Abstract
Patients with complex chronic disorders, such as asthma, present clinicians with important management problems. The identification of a clinical syndrome usually leads to the diagnosis of the disease entity. The next concern involves classification and a choice as to whether to use a few inclusive categories or multiple exclusive categories. Patients with asthma have multiple clinical syndromes, and these can be described as phenotypes. The use of cluster analysis allows investigators to identify phenotypes with less clinical bias. However, the identification of a particular phenotype does not necessarily provide much insight into the underlying pathogenesis. In asthma, the pathogenetic events are complex and multiple and require a classification based on endotypes. This difficulty introduces the idea of causation and models for causation. Asthma probably does not have a single universal necessary cause. However, it does have multiple sufficient component causes. Understanding these components and their interactions potentially leads to better treatment trials and more focused drug therapy. Clinicians need to identify asthmatic patients and classify them into particular phenotypes; they should also wonder about causation. Clinical investigators need to use these phenotypes to identify more homogenous groups of patients to study the underlying pathogenesis and establish endotypes. Focusing on causation can improve our understanding of disease entities, disease classification, and disease causation. This review outlines ideas relevant to causation in nearly all diseases.
Keywords: Asthma, causality, diagnosis, differential, endotype, multifactorial, phenotype
Clinicians use historical information, physical examination details, and routine laboratory tests to evaluate patients and initiate management plans. A collection of signs and symptoms may suggest a clinical syndrome, which potentially narrows the differential diagnosis and can lead to exact diagnoses in some cases. For practical purposes, the diagnosis represents the end goal in patient evaluation, and clinicians may not consider the various philosophical definitions and pathogenetic events (causation) related to the understanding of a disease entity. In this review, we discuss disease entities and various presentations (syndromes and phenotypes) and approaches to understanding causation (endotypes and pathogenesis). We use selected information from asthma studies to illustrate some of these ideas. The main effort is to discuss ideas about causation, which are potentially applicable to almost all chronic diseases, and not to provide a comprehensive review of the pathogenesis of asthma.
DISEASE ENTITY
In “Causation in medicine: the disease entity model,” Caroline Whitbeck explored the understanding of causal relationships in contemporary medicine and suggested that rather than defining disease entities as “invading bodies or … a static configuration of abnormal cells, tissues or organs,” a disease entity should be considered a “complex of processes.” 1 Whitbeck offered several criticisms of the “monocausal” conception of disease entity. Chief among these is the fact that “a single disease entity may have several pathological pictures,” but at the same time, “different [disease entities] may have clinical pictures which are indistinguishable relative to the observational distinctions available at a given time.” Due to these factors, older models of causation “fail to do justice to the process character of disease.”
Whitbeck explained the concept of disease entity as a complex of processes by noting that medical professionals “assume that for every clinical disease entity there must be a … disease entity.” It would be easy to conclude that there is a clinical process and a pathological process inherent to any disease, and one must logically follow the other. However, this is not always the case, as she noted that there is “no clear division between the clinical and pathological level.” As an example, Whitbeck listed a fever of 106°F, which is a clinical sign but is simultaneously a “significant factor in processes even at the cellular level.”
This “complex of processes” is most frequently identified or recognized as a clinical syndrome. At a basic level, this helps clinicians organize their thinking and evaluation of patients. As clinical information develops, some clinical syndromes may be divided into phenotypes. Deeper analysis leads to questions and information about causation, pathogenesis, and factors that contribute to the presentation of complicated chronic disorders.
CLINICAL SYNDROMES
Patients with asthma characteristically have episodic symptoms, such as dyspnea, cough, and wheezing, on physical examination. This constitutes a clinical syndrome, which has a broad differential diagnosis. Adding additional requirements to characterize the clinical syndrome, such as symptom triggers, pulmonary function testing, and responses to specific treatment, can increase the certainty of the diagnosis of asthma but also potentially increase the exclusion of patients with asthma who do not have all the listed criteria. This circumstance creates unavoidable tradeoffs in disease definition and ideally should lead to clinical studies characterizing patients using multiple observable parameters.
PHENOTYPES
Clinicians often classify patients into phenotypes (from the Greek words phainein, meaning “to show” and typos meaning “type”). The dictionary defines phenotype as the “observable properties of an organism that are produced by the interaction of the genotype and the environment.” 2 , 3 Hence, phenotype is the expression of genetic information. Consider Marfan syndrome, which has two alleles that we will designate M and m for the dominant and recessive alleles, respectively. There are three possible genotypes for this allele: MM, Mm, and mm. There are two possible phenotypes for this allele: positive for Marfan syndrome or negative for Marfan syndrome. The rare MM and Mm genotypes translate into the rare Marfan syndrome, while the far more common mm genotype translates into the negative or normal state.
The environment can have a role in a legitimate phenotype. Consider certain forms of cryoglobulinemia. The cryoglobulins are present all the time, but the clinical syndrome will not appear unless the ambient temperature is sufficiently cold. The cryoglobulin protein is a transcription product of the genotype, but the solubility of the cryoglobulin protein in blood changes abruptly at cold temperature to cause a physical sign and patient symptoms characteristic of a disease. It is proper to consider cryoglobulinemia as a phenotype, but proper understanding of the clinical syndrome requires knowledge of both the genetic and environmental components.
Asthma was defined by clinical criteria before any mechanisms were known. The World Health Organization definition of asthma starts with clinical criteria: “Asthma attacks all age groups but often starts in childhood. It is a disease characterized by recurrent attacks of breathlessness and wheezing, which vary in severity and frequency from person to person. In an individual, they may occur from hour to hour and day to day.” 3 This clinical definition is vague and not suitable as an inclusion criterion for studies. We rely on clinical experts to decide whether individual patients have asthma or not. The World Health Organization definition of asthma continues with a basis in pathophysiology: “This condition is due to inflammation of the air passages in the lungs and affects the sensitivity of the nerve endings in the airways so they become easily irritated. In an attack, the lining of the passages swell, causing the airways to narrow and reducing the flow of air in and out of the lungs.” 3 , 4 This mechanistic definition is not very useful without objective criteria for what constitutes “inflammation of the air passages” or thresholds for “reducing the flow of air.”
For any objective criterion, a patient will either meet the criterion (C+) or not (C–). These patients will be classified by an expert as either having asthma (A+) or not (A–). Four groups are defined: true positives (C+A+), false positives (C+A–), false negatives (C–A+), and true negatives (C–A–). The goal of the division of asthma into phenotypes is that the criterion C should have zero false-negatives and zero false-positives. This goal might be achieved by choosing phenotypes based on the presence or absence of genetically determined traits thought to contribute to the pathogenesis of asthma. Unfortunately, clinical asthma has turned out to be very heterogeneous with respect to mechanism. As such, all of the proposed phenotypes have significant false-positives and -negatives. Furthermore, some of the proposed phenotypes were not truly based on the expression of genotype and should not have been labeled as phenotypes.
A new term, endotype, has emerged based on biologic mechanisms that can include mechanisms that have no genetic basis. Division into endotypes should aid the development of studies with objective inclusion criteria, and the results of these studies should help identify mechanistic targets suitable for therapeutic intervention. Consider carbon monoxide poisoning. We understand the molecular basis for carbon monoxide poisoning, so defining an endotype based on a percentage of carboxy-hemoglobin found in the blood is useful for studying treatments even though the mechanism has no obvious genetic basis, ignoring the possibility that unidentified mutations could have subtle effects on hemoglobin binding capacities.
PHENOTYPE CLASSIFICATION BASED ON CLUSTER ANALYSIS
Cluster analysis is a statistical methodology that identifies groups of subjects (clusters) within a population that cannot be attributed to normal variation. It is presumed that these clusters identify a basis for understanding disease.
Haldar et al used cluster analysis to map asthmatics onto a plane with a measure of eosinophilic inflammation as one axis and self-reported clinical symptoms as the other axis. 4 Three groups of asthmatics with a total of 439 patients were analyzed using k-means cluster analysis to identify five clusters of patients. Two concordant clusters were identified—early onset atopic asthma and benign asthma—where eosinophilic inflammation and symptoms were present to a proportionate degree. A single cluster was identified—inflammation predominant—where eosinophilic inflammation was present out of proportion to clinical symptoms. Two clusters were identified—early symptom predominant and obese noneosinophilic—in which clinical symptoms were present out of proportion to eosinophilic inflammation. These clusters were called phenotypes; no evidence was presented to show that the basis for clustering was genetic expression. A genetic contribution to the development of these clusters might be assumed.
Moore et al analyzed 726 asthmatics using a tree-based (dendogram) cluster analysis methodology to identify five clusters of patients. 5 The variables used to distinguish the clusters included age, body mass index, asthma duration, baseline forced expiratory volume in 1 second, and atopy status. Like Haldar, Moore et al asserted, without any evidence, that the statistical clusters were separate phenotypes. Moore et al explicitly acknowledged the misuse of the word phenotype in their definition: “Identification of asthma subphenotypes has generally been accomplished in two ways: (1) through a priori definitions of a phenotype based on clinical characteristics of subjects or (2) through pathobiologic differences in sputum or bronchoscopy specimens.”
Siroux et al analyzed two groups of asthmatics with a total of 2536 patients by a latent class analysis method of cluster analysis to identify four clusters of asthmatics. 6 The four clusters were labeled as phenotypes E (active treated allergic childhood-onset asthma), F (active treated adult-onset asthma), G (inactive/mild untreated allergic childhood-onset asthma), and H (inactive/mild untreated adult-onset asthma).
Wu et al recently reported a multiview k-means cluster analysis of 100 clinical, physiological, inflammatory, and demographic variables recorded in 346 adult asthmatics with severe asthma. 7 This study included both static variables and dynamic variables based on changes in 15 factors following the administration of triamcinolone. This analysis identified four clusters, including two clusters of young patients with allergic asthma and relatively normal lung function, one cluster with late-onset asthma and reduced lung function, and one cluster of young obese asthmatics with severe airflow limitation and little response to triamcinolone. However, analysis of the figures in this paper indicates that there was a significant overlap in the factors used in this analysis in all four cluster groups. One group had a significant increase in the percentage of sputum neutrophils following the administration of triamcinolone, one group had a significant reduction in the percentage of sputum macrophages, and one group had a significant decrease in the percentage of sputum eosinophils. However, it is unclear how stable these dynamic changes are, and it is unclear whether or not these dynamic changes provide significant insight into the underlying pathogenesis of asthma in these patients.
These four studies all conflate a statistical difference between (among) clusters within a population with a genetic mechanism. The use of cluster analysis to identify subgroups of asthmatics may lead to a better understanding of the taxonomy of asthma, the underlying mechanisms, and approaches to treatment. All statistical analyses, however, can lead to type 1 and type 2 errors in which patterns are perceived where they do not exist and other patterns that do exist are not perceived. Even when statistical analysis leads to a perception of a real subgroup, we cannot conclude in the absence of additional evidence that the mechanism for the statistical cluster was genetic. An important issue is the uniformity of the group identified by observable difference(s).
ENDOTYPES/PATHOGENESIS
Anderson introduced the term endotype (a contraction of endophenotype) in a review of the pathogenetic mechanisms in asthma in 2008. 8 He defined an endotype as a subtype of disease defined functionally or pathologically by a particular molecular mechanism or by a treatment response. This word is a combination of the prefix endo-, from the Greek endon, “within,” and type, from Greek tupos, “impression, figure, type.” The study of endotypes helps focus clinical investigators on the underlying pathogenesis of various asthma subtypes or phenotypes. The identification of endotypes has the potential to create more homogeneous patient groups for clinical studies, such as genetic studies and drug trials. This has the long-term potential to help clinicians prescribe more effective drugs in particular endotypes and limit the use of ineffective drugs in these endotypes.
Lötvall and colleagues developed an approach to asthma classification based on endotypes and discussed the possible relationship between various phenotypes and endotypes. 9 They noted that a particular phenotype could have several endotypes, and that a single endotype could be associated with several phenotypes. Their table of proposed endotypes includes six examples and demonstrates the complexity and variability in asthma syndromes. An abbreviated version of their table is reproduced in Table 1.
Table 1.
Relationships between endotypes and phenotypes*
| Phenotype | Endotypes |
|---|---|
| Eosinophilic asthma | Allergic asthma; aspirin-sensitive asthma; late-onset hypereosinophilic asthma |
| Adult-onset asthma | Aspirin-sensitive asthma; infection-induced asthma; late-onset hypereosinophilic asthma |
| Poorly steroid-responsive asthma | Noneosinophilic, neutrophilic asthma; steroid-insensitive eosinophilic asthma; airflow obstruction caused by obesity |
Modified from Lötvall et al. 9
The following three endotypes illustrate this problem. Adults with allergic asthma have symptoms related to allergen exposure and typically have upper airway symptoms (allergic rhinitis). These patients have positive skin prick responses to aeroallergens and elevated IgE levels. Histopathologic studies of the airways reveal eosinophilic inflammation and basement membrane thickening. These patients usually respond to glucocorticoids and medications that inhibit the IL-4/13 pathways. The pathogenesis involves type 1 T-helper lymphocyte–mediated inflammation and single nucleotide polymorphisms in type 2 T-helper pathways.
Patients with aspirin-sensitive asthma often have chronic rhinosinusitis, nasal polyposis, and more severe asthma. They respond to aspirin challenges and have increased levels of urinary leukotrienes. The histopathology may reveal eosinophilic inflammation. These patients respond to leukotriene inhibitors. The proposed mechanism involves the increased production of leukotrienes and has been associated with leukotriene-related gene polymorphisms, which result in overexpression of proteins in leukotriene pathways.
Some elite cross-country skiers (15%–25%) develop asthma during intense training in cold environments. Their symptoms frequently develop following upper respiratory tract infection. They respond to exercise challenges with bronchospasm. Histopathology reveals low-grade noneosinophilic inflammation with increased numbers of neutrophils in the sputum, possibly correlating with training intensity. These skiers respond poorly to inhaled glucocorticoids and improve with reductions in training intensity. The pathogenesis probably involves cold dry air–induced cellular stress in airways, and there is no definite genetic component.
These three phenotypes clearly have different clinical presentations and likely represent different endotypes. However, our understanding of causation and the mechanism(s) of disease is clearly incomplete.
CAUSATION
Epidemiologists, scientists, and philosophers study causation. 10 Epidemiologists want to determine critical factors in the development of a disease entity to introduce interventions that might prevent the disease. Their approach requires the identification of necessary causes (NC) and sufficient causes (SC). This analysis creates a four-category classification that can explain the association between causal factors and outcomes (diseases): N+S+, N+S–, N–S+, and N–S–. These models best explain infectious diseases in which microbial pathogens, such as the tuberculous bacillus, are necessary causes for the development of tuberculous infections. However, tuberculous exposure or even tuberculous infection may not be sufficient to cause pulmonary tuberculosis, a disease entity. Consequently, there are situations in which there is an absolute necessary cause requirement and a requirement for a sufficient cause or causes that allow the disease entity to develop. In addition, sufficient causes may contribute to different pathways in the development of a disease; this is particularly true in chronic medical disorders.
Consider the idea of necessary causes and sufficient causes in the development of asthma phenotypes. The monocausal model argues that a single event causes every case of a particular disease entity. 11 It should also be true that this event does not cause an alternative disease. This model does not consider the possibility that other factors likely contribute to the development of the disease entity, and this concern leads to the idea of multicausal or multifactorial models. For example, in asthma, environmental exposures to high concentrations of aeroallergens or secondhand smoke could contribute to the development of a clinical syndrome in patients with a genetic predisposition to asthma. This modification of the monocausal model by Broadbent has been called the monocausal model with an additional circumstantial sufficiency requirement. 11
Broadbent also used a contrastive model to help explain disease causation. This model makes use of information from control or healthy subjects without the disease in question to help understand causation. For example, one child in the family may have asthma but the other children do not. To understand this outcome, the investigator needs to compare the asthmatic child with the “normal” children to determine differences that help explain causation.
Rothman introduced the sufficient component cause model to explain complex chronic diseases. 12 This model argues that often several components are necessary for the development of the disease and that these components do not need to be temporally associated. For example, an athlete with allergic asthma may develop symptoms only during seasons with high levels of allergens and significant training. Consequently, a cause is usually not a single component but rather a minimal set of conditions that inevitably produces the disease outcome. Each component in a sufficient cause is then called a component cause. A cause that is in every model is a necessary cause.
Diseases, such as asthma, almost certainly have multiple sufficient causes. Fuller has used the constitutive model for disease classification (or definition). 13 According to this model, a disease is present only in situations in which a certain characteristic is present. For example, patient A has asthma only if he or she also has a reversible obstructive process based on spirometric measurements. This approach works better with chronic diseases and helps investigators organize their thinking about the classification or definition of the disease, which in turn should lead to more focused clinical studies. Based on our current understanding of the pathogenesis of asthma, there is no obvious necessary cause, and the components in the sets of sufficient causes will clearly depend on the underlying endotype.
FACTORS INFLUENCING THE DEVELOPMENT OF A DISEASE ENTITY
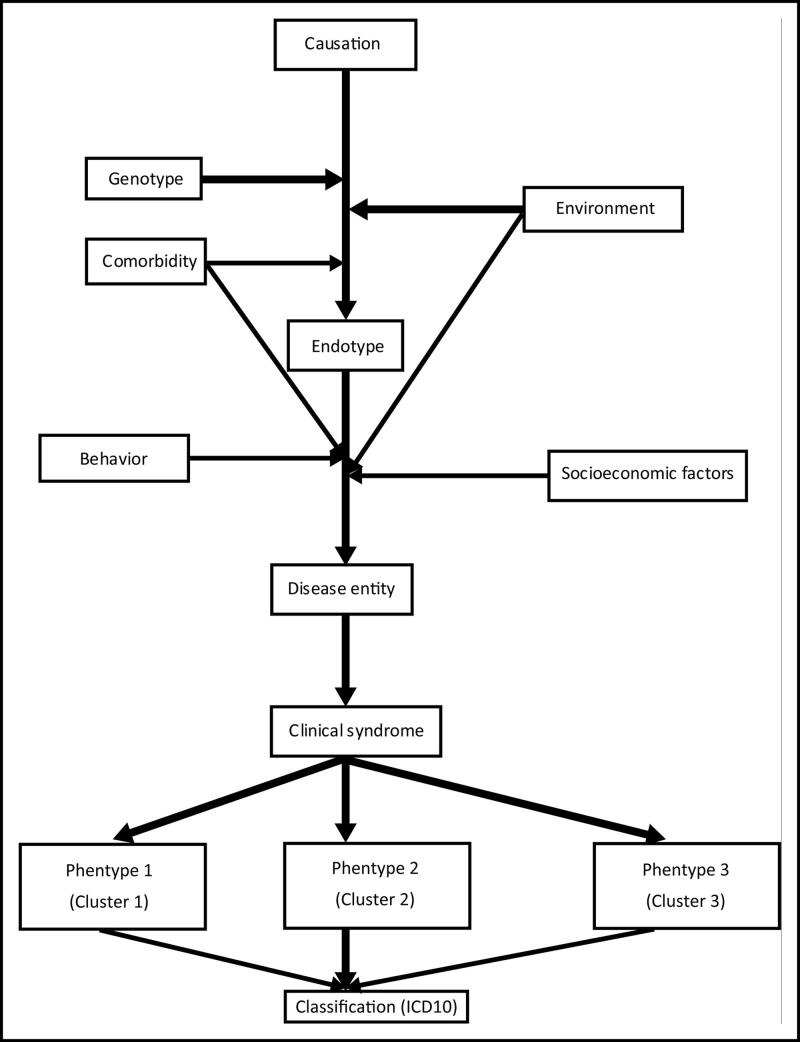
Understanding the cause and the relevant causation model in a particular endotype can lead to a better description of the pathogenesis (Figure 1). However, as discussed in the section on causation, identifying necessary causes and sufficient causes in chronic diseases can be very difficult. Multiple factors potentially contribute to the development of the endotype, even after the necessary cause has been identified. For example, repeated exposure to cold dry air is a necessary cause of asthma in some skiers. However, is it a sufficient cause? Is an underlying genetic predisposition necessary for these skiers to develop this endotype? Do other comorbidities, such as upper respiratory viral infections, or environmental exposures, such as smoke from indoor fireplaces, contribute to the development of this endotype? Finally, after the development of pathogenetic processes in the airway (i.e., the endotype), what other factors contribute to the development of the disease entity or clinical phenotype? Does comorbidity, such as an allergic diathesis, contribute to this, or do other environmental exposures contribute to it? Does the behavior of the individual patient contribute to it? For example, poor compliance with medication could contribute to the clinical presentation. Do social/economic factors contribute to it? For example, living in a remote area with limited access to medical care and treatment could reduce management opportunities.
Figure 1.
One possible pathway for the development of disease entities and phenotypes from endotypes.
Studies on endotypes are complex and difficult. 14 Important questions include the basis for the endotype classification, the timeframe for the development of a particular endotype, the stability of the endotype over time, and the clinical implications for treatment and prognosis. In patients with asthma, a single endotype may have several phenotypes and a single phenotype may have several endotypes. This clearly complicates the identification of study cohorts and management of drug trials.
APPROACHES TO STUDYING PATHOGENESIS
Studies on the pathogenesis of asthma present multiple problems. Table 2 provides examples of possible approaches to studying the pathogenic basis for asthma syndromes. 15–19 Some of these methods require complex biochemical analyses, and some require biopsy specimens that are invasive and potentially dangerous, especially in patients with severe asthma. Studies using exhaled breath condensates provide an opportunity to characterize chemical compounds originating either in the airways or alveolar spaces. These collections can be easily repeated in all patients and do not require invasive procedures. Studies on the natural history of asthma require well-characterized subgroups and prolonged follow-up. Studies on responses to drug therapy have two potential advantages. They provide immediate information about the benefit of drugs at least in certain subgroups. In addition, the mechanism of action may provide insight into the underlying pathogenesis of asthma. Drug studies that include a genetic analysis of the participants have the potential to provide information about the underlying genotype leading to the development of asthma or the genotype associated with a response to a particular drug.
Table 2.
Experimental studies on mechanism in asthma
| First author, year | Biological sample | Assay/test | Result | Comment |
|---|---|---|---|---|
| Zoratti, 2014 15 | Whole blood | Interleukins, interferon γ | TH2 polarized responses seen in allergic asthma, TH1 responses in nonallergic asthma | Distinct differences in immune endotypes |
| Meyer, 2014 16 | Exhaled air | VOC, clinical data | VOC can identify asthmatics | Asthma clusters with different VOC, similar VOC endotypes but different clinical characteristics |
| George, 2015 17 | Blood | Gene expression, biomarkers | Four distinct endotypes | Integrated approach necessary for complex diseases |
| Wesolowska- Andersen, 2015 18 | Airway mucosa, sputum | Biomarkers for TH2 inflammation | Distinct but related TH2 endotypes | Multiple non-TH2 molecular endotypes |
| Tse, 2011 19 | Blood | Genetic analysis | Genetic determinants of drug response | See text for discussion |
TH1 indicates T-helper type 1 cells; TH2, T-helper type 2 cells; VOC, volatile organic compounds.
CONCLUSIONS
Patients with complex chronic disorders, such as asthma, present clinicians with important management problems. The initial concern involves classification and a choice as to whether to use a few inclusive categories or multiple exclusive categories. In patients with asthma, this classification can use observable phenotypes based on clinical data, laboratory information, pulmonary function testing, and responses to treatment. However, this classification scheme provides minimal insight into the underlying pathogenesis of asthma in most patients. It also compromises treatment trials since study groups almost certainly include patients with different pathogenetic processes and potentially different responses to treatment. The use of a cluster analysis helps identify more homogeneous groups of patients with asthma and potentially improves classification. Using endotypes for classification helps clinical investigators focus on the underlying pathogenesis that can lead to better understanding of the disease process and possible treatment approaches. However, causation in asthma is almost certainly complex, and it is unlikely that there is any single necessary cause. Finally, studying and treating patients based on endotypes should lead to more focused studies on pathogenesis and possibly treatment and is likely the basis for future work with this disease. In addition, studies in patients with asthma provide a framework for studying acute disorders such as sepsis and chronic disorders such as obesity.
References
- 1. Whitbeck C. Causation in medicine: the disease entity model. Philos Sci. 1977;44(4):619–637. doi: 10.1086/288771. [DOI] [Google Scholar]
- 2. Phenotype. In: Merriam-Webster Ninth New Collegiate Dictionary. Springfield, MA: Merriam-Webster Inc; 1986. [Google Scholar]
- 3. World Health Organization . Asthma: Definition. https://www.who.int/respiratory/asthma/definition/en/. Accessed March 20, 2020.
- 4. Haldar P, Pavord ID, Shaw DE, et al. . Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178(3):218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moore WC, Meyers DA, Wenzel SE, et al. . Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181(4):315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siroux V, Basagaña X, Boudier A, et al. . Identifying adult asthma phenotypes using a clustering approach. Eur Respir J. 2011;38(2):310–317. doi: 10.1183/09031936.00120810. [DOI] [PubMed] [Google Scholar]
- 7. Wu W, Bang S, Bleecker ER, et al. . Multiview cluster analysis identifies variable corticosteroid response phenotypes in severe asthma. Am J Respir Crit Care Med. 2019;199(11):1358–1367. doi: 10.1164/rccm.201808-1543OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372(9643):1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 9. Lötvall J, Akdis CA, Bacharier LB, et al. . Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127(2):355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 10. Broadbent A. Conceptual and methodological issues in epidemiology: an overview. Prev Med. 2011;53(4–5):215–216. doi: 10.1016/j.ypmed.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 11. Broadbent A. Causation and models of disease in epidemiology. Stud Hist Philos Biol Biomed Sci. 2009;40(4):302–311. doi: 10.1016/j.shpsc.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 12. Rothman KJ, Greenland S.. Causation and causal inference in epidemiology. Am J Public Health. 2005;95(1):S144–S150. doi: 10.2105/AJPH.2004.059204. [DOI] [PubMed] [Google Scholar]
- 13. Fuller J. Universal etiology, multifactorial diseases and the constitutive model of disease classification. Stud Hist Philos Biol Biomed Sci. 2018;67:8–15. doi: 10.1016/j.shpsc.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 14. Donovan GM, Tawhai MH.. Phenotype, endotype and patient-specific computational modelling for optimal treatment design in asthma. Drug Discov Today Dis Models. 2015;15:23–27. doi: 10.1016/j.ddmod.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zoratti E, Havstad S, Wegienka G, et al. . Differentiating asthma phenotypes in young adults through polyclonal cytokine profiles. Ann Allergy Asthma Immunol. 2014;113(1):25–30. doi: 10.1016/j.anai.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meyer N, Dallinga JW, Nuss SJ, et al. . Defining adult asthma endotypes by clinical features and patterns of volatile organic compounds in exhaled air. Respir Res. 2014;15:136. doi: 10.1186/s12931-014-0136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. George BJ, Reif DM, Gallagher JE, et al. . Data-driven asthma endotypes defined from blood biomarker and gene expression data. PLoS One. 2015;10(2):e0117445. doi: 10.1371/journal.pone.0117445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wesolowska-Andersen A, Seibold MA.. Airway molecular endotypes of asthma: dissecting the heterogeneity. Curr Opin Allergy Clin Immunol. 2015;15(2):163–168. doi: 10.1097/ACI.0000000000000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tse SM, Tantisira K, Weiss ST.. The pharmacogenetics and pharmacogenomics of asthma therapy. Pharmacogenomics J. 2011;11(6):383–392. doi: 10.1038/tpj.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]