ABSTRACT
Enterococcus faecalis is one of the important causative agents of nosocomial and life-threatening infections in human. Several studies have demonstrated that the presence of CRISPR-cas is associated with antibiotic susceptibility and lack of virulence traits. In this study, we aimed to assess the phenotypic and genotypic virulence determinants in relation to CRISPR elements from the dental-root canals and hospital-acquired isolates of E. faecalis. Eighty-eight hospital-acquired and 73 dental-root canal isolates of E. faecalis were assessed in this study. Phenotypic screening of the isolates included biofilm formation, and gelatinase and hemolysis activities. Genotypical screening using PCR was further used to evaluate the presence of CRISPR elements and different virulence-associated genes such as efaA, esp, cylA, hyl, gelE, ace, ebpR, and asa1. Biofilm formation, gelatinase, and hemolysis activities were detected in 93.8%, 29.2%, and 19.2% of the isolates, respectively. The most prevalent virulence-associated gene was ace, which was followed by efaA, whereas cylA was the least identified. The presence of CRISPR1-cas, orphan CRISPR2, and CRISPR3-cas was determined in 13%, 55.3%, and 17.4% of the isolates, respectively. CRISPR elements were significantly more prevalent in the dental-root canal isolates. An inverse significant correlation was found between CRISPR-cas loci, esp, and gelE, while direct correlations were observed in the case of cylA, hyl, gelE (among CRISPR-loci 1 and 3), asa1, ace, biofilm formation, and hemolysis activity. Findings, therefore, indicate that CRISPR-cas might prevent the acquisition of some respective pathogenicity factors in some isolates, though not all; so selective forces could not influence pathogenic traits.
Abbreviations: BHI: brain-heart infusion agar; CRISPRs: Clustered regularly interspaced short palindromic repeats; Esp: Cell wall-associated protein; ENT: ear-nose-throat; ICU: intensive care units; OD: optical densities; PCR: polymerase chain reaction; SDS: sodium dodecyl sulfate; UTI: urinary tract infection
KEYWORDS: Enterococcus faecalis, CRISPR-Cas system, virulence genes, phenotypic characteristics, hospital-acquired bacteria, dental-root canal bacteria
Introduction
Enterococcus faecalisis a Gram-positive natural inhabitant of the mammalian digestive tract, including those of humans. It is also found in soil, plants, and dairy food products [1]. E. faecalisalso behaves as an opportunistic pathogen causing life-threatening infections in humans, such as endocarditis, meningitis, septicemia, urinary tract infections, and others [2,3]. E. faecalis is one of the frequent isolates of the endodontic pathogens ranging in terms of prevalence from 30% to 90% of the cases [4,5]. The restriction system of E. faecalisenables the bacterium to acquire, accumulate, and further transfer genetic elements potentially encoding antibiotic resistance genes and virulence factors. These virulence factors include exoenzymes and adhesins. Cytolysin is encoded by cyl operon, which is carried by a plasmid or integrated into the chromosome, with both hemolysin and bacteriocin activity [6,7]. Gelatinaseis is encoded by the chromosomal gelE gene, which is a zinc metalloprotease; it can hydrolyze gelatin, fibrinogen, collagen, casein, and insulin [8]. Another secreted factor is hyaluronidase, which is encoded by the hyl gene [7]. E. faecalis endocarditis antigen is encoded by theefaAgene that affects pathogenicity [9]. Cell wall-associated protein (Esp), encoded by pheromone-responsive plasmids or the chromosomal esp gene, is involved in biofilm formation and immune evasion [9]. Aggregation substance, encoded by theasa1 gene on the sex pheromone-plasmid pAD1, is a surface-bound glycoprotein which mediates the conjugative transfer of plasmids through the clumping of one E. faecalis to another and induces the formation of the cell-cell contact [10]. In addition, ebpRencodes an endocarditis- and biofilm-associated pilus regulator, which activates the ebpABC operon [11]. Another adhesion factor is a collagen-binding protein encoded by the ace gene, which mediates binding to collagen type I, collagen type IV, and laminin [12].
Clustered regularly interspaced short palindromic repeats (CRISPRs) loci and CRISPR-associated (Cas) protein-encoding genes are present in approximately 45% of eubacterial genomes sequenced [13–15]. There are three types of CRISPR loci in E. faecalis genome: CRISPR1-cas, orphan CRISPR2, and CRISPR3-cas [16–18]. CRISPR1-cas and orphan CRISPR2 were first found in the E. faecalis OG1RF strain: CRISPR1 is located between the OG1RF homolog of EF0672 and EF0673, which has the associated cas genes. CRISPR2 is located between the OG1RF homolog of EF2062 and EF2063, which is an orphan consisting only of spacers and palindromes, without any cas genes [16]. CRISPR3 was found in two genomes of the strains Fly1, as a fruit fly E. faecalis, and T11, as a urine E. faecalis isolate. CRISPR3 is located between the homologs of the E. faecalis V583 open reading frames EF1760 and EF1759 [18]. CRISPR1 possesses Nmeni subtype-specific genes csn1 and csn2 [16,18], while CRISPR3 only possesses csn1, not csn2 [18]. Both CRISPR1 and CRISPR2 contain seven repeats of a 37 bp palindromic sequence with no homology to any sequences of the 29 bp spacer [16]. Nevertheless, due to small spacer sequences, it is likely that they are derived from the pheromone-responsive type plasmids, plasmids integrated within the E. faecalis V583 genome, and Enterococcal prophage and phage [18]. Recent studies have demonstrated that the CRISPR/Cas system has applications for genome engineering and exerts a strong selective pressure for the acquisition of virulence factors and antibiotic resistance in pathogenic bacteria [18–21]. Mojica et al., for instance, have suggested that the pathogenicity of bacteria is largely controlled by conjugative plasmids and bacteriophages on an evolutionary timescale. As well, those CRISPR spacers that target these mobile elements might affect bacterial pathogenicity and virulence traits [22].
In this study, we aimed to assess the phenotypic and genotypic virulence determinants in relation to CRISPR elements from the dental-root canals and hospital-acquired isolates of E. faecalis.
Methods and materials
Bacterial strains
This study was approved by the Regional Ethics Committee of Tabriz (Tabriz University of Medical Sciences, Tabriz, Iran, No. IR.TBZMED.REC.1397.188). A total of 88 isolates of E. faecalis were collected from EmamReza Teaching and Treatment Hospital and pediatric hospitals of Tabriz, Iran. The specimen sources of hospital-acquired isolates included urinary tract infection (UTI) (78, 88.6%), wound (7, 7.9%), and blood (3, 3.4%). The specimens were obtained from different wards including outpatients (35, 39.8%), intensive (23, 26.1%), intensive care units (ICU) (12, 13.6%), infectious ward (13, 14.8%), emergency ward (3, 3.4%), ear-nose-throat (ENT) (1, 1.1%), urology, and nephrology (1, 1.1%). Forty-two (47.7%) isolates were from male and 46 (52.3%) were from female cases. The age range of patients was from 2 months to 86 y, with a mean of 39.04 y. At the same time, in order to collect 73 dental-root canal isolates of E. faecalis, patients in need of endodontic treatment were referred to the clinic of the Faculty of Dentistry at Tabriz University of Medical Sciences, Tabriz, Iran. Forty-nine (67.1%) of the isolates were obtained from the males and 24 (32.9%) from the females. The age range of endodontic treatment patients was 12–66 y, with a mean of 32.41 y. Briefly to collect the isolates, after stages of access cavity preparation by the dentist, tooth, and its surroundings were washed by sterile saline solutions and disinfected with 30% hydrogen peroxide followed by 2.5% sodium hypochlorite. Root canal of teeth with no prior endodontic treatment and teeth with previous root canal treatment that showed secondary infection was removed by drill and endodontic K-files without using any chemical solvents. After sampling the single root canal and multi-root canal of the teeth, paper points were transferred to a tube containing Enterococcal broth (Becton Dickenson microbiology systems, Cockeysville, MD) and cultured on a bile esculin azide agar (Himedia, India) and incubated at 37°C for 24–48 h [4]. Suspected colony was identified by the standard procedures of microbiology [23,24] and genotype detection was performed by ddlE primer [25,26], as shown in Table 1. Both clinical and tooth identified isolates for further studies were stored in a trypticase soy broth containing 10% glycerol at −70°C.
Table 1.
Primers used for the detection of virulence genes and CRISPR-associated genes.
| Gene | Primer | Sequence (5ʹ–3ʹ) | PCR product length (bp) | References |
|---|---|---|---|---|
| esp | espF | GGAACGCCTTGGTATGCTAAC | 95 | [46] |
| espR | GCCACTTTATCAGCCTGAACC | |||
| cylA | cylF | ACTCGGGGATTGATAGGC | 688 | [47] |
| cylR | GCTGCTAAAGCTGCGCTT | |||
| hyl | hylF | ACAGAAGAGCTGCAGGAAATG | 276 | [11] |
| hylR | GACTGACGTCCAAGTTTCCAA | |||
| efaA | efaF | TGGGACAGACCCTCACGAATA | 101 | [48] |
| efaR | CGCCTGTTTCTAAGTTCAAGCC | |||
| gelE | gelF | TATGACAATGCTTTTTGGGAT | 213 | [47] |
| gelR | AGATGCACCCGAAATAATATA | |||
| ace | aceF | GGAGAGTCAAATCAAGTACGTTGGTT | 101 | [49] |
| aceR | TGTTGACCACTTCCTTGTCGAT | |||
| ebpR | ebpF | AAAAATGATTCGGCTCCAGAA | 101 | [11] |
| ebpR | TGCCAGATTCGCTCTCAAAG | |||
| asa1 | asaF | GCACGCTATTACGAACTATGA | 375 | [47] |
| asaR | TAAGAAAGAACATCACCACGA | |||
| CRISPR1-cas csn1 | For | CAGAAGACTATCAGTTGGTG | 783 | [18] |
| Rev | CCTTCTAAATCTTCTTCATAG | |||
| CRISPR1-cas loci | For | GCGATGTTAGCTGATACAAC | 315 | [18] |
| Rev | CGAATATGCCTGTGGTGAAA | |||
| CRISPR2 loci | For | CTGGCTCGCTGTTACAGCT | variable | [18] |
| Rev | GCCAATGTTACAATATCAAACA | |||
| CRISPR3-cas csn1 | For | GCTGAATCTGTGAAGTTACTC | 258 | [18] |
| Rev | CTGTTTTGTTCACCGTTGGAT | |||
| CRISPR3-cas loci | For | GATCACTAGGTTCAGTTATTTC | 224 | [18] |
| Rev | CATCGATTCATTATTCCTCCAA |
Biofilm formation
Assessment of biofilm formation was done by quantitative biofilm formation in 96-well flat-bottom polystyrene microplates under static conditions for 48 h, as previously described [27,28]. Briefly, for each isolate, afresh colony cultured on a Muller-Hinton agar (Merck, Germany) containing 1% glucose was suspended in sterile saline and adjusted to 0.5 McFarland. Twenty microliters of the adjusted isolates was cultured in a 180-µl trypticase soy broth containing 1% glucose. After incubation for 48 h at 37°C, each well was washed by the 1X phosphate buffer saline (PBS; pH 7.4), fixed by methanol, and stained by 200 µl 0.1% crystal violet for 30 min at room temperature. The excess crystal violet was discarded and washed by water flow. Biofilm formation was measured by the absorbance of the supernatant after being solubilized in 33% acetic acid at 570 nm by using a microtiter plate reader (BioTeck, Winooski, USA). The biofilm formation of each isolate was tested in three independent 96-well microplates and the average of three optical densities (OD) was used as the final biofilm formation value. The cutoff absorbance for biofilm formation was considered higher than OD = 0.524, which was the absorbance of the biofilm produced byE. faecalisATCC® 29,212™. The mean of the biofilm formation of each isolate was grouped based on their level of distribution (OD570nm values) and categorized in quartiles higher than the cutoff absorbance and lower than the highest absorbance. Isolates whose absorbance of OD570nm fell below 0.524 were classed as non-biofilm formation, while those with 0.525–1.087 and 1.088–1.650 were grouped as low and moderate biofilm formation, respectively. Isolates with a biofilm formation greater than 1.651 were also considered with high biofilm formation.
Gelatinase production and hemolysis test
Hemolysis activity was assessed by blood agar plates prepared by brain–heart infusion agar (BHI, biomerieux, Poland, Ltd) containing 5% of the group ORh+ human blood. Cleared or green zone around the colonies was defined as hemolysis following incubation for 24 h at 37°C [29].
Production of gelatinase was assessed by the degradation of gelatin on the X-ray radiographic film, as described by Pickett et al. [30]. The heavy inoculum of individual isolates was cultured in the tubes containing 3 ml MHB and a strip of the X-ray radiographic film which had been cut into small strips (approximately 6 by 30 mm). The tubes were incubated for 24 h at 37°C and the cleared strip was defined as the production of gelatinase.
Genotype detection of virulence and cas genes
Total DNA for each isolate was extracted by the tissue buffer boiling method. Briefly, 20 µl tissue buffer (0.25% sodium doedecyl sulfate (SDS) and 0.05 M NaOH) were mixed with one colony of bacterial isolate and incubated at 95°C for 10 min. The suspension was centrifuged at 13,000 g for 1 min, and 180 µl DNase free water was added. Genotype analysis for each isolate was accomplished based on the multiplex polymerase chain reaction (PCR) of virulence determinants encoding the cytolysin activator cylA, hyl, esp, gelE, efaA, asa1, ace, ebpR, CRISPR1-cas, CRISPR1-cascsn1, CRISPR2, CRISPR3-cas, and CRISPR3-cascsn1. Each of the primer sequences and the amplified size are shown in Table 1. Two microliters of total DNA was used for the multiplex PCR in a 25 µl reaction mixture. The mix for the detection of esp, cyl, hyl genes contained 12.5 µl of the PCR master mix (Yekta Tajhiz Azma, Iran), with 0.5 µM of each primer. The mix for ebp, asa1, and efaA had the same condition. The mix for the detection of gelE and ace contained 12.5 µl of the PCR master mix (Yekta Tajhiz Azma, Iran), 1.5 mM-additional MgCl2 and 0.5 µM of each primer. The mix for CRISPR1-cascsn1, CRISPR3-cascsn1, CRISPR1-cas, CRISPR3-cas, and CRISPR2 contained 12.5 µl of the PCR master mix (Yekta Tajhiz Azma, Iran), 1 mM additional MgCl2, and 10 mM of each primer. The amplification condition was carried out with the following thermal cycling conditions: initial denaturation at 95°C for 10 min, 34 cycles of amplification consisting of 95°C for 30 s, 30 s at 58°C for esp, cylA, hyl, 58°C for efaA, 56°C for gel, ace, 52°C for ebpR, asa1, 60°C for all cas genes, and 72°C for 45 s, with 72°C for 5 min in the final polymerization. PCR products were analyzed by electrophoresis in a 1% agarose gel at 100 V for 1 h in a 1X TBE buffer containing the DNA safe stain. The size of the PCR product was correlated with a 100 based-pair DNA ladder (YektaTajhizAzma, Iran) to confirm the conjunction with their expected PCR amplicon size. In addition, the PCR procedure for each isolate was carried out twice in the case of each primer in order to check the consistency and reproducibility.
Statistical analysis
SPSS software, version 17.0 (Chicago, IL, USA) was used for statistical analysis. One-tailed Fisher’s exact test was used to compare the occurrence of CRISPR-cas loci in hospital-acquired and dental-root canal isolates and to evaluate the distribution of biofilm formation, gelatinase and hemolysin activities, and virulence genes among strains with CRISPR-cas. Student’s t-test was used to compare OD values among hospital-acquired and dental-root canal isolates. In addition, Spearman’s rank correlation was calculated between the presence of different virulence genes and CRISPR-cas loci among isolates. Significance was set at P ≤ 0.05.
Results
All isolates were investigated for the biofilm formation, in which the minimum, maximum, and average of biofilm formation (OD570 nm) were 0.054, 2.325, and 1.611, respectively. Most isolates showed strong biofilm formation (94, 58.4%), while 10 (6.2%) displayed no biofilm formation. Biofilm formation of hospital-acquired isolates was significantly higher than the dental-root canal isolates (P = 0.023). The biofilm formation absorbance according to the presence of virulence factors and CRISPR loci among E. faecalis isolates shown in Figure 1. Most of the isolates showed no gelatinase activity (70.8%), while hospital-acquired isolates significantly displayed the most gelatinase activity (P = 0.001). In addition, most isolates showed no hemolysis activity (80.7%), and all hemolysis activity was found in hospital-acquired isolates (19.2%). The most presence of the virulence genes among isolates were ace andefaAgenes (88.8% and 85.1%, respectively), and the lowest one belonged tocylA and asa1 (7.5% and 14.9%, respectively). The presence of gelE (contributing to gelatinase activity) and cylA (contributing to hemolysis activity) was significantly associated with phenotype gelatinase and hemolysis activity, respectively (P < 0.001, P = 0.013). In addition, the presence of efaA, cylA, and gelE was significantly more in hospital-acquired isolates, as compared to dental-root canal (P = 0.002, P < 0.001 and P = 0.008, respectively). Genotypic, and phenotypic determinants of hospital-acquired and dental-root canal isolates are shown in Table 2. TheefaA and gelEharboring isolates had a higher biofilm formation than negative isolates in all isolates (P = 0.017 and P = 0.042, respectively). The biofilm formation absorbance association to virulence genes and CRISPR loci among E. faecalis isolates is shown in Figure 2. By comparing the presence of virulence genes among isolates, it was found that hospital-acquired isolates had higher virulence genes than dental-root canal isolates (P = 0.007), such that all isolates had at least one virulence gene. The distribution of virulence gene counts among E. faecalis isolates is presented in Figure 3. The number of virulence genes was 1–7 among hospital-acquired isolates and 16 in the case of dental-root canal isolates. Among hospital-acquired isolates, the presence of five and four virulence genes was the highest (36.4% and 30.7%, respectively); also, the presence of 4 and 3 virulence genes was the highest among isolates of the dental-root canal (39.7% and 31.5%, respectively).
Figure 1.
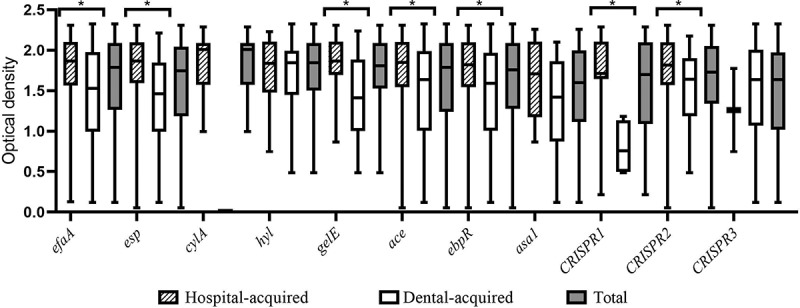
Biofilm formation absorbance by E. faecalis isolates according to the presence of virulence factors and CRISPR loci. (Error bars illustrate the minimum and maximum of three replicates of absorbance of the biofilm formation; *P-value was significant (P-value<0.05.)
Table 2.
Genotypic and phenotypic determinants of hospital-acquired and dental-root canal isolates.
|
esp |
cylA |
hyl |
efaA |
gelE |
ace |
ebpR |
asa1 |
Gelatinase activity |
Hemolysis |
Biofilm formation |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | P (n) | P (n) | P (n) | P (n) | P (n) | P (n) | P (n) | P (n) | Positive (n) | α | β | γ | N | + | ++ | +++ |
| Hospital-acquired (88) | 73.9% (65) | 13.6% (12) | 15.9% (14) | 93.2% (82) | 37.5% (33) | 88.6% (78) | 79.5% (70) | 15.9% (14) | 39.8% (35) | 29.5% (26) | 5.7% (5) | 64.8% (57) | 4.5% (4) | 6.8% (6) | 20.5% (18) | 68.2% (60) |
| UTIs (78) | 78.2% (61) | 15.4% (12) | 14.1% (11) | 93.6% (73) | 34.6% (27) | 89.7% (70) | 78.2% (61) | 16.7% (13) | 39.7% (31) | 29.5% (23) | 6.4% (5) | 64.1% (50) | 5.1% (4) | 3.8% (3) | 21.8% (17) | 69.2% (54) |
| Non-UTIs (10) | 40% (4) | 0 | 30% (3) | 90% (9) | 60% (6) | 80% (8) | 90% (9) | 10% (1) | 40% (4) | 30% (3) | 0 | 70% (7) | 0 | 30% (3) | 10% (1) | 60% (6) |
| Dental root (73) | 68.5% (55) | 0 | 24.7% (18) | 75.3% (55) | 19.2% (14) | 89% (65) | 84.9% (62) | 13.7% (10) | 16.4% (12) | 0 | 0 | 100% (73) | 8.2% (6) | 19.2% (14) | 26% (19) | 46.6% (34) |
| P-value§ | 0.282 | <0.001 | 0.118 | 0.002 | 0.008 | 0.570 | 0.249 | 0.435 | 0.001 | <0.001 | 0.023 | |||||
| Total (161) | 71.4% (115) | 7.5% (12) | 19.9% (32) | 85.1% (137) | 29.2% (47) | 88.8% (143) | 82% (132) | 14.9% (24) | 29.2% (47) | 16.1% (26) | 3.1% (5) | 80.7% (130) | 6.2% (10) | 12.4% (20) | 23% (37) | 58.4% (94) |
UTIs, urinary tract infections; non-UTIs: other isolates containing wound and blood; P: presence; N: negative; α: alpha hemolysin; β: beta hemolysin; γ: none hemolysin; +: low; ++: moderate; +++: strong
§ One-tailed Fisher’s exact test was used for comparison of hospital-acquired and dental-root canal groups.
Figure 2.
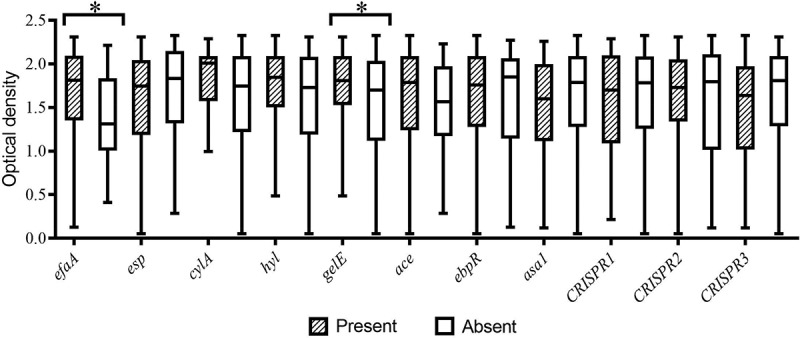
Biofilm formation absorbance association to virulence genes and CRISPR loci among E. faecalis isolates. (Error bars illustrate the minimum and maximum of three replicates of absorbance of the biofilm formation; *P-value was significant (P-value<0.05.)
Figure 3.
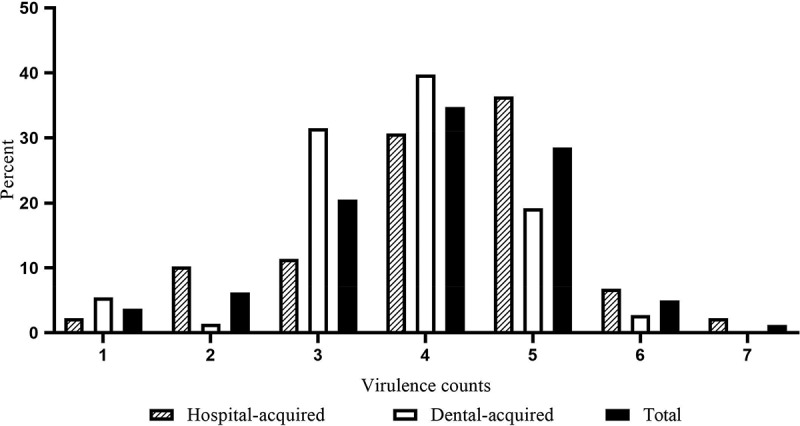
Distribution of virulence gene counts among E. faecalis isolates.
The occurrence of CRISPR-cas is shown in Table 3. Overall, the presence of CRISPR1-cas loci in dental-root canal isolates (4 of 73) was lower than that of hospital-acquired isolates (17 of 88) (P = 0.008), whereas the presence of CRISPR3-cas in dental-root canal isolates (26 of 73) was higher than that of hospital-acquired isolates (2 of 88) (P < 0.001); also, orphan CRISPR2 made no difference between hospital-acquired and dental-root canal isolates. None of the isolates had, however, both of CRISPR1-cas and CRISPR3-cas, as well as CRISPR1-cas, orphan CRISPR2, and CRISPR3-cas, at the same time. The isolates were more likely to harbor orphan CRISPR2 than CRISPR1-cas and CRISPR3-cas.In addition, the presence of orphan CRISPR2 was significantly correlated with CRISPR1-cas (P = 0.031, correlation coefficient = 0.163), whereas it was not significant with CRISPR3-cas. At least one CRISPR-cas locus was found in 106 (65.8%) of all isolates. The results, therefore, showed the isolates containing high virulence genes tended to have more frequently investigated cas genes. The presence of CRISPR1 and CRISPR 2 was significantly correlated with high distribution of virulence gene numbers (P = 0.010 and P = 0.011, respectively). The virulence gene counts association to CRISPR loci among E. faecalis isolates is shown in Figure 4. Overall, the absence of CRISPR1-casand one of CRISPR1 or CRISPR3weresignificantly correlated with the absence of the esp gene (P = 0.005, correlation coefficient = 0.204 andP = 0.033, correlation coefficient = 0.157, respectively). In addition, the presence of either CRISPR1-cas or orphan CRISPR2 and either CRISPR3-cas or orphan CRISPR2 was significantly correlated with the presence of ace and the absence of gelE, respectively (P = 0.019, correlation coefficient = 0.185 and P = 0.014, correlation coefficient = 0.184, respectively). In addition, presence of CRISPR1-cas was significantly correlated with the absence of hyl (P = 0.048, correlation coefficient = −0.147). Other significant correlations were found between the absence of CRISPR1 and the absence of cylAandasa1 (P < 0.05, correlation coefficient = 0.171 and 0.149, respectively), and between the absence of CRISPR2 and the absence of gelE (P = 0.001, correlation coefficient = 0.248). In hospital-acquired isolates, a significant correlation was found between the absence of CRISPR loci and the absence of gelE, asa1, gelatinase and hemolysis activity (P < 0.05); in dental-root canal isolates, a significant correlation was found between the absence of CRISPR3-cas and the absence of gelatinase (P = 0.003, correlation coefficient = 0.365), between the absence of either CRISPR1-cas or CRISPR2-cas and the absence of gelE (P = 0.021, correlation coefficient = 0.265), and between the presence of orphan CRISPR2 and either orphan CRISPR2 or CRISPR3-cas and biofilm production (P = 0.046, correlation coefficient = 0.247 andP = 0.044, correlation coefficient = 0.263, respectively) (see Table 4).
Table 3.
The presence of CRISPR-cas type in hospital-acquired and dental-root isolates of E. faecalis.
| CRISPR | CRISPR1-cas | CRISPR2 | CRISPR3-cas | CRISPR1-cas or CRISPR2 | CRISPR1-cas or CRISPR3-cas | CRISPR2 or CRISPR3-cas | CRISPR1-cas and CRISPR2 | CRISPR1-cas and CRISPR3-cas | CRISPR2 and CRISPR3-cas | CRISPR1-cas and CRISPR2 and CRISPR3-cas |
|---|---|---|---|---|---|---|---|---|---|---|
| Hospital-acquired isolates (88) | 19.3% (17) | 53.4% (47) | 2.3% (2) | 59.1% (52) | 21.6% (19) | 54.5% (48) | 13.6% (12) | 0 | 1.1% (1) | 0 |
| Dental root isolates (73) | 5.5% (4) | 57.5% (42) | 35.6% (26) | 58.9% (43) | 42.5% (31) | 72.6% (53) | 5.5% (4) | 0 | 20.5% (15) | 0 |
| P-value§ | 0.008 | 0.358 | <0.001 | 0.554 | 0.004 | 0.014 | 0.070 | NS | <0.001 | NS |
| Total (161) | 13% (21) | 55.3% (89) | 17.4% (28) | 59% (95) | 31.1% (50) | 62.7% (101) | 9.9% (16) | 0 | 9.9% (16) | 0 |
NS, not significant.
§ One-tailed Fisher’s exact test was used for comparison of hospital-acquired and dental-root canal groups.
Figure 4.
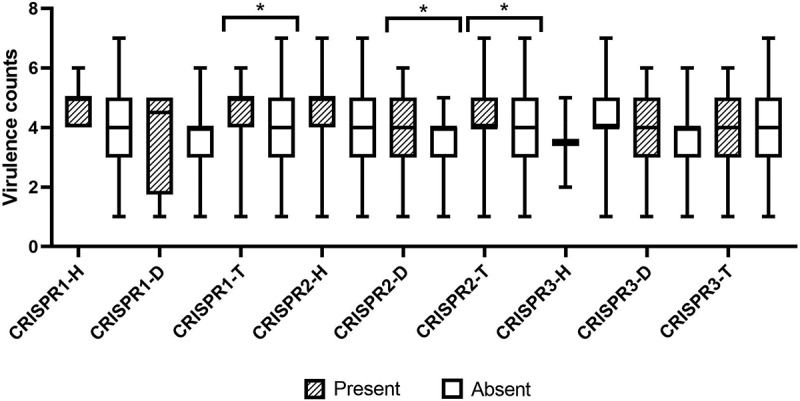
Virulence gene counts association to CRISPR loci among E. faecalis isolates. (Error bars illustrate the minimum and maximum of virulence gene counts; *P-value was significant (P-value <0.05; H: Hospital-acquired; D: Dental-acquired; T: Total).
Table 4.
Association between genotypic and phenotypic characteristics and the occurrence of CRISPR-cas in E. faecalis.
| Gene | CRISPR1-present | CRISPR1-absent | P-value | CRISPR2-present | CRISPR2-absent | P-value | CRISPR3-present | CRISPR3-absent | P-value | CRISPR1 or CRISPR2-present | CRISPR1 or CRISPR2-absent | P-value | CRISPR1 or CRISPR3-present | CRISPR1 or CRISPR3-absent | P-value | CRISPR2 or CRISPR3-present | CRISPR2 or CRISPR3-absent | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| esp-present | 20 | 95 | 0.005 | 64 | 51 | 0.509 | 21 | 94 | 0.417 | 68 | 47 | 0.549 | 41 | 74 | 0.033 | 72 | 43 | 0.554 |
| esp-absent | 1 | 45 | 25 | 21 | 7 | 39 | 27 | 19 | 9 | 37 | 29 | 17 | ||||||
| cylA-present | 4 | 8 | 0.053 | 6 | 6 | 0.464 | 0 | 12 | 0.092 | 7 | 5 | 0.594 | 4 | 8 | 0.544 | 6 | 6 | 0.258 |
| cylA-absent | 17 | 132 | 83 | 66 | 28 | 121 | 88 | 61 | 46 | 103 | 95 | 54 | ||||||
| hyl-present | 1 | 31 | 0.048 | 20 | 12 | 0.237 | 6 | 26 | 0.500 | 21 | 11 | 0.260 | 7 | 25 | 0.149 | 23 | 9 | 0.161 |
| hyl-absent | 20 | 109 | 69 | 60 | 22 | 107 | 74 | 55 | 43 | 86 | 78 | 51 | ||||||
| efaA-present | 20 | 117 | 0.139 | 75 | 62 | 0.461 | 22 | 115 | 0.214 | 81 | 56 | 0.557 | 43 | 94 | 0.518 | 86 | 51 | 0.575 |
| efaA-absent | 1 | 23 | 14 | 10 | 6 | 18 | 14 | 10 | 7 | 17 | 15 | 9 | ||||||
| gelE-present | 7 | 40 | 0.415 | 35 | 12 | 0.001 | 5 | 42 | 0.108 | 36 | 11 | 0.003 | 12 | 35 | 0.217 | 36 | 11 | 0.014 |
| gelE-absent | 14 | 100 | 54 | 60 | 23 | 91 | 59 | 55 | 38 | 76 | 65 | 49 | ||||||
| ace-present | 20 | 123 | 0.282 | 84 | 59 | 0.012 | 26 | 117 | 0.358 | 89 | 54 | 0.019 | 46 | 97 | 0.285 | 94 | 49 | 0.027 |
| ace-absent | 1 | 17 | 5 | 13 | 2 | 16 | 6 | 12 | 4 | 14 | 7 | 11 | ||||||
| ebpR-present | 18 | 114 | 0.451 | 75 | 57 | 0.263 | 24 | 108 | 0.399 | 80 | 52 | 0.250 | 42 | 90 | 0.418 | 85 | 47 | 0.235 |
| ebpR-absent | 3 | 26 | 14 | 15 | 4 | 25 | 15 | 14 | 8 | 21 | 16 | 13 | ||||||
| asa1-present | 6 | 18 | 0.067 | 15 | 9 | 0.293 | 3 | 21 | 0.363 | 17 | 7 | 0.146 | 9 | 15 | 0.303 | 16 | 8 | 0.425 |
| asa1-absent | 15 | 122 | 74 | 63 | 25 | 112 | 78 | 59 | 41 | 96 | 85 | 52 | ||||||
| Biofilm-positive | 19 | 132 | 0.384 | 86 | 65 | 0.092 | 27 | 124 | 0.453 | 91 | 60 | 0.176 | 46 | 105 | 0.377 | 97 | 54 | 0.117 |
| Biofilm-negative | 2 | 8 | 3 | 7 | 1 | 9 | 4 | 6 | 4 | 6 | 4 | 6 | ||||||
| Gelatinase-positive | 4 | 43 | 0.203 | 29 | 18 | 0.190 | 10 | 37 | 0.268 | 30 | 17 | 0.268 | 14 | 33 | 0.490 | 32 | 15 | 0.236 |
| Gelatinase-negative | 17 | 97 | 60 | 54 | 18 | 96 | 65 | 49 | 36 | 78 | 69 | 45 | ||||||
| Hemolysis-positive | 4 | 27 | 0.623 | 19 | 12 | 0.293 | 0 | 31 | 0.001 | 19 | 12 | 0.469 | 4 | 27 | 0.010 | 19 | 12 | 0.504 |
| Hemolysis-negative | 17 | 113 | 70 | 60 | 28 | 102 | 76 | 54 | 46 | 84 | 82 | 48 |
Discussion
In this study, we determined the occurrence of CRISPR loci and the content of virulence factors in E. faecalis strains isolated from different infectious sources as a pathogenic organism and the dental-root canal of patients. We found that the presence of CRISPR1 and CRISPR3 loci was varied among E. faecalis strains. The abundance of CRISPR1 among the dental-root canal isolates was significantly lower than that of hospital-acquired ones, whereas the reverse was significantly true for CRISPR3. These results were consistent with those obtained by Burley et al. study [31], who found the presence of CRISPR3-caswas significantly more in endodontic strains, as compared to hospital-acquired strains, and the majority of strains had CRISPR3. While these results were interesting, the reasons were not clear. In addition, we found that the presence of orphan CRISPR2 was more among E. faecalis strains in comparison to CRISPR1-cas and CRISPR3-cas, while CRISPR2 Lacks of cas genes. Palmer et al. [18] and Hullahalli et al. [32]suggested that CRISPR2 is functional for sequence interference and is functionally linked to CRISPR1-Cas or CRISPR3-Cas.
The results revealed that the presence of CRISPR loci was not significantly associated with a less number of virulence factors. There are several virulence factors in E. faecalis which play such roles as antiphagocytosis, adherence, biofilm formation, exoenzyme, toxin, and quorum sensing system. Although several studies have reported that there is no clear relation between origin isolation or a single gene and pathogenicity, and perhaps the surface proteins of E. faecaliscannot be considered as virulence factors [9,33,34], we found a correlation between the absence of CRISPR1-cas and the absence of theesp gene (P-value = 0.009, coefficient correlation = 0.204) and a correlation between the absence of CRISPR1-cas and the absence of cylA (P-value = 0.03, coefficient correlation = 0.171) and asa1 (P-value = 0.06, coefficient correlation = 0.149) genes. In addition, there was a correlation between the absence of single or multi-CRIPSR loci and the absence of some virulence factors. The cytolysin operon, cob and esp genes reside in the same pathogenicity island, which are located on either the chromosome or on large pheromone-responsive plasmids such as pAD1 [35,36]. The esp gene encodes a large surface protein with a variable number of highly conserved 82 amino acids repeats, contributing to the promotion of primary attachment, colonization and biofilm formation ofE. faecalis [36]. Our results, therefore, showed that the presence ofefaA,esp, gelE, ace, and ebpR genes were significantly associated with biofilm formation among the hospital-acquired isolates and efaA and gelE genes were significantly associated with biofilm formation in all E. faecalis isolates. Conflict outcomes have been, however, published regarding the role of the genes of biofilm formation. Duggan et al., for example, suggested that asa1, cylA, esp and gelEwere not associated with biofilm formation in the oral and endodontic isolates of E. faecalis [37], which is compatible with our results. In addition, the results revealed that 13.6% of hospital-acquired isolates carried thecylA gene, but only 35.2% of the isolates expressed hemolysin activity (both alpha and beta hemolysis). Several studies such as Sun et al. [38], Sedgley et al. [39] and Lindenstrauß et al. [40] have also determined 38%, 36% and 33.3% of the chronic periodontitis, endodontic, and clinical and food isolates of E. faecalis to be capable of producing hemolysis, respectively. These differences may be due to the differences in the types of blood used for the determination of the hemolysis activity, while we used human blood, others have employed horse and sheep blood. In addition, Sun et al. [38] and Sedgley et al. [39] reported the distribution of the cylA gene was detected only in 17% and 18.18% of the isolates, respectively; this was compatible with our results. These results may be due to such environmental factors as in vitro and in vivo conditions used to test for phenotypic characters, which could strongly influence gene expression [41] and can be the cause of the differences between our results and those obtained by others in the case of hemolysis activity. In addition, hemolysin activity was encoded by cyl operon in E. faecalis, where cylA is the only reading frame required for the expression of component A, a serine protease. As well, there is no association between CRISPR1-cas, biofilm-formation, and hemolysis activity. Several studies have reported that CRISPR loci play an inverse role in some virulence factors and acquisition of antibiotic resistance [18,31,40], such as Palmer and Gilmore’s study [42] and Burley et al.’s study [31], reporting that CRISPR loci were inversely associated with antibiotic resistance and some virulence factors in E. faecalis strains. In addition, similar to our results, Toro et al. [43] and Touchon et al. [44] reported that there was no significant association with CRISPR-cas and acquisition of integrons, plasmids, antibiotic resistance and virulence genes in Escherichia coli. However, an analysis of 370 other Archaeal and Eubacteria genomes showed that there was potential evidence for the propagation of CRISPR-cas genes to occur via horizontal gene transfer [45]. These findings, therefore, suggested that CRISPR loci could potentially inhibit or prevent some or part of the virulence factors and Pathogenicity Island could not serve as the selective forces to influence the pathogenic traits of E. faecalis.
Conclusion
The findings of this study indicated that CRISPR-casmightprevent the acquisition of some respective pathogenicity factors in some isolates, though not all; significant inverse correlations were found between CRISPR-cas loci, esp and gelE, while direct ones were found in cylA, hyl, gelE (between some CRISPR-loci), asa1, ace, biofilm formation, gelatinase, and hemolysis activities. However, other studies demonstrated that CRISPR-cas could prevent the acquisition of antibiotic resistance genes in E. faecalis and other bacteria. Further studies can determine the exact role of CRISPR-cas in the pathogenesis of Enterococcal infections.
Acknowledgments
We thank Antwerp University, Belgium, for providing us with the reference strains for study.
Funding Statement
This study was supported by Iran National Science Foundation (INSF) with the grant number 97015174 and Faculty of Medicine, Tabriz University of Medical Sciences.
Disclosure statement
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
This study was approved by the Regional Ethics Committee of Tabriz (Tabriz University of Medical Sciences, Tabriz, Iran, No. IR.TBZMED.REC.1397.188).
Consent to publish
For this study, the formal consent was filled by all participants before any procedure according to the ethics committee approved procedure.
Authors’ contributions
HSK, MY, MB and KG designed this study and discussed related methods; PG did experimental analysis, performed the statistical analysis, and was a major contributor in writing the manuscript. TP, MAR collected the data; PL reviewed the manuscript and put forward the comments; RQ, MA suggested and revised the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
The datasets used in this study are available from the corresponding author on reasonable request.
References
- [1].Tannock GW, Cook G.. Enterococci as members of the intestinal microflora of humans. In: Gilmore MS, Clewell DB, Courvalin P, et al., editors. The enterococci. [Google Scholar]
- [2].Baldassarri L, Creti R, Recchia S, et al. Virulence factors in enterococcal infections of orthopedic devices. Int J Artif Organs. 2006;29:402–406. [DOI] [PubMed] [Google Scholar]
- [3].Mohamed JA, Huang W, Nallapareddy SR, et al. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72:3658–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gomes BPFA, Pinheiro ET, Sousa ELR, et al. Enterococcus faecalis in dental root canals detected by culture and by polymerase chain reaction analysis. Oral Sur Oral Med Oral Pathol Oral Radiol Endodontol. 2006;102:247–253. [DOI] [PubMed] [Google Scholar]
- [5].Hidron AI, Edwards JR, Patel J, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. [DOI] [PubMed] [Google Scholar]
- [6].Gilmore MS, Segarra RA, Booth MC, et al. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol. 1994;176:7335–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Su Y, Sulavik M, He P, et al. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect Immun. 1991;59:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Galli D, Lottspeich F, Wirth R. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol Microbiol. 1990;4:895–904. [DOI] [PubMed] [Google Scholar]
- [11].Bourgogne A, Singh KV, Fox KA, et al. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol. 2007;189:6490–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nallapareddy SR, Qin X, Weinstock GM, et al. Enterococcus faecalis adhesin, ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun. 2000;68:5218–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR–Cas systems. Nature Rev Microbiol. 2011;9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bourgogne A, Garsin DA, Qin X, et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Haft DH, Selengut J, Mongodin EF, et al. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1:e00227–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gholizadeh P, Aghazadeh M, Asgharzadeh M, et al. Suppressing the CRISPR/Cas adaptive immune system in bacterial infections. Eur J Clin Microbiol Infect Dis. 2017;36:2043–2051. [DOI] [PubMed] [Google Scholar]
- [20].Marraffini LA, Sontheimer EJ. CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science. 2008;322:1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. [DOI] [PubMed] [Google Scholar]
- [22].Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic Elements. J Mol Evol. 2005;60:174–182. [DOI] [PubMed] [Google Scholar]
- [23].Facklam R, Collins M. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kafil HS, Mobarez AM, Moghadam MF, et al. Gentamicin induces efaA expression and biofilm formation in Enterococcus faecalis. Microb Pathog. 2016;92:30–35. [DOI] [PubMed] [Google Scholar]
- [25].Kafil HS, Mobarez AM, Moghadam MF. Adhesion and virulence factor properties of Enterococci isolated from clinical samples in Iran. Indian J Pathol Microbiol. 2013;56:238. [DOI] [PubMed] [Google Scholar]
- [26].Kariyama R, Mitsuhata R, Chow JW, et al. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 2000;38:3092–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Aghazadeh M, Zahedi Bialvaei A, Kabiri F, et al. Survey of the antibiofilm and antimicrobial effects of zingiber officinale (in Vitro Study). Jundishapur J Microbiol. 2016;9:e30167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kafil HS, Mobarez AM. Assessment of biofilm formation by enterococci isolates from urinary tract infections with different virulence profiles. J King Saud Univ Sci. 2015;27:312–317. [Google Scholar]
- [29].Gaspar F, Crespo M, Lopes MS. Proposal for a reliable enterococcal cytolysin production assay avoiding apparent incongruence between phenotype and genotype. J Med Microbiol. 2009;58:1122–1124. [DOI] [PubMed] [Google Scholar]
- [30].Pickett M, Greenwood J, Harvey S. Tests for detecting degradation of gelatin: comparison of five methods. J Clin Microbiol. 1991;29:2322–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Burley KM, Sedgley CM. CRISPR-Cas, a prokaryotic adaptive immune system, in endodontic, oral, and multidrug-resistant hospital-acquired enterococcus faecalis. J Endod. 2012;38:1511–1515. [DOI] [PubMed] [Google Scholar]
- [32].Hullahalli K, Rodrigues M, Palmer KL. Exploiting CRISPR-Cas to manipulate Enterococcus faecalis populations. eLife. 2017;6:e26664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hew CM, Korakli M, Vogel RF. Expression of virulence-related genes by Enterococcus faecalis in response to different environments. Syst Appl Microbiol. 2007;30:257–267. [DOI] [PubMed] [Google Scholar]
- [34].Martin B, Garriga M, Hugas M, et al. Genetic diversity and safety aspects of enterococci from slightly fermented sausages. J Appl Microbiol. 2005;98:1177–1190. [DOI] [PubMed] [Google Scholar]
- [35].Shankar N, Coburn P, Pillar C, et al. Enterococcal cytolysin: activities and association with other virulence traits in a pathogenicity island. Int J Med Microbiol. 2004;293:609–618. [DOI] [PubMed] [Google Scholar]
- [36].Shankar N, Lockatell CV, Baghdayan AS, et al. Role of Enterococcus faecalissurface protein ESP in the pathogenesis of ascending urinary tract infection. Infect Immun. 2001;69:4366–4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Duggan JM, Sedgley CM. Biofilm formation of oral and endodontic enterococcus faecalis. J Endod. 2007;33:815–818. [DOI] [PubMed] [Google Scholar]
- [38].Sun J, Sundsfjord A, Song X. Enterococcus faecalis from patients with chronic periodontitis: virulence and antimicrobial resistance traits and determinants. Eur J Clin Microbiol Infect Dis. 2012;31:267–272. [DOI] [PubMed] [Google Scholar]
- [39].Sedgley CM, Molander A, Flannagan SE, et al. Virulence, phenotype and genotype characteristics of endodontic Enterococcus spp. Oral Microbiol Immunol. 2005;20:10–19. [DOI] [PubMed] [Google Scholar]
- [40].Lindenstrauß AG, Pavlovic M, Bringmann A, et al. Comparison of genotypic and phenotypic cluster analyses of virulence determinants and possible role of CRISPR elements towards their incidence in Enterococcus faecalis and Enterococcus faecium. Syst Appl Microbiol. 2011;34:553–560. [DOI] [PubMed] [Google Scholar]
- [41].Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Palmer KL, Carniol K, Manson JM, et al. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J Bacteriol. 2010;192:2469–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Toro M, Cao G, Ju W, et al. Association of clustered regularly interspaced short palindromic repeat (CRISPR) elements with specific serotypes and virulence potential of Shiga toxin-producing Escherichia coli. Appl Environ Microbiol. 2014;80:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Touchon M, Charpentier S, Pognard D, et al. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology. 2012;158:2997–3004. [DOI] [PubMed] [Google Scholar]
- [45].Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–729. [DOI] [PubMed] [Google Scholar]
- [46].Shankar V, Baghdayan AS, Huycke MM, et al. Infection-derived Enterococcus faecal strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vankerckhoven V, Van Autgaerden T, Vael C, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42:4473–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lowe A, Lambert P, Smith A. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nallapareddy SR, Murray BE. Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host-E. faecalis interaction. Infect Immun. 2006;74:4982–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in this study are available from the corresponding author on reasonable request.


