for NR_003529 vs. pcDNA vector as quantified by qRT-PCR. (B) Expression of the DQ485454 transcript of ANRIL in HCAECs, HUVECs, and EA.hy926 cells transiently transfected with an expression plasmid for NR_003529 vs. pcDNA vector as quantified by qRT-PCR. (C) Relative adhesion of monocytes to HCAECs, HUVECs, and EA.hy926 cells transiently transfected with an expression plasmid for NR_003529 vs. pcDNA vector. (D) Relative transmigration of monocytes across a layer of HCAECs, HUVECs, and EA.hy926 cells transiently transfected with an expression plasmid for NR_003529 vs. pcDNA vector. *p < 0.05 and **p < 0.01 vs. vector, N = 5.
Knockdown of NR_003529 expression in endothelial cells reduces monocyte adhesion and transmigration.
To validate the finding of NR_003529 overexpression on EC functions involved in CAD, we performed similar functional assays but using ECs with siRNA knockdown of NR_003529 expression. Our qRT-PCR analysis showed that transient transfection of NR_003529-specific siRNA reduced its expression by 75–80% in HCAECs, HUVECs, and EA.hy926 cells (Fig. 3A). We also ensured that the NR_003529 knockdown did not affect the expression level of ANRIL transcript DQ485454 (Fig. 3B). In the assays for THP-1 monocyte adhesion to ECs and transendothelial migration of monocytes, we found that knockdown of NR_003529 expression in HCAECs, HUVECs, and EA.hy926 cells significantly reduced the function of ECs in monocyte adhesion (Fig. 3C) and transmigration (Fig. 3D). These data further show that ANRIL transcript NR_003529 plays an important role in EC functions involved in the pathogenesis of CAD.
Figure 3.
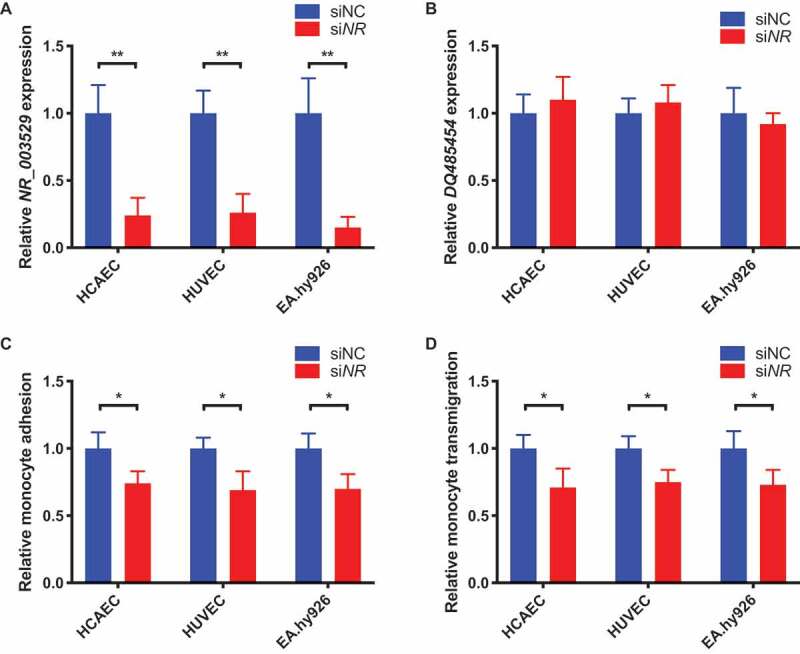
Knockdown of NR_003529 in HCAEC, HUVECs and EA.hy926 cells reduces monocyte adhesion and transmigration. (A) Expression of the NR_003529 transcript of ANRIL in HCAECs, HUVECs, and EA.hy926 cells transiently transfected with NR_003529-specific siRNA (siNR) vs. negative control siRNA (siNC) as quantified by qRT-PCR. (B) Expression of the DQ485454 transcript of ANRIL in HCAECs, HUVECs, and EA.hy926 cells transiently transfected with NR_003529-specific siRNA (siNR) vs. negative control siRNA (siNC) as quantified by qRT-PCR. (C) Relative adhesion of monocytes to HCAECs, HUVECs, and EA.hy926 cells transiently transfected with NR_003529-specific siRNA (siNR) vs. negative control siRNA (siNC). (D) Relative transmigration of monocytes across a layer of HCAECs, HUVECs, and EA.hy926 cells transiently transfected with NR_003529-specific siRNA (siNR) vs. negative control siRNA (siNC). *p < 0.05 and **p < 0.01 vs. vector, N = 5.
Combination of NR_003529 overexpression and DQ485454 knockdown in HCAECs dramatically increases monocyte adhesion and transmigration.
In human coronary artery tissue samples from CAD patients, we showed that the NR_003529 transcript of ANRIL was significantly upregulated (Fig. 1A), whereas the expression level of the DQ485454 transcript was significantly downregulated [13]. Therefore, we were interested in determining the effect of NR_003529 overexpression and DQ485454 knockdown on EC functions relevant to CAD. We co-transfected HCAECs with an expression plasmid for NR_003529 and two different siRNAs. Our qRT-PCR analysis showed that the co-transfection led to the overexpression of NR_003529 by 10–12 fold in HCAECs (Fig. 4A), and specific knockdown of DQ485454 by 70–75% with two separate siRNAs (Fig. 4B). Then, we utilized the transfected HCAECs for assays for THP-1 monocyte adhesion to ECs and transendothelial migration of monocytes. As shown in Fig. 4C, NR_003529 overexpression as well as siDQ1 and siDQ2 all significantly increased monocyte adhesion to HCAECs by about ~1.5 fold, however, the combination of NR_003529 overexpression with siDQ1 or siDQ2 dramatically increased monocyte adhesion to HCAECs by about 2.7 fold. Similar observations were made for transendothelial migration of monocytes (Fig. 4D). These data suggest that there is a synergistic effect between NR_003529 overexpression and DQ485454 knockdown on EC functions involved in the pathogenesis of CAD.
Figure 4.
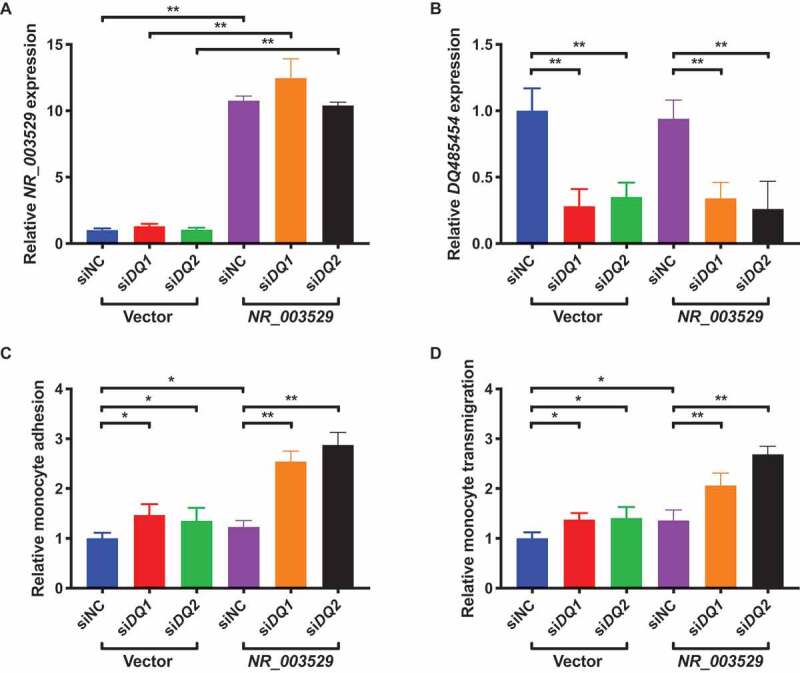
Overexpression of NR_003529 together with knockdown of DQ485454 in HCAECs dramatically increases monocyte adhesion and transmigration. (A) Expression of the NR_003529 transcript of ANRIL in HCAECs transiently co-transfected with DQ485454-specific siRNAs (siDQ1, siDQ2) vs. negative control siRNA (siNC) and the overexpression plasmid for NR_003529 vs. pcDNA vector as quantified by qRT-PCR. (B) Expression of the DQ485454 transcript of ANRIL in HCAECs transiently co-transfected with DQ485454-specific siRNAs (siDQ1, siDQ2) vs. negative control siRNA (siNC) and the overexpression plasmid for NR_003529 vs. pcDNA vector as quantified by qRT-PCR. (C) Relative adhesion of monocytes to HCAECs transiently co-transfected with DQ485454-specific siRNAs (siDQ1, siDQ2) vs. negative control siRNA (siNC) and the overexpression plasmid for NR_003529 vs. pcDNA vector. (D) Relative transmigration of monocytes across a layer of HCAECs transiently co-transfected with DQ485454-specific siRNAs (siDQ1, siDQ2) vs. negative control siRNA (siNC) and the overexpression plasmid for NR_003529 vs. pcDNA vector. *p < 0.05 and **p < 0.01 vs. vector, N = 5.
Combination of NR_003529 knockdown and DQ485454 overexpression in HCAECs dramatically decreases monocyte adhesion and transmigration.
We further investigated the effects of NR_003529 knockdown and DQ485454 overexpression on functions of HCAECs (Fig. 5A and 5B). As shown in Fig. 5C, knockdown of NR_003529 expression using siRNAs siNR1 and siNR2 as well as overexpression of DQ485454 all significantly decreased monocyte adhesion to HCAECs by about 25%, however, the combination of overexpression of DQ485454 with either NR_003529 siNR1 or siNR2 dramatically decreased monocyte adhesion to HCAECs by about 80%. Similar observations were made for transendothelial migration of monocytes (Fig. 5D). These data suggest that there is a synergistic effect between NR_003529 knockdown and DQ485454 overexpression on EC functions involved in the pathogenesis of CAD.
Figure 5.
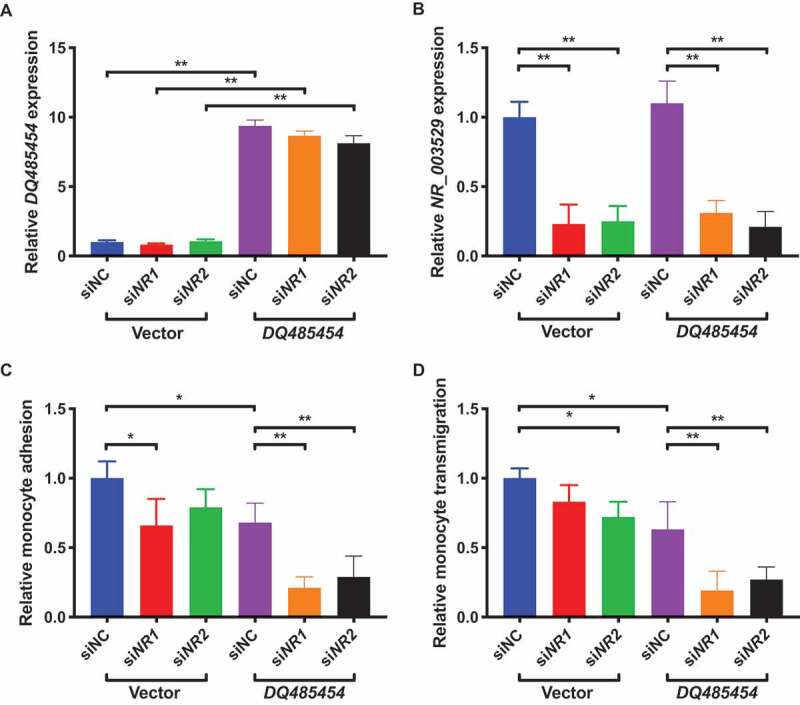
Overexpression of DQ485454 together with knockdown of NR_003529 in HCAECs dramatically reduces monocyte adhesion and transmigration. (A) Expression of the NR_003529 transcript of ANRIL in HCAECs transiently co-transfected with NR_003529-specific siRNAs (siNR1, siNR2) vs. negative control siRNA (siNC) and the overexpression plasmid for DQ485454 vs. pcDNA vector control as quantified by qRT-PCR. (B) Expression of the DQ485454 transcript of ANRIL in HCAECs transiently co-transfected with NR_003529-specific siRNAs (siNR1, siNR2) vs. negative control siRNA (siNC) and the overexpression plasmid for DQ485454 vs. pcDNA vector as quantified by qRT-PCR. (C) Relative adhesion of monocytes to HCAECs transiently co-transfected with NR_003529-specific siRNAs (siNR1, siNR2) vs. negative control siRNA (siNC) and the overexpression plasmid for DQ485454 vs. pcDNA vector. (D) Relative transmigration of monocytes across a layer of HCAECs transiently co-transfected with NR_003529-specific siRNAs (siNR1, siNR2) vs. negative control siRNA (siNC) and the overexpression plasmid for DQ485454 vs. pcDNA vector. *p < 0.05 and **p < 0.01 vs. vector, N = 5.
Transcript-specific regulation of downstream target genes by ANRIL transcripts NR_003529 and DQ485454
To determine why ANRIL transcript NR_003529 regulates EC functions in an opposite direction from DQ485454, we assessed their effects on the expression of downstream target genes, including CLIP1, EZR, LYVE1, TMEM100, TMEM106B, CXCL11, and ENC1 identified by global gene expression arrays [13]. As previously reported [13], our qRT-PCR analysis showed that overexpression of DQ485454 significantly increased the expression of CLIP1, EZR, LYVE1, CXCL11, and ENC1, but reduced the expression of TMEM100 and TMEM106B in HCAECs (Fig. 6A). However, overexpression of NR_003529 significantly decreased the expression of EZR and CXCL11, but increased the expression of LYVE1 and TMEM106B in HCAECs (Fig. 6A). Knockdown of NR_003529 expression significantly increased the expression of EZR and CXCL11 in HCAECs (Fig. 6B). Together, these data suggest that ANRIL transcripts NR_003529 and DQ485454 affect EC functions involved in CAD in an opposite direction by differentially regulating the expression of EZR, CXCL11, and probably TMEM106B.
Figure 6.
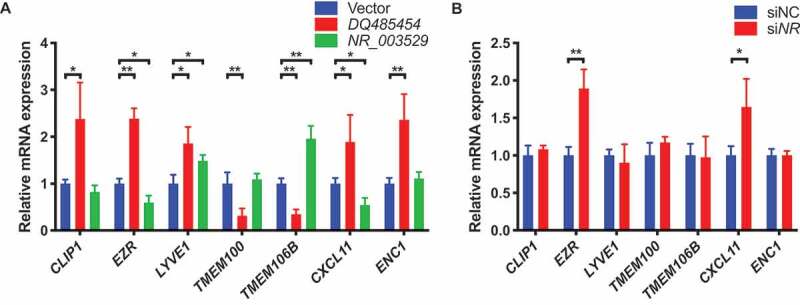
Expressions of EZR, CXCL11, and TMEM106B are differentially regulated by different transcripts of ANRIL. (A) Relative mRNA expressions of seven downstream genes of ANRIL (CLIP1, EZR, LYVE1, TMEM100, TMEM106B, CXCL11, and ENC1) in HCAECs transfected with the overexpression plasmid for either DQ485454 or NR_003529 transcripts of ANRIL vs. pcDNA3.1 vector control as determined by qRT-PCR. (B) Relative mRNA expressions of the seven downstream genes of ANRIL in HCAECs transfected with the NR_003529-specific siRNA (siNR) vs. negative control siRNA (siNC) as determined by qRT-PCR. *p < 0.05 and **p < 0.01 vs. vector, N = 5.
Discussion
Genomic variants in and near ANRIL on chromosome 9p21 were significantly associated with multiple diseases, including CAD, type 2 diabetes, several forms of cancer, aortic aneurysms, and others. For CAD, the 9p21 locus is the first locus identified by GWAS in 2007, and appears to be the most robust CAD locus with a recent association P value of 10−223 [21]. ANRIL shows a complex pattern of alternative splicing, resulting in more than 20 different isoforms of transcripts (Fig. 1A). There are two major groups of transcripts for ANRIL [22], one group ending with exon 19 such as the longest transcript NR_003529, and the other group ending with exon 13 such as DQ485454 (Fig. 1A). However, little is done with the characterization of different transcripts for their disease-associated or tissue-specific expression and functions. Such studies are essential for full understanding of the functionality of ANRIL and its role in the pathogenesis of human diseases. In this study, we focused on two major ANRIL transcripts in endothelial cells, NR_003529 and DQ485454. We found that the two transcripts showed opposite expression patterns in coronary arteries from CAD patients as compared with non-CAD tissue samples. Our qRT-PCR analysis showed that NR_003529 expression was significantly upregulated in CAD arteries (Fig. 1B), whereas DQ485454 expression was significantly downregulated in CAD arteries [13]. Similarly, we found that the expression of all transcripts ending with exon 19 were significantly upregulated in CAD arteries, where the expression of all transcripts ending with exon 13 were significantly downregulated (Fig. 1D and 1E). These data suggest that different ANRIL transcripts exhibit disease-specific expression patterns in CAD.
Consistent with their opposite expression patterns, NR_003529 and DQ485454 antagonize each other functionally (Fig. 2–5). In two endothelial cell assays relevant to the pathogenesis of CAD, NR_003529 increased monocytes adhesion to ECs (Fig. 2C) and transendothelial migration of monocytes (Fig. 2D) when overexpressed in ECs. Knockdown of NR_003529 expression reduced monocytes adhesion to ECs (Fig. 3C) and transendothelial migration of monocytes (Fig. 3D). Interestingly, the finding of upregulation of NR_003529 and downregulation of DQ485454 in CAD arteries showed a synergistic effect in dramatically increasing monocytes adhesion to ECs (Fig. 4C) and transendothelial migration of monocytes (Fig. 4D). Increased monocyte adhesion to ECs and transendothelial migration of monocytes are involved in promoting the initiation and development of atherosclerosis and CAD. These data further highlight the importance of molecular and functional characterization of different transcripts of ANRIL. More importantly, our data further support the hypothesis that ANRIL is the disease-causing gene at the 9p21 CAD locus.
We previously performed microarray global gene expression profiling with ECs transfected with ANRIL siRNA vs. scramble siRNA, and identified important downstream target genes of ANRIL. We showed that ANRIL can positively regulate the expression of ANHAK1, CLIP1, CXCL11, ENC1, EZR, LYVE1 and WASL, but negatively regulates expression of TMEM100 and TMEM106B [13]. Mechanistic studies showed that overexpression of ANRIL downstream target genes CLIP1, EZR, and LYVE1 reversed the effects of ANRIL KD on monocyte adhesion to ECs and TEM [13]. Recently, we showed that knockdown of TMEM106B, but not of TMEM100, reversed the increased monocyte adhesion to ECs and TEM induced by knockdown of DQ485454 [23]. Furthermore, overexpression of TMEM106B reversed the decreased monocyte adhesion to ECs and TEM induced by DQ485454 overexpression [23]. TMEM106B expression was significantly upregulated by >2-fold in CAD coronary arteries, and genomic variants in TMEM106B (but not in TMEM100) showed significant association with CAD (p = 1.9 × 10−8) in the UK Biobank samples [23]. We also identified significant gene-gene interaction between ANRIL variant rs2383207 and TMEM106B variant rs3807865 (p = 0.009) [23]. These studies suggest that ANRIL downstream target genes CLIP1, EZR, LYVE1, and TMEM106B are particularly important in regulating the functions of ANRIL in ECs involved in the development of CAD. To identify a molecular mechanism underlying the differential antagonizing functions of transcripts NR_003529 and DQ485454, we examined ANRIL downstream genes whose expression was affected by DQ485454 [13]. Overexpression of NR_003529 significantly reduced expression of EZR and CXCL11, and increased expression of TMEM106B, whereas overexpression of DQ485454 showed opposite expression patterns (Fig. 6A). On the other hand, knockdown of NR_003529 expression significantly increased the expression of EZR and CXCL11 (Fig. 6B). As CAD patient coronaries showed upregulation of NR_003529 and downregulation of DQ485454, we propose that reduced EZR expression and increased TMEM106B expression are more critically involved in EC functions regulated by ANRIL.
In addition to more than 20 different linear ANRIL transcripts, multiple circular forms of ANRIL (circANRIL) were reported [24–26]. Holdt et al showed that the expression of circANRIL with exons 5-6-7 was detected in vascular smooth muscle cells and macrophages in human atherosclerotic plaques, and a CAD protective haplotype was associated with increased expression of circANRIL in peripheral blood mononuclear cells, whole blood and endarterectomy samples [25]. Interestingly, we showed that the expression level of circANRIL was significantly less expressed in CAD coronary arteries than in non-CAD tissue samples (Fig. 1). Together, all these data suggest that reduced circANRIL expression is associated with risk of CAD, whereas increased circANRIL expression is associated with a protective role in CAD. Burd et al showed that the expression of both linear and circular ANRIL at the proximal end was correlated with CAD risk genotype of rs10757278, whereas distal ANRIL transcripts with exons 18 and 19 were independent to the risk genotype [24]. Moreover, some SNPs near exon 15 were predicted to affect ANRIL splicing [24]. The interesting findings support the hypothesis that different CAD genotypes (risk, protective) may differentially affect the expression of transcripts NR_003529 and DQ485454 by affecting ANRIL expression and/or splicing, resulting in the opposite expression patterns of the two transcripts in CAD coronary arteries. Future studies with large sample sizes may be able to test the interesting possibility.
There are similarities and differences between ANRIL transcripts DQ485454 and NR_003529 (Fig. 1A). DQ485454 is 2,659 bp in length and contains 13 exons, whereas NR_003529 is 3,857 bp long and has 19 exons (Fig. 1A). DQ485454 and NR_003529 share exons 1 to 12. DQ485454 contains a unique exon, exon 13, whereas NR_003529 contains an alternatively spliced exon 13, exon 13b, and 6 other exons from exon 14 to 19 (Fig. 1A). Therefore, an alternative splicing event at and after exon 12 is a key for generation of two different transcripts DQ485454 and NR_003529. The molecular mechanism for the differential expression of NR_003529 and DQ485454 in coronary artery tissue samples from CAD patients is unknown. One possibility is that different CAD-relevant splicing regulatory proteins act at the exon 12/exon 13 junction and the exon 12/exon 13b junction, respectively, to either inhibit or enhance alternative splicing, resulting in differential expression of DQ485454 and NR_003529 in CAD tissue samples. Alternatively, as noted above, a risk CAD genotype at the junction between exon 12 and exon 13 may inhibit splicing, resulting in reduced DQ485454 expression, whereas a different risk CAD genotype at the junction between exon 12 and exon 13b enhances splicing, resulting in increased NR_003529 expression in CAD tissue samples.
The molecular mechanism by which different ANRIL transcripts NR_003529 and DQ485454 differentially regulate the expression of its downstream genes EZR, CXCL11, and TMEM106B in endothelial cells is not clear. ANRIL can regulate expression of adjacent genes by facilitating the recruitment of the chromatin-modifying complex to promoter/regulatory regions via interaction with the components of PRC1/2, such as EZH2, SUZ12, and CBX7 [27–30]. The unique sequences of NR_003529 (exon 13a, 14–19) and DQ485454 (exon 13) may affect the interaction between ANRIL and different components of PRC1/2, and thus, regulate the expression of downstream genes in an opposite direction. Alternatively, as with many other lncRNAs, ANRIL can act as a sponge for some microRNAs such as let-7a [31], miR-9 [32], miR-7-5p [33], miR-127 [34], miR-181b [35], miR-181a [36,37], and potentially many other unreported miRNAs. The unique sequences of NR_003529 (exon 13a, 14–19) and DQ485454 (exon 13) may bind different miRNAs, and differentially regulate the expression of downstream target genes in endothelial cells.
ANRIL appears to be a primate-specific lncRNA. Sequence analysis did not detect an ortholog with a high similarity in the mouse genome to date. A lncRNA gene Gm12610 in the mouse genome was previously referred to as Anril (https://www.ncbi.nlm.nih.gov/nuccore/NR_132431.1) because it is located at the Cdkn2a/Cdkn2b locus on mouse chromosome 4. However, we performed BLASTN analysis and found no significant similarity between Gm12610 and human ANRIL transcript NR_003529 or between Gm12610 and DQ485454. Therefore, no in vivo knockout studies can be performed in animals to investigate the causal effect of ANRIL on atherosclerosis. Although the two functional assays used in this study, monocytes adhesion to ECs and transendothelial migration of monocytes, mimic the initiation processes involved in the development of atherosclerosis, they are in vitro assays and care should be taken when extrapolating the data to human CAD in vivo. Nevertheless, the data from two other studies provide evidence to support that ANRIL transcripts NR_003529 and DQ485454 are relevant to the pathogenesis of CAD in humans. First, we studied human coronary artery tissues from CAD patients and control individuals that are directly relevant to the pathogenesis of CAD, and found that ANRIL transcripts NR_003529 and DQ485454 were significantly upregulated and downregulated in coronary artery tissue samples from CAD patients, respectively (Fig. 1). Second, our genome-wide association studies (GWAS) showed that TMEM106B, one of the downstream genes of ANRIL, is a new susceptibility gene for CAD, and interacts with ANRIL to modulate the risk of CAD [23]. We further showed that overexpression of NR_003529 significantly increased the expression of TMEM106B in HCAECs (Fig. 6A), whereas overexpression of DQ485454 significantly reduced the expression of TMEM106B in HCAECs (Fig. 6A). The data suggest that the ANRIL-TMEM106B regulatory axis is involved in the development of CAD.
NR_003529 knockdown significantly reduced THP-1 monocyte adhesion (Fig. 3C) and transendothelial migration of monocytes (Fig. 3D) in all three different types of ECs, including HCAECs, HUVECs, and EA.hy926 cells. However, overexpression of NR_003529 significantly increased monocyte adhesion to ECs (Fig. 2C) and transendothelial migration of monocytes (Fig. 2D) in HCAECs and EA.hy926 cells, but not in HUVECs. The lower transfection efficiency of HUVECs for plasmid DNA (40% vs. 50% for HCAEs and 70% for EA.hy926) may be an underlying cause. Moreover, the transfection efficiency of plasmid DNA into ECs is lower than of siRNAs.
In summary, this study reveals an interesting finding that two different transcripts of ANRIL, NR_003529 and DQ485454, show an opposite expression pattern in coronary arteries from CAD patients, and exhibit an antagonizing role in EC functions involved in the initiation and development of CAD. Functionally, NR_003529 regulates the expression of ANRIL downstream genes EZR, CXCL11, and TMEM106B in an opposite direction against DQ485454, which may explain their antagonizing roles in EC functions. Our data further support the hypothesis that ANRIL is the disease-causing gene at the 9p21 CAD locus, and provide important insights into the biology of lncRNA ANRIL. The results highlight the importance of molecular characterization of different transcripts of a lncRNA for their expression and functions in human disease.
Materials and methods
Human subjects
This study involved human coronary artery tissue samples from six CAD patients and six age-matched non-CAD study subjects reported by us previously [13,23,38]. The mean age was 51 ± 4 years for both CAD and control groups. This study was approved by the Cleveland Clinic Institutional Review Board (IRB) on Human Subject Research and written consent was obtained from the subjects. This study abides by the Declaration of Helsinki principles.
Plasmids and siRNAs
A mammalian expression plasmid for ANRIL transcript DQ485454, referred to as pcDNA3.1-ANRIL (DQ485454), was described previously [13]. The pcDNA3.1-ANRIL (NR_003529) plasmid was originally created by cloning the full-length ANRIL transcript into Xho I and Hind III restriction sites of the pcDNA3.1 vector as previously described by us [39].
The siRNAs used in the study were ordered from Integrated DNA Technologies (Skokie, IL) and Lincode control siRNA was ordered from GE Dharmacon (Lafayette, CO). The specific sequences of the ANRIL siRNAs are as follows:
siDQ1: AUCUGUGUUUUUGUCCAAUGUCCTT;
siDQ2: GAAUGAUACCUGCUUCUCUAUCUTG;
siNR1: CAUGCUCCCUCCCCUCAUUGAGGTT;
siNR2: CACAGAUGCCUAACGCACUAUGGTA.
Cell culture and transfection
HCAECs (ATCC, USA) and HUVECs (Lonza, USA) were cultured in a phenol-red-free endothelial growth medium (ATCC, USA) supplemented with 2% foetal bovine serum (FBS), 0.4% human fibroblast growth factor, 0.1% human epidermal growth factor, 0.1% vascular endothelial growth factor, 0.1% insulin-like growth factor, 0.1% ascorbic acid, 0.1% heparin, 0.1% gentamicin/amphotericin-B and 0.04% hydrocortisone (all from ATCC, USA). EA.hy926, a human umbilical vascular EC line (ATCC, USA), was cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS. Human THP-1 monocytes were obtained from the ATCC and cultured in RPMI 1640 supplemented with 10% FBS. All cell cultures were maintained at 37°C in a humidified atmosphere with 5% CO2.
Transient transfection of ECs with mammalian expression plasmids and siRNAs was performed using non-liposomal TransIT-X2 Dynamic Delivery System (Mirus Bio, Madison, WI) according to the manufacturer’s instructions.
Monocyte adhesion to EC assays
Monocyte adhesion to ECs assays were performed as previously described [13]. Five independent experiments were performed.
Assays for transendothelial migration of monocytes
TEM was performed as previously described [13]. Five independent experiments were performed.
Real-time RT–PCR analysis
Total RNA samples were isolated from the transfected cells and coronary arteries using TRIzol reagent (Thermo Fisher Scientific) according to the Manufacturer’s instructions. cDNA synthesis and RT-PCR analysis were performed as described by us previously [19,20]. The RT-PCR analysis was performed in a final volume of 20 µl containing 10 µl Power SYBR Green PCR Master Mix kit (Applied Biosystems, USA), 0.4 µl of each primer at 10 pmol/µl, and 2 µl of the cDNA solution using an Applied Biosystems Prism 7900HT (Applied Biosystems, USA) system. Divergent primers were used to detect circANRIL. qRT-PCR primer sequences for CLIP1, EZR, LYVE1, TMEM100, TMEM106B, CXCL11, and ENC1 genes were provided previously [13] and primer sequences for the remaining genes are as follows:
ANRIL 12 F: AACCTGAGCAGCTGGGACTA
ANRIL 13bR: TGTGTCCATAGCACCTTCCA
ANRIL 5 F: TGAAAAACACACATCAAAGGAGA
ANRIL 7 R: TCACGAGGTCGAGAGTTCAA
ANRIL 11 F: CAGAAACCACATCCCTTGGA
ANRIL 13 R: CCTTTTATCACCCAGCTTCG
ANRIL 18 F: CTGCTACATGGAGGCTAGGG
ANRIL 19 R: GGTTCTGCCACAGCTTTGAT
circANRIL 7 F: CTCCCAAAGTGCTGGGATTA
circANRIL 5 R: ATCTGGTGGCCAGAAAACAG
Raw data were analysed using Sequence Detection System (SDS) Software v2.4 (Applied Biosystems, USA) and compared by the ΔΔCt method as previously described by us [19,20]. Results are expressed relative to the housekeeping GAPDH transcript and normalized to untreated cells. Each sample was analysed in triplicate. Each experiment was repeated at least three times.
Statistical analysis
Data are presented as mean ± standard deviation (SD). Statistical analysis was performed using a Student’s t-test (paired and two-tailed) or using analysis of variance (ANOVA) using GraphPad Prism 8.3.1 software. p < 0.05 was used as a cut-off value to determine statistical significance of the data.
Funding Statement
This work was supported by NIH/NHLBI grants R01 HL121358 (Q.K.W), R01 HL126729 (Q.K.W), and R01 HL138465 (Q.K.W. and Q.C.).
Author contributions
Study design: Q.K.W., Q.C., and H.C.; Experiments: H.C.; Data analysis and critical scientific inputs: H.C., Y.L., S.A., F.W., G.Y., S.C., Y.G., Q.C., and Q.K.W.; Manuscript preparation: H.C., Q.C., and Q.K.W.; Comments and revision of the manuscript: all authors; Study supervision: Q.C. and Q.K.W.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Benjamin EJ, Virani SS, Callaway CW, et al. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137:e67–e492. [DOI] [PubMed] [Google Scholar]
- [2].Lusis AJ. Atherosclerosis. Nature. 2000;407(6801):233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shen GQ, Li L, Seidelmann DG, et al. An LRP8 variant is associated with familial and premature coronary artery disease and myocardial infarction. Am J Hum Genet. 2007;81(4):780–791. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang Q, Rao S, Shen GQ, et al. Premature myocardial infarction novel susceptibility locus on chromosome 1P34-36 identified by genomewide linkage analysis. Am J Hum Genet. 2004;74(2):262–271. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Archacki SR, Angheloiu G, Moravec CS, et al. Comparative gene expression analysis between coronary arteries and internal mammary arteries identifies a role for the TES gene in endothelial cell functions relevant to coronary artery disease. Hum Mol Genet. 2012;21(6):1364–1373. [DOI] [PubMed] [Google Scholar]
- [6].Clarke SL, Assimes TL. Genome-wide association studies of coronary artery disease: recent progress and challenges ahead. Curr Atheroscler Rep. 2018;20(9):47. [DOI] [PubMed] [Google Scholar]
- [7].Wolin SL, Maquat LE. Cellular RNA surveillance in health and disease. Science. 2019;366(6467):822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sallam T, Sandhu J, Tontonoz P. Long noncoding RNA discovery in cardiovascular disease: decoding form to function. Circ Res. 2018;122(1):155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol. 2020;21(3):123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kopp F. Molecular functions and biological roles of long non-coding RNAs in human physiology and disease. J Gene Med. 2019;21(8):e3104. [DOI] [PubMed] [Google Scholar]
- [11].Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ribeiro DM, Zanzoni A, Cipriano A, et al. Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. 2018;46(2):917–928. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cho H, Shen GQ, Wang X, et al. Long noncoding RNA ANRIL regulates endothelial cell activities associated with coronary artery disease by up-regulating CLIP1, EZR, and LYVE1 genes. J Biol Chem. 2019;294(11):3881–3898. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sallam T, Jones M, Thomas BJ, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018;24(3):304–312. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gast M, Rauch BH, Nakagawa S, et al. Immune system-mediated atherosclerosis caused by deficiency of long non-coding RNA MALAT1 in ApoE−/− mice. Cardiovasc Res. 2019;115(2):302–314. . [DOI] [PubMed] [Google Scholar]
- [16].Kong Y, Hsieh CH, Alonso LC. ANRIL: a lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front Endocrinol (Lausanne). 2018;9:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Holdt LM, Teupser D. Long noncoding RNA ANRIL: lnc-ing genetic variation at the chromosome 9p21 locus to molecular mechanisms of atherosclerosis. Front Cardiovasc Med. 2018;5:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lo Sardo V, Chubukov P, Ferguson W, et al. Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing. Cell. 2018;175(7):1796–810 e20. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Luo C, Pook E, Tang B, et al. Androgen inhibits key atherosclerotic processes by directly activating ADTRP transcription. Biochim Biophys Acta Mol Basis Dis. 2017;1863(9):2319–2332. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Luo CY, Wang F, Ren X, et al. Identification of a molecular signaling gene-gene regulatory network between GWAS susceptibility genes ADTRP and MIA3/TANGO1 for coronary artery disease. Bba-Mol Basis Dis. 2017;1863(6):1640–1653. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122(3):433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hubberten M, Bochenek G, Chen H, et al. Linear isoforms of the long noncoding RNA CDKN2B-AS1 regulate the c-myc-enhancer binding factor RBMS1. Eur J Hum Genet. 2019;27(1):80–89. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Y, Cho H, Wang F, et al. Statistical and functional studies identify epistasis of cardiovascular risk genomic variants from genome-wide association studies. J Am Heart Assoc. 2020;9(7):e014146. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7(1):12429. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sarkar D, Oghabian A, Bodiyabadu PK, et al. Multiple isoforms of ANRIL in melanoma cells: structural complexity suggests variations in processing. Int J Mol Sci. 2017;18(7):1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5(8):2276–2292. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sarkar D, Leung EY, Baguley BC, et al. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10(2):103–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Meseure D, Vacher S, Alsibai KD, et al. Expression of ANRIL-polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. 2016;14(7):623–633. . [DOI] [PubMed] [Google Scholar]
- [30].Chi JS, Li JZ, Jia JJ, et al. Long non-coding RNA ANRIL in gene regulation and its duality in atherosclerosis. J Huazhong Univ Sci Technolog Med Sci. 2017;37(6):816–822. [DOI] [PubMed] [Google Scholar]
- [31].Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. 2017;31(7). doi:10.1002/jbt.21904. [DOI] [PubMed] [Google Scholar]
- [32].Deng W, Chen K, Liu S, et al. Silencing circular ANRIL protects HK-2 cells from lipopolysaccharide-induced inflammatory injury through up-regulating microRNA-9. Artif Cells Nanomed Biotechnol. 2019;47(1):3478–3484. [DOI] [PubMed] [Google Scholar]
- [33].Shu L, Zhang W, Huang C, et al. lncRNA ANRIL protects H9c2 cells against hypoxia-induced injury through targeting the miR-7-5p/SIRT1 axis. J Cell Physiol. 2019;235(2):1175–1183. [DOI] [PubMed] [Google Scholar]
- [34].Liu B, Cao W, Xue J. LncRNA ANRIL protects against oxygen and glucose deprivation (OGD)-induced injury in PC-12 cells: potential role in ischaemic stroke. Artif Cells Nanomed Biotechnol. 2019;47(1):1384–1395. [DOI] [PubMed] [Google Scholar]
- [35].Guo F, Tang C, Li Y, et al. The interplay of LncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-kappaB signalling pathway. J Cell Mol Med. 2018;22(10):5062–5075. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tan P, Guo YH, Zhan JK, et al. LncRNA-ANRIL inhibits cell senescence of vascular smooth muscle cells by regulating miR-181a/Sirt1. Biochem Cell Biol. 2019;97(5):571–580. [DOI] [PubMed] [Google Scholar]
- [37].He ZY, Wei TH, Zhang PH, et al. Long noncoding RNA-antisense noncoding RNA in the INK4 locus accelerates wound healing in diabetes by promoting lymphangiogenesis via regulating miR-181a/Prox1 axis. J Cell Physiol. 2019;234(4):4627–4640. [DOI] [PubMed] [Google Scholar]
- [38].Archacki SR, Angheloiu G, Tian XL, et al. Identification of new genes differentially expressed in coronary artery disease by expression profiling. Physiol Genomics. 2003;15(1):65–74. . [DOI] [PubMed] [Google Scholar]
- [39].Bai Y, Nie S, Jiang G, et al. Regulation of CARD8 expression by ANRIL and association of CARD8 single nucleotide polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke. 2014;45(2):383–388. . [DOI] [PMC free article] [PubMed] [Google Scholar]


