ABSTRACT
During the last years, the popularity of saliva has been increasing for its applicability as a diagnostic fluid. Blood biomarker molecules have to cross the blood-saliva barrier (BSB) in order to appear in saliva. The BSB consists of all oral and salivary gland epithelial barriers. Within this context, the optimization of in vitro models for mechanistic studies about the transport of molecules across the oral mucosa is an important task. Here, we describe the optimization and comprehensive characterization of a Transwell model of the oral mucosa based on the epithelial cell line TR146. Through systematic media optimization investigating 12 different set-ups, a significant increase of barrier integrity upon airlift cultivation was achieved for TR146 cell layers. The distinct improvement of the paracellular barrier was shown by measurements of transepithelial electrical resistance (TEER) and carboxyfluorescein permeability assays. Histological characterization supported TEER data and showed a stratified, non-keratinized multilayer of the optimized TR146 model. High-Throughput qPCR using 96 selected markers for keratinization, cornification, epithelial–mesenchymal transition, aquaporins, mucins, tight junctions, receptors, and transporter proteins was applied to comprehensively characterize the systematic optimization of the cellular model and validate against human biopsy samples. Data revealed the expression of several genes in the oral mucosa epithelium for the first time and elucidated novel regulations dependent on culture conditions. Moreover, functional activity of ABC transporters ABCB1 and ABCC4 was shown indicating the applicability of the model for drug transport studies. In conclusion, a Transwell model of the oral mucosa epithelium was optimized being suitable for transport studies.
KEYWORDS: Biomarker, oral cancer, sjörgen’s syndrome, cytokeratin, HIV
1. Introduction
The mouth, often referred as the mirror of the body, is a well-organized system that reflects and supports human health. It is continuously bathed in saliva, communicates with the external environment and fosters characteristic microorganisms. The mouth is organized into different kinds of tissues, each performing a unique function, and multiple structures that work in concert. Overall, the tissues comprise a stratified squamous epithelium that serves as a protective layer, prevents the passage of foreign particulate matter, deleterious molecules, viruses, bacteria, and irritants and allows exchange of gases and nutrients.1 The epithelium is coated with saliva that consists of water (99.5%), salts, enzymes, and proteins. Saliva is produced in the salivary glands by acinus cells, collected in the duct network and expelled into the mouth. Due to its viscoelastic properties, saliva retains on the surface and provides lubrication of the inner lining. The salivary glands are highly permeable and allow free entrance of blood derived-molecules via transcellular or paracellular routes.2 Thus, also biomarkers of local or systemic disorders can be absorbed and detected in this highly relevant physiological liquid.3 Compared to blood, which is currently the most frequently used diagnostic medium, saliva shows the advantage that the collection of samples is noninvasive, procurement is painless, and samples can be safely handled and easily stored.4 For example, Jasim et al. (2018) showed that the glutamate level, which plays an important role in the pathophysiology of chronic pain conditions, determined in stimulated whole saliva correlated with plasma.5 Besides saliva also the oral mucosa can be used for potential biomarker diagnosis. Studies with regard to Alzheimer's Disease (AD) demonstrated that potential biomarkers (e.g., Tau protein, CK14 expression) for early diagnosis of AD were detectable in the buccal mucosa.6,7 In addition to its function for biomarker diagnosis, the epithelium also provides an interesting target site for local and systemic drug delivery. Thereby, drug permeability is higher in the non-keratinized tissues (i.e., buccal, sublingual) than in the keratinized tissues (i.e., gingival, palatal) and strongly depends on the compound´s physicochemical properties such as molecular weight, hydrophobicity, or level of ionization. Methods commonly used to study drug interactions with molecules or their transport across the oral mucosa include in vivo and in vitro systems. Although the usage of animal models is still perceived as the “gold standard” (and also not to be excluded), the development and implementation of 3R models to Replace, Reduce, and Refine animal models have gained increased attention. To mimic the lining mucosa, Transwell systems are usually to be used, where TR146 cells are cultured on filters. The TR146 cells, which originate from a neck node metastasis of human buccal carcinoma, have been reported to be appropriate to study the transport of selected markers and drugs.8 Jacobson et al. (1995) showed that TR146 cells present ultrastructural similarity to the normal human buccal epithelium, resulting in four to eight cell layers and exhibit the formation of desmosomes during prolonged incubation time.9 The maximum integrity is obtained after 30 days. Stratification and the formation of a tight permeability barrier are reached after submerged culture for 23 days. The cells express keratins, involucrin, plasma membrane-associated transglutaminase, and EGF receptor and enzyme activity is comparable between porcine and human epithelium.10 Upon culturing at air-liquid interface, no differences with regard to differentiation markers were found for the two cultivation methods.11 Permeability studies revealed that for specific drugs (e.g., adrenoceptive drugs) the results correlated for both models.12 However, still there is a factor of difference in the transport of other substances. In order to clarify this, investigations are necessary that provide more detailed information about the physical barrier.
Here, we carefully studied the physical barrier of TR146 cells at the functional level by measuring transepithelial electrical resistance (TEER) and the permeability of paracellular marker carboxyfluorescein. To this end, cells were treated on airlift and submerged cultivation. The medium was complemented with supplements to create optimum conditions which resulted in a strong paracellular barrier showing significant effects of air-lift conditions for this cell line. High-Throughput qPCR was performed with selected markers to study keratinization, cornification, epithelial–mesenchymal transition, aquaporins, mucins, cell junctions, receptors, and transporter proteins and results were compared to human biopsies to select culture conditions nearest to the human in vivo situation. Moreover, activity of ABC transporters was proven indicating the applicability of the buccal epithelial model for drug transport studies.
2. Materials and methods
2.1. Cell culture
The buccal carcinoma cell line TR146 (Sigma-Aldrich, #10032305) was cultivated in T25 or T75 TC-treated cell culture flasks (Greiner, CELLSTAR, #690175, #658175) at 37°C, 5% CO2 and 95% humidity in Dulbecco´s Modified Eagle Medium (DMEM, Sigma-Aldrich, #D5796) supplemented with 1% Penicillin/Streptomycin (P/S, Merck, #A2213) and 10% Fetal Calf Serum (FCS, Sigma-Aldrich, #F9665), in the following termed as DMEM media. Cells were also cultivated in EpiLife (Gibco, #M-EPI-500-CA) supplemented with 1% Human Keratinocytes Growth Supplements (HKGS, Gibco, #S0015) and 1% P/S, termed as EpiLife media (E1). Cells were seeded at a concentration of 9.33 × 104 cells/cm2 for subcultivation once a week. Media change was performed every 2–3 days.
2.2. Transepithelial electrical resistance (TEER) measurements
ThinCert Inserts (Greiner, #662610) with a membrane (PET) area of 0.336 cm2 and 0.4 µm pore diameter were selected for 24-well experiments. The cells were seeded at a concentration of 4.29 × 104 cells/cm2 in 300 µL, the basolateral side was provided with 900 µL media at all times. For Transwell experiments, TR146 from passage 11–38 were used. EpiLife media (E1) additionally contained 1.44 mM CaCl2 (Sigma-Aldrich, #C7902) for seeding, whereby upon confluency on the inserts the media was additionally supplemented with 10 ng/mL Keratinocytes Growth Factor (KGF, Sigma-Aldrich, #K1757) and 252 nM Ascorbate-2-Phosphate (A2P, Sigma-Aldrich, #A8960), described as the final EpiLife media (E3). Cells seeded in DMEM media were supplemented with a variety of growth factors such as HKGS, KGF, A2P (applied at the same concentration as EpiLife) or hydrocortisone (HC, 10 nM, 100 nM or 1000 nM) or retinoic acid (RA, 3 µM or 10 µM) upon reaching confluency. Media change was performed every 2–3 days. The cells were either cultivated under submerged conditions for the entire time period or switched to airlift condition on the fifth day. First TEER measurements took place on day 5 upon seeding and were performed every 2–3 days until reaching the maximum of TEER values. Prior to measurement, media of the 24-well plates was exchanged and equilibrated for at least 30 min at room temperature under sterile conditions. The chopstick electrode (WPI, #STX2) was disinfected for a maximum of 10 min in 70% EtOH and subsequently equilibrated in media for at least 10 min. Ohmic resistance values were displayed as Ω upon measurement with Millicell ERS-1 Voltohmeter (Merck, #MERS00001) and multiplied with the surface area of the membrane for data analysis after subtraction of mean values from blanks without cells to obtain TEER values (Ω x cm2).
2.3. Permeability studies
To examine the paracellular barrier with a second parameter, transport assays with carboxyfluorescein (Fluka, #21877) were performed. The apical media was replaced with 300 µL DMEM media or final EpiLife media (E3) containing 10 µM carboxyfluorescein. After incubation for 2 h at 37°C the apical and basolateral media was collected and the carboxyfluorescein content was given as relative fluorescence units (RFU) upon measurement at 488–520 nm with the Enspire Multimode Plate Reader (PerkinElmer). Media without carboxyfluorescein were used as negative controls and stock solutions were used to determine the permeated content of carboxyfluorescein. Through linear regression analysis, the slope of the cleared volume against time was estimated and calculated with a factor considering the growth surface area of 0.336 cm2. The permeability coefficient was calculated as the inverse of the permeability [µm/min] after subtraction of the permeability of the blank as shown in the following equation, whereby PEall refers to the overall permeability, PEblank to the permeability of the blank and PEcell gives the permeability coefficient of the cell layer as described recently.13,14
2.4. RNA isolation and cDNA synthesis
The NucleoSpin RNA kit (Machery Nagel, #740955.250) was used according to manufacturer´s instruction and RNA was eluted with 40 µL nuclease-free water (Invitrogen, #AM9937). The RNA concentration [ng/µL] and the 260/280, 260/230 ratio were determined with a Nanodrop 2000 C (Thermo Scientific Peqlab) using 1 µL sample. Subsequent cDNA synthesis was performed with the Multiscribe Reverse Transcriptase Kit (Applied Biosystem Thermo Scientific, #4311235) applying 1 µg RNA for 20 µL cDNA. For RNA isolation after cultivation on 24-well inserts, two inserts cultivated under the same conditions were lysed and pooled as one sample. TR146 cultivated on 6-well plates (Becton Dickinson, #353502) were seeded at a density of 10,000 cells/cm2 (passage 12) in 3 mL DMEM media to obtain samples for PCR primer evaluation and lysed on day 9 for RNA isolation.
2.5. PCR and qPCR
PCR was performed with 10 ng cDNA per reaction. One reaction included 5 µM primer pair (Eurofins), dNTPs (ThermoScientific, #R0181) with a final concentration of 160 µM, PCR buffer containing 15 mM MgCl2 (QIAGEN, #154040672) and 0.6 units of Hot Star Plus Taq polymerase (QIAGEN, #154045537, 250 U) or Hot Star Taq Polymerase (QIAGEN, #1007837). PCR was performed with an activation time of 5 min for HotStarTaq Plus Polymerase or 15 min for HotStarTaq Polymerase at 95°C, continued with 35 cycles at 95°C for 40 s, 60°C for 40 s, and 72°C for 1 min 20 s. Final extension was performed at 72°C for 7 min with a subsequent cooling cycle at 4°C. The PCR program was performed with the Thermocycler (VWR Duocycler) or GeneAmp PCR system 2700 (Applied biosystems). The PCR products were diluted 1:6 with 6x DNA loading dye (Fermentas, #R0611) and separated on a 2% agarose gel containing 0.08% SYBR Safe (Invitrogen, #533102) in 1x Tris-Borate-EDTA (TBE) buffer for 1 h and 40 min at 120 V and 120 mA. Images were taken with the UVP Biospectrum 310 imaging System.
Real-time PCR was performed with 20 ng cDNA and 3 µM primer pairs as triplicates on white 96-well PCR plates (Framestar, #4ti-0951). One reaction of 20 µL contained 10 µL PowerUp Sybr Green Kit (Life technologies, #A25742), 2.8 µL 3 µM primer pair, 4 µL cDNA and 3.2 µL nuclease-free water. The qPCR program ran for 20 s at 95°C for polymerase activation and for 40 cycles for 3 s at 95°C and for 30 s at 60°C, melting stage for one cycle for 15 s at 95°C, 1 min at 60°C, 15 s at 95°C continuous using the Light Cycler480 II (Roche).
Prior to the 96.96 High-Throughput qPCR Chip TR146 cells grown on 6-well plates in DMEM media were compared against biopsy samples on the transcription level by performing PCR or qPCR using housekeeping genes, tight junction proteins, transporter proteins, receptors, mucins, and aquaporins. Epithelial–mesenchymal transition markers, cornification proteins, and cytokeratin were verified as well. Table 1 lists targets, which were expressed in TR146 cells as well as in the biopsy samples and were analyzed by 96.96 High-Throughput qPCR Chip.
Table 1.
Overview of targets expressed by both the TR146 cell line and the biopsy samples upon PCR or real-time PCR showing nine different categories of markers. Abbreviations are explained below the table (all biopsy samples additionally expressed tight junction protein CLDN5, 5 out of 8 samples expressed MUC19).
| Endogenous controls | 18sRNA | PPIA | GAPDH | ß-actin | B2M |
|---|---|---|---|---|---|
| Claudins | CLDN1 | CLDN2 | CLDN3 | CLDN4 | CLDN6 |
| CLDN7 | CLDN8 | CLDN9 | CLDN10 tva | CLDN10 tvb | |
| CLDN11 | CLDN12 tv1 | CLDN12 tv2 | CLDN12 tv3 | CLDN14 | |
| CLDN15 | CLDN16 | CLDN17 | CLDN18 tv1b | CLDN18 tv2b | |
| CLDN20 | CLDN22 | CLDN24 | CLDN 25 | ||
| Other Tight Junctions | ZO-1 | ZO-2 | ZO-3 | JAM-1 | JAM-3 |
| occludin | tricellulin | ||||
| Transporter proteins | P-gp (ABCB1) | MRP1 (ABCC1) | MRP2 (ABCC2) | MRP3 (ABCC3) | MRP4 (ABCC4) |
| MRP5 (ABCC5) | BCRP (ABCG2) | CAT1 (SLC7A1) | GLUT1 (SLC2A1) | LAT1 (SLC5A7) | |
| MCT1 (SLC16A1) | MCT8 (SLC16A2) | ||||
| Receptors | TfR | InR | LRP1 | LRP8 | |
| EMT | E-cadherin | ß-catenin | vimentin | VEFG-A | fibronectin |
| Cornification | filaggrin | involucrin | loricrin | ||
| Cytokeratins | CK1 | CK4 | CK5 | CK8 | CK10 |
| CK13 tv1 | CK13 tv2 | CK14 | CK16 | CK18 | |
| CK19 | |||||
| Aquaporins | AQP1 | AQP3 | AQP4 | AQP7 | AQP9 |
| AQP10 | AQP11 | ||||
| Mucins | MUC1A | MUC1B | MUC1 tv9 | MUC2 | MUC3A |
| MUC4 | MUC7 | MUC13 | MUC15 tv1,3 | MUC15 tv2 | |
| MUC16 | MUC18 | MUC19 | MUC20 | MUC21 |
ABC … ATP-binding cassette, AQP … Aquaporin, B2M … β2-Microglobulin, BCRP … Breast Cancer Resistance Protein, CK … Cytokeratin, CLDN … Claudin, EMT … Epithelial–mesenchymal transition, GAPDH … Glyceraldehyde-3-phosphate dehydrogenase, InR … Insulin receptor, JAM … Junctional Adhesion Molecule, LRP …Low density lipoprotein (LDL) receptor-related protein, MCT … Monocarboxylate-transporter, MRP … Multidrug Resistance-Associated Protein, MUC … Mucin, P-gp … P-glycoprotein, PPIA … Peptidylprolyl isomerase A, SLC … Solute carrier, TfR … Transferrin receptor, tv … transcript variant, VEFG … Vascular endothelial growth factor, ZO … Zonula occludens.
2.6. 96.96 high-throughput qPCR-chip
For cDNA synthesis 350 ng RNA of TR146 samples cultivated on 24-well inserts and 1 µg of eight biopsy samples were transcribed with the Multiscribe Reverse Transcription Kit as described above. The eight biopsy samples were obtained from five females and three males in the age range 21–78. Subjects without prior history of transplanted tissue, tumor, radiation, or diseases affecting the oral cavity were selected to avoid any influence of pathologically altered epithelial cells. Biopsy samples were taken during cystectomies, removal of metal subjects or tooth and uni/bimaxillary corrective osteotomies. With regard to the biopsy samples, all methods were carried out in accordance with relevant guidelines and regulations, all experimental protocols were approved by the ethical committee of the University of Würzburg (Ethic number AZ-182/10, University of Würzburg, Germany) and informed consent was obtained from all subjects. Each sample was preamplified for 18 cycles as previously described15 with two different 10x primer pools containing 500 nM of 43 or 56 primers. The target list of each primer pool is shown in the supplementary Table S3 and includes primers representing nine different target groups (housekeeping genes as endogenous controls; cytokeratins, cornification, and EMT transition markers for characterization of epithelial cells; mucins and aquaporins to evaluate mucosal properties; tight junction proteins for verification of the paracellular barrier; transporter and receptors to evaluate the possibility of transcellular transport). The preamplification was performed with 15 min at 95°C for HotStarTaq Polymerase and 5 min at 95°C for HotStarTaq Plus Polymerase, 18 cycles with 40 s at 95°C, 40 s at 60°C and 1 min 20 s at 72°C and one cycle for 7 min at 72°C. Then, 1.5 µL of 1:8 diluted preamplified cDNA containing preamplified products of both primer pools were mixed with 4.5 µL DNA Mix, additionally, 10 µM dilutions of 96 selected targets were prepared, whereby 2.7 µL of each target was mixed with 3.3 µL Assay Mix as previously described.15 The 96.96 Dynamic Array IFC for Gene Expression (Fluidigm, BMK-M-96.96) was prepared according to the manufacturer´s instruction and primed with Control Line Fluid (Fluidigm, #89000021) prior to loading 5 µL of the prepared sample (as duplicates) and target solutions on the inlets. The 96.96 Dynamic Array IFC was accomplished with Biomark System running the program for 120 s at 50°C, 30 min at 70°C and 10 min at 25°C for thermal mix, 2 min at 50°C and 10 min at 95°C for activation, 40 cycles at 95°C for 15 s and 60 s at 60°C following a melting cycle at 60°C continuously with 1°C/3 s until 95°C was reached. Data were collected with the Biomark and EP1 Systems (Fluidigm) and mean values of ΔCt were normalized to the endogenous control (PPIA) using the 2∆Ct method and referred to a standardized condition for each experiment.
2.7. Western blot
Western blotting was performed as recently described in Gerhartl et al. (2019).16 In brief 20 µg protein was loaded on 10% SDS gels upon determining the protein concentration using the BCA assay. Primary antibodies [P-gp (ABCB1, clone C219, ENZO, #ALX-801-002, mouse, 1:30), MRP4 (ABCC4, clone M4I-80, ENZO, #ALX-801-039, rat, 1:100), BCRP (ABCG2, clone BXP-53, Abcam, #ab24115, rat, 1:100)] and secondary antibodies [anti-mouse IgG HRP-linked (Cell signaling, #7076, goat, 1:5000) or anti-rat IgG HRP-linked (Invitrogen, #61-9520, rabbit 1:5000)] were diluted in 5% dry milk. For the loading control, an anti-β-actin−peroxidase antibody was used (Sigma-Aldrich, #A3584, mouse, 1:25,000). Incubation with antibodies was described in detail previously.17 Images were recorded with the ChemiDoc Touch Imaging System (170–8370, Bio-Rad Laboratories Ges.m.b.H), analysis was performed with the Imagelab 5.2.1 software.
2.8. ABC transporter uptake assays
Functionality of P-glycoprotein (P-gp, ABCB1 – ATP-binding cassette sub-family B member 1), Multidrug Resistance-Associated Protein 4 (MRP4, ABCC4), and Breast Cancer Resistance Protein (BCRP, ABCG2) in TR146 were assessed by measuring the uptake of specific transporter substrates. For that, the cells were either cultivated in DMEM media and E1 on transparent 96-well plates (Greiner, #655180) until reaching confluency (day 9–11) after seeding at a density of 9.33 × 103 cells/cm2. Media exchange was performed every 2–3 days. The cells were washed twice with 200 µL Hank´s Balanced Salt Solution (HBSS, Sigma-Aldrich, #H6648) per well and incubated with 100 µL serum-free DMEM media or EpiLife media (E1) containing specific inhibitors (P-gp: 10 µM and 100 µM verapamil (100 mM stock in DMSO), 5 µM Ko143 (5 mM stock in DMSO), 10 µM MK571 (10 mM stock in DMSO); MRP4: 10 µM MK571; BCRP: 5 µM and 10 µM Ko143) for 15–30 min at 37°C. Stock solution of substrates (P-gp: 1 mM Calcein-AM in DMSO; MRP4: 2 mM fluo-cAMP in DMSO; BCRP: 500 µM Bodipy-FL-Prazosin in DMSO) were thawed in the dark shortly before usage and diluted in serum-free media. 100 µL of 2-folded substrate solutions (end concentrations: 1 µM Calcein-AM, 10 µM fluo-cAMP, 500 µM Bodipy-FL-Prazosin) were applied for uptake and incubated for 45–120 min at 37°C as described previously.18 For lysis, the cells were washed twice with 100 µL precooled 1% Triton X-100/HBSS and incubated for 1 h under agitation. Fluorescence was measured at 480/522 nm excitation/emission wavelength with the EnSpire Multimode Plate Reader (PerkinElmer). Protein content was measured with the Pierce BCA Protein Assay (ThermoFisher, #23227) at 562 nm wavelength. Functionality of the ABC transporters was determined after deducting control values (serum-free media) by calculating the ratio of RFU/protein concentration. To assess the uptake of substrate upon the inhibition, the ratio without additional inhibitor was set as 100% for comparison.
2.9. Data analysis
Arithmetic means and standard deviations (SD) or standard error of the means (SEM) for Figure 1(A,C) to provide a better overview were plotted using SigmaPlot Version 14.0 (Systat Software, San Jose, CA). Heatmaps of the 96 × 96 High-Throughput Chip were drawn with Qlucore Omics Explorer 3.6. Statistical analysis was performed with Student’s t-test, and one-way or two-way ANOVA for comparison of multiple groups followed by post hoc testing using Tukey’s test, Holm–Sidak test, or Dunn´s test with SigmaPlot Version 14.0 or R Core Team (2018) [R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/]. For the data analysis, the post hoc tests upon ANOVA were chosen as conservative as possible depending on the suitability of the data set. Significant values were set as p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) with α = 5%.
Figure 1.

(A) TEER value progressions of submerged and airlift cultivation in final EpiLife media (E3) shown as Ω x cm2 (mean ± SEM, n = 3–12 inserts from four independent experiments). Statistical analysis was performed with two-way ANOVA following post hoc Tukey's test, with α = 0.05, ***p < 0.001. (B) Permeability coefficient values [µm/min] of carboxyfluorescein on the last day of experiments after cultivation in final EpiLife media (E3) (mean ± SD, n = 8–9 inserts from three independent experiments). Statistical analysis was performed with Student’s t-test (airlift vs submerged), α = 0.05, ***p < 0.001. (C) TEER value progressions of submerged and airlift cultivation in DMEM media shown as Ω x cm2 over time (mean ± SEM, n = 3–18 inserts from six independent experiments). Statistical analysis was performed with two-way ANOVA following Holm–Sidak as post hoc test, with α = 0.05, *p <0.05, **p < 0.01 ***p < 0.001. (D) Permeability coefficient values [µm/min] of carboxyfluorescein on the last day of experiments after cultivation in DMEM media under submerged and airlift condition (mean ± SD, n = 15 inserts from five independent experiments).
3. Results
3.1. Optimization of paracellular barrier properties
3.1.1. Effect of airlift cultivation in comparison to submerged cultivation
As soon as the cells reached confluency in the final EpiLife media (E3) or DMEM media one part of the inserts was switched to airlift condition, while others stayed under submerged cultivation. For cells cultivated in final EpiLife media (E3) the highest TEER value for submerged cultivation was reached on day 43 with 61.94 ± 0.85 Ω x cm2 (mean ± SEM, n = 3), while airlift cultivation showed a significantly higher TEER value of 158.14 ± 9.34 Ω x cm2 (mean ± SEM, n = 3) on the same day in final EpiLife media (E3) (p < 0.001, shown in Figure 1(A)). In case of cultivation in DMEM the highest TEER value was measured on day 10 under submerged cultivation with 51.28 ± 1.18 Ω x cm2 (mean ± SEM, n = 18) and on day 29 for airlift cultivation with 45.92 ± 0.67 Ω x cm2 (mean ± SEM, n = 3) (Figure 1(C)).
These figures illustrated that airlift cultivation in final EpiLife media (E3) showed a steady increase leading to a maximum of 2.57-fold value on day 44 compared to submerged cultivation (p < 0.05). Airlift cultivation in DMEM media on the other hand showed only partly moderate ameliorations over short time periods compared to submerged condition reaching a maximum of TEER differences on day 31, but failed to reach a steady increase.
Measurement of carboxyfluorescein showed a significant lower permeability coefficient under airlift conditions in the final EpiLife media (E3) with 4.18 ± 0.75 µm/min (mean ± SD, n = 9, p < 0.01) compared to submerged cultivation with 12.12 ± 2.57 µm/min (mean ± SD, three independent experiments with n = 8). Under DMEM cultivation a permeability coefficient of 16.24 ± 3.26 µm/min (mean ± SD, five independent experiments with n = 3) for submerged cultivation and 13.38 ± 4.85 µm/min (mean ± SD, n = 15) for airlift cultivation was calculated (Figure 1(B,D)).
3.1.2. Effect of supplements in DMEM medium on barrier properties
As cultivation of TR146 in final EpiLife media (E3) showed enhancing effects on the paracellular tightness, it was investigated whether supplements for the final EpiLife media (E3) showed a similar effect in DMEM media. As soon as the cells seeded in DMEM media reached confluency, the media was supplemented with 1% HKGS or HKGS/KGF/A2P. Hydrocortisone (HC), a compound contained in HKGS was also tested in DMEM media by supplementing DMEM media with 10, 100 and 1000 nM HC. All set-ups with additional supplements were carried out under airlift condition and compared to cells cultivated under submerged and airlift in DMEM media.
Data showed that HC increased TEER in a concentration-dependent manner (displayed in Figure 2(A) as mean ± SD, n = 9). In detail, 10 nM HC led to a maximum increase of 1.82-fold (± 0.49, p < 0.001, equivalent to TEER values of 42.67 ± 6.95 Ω x cm2) on day 22, 100 nM HC to an increase of 3.47-fold (± 0.84, p < 0.001, 86.39 ± 20.15 Ω x cm2) on day 24 and 1000 nM HC to the highest increase of TEER value with 3.73-fold (± 1.03, p < 0.001, 99.75 ± 28.61 Ω x cm2) on day 24 in comparison to submerged cultivation in DMEM (Figure 2(A)). This concentration-dependent enhancement of the paracellular barrier by addition of HC was confirmed by a concentration-dependent decrease of the permeation of carboxyfluorescein. More precisely, 10 nM HC reduced the permeability coefficient significantly to 8.83 ± 3.52 µm/min, 100 nM HC to 4.78 ± 1.21 µm/min and 1000 nM HC to 3.80 ± 1.55 µm/min in comparison to the permeability coefficient of airlift and submerged cultivation of 15.77 ± 4.85 µm/min and 16.51 ± 2.85 µm/min (Figure 2(B) as mean ± SD, n = 9).
Figure 2.

Cultivation of TR146 cells on inserts in DMEM with supplements monitored with TEER and permeability of carboxyfluorescein (A) Progression of TEER values versus time as x-fold. Ω x cm2 values of submerged cultivation in DMEM media (●) was used as reference for airlift cultivation in DMEM media (○), supplemented with 10 (▼), 100 (∆) or 1000 nM (□) HC. Statistical analysis was performed as two-way ANOVA with Holm–Sidak test (mean ± SD, n = 9 inserts from three independent experiments, α = 0.05, *p < 0.05, **p < 0.01, ***p < 0.001). (B) Corresponding permeability coefficients of carboxyfluorescein on the last day of experiments (mean ± SD, n = 9 inserts from three independent experiments, one-way ANOVA, Tukey test) to 2A. (C) Comparison of submerged cultivation in DMEM (●) to airlift in DMEM (○), supplemented with 1% HKGS (▼) or HKGS/KGF/A2P (∆) (mean ± SD, n = 9–12 inserts from three independent experiments, two-way ANOVA with Holm–Sidak test). (D) Permeability values of carboxyfluorescein on the last day of experiments corresponding to 2C (mean ± SD, n = 8 inserts from two independent experiments, one-way ANOVA, Dunn´s test).
Supplementing DMEM with 1% HKGS led to a 4.64-fold (± 1.53, n = 12, p < 0.001, 112.17 ± 36.81 Ω x cm2) increase of TEER on day 22 compared to submerged cultivation in DMEM (23.82 ± 4.95 Ω x cm2, n = 12), while HKGS/KGF/A2P led to a maximum increase of 2.64-fold (± 0.52, n = 9, p < 0.001, 62.38 ± 13.50 Ω x cm2). (Figure 2(C)).
The permeation of carboxyfluorescein after cultivation in DMEM media supplemented with 1% HKGS was significantly decreased (4.78 ± 1.53 µm/min), similarly to the permeability coefficient value of DMEM supplemented with HKGS/KGF/A2P (3.90 ± 0.69 µm/min). No difference could be detected between submerged (17.58 ± 2.45 µm/min) and airlift (17.53 ± 5.02 µm/min) cultivation in DMEM media (Figure 2(D), shown as mean ± SD, n = 6).
The cultivation duration using DMEM media supplemented with HKGS was shortened by approximately 2 weeks and showed a highly significant increase in barrier tightness. Hence, DMEM media supplemented with 1% HKGS was termed as optimized media.
3.1.3. Effect of serum on paracellular barrier properties
To determine the influence of serum on the model, the cells were also cultivated in DMEM media without serum, supplemented with HKGS or HKGS/KGF/A2P. To visualize the effect of serum deprivation, the respective TEER values were normalized to cultivation with the optimized media (DMEM media containing 10% serum supplemented with 1% HKGS) as shown in Figure 3(A) Interestingly, DMEM without serum supplemented with HKGS/KGF/A2P reached a 2.74 ± 2.34-fold normalized TEER value (equivalent to 157.14 ± 84.24 Ω x cm2) on day 17. The corresponding permeability coefficient values of carboxyfluorescein seemed to be in the same range as the optimized media with 4.34 ± 0.90 µm/min for DMEM without serum, 4.93 ± 0.56 µm/min for supplementing with 1% HKGS and 3.30 ± 1.42 µm/min for supplementing with HKGS/KGF/A2P (Figure 3(B), mean ± SD, n = 3–6).
Figure 3.
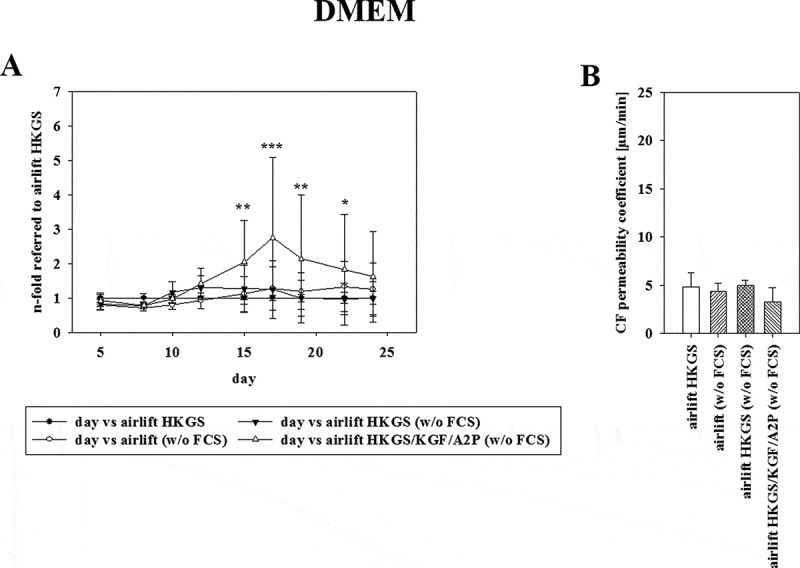
(A) Effect of serum deprivation on TEER using corresponding Ω x cm2 values of airlift cultivation in DMEM supplemented with HKGS (●) containing 10% serum as reference. DMEM without serum (○), supplemented with HKGS (▼) or HKGS/KGF/A2P (∆) was tested (mean ± SD, n = 3–6 inserts from three independent experiments, two-way ANOVA with Holm–Sidak test). (B) Permeability values of carboxyfluorescein on the last day of experiments corresponding to 3A (mean ± SD, n = 3–6 inserts from three independent experiments, one-way ANOVA).
3.2. High-throughput qPCR chip
For further comparison, the expression of selected markers of TR146 cultured on the Transwell model using various media supplements was compared against biopsy samples from the oral mucosa using a High-Throughput qPCR chip. Ninety-six targets of 9 target groups (endogenous control, tight junctions, transporter proteins, receptors, cytokeratins, cornification markers, epithelial–mesenchymal transition markers, aquaporins, and mucins) were tested for 47 TR146 samples, whereby for each analyzed sample cells from two inserts were pooled together. Twelve different cultivation conditions of TR146 were tested including submerged and airlift cultivation using DMEM media, airlift cultivation with DMEM media supplemented with 10, 100, or 1000 nM HC and HKGS or HKGS/KGF/A2P. The effect of serum-free media was tested under airlift with DMEM without FCS, supplemented with HKGS or HKGS/KGF/A2P next to cultivation with the serum-free final EpiLife media (E3) under airlift and submerged condition.
First analysis of ΔCt values, referring to PPIA as endogenous control, indicated most similar expression patterns of biopsy samples to TR146 samples cultivated under airlift conditions in optimized media (DMEM, supplemented with 10 % FCS, 1 % HKGS) or in final EpiLife media (E3) (supplementary Figure S1). Airlift cultivation in DMEM media supplemented with 10% FCS and 1% P/S was chosen as the reference condition for further data analysis. For a better overview, the results are divided into target groups shown in Figures 4 and 5, using the x-fold values upon referring to the reference condition. The corresponding values are shown as mean ± SD for the twelve cultivation conditions in supplementary Table S1. Expression values of biopsy samples normalized to reference condition of TR146 cultivated in DMEM media are shown in supplementary Table S2 (mean ± SD).
Figure 4.
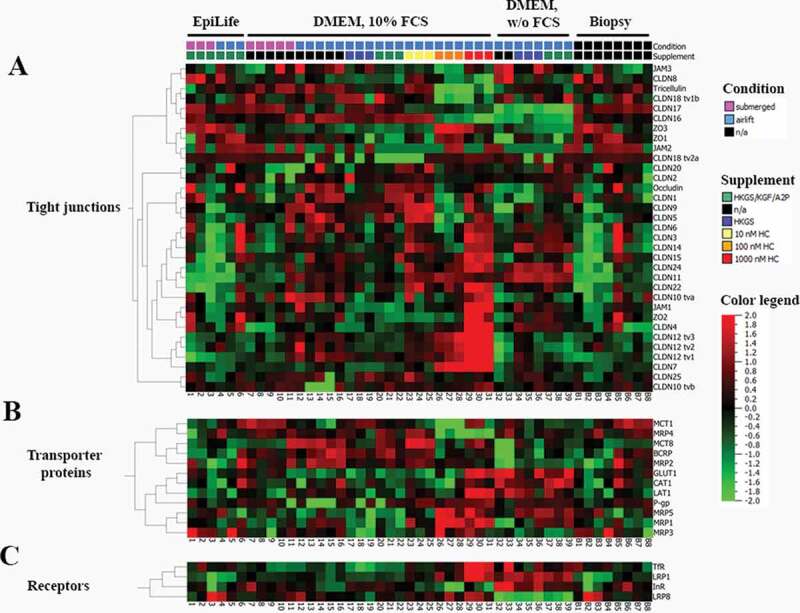
Expression values of (A) tight junction proteins, (B) transporter proteins, and (C,) receptors of the High-Throughput qPCR Chip shown as ∆Ct values, referred to PPIA (peptidylprolyl isomerase A) as endogenous control. TR146 samples [Samples 1–39, 1–3: cultivated in EpiLife media (E3), submerged; 4–6: EpiLife (E3), airlift; 7–11: DMEM media, submerged; 12–16: DMEM media, airlift; 17–19: DMEM media supplemented with 1% HKGS, airlift; 20–22: DMEM media supplemented with HKGS/KGF/A2P, airlift; 23–25: DMEM media supplemented with 10 nM HC, airlift; 26–28: DMEM media supplemented with 100 nM HC, airlift: 29–31: DMEM media supplemented with 1000 nM HC, airlift: 32–33: DMEM media w/o serum, airlift; 34–36: DMEM media w/o serum supplemented with 1% HKGS, airlift: 36–39: DMEM media w/o serum supplemented with HKGS/KGF/A2P, airlift] plotted against 8 biopsy samples (B1-8) of the oral mucosa.
Figure 5.
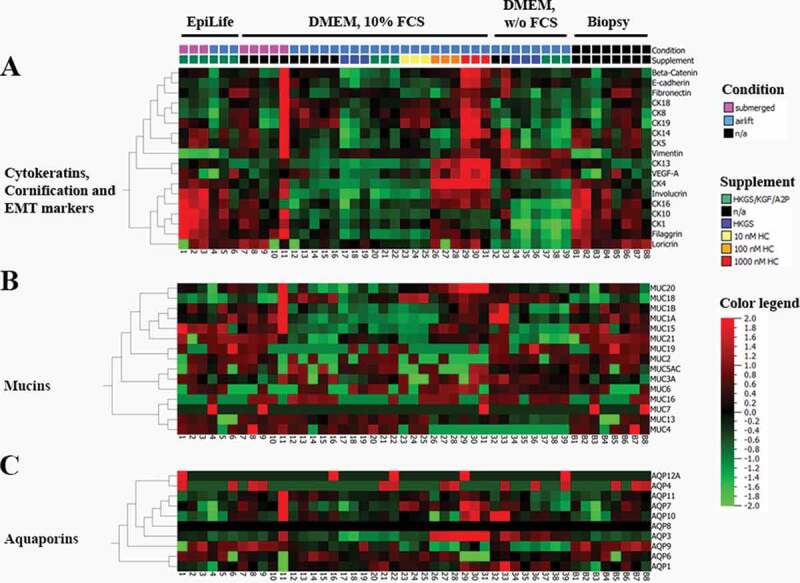
Expression values of (A) cytokeratins, cornification, and EMT (epithelial–mesenchymal transition) markers (B) mucins and (C) aquaporins of the High-Throughput qPCR Chip shown as ∆Ct values, referred to PPIA (peptidylprolyl isomerase A) as endogenous control. TR146 samples [Samples 1–39, 1–3: cultivated in EpiLife media (E3), submerged; 4–6: EpiLife (E3), airlift; 7–11: DMEM media, submerged; 12–16: DMEM media, airlift; 17–19: DMEM media supplemented with 1% HKGS, airlift; 20–22: DMEM media supplemented with HKGS/KGF/A2P, airlift; 23–25: DMEM media supplemented with 10 nM HC, airlift; 26–28: DMEM media supplemented with 100 nM HC, airlift: 29–31: DMEM media supplemented with 1000 nM HC, airlift: 32–33: DMEM media w/o serum, airlift; 34–36: DMEM media w/o serum supplemented with 1% HKGS, airlift: 36–39: DMEM media w/o serum supplemented with HKGS/KGF/A2P, airlift] plotted against 8 biopsy samples (B1-8) of the oral mucosa.
Thus, airlift cultivation in DMEM media supplemented with 10% FCS and 1% Pen/Strep was chosen as the reference condition for further data analysis. For a better overview, the results are shown divided into target groups in Figure 4, using the x-fold values upon referring to the reference condition. The corresponding values are shown as mean ± SD, in the supplementary file, Table S1 and the expression values of biopsy samples are shown in the supplementary Table S2.
Targets showing a significant change in expression (*p < 0.05, **p < 0.01, ***p < 0.001) were classified in groups and described in detail below as expression values normalized to the reference condition.
3.2.1. Tight junctions
Twenty-one different claudins, ZO1-ZO3, JAM1-JAM3, and tricellulin were tested as tight junction markers (Figure 4(A)). CLDN19, CLDN21, and CLDN23 were not tested in this set-up, as TR146 showed no expression in preliminary results. Cultivation in final EpiLife media (E3) under airlift resulted in the significant lowest expression of all three transcript variants of CLDN12 with tv1 showing a 0.29-fold (± 0.0059, n = 3), tv2 a 0.46-fold (± 0.0045, n = 3) and tv3 a 0.27-fold (± 0.0038, n = 3) expression in comparison to other cultivation conditions in DMEM media (see supplementary file, Table S1). Airlift cultivation in final EpiLife media (E3) also led to a significant downregulation of CLDN15 (0.41 ± 0.07, n = 3), CLDN22 (0.34 ± 0.14, n = 3) and JAM1 (0.47 ± 0.052, n = 3) in comparison to cultivation in DMEM media but showed a significantly increased expression of tricellulin (1.65 ± 0.13, n = 3). Submerged cultivation in final EpiLife media (E3) led to a similar regulation by showing significantly lower expression values of two transcript variants of CLDN12 (tv2: 0.65 ± 0.12, n = 3 and tv3: 0.48 ± 0.15, n = 3) and by achieving the significantly highest expression of tricellulin of 2.02-fold (± 0.12, n = 3) of all 12 tested cultivation conditions.
Submerged cultivation in DMEM media led to a significantly increased expression of CLDN11 (4.81 ± 1.94, n = 5), CLDN12 tv1 (1.6 ± 0.54, n = 5), CLDN15 (2.07 ± 0.9, n = 5) and CLDN24 (2.17 ± 0.69, n = 5). Airlift cultivation in DMEM media supplemented with HKGS showed a decreased expression of CLDN12 tv2 (0.61 ± 0.23, n = 2), indicating that HKGS is involved in the downregulation of CLDN12 in final EpiLife media (E3) and DMEM media. Supplementing DMEM with 10 nM HC under airlift led to a significantly increased expression of JAM1 of 1.2-fold (± 0.07, n = 3), while 100 nM HC even showed a 1.27-fold increase (± 0.049, n = 3). However, DMEM media deprived of serum and supplemented with HKGS/KGF/A2P led to a significantly higher expression of CLDN12 tv2 of 1.2-fold (± 0.13, n = 2), showing a supplement dependent regulation of CLDN12 and its transcript variants.
3.2.2. Transporter proteins
Only few out of the 12 tested transporter proteins showed a significant cultivation dependent regulation (Figure 4(B)). Under the reference cultivation condition, DMEM media (= DMEM, supplemented with 10% FCS, 1% P/S) under airlift, TR146 expressed MCT8 and GLUT1 significantly higher compared to other cultivation conditions, indicating that airlift cultivation in DMEM media induced the expression of transporter proteins in contrast to airlift cultivation in the final EpiLife media (E3). In this context, airlift cultivation in final EpiLife media (E3) led to the lowest expression of LAT1 (0.38 ± 0.045, n = 3) and GLUT1 (0.53 ± 0.21, n = 3) of all cultivation conditions. Submerged cultivation in final EpiLife media (E3) on the other hand led to one of the highest expression values of MCT8 with 0.93-fold (± 0.24, n = 3) as the transporter protein was weakly expressed by the majority of the tested conditions. MCT1 was also significantly upregulated upon submerged cultivation in E3 with a 2.69-fold (± 0.15, n = 3) expression. Due to the weak expression of MCT8 over all cultivation conditions, submerged cultivation in DMEM media showed one of the highest expression values with 0.94-fold (± 0.2, n = 5). Interestingly, cultivation of TR146 in DMEM media under submerged condition led to the significantly highest expression of BCRP (3.13-fold ±1.29, n = 5).
3.2.3. Receptors
Supplement- and media-specific regulation effects were observed for LRP1, LRP8, and TfR (Figure 4(C)). Cultivation in final EpiLife media (E3) under airlift led to the significantly lowest expression of LRP1 (0.16 ± 0.1, n = 3) and TfR (0.33 ± 0.12, n = 3) over all tested cultivation conditions. While neither submerged cultivation in final EpiLife (E3) or DMEM media showed a significant regulation of the receptors, DMEM media supplemented with 10 nM HC under airlift showed the highest significant expression of TfR (1.37 ± 0.2, n = 3) for all tested cultivation conditions. Deprivation of serum in DMEM media, supplemented with HKGS even led to the highest expression of LRP8 of 2.4-fold (± 0.42, n = 2), whereby supplementing the serum-free DMEM with HKGS/KGF/A2P also showed a significant increase of LRP8 (2.23 ± 0.52, n = 2).
3.2.4. Cytokeratins
From the tested cytokeratins, CK1, CK13, and CK18 were significantly regulated dependent on the different model set-ups (Figure 5(A)). Airlift cultivation in final EpiLife media (E3) showed the highest decrease in CK13 with an expression value of 0.0042-fold (± 0.0023, n = 3). Submerged cultivation in DMEM led to a lower expression of CK1 (i.e., 0.26 ± 0.15-fold, n = 5), but showed a highly significant increased expression of CK18 of 1.93-fold (± 0.44, n = 5). Interestingly, addition of hydrocortisone revealed a concentration-dependent inverse regulation. Supplementing 10 nM HC to DMEM media under airlift led to a significantly increased expression of CK18 of 1.35-fold (± 0.43, n = 3), whereby addition of 100 nM HC only showed a 0.9-fold (± 0.33, n = 3) and 1000 nM HC a 0.53-fold (± 0.33, n = 3) expression (see supplementary file, Table S1).
3.2.5. EMT-epithelial mesenchymal transition
Five markers for epithelial and mesenchymal transition were tested, of which E-cadherin and vimentin showed a significant regulation (Figure 5(A)). Airlift cultivation in final EpiLife media (E3) showed the significantly lowest expression of E-cadherin (0.34 ± 0.055, n = 3). On the other hand, submerged cultivation in DMEM media led to the highest expression of vimentin with a 7.79-fold expression (± 4.88, n = 5) (see supplementary file, Table S1) in comparison to the airlift cultivation in DMEM with 10% FBS and 1% P/S.
3.2.6. Mucins
Various mucin markers were tested from which MUC13, MUC15, and MUC21 showed a significant regulation upon cultivation in DMEM media supplemented with hydrocortisone or other supplements containing hydrocortisone and also upon cultivation in DMEM media deprived of serum (Figure 5(B)). Supplementing HKGS/KGF/A2P to DMEM media under airlift led to one of the highest upregulations for a marker by 725.6-fold (± 84.07, n = 2) for MUC21. Addition of 100 nM HC to DMEM media under airlift showed an upregulation of MUC15 (9.21 ± 3.05, n = 3) and MUC21 (21.63 ± 6.95, n = 3). Supplementing 1000 nM HC to DMEM media under airlift showed a similar effect with a 10.97-fold (± 1.04, n = 3) expression for MUC15 and a 218.85-fold (± 51.66, n = 3) expression for MUC21. This suggested a correlation between the concentration of supplemented hydrocortisone and the expression of MUC21.
Serum-free cultivation in DMEM supplemented with HKGS also showed a significant upregulation of MUC15 (10.03 ± 5.38, n = 2) and MUC21 (320.97 ± 28.24, n = 2). Supplementing the serum-free DMEM media with HKGS/KGF/A2P even showed higher significant upregulations for MUC15 (13.06 ± 1.33, n = 2) and MUC21 (1292.78 ± 323.49, n = 2) and also led to an increased expression of MUC13 of 21.02-fold (± 11.83, n = 2).
3.2.7. Aquaporins
Out of the 10 tested aquaporins, AQP4, AQP8, and AQP12A showed very weak or no expression across all cultivation conditions, whereby others showed a significant regulation (Figure 5(C)). Submerged cultivation in final EpiLife media (E3) led to the highest increase of all tested aquaporins for AQP11 (113.05 ± 33.45-fold, n = 3). On the other hand, submerged cultivation in DMEM led to an increased expression of AQP1 (2.24 ± 0.92-fold, n = 5). Airlift cultivation in DMEM supplemented with 10 nM HC showed a significantly increased expression of 2.61-fold (± 0.23, n = 3) for AQP3, whereby a significant downregulation upon supplementing DMEM with HKGS was reached for AQP11 (0.22 ± 0.31, n = 2). As the aquaporins were weakly or irregularly expressed (see supplementary file, Table S1), no favorable cultivation condition or supplement could be defined.
In order to compare the optimized Transwell TR146 model with the biopsy samples, Table 2 gives an overview of the highest expressed targets in the respective marker panels. Twelve of 14 targets revealed no significant difference (tight junction markers: CLDN1, CLDN4, CLDN7, JAM1; cytokeratins: CK4, CK5, CK10, CK14, CK18; cornification marker involucrin, aquaporin AQP3; mucin MUC21) whereas transporter protein LAT1 was significantly higher and CK13 was significantly lower expressed.
Table 2.
Comparison of highly expressed targets in human oral biopsy samples (n = 8) versus TR146 cells cultivated under optimized conditions (DMEM media, supplemented with HKGS, airlift, n = 3, final TEER of 115.39 ± 38.48 Ω x cm2). Ct values were obtained with the 96.96 High-Throughput qPCR-Chip, referred to PPIA (peptidylprolyl isomerase A) as housekeeping gene, normalized to the expression values of biopsy samples and listed as x-fold values (mean ± SEM). Statistical analysis was performed using the Student´s t-test with α = 0.05, *p <0.05, **p < 0.01, ***p < 0.001.
| Target groups | Targets | TR146 | Biopsies | p-value |
|---|---|---|---|---|
| Tight Junctions | CLDN1 | 1.69 ± 0.23 | 1 ± 0.42 | |
| CLDN4 | 0.81 ± 0.08 | 1 ± 0.22 | ||
| CLDN7 | 0.76 ± 0.07 | 1 ± 0.40 | ||
| JAM1 | 1.61 ± 0.22 | 1 ± 0.16 | ||
| Transporters | LAT1 | 4.57 ± 0.38 | 1 ± 0.21 | ** |
| Cytokeratin | CK4 | 0.08 ± 0.05 | 1 ± 0.59 | |
| CK5 | 0.47 ± 0.21 | 1 ± 0.18 | ||
| CK10 | 2.58 ± 1.46 | 1 ± 0.36 | ||
| CK13 | 0.11 ± 0.05 | 1 ± 0.20 | ** | |
| CK14 | 0.52 ± 0.14 | 1 ± 0.20 | ||
| CK18 | 1.42 ± 0.77 | 1 ± 0.42 | ||
| Cornification | Involucrin | 3.99 ± 1.75 | 1 ± 0.23 | |
| Aquaporins | AQP3 | 0.63 ± 0.26 | 1 ± 0.17 | |
| Mucins | MUC21 | 0.003 ± 0.002 | 1 ± 0.56 |
3.3. Functionality of ABC transporters
Since ABC transporters regulate the transport of a broad range of drugs, and their presence in our models was confirmed on the mRNA and protein level (Figures 4(B) and 6), we tested the functionality of three selected ABC transporters in cell line TR146. The assays were performed with cell layers cultivated either in EpiLife media (E1) or DMEM media to assess the functionality of P-gp (ABCB1), MRP4 (ABCC4), or BCRP (ABCG2). The results are shown below in Table 3 as mean ± SD. Addition of 100 µM P-gp inhibitor verapamil increased the uptake of the P-gp substrate Calcein-AM (1.5 ± 0.16, n = 4, p < 0.001 for DMEM, 1.45 ± 0.26, n = 5, p < 0.001 for EpiLife) significantly, whereas 10 µM verapamil, 5 µM Ko143 or 10 µM MK571 did not change the uptake of 1 µM Calcein-AM in comparison to the control. With regard to MRP4, 10 µM MK571 increased the uptake of MRP4 substrate fluo-cAMP (10 µM) 1.46-fold (± 0.58, n = 4) in DMEM and 1.35-fold (± 0.24, p < 0.01, n = 4) in EpiLife media (E1) in comparison to the control. While 5 µM and 10 µM Ko143 did not show an inhibitory effect on BCRP in DMEM, a decreased uptake of Bodipy-FL-Prazosin was measured upon addition of the inhibitor in EpiLife media (E1). In summary, efflux activity of P-gp and MRP4 was proven for TR146 cells.
Figure 6.
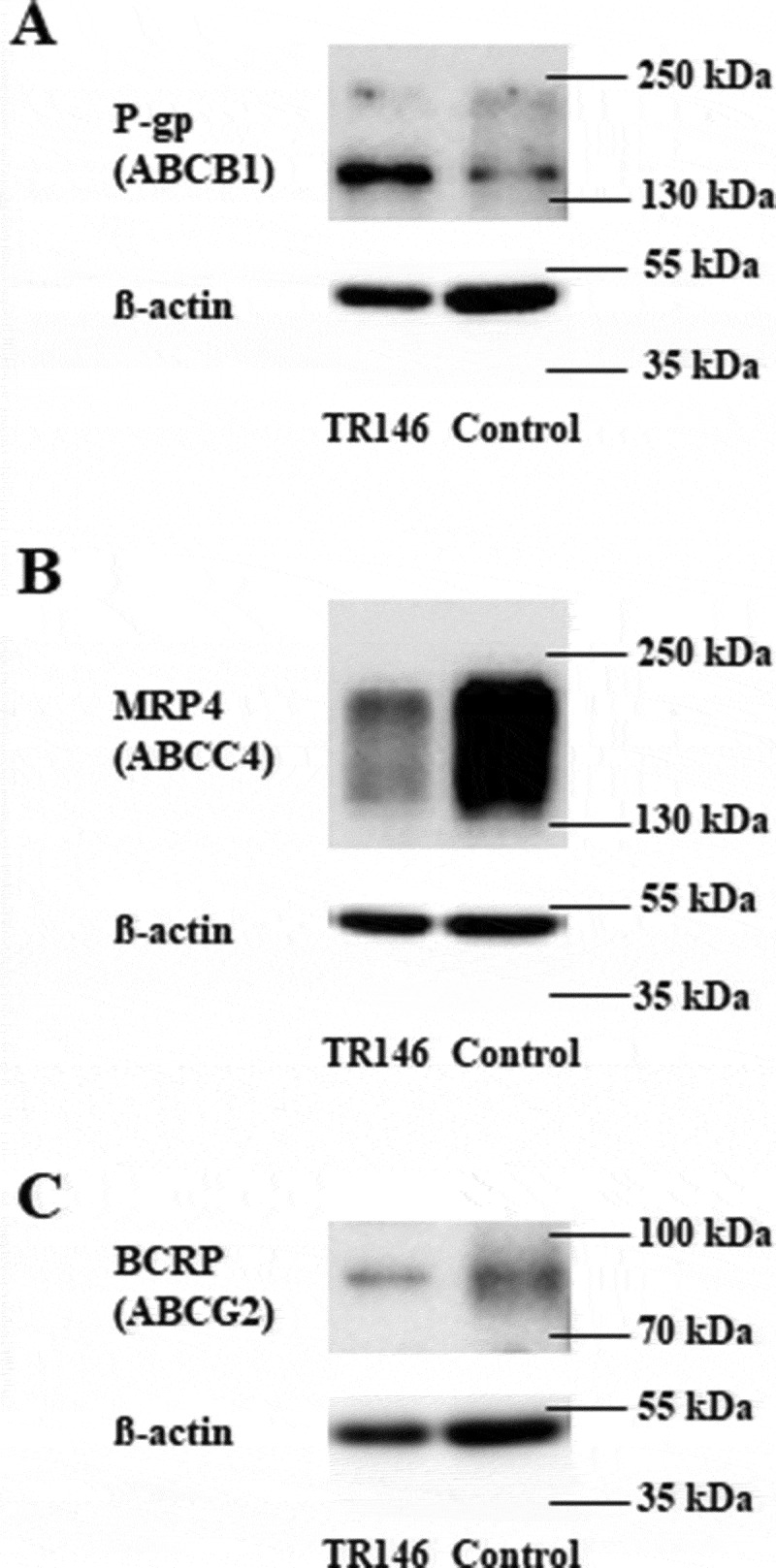
Representative western blots of TR146 cells cultivated in optimized media under airlift conditions in comparison to a control sample (immortalized mouse capillary endothelial cell line cerebEND cultivated as previously published18) showing the expression of ABC transporters (A) P -gp (ABCB1, 170 kDa), (B) MRP4 (ABCC4, 140–200 kDa), (C) BCRP (ABCG2, 72-75 kDa); corresponding ß-actin (42 kDa) was used as loading control.
Table 3.
Results of ABC transporter functionality tests in TR146 cells. For each assay at least 6 values were obtained for each condition (control, control with fluorescent substrate, samples of fluorescent substrate with or without inhibitor). The calculated ratios of fluorescent signal to protein content were normalized to samples only exposed to the fluorescent substrate (set as 100%). An active inhibition is noticeable by a significantly increased uptake of the substrate (>100%). Values are shown for both DMEM and EpiLife media as mean ± SD. Data were analyzed with Student’s t-test or with one-way ANOVA with post hoc comparison using Dunn´s method and Holm–Sidak test for multigroup comparison, **p < 0.01, ***p < 0.001, SC = statistical significance, n/a = not analyzed, n.s. = not significant, n = number of independently conducted experiments.
| DMEM media |
EpiLife media |
|||||||
|---|---|---|---|---|---|---|---|---|
| ABC transporter | Substrate | Inhibitor | mean ± SD [%] | SC | mean ± SD [%] | SC | ||
| P-gp (ABCB1) | Calcein-AM [1 µM] | - | 100.00 ± 3.95 | n/a | n = 4 | 100.00 ± 7.55 | n/a | n = 5 |
| Calcein-AM [1 µM] | Verapamil [100 µM] | 150.67 ± 16.26 | *** | n = 4 | 144.65 ± 26.26 | *** | n = 5 | |
| Calcein-AM [1 µM] | Verapamil [10 µM] | 107.84 ± 7.39 | n.s. | n = 4 | Not tested | |||
| Calcein-AM [1 µM] | Ko143 [5 µM] | 99.95 ± 6.80 | n.s. | n = 2 | Not tested | |||
| Calcein-AM [1 µM] | MK571 [10 µM] | 112.63 ± 9.40 | n.s. | n = 2 | Not tested | |||
| MRP4 (ABCC4) | Fluo-cAMP [10 µM] | - | 100.00 ± 24.48 | n/a | n = 4 | 100.00 ± 4.68 | n/a | n = 4 |
| Fluo-cAMP [10 µM] | MK571 [10 µM] | 145.66 ± 57.74 | n.s. | n = 4 | 134.84 ± 23.91 | ** | n = 4 | |
| BCRP (ABCG2) | Bodipy-FL-Prazosin [0.5 µM] | - | 100.00 ± 7.31 | n/a | n = 8 | 100.00 ± 5.13 | n/a | n = 8 |
| Bodipy-FL-Prazosin [0.5 µM] | Ko143 [5 µM] | 96.94 ± 11.53 | n.s. | n = 4 | 87.92 ± 7.53 | *** | n = 4 | |
| Bodipy-FL-Prazosin [0.5 µM] | Ko143 [10 µM] | 96.99 ± 23.03 | n.s. | n = 3 | 81.55 ± 10.96 | *** | n = 3 | |
4. Discussion
Even though over 50 tumor-derived cell lines of the oral mucosa have already been described, the majority of those is used for applications such as cytotoxicity testing, drug sensitivity, or migration studies as reported by Bierbaumer et al. recently.19 The oral mucosa is built by three different kinds of mucosa depending on the location (keratinized: gingiva, non-keratinized: buccal, specialized: tongue). Several cancer cell lines or immortalized cell lines from the oral cavity have been decribed previously. 20–24 However, most of the commonly used cell lines originate from the floor of the mouth or from the gingival area, and a standardized cell line for the buccal mucosa is still missing. Even though primary models of the buccal mucosa are commercially available, they are mainly used for disease modeling or viability tests upon short-time exposure of certain molecules.25,26
EpiSkin commercialized the buccal carcinoma cell line TR146 as an airlift model described as RHE, reconstituted human epithelium (SkinEthic). However, the cultivation conditions are not published for how to arrive at the differentiation status of this model and it has mainly been used for infection studies with candida strains.27,28 Hence, an extensive characterization of the model for transport studies of biomarkers over a longer time period is missing. Thus, our aim was to develop a human cell-based model of the oral mucosa based on TR146 and optimize it for transport studies for biomarkers. Since most molecules are assumed to pass from blood to saliva using the paracellular transport route, paracellular integrity is of high importance for the model.29,30 In addition, strong tight junctions enhance cell polarization and transporter localization either to the apical or basolateral side.
TR146 has already been described with regards to its paracellular barrier integrity by TEER measurements and permeability assays with FITC-dextran previously.8,9,11,31 In those studies, the cells were cultivated in DMEM with supplements (serum with P/S or serum with gentamicin and p- hydroxybenzoic acid n-butyl) on Transwell inserts in a 6- or 12-well format (Becton Dickinson (BD) or Corning Costar). Results of these studies were quite diverse and not conclusive on which insert type the highest paracellular barrier could be reached. For example, Portero et al. (2002) published TEER values of 339 ± 89 Ω x cm2 after cultivating for 28–30 days using inserts from BD in a 12-well format, while Teubl et al. (2013) only reached TEER values of 50.02 ± 2.87 Ω x cm2 after 27–28 days using inserts from Corning Costar inserts in the same format.32,33 Therefore, we tested several 24-well inserts from different companies (BD, Corning Costar, GreinerBioOne, Brand) during preliminary studies. Since TR146 layers on Greiner ThinCert showed the highest TEER values in DMEM media under submerged cultivation (data not shown), it was decided to use these for further studies. For the next experiments, two different media (i.e., EpiLife and DMEM) and two experimental set-ups (i.e., airlift and submerged) were tested. Airlift cultivation of TR146 in final EpiLife media (E3) showed a continuous increase of TEER, whereas cultivation in DMEM media led to a smaller increase of TEER values. However, even though the HE stainings showed multilayered cells as a result of airlift cultivation in DMEM media (supplementary Figure S2C), the corresponding permeability coefficient for carboxyfluorescein did not indicate a tighter paracellular barrier. These results coincided with Jacobsen et al. (1999), who showed that airlift cultivation did not enhance the integrity of the paracellular barrier of TR146 using DMEM.11 Thus, it was hypothesized that the supplements of the final EpiLife media (E3) might be necessary to induce an increase of the tightness during airlift cultivation.
Supplementing HKGS to DMEM under airlift showed a twofold higher effect compared to EpiLife under airlift and reduced the cultivation period by 2 weeks. HKGS contains human epidermal growth factor (EGF) and hydrocortisone (HC) among other substances, potentially having an influence on the differentiation. EGF has shown to enhance differentiation of other epidermal barriers,34,35 while HC is known for its promoting effects on the tightness of brain endothelial cells.36-38 Addition of hydrocortisone alone at a physiological concentration to DMEM media (supplemented with 10% FCS, 1% P/S) improved the barrier upon airlift cultivation significantly in a concentration-dependent manner.39,40
As further supplement, we tested retinoic acid (RA) in DMEM without serum as RA is often described to enhance the barrier.41 However, RA did not show any enhancement in our model (supplementary Figure S2). In concordance to this, Reiss et al. (1985) discovered RA to be an inhibitor of terminal differentiation for cell line SqCC/Y1 derived from a squamous carcinoma of the buccal mucosa.42
Deprivation of serum and supplementing DMEM with HKGS, KGF, and A2P under airlift led to the highest increase of TEER values and accordingly to the lowest permeability coefficient of all tested media compositions. However, the TEER values showed very high variances, probably caused by the non-stratified morphological phenotype typical for deprivation of serum using this cell line (see supplementary Figure S3). Therefore, we have defined the DMEM media supplemented with 1% HKGS and containing 10% serum as the optimized media.
The 96.96 High-Throughput qPCR Chip data provided the possibility to investigate the influence of the different growth media treatments at a molecular level. Data of TR146 cells cultivated in different set-ups revealed significant changes in mRNA expression in the target groups dependent on the supplements or cultivation conditions (supplementary file, Table 1S).
One major target group included tight junction proteins that define the integrity of paracellular barriers and are composed of a network of transmembrane proteins (claudins, occludin, tricellulin, JAM), linked to intracellular scaffolding proteins such as ZO.43-46 However, only sparse information is available about the expression of this target group, e.g. regarding the expression of claudins in oral mucosa epithelial cells. In case of CLDN1, it was shown to be expressed in squamous cell carcinoma of the oral cavity and was linked to the advanced disease stage.47,48 However, no regulation upon supplementation was detected in our TR146 set-ups. Other tight junction proteins such as CLDN11 and CLDN24 were higher expressed under submerged conditions in DMEM media compared to airlift cultivation. Anyhow, since no data about expression of CLDN11 and CLDN24 are available in the oral epithelium, further studies are needed to evaluate their role in the oral mucosa.
Interestingly, cultivation in final EpiLife media under airlift led to a reduced mRNA expression of the majority of tested tight junction markers after normalization to airlift cultivation of TR146 in DMEM, even though the Epilife media was successfully used for a model based on primary cells or for 3D modeling of oral mucosa constructs.49,50 Results of the High-Throughput qPCR Chip indicated similarity of biopsy samples to TR146 cells cultivated under airlift conditions in DMEM media supplemented with HKGS. Using HKGS as a supplement also showed to be of advantage regarding the permeability studies as it led to a significant improvement of the integrity of the paracellular barrier.
In order to confirm the presence of continuous tight junctions, the localization of tight junction marker occludin was morphologically characterized using immunofluorescent staining on microscope slides (see supplementary Figure S4A). To confirm the presence of cytokeratins, differentiation markers for epithelial cells, at the protein level additionally to the mRNA level, immunofluorescent staining (K5/K8) was accomplished on microscope slides and inserts (see supplementary Figure S4A-B). In general, HE stainings showed a stratified non-keratinized epithelium, which corresponds to the phenotype of the cell line of the buccal region. Interestingly, the cornification markers (loricrin > filaggrin > involucrin) showed a high upregulation under the presence of hydrocortisone or other supplement compositions containing hydrocortisone such as HKGS and HKGS/KGF/A2P. This confirmed the enhanced differentiation upon hydrocortisone in epithelial cell layers, which was in concordance to higher TEER values of the accordingly treated TR146 inserts. In the oral epithelium, desmosomes play a pivotal role as intercellular junctions.51 Although one focus of the presented study is the comprehensive investigation of the expression of tight junction proteins, the expression of desmoglein 3, a desmosome formation facilitating protein, was evaluated with conventional qPCR (see supplementary Figure S5). Interestingly, DMEM media supplemented with 1000 nM hydrocortisone, HKGS (the optimized media) and with HKGS/KGF/A2P showed an upregulation to a similar scale compared to the biopsy samples of healthy donors.
The defensive role and expression of mucin glycoproteins in the oral cavity have been described before.52 High-throughput qPCR data revealed that some of the mucins (MUC6, MUC16, and MUC19) were only detected in half of the TR146 samples, while MUC7 was only found in four of the tested samples. In this regard, the oral cavity was described to express MUC7 and MUC19 in general,53 but MUC19 was not found in primary oral mucosal epithelial cells54 and MUC7 (important for bacterial clearance in saliva) was only weakly expressed in mucoepidermoid carcinoma.55 To our best knowledge, the presence of MUC6 has not been shown in the oral mucosa until now, whereby the expression of MUC16 in saliva samples of healthy patients and patients with oral squamous carcinoma showed no correlation to the prognosis as described by Suh et al. (2017).56 On the other hand, all samples expressed MUC1A, MUC1B, MUC3A, MUC15, MUC18, and MUC20. The expression of MUC1 in the buccal area and salivary glands has been described before.57-59 MUC3 has shown to be upregulated in mucosal tissue (intestine) in a stress response,60 but no data about expression in the oral mucosa has been described yet. MUC15 was shown to be expressed in explants of human oral mucosal epithelial cells.61 The cell surface glycoprotein MUC18, also described as MCAM (melanoma cell adhesion molecule),62 has also shown to correlate with epithelial carcinoma or bacterial infections in various epithelial tissues.63,64 Even though MUC20 has been expressed in corneal, respiratory epithelia and in esophageal squamous cell carcinoma, no data about the expression in the buccal mucosa was found.65-67 Interestingly a strong regulation of MUC21 in correlation with increasing hydrocortisone (>300 fold) was shown in our model. MUC21 was identified in 2008 by Itoh et al. as a membrane bound protein.68 An upregulation of over 30-fold of MUC21 was described during surgical wound healing of the gingiva upon a study with 10 volunteers69 suggesting a similar underlying pathway for MUC21 during wound healing and treatment with hydrocortisone.
Aquaporins (AQP), integral membrane proteins forming water channels for rapid fluid transport, are very strongly expressed by, e.g., the salivary glands. Therefore, characterization of aquaporins is mostly referred to mouse, rat, or human salivary glands, but a thorough characterization for the human oral mucosa is still missing.70 In our oral mucosa model, we demonstrated the expression of AQP1, AQP3, AQP6, AQP7, AQP9, AQP10, and AQP11. The biopsy samples of the human oral mucosa showed a similar expression pattern as our model. A consistent expression for AQP1, AQP3, AQP7, AQP9, AQP10, and AQP11, a weak expression for AQP4 and no expression for AQP6, AQP8 and AQP12A was obtained. This confirms the similarity of our cell line-based model to the human model. Even though the expression of aquaporins was mostly described in the salivary glands, AQP3 and AQP9 were also found in rat buccal mucosa epithelium at the mRNA and protein level. Moreover, a correlation was suggested between AQP3 localization and the differentiation of keratinocytes.71 To understand the expression of aquaporins in our human buccal mucosa model, further investigations on their functionality in the oral mucosa are necessary.
ABC transporters play a pivotal role in the transport of physiological substrates and efflux of drugs.72 From the 48 ABC transporters proteins nine are involved in multidrug resistance transport (P-glycoprotein, seven multidrug resistance-associated proteins and Breast Cancer Resistance Protein) with the ability to extrude a large variety of xenobiotics.73 For the first time, we were able to show the functionality of P-gp and MRP4 in TR146 cells. The expression of BCRP was confirmed at the mRNA level (see Table 1), but no BCRP efflux activity was detectable in TR146 cells. This corresponded well with the relatively low BCRP protein expression compared to the control cell line (Figure 6). With P-gp being one of the most investigated efflux transporters,74 the established TR146 model could be used to test the role of this ABC transporter on the pharmacokinetics of oral drugs, but also for biomarkers. For example, P-gp was described to actively transport the steroid hormone cortisol, a biomarker often used to evaluate the stress level of patients.75-77 Studies showed a correlation between the concentrations of cortisol in serum and saliva, with one-tenth of the serum concentration detected in saliva. Assuming that a maximum 1% of blood molecules can be detected in saliva due to commingling of saliva with the crevicular gingival fluid, the cortisol data suggested a directed transport of cortisol from blood into saliva via the blood-saliva barrier.30,78-80 With the uprising use of saliva for biomarker detection in the past 60 years, several biomarkers have been investigated and described in saliva for diagnosis purposes. For example, C-reactive protein (CRP) has commonly been used as a marker for systemic inflammation in serum and there is a significant number of studies suggesting applying salivary CRP as a noninvasive biomarker. In this context, Iyengar et al. (2014) described a correlation between serum and salivary concentration of CRP in neonates, offering a stress-free detection method of abnormal serum levels for newborns.81 Especially, high CRP concentrations in saliva under disease conditions suggested active transport from blood into saliva by yet unknown transport mechanisms across the blood-saliva barrier. But there are also several studies that did not show sufficient correlations between serum and salivary CRP underlining the need for a better understanding of the origin of the measured CRP in saliva, the degradation, and clearance half-life of CRP in saliva and the influence of the oral microenvironment on salivary CRP concentration.74
In summary, the transport mechanisms of biomarkers from blood to saliva are often not understood emphasizing the necessity of validated and optimized models representing the blood-saliva barrier in order to elucidate if and how these biomarkers can permeate from blood into saliva.
5. Conclusion
We showed that a Transwell model based on human buccal carcinoma cell line TR146 displayed improved barrier properties upon airlift cultivation. Different supplements (hydrocortisone, HKGS, KGF, A2P, FCS) and their influence on the barrier properties and morphology were tested and comprehensively characterized by TEER measurement and the assessment of carboxyfluorescein permeation, supported by comprehensive mRNA expression data and its comparison to the expression pattern of biopsy samples from the oral mucosa. Future studies could include the influence of the microenvironment on the model characteristics by co-cultivating with other cell types present in the oral mucosa such as fibroblasts and endothelial cells.
Supplementary Material
Acknowledgments
We are very grateful to Lisa Bierbaumer and Lisa Koch (AIT – Austrian Institute of Technology, GmbH, Vienna, Austria) who helped with the first steps during the establishment of the model. Moreover, we are thankful to Julia Gausterer, Michael Wirth, and Franz Gabor for supporting the project with the immunofluorescent microscopy equipment and expertise in the Department of Pharmaceutical Technology and Biopharmaceutics, University of Vienna, Austria. Additionally, we want to thank Silvia Schönthaler, Manuela Hofner, and Walter Pulverer (AIT – Austrian Institute of Technology, GmbH, Vienna, Austria) for their expertise and support developing the High-Throughput qPCR-Chip.
Competing interests
The authors declare no competing interests.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Squier CA, Kremer MJ.. Biology of oral mucosa and esophagus. J. Natl. Cancer Inst. Monogr. 2001;52242:1–22. [DOI] [PubMed] [Google Scholar]
- 2.Drobitch RK, Svensson CK.. Therapeutic drug monitoring in saliva. Clin Pharmacokinet. 1992;23:365–379. doi: 10.2165/00003088-199223050-00003. [DOI] [PubMed] [Google Scholar]
- 3.Haeckel R, Hänecke P. The application of saliva, sweat and tear fluid for diagnostic purposes. Ann Biol Clin. 1993;51:903–910. [PubMed] [Google Scholar]
- 4.Yoshizawa JM, Schafer CA, Schafer JJ, Farrell JJ, Paster BJ, Wong DTW. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013;26:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jasim H, Carlsson A, Hedenberg-Magnusson B, Ghafouri B, Ernberg M. Saliva as a medium to detect and measure biomarkers related to pain. Sci Rep. 2018;8:1–9. doi: 10.1038/s41598-018-21131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francois M, Leifert W, Martins R, Thomas P, Fenech M. Biomarkers of Alzheimer’s disease risk in peripheral tissues; focus on buccal cells. Curr Alzheimer Res. 2014;11:519–531. doi: 10.2174/1567205011666140618103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.François M, F. Fenech M, Thomas P, Hor M, Rembach A, N. Martins R, R. Rainey-Smith S, L. Masters C, Ames D, C. Rowe C, et al. High content, multi-parameter analyses in buccal cells to identify Alzheimer’s disease. Curr. Alzheimer Res. 2016;13:787–799. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen HM, Rassing MR. TR146 cells grown on filters as a model of human buccal epithelium: III. Permeability enhancement by different pH values, different osmolality values, and bile salts. Int J Pharm. 1999;185:215–225. doi: 10.1016/S0378-5173(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 9.Jacobsen J, van Deurs B, Pedersen M, Rassing MR. TR146 cells grown on filters as a model for human buccal epithelium: I. Morphology, growth, barrier properties, and permeability. Int J Pharm. 1995;125:165–184. doi: 10.1016/0378-5173(95)00109-V. [DOI] [Google Scholar]
- 10.Mørck Nielsen H, Rømer Rassing M. TR 146 cells grown on filters as a model of human buccal epithelium: V. Enzyme activity of the TR146 cell culture model, human buccal epithelium and porcine buccal epithelium, and permeability of leu-enkephalin. Int J Pharm. 2000;200:261–270. doi: 10.1016/S0378-5173(00)00394-X. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen J, Nielsen EB, Brondum-Nielsen K, Christensen ME, Olin HBD, Tommerup N, Rassing MR. Filter-grown TR146 cells as an in vitro model of human buccal epithelial permeability. Eur J Oral Sci. 1999;107:138–146. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen HM, Rassing MR. TR146 cells grown on filters as a model of human buccal epithelium: IV. Permeability of water, mannitol, testosterone and β-adrenoceptor antagonists. Comparison to human, monkey and porcine buccal mucosa. Int J Pharm. 2000;194:155–167. doi: 10.1016/S0378-5173(99)00368-3. [DOI] [PubMed] [Google Scholar]
- 13.Novakova I, Subileau E-A, Toegel S, Gruber D, Lachmann B, Urban E, Chesne C, Noe CR, Neuhaus W, et al. Transport rankings of non-steroidal antiinflammatory drugs across blood-brain barrier in vitro models. PLoS One. 2014;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neuhaus W, Plattner VE, Wirth M, Germann B, Lachmann B, Gabor F, Noe CR. Validation of in vitro cell culture models of the blood–brain barrier: tightness characterization of two promising cell lines. J Pharm Sci. 2008;97:5158–5175. [DOI] [PubMed] [Google Scholar]
- 15.Ramme AP, Koenig L, Hasenberg T, Schwenk C, Magauer C, Faust D, Lorenz AK, Krebs A, Drewell C, Schirrmann K, et al. Towards an autologous iPSC-derived patient-on-a-chip. bioRxiv 376970. 2018. doi: 10.1101/376970. [DOI] [Google Scholar]
- 16.Gerhartl A, Hahn K, Neuhoff A, Friedl H-P, Förster CY, Wunder C, Schick M, Burek M, Neuhaus W. Hydroxyethylstarch (130/0.4) tightens the blood-brain barrier in vitro. Brain Res. 2020;1727:146560. [DOI] [PubMed] [Google Scholar]
- 17.Neuhaus W, Samwer F, Kunzmann S, Muellenbach RM, Wirth M, Speer CP, Roewer N, Förster CY. Lung endothelial cells strengthen, but brain endothelial cells weaken barrier properties of a human alveolar epithelium cell culture model. Differentiation. 2012;84:294–304. doi: 10.1016/j.diff.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Neuhaus W, Gaiser F, Mahringer A, Franz J, Riethmüller C, Förster C. The pivotal role of astrocytes in an in vitro stroke model of the blood-brain barrier. Front Cell Neurosci. 2014;8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bierbaumer L, Schwarze UY, Gruber R, Neuhaus W. Cell culture models of oral mucosal barriers: A review with a focus on applications, culture conditions and barrier properties. Tissue Barriers. 2018;1–42. doi: 10.1080/21688370.2018.1479568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colley HE, Hearnden V, Jones AV, Weinreb PH, Violette SM, MacNeil S, Thornhill MH, Murdoch C. Development of tissue-engineered models of oral dysplasia and early invasive oral squamous cell carcinoma. Br J Cancer. 2011;105:1582–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buskermolen JK, Reijnders CMA, Spiekstra SW, Steinberg T, Kleverlaan CJ, Feilzer AJ, Bakker AD, Gibbs S. Development of a full-thickness human gingiva equivalent constructed from immortalized keratinocytes and fibroblasts. Tissue Eng Part C Methods. 2016;22:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jennings LR, Colley HE, Ong J, Panagakos F, Masters JG, Trivedi HM, Murdoch C, Whawell S. Development and characterization of in vitro human oral mucosal equivalents derived from immortalized oral keratinocytes. Tissue Eng Part C Methods. 2016;22:1108–1117. doi: 10.1089/ten.tec.2016.0310. [DOI] [PubMed] [Google Scholar]
- 23.Dalley AJ, Abdulmajeed AA, Upton Z, Farah CS. Organotypic culture of normal, dysplastic and squamous cell carcinoma-derived oral cell lines reveals loss of spatial regulation of CD44 and p75NTR in malignancy. J Oral Pathol Med. 2013;42:37–46. doi: 10.1111/j.1600-0714.2012.01170.x. [DOI] [PubMed] [Google Scholar]
- 24.Dongari-Bagtzoglou A, Kashleva H. Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nat Protoc. 2006;1:2012–2018. doi: 10.1038/nprot.2006.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanetti F, Sewer A, Mathis C, Iskandar AR, Kostadinova R, Schlage WK, Leroy P, Majeed S, Guedj E, Trivedi K, et al. Systems toxicology assessment of the biological impact of a candidate modified risk tobacco product on human organotypic oral epithelial cultures. Chem Res Toxicol. 2016;29:1252–1269. [DOI] [PubMed] [Google Scholar]
- 26.Koschier F, Kostrubsky V, Toole C, Gallo MA. In vitro effects of ethanol and mouth rinse on permeability in an oral buccal mucosal tissue construct. Food Chem Toxicol. 2011;49:2524–2529. doi: 10.1016/j.fct.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Lu Q, Jayatilake JAMS, Samaranayake LP, Jin L. Hyphal invasion of Candida albicans inhibits the expression of human β-defensins in experimental oral candidiasis. J Invest Dermatol. 2006;126:2049–2056. doi: 10.1038/sj.jid.5700346. [DOI] [PubMed] [Google Scholar]
- 28.Green CB, Cheng G, Chandra J, Mukherjee P, Ghannoum MA, Hoyer LL. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology. 2004;150:267–275. [DOI] [PubMed] [Google Scholar]
- 29.Wang A, Wang CP, Tu M, Wong DTW. Oral biofluid biomarker research: current status and emerging frontiers. Diagnostics. 2016;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic potential of saliva: current state and future applications. Clin Chem. 2011;57:675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- 31.Klemetsrud T, Kjøniksen AL, Hiorth M, Jacobsen J, Smistad G. Polymer coated liposomes for use in the oral cavity–a study of the in vitro toxicity, effect on cell permeability and interaction with mucin. J Liposome Res. 2018;28:62–73. doi: 10.1080/08982104.2016.1255640. [DOI] [PubMed] [Google Scholar]
- 32.Teubl BJ, Absenger M, Fröhlich E, Leitinger G, Zimmer A, Roblegg E. The oral cavity as a biological barrier system: design of an advanced buccal in vitro permeability model. Eur J Pharm Biopharm. 2013;84:386–393. [DOI] [PubMed] [Google Scholar]
- 33.Portero A, Remuñán-López C, Nielsen HM. The potential of chitosan in enhancing peptide and protein absorption across the TR146 cell culture model - An in vitro model of the buccal epithelium. Pharm Res. 2002;19:169–174. doi: 10.1023/A:1014220832384. [DOI] [PubMed] [Google Scholar]
- 34.Kim YJ, Choi MJ, Bak DH, Lee BC, Ko EJ, Ahn GR, Ahn SW, Kim MJ, Na J, Kim BJ. Topical administration of EGF suppresses immune response and protects skin barrier in DNCB-induced atopic dermatitis in NC/Nga mice. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang X, Liu H, Yang S, Li Z, Zhong J, Fang R. Epidermal growth factor and intestinal barrier function. Mediators Inflamm. 2016;2016:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barabutis N, Khangoora V, Marik PE, Catravas JD. Hydrocortisone and ascorbic acid synergistically prevent and repair lipopolysaccharide-induced pulmonary endothelial barrier dysfunction. Chest. 2017;152:954–962. doi: 10.1016/j.chest.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kürti L, Veszelka S, Bocsik A, Ózsvári B, Puskás LG, Kittel Á, Szabó-Révész P, Deli MA. Retinoic acid and hydrocortisone strengthen the barrier function of human RPMI 2650 cells, a model for nasal epithelial permeability. Cytotechnology. 2013;65:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrot S, Weidenfeller C, Schäffer TE, Robenek H, Galla H-J. Influence of hydrocortisone on the mechanical properties of the cerebral endothelium in vitro. Biophys J. 2005;89:3904–3910. doi: 10.1529/biophysj.104.058750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kılıc Y, Cetin HN, Sumlu E, Pektas MB, Koca HB, Akar F. Effects of boxing matches on metabolic, hormonal, and inflammatory parameters in male elite boxers. Med. 2019;55:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cannizzaro E, Cirrincione L, Mazzucco W, Scorciapino A, Catalano C, Ramaci T, Ledda C, Plescia F. Night-time shift work and related stress responses: A study on security guards. Int J Environ Res Public Health. 2020;17:562. doi: 10.3390/ijerph17020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckert RL, Rorke EA. Molecular biology of keratinocyte differentiation. Environ Health Perspect. 1989;80:109–116. doi: 10.1289/ehp.8980109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiss M, Pitman SW, Sartorelli AC. Modulation of the terminal differentiation of human squamous carcinoma cells in vitro by all-trans-retinoic acid1, 2. J Natl Cancer Inst. 1985;74:1015–1023. [PubMed] [Google Scholar]
- 43.Buckley A, Turner JR. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb Perspect Biol. 2018;10:a029314. doi: 10.1101/cshperspect.a029314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber CR. Dynamic properties of the tight junction barrier. Ann N Y Acad Sci. 2012;1257:77–84. doi: 10.1111/j.1749-6632.2012.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Itallie CM, Anderson JM. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol. 2014;36:157–165. doi: 10.1016/j.semcdb.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsukita S, Tanaka H, The Claudins: TA. From tight junctions to biological systems. Trends Biochem Sci. 2019;44:141–152. doi: 10.1016/j.tibs.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Dos Reis PP, Bharadwaj RR, Machado J, MacMillan C, Pintilie M, Sukhai MA, Perez-Ordonez B, Gullane P, Irish J, Kamel-Reid S, et al. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer. 2008;113:3169–3180. [DOI] [PubMed] [Google Scholar]
- 48.Lourenço SV, Coutinho-Camillo CM, Buim MEC, Pereira CM, Carvalho AL, Kowalski LP, Soares FA. Oral squamous cell carcinoma: status of tight junction claudins in the different histopathological patterns and relationship with clinical parameters. A tissue-microarray-based study of 136 cases. J Clin Pathol. 2010;63:609–614. [DOI] [PubMed] [Google Scholar]
- 49.Bayar GR, Aydintug YS, Gulses A, Elci P, Sarper M. A pilot study of the primary culture of the oral mucosa keratinocytes by the direct explant technique. Oral Health and Dental Management in Black Sea Countries. 2011;10:88–92. [Google Scholar]
- 50.Peramo A, Marcelo CL, Feinberg SE. Tissue engineering of lips and muco-cutaneous junctions: in vitro development of tissue engineered constructs of oral mucosa and skin for lip reconstruction. Tissue Eng Part C Methods. 2012;18:273–282. doi: 10.1089/ten.tec.2011.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donetti E, Bedoni M, Boschini E, Dellavia C, Barajon I, Gagliano N. Desmocollin 1 and desmoglein 1 expression in human epidermis and keratinizing oral mucosa: a comparative immunohistochemical and molecular study. Arch Dermatol Res. 2005;297:31–38. [DOI] [PubMed] [Google Scholar]
- 52.Slomiany BL, Murty VLN, Piotrowski J, Slomiany A. Salivary mucins in oral mucosal defense. Gen Pharmacol. 1996;27(5):761–771. doi: 10.1016/0306-3623(95)02050-0. [DOI] [PubMed] [Google Scholar]
- 53.Frenkel ES, Ribbeck K. Salivary mucins in host defense and disease prevention. J Oral Microbiol. 2015;7(1):29759. doi: 10.3402/jom.v7.29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sen S, Sharma S, Gupta A, Gupta N, Singh H, Roychoudhury A, Mohanty S, Sen S, Nag TC, Tandon R, et al. Molecular characterization of explant cultured human oral mucosal epithelial cells. Investig Ophthalmol Vis Sci. 2011;52:9548–9554. [DOI] [PubMed] [Google Scholar]
- 55.Alos L, Lujan B, Castillo M, Nadal A, Carreras M, Caballero M, de Bolos C, Cardesa A. Expression of Membrane-Bound Mucins (MUC1 and MUC4) and Secreted Mucins (MUC2, MUC5AC, MUC5B, MUC6 and MUC7) in mucoepidermoid carcinomas of salivary glands. Am J Surg Pathol. 2005;29:806–813. doi: 10.1097/01.pas.0000155856.84553.c9. [DOI] [PubMed] [Google Scholar]
- 56.Suh H, Valle S, Morris DL. Targeting MUC16 in cancer therapy. Chemother Open Access. 2017;06:6–10. [Google Scholar]
- 57.Ukkonen H, Pirhonen P, Herrala M, Mikkonen JJW, Singh SP, Sormunen R, Kullaa AM. Oral mucosal epithelial cells express the membrane anchored mucin MUC1. Arch Oral Biol. 2017;73:269–273. [DOI] [PubMed] [Google Scholar]
- 58.Kho HS. Oral epithelial MUC1 and oral health. Oral Dis. 2018;24:19–21. doi: 10.1111/odi.12713. [DOI] [PubMed] [Google Scholar]
- 59.Thakur A, Tupkari J, Joy T, Kende P, Siwach P, Ahire M. Expression of mucin-1 in oral squamous cell carcinoma and normal oral mucosa: an immunohistochemical study. J Oral Maxillofac Pathol. 2018;22:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Louis NA, Hamilton KE, Canny G, Shekels LL, Ho SB, Colgan SP. Selective induction of mucin-3 by hypoxia in intestinal epithelia. J Cell Biochem. 2006;99:1616–1627. [DOI] [PubMed] [Google Scholar]
- 61.Agrawal S, Bhattacharya A, Manhas J, Kholakiya Y, Khera N, Roychoudhury A, Sen S. Increased mucin expression in oral mucosal epithelial cells in vitro: a potential new role of mycophenolate mofetil. Tokai J Exp Clin Med. 2018;43:132–138. [PubMed] [Google Scholar]
- 62.Bai Q, Liu L, Long Q, Xia Y, Wang J, Xu J, Guo J. Decreased expression of mucin 18 is associated with unfavorable postoperative prognosis in patients with clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2015;8:11005–11014. [PMC free article] [PubMed] [Google Scholar]
- 63.Simon GC, Martin RJ, Smith S, Thaikoottathil J, Bowler RP, Barenkamp SJ, Chu HW. Up-regulation of MUC18 in airway epithelial cells by IL-13 implications in bacterial adherence. Am J Respir Cell Mol Biol. 2011;44:606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Q, Case SR, Minor MN, Jiang D, Martin RJ, Bowler RP, Wang J, Hartney J, Karimpour-Fard A, Chu HW, et al. A novel function of MUC18: amplification of lung inflammation during bacterial infection. Am J Pathol. 2013;182:819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma J, Rubin BK, Voynow JA. Mucins, mucus, and goblet cells. Chest. 2018;154:169–176. doi: 10.1016/j.chest.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 66.Woodward AM, Argüeso P. Expression analysis of the transmembrane mucin MUC20 in human corneal and conjunctival epithelia. Investig Ophthalmol Vis Sci. 2014;55:6132–6138. doi: 10.1167/iovs.14-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H, Wang H, Shen L, Lin Y, Shi Q, Yang Y. The expression and prognostic significance of Mucin 13 and Mucin 20 in esophageal squamous cell carcinoma. J Cancer Res Ther. 2015;11:74–79. [DOI] [PubMed] [Google Scholar]
- 68.Itoh Y, Kamata-Sakurai M, Denda-Nagai K, Nagai S, Tsuiji M, Ishii-Schrade K, Okada K, Goto A, Fukayama M, Irimura T, et al. Identification and expression of human epiglycanin/MUC21: a novel transmembrane mucin. Glycobiology. 2008;18:74–83. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Tatakis DN. Human gingiva transcriptome during wound healing. 2016. doi: 10.1111/jcpe.12669. [DOI] [PubMed] [Google Scholar]
- 70.Delporte C, Bryla A, Perret J. Aquaporins in salivary glands: from basic research to clinical applications. Int J Mol Sci. 2016;17:1–13. doi: 10.3390/ijms17020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poveda M, Hashimoto S, Matsuki-Fukushima M, Sasaki H, Sakurai K, Masaki S. Expression and localization of aqua-glyceroporins AQP3 and AQP9 in rat oral epithelia. Bull Tokyo Dent Coll. 2014;55:1–10. [DOI] [PubMed] [Google Scholar]
- 72.Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015;7:1–9. doi: 10.12703/P7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marquez B, Van Bambeke F. ABC multidrug transporters: target for modulation of drug pharmacokinetics and drug-drug interactions. Curr Drug Targets. 2011;12:600–620. doi: 10.2174/138945011795378504. [DOI] [PubMed] [Google Scholar]
- 74.Lin DJH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics. Clin Pharmacokinet. 2012;42:59–98. doi: 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- 75.Peng R, Zhang H, Zhang Y, Wei DY. Impacts of ABCB1 (G1199A) polymorphism on resistance, uptake, and efflux to steroid drugs. Xenobiotica. 2016;46:948–952. doi: 10.3109/00498254.2016.1138249. [DOI] [PubMed] [Google Scholar]
- 76.Paitz RT, Bukhari SA, Bell AM. Stickleback embryos use ATP-binding cassette transporters as a buffer against exposure to maternally derived cortisol. Proc R Soc B Biol Sci. 2016;283:1–7. doi: 10.1098/rspb.2015.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burns VE, Kerppola TK. ATR-101 inhibits cholesterol efflux and cortisol secretion by ATP-binding cassette transporters, causing cytotoxic cholesterol accumulation in adrenocortical carcinoma cells. Br J Pharmacol. 2017;174:3315–3332. doi: 10.1111/bph.13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neary JP, Malbon L, McKenzie DC. Relationship between serum, saliva and urinary cortisol and its implication during recovery from training. J Sci Med Sport. 2002;5:108–114. doi: 10.1016/S1440-2440(02)80031-7. [DOI] [PubMed] [Google Scholar]
- 79.Turpeinen U, Hämäläinen E. Determination of cortisol in serum, saliva and urine. Best Pract Res Clin Endocrinol Metab. 2013;27:795–801. doi: 10.1016/j.beem.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 80.Bicakci T, Ozcaka O, Tuzunsoy-aktas R, Nalbantsoy A, Akcali A, Bicakci N, Kose T. Effects of potassium aluminum sulfate on TNF-α, MMP −1 and MMP-8 levels at gingival crevicular fluid in periodontally healthy subjects: a pilot study. Turkish J Biochem. 2012;37:315–321. [Google Scholar]
- 81.Iyengar A, Paulus JK, Gerlanc DJ, Maron JL. Detection and potential utility of C-reactive protein in saliva of neonates. Front Pediatr. 2014;2:1–6. doi: 10.3389/fped.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


