ABSTRACT
Atopic dermatitis (AD) is a chronic inflammatory disease that affects approximately 2-5% of adults worldwide. The pathogenesis of AD continues to be a well-debated point of conjecture, with numerous hypotheses having been proposed. AD conditions are associated with increased populations of Staphylococcus aureus and reduced skin lipids. In this study, we evaluate the ability of S. aureus to permeate across human stratum corneum (SC) exhibiting both normal and depleted lipid conditions consistent with AD. This permeation would enable bacteria to interact with underlying viable epidermal cells, which could serve as a trigger for inflammation and disease onset. Our results indicate that permeation of S. aureus through SC exhibiting normal lipid conditions is not statistically significant. However, bacteria can readily permeate through lipid depleted tissue over a 9-d period. These findings suggest that S. aureus may potentially act as the mechanistic cause, rather than merely the result of AD.
Abbreviations
AD: Atopic dermatitis; SC: Stratum Corneum; AMP: Antimicrobial peptide; DIW: Deionized water; PDMS: Polydimethylsiloxane; GFP: Green fluorescent protein; BHI: Brain heart infusion medium
KEYWORDS: Stratum Corneum, Staphylococcus aureus, lipids, penetration, cisternae
Introduction
Atopic dermatitis (AD) is a chronic inflammatory skin disease, characterized by erythema, pruritis, and the formation of exudative lesions.1 AD affects approximately 2–5% of adults and 15–20% of children worldwide.2,3 This skin disease is associated with a depletion of stratum corneum (SC) lipids,4 an increase in trans-epidermal water loss,5 and a shift of the skin microbiome to predominantly Staphylococcus aureus.6 Currently, it remains unclear if these observed changes are merely the result of AD, or contribute to the pathogenesis of the disease.7,8 To date, numerous hypotheses for AD pathogenesis have been proposed. The inside-out hypothesis posits that a dysfunctional immune system drives inflammation, which precedes and triggers barrier dysfunction.9,10 Of the many immune-associated genes (adaptive or innate) that produce response proteins related to AD,11 the most reported include IL4, IL13, RANTES, CD14, and NOD1.9,11 To date, however, the role of these immunological changes as the pathogenic cause of AD has not been clinically identified.8 More recently, the outside-in model proposes that an impaired skin barrier enables external allergens or microbials to penetrate into the skin and cause inflammation.12 A major gene mutation that contributes to AD-associated impaired barrier function is FLG, which encodes filaggrin; a structural protein that aggregates keratin intermediate filaments into a tight matrix of bundles within keratinocytes during cornification.13,14 Barrier dysfunction from impaired FLG formation however only accounts for up to 66% of AD cases.11,13,15 Moreover, FLG mutation by itself has been demonstrated not to change SC hydration or trans-epidermal water loss in mice models,16 affect lipid organization and composition in SC of human skin equivalents,17 and alter the penetrative ability of allergens in ex vivo skin.18 As such, other factors are likely to be contributing to the loss of barrier function that would enable the outside-in model to be viable.
We propose that decreases in lipid populations on the SC may cause sufficient barrier dysfunction to enable S. aureus to permeate across the SC, and act as a potential inflammatory trigger of the disease. Previous studies have demonstrated that bacteria reside in healthy human subepidermal components.19 Moreover, it has previously been demonstrated that S. aureus (isolated from corneal ulcers) can penetrate into organotypic skin and the skin of mice, while at the same time, in the latter model, triggering an immune response similar to human AD patients.20 This bacterial penetration was found to be dependent on bacterial viability, antimicrobial peptide (AMP) activity, S. aureus proteolytic activity, and ceramide levels.20 Furthermore, prior studies have also indicated that SC ceramides can influence S. aureus bacterial growth patterns on ex vivo human SC;21 a depletion enabling the bacteria to employ topographical microchannels as conduits for pervasive biofilm growth. Depleted skin lipid conditions can be caused by abnormalities in lipid-processing proteins,22 occupational hazards such as repeated hand washing required for sterile environments,23,24 or contact with metalworking fluids, solvents and caustic chemicals.25 Yet to date, the ability of physiologically relevant S. aureus to penetrate across the SC barrier of human skin has yet to be demonstrated. As such, in this study, we characterize the ability of S. aureus (isolated from human skin lesions) to penetrate SC under control and lipid reduced conditions.
Materials and methods
SC isolation
Full thickness (32 and 41 y) female breast skin samples were obtained from Yale Pathology Tissue Services (New Haven, CT) within 24 hr of elective surgery. In accordance with the Department of Health and Human Services regulations, 45 CFR 46.101:b:4, an exempt approval (3002–13) was attained to perform research using de-identified tissue samples. SC was isolated using standard heat bath and trypsin techniques.26 After isolation, SC sheets were placed on plastic mesh, rinsed in deionized water (DIW), and dried at room temperature and humidity (23 ± 2°C, 29 ± 3% RH).
SC lipid depletion
SC sheets were divided equally, with one half immersed in DIW for 60 min; a treatment that does not deplete lipids or irreversibly alter the intercellular lipid structure.27 This tissue was used for control experiments. The remaining tissue was immersed in a mixture of chloroform and methanol (2:1 by volume, Sigma-Aldrich, St. Louis, MO) for 60 min, followed by DIW for 60 min (lipid depleted). This treatment partially depletes intercellular ceramides, cholesterol, and free fatty acids found in SC.28 From estimates of human SC lipid concentrations and composition,29-31 treatments using similar solvent extraction protocols on human and porcine SC31-34 reduce lipids by 54 ± 30%. SC samples were then cut from both control and lipid depleted SC tissue sheets using a 6 mm diameter circular hole punch (Harris Uni-Core, Redding, CA).
Substrate preparation
Polydimethylsiloxane (PDMS) silicone elastomer (Sylgard 184, Dow Corning, Midland, MI) was prepared by mixing a 10:1 ratio of base to curing agent by weight. After mixing and degassing, the mixture was spin coated (WS-400B-6NNP/LITE, Laurell Technologies Corporation, North Wales, PA) on to a glass coverslip sequentially at 500, 1000, 1500, 3000, 5000, and 6000 rpm, each for 15 s. This produced a uniform silicone elastomer film 14 μm in thickness. Control and lipid depleted SC samples were alternately embedded in the uncured elastomer along the centerline of the coverslip, leaving only their outermost face exposed. This embedding process occludes the sides and underside of the SC sample, preventing bacterial growth in these regions. Substrates were then placed under vacuum in a vacuum desiccator (5310–0250, Nalgene®, ThermoFisher Scientific, Waltham, MA) with an attached vacuum pump (ME4 NT Vacuubrand, BrandTech, Essex, CT) for 4 hr to eliminate microbubbles between the SC and PDMS. The elastomer was then cured at room temperature and humidity for 48 hr. For each substrate tested, the order of the conditioned SC samples deposited was randomized.
Bacterial strains
All bacterial studies used Staphylococcus aureus ATCC 6538 (Rosenbach, American Type Culture Collection (ATCC), Manassas, VA) isolated from human lesions. This strain was modified with pALC2084 and edited to constitutively express green fluorescence protein (GFP). Overnight cultures were grown in brain heart infusion media (BHI, Becton, Dickinson, Sparks, MD) supplemented with 10 mg/L chloramphenicol (Mediatech, Corning Life Sciences, Corning, NY) for plasmid maintenance, and 250 ng/mL tetracycline (Amresco, Solon, OH) for induction of GFP, in Erlenmeyer flasks at 37°C with agitation (220 rpm).
Flow cell setup and inoculation
Embedded SC substrates were sterilized under ultraviolet light for 15 min, then mounted in an anodized aluminum flow cell reactor (FC81, BioSurface Technologies Corp., Bozeman, MN). The flow cell was connected to an inlet carboy containing 20% BHI medium (Franklin Lakes, NJ), as well as an outlet waste carboy using silicone tubing. The medium was supplemented with 250 ng/mL tetracycline hydrochloride (Amresco, Solon, OH) for induction of GFP. The system was maintained at atmospheric pressure using a gas permeable filter (200 nm pore size) fitted to each vessel. The flow cell was first filled with media at 8 mL/hr, stopped, and subsequently injected with a fatty acid dye (D3835, ThermoFisher, Waltham, MA) at a 10 μM concentration in 10% BHI medium. The lipid stain was allowed to bind to all SC samples for 30 min under static conditions before laminar flow was reinstated for 30 min to remove excess dye. For initial experiments ( SC samples per flow cell, SC samples for each lipid condition, flow cells) flow cells were again stopped to inoculate with 5 mL of a stationary phase culture of S. aureus (108 CFU/mL), grown in tetracycline (250 mg/L) and chloramphenicol (10 mg/L) supplemented BHI. Flow cells were inverted to prevent gravitational sedimentation of bacteria onto the SC samples. For delayed experiments ( SC samples per flow cell, SC samples for each lipid condition, flow cells) media was perfused through the flow cell for 48 hr prior to inoculation. In both experiment types, S. aureus was allowed to attach to the substrate for 2 hr under static conditions before laminar flow was reinstated and flow cells were turned right side up.35,36 The fluorescent fatty acid stain was found not to alter the growth behavior of the bacteria.
Microscopic imaging of bacterial colonization of SC
S. aureus development was monitored every 24 hr using confocal fluorescence microscopy (Leica SP5, Wetzlar, Germany) over a 10-d period. Images with a spatial resolution of 0.38 μm/pixel (1024 x 1024 pixels) were acquired using a 40x objective lens (400x magnification) with numerical aperture of 1.25. SC samples were illuminated sequentially with transmitted light, then at 455 and 543 nm. Images for the latter two excitation wavelengths were captured, respectively, across a bandwidth of 500–540 and 560–590 nm. Transmitted light images were used to distinguish topographical regions of the SC in order to image the same position every day (to within ~5 μm spatial accuracy). The SC position imaged was carefully chosen as to not include apocrine pores. Supplemental Figure S1 distinguishes that an apocrine pore is approximately 100 ± 40 μm wide in diameter, in agreement with previous studies.37 The 455 nm illumination was used to excite GFP-tagged S. aureus. The 543 nm illumination was used to excite the fluorescent BODIPY stained SC. At each recorded time point, z-stack images were taken across the full depth of the SC sample and substrate at height increments of 0.15 μm.
Identification of SC thickness using BOPIDY lipid stained images
Composite fluorescent z-stack images for each time point were used to establish vertical cross-sectional profiles of the SC, as shown in supplemental Figure S2(a,b). Employing only the channel revealing the BODIPY stain, the peak intensity, , was established at each lateral position across the cross section. Half peak intensities, were then employed as a minimum threshold to define the upper and lower extents of the SC, as shown in supplemental Figure S2(c,d). If intensity values did not extend above along the lateral position, the upper and lower positions were left undefined. A smooth curve fitting function based on a moving average filter was then applied to the top and bottom regions profiles, as shown in supplemental Figure S2(e). The profile defining the midpoints of the cross section was then established from the upper and lower profiles.
Quantifying bacteria at different SC depths
With the upper and lower interface of the SC identified, the positions of bacteria above and within the SC are established from the channel revealing GFP emission. For each cross-sectional profile and timepoint, a background subtraction was first applied to reduce noise. The average fluorescent intensity of the image was then quantified. Cross-sectional profiles were then separated into four regions based on the upper, lower and midpoint profiles of the SC: above the SC (Above), upper-half of the SC (Middle 1), lower-half of the SC (Middle 2) and below the SC (Below). A centroid tracking algorithm was used to count the number of bacteria in each region, as shown in supplemental Figure S2(f). S. aureus bacterium has a maximum diameter of approximately 1 μm,38 therefore two bacterial point spread functions were considered distinct only if separated by greater than 4 pixels (1.52 μm). Moreover, to prevent counting bacteria more than once, all cross-sectional profiles analyzed were separated by a minimum distance of 1.14 μm.
Statistical analysis
All statistical analyses were performed using R (version 3.4.2). A 1-way ANOVA was used to test for statistical significance in Figure 2(b), where average values in each region (Middle 1, Middle 2, Below) were compared to their respective d 0 values. Levene’s and Shapiro-Wilk’s tests were, respectively, used to determine equality of variances and normality. Results in Figure 2(a) (Middle 1 d 5; Middle 2 d 2; Below d 3), and Figure 2(b) (Middle 1 d 1 and 6; Middle 2 d 1, 3 and 6; Below d 3, 6 and 9) were found to exhibit non-normal distributions and unequal variances. Here a Kruskal-Wallace analysis was performed. The remainder of the results was found to exhibit normal distributions, but un-equal variances. Here a 1-way ANOVA with Welch correction was performed. Post-hoc analyses were performed if statistical significance levels below 5% were established. A one-tail unpaired t-test was used to test for statistical significance in Figure 3, where selective regions (Middle 1, Middle 2, Below) within control and lipid depleted SC samples were compared for each day. The null hypothesis () was lipid depleted > control . F-test and Shapiro-Wilk’s tests were, respectively, used to determine equality of variances and normality. Results in Figure 3(a) (Middle 1 d 6), 3(b) (Middle 2 d 1, 2, 3, and 6), and 3(c) (Below d 6 and 8) were found to exhibit non-normal distributions and unequal variances. Here a Wilcoxon rank-sum test with Welch correction was performed. Results in Figure 3(a) (Middle 1 d 0, 7, 8, and 10), 3(b) (Middle 2 d 4, 5, 7, 8, and 10), and 3(c) (Below d 4 and 10) were found to exhibit normal distributions, but un-equal variances. Here a student t-test with Welch correction was performed. Results in Figure 3(a) (Middle 1 d 1 and 5) and 3(c) (Below d 3, 5, and 7) were found to exhibit non-normal distribution, but equal variances. Here a Wilcoxon rank-sum test was performed. Results in Figure 3(a) (Middle 1 d 2, 3, 4, and 9), 3(b) (Middle 2 d 0 and 9), and 3(c) (Below d 0, 1, 2, and 9) were found to exhibit normal distribution and equal variances. Here a standard student t-test was performed. A 1-way ANOVA was used to test for statistical significance in Figure 5, where average values in each region (Middle 1, Middle 2, Below) were compared to their respective d 3 values. Levene’s and Shapiro-Wilk’s tests were respectively used to determine equality of variances and normality. Results in Figure 5(a) (Middle 1 d 7; Middle 2 d 5 and 7; Below d 4–7), and Figure 5(b) (Middle 2 d 4, 5, 7, 8 and 10; Below d 4, 5, 6, 7, and 11) were found to exhibit non-normal distributions and unequal variances. Here a Kruskal-Wallace analysis was performed. The remainder of the results was found to exhibit normal distributions, but un-equal variances. Here a 1-way ANOVA with Welch correction was performed. Post-hoc analyses were performed if statistical significance levels below 5% were established. In the figures, * denotes , ** denotes , and *** denotes .
Figure 2.
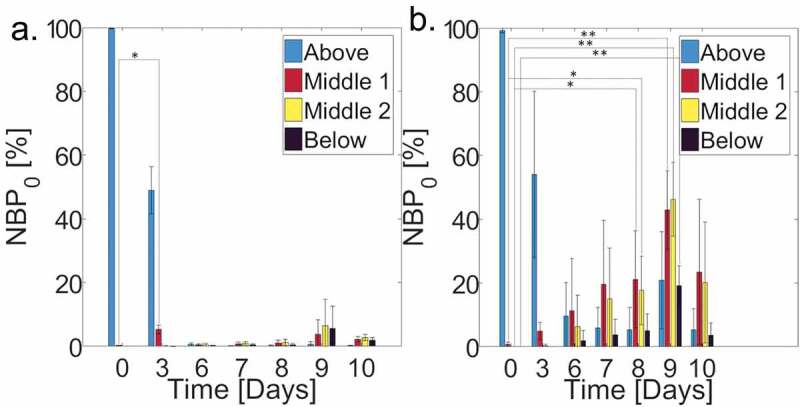
Average normalized bacterial count in each of the 4 SC regions, scaled by the total bacterial count on d 0 () for (a) control ( individual samples; cross-sections per sample) and (b) lipid depleted ( individual samples; cross-sections per sample) SC samples over a 10-d period. (Blue bars, Above) Above the SC, (Red bars, Middle 1) upper half of SC, (Yellow bars, Middle 2) lower half of the SC, and (Black bars, Below) below the SC. Error bars denote standard deviations.
Figure 3.
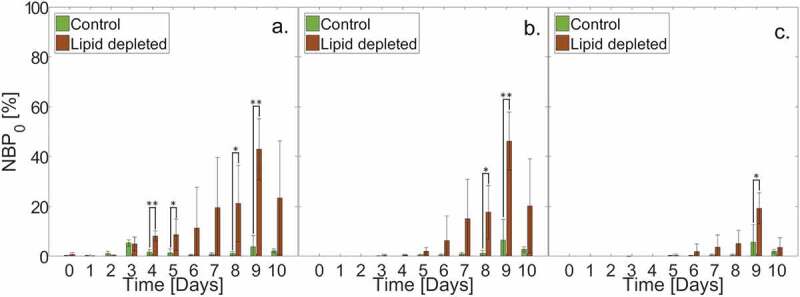
Average normalized bacterial count within or beneath the SC, scaled by the total amount of bacteria on d 0 () for (Green bars) control ( individual SC samples; cross-sections per sample) and (Brown bars) lipid depleted ( individual SC samples; cross-sections per sample) samples over a 10-d period. (a) Upper half of the SC (Middle 1). (b) Lower half of the SC (Middle 2). (c) Below the SC (Below). Error bars denote standard deviations.
Figure 5.
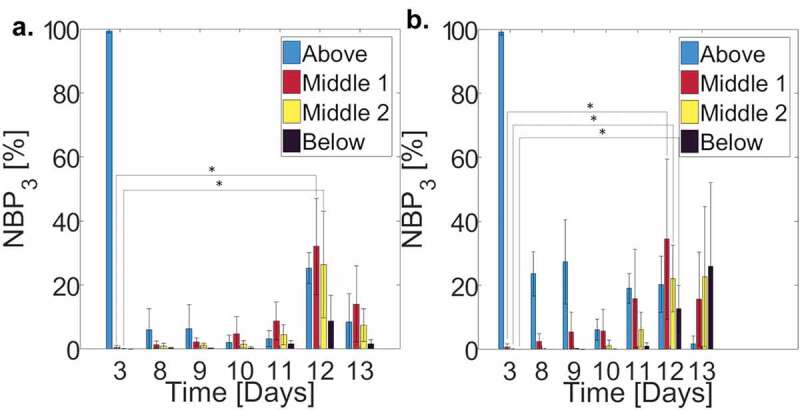
Average normalized bacterial count in each of the 4 SC regions, scaled by the total amount of bacteria on d 3 () for (a) control ( individual samples; cross-sections per sample) and (b) lipid depleted ( individual samples; cross-sections per sample) SC samples over a 10-d period. (Blue bars) Above the SC, (Red bars) upper half of SC, (Yellow bars) lower half of the SC, and (Black bars) below the SC. Error bars denote standard deviations.
Results and discussion
S. aureus permeates into human SC
Composite fluorescent side profile (XZ) confocal images in Figure 1 provide representative slices through the SC of S. aureus (green) as well as fatty acids (red) in and within the SC. Figure 1(a–d) shows bacteria progression over a 9-d period for a lipid depleted SC sample, whose lipid content is consistent with AD conditions.4,31,33,34 Individual bacterium begins to enter the tissue by d 3, as shown in Figure 1(b) (white arrow). By d 6 (Figure 1(c)), bacterium is located within the lipid depleted SC, with the number of bacteria increasing dramatically up until d 9 (Figure 1(d)). Figure 1(e–h) shows the bacterial progression through a control SC sample over a 9-d period. Similar to the lipid depleted SC sample, some bacterium begin to enter the tissue by d 3 (Figure 1(f)) (white arrows). However, bacterium is not seen at all depths within the SC, as observed for the lipid depleted conditions on d 6–9 (Figure 1(g,h)). While bacterial progression in Figure 1 is representative of all lipid depleted conditions, only half of the control SC samples displayed bacterium fully within the tissue. Supplemental Figure S3 provides an example of a control SC sample showing full bacterial entry.
Figure 1.
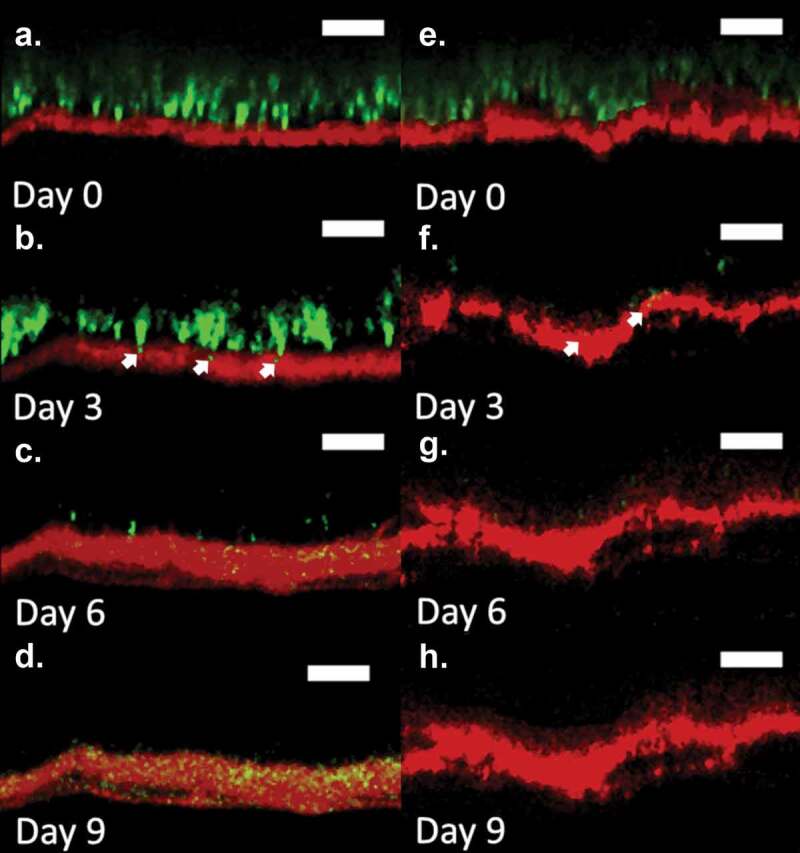
Fluorescent confocal cross-sectional profiles of S. aureus permeation into a lipid depleted (a–d) and control (e–h) SC over a 9-d period. Images show representative fluorescent cross-sectional profiles of BODIPY lipid stained SC (red) inoculated with GFP labeled S. aureus bacteria (green) after (a) 0, (b) 3, (c) 6, and (d) 9 d. Arrows indicate S. aureus initial entry into the SC at d 3. An identical contrast change has been imposed on all images in each lipid condition to enhance visual clarity. Original unaltered images are used to establish quantitative results in Figures 2 and 3. (Scale bar) 15 µm.
We further quantify bacterial permeation by dividing the SC into four distinct regions: above the superficial SC surface (Above), the upper half of the SC (Middle 1), the lower half of the SC (Middle 2) and beneath the SC underside (Below). Figure 2 shows averaged ( samples for each lipid condition; cross-sections per sample) normalized bacterial percentages () in the four regions over a 10-d period for both control and lipid depleted samples. For each SC sample, denotes the total bacterial count quantified in each SC region and timepoint, normalized by the total bacterial count 2 hr after inoculation (d 0). The total bacterial count on d 0 therefore sums to 100%. Across all SC samples, the magnitude of bacteria at d 0 fluctuated by ± 28%, with lipid depleted samples exhibiting increased bacterial populations similar to what has been observed previously.21 This variation therefore necessitated normalization. While a statistically significant rise in is observed for lipid depleted samples in Figure 2(b) on d 8 for Middle 1 and Middle 2 regions of the SC, and d 9 for Middle 1, Middle 2, and Below regions of the SC, relative to d 0, no statistically significant bacterial populations are observed in these regions throughout the 10-d period (Figure 2(a)), except for a significant increase in in the Middle 1 region for control samples on d 3.
Figure 3 directly compares the across lipid conditions within and beneath the SC (Middle 1, Middle 2, Below). Relative to controls, Figure 3(a) shows that lipid depleted samples exhibit significantly higher values of within the Middle 1 region on d 4, 5, 8, and 9. Figure 3(b) shows that lipid depleted samples exhibit significantly higher values of within the Middle 2 region on d 8 and 9, relative to controls. Finally, Figure 3(c) shows that lipid depleted samples exhibit a significantly higher value of beneath the SC on d 9, relative to controls. These combined results indicate that loss of SC lipids not only enables increased bacterial populations to grow within the SC tissue, it also enables bacteria to fully permeate through the tissue.
Cisternae formation does not alter bacterial permeation rate
Due to prolonged liquid media immersion over the course of a 10 d period, delamination of the tissue started to occur, resulting in the formation of cisternae, water filled voids within the tissue.27 This structural change agrees with previous studies that highlight their presence after 4 hr of water exposure,27,39 and monotonic growth with subsequent immersion. Figure 4(a,b) shows that in comparison to d 0 cross-sectional SC profiles, cisternae are visibly present after 3 d immersion in media. The influence of these cisternae on bacterial permeation is subsequently quantified by performing delayed bacterial permeation studies on SC containing cisternae prior to bacterial inoculation; achieved by immersing the SC samples in liquid media for 3 d prior to the introduction of bacteria. Figure 5 shows averaged ( samples for each lipid condition; cross-sections per sample) normalized bacterial percentages () within the four regions of SC over a 10-d period. This percentage denotes the total bacterial count quantified in each SC region and timepoint normalized by the total bacterial count 3 d after liquid media immersion and 2 hr after bacterial inoculation (d 3). Figure 5(a,b) both highlight that while cisternae have already developed in both control and lipid depleted tissue samples, within the SC is greatest after d post-inoculation (d post-submergence). This aligns with the results of Figure 2, where bacterial levels within lipid depleted SC are also greatest d post-inoculation. This indicates that the presence of cisternae does not alter the rate of bacterial permeation through the SC. The results further verify that only complete bacterial permeation through SC samples occurs with the lipid depleted samples. However, statistically significant increases in bacterial populations are present within the control SC tissue (Middle 1, Middle 2) 9 d after inoculation, suggesting the ability of bacteria to penetrate into SC tissue is increased by the prolonged water exposure. We anticipate that this may be due to the disruption of intercellular lipid lamellae that can occur within 24 hr of water immersion.39
Figure 4.
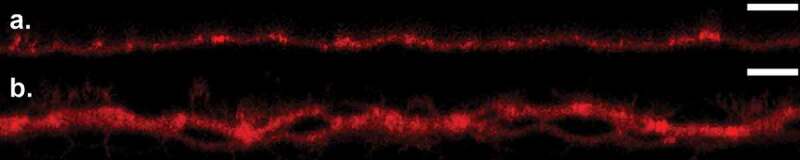
Cross-sectional confocal fluorescent images of BODIPY lipid stained (red) SC samples at (a) 0 and (b) 3 d after media incubation. An identical contrast change has been imposed on all images to enhance visual clarity. (Scale bar) 30 µm.
Mechanism of bacterial permeation
To date, the mechanistic process of non-motile bacterial permeation into and through biological tissue has not been revealed. We take a first step toward understanding this phenomenon by examining the ability of non-motile S. aureus bacteria40 to actively diffuse through the tissue via intercellular, or intercellular and intracellular pathways. For a bacterium located on the SC surface, after binary fission occurs, the centroid of a second adjacent bacterium will be arbitrarily located, relative to the first, by a distance equivalent to the bacterium diameter, , after the doubling timescale . The new bacterium position can therefore be modeled using a 3D random walk,41 , where denotes the mean square displacement after time, , and is the diffusion constant, equal to . For S. aureus, μm38,42 and hr for the media type used,43 resulting in m2 s−1. Currently, it is unclear if in order to fully traverse the SC tissue, bacteria must remain exclusively within intercellular regions, or can progress through both inter- and intracellular regions. If bacteria can pass into, and out of corneocytes, the diffusion length scale required to traverse the SC would be , where is the number of corneocyte layers in the tissue and is the corneocyte thickness. In contrast, if bacteria remain exclusively in intercellular regions, the brick and mortar organization of the tissue44 would require bacteria to diffuse an approximate length scale of , where is the shortest overlapping offset between vertically adjacent corneocytes.45,46 With µm,45-47 μm48 and cells,49,50 the timescale required for bacterial permeation through SC for exclusively intercellular diffusion ranges between d, whereas both inter- and intracellular diffusion timescales range between d. This suggests that permeation of S. aureus into and through SC tissue is unlikely to occur through the growth of bacteria entirely within intercellular regions, requiring the S. aureus bacteria to grow into corneocytes. There is however evidence of cellular internalization of S. aureus, with these bacteria observed both in vivo and in vitro within keratinocytes at various levels of differentiation up until the stratum granulosum,51,52 and within nasal corneocytes.52 While both apocrine and eccrine pores within the SC are larger (approximately 100 ± 40 µm37 and 40 ± 20 µm53 respectively) than S. aureus bacteria, and could act as a transport pathway through the tissue, apocrine pores were easily detectable during imaging and ignored (Supplemental Figure S1). Based on eccrine pore distributions, we further establish that the likelihood of imaging a region containing a pore within our studies was 6.5 ± 4.5%.54 Moreover, the distribution of bacteria within the tissue did not show any observable regions of heightened bacteria localization that would indicate pore transport.
Conclusions
In this article, we show that permeation of Staphylococcus aureus through isolated human stratum corneum exhibiting normal lipid conditions is not statistically significant. However, bacteria can readily permeate through stratum corneum over a 9-d period when depleted to lipids levels consistent with atopic dermatitis. Permeation of these non-motile bacteria can be adequately characterized by bacterial diffusion mediated by random walk binary fission. Permeation timescales equivalent to those observed in experiments suggest bacterium permeate via inter- and intra-cellular pathways. The results of this study align with findings by Nakatsuji et al. (2016) that shows the application of a moisturizer containing ceramides can cause a decrease in bacterial penetration in atopic dermatitis mice models,20 further reinforcing the importance of skin lipids in governing bacterial growth.
While previous articles have demonstrated that bacteria can permeate murine and organotypic skin, neither tissue model adequately characterizes human skin. Murine epidermal tissue contains fewer cell layers compared to the human epidermis, resulting in decreased barrier function and enhanced permeability.55 Moreover, organotypic skin only recreates part of human skin organization and function56 and can be altered both by the composition of culture media used57 and diversity of cell types in the culture.58 Organotypic stratum corneum has been shown to exhibit notable variations in lipid structure and composition compared to in vivo stratum corneum.59,60 Further, we note that while the bacteria strain used in the current study originates from skin lesions, prior studies such as Nakatsuji et al. (2016)20 employ a bacterial strain from corneal ulcers. We anticipate therefore that our study provides the closest physiological model to S. aureus interactions on human skin, and its relation to atopic dermatitis pathogenesis.
In order to further understand S. aureus’s role in the pathogenesis of atopic dermatitis, future work should expand upon these studies by examining the growth and permeation of mixed populations of bacteria on skin, such as S. aureus and S. epidermidis, and their competition on a surface that ordinarily exhibits heterogeneous distributions of lipids.21 Furthermore, while our work focused specifically on the removal of lipids to reduce stratum corneum barrier function, in the context of an outside-in atopic dermatitis model, alteration of other factors, such as tight junction proteins (claudin-1), have additionally been noted to diminish barrier function in the epidermis of atopic dermatitis patients.61,62 While tight junction-like structures have also been observed in the stratum corneum,63-65 the isolated effect these structures might have on bacteria, or vice-versa, has not to our knowledge been investigated ex vivo. However, Ohnemus et al. did show that S. aureus induced a time and concentration dependent structural and delocalization/downregulation to key TJ components (Cldn-1, ZO-1, and occludin) in ex vivo porcine skin, resulting in a reduction in transepithelial resistance.66 Moreover, tight junctions have been demonstrated to create resistance for intercellular travel of small molecules (~550-600 Da) between the stratum granulosum and stratum corneum when using subcutaneous injection.67,68 S. aureus is orders of magnitude larger than the tested molecules, suggesting that movement between these tight junctions could be difficult, which would further align with our argument for intracellular travel. To that end, internalization of S. aureus into ex vivo human stratum corneum should additionally be studied.
Limitations
The general consensus agrees that there is a reduction in SC lipids for AD patients,4,69 with Imokawa et al. citing an approximate 53 ± 43% decrease for lesional SC and 44 ± 41% for non-lesional SC compared to healthy control SC.4 Lipid depletion processes similar to our work show a comparable range of decrease, 54 ± 30%31,33,34 compared to healthy control SC. While the lipid depletion process employed in this work generally reduces lipids to similar levels found in human AD skin, the class and subclass of lipids that are altered, and to what degree, is ambiguous for AD patients from the literature.4,70-76 For example, while some studies show a decrease in SC ceramides for both lesional and non-lesional AD patients compared to healthy control patients,4 others only show a change for lesional patients,70 and some observe no difference altogether.71 Of the studies that observe a decrease in ceramides, a specific decrease in EO ceramides4,72,75 is detected, which our delipidation method also shows a bias toward.77 In addition to limitations in creating a comparable lipid profile to AD patients, the method of standard biofilm growth used36,78 does not simulate physiologically relevant conditions, as the skin is not continually immersed in liquid media throughout the day. In vivo, bacteria would naturally survive off of nutrients available such as keratin and lipids.79
Supplementary Material
Funding Statement
This material is based upon work supported by the National Science Foundation under Grant No. [1653071].
Disclosure of potential conflicts of interest
The authors state no conflict of interest.
Author contributions
G.K.G and C.N.H.M. conceived the idea. Z.W.L. carried out the experiments. Everyone contributed to writing the manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Leung DY, Bieber T.. Atopic dermatitis. Lancet [Internet]. 2003;361(9352):1–12; [accessed 2019 February14]. https://www.sciencedirect.com/science/article/pii/S0140673603121939?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 2.Barbarot S, Auziere S, Gadkari A, Girolomoni G, Puig L, Simpson EL, Margolis DJ, de Bruin-weller M, Eckert L.. Epidemiology of atopic dermatitis in adults: results from an international survey. Allergy Eur J Allergy Clin Immunol. 2018;73(6):1284–1293. doi: 10.1111/all.13401. [DOI] [PubMed] [Google Scholar]
- 3.Asher MI, Montefort S, Björkstén B, Lai CK, Strachan DP, Weiland SK, Williams H. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional surveys. Lancet. 2006;368(9537):733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 4.Imokawa G, Abe A, Jin K, Higaki Y, Kawashima M, Hidano A. Decreased level of ceramides in stratum corneum of atopic dermatitis: an etiologic factor in atopic dry skin? J Invest Dermatol. 1991;96(4):523–526. doi: 10.1111/1523-1747.ep12470233. [DOI] [PubMed] [Google Scholar]
- 5.Werner Y, Lindberg M. Transepidermal water loss in dry and clinically normal skin in patients with atopic dermatitis. Acta Derm Venereol. 1985;65(2):102–105. [PubMed] [Google Scholar]
- 6.Chiller K, Selkin BA, Murakawa GJ, Microflora S. Bacterial infections of the skin. J Invest Dermatol Symp Proc. 2001;6:170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 7.Silverberg NB, Silverberg JI. Inside out or outside in: does atopic dermatitis disrupt barrier function or does disruption of barrier function trigger atopic dermatitis? Cutis. 2015;96(6):359–61. [PubMed] [Google Scholar]
- 8.Sullivan M, Silverberg NB. Current and emerging concepts in atopic dermatitis pathogenesis. Clin Dermatol. 2017;35(4):349–353. doi: 10.1016/j.clindermatol.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Hanifin JM. Evolving concepts of pathogenesis in atopic dermatitis and other eczemas. J Invest Dermatol. 2009;129(2):320–322. doi: 10.1038/jid.2008.252. [DOI] [PubMed] [Google Scholar]
- 10.Bieber T. Atopic dermatitis. Ann Dermatol [Internet]. 2010;22(2):125–137. https://www.ncbi.nlm.nih.gov/pubmed/20548901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010;125(1):16–29.e11. doi: 10.1016/j.jaci.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillespie RM. From the outside-in: epidermal targeting as a paradigm for atopic disease therapy. World J Dermatol. 2015;4(1):16. doi: 10.5314/wjd.v4.i1.16. [DOI] [Google Scholar]
- 13.Palmer CNA, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJD, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38(4):441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 14.Irvine AD, McLean WHI, Leung DYM. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med. 2011;365(14):1315–1327. doi: 10.1056/NEJMra1011040. [DOI] [PubMed] [Google Scholar]
- 15.Morar N, Cookson WOCM, Harper JI, Moffatt MF. Filaggrin mutations in children with severe atopic dermatitis. J Invest Dermatol. 2007;127(7):1667–1672. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H, Nagao K, Kubo A, Hata T, Shimizu A, Mizuno H, Yamada T, Amagai M. Altered stratum corneum barrier and enhanced percutaneous immune responses in filaggrin-null mice. J Allergy Clin Immunol. 2012;129(6):1538–1546.e6. doi: 10.1016/j.jaci.2012.01.068. [DOI] [PubMed] [Google Scholar]
- 17.van Drongelen V, Alloul-Ramdhani M, Danso MO, Mieremet A, Mulder A, van Smeden J, Bouwstra JA, El Ghalbzouri A. Knock-down of filaggrin does not affect lipid organization and composition in stratum corneum of reconstructed human skin equivalents. Exp Dermatol. 2013;22(12):807–812. doi: 10.1111/exd.12271. [DOI] [PubMed] [Google Scholar]
- 18.Newell L, Polak ME, Perera J, Owen C, Boyd P, Pickard C, Howarth PH, Healy E, Holloway JW, Friedmann PS, et al. Sensitization via healthy skin programs Th2 responses in individuals with atopic dermatitis. J Invest Dermatol. 2013;133(10):2372–2380. doi: 10.1038/jid.2013.148. [DOI] [PubMed] [Google Scholar]
- 19.Nakatsuji T, Chiang H-I, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun [Internet]. 2013;4(1):1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, Hata TR, Gallo RL. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol. 2016;136(11):2192–2200. doi: 10.1016/j.jid.2016.05.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cleary JM, Lipsky ZW, Kim M, Marques CNH, German GK. Heterogeneous ceramide distributions alter spatially resolved growth of Staphylococcus aureus on human stratum corneum. J R Soc Interface. 2018;15(141):20170848. doi: 10.1098/rsif.2017.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin J, Friedlander SF, Del Rosso JQ. Atopic dermatitis and the stratum corneum part 2: other structural and functional characteristics of the stratum corneum barrier in atopic skin. J Clin Aesthet Dermatol. 2013;6(11):49. [PMC free article] [PubMed] [Google Scholar]
- 23.Ananthapadmanabhan KP, Moore DJ, Subramanyan K, Misra M, Meyer F. Cleansing without compromise: the impact of cleansers on the skin barrier and the technology of mild cleansing. Dermatol Ther. 2004;17(Suppl 1):16–25. doi: 10.1111/j.1396-0296.2004.04S1002.x. [DOI] [PubMed] [Google Scholar]
- 24.Kirk JE. Hand washing. Quantitative studies on skin lipid removal by soaps and detergents based on 1500 experiments. Acta Derm Venereol. 1966;46(57):25–71. [PubMed] [Google Scholar]
- 25.Monteiro-Riviere NA. Toxicology of the skin. United Kingdom: CRC Press; 2010. [Google Scholar]
- 26.Kligman AM, Christophers E. Preparation of isolated sheets of human stratum corneum. Arch Dermatol [Internet]. 1963;88:702–705. doi: 10.1001/archderm.1963.01590240026005. [DOI] [PubMed] [Google Scholar]
- 27.Warner RR, Boissy YL, Lilly NA, Spears MJ, Mckillop K, Marshall JL, Stone KJ. Water disrupts stratum corneum lipid lamellae: damage is similar to surfactants1. J Invest Dermatol [Internet]. 1999;113:960–966. [accessed 2017 February14]. http://cat.inist.fr/?aModele=afficheN&cpsidt=1255154 [DOI] [PubMed] [Google Scholar]
- 28.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 29.Schaefer H, Rodelmeier TE. Skin Barrier: Principles of percutaneous absorption. Basel (Switz): Karger; 1996. [Google Scholar]
- 30.Anderson RL, Cassidy JM. Variations in physical dimensions and chemical composition of human stratum corneum. J Invest Dermatol [Internet]. 1973;61(1):30–32. http://linkinghub.elsevier.com/retrieve/pii/S0022202X15441089 [DOI] [PubMed] [Google Scholar]
- 31.Abrams K, Harvell JD, Shriner D, Wertz P, Maibach H, Maibach HI, Rehfeld SJ. Effect of organic solvents on in vitro human skin water barrier function. J Invest Dermatol. 1993;101(4):609–613. doi: 10.1111/1523-1747.ep12366068. [DOI] [PubMed] [Google Scholar]
- 32.Caussin J, Gooris GS, Janssens M, Bouwstra JA. Lipid organization in human and porcine stratum corneum differs widely, while lipid mixtures with porcine ceramides model human stratum corneum lipid organization very closely. Biochim Biophys Acta (BBA)-Biomembr. 2008;1778:1472–1482. doi: 10.1016/j.bbamem.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Lampe MA, Burlingame AL, Whitney J, Williams ML, Brown BE, Roitman E, Elias PM. Human stratum corneum lipids: characterization and regional variations. J Lipid Res [Internet]. 1983;24:120–130. http://www.ncbi.nlm.nih.gov/pubmed/6833889. [PubMed] [Google Scholar]
- 34.Wertz PW, Kremer M, Squier CA. Comparison of lipids from epidermal and palatal stratum corneum. J Invest Dermatol. 1992;98(3):375–378. doi: 10.1111/1523-1747.ep12499809. [DOI] [PubMed] [Google Scholar]
- 35.Marques CNH, Craver SA. Quantification of respiratory activity in biofilms. Bio-protocol. 2015;5(18):e1591. doi: 10.21769/BioProtoc.1591. [DOI] [Google Scholar]
- 36.Davies DG, Marques CNH. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191(5):1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otberg N, Richter H, Schaefer H, Blume-Peytavi U, Sterry W, Lademann J. Variations of hair follicle size and distribution in different body sites. J Invest Dermatol. 2004;122(1):14–19. doi: 10.1046/j.0022-202X.2003.22110.x. [DOI] [PubMed] [Google Scholar]
- 38.Richardson H, Smaill F. Medical microbiology. BMJ. 1998;317(7165):1060–1062. doi: 10.1136/bmj.317.7165.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner RR, Stone KJ, Boissy YL. Hydration disrupts human stratum corneum ultrastructure. J Invest Dermatol. 2003;120(2):275–284. doi: 10.1046/j.1523-1747.2003.12046.x. [DOI] [PubMed] [Google Scholar]
- 40.Kloos WE, Schleifer KH. Simplified scheme for routine identification of human Staphylococcus species. J Clin Microbiol. 1975;1(1):82–88. doi: 10.1128/JCM.1.1.82-88.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson P. Biological physics-energy, information, life. New York: WH Freeman. 2004;. 98–137. [Google Scholar]
- 42.Tortora GJ, Funke BR, Case CL, Johnson TR. Microbiology: an introduction. San Francisco (CA): Benjamin Cummings; 2004. [Google Scholar]
- 43.Lindqvist R. Estimation of Staphylococcus aureus growth parameters from turbidity data: characterization of strain variation and comparison of methods. Appl Environ Microbiol. 2006;72(7):4862–4870. doi: 10.1128/AEM.00251-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elias PM, Grayson S, Lampe MA, Williams ML, Brown BE. The Intercorneocyte Space. In: Marks R, Plewig G, editors. Stratum Corneum. Berlin (Heidelberg): Springer Berlin Heidelberg; 1983. p. 53–67. [Google Scholar]
- 45.Frasch HF, Barbero AM. Steady-state flux and lag time in the stratum corneum lipid pathway: results from finite element models. J Pharm Sci. 2003;92(11):2196–2207. [DOI] [PubMed] [Google Scholar]
- 46.Talreja PS, Kasting GB, Kleene NK, Pickens WL, Wang T-F. Visualization of the lipid barrier and measurement of lipid pathlength in human stratum corneum. AAPS J. 2001;3(2):48–56. doi: 10.1208/ps030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen J, Koo SM, Lape N. How sensitive are transdermal transport predictions by microscopic stratum corneum models to geometric and transport parameter input? J Pharm Sci. 2018;107(2):612–623. doi: 10.1016/j.xphs.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Michael Licht MS, Heise HM. Surface ultra-structure and size of human corneocytes from upper stratum corneum layers of normal and diabetic subjects with discussion of cohesion aspects. J Diabetes Metab. 2015;06(9). doi: 10.4172/2155-6156.1000603. [DOI] [Google Scholar]
- 49.Cork MJ, Robinson DA, Vasilopoulos Y, Ferguson A, Moustafa M, MacGowan A, Duff GW, Ward SJ, Tazi-Ahnini R. New perspectives on epidermal barrier dysfunction in atopic dermatitis: gene-environment interactions. J Allergy Clin Immunol. 2006;118(1):3–21. doi: 10.1016/j.jaci.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 50.Silva CL, Topgaard D, Kocherbitov V, Sousa JJS, Pais AACC, Sparr E. Stratum corneum hydration: phase transformations and mobility in stratum corneum, extracted lipids and isolated corneocytes. Biochim Biophys Acta Biomembr. 2007;1768(11):2647–2659. doi: 10.1016/j.bbamem.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 51.Soong G, Paulino F, Wachtel S, Parker D, Wickersham M, Zhang D, Brown A, Lauren C, Dowd M, West E, et al. Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes. MBio. 2015;6(2). doi: 10.1128/mBio.00289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rigaill J, Morgene MF, Gavid M, Lelonge Y, He Z, Carricajo A, Grattard F, Pozzetto B, Berthelot P, Botelho-Nevers E, et al. Intracellular activity of antimicrobial compounds used for Staphylococcus aureus nasal decolonization. J Antimicrob Chemother. 2018;73(11):3044–3048. doi: 10.1093/jac/dky318. [DOI] [PubMed] [Google Scholar]
- 53.Wilke K, Martin A, Terstegen L, Biel SS. A short history of sweat gland biology. International Journal of Cosmetic Science. 2007;29(3):169–179. doi: 10.1111/j.1467-2494.2007.00387.x. [DOI] [PubMed] [Google Scholar]
- 54.Hwang K, Baik SH. Distribution of hairs and sweat glands on the bodies of korean adults: A morphometric study. Cells Tissues Organs. 1997;158(2):112–120. doi: 10.1159/000147920. [DOI] [PubMed] [Google Scholar]
- 55.Zomer HD, Trentin AG. Skin wound healing in humans and mice: challenges in translational research. J Dermatol Sci. 2018;90(1):3–12. doi: 10.1016/j.jdermsci.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Oh JW, Hsi TC, Fernando C, Ramos R, Plikus MV. Organotypic skin culture. J Invest Dermatol. 2013;133(11):1–4. doi: 10.1038/jid.2013.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stark H-J, Baur M, Breitkreutz D, Mirancea N, Fusenig NE. Organotypic keratinocyte cocultures in defined medium with regular epidermal morphogenesis and differentiation. J Invest Dermatol. 1999;112(5):681–691. doi: 10.1046/j.1523-1747.1999.00573.x. [DOI] [PubMed] [Google Scholar]
- 58.Sriram G, Bigliardi PL, Bigliardi-Qi M. Fibroblast heterogeneity and its implications for engineering organotypic skin models in vitro. Eur J Cell Biol. 2015;94(11):483–512. doi: 10.1016/j.ejcb.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Parenteau NL, Bilbo P, Nolte CJM, Mason VS, Rosenberg M. The organotypic culture of human skin keratinocytes and fibroblasts to achieve form and function. Cytotechnology. 1992;9(1–3):163–171. doi: 10.1007/BF02521744. [DOI] [PubMed] [Google Scholar]
- 60.Bouwstra JA, Gooris GS, Weerheim A, Kempenaar J, Ponec M. Characterization of stratum corneum structure in reconstructed epidermis by X-ray diffraction. J Lipid Res. 1995;36(3):496–504. [PubMed] [Google Scholar]
- 61.De Benedetto A, Rafaels NM, McGirt LY, Ivanov AI, Georas SN, Cheadle C, Berger AE, Zhang K, Vidyasagar S, Yoshida T, et al. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol. 2011;127(3):773–786.e7. doi: 10.1016/j.jaci.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bergmann S, von Buenau B, Vidal-y-Sy S, Haftek M, Wladykowski E, Houdek P, Lezius S, Duplan H, Bäsler K, Dähnhardt-Pfeiffer S, et al. Claudin-1 decrease impacts epidermal barrier function in atopic dermatitis lesions dose-dependently. Sci Rep. 2020;10(1). doi: 10.1038/s41598-020-58718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishida-Yamamoto A, Igawa S, Kishibe M. Molecular basis of the skin barrier structures revealed by electron microscopy. Exp Dermatol. 2018;27:841–846. doi: 10.1111/exd.13674. [DOI] [PubMed] [Google Scholar]
- 64.Kitajima Y. Desmosomes and corneodesmosomes are enclosed by tight junctions at the periphery of granular cells and corneocytes, suggesting a role in generation of a peripheral distribution of corneodesmosomes in corneocytes. J Dermatol Sci. 2016;83:73–75. doi: 10.1016/j.jdermsci.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 65.Ishida-Yamamoto A, Igawa S, Kishibe M, Honma M. Clinical and molecular implications of structural changes to desmosomes and corneodesmosomes. J Dermatol. 2018;45:385–389. doi: 10.1111/1346-8138.14202. [DOI] [PubMed] [Google Scholar]
- 66.Ohnemus U, Kohrmeyer K, Houdek P, Rohde H, Wladykowski E, Vidal S, Horstkotte MA, Aepfelbacher M, Kirschner N, Behne MJ, et al. Regulation of epidermal tight-junctions (TJ) during infection with exfoliative toxin-negative Staphylococcus strains. J Invest Dermatol. 2008;128(4):906–916. doi: 10.1038/sj.jid.5701070. [DOI] [PubMed] [Google Scholar]
- 67.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J Cell Biol. 2002;156(6):1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuki T, Hachiya A, Kusaka A, Sriwiriyanont P, Visscher MO, Morita K, Muto M, Miyachi Y, Sugiyama Y, Inoue S. Characterization of tight junctions and their disruption by UVB in human epidermis and cultured keratinocytes. J Invest Dermatol. 2011;131(3):744–752. doi: 10.1038/jid.2010.385. [DOI] [PubMed] [Google Scholar]
- 69.Janssens M, Van Smeden J, Puppels GJ, Lavrijsen APM, Caspers PJ, Bouwstra JA. Lipid to protein ratio plays an important role in the skin barrier function in patients with atopic eczema. Br J Dermatol. 2014;170(6):1248–1255. doi: 10.1111/bjd.12908. [DOI] [PubMed] [Google Scholar]
- 70.Farwanah H, Raith K, Neubert RHH, Wohlrab J. Ceramide profiles of the uninvolved skin in atopic dermatitis and psoriasis are comparable to those of healthy skin. Arch Dermatol Res. 2005;296(11):514–521. doi: 10.1007/s00403-005-0551-2. [DOI] [PubMed] [Google Scholar]
- 71.Angelova-Fischer I, Mannheimer A-C, Hinder A, Ruether A, Franke A, Neubert RHH, Fischer TW, Zillikens D. Distinct barrier integrity phenotypes in filaggrin-related atopic eczema following sequential tape stripping and lipid profiling. Exp Dermatol. 2011;20(4):351–356. doi: 10.1111/j.1600-0625.2011.01259.x. [DOI] [PubMed] [Google Scholar]
- 72.Di Nardo A, Wertz P, Giannetti A, Seidenari S. Ceramide and cholesterol composition of the skin of patients with atopic dermatitis. Acta Derm Venereol. 1998;78:27–30. doi: 10.1080/00015559850135788. [DOI] [PubMed] [Google Scholar]
- 73.Ishikawa J, Narita H, Kondo N, Hotta M, Takagi Y, Masukawa Y, Kitahara T, Takema Y, Koyano S, Yamazaki S, et al. Changes in the ceramide profile of atopic dermatitis patients. J Invest Dermatol. 2010;130:2511–2514. doi: 10.1038/jid.2010.161. [DOI] [PubMed] [Google Scholar]
- 74.Melnik B, Hollmann J, Plewig G. Decreased stratum corneum ceramides in atopic individuals–a pathobiochemical factor in xerosis? Br J Dermatol. 1988;119:547–549. doi: 10.1111/j.1365-2133.1988.tb03262.x. [DOI] [PubMed] [Google Scholar]
- 75.Yamamoto A, Serizawa S, Ito M, Sato Y. Stratum corneum lipid abnormalities in atopic dermatitis. Arch Dermatol Res. 1991;283(4):219–223. doi: 10.1007/BF01106105. [DOI] [PubMed] [Google Scholar]
- 76.Janssens M, van Smeden J, Gooris GS, Bras W, Portale G, Caspers PJ, Vreeken RJ, Hankemeier T, Kezic S, Wolterbeek R, et al. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients. J Lipid Res. 2012;53:2755–2766. doi: 10.1194/jlr.P030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boiten W, Absalah S, Vreeken R, Bouwstra J, van Smeden J. Quantitative analysis of ceramides using a novel lipidomics approach with three dimensional response modelling. Biochim Biophys Acta - Mol Cell Biol Lipids. 2016;1861(11):1652–1661. doi: 10.1016/j.bbalip.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Sauer K, Camper A, Ehrlich G, Costerton J, Davies D. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184(4):1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fredricks DN. Microbial ecology of human skin in health and disease. J Invest Dermatol Symp Proc. 2001. Elsevier;6(3):167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


