ABSTRACT
With the outbreak of the recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019, coronaviruses have become a global research hotspot in the field of virology. Coronaviruses mainly cause respiratory and digestive tract diseases, several coronaviruses are responsible for porcine diarrhea, such as porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), and emerging swine acute diarrhea syndrome coronavirus (SADS-CoV). Those viruses have caused huge economic losses and are considered as potential public health threats. Porcine torovirus (PToV) and coronaviruses, sharing similar genomic structure and replication strategy, belong to the same order Nidovirales. Here, we developed a multiplex TaqMan-probe-based real-time PCR for the simultaneous detection of PEDV, PDCoV, PToV, and SADS-CoV for the first time. Specific primers and TaqMan fluorescent probes were designed targeting the ORF1a region of PDEV, PToV, and SADS-CoV and the ORF1b region of PDCoV. The method showed high sensitivity and specificity, with a detection limit of 1 × 102 copies/μL for each pathogen. A total of 101 clinical swine samples with signs of diarrhea were analyzed using this method, and the result showed good consistency with conventional reverse transcription PCR (RT-PCR). This method improves the efficiency for surveillance of these emerging and reemerging swine enteric viruses and can help reduce economic losses to the pig industry, which also benefits animal and public health.
KEYWORDS: Real-time PCR, swine diarrhea virus, coronavirus, detection
Introduction
Coronaviruses, belonging to the family Coronaviridae, order Nidovirales, are single-stranded, positive-sense RNA viruses with the largest genome among known RNA viruses [1,2]. According to genetic and antigenic characteristics, coronaviruses can be divided into four genera: α-coronaviruses, β-coronaviruses, γ-coronaviruses, and δ-coronaviruses [3–5]. Coronaviruses can cause respiratory and gastrointestinal diseases in animals and humans [6]. Generally, α- and β-coronaviruses only infect mammals, while γ- and δ-coronaviruses mainly infect birds, but some of them can also infect mammals [4]. Of note, coronaviruses exhibit a pronounced propensity for interspecies transmission as illustrated by important emerging viruses in humans such as SARS-CoV and Middle East respiratory syndrome-related coronavirus (MERS-CoV), as well as the recent SARS-CoV-2 that is causing a major human pandemic [7,8,9].
Compared to many other species, pigs are in frequent contact with both humans and other animals such as pets, livestock and wild animals, and theoretically possess a greater chance to promote cross-species viral transmission. There are currently six coronaviruses that can infect pigs: PEDV, transmissible gastroenteritis virus (TGEV), porcine respiratory coronavirus (PRCV), porcine hemagglutinating encephalomyelitis virus (PHEV), SADS-CoV, and PDCoV [10]. With the exception of PRCV and PHEV, the remaining four can all cause severe diarrhea, dehydration, and death in pigs [3,11]. PEDV is an α-coronavirus that causes long-lasting and extremely harmful swine diarrhea worldwide. PEDV may have originated from bat coronavirus [12]. It has high genome variability and different degrees of virulence among different strains [13]. PEDV strains can be divided into genotype G1 and genotype G2 with high genetic diversity. Both genotypes can cause catastrophic herd harm, of which the G2 variant has spread rapidly worldwide since 2010 [14–17]. SADS-CoV is also an α-coronavirus and is regarded to share a relationship with rhinolophus bat coronavirus HKU2 [18]. It was first discovered in January, 2017 in Guangdong, China. Afterward, there were no other reemerging SADS-CoV strains detected until it appeared again in Guangdong, China causing devastating damage to the local pig industry in 2019 [11]. This suggests possible periodic outbreaks of SADS-CoV could be observed in years to come. PDCoV, a δ-coronavirus, was first detected in 2012 and caused PEDV-like signs in pigs [19–21,22]. Birds can be considered as the natural hosts of δ-coronaviruses. Based on their ability to spread across species, δ-coronaviruses may “jump” the species barrier and adapt to mammals [23]. It has been reported that δ-coronaviruses have also been detected in Asian leopard cats and Chinese ferret-badgers [10,24]. PToV was first discovered by Kroneman et al. in 1998 [2]. PToV was considered as a coronavirus for a long time before the ICTV (International Committee on Taxonomy of Viruses) classified it into family Tobaniviridae, order Nidovirales. PToV has a high positive rate in swine diarrhea samples [25]. However, it remains to be a potential pathogen in pigs [25–27]. It should be noted that frequent recombination events involving PToV have been discovered, some of which, especially those on the Hemagglutinin-esterases (HE) or Spike (S) genes, which encode proteins relating to attachment and invasion, could lead to changes in pathogenicity or host-specificity [28–32].
So far, PEDV, PDCoV, SADS-CoV, and the coronavirus-like virus PToV have caused huge economic losses to the pig industry worldwide. In addition, their cross-species transmission ability may pose a threat to public health. In order to monitor these four viruses more efficiently, it is extremely important to develop a fast, simple, and accurate method for the detection and differentiation of those viral swine diarrhea pathogens. In clinical testing, multiplex real-time PCR has excellent performance. It has a larger detection capacity with higher speed and lower labor costs. However, the main reason hindering the application of multiplex real-time PCR on pathogen detection is the difficulty in assay design. Here, we developed a multiplex TaqMan-probe-based real-time PCR method for these four emerging and reemerging swine enteric viruses.
Experimental section
Primers and probes
To ensure the detection performance of primers used in the multiplex real-time PCR method, all available sequences of PEDV, PDCoV, PToV, and SADS-CoV from GenBank were obtained and analyzed. The conserved region of ORF1a was chosen for designing primers and probes for PEDV, PToV, and SADS-CoV, and the ORF1b region was chosen for PDCoV. Four sets of primers and probes were designed using the Oligo 7 (Version 7.60) software (Table 1). Primers and probes were synthesized by Sangon Biotech (Shanghai) Co., Ltd. Primers were also used for the construction of plasmid standards.
Table 1.
Primers and probes*.
| Pathogens | Primers and Probes | Sequences (5ʹ end to 3ʹ end) | Length (bp) | Gene | Position |
|---|---|---|---|---|---|
| PEDV | PE-Detection(F) | CTCCCTTGAATTTGAGTTCG | 85 | ORF1a | 3041–3125b |
| PE-Detection(R) | ACCACCTGTAACCTTGATAC | ||||
| PE-Detection(Probe) | FAM-TTACCAACAGCCTTATTAAGCAC-MGB | ||||
| PDCoV | PD-Detection(F) | AAAGCTTTCAAGACAATACCT | 87 | ORF1b | 15,130–15,216c |
| PD-Detection(R) | TACGACAAACTCCTGAAAGCA | ||||
| PD-Detection(Probe) | Texas Red-TACGATACGACTGCATTGGCCTAC-BHQ2 | ||||
| PToV | PT-Detection(F) | TCATCCACCCAGTTCAAAT | 73 | ORF1a | 1024–1096d |
| PT-Detection(R) | TGCACAATTCTCTCTCCAAAT | ||||
| PT-Detection(Probe) | VIC-CCTCAGaTTTCGaAGATAGaACC-BHQ1 | ||||
| SADS-CoV | SA-Detection(F) | CATTTGCCGTTCTTGACCAT | 95 | ORF1a | 5269–5363e |
| SA-Detection(R) | AACCCAGCAATTGTTATCTGAA | ||||
| SA-Detection(Probe) | Cy5-CAAGTGCACGCTTACCATCAACTACT-BHQ3 |
aLNA, Locked Nucleic Acid.
bGenBank accession No. AF353511.1.
cGenBank accession No. JQ065042.2.
dGenBank accession No. NC_022787.1.
eGenBank accession No. MF167434.1.
*All primers and probes are protected by patent. For any commercial use please contact the corresponding author.
All primers for conventional RT-PCR in this study referred to former reports [33–42].
Virus strains and field samples
Clinical samples collected during 2017–2019 were preserved at −80°C in our laboratory. Those samples were mainly from Henan, Jiangsu, Anhui and Guangdong provinces in China. PCR templates were DNA or cDNA, preserved at −20°C. All positive samples were identified by singleplex conventional RT-PCR in our laboratory and confirmed with DNA sequencing by Sangon Biotech (Shanghai) Co., Ltd.
RNA extraction and reverse transcription
Intestinal tissues or feces samples were treated with 3 to 5 volumes of PBS, mixed by shaking or vortex and supernatant was collected after centrifuged at 12,000 × g at 4°C for 15 minutes. Nucleic acids were extracted using the RNApure Virus Kit (Beijing ComWin Biotech Co., Ltd.) following the manufacturer’s instructions. Reverse transcription was performed using the HiScript III RT SuperMix for qPCR (+gDNA wiper) Kit (Nanjing Vazyme biotechnology Co., Ltd.).
Construction of plasmid standards
The target fragments of PEDV, PDCoV, PToV, and SADS-CoV were amplified separately via PCR using the cDNA obtained in the previous step with the Phanta Max Super-Fidelity DNA Polymerase (Nanjing Vazyme biotechnology Co., Ltd.) following the manufacturer’s instructions. Primers used in the amplification were the same as used in multiplex real-time PCR method. The PCR fragments were then cloned into the pMD18-T vector (Takara Biomedical Technology (Beijing) Co., Ltd.) through TA colony and confirmed by DNA sequencing.
The plasmid copy number was calculated and the plasmids were diluted from 1 × 107 copies/μL to 1 × 101 copies/μL. Singleplex real-time PCR was performed for each virus using the 10-fold diluted plasmids to generate standard curves, based on which the E value (amplification efficiency), R2 (correlation coefficient), and the standard equation were calculated.
Reaction conditions of the singleplex real-time PCR
As shown in Table S1, the total volume of the singleplex real-time PCR reaction was 20 μL, consisting of 10 μL of 2× AceQ qPCR Probe Master Mix (AceQ qPCR Probe Master Mix kit, Nanjing Vazyme biotechnology Co., Ltd.), 0.4 μL of each forward and reverse primer (10 μM), 0.2 μL of TaqMan probe (10 μM), 2 μL of template, and the remaining volume of nuclease-free water.
Amplification was carried out on a Roche LightCycler® 96 Instrument (Roche Life Science) using the following program: 95°C for 600 s; 40 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 20 s. Fluorescence signal was automatically collected at the end of each cycle.
Reaction condition optimization for multiplex real-time PCR
The singleplex real-time PCR assays for PEDV, PDCoV, PToV, and SADS-CoV described above were multiplexed into one reaction system consisting of 2× AceQ qPCR Probe Master Mix (AceQ qPCR Probe Master Mix kit, Nanjing Vazyme biotechnology Co., Ltd.), primers and probes for all four viruses, and templates. The multiplex reaction system was then optimized using different volumes of primers (10 μM) and probes (10 μM), and the optimal volumes of templates were determined. In the optimization stage, the final concentration of primers and probes in the system ranged from 1200 nM to 2400 nM and 200 nM to 1000 nM, respectively. The plasmid standards containing 1 × 103 copies/μL were chosen as templates. The same instrument and real-time PCR program were used as described above.
Sensitivity of the multiplex real-time PCR assay
To determine the limit of detection (LOD) of the multiplex detection method, we performed real-time PCR reactions for each virus separately, using 10-fold serial dilutions of standard plasmid templates ranging from 1 × 107 copies/μL to 1 × 101 copies/μL. To confirm the detection limit, a multiplex real-time PCR was performed using plasmid templates of all four viruses at the concentration of the presumable detection limit with 23 replicates for each concentration. The lowest concentration that met the positive detection rate of 95% was considered as the reliable LOD.
Specificity of the multiplex real-time PCR assay
To rule out potential false positives caused by other viruses that may present in the samples, positive samples for PEDV, PDCoV, PToV, SADS-CoV, TGEV, porcine kobuvirus (PKV), classical swine fever virus (CSFV), porcine sapelovirus (PSV), porcine teschenvirus (PTV), and porcine rotavirus (PoRV) were tested using the multiplex real-time PCR detection method. All the cDNA samples were previously synthesized and stored in our laboratory.
Repeatability of the multiplex real-time PCR assay
The assay was repeated three times with a 7-days interval, using 10-fold dilutions of the standard plasmid of each pathogen ranging from 1 × 107 copies/μL to the LOD with three replicates per reaction. Each template was a mixture of standard plasmid of four pathogens at the same concentration. The coefficient of variation (CV) of the Cq values of the samples at each concentration in the three experiments was calculated to estimate repeatability.
Simulation of co-infection by combining same concentration of standard samples
Plasmid standards of two, three or four target pathogens at the same concentration were randomly chosen and mixed as templates and detected using our new method. Three concentrations (1 × 107 copies/μL, LOD and 10 times the LOD) of the plasmid standards were tested.
Simulation of co-infection by combining different concentrations of standard samples
To simulate actual co-infection events, we mixed the plasmid standards of the four target pathogens with one at 1 × 107 copies/μL and the other three at the LOD and then detected the template mixture using our multiplex detection method.
Clinical sample detection
We tested 45 newly-collected samples from pigs showing signs of diarrhea and 56 previously-confirmed positive samples (31 with PEDV only, 16 with PDCoV only, 3 with PToV only, 1 with SADS-CoV only, 4 with PEDV and PToV, and 1 with PEDV, PDCoV and PToV) using our multiplex detection method. The clinical performance of our established methods was evaluated by comparing the results with those of singleplex conventional RT-PCR. Positive samples detected by either method were then confirmed through DNA sequencing by Sangon Biotech (Shanghai) Co., Ltd.
Results
Preparation of primers, probes and plasmid standards
As shown in Table 1, the sequences of the primers and probes designed in this study are presented. PEDV, PDCoV, PToV, and SADS-CoV probes were labeled with FAM, VIC, Texas Red, and Cy5 separately. Plasmid standards with concentrations ranging from 1 × 107 copies/μL to 1 × 101 copies/μL of each pathogen were selected to perform a singleplex real-time PCR (Figure 1). The standard curves showed an acceptable amplification efficiency and correlation coefficient: PEDV R2 = 0.9957, E value = 99%; PDCoV R2 = 0.9990, E value = 100%; PToV R2 = 0.9943, E value = 94%; and SADS-CoV R2 = 0.9982, E value = 99%, indicating that our plasmid standards were qualified, and the primers and probes designed were efficient.
Figure 1.
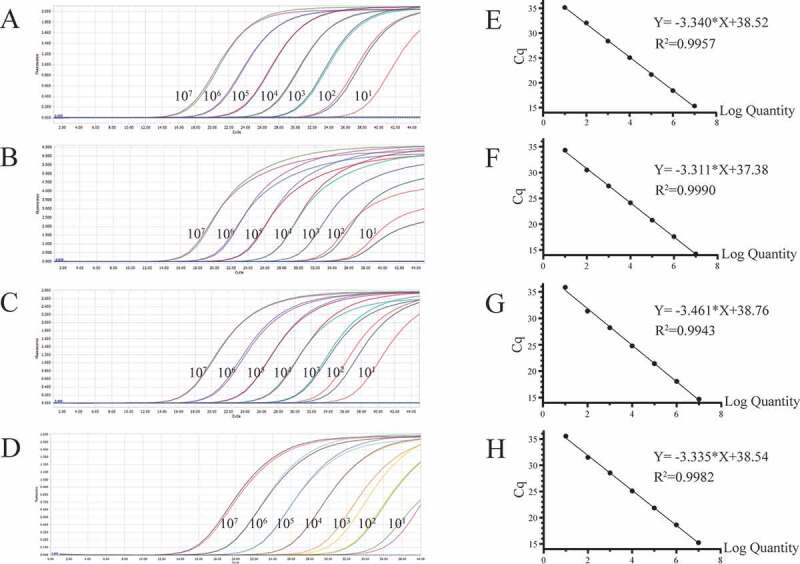
Preparation of plasmid standards. A-D: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PEDV, PDCoV, PToV, and SADS-CoV for each plasmid standard of concentrations with 1 × 107 copies/μL to 1 × 101 copies/μL; E-H: standard curves of plasmid standards of PEDV, PDCoV, PToV, and SADS-CoV. All standard curves were conducted with software GraphPad Prism 8.
Optimization of the multiplex reaction system
Among the four fluorophores used in the multiplex detection method, the fluorescence of Cy5 was the weakest and very susceptible to interference from other fluorophores due to its own physical properties. Therefore, the main purpose of the reaction optimization system was to improve the performance of the Cy5 fluorophores without hindering other fluorophores, and to achieve the best amplification efficiency (i.e. the lowest Cq value). We performed multiplex real-time PCR with final concentration of probes ranging from 200 nM to 1000 nM, and of primers ranging from 1200 nM to 2400 nM and compared the fluorescence intensity and Cq values of each possible combination. We concluded that the best final concentrations for probes and primers are 1000 nM and 2400 nM, respectively (Table 2 and Figure 2). The maximum volume of template that can be loaded in this system is 4.8 μL as shown in Table S2.
Table 2.
Cq values of PEDV, PDCoV, PToV, and SADS-CoV detected by this multiplex real-time PCR assay with different probe and primer concentrations.
| PEDV |
PDCoV |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe concentration (nM) |
Primer concentration (nM) |
Probe concentration (nM) |
Primer concentration (nM) |
||||||
| 1200 | 1600 | 2000 | 2400 | 1200 | 1600 | 2000 | 2400 | ||
| 200 | 31.14 | 29.75 | 29.46 | 29.58 | 200 | 30.97 | 29.07 | 28.54 | 28.29 |
| 400 | 30.50 | 29.49 | 28.47 | 29.40 | 400 | 30.20 | 28.60 | 27.36 | 28.09 |
| 600 | 29.78 | 29.19 | 29.04 | 29.02 | 600 | 29.73 | 28.31 | 28.01 | 27.92 |
| 800 | 29.43 | 29.18 | 28.91 | 28.91 | 800 | 29.14 | 28.21 | 27.90 | 27.93 |
| 1000 |
29.55 |
29.08 |
29.11 |
28.84 |
1000 |
28.42 |
27.94 |
28.13 |
27.83 |
| PToV |
SADS-CoV |
||||||||
| Probe concentration (nM) |
Primer concentration (nM) |
Probe concentration (nM) |
Primer concentration (nM) |
||||||
| 1200 | 1600 | 2000 | 2400 | 1200 | 1600 | 2000 | 2400 | ||
| 200 | 31.63 | 29.35 | 29.48 | 29.24 | 200 | 29.82 | 29.10 | 29.08 | 28.99 |
| 400 | 30.87 | 29.57 | 28.04 | 28.77 | 400 | 30.56 | 29.51 | 29.26 | 29.35 |
| 600 | 30.17 | 29.05 | 28.81 | 28.72 | 600 | 31.47 | 29.77 | 29.68 | 29.51 |
| 800 | 29.49 | 28.73 | 28.65 | 28.40 | 800 | 31.66 | 30.14 | 29.42 | 29.59 |
| 1000 | 28.57 | 28.40 | 28.17 | 28.05 | 1000 | 33.66 | 30.58 | 30.21 | 29.96 |
Figure 2.
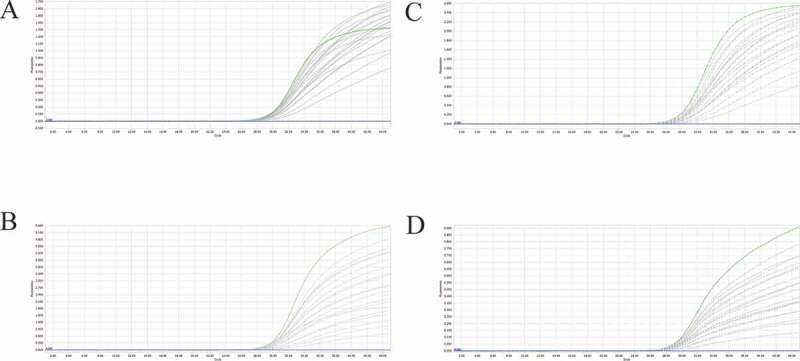
A-D: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PEDV, PDCoV, PToV, and SADS-CoV detected by multiplex real-time PCR with different probe and primer concentrations. Plasmid standards with concentration of 1 × 103 copies/μL were chosen as templates for the reactions. The four green lines are the amplification curves of four fluorescence of the most suitable reaction tube.
Sensitivity of the multiplex real-time PCR assay
Using the optimized system and the plasmid standards of each pathogen with concentrations ranging from 1 × 107 copies/μL to 1 × 101 copies/μL, we found that the method could identify positive samples with the concentrations as low as 1 × 101 copies/μL (Figure 3a and Table S3). However, follow-up experiments indicated that the detection rate of samples at 1 × 101 copies/μL was less than 95% of replicates (Table 3). Thus, the reliable LOD of this method is 1 × 102 copies/μL. In this experiment, the cutoff line of positivity was automatically decided by the LightCycler® 96 Instrument.
Figure 3.
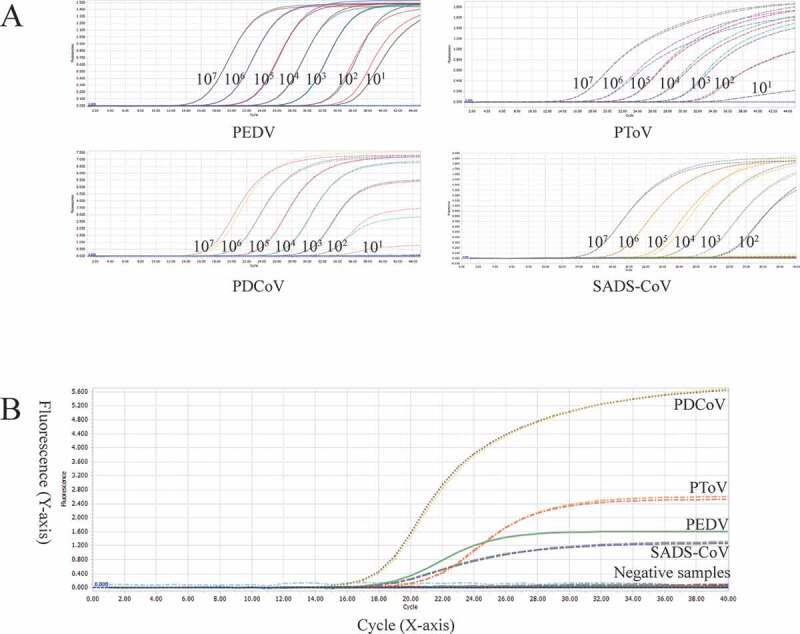
A: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of 10-fold serial dilutions (1 × 107–1 × 101 copies/μL) of plasmid standards of PEDV, PDCoV, PToV, and SADS-CoV detected by multiplex real-time PCR. B: four amplification curves represent samples positive for PEDV, PDCoV, PToV, and SADS-CoV detected by our multiplex real-time PCR assay; negative samples include TGEV, PoRV, PSV, PTV, CSFV, PKV, and negative control.
Table 3.
Sensitivity of the multiplex real-time PCR assay.
| Pathogens | Concentration | Total | Positive | Detection rate | 95% detection rate |
|---|---|---|---|---|---|
| PEDV | 1000 copies/μL | 23 | 23 | 100.0% | > 95% |
| 100 copies/μL | 23 | 23 | 100.0% | > 95% | |
| 10 copies/μL | 23 | 20 | 87.0% | < 95% | |
| PDCoV | 1000 copies/μL | 23 | 23 | 100.0% | > 95% |
| 100 copies/μL | 23 | 23 | 100.0% | > 95% | |
| 10 copies/μL | 23 | 14 | 60.9% | < 95% | |
| PToV | 1000 copies/μL | 23 | 23 | 100.0% | > 95% |
| 100 copies/μL | 23 | 23 | 100.0% | > 95% | |
| 10 copies/μL | 23 | 17 | 73.9% | < 95% | |
| SADS-CoV | 1000 copies/μL | 23 | 23 | 100.0% | > 95% |
| 100 copies/μL | 23 | 23 | 100.0% | > 95% | |
| 10 copies/μL | 23 | 12 | 52.2% | < 95% |
The cutoff line of positivity is automatically decided by Roche LightCycler® 96 Instrument.
We set the cutoff line of positivity of our method at 35, which means samples with a Cq value less than or equal to 32 (≤ 32) are regarded as positive, higher than 32 but less than or equal to 35 (32 < and ≤ 35) are invalid, higher than 35 (>35) are negative. The criteria were set based on two reasons. First, the LOD of our detection method was 1 × 102 copies/μL, of which the Cq value was around 32. Second, some samples at 1 × 101 copies/μL were detectable, however, the detection rate was unqualified, and the Cq values of those detectable samples were around 35. All following experiments complied with these criteria.
Specificity of the multiplex real-time PCR assay
The optimized method was used to detect PEDV, PDCoV, PToV, and SADS-CoV in positive samples and some other samples derived from pigs with diarrhea positive for TGEV, PKV, CSFV, PSV, PTV, and PoRV. As shown in Figure 3b and Table S4, the target pathogens were detected while the other pathogens were negative, indicating good specificity.
Repeatability of the multiplex real-time PCR assay
As is shown in Table S5, most %CV values of the Cq values of the plasmid standard were less than 1% (81/96) with only a few %CV values ranging from 1% to 5% (15/96), indicating that this multiplex detection method is stable.
Co-infection simulation experiment
We selected plasmid standards with concentrations of 1 × 107 copies/μL, 1 × 103 copies/μL, and 1 × 102 copies/μL of different pathogens as templates to perform a co-infection simulation experiment. As shown in Figures 4, 5, and 6, the multiplex detection method could detect duplex, triplex, or quadruplex simulation co-infections of the target pathogens, even pathogens with different concentrations (Figure 7).
Figure 4.
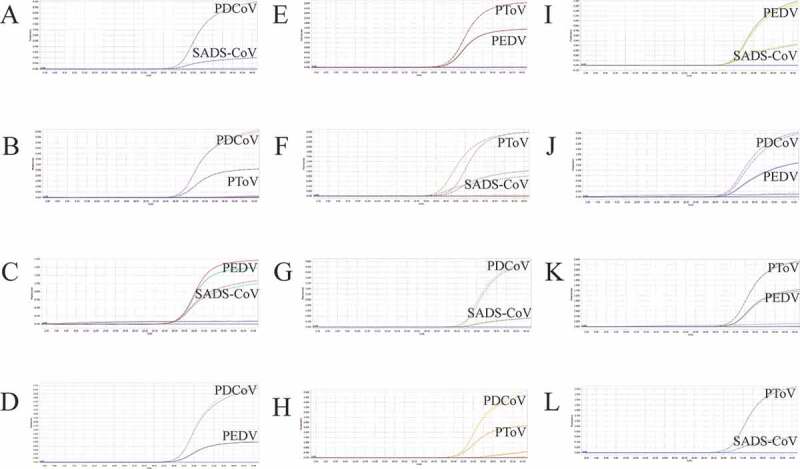
Co-infection simulation experiments with two pathogens. A-F: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PDCoV + SADS-CoV, PDCoV + PToV, PEDV + SADS-CoV, PEDV + PDCoV, PEDV + PToV, PToV + SADS-CoV at concentrations of 1 × 103 copies/μL; G-L: amplification curves of PDCoV + SADS-CoV, PDCoV + PToV, PEDV + SADS-CoV, PEDV + PDCoV, PEDV + PToV, PToV + SADS-CoV at concentrations of 1 × 102 copies/μL. Two replicates were set per reaction.
Figure 5.
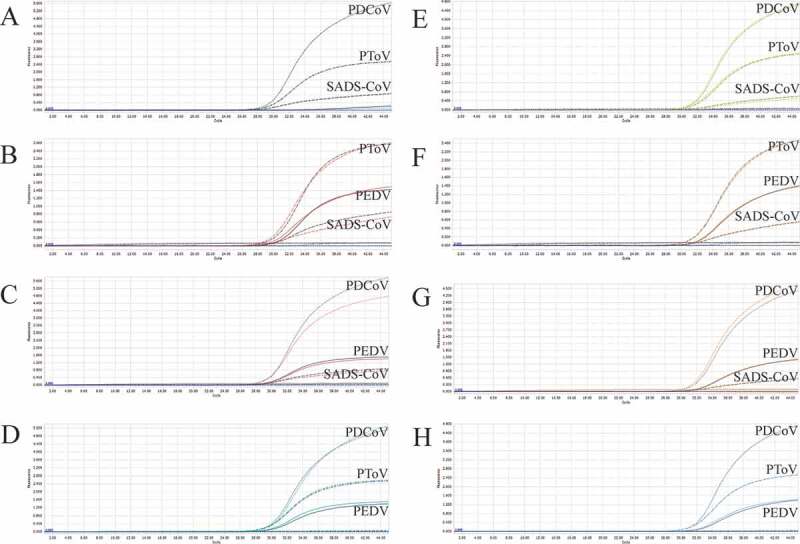
Co-infection simulation experiments with three pathogens. A-D: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PDCoV + PToV + SADS-CoV, PEDV + PToV + SADS-CoV, PEDV + PDCoV + SADS-CoV, PEDV + PDCoV + PToV with concentration of 1 × 103 copies/μL; E-H: amplification curves of PDCoV + PToV + SADS-CoV, PEDV + PToV + SADS-CoV, PEDV + PDCoV + SADS-CoV, PEDV + PDCoV + PToV with concentrations of 1 × 102 copies/μL. Two replicates were set per reaction.
Figure 6.
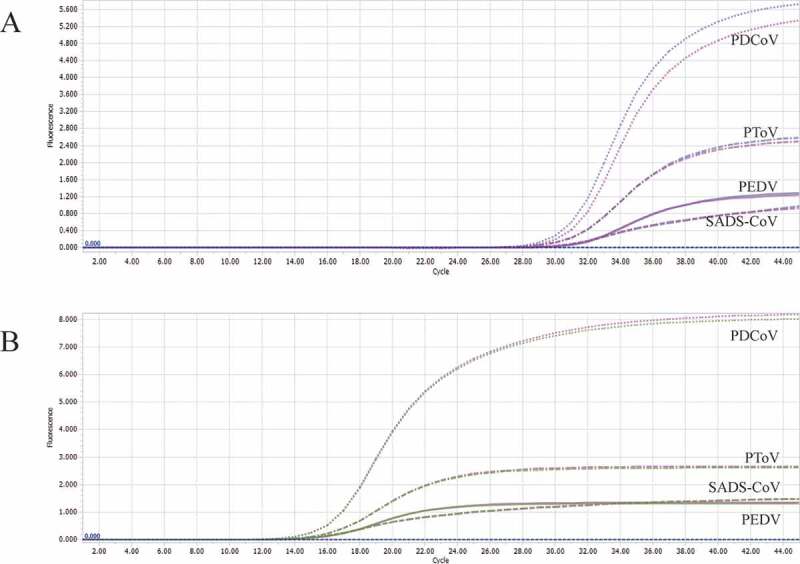
Co-infection simulation experiments with four pathogens. A-B: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PEDV + PDCoV + PToV + SADS-CoV at concentrations of 1 × 102 copies/μL and 1 × 107 copies/μL. Two replicates were set per reaction.
Figure 7.
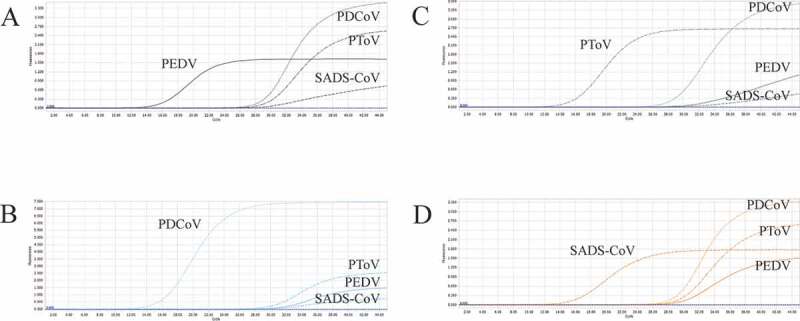
Co-infection of all the four pathogens at different concentrations. A: the concentration of plasmid standard of PEDV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL; B: the concentration of plasmid standard of PDCoV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL; C: the concentration of plasmid standard of PToV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL; D: the concentration of plasmid standard of SADS-CoV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL. Two replicates were set per reaction. X-axis: Cycle, Y-axis: Fluorescence.
Clinical sample detection
Two batches of clinical samples were tested by our established method to validate its performance in clinical use. The first batch consisted of 45 digestive tract samples recently collected from pig farms in China. The results of our method were 100% consistent with the results of singleplex conventional RT-PCR (Table S6). The second batch consisted of 56 positive samples stored in our laboratory, of which 31 were positive for PEDV alone, 16 were positive for PDCoV alone, 1 was positive for SADS-CoV alone, and 8 were positive for PToV (not necessarily alone). Our method was 100% consistent with singleplex conventional RT-PCR tests. It should be noted that among the 8 PToV positive samples, 3 samples were positive only for PToV, while the remaining 5 were also positive for other viruses, including 4 samples which were also positive for PEDV, and one positive for PEDV, PDCoV, and PToV (Table S6).
Discussion
Virus cross-species transmission from wildlife reservoirs poses a remarkable threat to human and domestic animal health [18,43]. Coronaviruses can cross the species barrier and gradually adapt to new hosts [44,45]. For example, the recently emerged SARS-CoV-2 was estimated to have originated from bats spreading to wild animals or livestock and then to humans [46]. Animals may potentially serve as mixing vessels for the generation of novel recombinant coronaviruses and facilitate the viruses to expand their host tropism to humans [47]. In current swine breeding practices, humans have close contact with pigs, which further increases the possibility of viral transmission to humans, posing potential threat to human health [10]. Therefore, the detection of emerging and reemerging coronaviruses is of significance for farming and public health.
Among the four target pathogens in this study, PEDV is a coronavirus with considerably high prevalence and frequent recombination and rapid evolution rate [13]. SADS-CoV and PDCoV are two emerging coronaviruses and have been reported to be spreading and causing economic losses [11,23]. PToV is regarded as a swine diarrhea virus that has not caused huge economic losses [25]. However, given the high recombination rate and evolutionary characteristics of coronavirus and torovirus, we cannot underestimate their potential threat [28–30]. It is very likely that a virulent strain could emerge and cause unpredictable damage to the pig industry [13]. Thus, monitoring these currently less harmful viruses, such as SADS-CoV, PDCoV, and PToV, is of great significance.
Pathogen monitoring nowadays largely relies on laboratory detection methods. Singleplex conventional RT-PCR has been widely applied for pathogen detection [34–36,38]. However, such kind of assays are not convenient for the simultaneous detection of co-infection of multiple pathogens, which hinders further study of viral recombination events [3]. Given that co-infections of different coronaviruses and torovirus are common in the field and the high recombination frequency of coronavirus and torovirus, it is necessary to develop a fast, convenient detection method (e.g. multiplex real-time PCR) for the diagnosis of co-infections. PCR is fast, accurate and convenient for clinical sample screening. Real-time PCR is better than conventional PCR due to its faster, more sensitive and accurate detection capacity [48]. The use of probes is the most obvious and critical difference [49,50]. In general, conventional RT-PCR is less sensitive than real-time PCR. The LOD of our multiplex real-time PCR detection method can reach as low as 1 × 102 copies/μL for each pathogen, while that of the singleplex conventional RT-PCR is generally around 1 × 103 copies/μL to 1 × 104 copies/μL [51]. Likewise, multiplex conventional RT-PCR shows no noticeable advantage in terms of sensitivity [34,52]. Hui et al. developed a multiplex conventional RT-PCR assay for pancoronaviruses, in which the limits of detection were no less than 1 × 103 copies/μL [40]. Zhao et al. developed a multiplex RT-PCR detection for CSFV, porcine reproductive and respiratory syndrome virus (PRRSV), PEDV, and TGEV. The limit of detection of this method was 1 × 103 copies/μL [52]. On the other hand, although multiplex real-time PCR combines high sensitivity and high detection efficiency, the design of qualified multiplex real-time PCR assays, especially of quadruplex quantitative real-time PCR assays, is challenging [48,49,53]. In multiplex real-time PCR assays, multiple sets of oligonucleotides exist simultaneously in the reaction system, increasing the possibility for nonspecific amplification, which poses high demands on the specificity of primers and probes.
Improvements in detection method can bring many benefits. Accurate detection of virus at lower concentration enables the diagnosis and prevention of porcine diarrhea at an early stage. Some virulent strains may cause severe signs at a low titer, and thus a sensitive detection method is indispensable in such situations. Due to its lower false negative rate caused by lower LODs, our new detection method enables more powerful surveillance over those four swine diarrhea viruses and, hopefully, can benefit the construction of pig farms with higher biosecurity. However, the improvement in sensitivity also increases the possibility of false positive results, which imposes higher requirements on the prevention of contamination during sampling and assay set up. Good practice in the laboratory is necessary in order to get credible results.
In short, an excellent and efficient detection method should accurately reflect the epidemiological data (e.g. the scale of the disease, the rate of transmission, and the severity of the epidemic), so as to be beneficial to the monitoring and prevention of the disease [53]. Multiplex real-time PCR achieves better detection capability in less time and with lower labor cost, but its development is still challenging compared to singleplex conventional RT-PCR due to technical difficulties at the development stage. To the best of our knowledge, this is the first multiplex real-time PCR detection method for PEDV, PDCoV, PToV, and emerging SADS-CoV, which often present simultaneously among pigs. The multiplex detection method developed here can detect multiple pathogens in a single reaction, making detection for co-infection more convenient. This method can ensure good specificity and sensitivity, which will undoubtedly save labor and material costs.
Conclusion
Here, we developed a TaqMan-probe-based multiplex real-time PCR method for the simultaneous detection of emerging and reemerging PEDV, PDCoV, PToV and SADS-CoV of swine. The limit of detection can reach as low as 1 × 102 copies/μL for each pathogen with good specificity and repeatability. The application of this method in clinical detection will not only improve the detection capacity, but also reduce workload and cost, benefiting clinicians and epidemiologists.
Supplementary Material
Acknowledgments
We thank Professor Huang Yaowei (College of Animal Sciences, Zhejiang University) and Professor Zhou Jiyong (College of Animal Sciences, Zhejiang University) for their guidance on our coronavirus research and the help they provided on biological materials. We thank Haitao Qi (New Hope Liuhe Company) and Ao Liao (Han Shiwei Company) for providing pig samples. We thank Jiexiong Xie (Laboratory of Virology, Faculty of Veterinary Medicine, Ghent University) for helping us improve the paper.
Funding Statement
This work was financially supported by the National Natural Science Foundation of Outstanding Youth Fund in China (NSFC Grant no. 31922081), the National Key Research and Development Program of China (Grant no. 2016YFD0500402), National Ten Thousand Talent Program, the Fundamental Research Funds for the Central Universities (Grant no. Y0201900459), Six Talent Peaks Project of Jiangsu Province of China (Grant no. NY-045) and the Bioinformatics Center of Nanjing Agricultural University.
Disclosure statement
The authors declare no competing financial interest.
Author contributions
SS provided the study design and experiment, other authors contributed to the experiment. All authors contributed to writing the manuscript. All authors have approved the final version of the manuscript.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Brian DA, Baric RS.. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kroneman A, Cornelissen LA, Horzinek MC, et al. Identification and characterization of a porcine torovirus. J Virol. 1998;72(5):3507–3511.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huang X, Chen J, Yao G, et al. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl Microbiol Biotechnol. 2019;103(12):4943–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Woo PC, Huang Y, Lau SK, et al. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Banerjee A, Kulcsar K, Misra V, et al. Bats and Coronaviruses. Viruses. 2019;11(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gralinski LE, Menachery VD. Return of the Coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. https://doi.org/10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sun J, He W-T, Wang L, et al. COVID-19: epidemiology, Evolution, and Cross-Disciplinary Perspectives. Trends Mol Med. 2020;26(5):483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhai X, Sun J, Yan Z, et al. Comparison of SARS-CoV-2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. Journal of virology, JVI.00831-20. Advance online publication. 10.1128/JVI.00831-20 [DOI] [PMC free article] [PubMed]
- [10].Wang L, Su S, Bi Y, et al. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol. 2018;26(6):466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhou L, Li QN, Su JN, et al. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound Emerg Dis. 2019;66(5):2180–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Huang Y-W, Dickerman AW, Pineyro P, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4(5):e00737–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lin C-M, Saif LJ, Marthaler D, et al. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44(2):167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bi J, Zeng S, Xiao S, et al. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J Virol. 2012;86(19):10910–10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pan Y, Tian X, Li W, et al. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol J. 2012;9(1):195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li W, Li H, Liu Y, et al. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. 2012;18(8):1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou P, Fan H, Lan T, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chu DK, Leung CY, Gilbert M, et al. Avian coronavirus in wild aquatic birds. J Virol. 2011;85(23):12815–12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Woo PC, Lau SK, Lam CS, et al. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J Virol. 2009;83(2):908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].W T He, Ji X, He W, et al. Genomic epidemiology, evolution, and transmission dynamics of porcine deltacoronavirus. Molecular biology and evolution, msaa117. Advance online publication. 10.1093/molbev/msaa117 [DOI] [PMC free article] [PubMed]
- [23].Ma Y, Zhang Y, Liang X, et al. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6(2):e00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dong BQ, Liu W, Fan XH, et al. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J Virol. 2007;81(13):6920–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hu Z-M, Yang Y-L, Xu L-D, et al. Porcine Torovirus (PToV)—a brief review of etiology, diagnostic assays and current epidemiology. Front Vet Sci. 2019;6:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Weiss M, Steck F, Horzinek MC. Purification and partial characterization of a new enveloped RNA virus (Berne virus). J Gen Virol. 1983;64(9):1849–1858. [DOI] [PubMed] [Google Scholar]
- [27].Woode GN, Reed DE, Runnels PL, et al. Studies with an unclassified virus isolated from diarrheic calves. Vet Microbiol. 1982;7(3):221–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Conceicao-Neto N, Theuns S, Cui T, et al. Identification of an enterovirus recombinant with a torovirus-like gene insertion during a diarrhea outbreak in fattening pigs. Virus Evol. 2017;3(2):vex024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Knutson TP, Velayudhan BT, Marthaler DG. A porcine enterovirus G associated with enteric disease contains a novel papain-like cysteine protease. J Gen Virol. 2017;98(6):1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee S, Lee C. First detection of novel enterovirus G recombining a torovirus papain-like protease gene associated with diarrhoea in swine in South Korea. Transbound Emerg Dis. 2019;66(2):1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shang P, Misra S, Hause B, et al. A naturally occurring recombinant enterovirus expresses a torovirus deubiquitinase. J Virol. 2017;91(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang Y, Zhang W, Liu Z, et al. Full-length and defective enterovirus G genomes with distinct torovirus protease insertions are highly prevalent on a Chinese pig farm. Arch Virol. 2018;163(9):2471–2476. [DOI] [PubMed] [Google Scholar]
- [33].Pan Y, Tian X, Qin P, et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol. 2017;211:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ma L, Zeng F, Huang B, et al. Development of a conventional RT-PCR assay for rapid detection of porcine deltacoronavirus with the same detection limit as a SYBR green-based real-time RT-PCR assay. Biomed Res Int. 2018;2018:5035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen M, Tang W, Hua X. Molecular characterization of a porcine teschovirus HuN-1 isolate proliferating in PK-15 cell. BMC Vet Res. 2018;14(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jiang C, He H, Zhang C, et al. One-step triplex reverse-transcription PCR detection of porcine epidemic diarrhea virus, porcine sapelovirus, and porcine sapovirus. J Vet Diagn Invest. 2019;31(6):909–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhou L, Wei H, Zhou Y, et al. Molecular epidemiology of Porcine torovirus (PToV) in Sichuan Province, China: 2011–2013. Virol J. 2014;11(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ogawa H, Taira O, Hirai T, et al. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J Virol Methods. 2009;160(1–2):210–214. [DOI] [PubMed] [Google Scholar]
- [39].Aguero M, Fernandez J, Romero LJ, et al. A highly sensitive and specific gel-based multiplex RT-PCR assay for the simultaneous and differential diagnosis of African swine fever and Classical swine fever in clinical samples. Vet Res. 2004;35(5):551–563. [DOI] [PubMed] [Google Scholar]
- [40].Hu H, Jung K, Wang Q, et al. Development of a one-step RT-PCR assay for detection of pancoronaviruses (α-, β-, γ-, and δ-coronaviruses) using newly designed degenerate primers for porcine and avian `fecal samples. J Virol Methods. 2018;256:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou X, Zhang T, Song D, et al. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol Cell Probes. 2017;33:36–41. [DOI] [PubMed] [Google Scholar]
- [42].Ding G, Fu Y, Li B, et al. Development of a multiplex RT-PCR for the detection of major diarrhoeal viruses in pig herds in China. Transbound Emerg Dis. 2020;67(2):678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Olival KJ, Hosseini PR, Zambrana-Torrelio C, et al. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546(7660):646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84(7):3134–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus Spike Protein and Tropism Changes. Adv Virus Res. 2016;96:29–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Singh C, Roy-Chowdhuri S. Quantitative real-time PCR: recent advances. Methods Mol Biol. 2016;1392:161–176. [DOI] [PubMed] [Google Scholar]
- [49].Heid CA, Stevens J, Livak KJ, et al. Real time quantitative PCR. Genome Res. 1996;6(10):986–994. [DOI] [PubMed] [Google Scholar]
- [50].Arya M, Shergill IS, Williamson M, et al. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn. 2005;5(2):209–219. [DOI] [PubMed] [Google Scholar]
- [51].Zhao P-D, Bai J, Jiang P, et al. Development of a multiplex TaqMan probe-based real-time PCR for discrimination of variant and classical porcine epidemic diarrhea virus. J Virol Methods. 2014;206:150–155. [DOI] [PubMed] [Google Scholar]
- [52].Zhao Y, Liu F, Li Q, et al. A multiplex RT-PCR assay for rapid and simultaneous detection of four RNA viruses in swine. J Virol Methods. 2019;269:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kurkela S, Brown DWG. Molecular diagnostic techniques. Medicine (Abingdon). 2009;37(10):535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


