Highlights
-
•
Non-invasive detection of copy number variations (CNVs) was firstly reported in patients with biliary tract cancer.
-
•
Biliary tract cancer harbors rich information of CNVs based on plasma cell-free DNA analyses.
-
•
The sensitivity and specificity of CNVs for diagnosing biliary tract cancer were 89.7% and 88.9%, respectively. The diagnostic performance of CNVs significantly outperformed CA 19-9 and CEA.
Keywords: Copy number variation, Cell-free DNA, Low-coverage whole genome, Biliary tract cancer, Diagnosis
Abbreviations: BTC, biliary tract cancer; CEA, carcinoembryonic antigen; CC, cholangiocarcinoma; cfDNA, cell-free DNA; CIN, chromosomal instability; CNV, copy number variation; GBC, gallbladder cancer; LCWG, low-coverage whole genome; WGS, whole genome sequencing
Abstract
Background
The diagnosis of biliary tract cancer (BTC) is challenging in clinical practice. We performed a prospective study to evaluate the value of plasma copy number variation (CNV) assays in diagnosing BTC.
Methods
47 treatment-naïve patients with suspicious biliary lesions were recruited. Plasma samples were collected at admission. Cell-free DNA was analyzed by low coverage whole genome sequencing, followed by CNV analyses via a customized bioinformatics workflow, namely the ultrasensitive chromosomal aneuploidy detector.
Results
29 patients were pathologically diagnosed as BTC, including 8 gallbladder cancers (GBCs) and 21 cholangiocarcinomas (CCs). Cancer patients had more CNV signals as compared with benign patients (26/29 vs. 2/18, P < 0.001). The most frequent copy number gains were chr3q (7/29) and chr8q (6/29). The most frequent copy number losses were chr7p (6/29), chr17p (6/29), and chr19p (6/29). The sensitivity and specificity of plasma CNV assays in diagnosing BTC were 89.7% and 88.9%, respectively. For CA 19-9 (cutoff: 37 U/ml), the sensitivity was 58.6% and the specificity was 72.2%. The diagnostic accuracy of CNV assays significantly outperformed CA 19-9 (AUC 0.91 vs. 0.62, P = 0.004). Compared with CA 19-9 alone, the adding of CNV profiles to CA 19-9 increased the sensitivity in diagnosing GBC (75.0% vs. 25.0%) and CC (100% vs. 52.4%). Higher CNV burden was also associated with decreased overall survival (Hazard ratio = 4.32, 95% CI 2.06–9.08, P = 0.033).
Discussion
Our results suggest that BTC harbors rich plasma CNV signals, and their assays might be useful for diagnosing BTC.
Introduction
Biliary tract cancers (BTCs) include intrahepatic, perihilar, and distal cholangiocarcinomas as well as gallbladder cancer. BTCs are a group of devastating malignancies with dismal prognosis, and the prevalence has been increased globally over the past 20 years [1]. The diagnosis of BTC mainly relies on serum tumor biomarkers, radiographic imagings, and endoscopic examinations. However, hepatobiliary surgeons always face the dilemma of diagnosing indeterminate biliary strictures or uncertain gallbladder masses. In one study of 707 suspected cases of perihilar cholangiocarcinoma, 22 (3.1%) patients had a final pathology of benign hilar stricture [2]. Another study showed a misdiagnosis rate as high as 15% in 323 patients resected for presumed perihilar cholangiocarcinoma [3]. As for gallbladder lesions, extended resection for xanthogranulomatous cholecystitis that masquerades as cancer was not unusual [4]. Meanwhile, incidental cancer detected after cholecystectomy occurred in about 1% of all cholecystectomies [5].
CA 19-9 and carcinoembryonic antigen (CEA) are the most commonly used serum biomarkers for diagnosing BTC; however, both have only moderate diagnostic accuracy [6]. CA 19-9 may be elevated in patients with acute cholangitis, obstructive jaundice, or xanthogranulomatous cholecystitis [4, 7, 8]. Meanwhile, CA 19-9 could be undetected in cancer patients with Lewis (-) phenotypes [9]. Multidetector-row computed tomography (MDCT) and magnetic resonance imaging (MRI) are baseline imaging modalities, whereas some patients with either early cholangiocarcinoma or IgG4-related cholangitis may display equivocal or negative findings on conventional imaging [10]. Although PET-CT is a valuable complimentary tool, it is costly and may result in false-positive findings in inflammatory biliary conditions [11]. Unlike gastrointestinal cancers, it is an arduous undertaking to perform biopsy for BTC. Endoscopic ultrasound (EUS) and endoscopic retrograde cholangiopancreatography (ERCP) with cytological brushing or biopsy, along with avant-garde visualization techniques, are increasingly used to improve the diagnostic yield. Regretfully, they are hampered by low sensitivity, accessibility, poor cost-effectiveness and potentially severe complications [12].
The past decade witnessed exponential progress in non-invasive liquid biopsy for cancer, which is epitomized by the testing of circulating tumor DNA (ctDNA), namely the fraction of cell-free DNA (cfDNA) that is shed from tumor cells into bloodstream [13, 14]. BTC is notoriously known as a heterogeneous cancer entity with complex cell origins and mutation profiles [15]. Plasma ctDNA sampling is attractive in capturing the tumor heterogeneity harbored by multiple clonal populations [13]. Either analyzed by polymerase-chain-reaction (PCR) or ultra-deep next-generation sequencing (NGS), ctDNA has been tentatively examined in patients with BTC [16, 17]. However, the majority of studies focused on gene mutations that were used for predicting prognosis or guiding therapy decisions [18]. Supplementary Table S1 systematically reviewed previous studies on cfDNA analysis in BTC.
Chromosomal instability (CIN) is a promising alternative diagnostic tool. CIN refers to ongoing aberrant chromosome segregation during cell divisions, generally exhibiting as somatic copy number variations (CNVs). As a hallmark of cancer, CIN pervades in almost 90% of solid malignancies. It also underpins much of the tumor heterogeneity and is central to cancer evolution [19, 20]. The effect of CNVs exerting on the cancer genome far outweighs certain genetic mutations [21]. Previous studies based on tumor tissues proved that copy number signals were frequently detected in BTC [22, 23].
Although ctDNA is a potentially rich reservoir to deduce CNV profiles, the fraction of ctDNA in plasma is basically at a low level [24]. It is intriguing to uncover CNVs by analyzing ctDNA with whole genome sequencing (WGS). The feasibility of this approach has been verified, mostly relating to therapy efficacy or disease monitoring [25], [26], [27]. It remains an untapped territory for detecting plasma CNVs in biliary malignancies. Consequently, we conducted this prospective cohort study to identify CNVs by using low-coverage whole genome sequencing of plasma cell-free DNA in patients with BTC. We aimed to evaluate whether plasma CNV profiles have a diagnostic role for discriminating BTC from benign biliary lesions. The prognostic value of CNV burden was also investigated.
Materials and methods
Our study adhered to the Standards for Reporting Diagnostic Accuracy Studies (STARD) 2015 guideline [28]. This prospective study was approved by the Institutional Review Board of the Eastern Hepatobiliary Surgery Hospital (EHBH) (NO. EHBHKY2019-01-004). Informed consent was obtained from all patients. We enrolled consecutive non-overlapping patients who underwent surgical resection for biliary diseases between March 2019 and January 2020 at the Second Department of Biliary Surgery in EHBH.
Inclusion and exclusion criteria
All patients were over 18 years of age. We defined a prior the inclusion and exclusion criteria. Two groups of patients were eligible for inclusion: (1) highly suspicious group: patients were diagnosed as BTC based on preoperative clinical data as judged by radiologists and senior authors (W.L.Y and Y.J.Z); (2) moderately suspicious group: patients had long-term (> 5 years) precancerous biliary lesions or suspicious clinical signs (e.g. irregular thickness of the gallbladder wall, indeterminate enhancement on CT/MRI, elevated serum tumor markers); precancerous lesions may include hepatolithiasis, choledochal cysts, gallbladder polypoid lesions, gallstones, primary sclerosing cholangitis, and IgG4-related cholangitis. Exclusion criteria were defined as follows: (1) benign biliary lesions with very low risk of malignancy (e.g. gallstones of diameter ≤ 2 cm, gallbladder polypoid lesions of diameter < 1 cm, choledocholithiasis, or bile duct injury after cholecystectomy); (2) postoperative pathological information was unavailable; (3) history of non-biliary malignancies; (4) hepatocellular, pancreatic, or duodenal carcinomas; (5) testing for postoperative surveillance; (6) unwillingness to participate in the study. All participants underwent standard collection of blood samples at admission to examine CNV profiles and tumor markers. We focused on CA 19-9 and CEA examinations as recommended by the NCCN guideline [6]. The cutoff values for tumor biomarkers were: CA19-9, 37.0 U/mL; CEA, 5 ng/mL [29]. We also tested a cutoff of 90 U/ml for CA 19-9 given that obstructive jaundice or cholangitis might affect CA 19-9 levels [8]. Tumor stages were classified according to the 8th edition of the American Joint Committee on Cancer (AJCC) manual [30].
DNA extraction
Total genomic DNA and cfDNA were isolated from plasma by using the Amp Genomic DNA Kit (TIANGEN) and QIAseq cfDNA Extraction kit (Qiagen), respectively. Next generation sequencing was performed as previously described. DNA was fragmented into an average size of 300 bp (cfDNA without fragmentation), and then 100 ng of fragmented genomic DNA (cfDNA 10 ng) was used for the preparation of sequencing libraries (NEBnext Ultra II). 8 bp barcoded sequencing adaptors were then ligated with DNA fragments and amplified by PCR. Purified sequencing libraries were massively parallel sequenced by Illumina HiSeq Xten platform. About 4G sequencing raw data per sample was filtered and aligned to the human reference genome.
Low-coverage WGS
For low-coverage WGS, libraries were prepared using the Kapa Hyper Prep kit with custom adapters (IDT and Broad Institute). Low-pass whole-genome sequencing was started with 3–20 ng of cfDNA input (median, 5 ng), or approximately 1000 to 7000 haploid genome equivalents. Up to 22 libraries were pooled and sequenced using 150 bp pair-end runs over 1 × lane on a HiSeq X10 (Illumina). Segment copy numbers were derived via the customized workflow, namely ultrasensitive chromosomal aneuploidy detector (UCAD). Samples were excluded if the median absolute deviation of copy ratios (log2 ratio) between adjacent bins, genome-wide, was > 0.38, suggesting poor-quality sequence data. Reporters of UCAD test were blinded to the clinical information.
Statistical analysis
Plasma cell free DNA was extracted and analyzed by Illumina X10. At least 10M paired reads were collected for each sample. The reads were mapped to human reference genome hg19. Genomic coverage was counted by using the software SAMtools mpileup [31]. Then we calculated average coverage for each 200k bin. Z-scores for each bin was then normalized by Z-score by using the formula as below:
Circular binary segmentation (CBS) algorithm was then used to find significant genomic breakpoints and genomics segments with changed copy number, by using the R package ‘DNACopy’ [32]. A previous study showed that the areas under the curve (AUC) was 0.53 for CA 19-9 to diagnose BTC [33]. We estimated the required sample size a prior at a clinically useful AUC of 0.75 and a null hypothesis value of 0.53, corresponding with a 5% alpha error and a 20% beta error. The calculated sample size was 50. The sensitivity and specificity of UCAD and tumor markers (CA 19-9 and CEA) were calculated by the receiver operating characteristic (ROC) method. The AUCs were compared by using the Delong method [34].
Categorical variables were reported as frequencies and percentages, and continuous variables were described as mean and standard deviation (SD), or median with interquartile range (IQR), as appropriate. Continuous variables were analyzed using the Mann–Whitney U test, and categorical variables were analyzed by the Pearson chi-square test or Fisher's exact test, as appropriate. Missing data were discarded from analyses. Kaplan–Meier survival curves were used to estimate the association between CNV burden and overall survival in patients with BTCs. All analyses were performed by using SPSS 18.0 (SPSS Inc., Chicago, IL, USA), R software (version 3.4.3; R Foundation for Statistical Computing), and MedCalc software (version 19.1, Mariakerke, Belgium). P < 0.05 was considered as statistically significant. Original data and R code used in the statistical analyses would be available on request.
Results
Patient characteristics
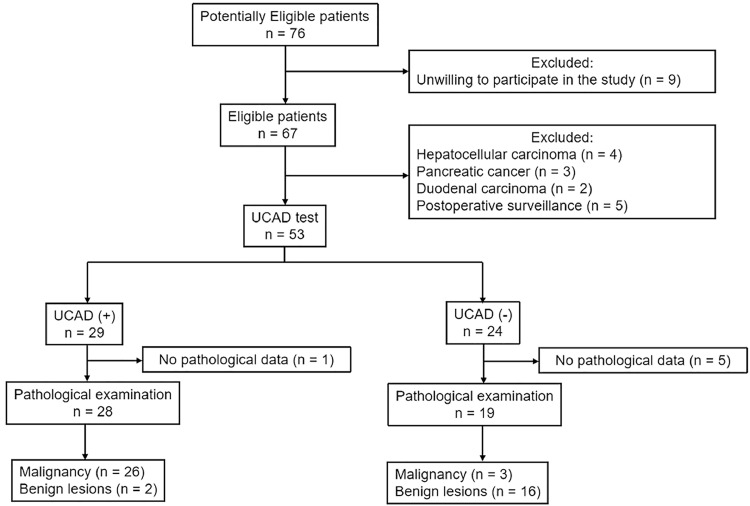
Between Mar 2019 and Jan 2020, 76 consecutive patients, moderately to highly suspicious for biliary malignancies, were admitted to our department. We excluded 9 patients unwilling to participate in the study, 4 patients with histologically proven hepatocellular carcinoma, 3 patients with pancreatic cancer, 2 patients with duodenal carcinoma, 5 patients of postoperative cancer surveillance, and 6 patients without histological examinations. The STARD flowdiagram was shown in Fig. 1. Finally, a total of 47 patients with biliary diseases were eligible for analysis. The median age was 59 years and 24 (51.1%) patients were female. On pathological examination, 29 (61.7%) patients had biliary tract cancer. Primary disease site was intrahepatic cholangiocarcinoma in 5 patients, perihilar cholangiocarcinoma in 10 patients, distal cholangiocarcinoma in 6 patients, and gallbladder cancer in 8 patients. Eighteen patients had postoperative benign lesions: hepatolithiasis (n = 4), gallbladder polyps (n = 4), xanthogranulomatous cholecystitis (n = 3), choledochal cyst (n = 1), inflammatory biliary stricture (n = 5), and IgG4-related biliary stricture (n = 1). Compared with the benign group, the malignancy group had more patients with hyperbilirubinemia and higher levels of alkaline phosphatase (AKP) as well as alanine transaminase (ALT). The baseline characteristics of these patients were shown in Table 1. The STARD checklist was provided in Supplementary Table S2.
Fig. 1.
The STARD flowdiagram for participants’ recruitment.
Table 1.
Baseline characteristics of the included patients.
| Total | Malignancy | Benign | P value | |
|---|---|---|---|---|
| Number of patients | 47 | 29 (61.7%) | 18 (38.3%) | – |
| Female | 24 (51.1%) | 16 (55.2%) | 8 (44.4%) | 0.474 |
| Age (year) | 59.0 (50.0–63.0) | 61.0 (52.5–66.0) | 56.5(41.3–60.0) | 0.074 |
| Age > 70 | 7 (14.9%) | 5 (17.2%) | 2 (11.1%) | 0.692a |
| Diabetes mellitus | 3 (6.4%) | 1 (3.5%) | 2 (11.1%) | 0.549a |
| Smoking | 1 (2.1%) | 1 (3.5%) | 0 (0%) | > 0.999a |
| History of biliary surgery | 6 (12.8%) | 3 (10.3%) | 3 (16.7%) | 0.662a |
| Hepatitis virus infection | 2 (4.3%) | 2 (6.9%) | 0 (0%) | 0.517a |
| Acute biliary infection* | 20 (42.6%) | 9 (31.0%) | 11 (61.1%) | 0.069a |
| Hyperbilirubinemia (≥ 2 mg/dl) | 17 (36.2%) | 16 (55.2%) | 1 (5.6%) | 0.007a |
| C-reactive protein level (μg/ml) | 0.50 (0.50–4.75) | 0.50 (0.50–3.65) | 0.50 (0.50–9.02) | 0.802 |
| WBC count (× 109/L) | 5.91 (4.58–7.52) | 5.95 (5.30–7.96) | 5.52 (4.28–6.60) | 0.175 |
| ALT (U/L) | 34.0 (19.0–65.0) | 49.0 (21.0–92.5) | 29.0 (12.75–45.00) | 0.037 |
| AKP (U/L) | 140.0 (79.0–268.0) | 164.0 (109.50–387.50) | 91.5 (46.5–158.25) | 0.004 |
| GGT (U/L) | 172.0 (30.0–309.0) | 201.0 (43.0–311.50) | 68.50 (20.75–257.25) | 0.090 |
| CA 19-9 level (U/ml) | 35.0 (10.40–110.00) | 61.30 (12.20–141.00) | 15.45 (8.65–41.73) | 0.088 |
| CEA level (ng/ml) | 1.90 (1.20–3.10) | 1.90 (1.35–3.75) | 1.90 (0.90–2.95) | 0.341 |
| Elevated CA 19-9 level (> 37 U/ml) | 24 (51.1%) | 18 (62.1%) | 6 (33.3%) | 0.055 |
| Elevated CA 19-9 level (> 90 U/ml) | 14 (29.8%) | 11 (37.9%) | 3 (16.7%) | 0.104 |
| Elevated CEA level (> 5 ng/ml) | 7 (14.9%) | 6 (20.7%) | 1 (5.6%) | 0.225a |
| Elevated CEA level (> 10 ng/ml) | 1 (2.1%) | 1 (3.45%) | 0 (0%) | > 0.999a |
| Positive UCAD testing | 28 (59.6%) | 26 (89.7%) | 2 (11.1%) | < 0.001 |
| Intrahepatic cholangiocarcinoma | 5 (10.6%) | 5 (17.2%) | – | – |
| Perihilar cholangiocarcinoma | 10 (21.3%) | 10 (34.5%) | – | – |
| Distal cholangiocarcinoma | 6 (12.8%) | 6 (20.7%) | – | – |
| Gallbladder cancer | 8 (17.0%) | 8 (27.6%) | – | – |
| Hepatolithiasis | 4 (8.5%) | – | 4 (22.2%) | – |
| Choledochal cyst | 1 (2.1%) | – | 1 (5.6%) | – |
| Gallbladder polyps | 4 (8.5%) | – | 4 (22.2%) | – |
| Xanthogranulomatous cholecystitis | 3 (6.4%) | – | 3 (16.7%) | – |
| IgG4-related cholangitis (≥ 135 mg/dl) | 1 (2.1%) | – | 1 (5.6%) | – |
| Benign biliary stricture | 5 (10.6%) | – | 5 (27.8%) | – |
Including acute cholecystitis and cholangitis.
AKP, alkaline phosphatase; ALT, alanine transaminase; BTC, biliary tract cancer; GGT, gamma-glutamyl transferase; WBC, white blood cell.
Fisher exact test was used.
Bold values indicate statistical significance (P < 0.05).
CNV profiles
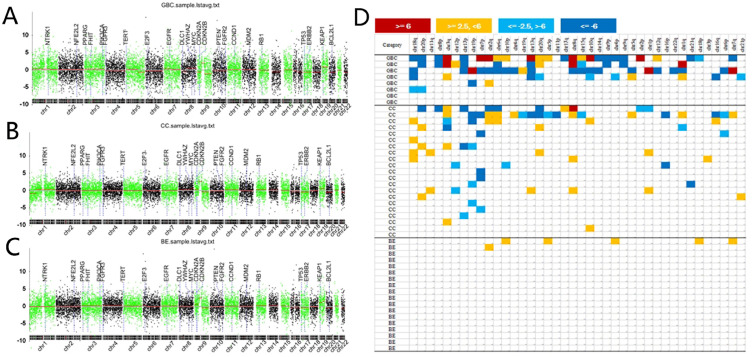
All plasma samples passed sequencing data quality assessment. The positive rate of CNV assays was 89.7% (26/29) in patients with BTC, and was 11.1% (2/18) in patients with benign biliary disease. The genome-wide landscape of CNVs in all patients was shown in Fig. 2A-C. Representative chromosome CNVs included 13q loss (n = 3), 17p loss (n = 3), 18q loss (n = 4), and 20p loss (n = 3), and 3q gain (n = 3) in gallbladder cancer (Fig. 2A), and 7p loss (n = 5), 19p loss (n = 5), 3q gain (n = 4), 8q gain (n = 4), and 18q gain (n = 4) in cholangiocarcinoma (Fig. 2B). None of these CNVs were found in benign lesions (Fig. 2C). Supplementary Fig. S1 showed the average chromosome CNVs in plasma samples for cholangiocarcinoma and gallbladder cancer. Z-scores for chromosomal arms (1p, 1q, 2p, 2q, 3p, 3q, 4p, 4q, 5p, 5q, 6p, 6q, 7p, 7q, 8p, 8q, 9p, 9q, 10p, 10q, 11p, 12q, 13q, 14q, 15q, 16p, 16q, 17p, 17q, 18p, 18q, 19p, 19q, 20p, 20q, 21p, 21q, 22q) were calculated normalized to benign controls. A heatmap of Z-scores was generated (Fig. 2D). The cancer group had more genome aberrations compared with the benign group. Chr3q and 8q gains were found in both gallbladder cancer and cholangiocarcinoma patients. With respect to Chr18q and 20p, more frequent gains were shown for cholangiocarcinoma, whereas more frequent losses were shown for gallbladder cancer (P < 0.05). Chr3q, 12p, and 19p showed more frequent losses in cholangiocarcinoma, but showing more frequent gains in gallbladder cancer (P < 0.05). These different CNV patterns indicated that gallbladder cancer and cholangiocarcinoma had potentially different cell origins and tumorigenesis processes. The overview of CNV profiles and potentially relevant genes described in literature was shown in Supplementary Table S3.
Fig. 2.
Overview of copy number variations via plasma cell-free DNA analysis in the included patients. (A) copy number variation genome of gallbladder cancer. (B) copy number variation genome of cholangiocarcinoma. (C) Copy number changes of benign biliary lesions. (D) heatmap of copy number variation quantified by chromosome Z-scores for all patients. GC, gallbladder cancer; CC, cholangiocarcinoma; BE, benign lesions.
Z-scores between malignant and benign lesions
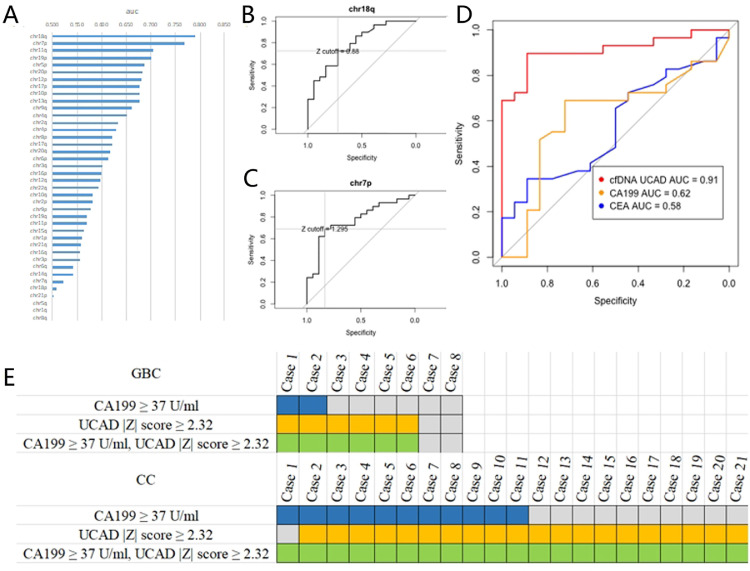
We explored the value of Z-score of each chromosome arm in differentiating biliary malignancies from benign lesions. The AUCs ranged from 0.45 to 0.79 (median AUC = 0.60, Fig. 3A). Chr18q and 7p showed high diagnostic accuracy, with AUCs of 0.79 and 0.77, respectively (Fig. 3B–C). We combined all the chromosomes information to build a diagnostic model for BTC. The optimal Z-score cutoff |Z| ≥ 2.32 was calculated by Youden Index. At this cutoff, UCAD test showed a sensitivity of 89.7% and a specificity of 88.9% (Table 2). The AUC was 0.91 (0.83–0.99) (Fig. 3D), which was better than the result from any single chromosome. A higher cutoff (|Z| = 3) showed better specificity (100.0%), whereas compromising the sensitivity (62.1%). We used CA 19-9 > 37 U/ml and CEA > 5 ng/ml as cutoffs. Compared with traditional tumor biomarkers, the overall diagnostic accuracy of UCAD test significantly outperform CA 19-9 (AUC 0.91 vs 0.62, P = 0.004) as well as CEA (AUC 0.91 vs 0.58, P = 0.001). When using a cutoff of 90 U/ml for CA 19-9, the specificity was 83.3% and the sensitivity was only 34.5% (Table 2). As shown in Fig. 3E, the diagnosis model identified 92.3% (12/13) of the CA 19-9 positive cancers, and 87.5% (14/16) of the CA 19-9 negative cancers. Compared with CA 19-9 alone, the adding of CNVs to CA 19-9 increased the sensitivity from 25.0% to 75.0% for GBC, and from 52.4% to 100% for CC. The AUC for combining CA 19-9 with UCAD test was 0.91 (95% CI: 0.80–0.98), which was not marked changed compared with UCAD test alone (P = 0.48). Correlations between UCAD test positivity and patient clinicopathological features were explored (Table 3). Cancer types, TNM stages, perineural invasion, vascular invasion, lymph node metastasis, histological grade, or distant metastasis were not significantly associated with UCAD test positivity.
Fig. 3.
Diagnostic performance of chromosome Z-scores. (A) calculation of area under the curve for each chromosome aberration. (B) ROC curve for chr18q. (C) ROC curve for chr7p. (D) Comparison of ROC curves between chromosome Z-scores incorporating all information of copy number aberrations, CA 19-9 and CEA. (E) the adding of Z-scores to CA 19-9 increased the detection rate of bile duct cancer. GBC, gallbladder cancer. CC, cholangiocarcinoma.
Table 2.
Diagnostic performance of |Z|-score in UCAD test by incorporating all chromosomes, CA 19-9 and CEA for diagnosing biliary tract cancer.
| Cutoff | Specificity | Sensitivity | Accuracy | TN | TP | FN | FP | NPV | PPV | |
|---|---|---|---|---|---|---|---|---|---|---|
| |Z|-score in UCAD test | 2 | 55.6% | 93.1% | 78.7% | 10 | 27 | 2 | 8 | 84.6% | 77.1% |
| 2.32 | 88.9% | 89.7% | 89.4% | 16 | 26 | 3 | 2 | 84.2% | 93.1% | |
| 3 | 100.0% | 62.1% | 76.6% | 18 | 18 | 11 | 0 | 62.1% | 100.0% | |
| CA 19-9 | 37 U/ml | 72.2% | 58.6% | 63.8% | 13 | 17 | 12 | 5 | 52.0% | 77.8% |
| CA 19-9 | 90 U/ml | 83.3% | 34.5% | 53.2% | 15 | 10 | 19 | 3 | 44.1% | 77.8% |
| CEA | 5 ng/ml | 94.4% | 20.7% | 48.9% | 17 | 6 | 23 | 1 | 42.5% | 87.5% |
TN, true negative; TP, true positive; FN, false negative; FP, false positive; NPV, negative predictive value; PPV, positive predictive value.
Table 3.
Correlation between clinicopathological features and UCAD results in 29 patients with biliary tract cancer.
| N | % | UCAD (+) | UCAD (-) | P valuea | |
|---|---|---|---|---|---|
| No. patients | 29 | 26 | 3 | ||
| Cancer type | 0.176 | ||||
| Gallbladder cancer | 8 | 27.6 | 6 | 2 | |
| Cholangiocarcinoma | 21 | 72.4 | 20 | 1 | |
| TNM Stage | > 0.999 | ||||
| Stage I | 3 | 13.8 | 3 | 0 | |
| Stage II | 4 | 10.3 | 4 | 0 | |
| Stage III | 14 | 48.3 | 12 | 2 | |
| Stage IV | 8 | 27.6 | 7 | 1 | |
| Perineural invasion | 21 | 72.4 | 19 | 2 | > 0.999 |
| Vascular invasion | 7 | 24.14 | 7 | 0 | 0.557 |
| Lymph node metastasis | 16 | 55.17 | 15 | 1 | 0.573 |
| Poor histological grade | 3 | 10.34 | 3 | 0 | > 0.999 |
| Distant metastasis | 2 | 6.90 | 2 | 0 | > 0.999 |
BTC, biliary tract cancer.
Bold values indicate statistical significance (P < 0.05).
Fisher exact test.
CNV burden and overall survival
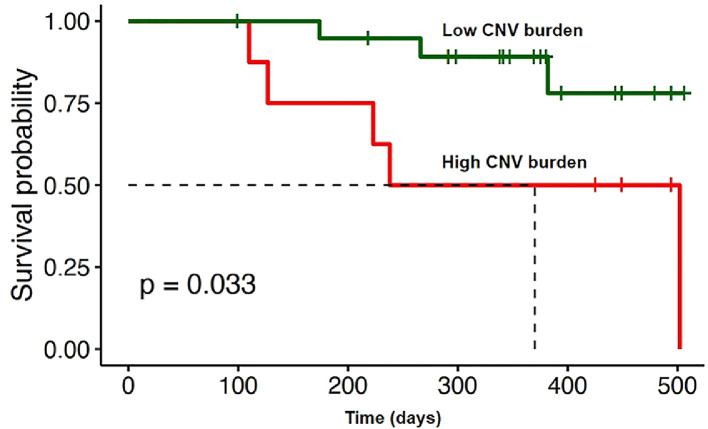
One patient died 3 days after surgery due to acute liver failure and was excluded from survival analysis. We completed follow-up for 28 patients with BTC. Ten patients had 4 or more detectable chromosomal aberrations in plasma. Five patients (50.0%) died within 8 months after surgery. In comparison, among 17 patients with less than 4 detectable chromosomal aberrations in plasma, 2 (11.8%) of them died within 8 months. High CNV burden (4 or more detectable chromosomal aberrations) in plasma was significantly associated with worse survival (HR = 4.32, 95% CI 2.06–9.08, P = 0.033) (Fig. 4).
Fig. 4.
Kaplan-Meier estimates of overall survival for 28 patients with BTC stratified by CNV burden.
Discussion
This pilot prospective study enrolled 29 biliary tract cancers and 18 benign biliary diseases. It provided the first evidence that plasma ctDNA-based CNV signals across the whole genome were rich in BTC. The sensitivity and specificity of plasma CNV assays (UCAD test) for diagnosing BTC were 89.7% and 88.9%, respectively (|Z| score cutoff: 2.32). The diagnostic accuracy of this test significantly outperformed CA 19-9 and CEA. In addition, higher CNV burden was useful to identify the subgroup of patients with unfavorable prognosis. This study represented a major step forward from previous work, which generally focused on panel-based gene mutations via tumor tissue or ctDNA analyses [16]. By contrast, plasma CNVs could play as a black box with abundant and mixed genetic aberrations, and might provide fuller picture of the genomic instability, tumor heterogeneity and clone evolution [35].
CA 19-9, an epitope on a complex oligosaccharide categorized as a Lewis blood group antigen, is regarded as the textbook biomarker for biliary malignancies. However, its diagnostic value is far from accurate. In our data regarding CA 19-9, the sensitivity and specificity, respectively, were 58.6% and 72.2%. Obstructive jaundice and biliary infection are commonly seen in cholangiocarcinoma. Hyperbilirubinemia and inflammation may promote the proliferation of biliary epithelial cells and subsequently increased systemic absorption of CA 19-9, and hence increasing its concentration [36]. Some authors suggested a higher cutoff level of 90 U/ml for CA 19-9 [8]. When we used this cutoff, the specificity increased to 83.3%, whereas the sensitivity decreased to 34.5%. In fact, falsely increased CA 19-9 levels misled us to perform extended multiple organ resection for a patient with xanthogranulomatous cholecystitis, who had negative UCAD test. Further complicating the situation is that approximately 5−10% of the population were Lewis (-). This subgroup lacks the enzyme 1,4-fucosyl transferase required for antigen epitope production, resulting in very low or even absent secretion of CA19-9 [36]. CEA is another widely used glycoprotein tumor marker, especially for gastrointestinal malignancies. Although recommended by the NCCN guideline [6], CEA had only limited value in diagnosing BTC. Our study showed that its sensitivity was only 20.7%.
In specialist centers, endoscopic procedures were always carried out for patients with indeterminate biliary strictures, along with histopathological examinations. Regretfully, the cumbersome and potentially risky endoscopic procedure could only acquire very small amount of brushings or biopsy tissues. Recently, fluorescence in situ hybridization (FISH) was introduced to amplify the faint signals captured by tiny forceps. Nevertheless, a meta-analysis showed a suboptimal diagnostic value of FISH in cholangiocarcinoma, with the sensitivity and specificity of 68% and 70%, respectively [37]. A study advised to perform serial polysomy FISH tests for patients with primary sclerosing cholangitis, given that nearly half (47%) of them did not have definitive malignancy evidence at the initial polysomy FISH test [38]. Vysis UroVysion is the most widely used FISH test, but it only detects polysomy of chromosomes 3, 7, and 17 and the deletion of chromosome locus 9p21 [39]. In comparison, the UCAD approach provided a panorama of the whole chromosomes. The alteration at a single chromosome might have insufficient power to discriminate BTC from benign lesions.
Our application of UCAD test is endorsed by that CNV profiles are particularly rich in BTC patients’ tumor tissues. Human cancers can be divided into two groups on the basis of oncogenic signatures: M class (primarily with mutations) and C class (primarily with copy number alterations). TP53 mutation was regarded as a typical feature of C-class cancer [40]. In BTC patients, TP53 mutation is most frequently detected, and is more predominant in gallbladder cancer (47.1−59%) and extrahepatic cholangiocarcinoma (40%), which mainly constituted our cohort [15]. The whole-genome study revealed that fluke-negative cholangiocarcinomas exhibited particularly high copy-number alterations [41]. None of our patients had history of fluke infection or fluke-associated findings on pathology. In fact, a series of chromosomal aberrations have been revealed among BTC patients [42], [43], [44]. A recent single-cell RNA sequencing analysis on cholangiocarcinoma has employed the CNV signals to identify the tumor cell cluster [45].
Our chromosomal findings were also in line with previous genetic studies on BTC. In our data, loss of chromosome 18q showed the highest diagnostic accuracy, with the sensitivity and specificity of 72.4% and 72.2%, respectively. Chromosome 18q harbors the SMAD gene, which is mutated in approximately 21% of patients with extrahepatic cholangiocarcinoma [15]. The DCC gene is also located herein. A Japanese study on GBC showed that reduced expression of DCC was associated with poorly differentiated histological type and increased proliferation and metastasis [46]. A study based on UroVysion test showed that increased chromosome 7 copy number was present in 26.5% (13/49) of patients with cholangiocarcinoma and was significantly associated with poor prognosis [47]. Chromosome 7 contains several oncogenes, including EGFR and MET genes. Our data showed that the sensitivity and specificity, respectively, were 69.0% and 83.3% for chromosome 7p gains in diagnosing BTC. Patients with GBC were likely to have more frequent 7p gains than those with CCs. This was in accordance with that ErbB signaling was the most extensively mutated pathway in GBC [48]. TP53 gene is one of the most famous tumor suppressor genes that located at chromosome 17p13.1. Mutational inactivation of TP53 is always accompanied with the loss of chromosome 17p. Our results showed that the aberration of chromosome 17 (17p-) did not differ significantly between cholangiocarcinoma and GBC, indicating that TP53 deletion might participate in the carcinogenesis for both subtypes. However, specific gene analysis was beyond the scope of our study. We failed to verify the mRNA expressions of specific candidate genes as no sufficient fresh frozen tissues were available. In fact, the association between gene expression and CNVs was far from clear. One study suggested that copy number only explained 12–40% of the variation in gene expression, given that other biological processes may participate in this complex process, such as histone modifications, mutations in DNA sequence, microRNA molecules and protein activity [49]. Our feasibility study laid the foundation for future work to depict the genomic and transcriptomic landscapes of BTCs via plasma CNV profiles.
A perfect marker is like the distant holy grail in tumor diagnosis. We have tried to optimize the UCAD workflow to reduce the interference of CNV results from normal cfDNA. The repetitive regions and genome duplication regions of human genome were excluded from analyses. Our workflow has previously sequenced a thousands of ‘non-cancer’ individuals to filter out the potential non-specific CNVs [50]. However, several other factors may add noise to our diagnostic accuracy. Firstly, the sensitivity of low pass WGS is not as high as deep-sequencing, and requires a relatively high tumor fraction. The fraction of ctDNA can range from 0.01% to 93% in the plasma [51]. A study on lung cancer showed that ctDNA was undetectable among the largest proportion of stage I patients by using the current NGS technique [52]. It is possible that in early BTC, the ctDNA level is below optimal level for the detection of CNVs. Nevertheless, our study showed that UCAD test was positive for all of the seven stage I-II patients. Chromosomal alterations could occur in biliary dysplastic tissue especially high-grade dysplasia, which may be similar to the aberrations detected in cholangiocarcinoma [53]. UCAD test was positive in one patient with gallbladder adenoma and low-grade dysplasia, both known as cancer precursors. He was closely followed up after cholecystectomy. A series of precancerous lesions may lead to BTC development, including IgG4-related cholangitis, hepatolithiasis, and choledochal cyst. Discriminating a precursor from true malignancy can be extremely difficult despite clinical, biochemical, and radiologic findings. Intriguingly, BTCs secondary to these high-risk lesions were all seen in our cohort.
Our study had several advantages. This study was prospectively designed, and the test was done blinded to the clinical data. Thus, we avoided the impact of hypothesis-generating data in most retrospective or non-blinded studies. Compared with panel-based high-depth sequencing, low-coverage genome-wide sequencing of cfDNA markedly decreased costs and shortened testing duration. By covering the whole genome, this method detected larger number of chromosomal rearrangements. Furthermore, our cohort included a group of patients with heterogeneous biliary etiologies and thus indicated the generalizability of UCAD test in the real world practice. Since many patients were reluctant to undergo invasive ERCP procedures and preferred blood tests, our test could fill a critical niche for strengthening surveillance among high-risk individuals with benign biliary lesions.
Despite the merits, several limitations should be acknowledged. Given the low incidence of biliary malignancy and the adverse impact of COVID-19 for patients’ recruitment, the sample size was not quite large and the enrolling process was slow recently. There may exist a chance for a type I error. The small number of cases also preluded us from verifying distinct CNV patterns for different BTC subtypes. Our favorable preliminary data may promote more biliary surgeons to participate in future multicenter validation. Notably, none of our patients had a history of primary sclerosing cholangitis, which is the major cause of cholangiocarcinoma in Europe or North America whereas is rare in Asia. Besides, fluke infection was absent in our cohort. Thus, the cohort representativeness may be compromised. Furthermore, due to cost concern, we only provided plasma results without tissue confirmation. Notably, large well-controlled studies have shown that the concordance rate of ctDNA levels between plasma and tissue samples was as high as 80–90% [13]. Moreover, the analyzed sliced tissue may only harbor a small sample of the entire cancer events and might not be as representative as plasma ctDNA [54]. As our study focused on diagnosing BTC at the chromosomal level, we did not further crack the genetic codes that delineate the formation of CNVs. In some patients with BTC, the carcinogenesis may be primarily driven by specific genetic mutations, and UCAD may incidentally miss positive findings in this subgroup.
In the future, large cohort is needed to validate whether UCAD test could be implemented as routine practice to aid diagnosis, monitor recurrence, and guide therapy efficacy among patients with BTC. It remains unknown whether CNV profiles differ according to different cancer etiologies or BTC subtypes. Mysteries relating to cancer cell origins or tumor heterogeneity may be unraveled by CNV signals. Deeper sequencing techniques are expected to improve the sensitivity among patient with low tumor fraction. It is also appealing to explore CNV profiles in the bile juice from BTC patients.
Conclusions
Our study demonstrates a proof of concept for employing the plasma CNV assays (UCAD test) to discriminate biliary tract cancers from benign biliary diseases. The diagnostic accuracy of UCAD test was significantly superior to CA 19-9 and CEA. Higher CNV burden was also associated with poor prognosis. As a non-invasive, cost-effective, and time-saving method, plasma CNV assays might be a promising diagnostic tool for patients with indeterminate biliary stricture, suspicious gallbladder thickening, or long-term precancerous biliary conditions.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100908.
Contributor Information
Yong-Jie Zhang, Email: yongjie_zh@outlook.com.
Wen-Long Yu, Email: Yu_wenlong@sina.com.
Appendix. Supplementary materials
References
- 1.Florio A.A., Ferlay J., Znaor A., Ruggieri D., Alvarez C.S., Laversanne M., Bray F., McGlynn K.A., Petrick J.L. Global trends in intrahepatic and extrahepatic cholangiocarcinoma incidence from 1993 to 2012. Cancer. 2020;126:2666–2678. doi: 10.1002/cncr.32803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otsuka S., Ebata T., Yokoyama Y., Igami T., Mizuno T., Yamaguchi J., Onoe S., Watanabe N., Shimoyama Y., Nagino M. Benign hilar bile duct strictures resected as perihilar cholangiocarcinoma. Br. J. Surg. 2019;106:1504–1511. doi: 10.1002/bjs.11257. [DOI] [PubMed] [Google Scholar]
- 3.Roos E., Hubers L.M., Coelen R.J.S., Doorenspleet M.E., de Vries N., Verheij J., Beuers U., van Gulik T.M. IgG4-associated cholangitis in patients resected for presumed perihilar cholangiocarcinoma: a 30-year tertiary care experience. Am. J. Gastroenterol. 2018;113:765–772. doi: 10.1038/s41395-018-0036-5. [DOI] [PubMed] [Google Scholar]
- 4.Spinelli A., Schumacher G., Pascher A., Lopez-Hanninen E., Al-Abadi H., Benckert C., Sauer I.M., Pratschke J., Neumann U.P., Jonas S. Extended surgical resection for xanthogranulomatous cholecystitis mimicking advanced gallbladder carcinoma: a case report and review of literature. World J. Gastroenterol. 2006;12:2293–2296. doi: 10.3748/wjg.v12.i14.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitt S.C., Jin L.X., Hall B.L., Strasberg S.M., Pitt H.A. Incidental gallbladder cancer at cholecystectomy: when should the surgeon be suspicious? Ann. Surg. 2014;260:128–133. doi: 10.1097/SLA.0000000000000485. [DOI] [PubMed] [Google Scholar]
- 6.A.B. Benson, M.I. D'Angelica, D.E. Abbott, T.A. Abrams, S.R. Alberts, D.A. Anaya, R. Anders, C. Are, D. Brown, D.T. Chang, Hepatobiliary Cancers, Version 2.2019, Accessed Febrary 13, 2020 To view the most recent version of these guidelines, visit NCCNorg. (2019).
- 7.Albert M.B., Steinberg W.M., Henry J.P. Elevated serum levels of tumor marker CA19-9 in acute cholangitis. Dig. Dis. Sci. 1988;33:1223–1225. doi: 10.1007/BF01536670. [DOI] [PubMed] [Google Scholar]
- 8.Marrelli D., Caruso S., Pedrazzani C., Neri A., Fernandes E., Marini M., Pinto E., Roviello F. CA19-9 serum levels in obstructive jaundice: clinical value in benign and malignant conditions. Am. J. Surg. 2009;198:333–339. doi: 10.1016/j.amjsurg.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Luo G., Liu C., Guo M., Cheng H., Lu Y., Jin K., Liu L., Long J., Xu J., Lu R. Potential biomarkers in Lewis negative patients with pancreatic cancer. Ann. Surg. 2017;265:800–805. doi: 10.1097/SLA.0000000000001741. [DOI] [PubMed] [Google Scholar]
- 10.Joo I., Lee J.M., Yoon J.H. Imaging diagnosis of intrahepatic and perihilar cholangiocarcinoma: recent advances and challenges. Radiology. 2018;288:7–13. doi: 10.1148/radiol.2018171187. [DOI] [PubMed] [Google Scholar]
- 11.Aljiffry M., Abdulelah A., Walsh M., Peltekian K., Alwayn I., Molinari M. Evidence-based approach to cholangiocarcinoma: a systematic review of the current literature. J. Am. Coll. Surg. 2009;208:134–147. doi: 10.1016/j.jamcollsurg.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 12.de Vries A.B., van der Heide F., Ter Steege R.W.F., Koornstra J.J., Buddingh K.T., Gouw A.S.H., Weersma R.K. Limited diagnostic accuracy and clinical impact of single-operator peroral cholangioscopy for indeterminate biliary strictures. Endoscopy. 2020;52:107–114. doi: 10.1055/a-1061-7067. [DOI] [PubMed] [Google Scholar]
- 13.Corcoran R.B., Chabner B.A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 14.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J. Clin. Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 15.Valle J.W., Lamarca A., Goyal L., Barriuso J., Zhu A.X. New horizons for precision medicine in biliary tract cancers. Cancer Discov. 2017;7:943–962. doi: 10.1158/2159-8290.CD-17-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zill O.A., Greene C., Sebisanovic D., Siew L.M., Leng J., Vu M., Hendifar A.E., Wang Z., Atreya C.E., Kelley R.K. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015;5:1040–1048. doi: 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mody K., Kasi P.M., Yang J., Surapaneni P.K., Bekaii-Saab T., Ahn D.H., Mahipal A., Sonbol M.B., Starr J.S., Roberts A. Circulating tumor DNA profiling of advanced biliary tract cancers. JCO Precis. Oncol. 2019;3:1–9. doi: 10.1200/PO.18.00324. [DOI] [PubMed] [Google Scholar]
- 18.Campos-Carrillo A., Weitzel J.N., Sahoo P., Rockne R., Mokhnatkin J.V., Murtaza M., Gray S.W., Goetz L., Goel A., Schork N. Circulating tumor DNA as an early cancer detection tool. Pharmacol. Ther. 2020;207 doi: 10.1016/j.pharmthera.2019.107458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakhoum S.F., Cantley L.C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 2018;174:1347–1360. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turajlic S., Sottoriva A., Graham T., Swanton C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 2019;20:404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 21.Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura H., Arai Y., Totoki Y., Shirota T., Elzawahry A., Kato M., Hama N., Hosoda F., Urushidate T., Ohashi S. Genomic spectra of biliary tract cancer. Nat. Genet. 2015;47:1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 23.Yoo K.H., Kim N.K., Kwon W.I., Lee C., Kim S.Y., Jang J., Ahn J., Kang M., Jang H., Kim S.T. Genomic alterations in biliary tract cancer using targeted sequencing. Transl. Oncol. 2016;9:173–178. doi: 10.1016/j.tranon.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3007094. 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen T.J., Goodman A.M., Kato S., Ellison C.K., Daniels G.A., Kim L., Nakashe P., McCarthy E., Mazloom A.R., McLennan G. Genome-wide sequencing of cell-free dna identifies copy-number alterations that can be used for monitoring response to immunotherapy in cancer patients. Mol. Cancer Ther. 2019;18:448–458. doi: 10.1158/1535-7163.MCT-18-0535. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z., Zhang C., Zhang M., Li B., Niu Y., Chen L., Yang J., Lu S., Gao J., Shen L. Chromosomal instability of circulating tumor DNA reflect therapeutic responses in advanced gastric cancer. Cell Death Dis. 2019;10:697. doi: 10.1038/s41419-019-1907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stover D.G., Parsons H.A., Ha G., Freeman S.S., Barry W.T., Guo H., Choudhury A.D., Gydush G., Reed S.C., Rhoades J. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative breast cancer. J. Clin. Oncol. 2018;36:543. doi: 10.1200/JCO.2017.76.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bossuyt P.M., Reitsma J.B., Bruns D.E., Gatsonis C.A., Glasziou P.P., Irwig L., Lijmer J.G., Moher D., Rennie D., de Vet H.C. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical practice guidelines for the use of tumor markers in breast and colorectal cancer. Adopted on May 17, 1996 by the American Society of Clinical Oncology. J. Clin. Oncol. 1996;14:2843–2877. doi: 10.1200/JCO.1996.14.10.2843. [DOI] [PubMed] [Google Scholar]
- 30.Amin M.B., Edge S.B. springer; 2017. AJCC Cancer Staging Manual. [Google Scholar]
- 31.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome S. Project data processing, the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. V.E., O. A, DNAcopy: DNA copy number data analysis. R package version 1.42.0., URL https://www.bioconductororg/packages/release/bioc/html/DNAcopyhtml.
- 33.Kim H.J., Kim M.H., Myung S.J., Lim B.C., Park E.T., Yoo K.S., Seo D.W., Lee S.K., Min Y.I. A new strategy for the application of CA19-9 in the differentiation of pancreaticobiliary cancer: analysis using a receiver operating characteristic curve. Am. J. Gastroenterol. 1999;94:1941–1946. doi: 10.1111/j.1572-0241.1999.01234.x. [DOI] [PubMed] [Google Scholar]
- 34.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 35.Ben-David U., Amon A. Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 2020;21:44–62. doi: 10.1038/s41576-019-0171-x. [DOI] [PubMed] [Google Scholar]
- 36.Ballehaninna U.K., Chamberlain R.S. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J. Gastrointest. Oncol. 2012;3:105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navaneethan U., Njei B., Venkatesh P.G., Vargo J.J., Parsi M.A. Fluorescence in situ hybridization for diagnosis of cholangiocarcinoma in primary sclerosing cholangitis: a systematic review and meta-analysis. Gastrointest. Endosc. 2014;79:943–950.e943. doi: 10.1016/j.gie.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Barr Fritcher E.G., Kipp B.R., Voss J.S., Clayton A.C., Lindor K.D., Halling K.C., Gores G.J. Primary sclerosing cholangitis patients with serial polysomy fluorescence in situ hybridization results are at increased risk of cholangiocarcinoma. Am. J. Gastroenterol. 2011;106:2023–2028. doi: 10.1038/ajg.2011.272. [DOI] [PubMed] [Google Scholar]
- 39.Kipp B.R., Stadheim L.M., Halling S.A., Pochron N.L., Harmsen S., Nagorney D.M., Sebo T.J., Therneau T.M., Gores G.J., de Groen P.C. A comparison of routine cytology and fluorescence in situ hybridization for the detection of malignant bile duct strictures. Am. J. Gastroenterol. 2004;99:1675–1681. doi: 10.1111/j.1572-0241.2004.30281.x. [DOI] [PubMed] [Google Scholar]
- 40.Ciriello G., Miller M.L., Aksoy B.A., Senbabaoglu Y., Schultz N., Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jusakul A., Cutcutache I., Yong C.H., Lim J.Q., Huang M.N., Padmanabhan N., Nellore V., Kongpetch S., Ng A.W.T., Ng L.M. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong N., Li L., Tsang K., Lai P.B., To K.F., Johnson P.J. Frequent loss of chromosome 3p and hypermethylation of RASSF1A in cholangiocarcinoma. J. Hepatol. 2002;37:633–639. doi: 10.1016/s0168-8278(02)00269-6. [DOI] [PubMed] [Google Scholar]
- 43.Kang M.J., Kim J., Jang J.Y., Park T., Lee K.B., Kim S.W. 22q11-q13 as a hot spot for prediction of disease-free survival in bile duct cancer: integrative analysis of copy number variations. Cancer Genet. 2014;207:57–69. doi: 10.1016/j.cancergen.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Eaton J.E., Barr Fritcher E.G., Gores G.J., Atkinson E.J., Tabibian J.H., Topazian M.D., Gossard A.A., Halling K.C., Kipp B.R., Lazaridis K.N. Biliary multifocal chromosomal polysomy and cholangiocarcinoma in primary sclerosing cholangitis. Am. J. Gastroenterol. 2015;110:299–309. doi: 10.1038/ajg.2014.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M., Yang H., Wan L., Wang Z., Wang H., Ge C., Liu Y., Hao Y., Zhang D., Shi G. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020 doi: 10.1016/j.jhep.2020.05.039. [DOI] [PubMed] [Google Scholar]
- 46.Cha P.C., Zembutsu H., Takahashi A., Kubo M., Kamatani N., Nakamura Y. A genome-wide association study identifies SNP in DCC is associated with gallbladder cancer in the Japanese population. J. Hum. Genet. 2012;57:235–237. doi: 10.1038/jhg.2012.9. [DOI] [PubMed] [Google Scholar]
- 47.Kato A., Naitoh I., Miyabe K., Hayashi K., Yoshida M., Hori Y., Natsume M., Jinno N., Asano G., Kato H. An increased chromosome 7 copy number in endoscopic bile duct biopsy specimens is predictive of a poor prognosis in cholangiocarcinoma. Dig. Dis. Sci. 2018;63:3376–3381. doi: 10.1007/s10620-018-5280-4. [DOI] [PubMed] [Google Scholar]
- 48.Li M., Zhang Z., Li X., Ye J., Wu X., Tan Z., Liu C., Shen B., Wang X.A., Wu W. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat. Genet. 2014;46:872–876. doi: 10.1038/ng.3030. [DOI] [PubMed] [Google Scholar]
- 49.Gu W., Choi H., Ghosh D. Global associations between copy number and transcript mRNA microarray data: an empirical study. Cancer Inform. 2008;6:17–23. doi: 10.4137/cin.s342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan M., Ziliang Q., Fang J., Zhu Z., Wu J., Guo X., Liu W., Qian H., Xu. iStopCancer Q. A database of 6016 low pass whole genome sequencing of minimal invasive samples from 21 cancer types of Chinese population. Am. Soc. Clin. Oncol. 2020 [Google Scholar]
- 51.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 52.Abbosh C., Birkbak N.J., Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 2018;15:577–586. doi: 10.1038/s41571-018-0058-3. [DOI] [PubMed] [Google Scholar]
- 53.DeHaan R.D., Kipp B.R., Smyrk T.C., Abraham S.C., Roberts L.R., Halling K.C. An assessment of chromosomal alterations detected by fluorescence in situ hybridization and p16 expression in sporadic and primary sclerosing cholangitis-associated cholangiocarcinomas. Hum. Pathol. 2007;38:491–499. doi: 10.1016/j.humpath.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Heitzer E., Ulz P., Geigl J.B., Speicher M.R. Non-invasive detection of genome-wide somatic copy number alterations by liquid biopsies. Mol. Oncol. 2016;10:494–502. doi: 10.1016/j.molonc.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.