ABSTRACT
The production of mRNA is a dynamic process that is highly regulated by reversible post-translational modifications of the C-terminal domain (CTD) of RNA polymerase II. The CTD is a highly repetitive domain consisting mostly of the consensus heptad sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7. Phosphorylation of serine residues within this repeat sequence is well studied, but modifications of all residues have been described. Here, we focus on integrating newly identified and lesser-studied CTD post-translational modifications into the existing framework. We also review the growing body of work demonstrating crosstalk between different CTD modifications and the functional consequences of such crosstalk on the dynamics of transcriptional regulation.
KEYWORDS: Transcription regulation, Gene regulation, RNA polymerase II, Crosstalk, Post-translational modification
Introduction
Precisely tuned gene expression is crucial for maintenance of organismal homeostasis and for response to external stimuli. RNA Polymerase II (RNAPII) is a large multi-subunit enzymatic complex that is essential for constitutive expression of mRNA and small nuclear RNAs as well as their rapid production in response to altering cell signaling events. While RNAPII is highly conserved across eukaryotes, the C-terminal domain (CTD) of its largest catalytic subunit, Rpb1, has evolved to harbor unique features across species [1,2]. The CTD is highly repetitive, unstructured, and of low diversity [3]. The Rpb1 CTD consensus heptad sequence (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7) is highly conserved as exemplified by the similarity of the Sacccharomyces cervisiae CTD to the first 26 proximal repeats of the human CTD. However, the CTD has varying divergence across eukaryotes with higher eukaryotes evolving a substantially longer CTD following the common ancestor of Metazoa [4]. The distal CTD in higher metazoans has maintained the heptad repeat structure, yet diverged from the consensus heptad sequence [1,5–8] (Figure 1). Recent work studying the ability of RNAPII to induce liquid phase separation suggest that organisms with increased CTD length harbor more non-consensus repeats to prevent protein aggregation of the CTD while still allowing phase separation creating transcriptional hubs [9–12].
Figure 1.
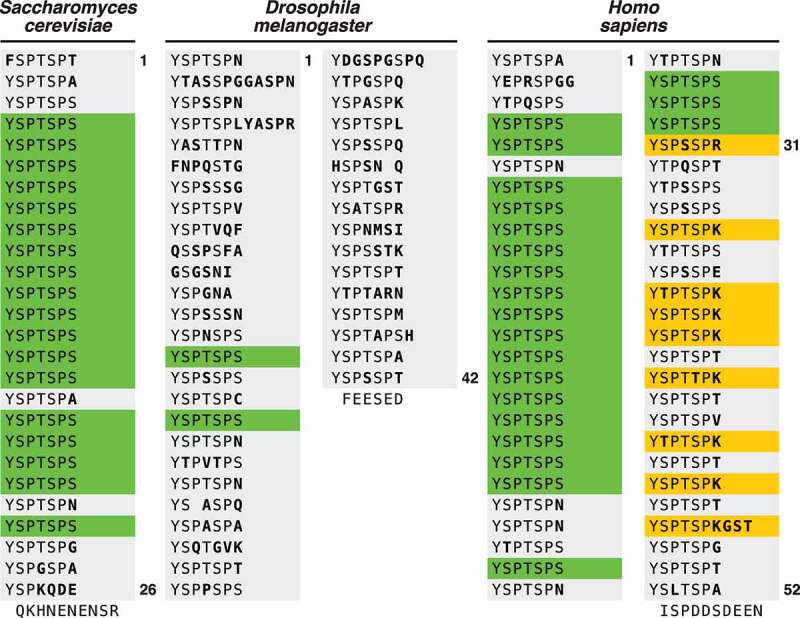
Schematic of the CTD. The RNAPII CTD of Saccharomyces cerevisiae, Drosophila melanogaster, and Homo sapiens are presented. Each heptad repeat is represented as a new line. Conserved repeats are in green, non-conserved amino acids are in bold, non-consensus repeats that can be uniquely modified are in yellow.
Although the CTD is a relatively small appendage of a massive molecular machine, it has a profound significance on the regulation of gene expression. The unstructured nature of the CTD hypothetically allows for increased protein-protein interactions with major transcriptional effector molecules, including the enzymes mediating or removing posttranslational modifications (PTMs; writers and erasers), and the proteins containing specific recognition domains for PTMs (readers). Many CTD-modifying or -recognizing proteins essential for regulating the process of transcription are highlighted in this review.
The most extensively studied modification of the CTD is the dynamic and reversible phosphorylation of serine residues, specifically of Ser2 and Ser5; the roles of phospho-Ser2 and -Ser5 in the regulation of transcription have been reviewed previously [5,7,13–16]. However, on-going research in the field has shown that every amino acid in the consensus heptad repeat, as well as arginine and lysine residues of non-consensus repeats, can be modified during transcription. Recognized CTD PTMs include phosphorylation of serine, tyrosine, and threonine, O-GlcNAcylation (O-GlcNAc), proline isomerization, arginine and lysine methylation, arginine citrullination, and lysine acetylation (Table 1). The repertoire of PTMs that have been attributed to the CTD is often referred to as a “CTD code”, in which certain modifications are attributed to particular functions during specific steps of the cycle (Figure 2).
Table 1.
CTD modifications, their functions and interacting partners.
| Modification | Function | Reader | Writer | Eraser | Reference |
|---|---|---|---|---|---|
| Tyrosine 1 phosphorylation | Prevention of pre-mature termination, CTD stability, enhancer transcription | c-Abl | [79,80,108,163,166] | ||
| Threonine 4 phosphorylation |
Transcription elongation, termination, processing of histone mRNA, chromatin remodeling | Plk1/3, CDK9 | [167–170] | ||
| Proline Isomerization | cis-trans isomerization of CTD prolines, regulates activity of serine 5 phosphatases | Ssu72, FCP1 | Pin1 | [149–151,153,154,171] | |
| Arginine citrullination | Promoter-proximal pause release | P-TEFb | PADI2 | [109] | |
| Arginine methylation | Transcription termination, terminal R-loop resolution, snRNA and snoRNA expression | SMN, TDRD3 | PRMT5, PTRM4/CARM1 | [27,28,172] | |
| Lys Ubiquitination | Degradation of RPB1 | rpfl/hNedd4 | pVHL | [173,174] | |
| O-GlcNAc | Assembly of PIC | OGT | OGA | [19–23,61] | |
| Serine 2 phosphorylation |
Promoter-proximal pause release, elongation, termination, splicing | PAF1, SPT6, TCERG1, U2AF65-Prp19, SET2, HDAC/HAT, SCAF8, SCAF4 | CDK9(P-TEFb), CDK12, CDK13 | Ssu72, FCP1 |
[30,37,42,103,122,152,157,175–190] |
| Serine 5 phosphorylation |
mRNA capping, promoter-proximal pausing, chromatin remodeling, ncRNA transcription termination, splicing, prevention of premature transcription termination | DYRK1a, SCP1, SCP4, CDC14, MLL1/2, Guanyltransferase, Pin1 | CDK7, CDK8, CDK9, CDK12, CDK13 | Ssu72, RPAP2 | [74,76,151,153,155,191–198] |
| Serine 7 phosphorylation |
snRNA expression, Interaction with Integrator, promoter-proximal pausing | CDK7, CDK9 | Ssu72 | [152,199–202] |
Figure 2.
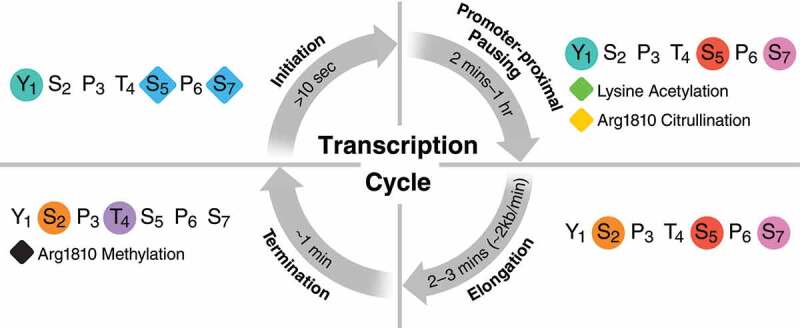
The cycle of CTD modifications. Key CTD post translational modifications of each major phase of transcription are shown, with the average duration of each phase noted. The rate of transcriptional initiation is not listed as a time range as rates of initiation vary dramatically depending on if the mediator complex is already resident at the gene promoter. Timing for phases is from eukaryotes in general [31,32,38–44,46-50]. Circles denote phosphorylation and diamonds indicate O-GlcNAc unless otherwise noted.
Although many modifications are highly conserved, their time of occurrence and associated functions can vary across species. For example, the role of Tyr1 and Ser7 phosphorylation vary considerably between mammals and yeast. In mammals, both modifications control early steps of transcription, but in yeast, Ser7 phosphorylation acts both early in transcription and as a termination signal and Tyr1 phosphorylation has primarily been linked to control of transcription termination [5,17,18]. Other modifications have only been described in mammals, including O-GlcNAcylation [19–23], lysine acetylation [4,24–26] and arginine methylation [27,28], and the extended PTM repertoire suggests a need for enhanced regulation in more complex organisms.
Increasing evidence indicates that individual PTMs can communicate and act to modulate each other’s presence and function. This crosstalk expands the regulatory capacity and complexity of the CTD and allows RNAPII to rapidly and transiently react to external stimuli. Both positive and negative crosstalk mechanisms have been described, either promoting or prohibiting the occurrence of modifications, or enhancing or impairing reading of the modifications’ “code”. In this review, we focus on PTMs of the mammalian CTD during mRNA synthesis, including those that are less studied, and discuss the emerging significance of PTM crosstalk within the CTD.
Timing of individual CTD modifications
Transcription of genes coding for mRNA takes place in four key phases: initiation, promoter-proximal pausing, elongation, and termination [29].The dynamic, reversible PTMs of the CTD mark the various stages of transcription (Figure 3), from an uninitiated polymerase to transcription termination [30]. This is key to allowing transcription to adapt to specific needs, for example transcription must progress at fast rates in response to external stimuli as well as during the rapid cell divisions of early development [31,32]. Although CTD modifications play a key role in the regulation of transcription rates, it is important to note that while not discussed in this review other features of genes, such as nucleosome position, DNA sequence, DNA structure, and co-transcriptional processes, are also involved [33–37]. Studies on bulk transcription rates in human cells indicate that the average gene can be transcribed on the scale of several minutes [38–44]. However, this varies dramatically with gene size; indeed, the largest gene in the human genome takes 16 hours to transcribe [45]. Development of new single molecule techniques for studying individual transcripts may allow further elucidation of the rate of individual phases of transcription and the role of PTMs in regulating these phases [46–51].
Figure 3.
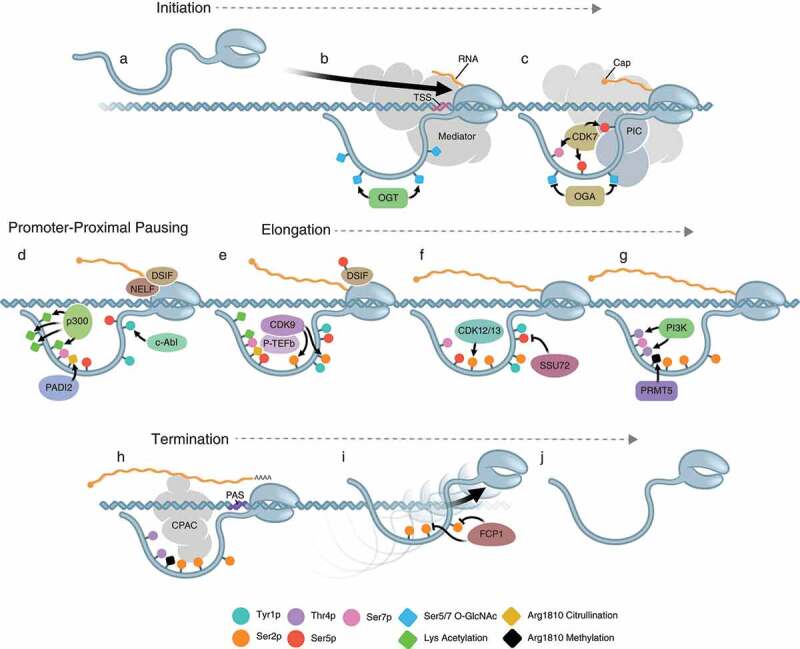
Key steps of transcription. A and B) An unmodified RNAPII is recruited to the transcription start site (TSS) by the Mediator complex. O-GlcNAcylation of Ser5 and 7 is performed by OGT. C) The pre-initiation complex (PIC) is formed and its CDK7 subunit phosphorylates Ser5 and 7 after O-GlcNAc is removed by OGA. D) Ser5 and 7 phosphorylation releases the polymerase from interactions with Mediator and the PIC. Following release, the polymerase pauses proximally to the promoter, marked by Tyr1 phosphorylation by c-Abl, acetylation of Lys7 by p300, and citrullination of non-consensus Arg1810 by PADI2. Pausing is facilitated by the recruitment of NELF and DSIF. E) The polymerase is released from pausing by phosphorylation of Ser2 by the CDK9 subunit of PTEF-b, as well as the phosphorylation of DSIF and removal of NELF. F) Once productive elongation begins, CDK12 and CDK13 maintain the phosphorylation of Ser2, and phosphorylation of Ser5 is removed by the Ssu72 phosphatase. G) As the polymerase reaches the 3ʹ end of the gene, Thr4 phosphorylation by PI3 K occurs, as well as methylation of Arg1810 by PRMT5. H) When the polymerase is in the proximity of a poly-adenylation site (PAS), it interacts with the cleavage and polyadenylation complex (CPAC) via Ser2 phosphorylation, allowing for cleavage of the mRNA from the polymerase. I) The polymerase is then removed from the DNA and the remaining Ser2 phosphorylation is removed by FCP1. J) A unmodified polymerase is free to reinitiate new rounds of transcription.
Transcriptional initiation
Initiation is the earliest stage of the transcription cycle and involves several key steps: initial recruitment of the polymerase to a promoter, proper formation of the transcription preinitiation complex (PIC), and promoter escape. Classically, transcriptional initiation was believed to be associated with an unmodified CTD [35]. However, an increasing body of work has shown that less-studied PTMs are critical to steps in early transcriptional initiation [17,20,26,52,53].
First, a polymerase with an unmodified CTD interacts with Mediator, the transcriptional co-activator complex [54,55]. This is thought to function as a bridge between general transcription factors and RNAPII, and facilitate the formation of the PIC [56–59]. Proper formation of the PIC requires specific CTD PTMs, including O-GlcNAcylation of Ser5 and Ser7 subsequent to polymerase binding to Mediator [20,23,52]. O-GlcNAcylation is a highly transient event mediated by the O-GlcNAc transferase (OGT) and rapidly reversed by the N-acetylglucos-aminidase (OGA), a member of the PIC [19]. The removal of O-GlcNAc after PIC formation is required for phosphorylation of Ser5 and Ser7, which mark the transition to elongation, supporting the model that O-GlcNAc of RNAPII is transient and likely only exists for the formation of the PIC [19,22]. Notably, O-GlcNAc modifications have only been described in mammals and are likely unique to vertebrates, as the O-GlcNAc enzymes are not present in lower eukaryotes, including yeast [60,61].
To begin early elongation, RNAPII must escape interactions with the Mediator complex and the PIC. Notably, many polymerases will fail to escape the promoter and will be turned over through abortive termination [35,62]. The interaction among RNAPII, the PIC, and Mediator are disrupted by phosphorylation of Ser5 and Ser7 by TFIIH, a member of the PIC, via its CDK7 subunit, which frees RNAPII to initiate the early steps of elongation [63–68]. Interestingly, the kinase activity of TFIIH is not required for initiation in vitro, although it is required for early elongation [69–73]. Ser5 phosphorylation is also critical for recruitment of enzymes involved in capping the nascent RNA, which protects it from degradation [74–76]. This function is supported by the recruitment of Dichloro-1-B-D-ribofuranosyl-benzimidazole Sensitivity-Inducing Factor (DSIF), which assists in RNA capping and recruitment of the Negative Elongation Factor (NELF) [42,57].
Two other less studied PTMs play a role during transcriptional initiation: phosphorylation of Tyr1, one of the most highly conserved CTD residues, and methylation of Lys7 [17,26,53,77–80]. Phosphorylation of Tyr1 has been associated with RNAPII occupying enhancers and promoters, and more recently attributed to driving specificity of CDK kinase activity on CTD repeats (discussed below). Tyr1-phosphorylated RNAPII is also involved in the production of enhancer RNAs that occur specifically in the anti-sense direction of gene bodies [17,77–80]. The mechanism and role of lysine methylation remain largely unknown, but initial work indicates that it is involved in preventing Lys7 acetylation, and may negatively regulate transcription [26,53]. Of note, Lys7 exists on only eight non-consensus repeats in the distal metazoan CTD, thus this lysine methylation is a unique feature of higher eukaryotic CTDs.
Promoter-proximal pausing
After promoter escape, RNAPII can pause proximal to the promoter, at an average of 25–50 bp downstream of the transcriptional start site (TSS) [57,81–85]. This phenomenon was initially believed to occur at a small number of genes or viruses such as heat shock genes in Drosophila [86–88] and the integrated HIV provirus [81]. It is now recognized that promoter-proximal pausing is a feature of RNAPII transcription of most genes, as disruption of the release from pausing abrogates transcription of nearly all genes [42,89–92]. Not all paused polymerases will continue into productive elongation, and some paused polymerases will be removed and replaced [42,93–96]. The turnover of paused polymerases may play a critical role in regulating the rate of transcription. Whether CTD PTMs play a role in RNAPII turnover remains to be determined.
Promoter-proximal pausing is established and maintained by the interaction of the CTD with negative elongation factors NELF and DSIF [92,97–100]. Paused polymerases are classically characterized by high levels of Ser5 and Ser7 phosphorylation, but newer research has highlighted the role of additional PTMs. The transition of the polymerase from a paused state to an actively elongating complex, called pause release, is catalyzed by the recruitment of the positive transcriptional elongation factor b (P-TEFb) [37,57,89,90,101–105]. P-TEFb is composed of a regulatory cyclin and catalytic CDK9 component, which phosphorylates Ser2 residues to enhance polymerase processivity [102–104,106]. The ability of P-TEFb to phosphorylate Ser2 is primed by previous phosphorylation events at Ser7 [107] and Tyr1 [108]. A recent study in a breast cancer cell line identified citrullination of Arg1810 by peptidyl arginine deiminase 2 (PADI2), a calcium-dependent enzyme, as a key CTD modification for recruitment of P-TEFb to the paused polymerase of genes involved in cell proliferation [109]. It is important to note the breast cancer cell line studied expressed high levels of PADI2 and the role of citrullination in cells with normal levels of PADI2 expression remains to be fully explored. Given that PADI2 is a calcium dependent enzyme [109], it is intriguing to hypothesize that citrullination may act in a gene-selective manner modulating promoter-proximal release in response to calcium-induced signaling cascades. To date, there is no known mechanism for reversal of arginine citrullination, leading to questions about what happens to citrullinated RNAPII following pause release [109–111]. P-TEFb also phosphorylates DSIF and NELF; phosphorylation of NELF leads to its dissociation from the polymerase complex, whereas phosphorylation of DSIF turns DSIF into a positive elongation factor [100–103,112-115].
Paused polymerases are also highly acetylated at Lys7 residues, mediated by the acetyltransferase p300 [24,25]. Lys7 acetylation serves to recruit the RPRD complex, which includes RPAP2, which serves as a Ser5 phosphatase, and HDAC1, which deacetylates Lys7 [24]. Phosphorylation of Ser5 has been shown to inhibit the ability of P-TEFb to phosphorylate Ser2 on the same heptad in vitro [107], so recruitment of RPRD may prime RNAPII for processive elongation. Furthermore, because Lys7 acetylation results in recruitment of its own deacetylase, Lys7 acetylation is limited to a very narrow window downstream of the TSS [24,25]. The fact that methylation of Lys7 may further restrict Lys7 acetylation underscores the notion that Lys7 acetylation acts at a very defined time during the transcription cycle [26,53].
Transcriptional elongation
Transcriptional elongation is characterized by a gradual loss of Ser5 and Ser7 phosphorylation and Lys7 acetylation [25,116]. In addition, there is an increase in Ser2 phosphorylation, beginning just downstream of promoter pausing and increasing gradually along the gene body, reaching maximal levels at the 3ʹ end of genes [117]. The rate of transcriptional elongation also increases along the body of the gene [32,42–44], and the accumulation of Ser2 phosphorylation along the gene appears to be regulated by the rate of elongation, as RNAPII mutants with slower elongation rates lead to increased Ser2 phosphorylation toward the 5ʹ end of genes [118].
Critical to pause release and transcriptional elongation is the recruitment of complexes mediating co-transcriptional processes, such as splicing or polyadenylation [31,119,120]. Interactions between the CTD of actively transcribing RNAPII and the spliceosome were thought to be mediated primarily through interactions with phosphorylated Ser5 [121]. However, the crystal structure of transcriptional elongation regulator 1 (TCERG1), which mediates interactions between RNAPII and the spliceosome, indicates that hyperphosphorylation of all three serine residues, Ser2, Ser5 and Ser7, are required for optimal interactions between RNAPII and the spliceosome during elongation [122–124]. Additionally, premature termination of transcription is inhibited by the interaction between the CTD phosphorylated on Ser2 and Ser5 and the human anti-termination proteins SCAF4 and SCAF8, members of the arginine/serine-rich splicing factor family [125]. Indeed, loss of Ser2 phosphorylation leads to increases in the usage of early alternative polyadenylation sites by RNAPII [126].
While the initial Ser2 phosphorylation is placed by P-TEFb/CDK9, maintenance of Ser2 phosphorylation throughout elongation is carried out by different kinases, specifically CDK12 and CDK13 [127–130]. Experiments knocking down CDK12 and CDK13 indicate that these kinases have individual as well as overlapping function, however, it is unlikely that they cooperate on individual genes [128,130–132]. How these kinases act to maintain Ser2 phosphorylation in a gene-specific manner is an area of ongoing research.
Transcriptional termination
Recent work has dramatically increased our understanding of the mechanism of transcriptional termination in mammalian systems. However, the exact role that CTD modifications play in termination in mammals remains elusive. There are two dominant models of mammalian termination; Allosteric, where conformational changes to RNAPII allow for termination, and the Torpedo model, where RNA polymerase continues transcribing until it is removed by XRN2 [133]. Which of these models is correct remains unclear, although recent work suggests that aspects of both occur together for efficient termination [134]. Termination occurs in two steps: cleavage/polyadenylation of mRNAs, and removal of RNAPII from DNA [133,135]. These steps occur separately, which suggests that identification of the correct polyadenylation site is necessary for efficient termination. Transcriptional termination is associated with a second pausing of the polymerase, this time at the 3ʹ end of the gene [136–140]. Whether 3ʹ pausing is required for termination remains controversial, and the cause remains elusive, although it may involve interactions of RNAPII with the cleavage and polyadenylation (CPA) complex and the polyadenylation signal [36,138,141].
Interactions between the CPA complex and RNAPII are mediated through the termination factor PCF11, which shows selective interaction with Ser2-phosphorylated RNAPII [142,143]. Additionally, Arg1810 di-methylated by the methyltransferase PRMT5 is recognized by SMN, a protein involved in spliceosome assembly and ribonucleosome biogenesis [27,144]. SMN in turn recruits Sentataxin (SETX), a DNA-RNA helicase required for cleavage of the mRNA from RNAPII [27]. Although the PRMT proteins needed for arginine methylation are conserved across eukaryotes, non-consensus repeats harboring arginine only exist in vertebrates [27,145]. Following cleavage of the mRNA, the exonuclease XRN2 removes RNAPII from chromatin [36,141]. Once the RNAPII is removed from the DNA, it is maintained in a hypo-phosphorylated state by the phosphatase FCP1, which allows efficient recycling of RNAPII into new rounds of transcription [146].
Crosstalk between individual CTD modifications
While CTD modifications have long been considered individually in a systematic “ON” and “OFF” exchange throughout the transcription cycle, recent examples of cooperativity between PTMs have uncovered new functions of individual PTMs and have increased the complexity of our model of transcriptional regulation (Figure 4). Analogous examples of PTM crosstalk, such as among histone modifications, demonstrate that the complex combinatorial nature of modifications can drive specificity and selectivity of interactions with reader proteins [147]. The CTD, similar to the histone tails, is structurally flexible and can support interactions between different PTMs within the same repeat or between different repeats. The CTD is, however, unique in its highly repetitive sequence, and it remains unclear how many repeats within the CTD carry actual modifications. Recent mass spectrometry studies to map phosphorylation of the CTD have started to address these issues. Interestingly, these studies show that the CTD is not heavily phosphorylated, with phosphorylation of Ser2 and Ser5 being predominant and that heptads with multiple phosphorylation events are rare [106,148]. One limitation of these heptad-specific studies is that only phosphorylation marks of the CTD have been mapped and do not address less studied CTD modifications. These studies are also performed on bulk RNAPII and modifications that may be very transient or unstable during sample processing may be difficult to detect and accurately quantify.
Figure 4.
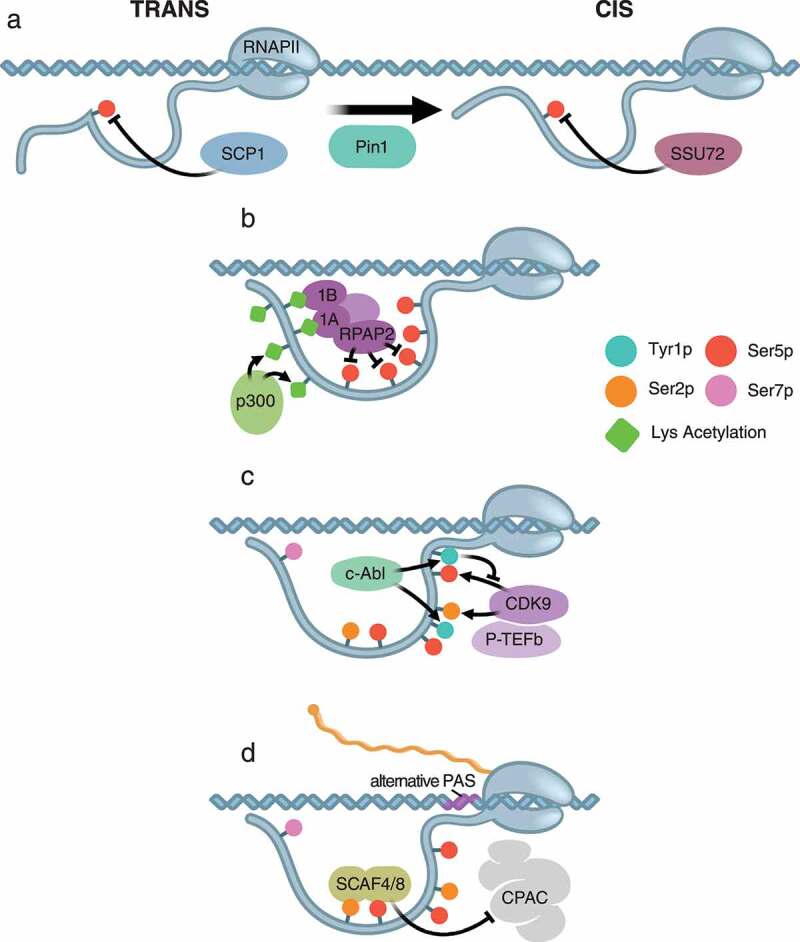
Cross-talk in the CTD. A) Proline isomerization influences the ability of CTD phosphatases to remove Ser5 phosphorylation. In the trans configuration, SCP1 is able to de-phosphorylate Ser5. While in the cis configuration mediated by the proline isomerase Pin1, SSU72 is able to de-phosphorylate Ser5. B) Acetyled non-consensus lysine residues interact with the reader proteins RPRD1A (1A) and RPRD1B (1B), which recruit the Ser5 phosphatase RPAP2. C) Phosphorylation of Tyr1 by c-Abl directs the kinase activity of the CDK9 subunit of PTEF-b to Ser2 by preventing it from phosphorylating Ser5 of nearby repeats. D) Premature termination is prevented by the recognition of Ser2 and Ser5 phosphorylation by SCAF4 and SCAF8, which block recruitment of CPAC to alternative polyadenylation sites (PAS).
Proline isomerization couples CTD phosphorylation to RNA processing events
A classical example of CTD crosstalk is the role of proline isomerization in directing serine/threonine phosphorylation. Prolines are highly conserved at position 3 and 6 of consensus repeats, and are also widely found in non-consensus repeats (Figure 1). Although the dominant state of the proline within the CTD is the more energetically stable trans-configuration, the peptidyl-prolyl isomerase Pin1 is able to recognize phosphorylated serine-proline motifs (Ser2-Pro3 or Ser5-Pro6) and isomerize them from the trans- to the cis-configuration [149–151].
In turn, the conformation of prolines can affect CTD phosphatases and influence the phosphorylation status of Ser2 and Ser5. Indeed, Pin1 peptidyl-prolyl isomerase activity influences the action of the human serine phosphatases Ssu72 and SCP1 [152–156] (Figure 4a). Molecular modeling studies predict that the presence of cis-proline significantly disrupts the active site of SCP1, thus making SCP1 preferentially act on serine residues proximal to trans-prolines [154]. Ssu72 is ubiquitously expressed and acts only on CTD repeats when prolines are in cis conformation [153,155,157].
While effects on serine phosphorylation are clear, it is unclear yet when isomerization occurs in the transcription cycle. Notably, Ssu72 functions as a Ser5 phosphatase only after promoter-proximal pause release [158–160], suggesting Pin1 functions during transcriptional elongation. Interestingly, overexpression of Pin1 leads to overall increased levels of Ser2 and Ser5 phosphorylation and a global shut-down of transcription [150]. These results support the hypothesis that the timing of proline isomerization plays a key role in the progression of transcription, but further studies are required to fully elucidate the function of proline isomerization in gene expression.
Reading of lysine acetylation leads to CTD de-phosphorylation
Higher eukaryotes have evolved a longer CTD and incorporated a varying number of non-consensus repeats carrying Lys7 residues [4]. Lys7 can be methylated and acetylated at different times during the transcription cycle [24–26,53]. Lys7 acetylation is functionally required for activation of signal response genes, e.g. in response to epidermal growth factor signaling, an essential function in the development of multicellular organisms [25].
A recent quantitative mass spectrometry analysis connected Lys7 acetylation with the preferred recruitment of the RPRD1 complex [24]. RPRD1A and RPRD1B and their yeast homologs were previously shown to interact with the Ser2-phosphorylated CTD, with increased affinity for repeats that are dually phosphorylated, and play a role in transcriptional termination [161,162]. However, in the more recent study Lys7 acetylation was shown to enhance RPRD1A and B interaction by providing new electrostatic interactions with the CTD [24] (Figure 4b).
As the RPRD1 protein complex also contains RPAP2, a serine 5 phosphatase, experiments using deacetylase inhibitors were performed to test the effect of increased Lys7 acetylation on Ser5 phosphorylation. Indeed, increased lysine acetylation led to decreased Ser5 phosphorylation, consistent with enhanced recruitment of RPAP2 to the acetylated CTD [24]. Interestingly, knockdown of RPRD1B also increased Lys7 acetylation, and indeed, the class I deacetylase HDAC1 was found associated with the RPRD1 complex [24]. This establishes not only a new link between Lys7 acetylation and Ser5 dephosphorylation, it also shows that certain modifications can autoregulate by recruiting an eraser protein as part of a reader complex. Notably, Lys7 acetylation occurs exclusively in distal repeats, while Ser5 phosphorylation is found across the CTD [26]. The crosstalk between Lys7 acetylation and Ser5 phosphorylation likely occurs between different repeats, in accordance with biochemical studies which showed that Ser5 phosphorylated repeats cannot be acetylated, and vice versa [24,25].
Tyrosine phosphorylation is required for RNAPII elongation via Ser2 phosphorylation
While Lys7 acetylation negatively regulates Ser5 phosphorylation, Tyr1 phosphorylation positively influences Ser2 phosphorylation. Tyr1 phosphorylation has been implicated in regulating transcriptional termination in yeast, specifically by preventing binding of the termination factor Nrd1 [18]. However, in mammals, Tyr phosphorylation occurs primarily at the 5ʹ end of genes [80,163]. A recent study showed that Tyr1 phosphorylation by c-Abl selectively directs P-TEFb/CDK9 to phosphorylate Ser2 [108] (Figure 4c). In biochemical studies, CDK9 can phosphorylate both Ser2 and Ser5 [107], but in vivo it specifically targets Ser2 [105]. Notably, the in vitro assessment of CDK9 function was performed using short CTD peptides, and thus may not account for spatially separated interactions between different repeats.
Prevention of early termination is mediated by dual-phosphorylation of Ser2 and Ser5
Transcriptional termination depends on precise polyadenylation of mRNAs, which is complicated by the existence of multiple early polyadenylation sequences in human genes [164,165]. Thus, extraneous polyadenylation sites must be ignored by the actively transcribing polymerase complex. Recent work showed that the proteins SCAF4 and SCAF8 prevent premature termination by inhibiting recognition of early alternative polyadenylation sites [125] (Figure 4d). In vivo, SCAF4 and SCAF8 co-immunoprecipitate with actively elongating RNAPII that is hyperphosphorylated at Ser2/5/7 and at Tyr1 and Thr4. In vitro experiments with purified CTD heptads showed that SCAF4 and SCAF8 had strong preference for heptads that are dually-phosphorylated at Ser2 and Ser5. This suggests that by recognizing dually-phosphorylated heptads, SCAF4 and SCAF8 identify polymerases early in the process of elongation, preventing premature recruitment of the CPA complex. This is supported by recent work on the function of CDK12, a kinase that supports Ser2 phosphorylation during elongation. Loss of CDK12, and thus loss of Ser2 phosphorylation, led to use of early polyadenylation sites [126].
Conclusions
CTD modifications are key regulators of each step of eukaryotic transcription, and our emerging understanding of the interplay between CTD modifications sheds light on how transcription can be rapidly and dynamically responsive to stimuli. Deciphering the interactions between modifications will lead to an improved understanding of their timing and distribution during the transcriptional cycle. It will also clarify the dynamics of reader protein recruitment, and could lead to new approaches to therapeutically interfere with these dynamics. New research is also beginning to reveal the role of modifications of the CTD at non-consensus sites, which underlie the enhanced transcriptional complexity in higher eukaryotes.
However, the field is hampered by technical challenges, such as antibodies with limited specificity, and the inability of mass spectrometry to provide resolution at the level of a single repeat. In addition, while recent advances such as cryo-EM have greatly improved the ability to capture structural information about RNAPII, the CTD structure in the various phases of transcription remains elusive because of its flexible nature. Future studies directly addressing these issues are necessary to fully capture the complexity of transcription regulation and the important role of CTD modifications in this process.
Funding Statement
This work was supported by the National Institute of Allergy and Infectious Diseases [1R61AI140465-01]; National Institute of Allergy and Infectious Diseases [2R37AI083139-10]; National Institute of Allergy and Infectious Diseases [1R61DA048444-01]; National Institute on Drug Abuse [5R01AI097552-03]; National Institute on Drug Abuse [1R01DA043142-01]; National Institutes of Health [5DP1DA038043-03]; Tobacco-Related Disease Research Program (US) [T30DT1006]; National Institute of Diabetes and Digestive and Kidney Diseases (US) [1R01DK123746-01].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Yang C, Stiller JW.. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci U S A. 2014;111(16):5920–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stump AD, Ostrozhynska K. Selective constraint and the evolution of the RNA polymerase II C-terminal domain. Transcription. 2013;4(2):77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Portz B, Lu F, Gibbs EB, et al. Structural heterogeneity in the intrinsically disordered RNA polymerase II C-terminal domain. Nat Commun. 2017;8:15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Simonti CN, Ostrozhynska K. Evolution of lysine acetylation in the RNA polymerase II C-terminal domain. BMC Evol Biol. 2015;15:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Harlen KM, Churchman LS. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol. 2017;18(4):263–273. [DOI] [PubMed] [Google Scholar]
- [6].Chapman RD, Heidemann M, Hintermair C, et al. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24(6):289–296. [DOI] [PubMed] [Google Scholar]
- [7].Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113(11):8456–8490. [DOI] [PubMed] [Google Scholar]
- [8].Stiller JW, Hall BD. Evolution of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci U S A. 2002;99(9):6091–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lu F, Portz B, Gilmour DS. The C-terminal domain of RNA polymerase II is a multivalent targeting sequence that supports drosophila development with only consensus heptads. Mol Cell. 2019;73(6):1232–1242 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boehning M, Dugast-Darzacq C, Rankovic M, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25(9):833–840. . [DOI] [PubMed] [Google Scholar]
- [11].Guo YE, Manteiga JC, Henninger JE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature; 2019;572(7770):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu H, Yu D, Hansen AS, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558(7709):318–323. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zaborowska J, Egloff S, Murphy S. The pol II CTD: new twists in the tail. Nat Struct Mol Biol. 2016;23(9):771–777. [DOI] [PubMed] [Google Scholar]
- [14].Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36(4):541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jeronimo C, Collin P, Robert F. The RNA Polymerase II CTD: the increasing complexity of a low-complexity protein domain. J Mol Biol. 2016;428(12):2607–2622. [DOI] [PubMed] [Google Scholar]
- [16].Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20(21):2922–2936. [DOI] [PubMed] [Google Scholar]
- [17].Yurko NM, Manley JL. The RNA polymerase II CTD “orphan” residues: emerging insights into the functions of Tyr-1, Thr-4, and Ser-7. Transcription. 2018;9(1):30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mayer A, Heidemann M, Lidschreiber M, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336(6089):1723–1725. [DOI] [PubMed] [Google Scholar]
- [19].Ranuncolo SM, Ghosh S, Hanover JA, et al. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem. 2012;287(28):23549–23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lewis BA, Hanover JA. O-GlcNAc and the epigenetic regulation of gene expression. J Biol Chem. 2014;289(50):34440–34448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lu L, Fan D, Hu CW, et al. Distributive O-GlcNAcylation on the highly repetitive C-terminal domain of RNA polymerase II. Biochemistry. 2016;55(7):1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268(14):10416–10424. [PubMed] [Google Scholar]
- [23].Lewis BA, Burlingame AL, Myers SA. Human RNA polymerase II promoter recruitment in vitro is regulated by O-linked N-Acetylglucosaminyltransferase (OGT). J Biol Chem. 2016;291(27):14056–14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ali I, Ruiz DG, Ni Z, et al. Crosstalk between RNA Pol II C-terminal domain acetylation and phosphorylation via RPRD proteins. Mol Cell. 2019;74(6):1164–1174 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schroder S, Herker E, Itzen F, et al. Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Mol Cell. 2013;52(3):314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Voss K, Forne I, Descostes N, et al. Site-specific methylation and acetylation of lysine residues in the C-terminal domain (CTD) of RNA polymerase II. Transcription. 2015;6(5):91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhao DY, Gish G, Braunschweig U, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016;529(7584):48–53. [DOI] [PubMed] [Google Scholar]
- [28].Sims RJ 3rd, Rojas LA, Beck DB, et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332(6025):99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shandilya J, Roberts SG. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim Biophys Acta. 2012;1819(5):391–400. [DOI] [PubMed] [Google Scholar]
- [30].Harlen KM, Trotta KL, Smith EE, et al. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Rep. 2016;15(10):2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Alpert T, Herzel L, Neugebauer KM. Perfect timing: splicing and transcription rates in living cells. Wiley Interdiscip Rev RNA. 2017;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Danko CG, Hah N, Luo X, et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell. 2013;50(2):212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15(3):163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aoi Y, Smith ER, Shah AP, et al. NELF regulates a promoter-proximal step distinct from RNA Pol II pause-release. Mol Cell. 2020;78(2):261–274.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):129–143. [DOI] [PubMed] [Google Scholar]
- [36].Porrua O, Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol. 2015;16(3):190–202. [DOI] [PubMed] [Google Scholar]
- [37].Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Darzacq X, Shav-Tal Y, de Turris V, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14(9):796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13(1):49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jiang Y, Gralla JD. Uncoupling of initiation and reinitiation rates during HeLa RNA polymerase II transcription in vitro. Mol Cell Biol. 1993;13(8):4572–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29(5):600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Veloso A, Kirkconnell KS, Magnuson B, et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24(6):896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fuchs G, Voichek Y, Benjamin S, et al. 4sUDRB-seq: measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biol. 2014;15(5):R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9(2):184–190. [DOI] [PubMed] [Google Scholar]
- [46].Li J, Dong A, Saydaminova K, et al. Single-molecule nanoscopy elucidates RNA polymerase II transcription at single genes in live cells. Cell. 2019;178(2):491–506 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tang GQ, Roy R, Bandwar RP, et al. Real-time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proc Natl Acad Sci U S A. 2009;106(52):22175–22180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang Z, Tjian R. Measuring dynamics of eukaryotic transcription initiation: challenges, insights and opportunities. Transcription. 2018;9(3):159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Larson DR, Zenklusen D, Wu B, et al. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332(6028):475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang Z, Revyakin A, Grimm JB, et al. Single-molecule tracking of the transcription cycle by sub-second RNA detection. Elife. 2014;3:e01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lis JT. A 50 year history of technologies that drove discovery in eukaryotic transcription regulation. Nat Struct Mol Biol. 2019;26(9):777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schilbach S, Hantsche M, Tegunov D, et al. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature. 2017;551(7679):204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dias JD, Rito T, Torlai Triglia E, et al. Methylation of RNA polymerase II non-consensus Lysine residues marks early transcription in mammalian cells. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tsai KL, Yu X, Gopalan S, et al. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544(7649):196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sato S, Tomomori-Sato C, Tsai KL, et al. Role for the MED21-MED7 hinge in assembly of the mediator-RNA polymerase II holoenzyme. J Biol Chem. 2016;291(52):26886–26898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Allen BL, Taatjes DJ. The mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16(3):155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Core L, Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33(15–16):960–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Grunberg S, Hahn S. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci. 2013;38(12):603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Naar AM, Taatjes DJ, Zhai W, et al. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 2002;16(11):1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jiang MS, Hart GW. A subpopulation of estrogen receptors are modified by O-linked N-acetylglucosamine. J Biol Chem. 1997;272(4):2421–2428. [DOI] [PubMed] [Google Scholar]
- [61].Lubas WA, Frank DW, Krause M, et al. O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem. 1997;272(14):9316–9324. [DOI] [PubMed] [Google Scholar]
- [62].Revyakin A, Zhang Z, Coleman RA, et al. Transcription initiation by human RNA polymerase II visualized at single-molecule resolution. Genes Dev. 2012;26(15):1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Meyer KD, Lin SC, Bernecky C, et al. p53 activates transcription by directing structural shifts in mediator. Nat Struct Mol Biol. 2010;17(6):753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Boeing S, Rigault C, Heidemann M, et al. RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. J Biol Chem. 2010;285(1):188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Glover-Cutter K, Larochelle S, Erickson B, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29(20):5455–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Rodriguez-Molina JB, Tseng SC, Simonett SP, et al. Engineered covalent inactivation of TFIIH-kinase reveals an elongation checkpoint and results in widespread mRNA stabilization. Mol Cell. 2016;63(3):433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wong KH, Jin Y, Struhl K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol Cell. 2014;54(4):601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kwon I, Kato M, Xiang S, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155(5):1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Serizawa H, Conaway JW, Conaway RC. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature. 1993;363(6427):371–374. [DOI] [PubMed] [Google Scholar]
- [70].Makela TP, Parvin JD, Kim J, et al. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci U S A. 1995;92(11):5174–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tirode F, Busso D, Coin F, et al. Reconstitution of the transcription factor TFIIH. Mol Cell. 1999;3(1):87–95. [DOI] [PubMed] [Google Scholar]
- [72].Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14(19):2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Payne JM, Laybourn PJ, Dahmus ME. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989;264(33):19621–19629. [PubMed] [Google Scholar]
- [74].Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3(3):405–411. [DOI] [PubMed] [Google Scholar]
- [75].Rodriguez CR, Cho EJ, Keogh MC, et al. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20(1):104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fabrega C, Shen V, Shuman S, et al. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11(6):1549–1561. [DOI] [PubMed] [Google Scholar]
- [77].Burger K, Schlackow M, Gullerova M. Tyrosine kinase c-Abl couples RNA polymerase II transcription to DNA double-strand breaks. Nucleic Acids Res. 2019;47(7):3467–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shah N, Maqbool MA, Yahia Y, et al. Tyrosine-1 of RNA polymerase II CTD controls global termination of gene transcription in mammals. Mol Cell. 2018;69(1):48–61 e6. [DOI] [PubMed] [Google Scholar]
- [79].Baskaran R, Escobar S, Wang J. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the mammalian RNA polymerase II: possible role in transcription elongation. Cell Growth Differ. 1999;6(10):387–396. [PubMed] [Google Scholar]
- [80].Descostes N, Heidemann M, Spinelli L, et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. Elife. 2014;3:e02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kao SY, Calman AF, Luciw PA, et al. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330(6147):489–493. [DOI] [PubMed] [Google Scholar]
- [82].Krumm A, Meulia T, Brunvand M, et al. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6(11):2201–2213. [DOI] [PubMed] [Google Scholar]
- [83].Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. Embo J. 1992;11(9):3307–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Min IM, Waterfall JJ, Core LJ, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25(7):742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Nat Acad Sci. 1993;90(17):7923–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6(11):3984–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54(6):795–804. [DOI] [PubMed] [Google Scholar]
- [89].Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276(34):31793–31799. [DOI] [PubMed] [Google Scholar]
- [90].Ni Z, Saunders A, Fuda NJ, et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28(3):1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Rahl PB, Lin CY, Seila AC, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Henriques T, Gilchrist DA, Nechaev S, et al. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52(4):517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 2015;29(1):39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Krebs AR, Imanci D, Hoerner L, et al. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol Cell. 2017;67(3):411–422 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Shao W, Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet. 2017;49(7):1045–1051. [DOI] [PubMed] [Google Scholar]
- [96].Erickson B, Sheridan RM, Cortazar M, et al. Dynamic turnover of paused Pol II complexes at human promoters. Genes Dev. 2018;32(17–18):1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Adelman K, Henriques T. Transcriptional speed bumps revealed in high resolution. Nature. 2018;560(7720):560–561. [DOI] [PubMed] [Google Scholar]
- [98].Vos SM, Farnung L, Boehning M, et al. Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature. 2018;560(7720):607–612. [DOI] [PubMed] [Google Scholar]
- [99].Yamaguchi Y, Takagi T, Wada T, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97(1):41–51. [DOI] [PubMed] [Google Scholar]
- [100].Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282(30):21901–21912. [DOI] [PubMed] [Google Scholar]
- [101].Yamada T, Yamaguchi Y, Inukai N, et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21(2):227–237. . [DOI] [PubMed] [Google Scholar]
- [102].Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20(8):2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. [DOI] [PubMed] [Google Scholar]
- [104].Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270(21):12335–12338. [DOI] [PubMed] [Google Scholar]
- [105].Marshall NF, Peng J, Xie Z, et al. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271(43):27176–27183. . [DOI] [PubMed] [Google Scholar]
- [106].Schuller R, Forné I, Straub T, et al. Heptad-specific phosphorylation of RNA polymerase II CTD. Mol Cell. 2016;61(2):305–314. [DOI] [PubMed] [Google Scholar]
- [107].Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3(1):842. [DOI] [PubMed] [Google Scholar]
- [108].Mayfield JE, Irani S, Escobar EE, et al. Tyr1 phosphorylation promotes phosphorylation of Ser2 on the C-terminal domain of eukaryotic RNA polymerase II by P-TEFb. Elife. 2019;8. DOI: 10.7554/eLife.48725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Sharma P, Lioutas A, Fernandez-Fuentes N, et al. Arginine citrullination at the C-terminal domain controls RNA polymerase II transcription. Mol Cell. 2019;73(1):84–96 e7. [DOI] [PubMed] [Google Scholar]
- [110].Wang Y, Wysocka J, Sayegh J, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–283. . [DOI] [PubMed] [Google Scholar]
- [111].Cuthbert GL, Daujat S, Snowden AW, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–553. . [DOI] [PubMed] [Google Scholar]
- [112].Guo S, Yamaguchi Y, Schilbach S, et al. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408(6810):366–369. . [DOI] [PubMed] [Google Scholar]
- [113].Wada T, Takagi T, Yamaguchi Y, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12(3):343–356. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Hartzog GA, Fu J. The Spt4–Spt5 complex: A multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829(1):105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Bernecky C, Plitzko JM, Cramer P. Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat Struct Mol Biol. 2017;24(10):809–815. [DOI] [PubMed] [Google Scholar]
- [116].Ni Z, Olsen JB, Guo X, et al. Control of the RNA polymerase II phosphorylation state in promoter regions by CTD interaction domain-containing proteins RPRD1A and RPRD1B. Transcription. 2011;2(5):237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Mayer A, Lidschreiber M, Siebert M, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17(10):1272–1278. . [DOI] [PubMed] [Google Scholar]
- [118].Fong N, Saldi T, Sheridan RM, et al. RNA Pol II dynamics modulate co-transcriptional chromatin modification, CTD phosphorylation, and transcriptional direction. Mol Cell. 2017;66(4):546–557 e3. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Coulon A, Ferguson ML, de Turris V, et al. Kinetic competition during the transcription cycle results in stochastic RNA processing. Elife. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Herzel L, Ottoz DSM, Alpert T, et al. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol. 2017;18(10):637–650. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Nojima T, Rebelo K, Gomes T, et al. RNA polymerase II phosphorylated on CTD serine 5 interacts with the spliceosome during co-transcriptional splicing. Mol Cell. 2018;72(2):369–379 e4. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Liu J, Fan S, Lee C-J, et al. Specific interaction of the transcription elongation regulator TCERG1 with RNA polymerase II requires simultaneous phosphorylation at Ser 2, Ser 5, and Ser 7 within the carboxyl-terminal domain repeat. J Biol Chem. 2013;288(15):10890–10901. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Pearson JL, Robinson TJ, Muñoz MJ, et al. Identification of the cellular targets of the transcription factor TCERG1 reveals a prevalent role in mRNA processing. J Biol Chem. 2008;283(12):7949–7961. . [DOI] [PubMed] [Google Scholar]
- [124].Sanchez-Hernandez N, Boireau S, Schmidt U, et al. The in vivo dynamics of TCERG1, a factor that couples transcriptional elongation with splicing. RNA. 2016;22(4):571–582. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Gregersen LH, Mitter R, Ugalde AP, et al. SCAF4 and SCAF8, mRNA anti-terminator proteins. Cell. 2019;177(7):1797–1813 e18. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Dubbury SJ, Boutz PL, Sharp PA. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature. 2018;564(7734):141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Bartkowiak B, Liu P, Phatnani HP, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24(20):2303–2316. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Blazek D, Kohoutek J, Bartholomeeusen K, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25(20):2158–2172. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Eifler TT, Shao W, Bartholomeeusen K, et al. Cyclin-dependent kinase 12 increases 3′ end processing of growth factor-induced c-FOS transcripts. Mol Cell Biol. 2015;35(2):468–478. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Greenleaf AL. Human CDK12 and CDK13, multi-tasking CTD kinases for the new millenium. Transcription. 2019;10(2):91–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kohoutek J, Blazek D. Cyclin K goes with Cdk12 and Cdk13. Cell Div. 2012;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Choi SH, Martinez TF, Kim S, et al. CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability. Genes Dev. 2019;33(7–8):418–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Proudfoot NJ. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science. 2016;352(6291):aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Cortazar MA, Sheridan RM, Erickson B, et al. Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 dephosphorylation facilitates termination by a “sitting duck torpedo” mechanism. Mol Cell. 2019;76(6):896–908 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23(11):1247–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Plant KE, Dye MJ, Lafaille C, et al. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human Ggamma-globin gene. Mol Cell Biol. 2005;25(8):3276–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26(10):3986–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Nag A, Narsinh K, Martinson HG. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol. 2007;14(7):662–669. [DOI] [PubMed] [Google Scholar]
- [139].Glover-Cutter K, Kim S, Espinosa J, et al. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15(1):71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Grosso AR, de Almeida SF, Braga J, et al. Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res. 2012;22(8):1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].West S, Gromak N, Proudfoot NJ. Human 5ʹ –> 3ʹ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432(7016):522–525. [DOI] [PubMed] [Google Scholar]
- [142].Kamieniarz-Gdula K, Gdula MR, Panser K, et al. Selective roles of vertebrate PCF11 in premature and full-length transcript termination. Mol Cell. 2019;74(1):158–172 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Volanakis A, Kamieniarz-Gdula K, Schlackow M, et al. WNK1 kinase and the termination factor PCF11 connect nuclear mRNA export with transcription. Genes Dev. 2017;31(21):2175–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Chaytow H, Huang YT, Gillingwater TH, et al. The role of survival motor neuron protein (SMN) in protein homeostasis. Cell Mol Life Sci. 2018;75(21):3877–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Sun L, Wang M, Lv Z, et al. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc Natl Acad Sci U S A. 2011;108(51):20538–20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Fuda NJ, Buckley MS, Wei W, et al. Fcp1 dephosphorylation of the RNA polymerase II C-terminal domain is required for efficient transcription of heat shock genes. Mol Cell Biol. 2012;32(17):3428–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. [DOI] [PubMed] [Google Scholar]
- [148].Suh H, Ficarro SB, Kang UB, et al. Direct analysis of phosphorylation sites on the Rpb1 C-terminal domain of RNA polymerase II. Mol Cell. 2016;61(2):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Xu YX, Hirose Y, Zhou XZ, et al. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17(22):2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Xu YX, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. 2007;21(22):2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Verdecia MA, Bowman ME, Lu KP, et al. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7(8):639–643. [DOI] [PubMed] [Google Scholar]
- [152].Zhang DW, Mosley AL, Ramisetty SR, et al. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem. 2012;287(11):8541–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Xiang K, Nagaike T, Xiang S, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467(7316):729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Zhang Y, Kim Y, Genoud N, et al. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell. 2006;24(5):759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Werner-Allen JW, Lee CJ, Liu P, et al. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem. 2011;286(7):5717–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Zhang M, Liu J, Kim Y, et al. Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein Sci. 2010;19(5):974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Irani S, Sipe SN, Yang W, et al. Structural determinants for accurate dephosphorylation of RNA polymerase II by its cognate C-terminal domain (CTD) phosphatase during eukaryotic transcription. J Biol Chem. 2019;294(21):8592–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Chen Y, Zhang L, Estaras C, et al. A gene-specific role for the Ssu72 RNAPII CTD phosphatase in HIV-1 Tat transactivation. Genes Dev. 2014;28(20):2261–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].He X, Khan AU, Cheng H, et al. Functional interactions between the transcription and mRNA 3ʹ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17(8):1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Dichtl B, Blank D, Ohnacker M, et al. A Role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell. 2002;10(5):1139–1150. [DOI] [PubMed] [Google Scholar]
- [161].Morales JC, Richard P, Rommel A, et al. Kub5-Hera, the human Rtt103 homolog, plays dual functional roles in transcription termination and DNA repair. Nucleic Acids Res. 2014;42(8):4996–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Ni Z, Xu C, Guo X, et al. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat Struct Mol Biol. 2014;21(8):686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Hsin JP, Li W, Hoque M, et al. RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. Elife. 2014;3:e02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Derti A, Garrett-Engele P, Macisaac KD, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22(6):1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. [DOI] [PubMed] [Google Scholar]
- [166].Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci U S A. 1993;90(23):11167–11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Hintermair C, Heidemann M, Koch F, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. Embo J. 2012;31(12):2784–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Hintermair C, Voss K, Forne I, et al. Specific threonine-4 phosphorylation and function of RNA polymerase II CTD during M phase progression. Sci Rep. 2016;6:27401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Schlackow M, Nojima T, Gomes T, et al. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell. 2017;65(1):25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3ʹ end processing. Science. 2011;334(6056):683–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Zhang M, Wang XJ, Chen X, et al. Structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the CTD code. ACS Chem Biol. 2012;7(8):1462–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Sikorsky T, Hobor F, Krizanova E, et al. Recognition of asymmetrically dimethylated arginine by TDRD3. Nucleic Acids Res. 2012;40(22):11748–11755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Mikhaylova O, Ignacak ML, Barankiewicz TJ, et al. The von Hippel-Lindau tumor suppressor protein and Egl-9-Type proline hydroxylases regulate the large subunit of RNA polymerase II in response to oxidative stress. Mol Cell Biol. 2008;28(8):2701–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Beaudenon SL, Huacani MR, Wang G, et al. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(10):6972–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Spector BM, Turek ME, Price DH. Functional interaction of human Ssu72 with RNA polymerase II complexes. PLoS One. 2019;14(3):e0213598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Clemente-Blanco A, Sen N, Mayan-Santos M, et al. Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat Cell Biol. 2011;13(12):1450–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Patturajan M, Wei X, Berezney R, et al. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol Cell Biol. 1998;18(4):2406–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Qiu H, Hu C, Gaur NA, et al. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. Embo J. 2012;31(16):3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].David CJ, Boyne AR, Millhouse SR, et al. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25(9):972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Yoh SM, Cho H, Pickle L, et al. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21(2):160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Li B, Howe L, Anderson S, et al. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278(11):8897–8903. [DOI] [PubMed] [Google Scholar]
- [182].Sun M, Lariviere L, Dengl S, et al. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J Biol Chem. 2010;285(53):41597–41603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Govind CK, Qiu H, Ginsburg DS, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39(2):234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Govind CK, Zhang F, Qiu H, et al. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25(1):31–42. [DOI] [PubMed] [Google Scholar]
- [185].Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol. 2009;29(24):6473–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Liang K, Gao X, Gilmore JM, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol Cell Biol. 2015;35(6):928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Bosken CA, Farnung L, Hintermair C, et al. The structure and substrate specificity of human Cdk12/Cyclin K. Nat Commun. 2014;5:3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Gu B, Eick D, Bensaude O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2013;41(3):1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Schaft D, Roguev A, Kotovic KM, et al. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31(10):2475–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Drouin S, Laramee L, Jacques PE, et al. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6(10):e1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Ganem C, Devaux F, Torchet C, et al. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. Embo J. 2003;22(7):1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Krishnamurthy S, He X, Reyes-Reyes M, et al. Ssu72 Is an RNA Polymerase II CTD Phosphatase. Mol Cell. 2004;14(3):387–394. [DOI] [PubMed] [Google Scholar]
- [193].Milne TA, Dou Y, Martin ME, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005;102(41):14765–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Ghosh A, Shuman S, Lima CD. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell. 2011;43(2):299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Hughes CM, Rozenblatt-Rosen O, Milne TA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the Hoxc8 locus. Mol Cell. 2004;13(4):587–597. [DOI] [PubMed] [Google Scholar]
- [196].Yaffe MB, Schutkowski M, Shen M, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278(5345):1957–1960. [DOI] [PubMed] [Google Scholar]
- [197].Di Vona C, Bezdan D, Islam AB, et al. Chromatin-wide profiling of DYRK1A reveals a role as a gene-specific RNA polymerase II CTD kinase. Mol Cell. 2015;57(3):506–520. [DOI] [PubMed] [Google Scholar]
- [198].Yu D, Cattoglio C, Xue Y, et al. A complex between DYRK1A and DCAF7 phosphorylates the C-terminal domain of RNA polymerase II to promote myogenesis. Nucleic Acids Res. 2019;47(9):4462–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Baillat D, Hakimi MA, Naar AM, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123(2):265–276. [DOI] [PubMed] [Google Scholar]
- [200].Egloff S, O'Reilly D, Chapman RD, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318(5857):1777–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Egloff S, Szczepaniak SA, Dienstbier M, et al. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem. 2010;285(27):20564–20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Egloff S, Zaborowska J, Laitem C, et al. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell. 2012;45(1):111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]


