Abstract
An urgent question of coronavirus disease 2019 (COVID-19) is population variation in susceptibility to SARS-CoV-2 infection and symptom severity. We explore the expression profiles of SARS-CoV-2 host genes, their population variations, associated genetic variants, age- and sex-dependency in normal individuals. SARS-CoV-2 host genes are provisionally defined as the human genes that are experimentally validated or bioinformatically predicted to interact with SARS-CoV-2 proteins. Genes exhibiting most variable expression include ACE2, CLEC4G, CLEC4M, CD209 (interact with the SARS-CoV-2 spike protein); REEP6 (a receptor accessory protein expressed in the olfactory epithelium); SLC27A2 and PKP2 (inhibit virus replication); and PTGS2 (mediates fever response). SNP rs4804803, associated with SARS severity, affects expression of CLEC4G and CD209. Genetic variants of proteases associated with SARS-CoV-2 entry (TMPRSS2, CTSB, and CTSL) are strongly associated with their expression variation, suggesting a genetic contribution to phenotypic variations in multiple organs upon virus attack. The most significant age-dependent gene is ACE2, the cellular receptor of SARS-CoV-2. Others include TGF-β family member GDF15, mediating inflammation, and VKORC1, possibly explaining vitamin K deficiency in COVID-19. TIMM10 and ERGIC1 exhibit significant sex differences. In summary, our results show genetic and multiple biological variables may underlie the population variation in SARS-CoV-2 infection and symptom severity.
Keywords: COVID-19, Expression variation, Genetic variants, Host genes, Perturbagens, SARS-CoV-2, Transcriptome
1. Introduction
The pandemic outbreak of coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 infection has affected more than 32 million people globally, with an average fatality rate of 3.0% (https://coronavirus.jhu.edu/map.html, as of September 25, 2020). The disease manifested through a broad spectrum attacking multiple tissue organs [[1], [2], [3], [4], [5]]. Besides respiratory symptoms (e.g., cough and pneumonia), some COVID-19 patients experience gastrointestinal (GI) symptoms (e.g., loss of appetite, nausea, vomiting, diarrhea, and abdominal pain) and/or neurological symptoms (e.g., loss of smell and taste, muscle weakness, tingling or numbness in hands and feet, dizziness, confusion, delirium, seizures, and stroke). Symptom severity varies widely among patients.
We do not yet know why or how SARS-CoV-2 infections lead to this wide range of outcomes. A large proportion of people who tested positive for SARS-CoV-2 infection were asymptomatic [[6], [7], [8]]. Even for symptomatic patients, reported illnesses ranged from very mild cold-like symptoms to severe illness and death [6,9]. Anecdotal evidence suggests not all people exposed to SARS-CoV-2 were infected. While the sources of these variations are undoubtedly multifactorial, one possibility lies with genetic variants and variable gene expression of host cells.
It is essential to know the causes of variations among the normal population regarding an individual's intrinsic susceptibility of being infected, their responses to virus infection, and disease progression. It is also critical to understand the mechanisms underlying tissues' variable vulnerability to SARS-CoV-2 virus attack. Knowledge about these variations will not only help develop effective prophylactic and therapeutic strategies but may also inform epidemiological and economic policies. Medical tests can be better developed to accommodate these variations in monitoring virus transmission and disease pathology, which helps guide mitigation and treatment options.
No effective treatment has been approved by FDA for COVID-19 as yet, though five treatments were authorized for emergency use and more than 590 drug development programs are in planning stages (https://www.fda.gov/drugs/coronavirus-covid-19-drugs/coronavirus-treatment-acceleration-program-ctap, as of September 25, 2020). Low-dose radiation therapy for COVID-19 pneumopathy was examined and may be worth investigating in the clinical setting [10]. There is an urgent need to pinpoint possible drug candidates amid the outbreak because vaccines will likely take over a year to develop. An effective drug could interfere with the SARS-CoV-2 life cycle. Since the virus hijacks host molecules for fusion, replication, packaging and release, targeted perturbation of virus host genes would be a reasonable strategy to slow and/or stop virus infection and disease development.
In this study, we investigate the expression profiles of known and predicted SARS-CoV-2 human host genes in tissues from the normal population. Host genes include those interacting with SARS-CoV-2 proteins in human cells [11] https://www.proteinatlas.org/humanproteome/sars-cov-2 and other predicted interactors based on homology with SARS [12] http://korkinlab.org/wuhan. Gordon et al. cloned and expressed 26 of the 29 SARS-CoV-2 proteins in human cells. Human proteins physically associated with each of the SARS-CoV-2 proteins were identified through affinity-purification mass spectrometry. They identified 332 human cell proteins and provisionally named these proteins the SARS-CoV-2 host proteins. Srinivasan et al. applied an integrated bioinformatics approach to identify SARS-CoV-2 host proteins based on structural genomics and interactomics. Although the function these interactions regarding viral activity will need to be validated, they serve a reasonable starting point to understand virus-host interactions. Here, we provisionally called all these proteins the SARS-CoV-2 host proteins.
We reasoned that individual expression variation of SARS-CoV-2 host genes may contribute to the variability of host response to SARS-CoV-2 and therefore a wide range of outcomes from no symptoms to death. Tissue expression variability of SARS-CoV-2 host genes may also underlie the different vulnerability of tissue organs to SARS-CoV-2 attacks. To understand population and tissues expression variation of SARS-CoV-2 host genes, we comprehensively analyzed the large-scale tissue transcriptome data in the Genotype-Tissue Expression (GTEx) project [13] https://gtexportal.org/. GTEx aims to characterize genetic effects on gene expression variation across different individuals (>900) and a variety of healthy tissues (>50). It provides genotype data and transcriptome data of multiple tissues from the same individuals. We examined host gene expression variation in this population and identified associated genetic variants. We derived tissue similarity in relation to the lung to understand tissue vulnerability based on the expression of SARS-CoV-2 host genes. Our results would help understanding the variation of individuals’ susceptibility to SARS-CoV-2.
2. Results
2.1. Tissue expression of SARS-CoV-2 host genes in the normal population
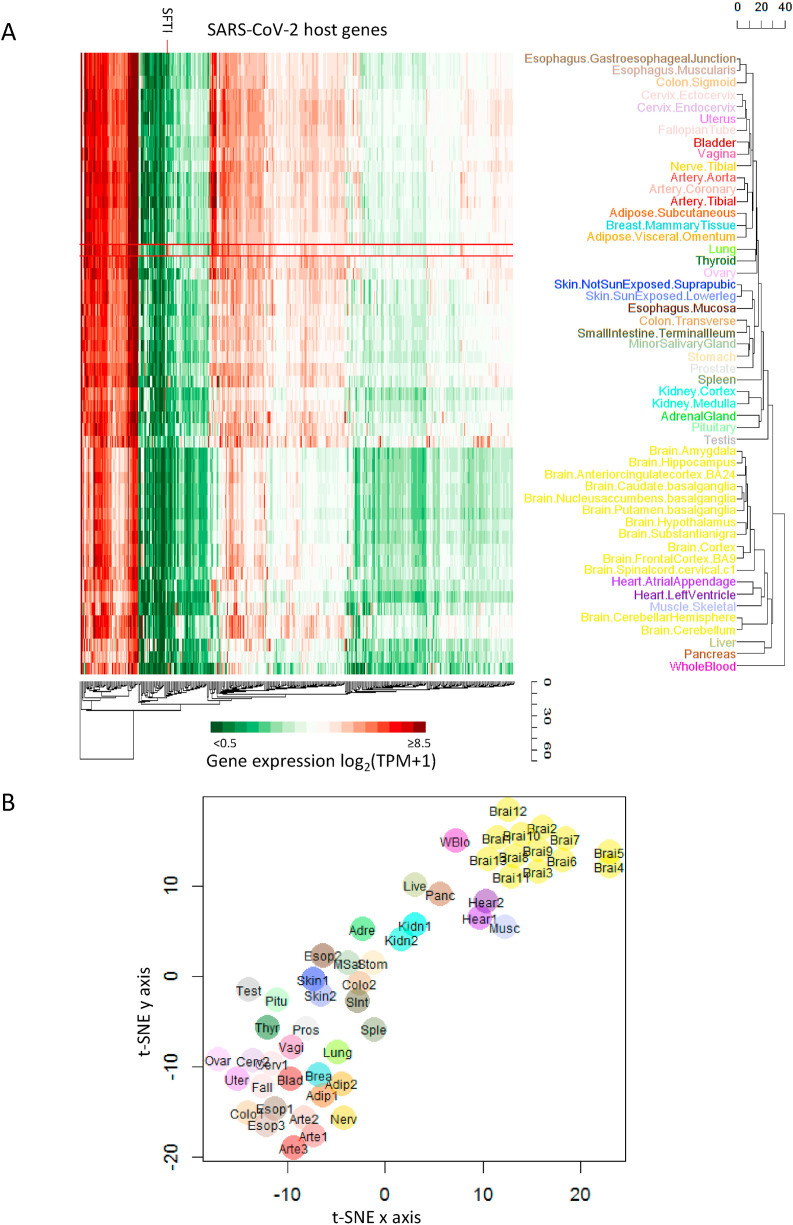
Herein, ‘normal’ means non-diseased tissues profiled by GTEx. We studied tissue expression of SARS-CoV-2 host genes in the normal population using transcriptomics data from non-diseased tissue sites profiled by GTEX. Unbiased clustering separated all profiled tissues into three groups (Fig. 1 A). Whole blood stands alone and is distinct from other tissues. Brain, heart, muscle, liver and pancreas belong to one cluster. Lung and others belong to the third cluster. The t-SNE visualization based on expression of SARS-CoV-2 host genes shows a similar grouping of tissues (Fig. 1B).
Fig. 1.
Expression profiles of SARS-CoV-2 host genes across different normal tissues. (A) Heatmap of host gene expression across tissues. We obtained the expression level of each gene in each sample quantified by GTEx RNA-seq. We then calculated the average expression level across multiple samples of the same tissue. The color warmness in the heatmap represents the expression level, as shown in the color scale. Hierarchical clustering based on average linkage was performed for both genes (columns) and tissues (rows). (B) t-SNE visualization of tissues based on their expression profiles of host genes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To further understand the differences among tissues for SARS-CoV-2 infection, we identified tissue-distinctive genes whose expression in that tissue displayed a minimum of 2-fold change compared to all other tissues. That is, the distinctive tissue exhibits the largest (or smallest) expression among all tissues with a difference ≥ 1 (log2 scale) from the second largest (or smallest) expression. In summary, we found 41 host genes exhibiting tissue-distinctive high expression and 37 tissue-distinctive low expression (Table S1).
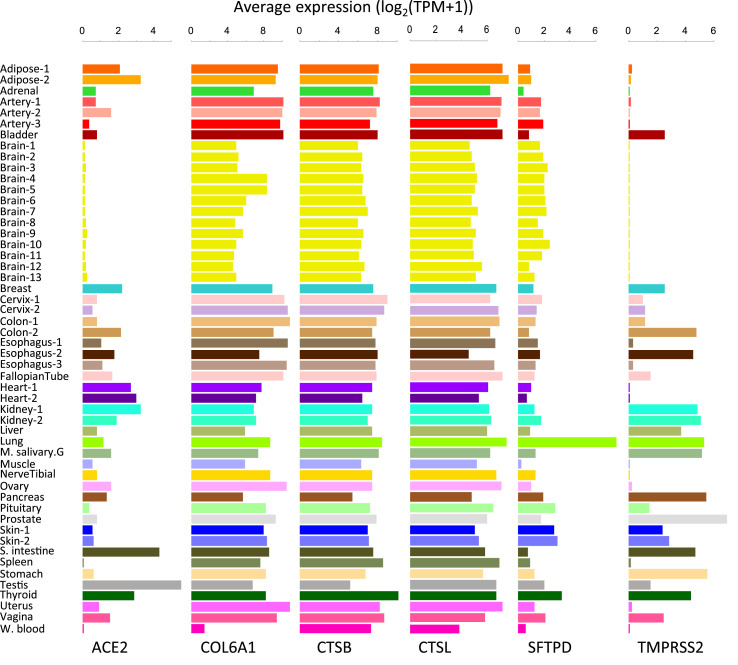
High expression of host genes in the lung may underlie lung's unique susceptibility to SARS-CoV-2. SFTPD is particularly highly expressed in the lung (Fig. 2 ). It is involved in the innate immune response, and protects the lung against inhaled microorganisms and chemicals [14]. Based on the Human Protein Atlas (HPA) [15], we confirmed SFTPD protein is highly expressed in pneumocytes of lung. SFTPD is predicted to interact with the spike protein of SARS-CoV-2 [12]. Given its distinctly high expression in the lung, future studies can prioritize SFTPD to understand its role in the lung's unique susceptibility and innate immune response to COVID-19.
Fig. 2.
Expression profiles of important SARS-CoV-2 host genes.
Although SARS-CoV-2 can be detected in blood, the positive rate was only 1% [16]. By contrast, bronchoalveolar lavage fluid exhibited a positive rate as high as 93% [16], suggesting that blood cells are not the common infection target. We found 33 of 420 host genes were lowly expressed specifically in the blood, representing 89% (33/37) of tissue-distinctive lowly expressed genes, and establishing blood as the most distinctive tissue from the lung. COL6A1 displays the most tissue-distinctive low expression (>8-fold change compared to other tissues, Fig. 2). COL6A1 encodes a collagen chain to maintain tissue integrity [14]. On the other hand, three genes (DEDD2, IL17RA, LCP1) are highly expressed in the blood. Whether the distinctive expression of these 36 genes in the blood spares blood cells from SARS-CoV-2 attack is an interesting question for experimental validation.
Male sex hormones are significantly changed with SARS-CoV-2 infection [17]. We found 16 of 420 genes are highly expressed in the testis, including ACE2, the cellular receptor of SARS-CoV-2 (Fig. 2). ACE2 is also highly expressed in the small intestine. We examined HPA data and confirmed the selective membranous ACE2 protein expression in testis, intestinal tract, renal tubules, and gallbladder. These data suggest an enhanced risk of infecting male gonads and gastrointestinal tissues compared to other tissues in Covid-19 patients.
SARS-CoV-2 entry-associated proteases include TMPRSS2, CTSB, and CTSL. GTEx expression profiles show that TMPRSS2 is distinctively high in the prostate (Fig. 2). HPA confirms the selective protein expression of TMPRSS2 in the prostate, gastrointestinal tract, kidney, and pancreas. CTSB is distinctively expressed in the thyroid but is generally expressed in all other tissues. HPA protein data also show CTSB protein expression in many tissues. CTSL is not particularly distinctive in any tissue based on the threshold (≥2-fold changes). These data suggest that TMPRSS2 expression is less ubiquitous than CTSB and CTSL and would be a more selective target to limit SARS-CoV-2 entry.
Caspases play an important role in the host cell death [18]. Although they are not on in the provisional host gene list, we profiled their expression across tissues (Fig. S1) and found that their expression levels in the lung were relatively high. Therefore, lung cells may succumb to cell death due to high caspase levels.
2.2. Alternative splicing patterns of SARS-CoV-2 host genes in tissues of normal population
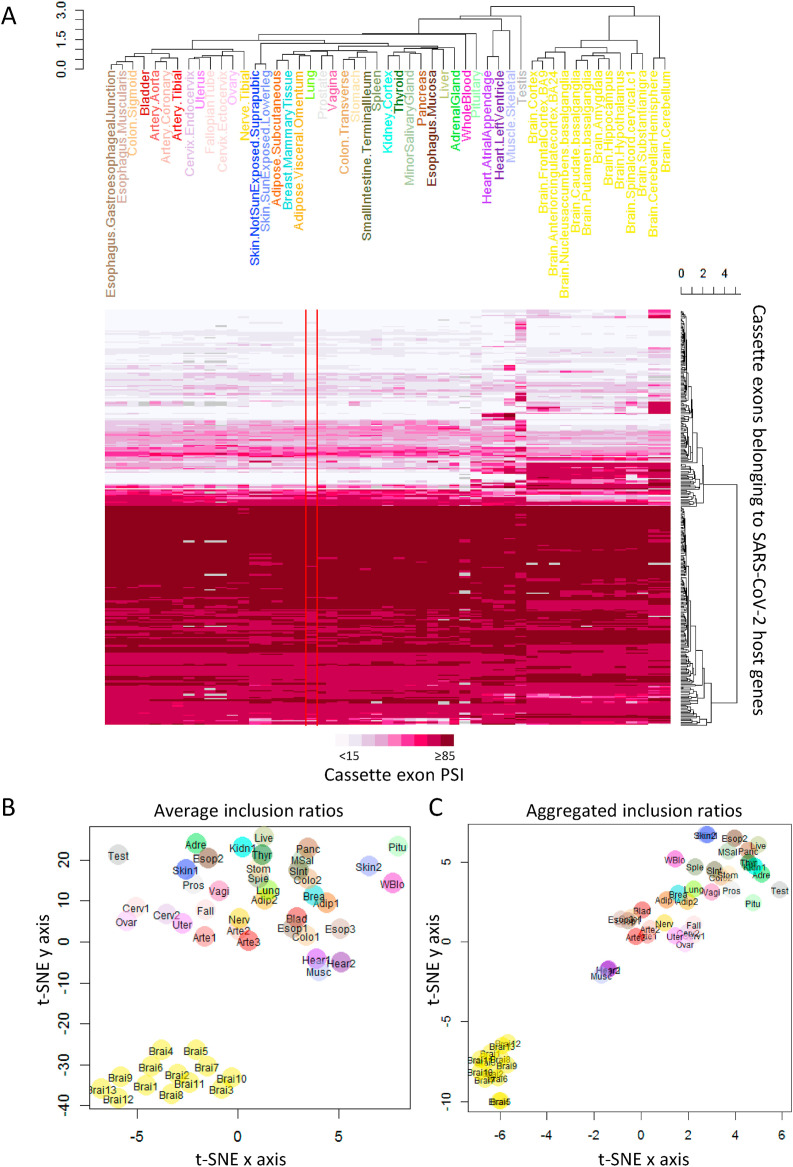
We studied alternative splicing of the SARS-CoV-2 host genes. GENCODE (V26) [19] annotates 944 host gene cassette exons (i.e., an internal exon included in some transcripts but skipped in others). We determined their alternative splicing levels using PSI (percent-spliced-in, in the range of 0–100, see Methods). Among them, 298 cassette exons exhibiting (maxPSI−minPSI) ≥ 10 across tissues were used for clustering analysis. The splicing pattern in the lung is the most similar to those in adipose, breast, sexual organs, spleen, and gastrointestinal tract tissues including stomach, colon, and small intestine (Fig. 3 ). Notably, some COVID-19 patients experienced digestive symptoms and weight loss. The overall similarity of gene expression and splicing patterns between lung, adipose, and gastrointestinal tissues may underlie their common tissue vulnerability to SARS-CoV-2.
Fig. 3.
Splicing profiles of cassette exons belonging to SARS-CoV-2 host genes across different normal tissues. (A) Heatmap of the average inclusion ratios (PSI) of cassette exons. Hierarchical clustering was performed on 298 cassette exons which exhibited PSI ranges of at least 10 across tissues (kidney.medulla excluded) and less than five missing ratios due to insufficient junction reads. (B) t-SNE visualization based on the average inclusion ratios of the 298 cassette exons. (C) t-SNE visualization based on the aggregated inclusion ratios of cassette exons. We pooled junction reads from samples of the same tissue to calculate the aggregated inclusion ratios. A total of 371 cassette exons exhibited ranges of at least 10 for aggregated PSI across tissues (kidney.medulla excluded) and less than five missing ratios due to zero junction reads. Missing values (0.6% for B and 0.1% for C) were imputed as the mean inclusion ratio across all tissues for a specific cassette exon.
We determined tissue-distinctive alternative exons whose PSI in the distinctive tissue is larger (or smaller) than those in all other tissues by 10 or more. We found 18 testis-distinctive cassette exons, 10 in the skeletal muscle, one in the cerebellum, one in the cerebellar hemisphere, and one in the pancreas (Table S1). The biological significance of these differential splicing events is unknown and need further experimental studies.
2.3. Population variation in expression and splicing of SARS-CoV-2 host genes
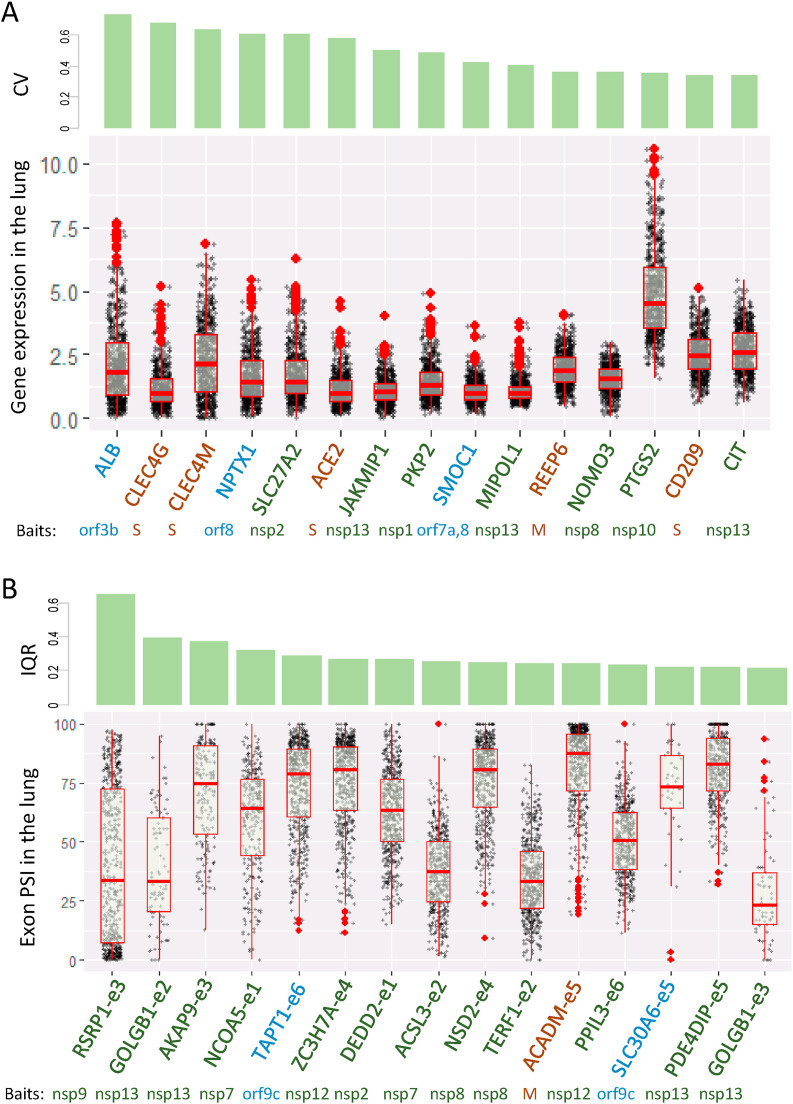
We explored expression variation of SARS-CoV-2 host genes in the normal population. We ranked the host genes by their coefficient of variation (CV, i.e., stochasticity) of expression levels in the lung across individuals after excluding seven lowly expressed host genes (average TPM (transcript per million) < 1). The top 15 host genes are presented in Fig. 4 A along with their CV (top panel) and gene expression values (bottom panel).
Fig. 4.
Population variation in expression and splicing of SARS-CoV-2 host genes. (A) Expression of top 15 genes with the largest coefficient of variation (CV) in the lung. Expression levels are at log2 (TPM+1). Genes with average TPM <1.0 were excluded. (B) Inclusion ratios (PSI) of top 15 cassette exons sorted by their interquartile ranges (IQR) in the lung. Baits are those SARS-CoV-2 proteins tested in Gordon et al.
The stochasticity of SARS-CoV-2 receptor ACE2 is high (ranked 6th). Although no data are available to correlate ACE2 expression with COVID-19 susceptibility and symptom severity, low ACE2 expression probably limits virus entry into host cells resulting in a different outcome from those with high ACE2 expression. ALB displays the largest lung expression stochasticity in the population. ALB acts as a carrier protein for a wide range of endogenous molecules, including hormones, fatty acids, and metabolites, as well as exogenous drugs [14]. The role of ALB in COVID-19 has yet to be reported.
Among the top six genes, CLEC4G, CLEC4M, and ACE2, interact with the spike (S) proteins and presumably regulate SARS-CoV-2 entry. CLEC4M (also known as DCSIGNR, CD209L) is a glycoprotein with a high affinity for HIV gp120 and has been shown to function as another receptor for SARS coronavirus to mediate virus entry [20,21]. Given the similarity of the S proteins between SARS and SARS-CoV-2, and that both viruses can use the same cellular receptor (ACE2), CLEC4M likely mediates SARS-CoV-2 entry, though this awaits experimental validation. CLEC4G (also known as LSECtin) is a C-type lectin domain protein also predicted to interact with the S protein of SARS, and was proposed as an attachment factor enhancing virus infection [22].
Another variably expressed gene (ranked 14th) is CD209. CD209, predicted to interact with the S protein, is involved in the innate immune system and recognizes numerous pathogens ranging from parasites to viruses such as Ebola and SARS [14]. Interestingly, a genetic variant (rs4804803) located in the promoter region of CD209 is associated with SARS severity [23]. We found this variant affects CD209 expression (see below), further suggesting the contribution of genetic and expression variations to disease severity.
REEP6 (ranked the 11th) was predicted to interact with the membrane protein M, another structural protein of SARS-CoV-2. REEP6 is a receptor accessory protein, whose gene family members have been shown to enhance cell surface expression of G protein-coupled receptors (e.g., odorant receptors, and taste receptors) [24,25]. Human mutations in REEP6 cause retinitis pigmentosa [26]. Interestingly, REEP6 appears to be specifically expressed in the supporting cells of the olfactory epithelium [24]. Whether SARS-CoV-2 affects the function of these supporting cells leading to temporary loss of smell would be an interesting topic for future studies.
Seven genes, including SLC27A2, PKP2, and PTGS2, interact with non-structural proteins. Interestingly, interferon alpha-mediated upregulation of SLC27A2 has been shown to suppress Dengue virus [27]. PKP2 interferes with the polymerase complex of influenza A viruses thereby inhibiting virus replication [28]. PTGS2 (COX2) is the major prostaglandin synthase catalyzing the rate-limiting step of prostaglandin production in response to injury and inflammation [29]. Prostaglandins are hormones of diverse function [30]. For example, prostaglandin E acts on the hypothalamus to trigger fever as a systemic response to infection [31,32]. Prostaglandins also function as vasodilators and inhibit platelet aggregation [33]; blood clotting has emerged as a serious complication of COVID-19 [34,35]. In summary, our results identified host genes exhibiting the highest expression variabilities among normal population. Their known and presumptive functions in mediating virus entry as well as in cellular and hormonal responses suggest their important roles underlying individual variability in response to SARS-CoV-2 infection.
We then determined population variation of host gene alternative splicing events. We ranked them by the interquartile range since the percent spliced-in (PSI) is between 0 and 100. The top 15 ranked cassette exons (of 14 genes) exhibit a large range of PSI among individuals (Fig. 4B). For example, the medium PSI of one ACADM cassette exon is 88 but in some individual the PSI is as low as 19. Another example is a cassette exon of ACSL3, whose PSI is as high as 75–100 in certain samples, but the medium is 25. ACADM interacts with structural protein M. It encodes the medium-chain specific acyl-Coenzyme A dehydrogenase to catalyze the initial step of the mitochondrial fatty acid beta-oxidation pathway [14]. Defects in this gene cause MCADD disease characterized by hepatic dysfunction, fasting hypoglycemia, encephalopathy, and sudden infant death syndrome (SIDS)-like illness [36]. ACSL3 interacts with a nonstructural protein, and is an isozyme of the long-chain fatty-acid-coenzyme A ligase family. It plays a key role in lipid biosynthesis and fatty acid degradation [14].
2.4. Genetic variants associated with expression variation of SARS-CoV-2 host genes
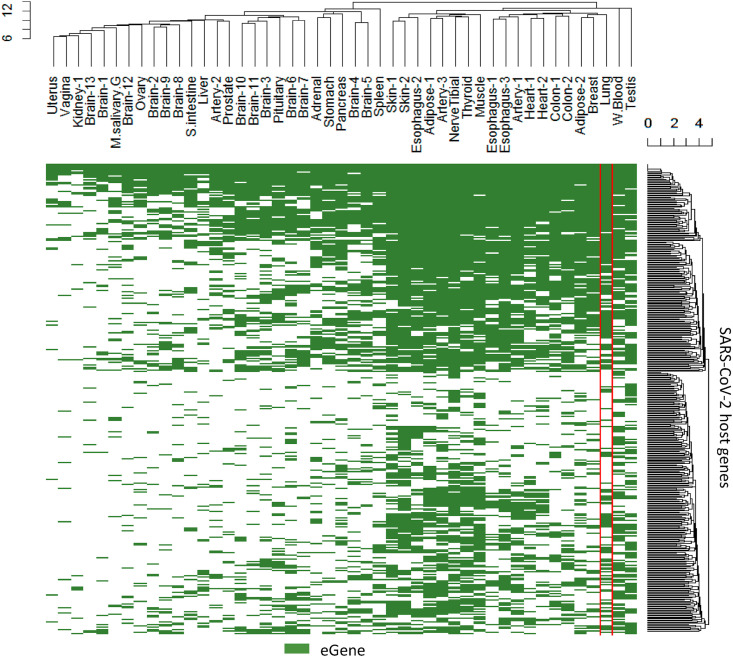
The variation of expression levels can be attributed to nearby genetic variants, which is the principal of expression quantitative trait loci (eQTL) studies. We therefore explored the extent to which SARS-CoV-2 host gene expression variation is associated with common genetic variants in the population. As defined by the GTEx project, if a gene has at least one nearby SNP that is significantly associated with expression differences of that gene, the gene is called an eGene. We found all but one SARS-CoV-2 host genes can be classified as an eGene in at least one tissue (Table S2). In the lung, 238 are qualified as eGenes (Fig. 5 ).
Fig. 5.
Tissues in which SARS-CoV-2 host genes are eGenes. The host genes have common genetic variants nearby to affect their expression levels in the green highlighted tissues. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
SARS-CoV-2 host genes have eQTLs in more tissues than other protein-coding genes (an average of 19 vs 16 tissues, p = 3.3 × 10−6, Wilcoxon test). When compared with all other genes, including non-coding genes, the difference is even larger and more significant (an average of 19 vs 8 tissues, p < 2.2 × 10−16, Wilcoxon test). In other words, common genetic variants in the population tend to perturb expression levels of SARS-CoV-2 host genes in more tissues than regular human protein-coding genes, suggesting a genetic contribution to phenotypic variations in multiple organs upon virus attack.
For example, ACE2 has effective common variants in 20 tissues, although not in the lung. All three SARS-CoV-2 entry associated proteases (TMPRSS2, CTSB, and CTSL) have effective common variants in the lung. Among them, CTSB is an eGene in 34 tissues; CTSL in 17 tissues; and TMPRSS2 in 5 tissues. The lung-distinctive gene SFTPD has effective common variants in 43 tissues including the lung.
SNP rs4804803, located in the promoter region of CD209 and associated with SARS severity, is an effective genetic variant for expression of CD209 in the esophagus (muscularis). It is also an effective genetic variant for expression of the nearby gene CLEC4G in the skin (sun exposed, lower leg). As previously mentioned, both CD209 and CLEC4G were predicted to interact with the spike protein of SARS-CoV-2.
2.5. Age and sex differences in SARS-CoV-2 host gene expression in the normal population
Age and sex are two known variables affecting vulnerabilities to Covid-19. Children and young people (<45 years old) show a lower chance of developing symptoms and, if they do, exhibit less severe symptoms than older people [[37], [38], [39]]. Men appear to at a higher risk for severe symptoms and death than women [40]. We therefore checked the possible sex and age dependency of SARS-CoV-2 host gene expression in normal lung tissues (total 578 samples, 515 of them with complete covariate information). We fit a regression model between gene expression versus sex, age, top five genotyping principal components, sequencing platform and protocol, and expression confounding factors corrected by Probabilistic Estimation of Expression Residuals (PEER) [41]. Note that GTEx projects only include people over 20 years old and stratify age in six groups every 10 years (’20–29’, ‘30–39’, …., ‘70–79’).
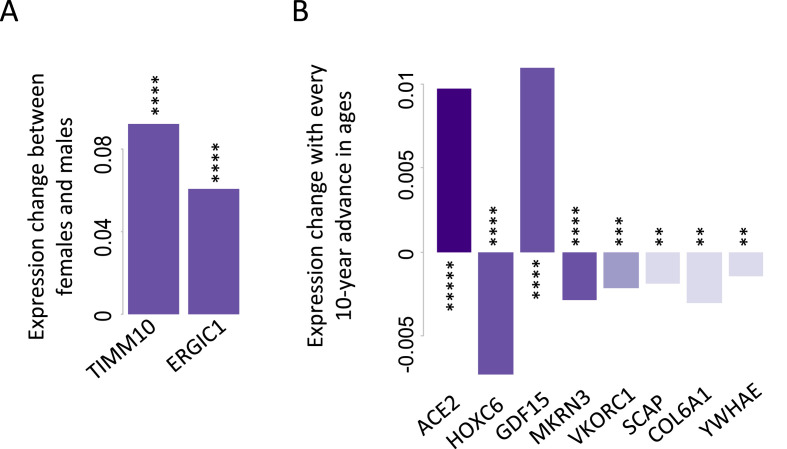
We found TIMM10 and ERGIC1 exhibit significant sex differences (P < 0.0005 and FDR<0.1, Fig. 6 A), with females expressing TIMM10 and ERGIC1 higher than males (the log2 scale TPM in females are 0.09 or 0.06 higher than those in males when keeping all other covariates unchanged). Note that the average log2 scale TPM values of TIMM10 and ERGIC1 are 5.2 and 5.9, respectively. TIMM10 is essential for mitochondria function as it acts as a chaperon protein to transport mitochondrial inner membrane proteins through the outer membrane and across the intermembrane space [42]. ERGIC1 is a membrane protein in the endoplasmic reticulum (ER) and Golgi that may control the secretory pathway [43]. It is conceivable that higher expression of these essential genes in females contributes to mitochondria integrity and homeostasis of protein secretion in combating virus infection.
Fig. 6.
Sex- (A) or age-dependent (B) expression for SARS-CoV-2 host genes. The expression changes were derived from the coefficients of fitted regression models. Expression levels are at log2 (TPM+1). FDR <0.1 was used as the cutoff for the multiple comparison adjustment. *****P ≤ 0.00005; ****P ≤ 0.0005; ***P ≤ 0.001; and **P ≤ 0.005.
Eight SARS-CoV-2 genes exhibit significant age effect (P < 0.005 and FDR<0.1, Fig. 6B). The most significant age-dependent gene is ACE2 (P = 2.2 × 10−5, R2 = 0.55). The average log2 scale TPM of ACE2 is 1.16. When ages advance every 10 years, the log2 scale TPM of ACE2 increases by 0.01. This small but statistically significant age-dependent expression increase in ACE2 may contribute to the higher prevalence of COVID-19 in elderly people.
GDF15 also exhibits increasing expression with ages (Fig. 6B). GDF15 is a secreted ligand of the TGF-beta superfamily [44]. Increased protein levels of GDF15 are associated with disease states such as tissue hypoxia, inflammation, acute injury, and oxidative stress [45]. Uncontrolled inflammation (cytokine storm) is often observed in rapidly deteriorating COVID-19 patients who are over-represented in aged population [46,47].
Genes decreasing expression with age includes VKORC1 (Fig. 6B). VKORC1 encodes the vitamin K epoxide reductase enzyme necessary for vitamin K biosynthesis [14]. Functional vitamin K insufficiency is more prevalent among elderly people and those with hypertension, type 2 diabetes, chronic kidney disease, and cardiovascular disease [48]. Dofferhoff et al. found that vitamin K was reduced in COVID-19 patients and was associated with poor prognosis [49]. Low levels of vitamin K are associated with coagulopathy [50], and coagulopathy is prevalent in severe COVID-19 [51]. Concomitant administration of heparin and vitamin K have therefore been proposed to benefit COVID-19 patients [49]. Our finding of age-dependent expression of VKORC1 provides direct insights to these COVID-related symptoms and proposed remedy.
2.6. Diseases related to SARS-CoV-2 host genes
We used DisGeNet [52], a public disease database, to understand the pathological relevance of SARS-Cov-2 host genes and infer their functional importance in general. DisGeNet integrates gene-disease associations from literature and a group of public data sources. Our search identified 365 out of 420 host genes that were related to 5181 diseases or phenotypes. We found SARS-CoV-2 host genes are more likely associated with diseases than other human protein-coding genes (odds ratio = 2.13 (95% C.I. 1.59–2.88), p = 2.3 × 10−8, Fisher's exact test). When comparing with all other human genes including non-coding ones, the odds ratio is 16.56 (95% C.I. 12.44–22.43, p < 2.2 × 10−16, Fisher's exact test), indicating the functional importance of SARS-CoV-2 host genes.
Among the 5181 disease terms, 18 were enriched with SARS-Cov-2 host genes (FDR ≤0.05, enriched with ≥4 host genes, Table 1 ). As expected, the ranked 1st disease is SARS. Interestingly, Alzheimer's disease ranked 4th, indicating malfunction of SARS-Cov-2 host genes could be related to Alzheimer's disease phenotypes. Notable neurologic features of severe COVID-19 illness include encephalopathy, prominent agitation and confusion, and diffuse corticospinal tract signs [53].
Table 1.
List of diseases enriched with SARS-CoV-2 host genes. FDR ≤0.05, enriched with ≥4 host genes. P-values were calculated based on hypergeometric tests.
| Disease | Host gene count | Total gene count | P | FDR |
|---|---|---|---|---|
| Severe Acute Respiratory Syndrome | 11 | 80 | 1.2 × 10−6 | 0.0038 |
| Muscle Spasticity | 23 | 337 | 1.5 × 10−6 | 0.0038 |
| Contracture | 15 | 162 | 2.7 × 10−6 | 0.0042 |
| Alzheimer's Disease | 70 | 1853 | 3.2 × 10−6 | 0.0042 |
| Flexion contracture | 13 | 133 | 6.8 × 10−6 | 0.0071 |
| Contracture of joint | 13 | 137 | 9.4 × 10−6 | 0.0081 |
| Respiratory Failure | 15 | 182 | 1.1 × 10−5 | 0.0082 |
| Presenile dementia | 21 | 342 | 2.1 × 10−5 | 0.014 |
| Congenital contractural arachnodactyly | 15 | 194 | 2.4 × 10−5 | 0.014 |
| Flexion contractures of joints | 12 | 132 | 3.2 × 10−5 | 0.016 |
| Dementia | 22 | 395 | 5.9 × 10−5 | 0.028 |
| Osteosarcoma of bone | 37 | 862 | 6.8 × 10−5 | 0.029 |
| Global developmental delay | 43 | 1068 | 7.6 × 10−5 | 0.030 |
| Ehlers-Danlos Syndrome | 10 | 103 | 8.3 × 10−5 | 0.031 |
| Osteosarcoma | 38 | 923 | 1.3 × 10−4 | 0.043 |
| Amelanotic Skin Melanoma | 4 | 13 | 1.4 × 10−4 | 0.043 |
| Infection | 24 | 479 | 1.4 × 10−4 | 0.043 |
| Optic Atrophy | 19 | 335 | 1.5 × 10−4 | 0.043 |
3. Discussion
In this study, we explored the transcriptome landscape of SARS-CoV-2 host genes in the normal population. We focused on tissue and population variation of gene expression to provide immediate insight to variable susceptibility to and manifestations of COVID-19. Particularly, the expression and splicing of SARS-CoV-2 host genes in normal lung tissue are probably associated with the chances of infection and symptom severity.
Distinctive gene expression associated with vulnerable tissues may shed light on the cellular response to SARS-CoV-2 infection. For example, we found SFTPD is highly expressed in the lung and predicted to interact with the spike protein of SARS-CoV-2, which warrants immediate attention for study of its role in COVID-19. On the other hand, blood, much less vulnerable to the infection, exhibits an expression profile substantially distinct from the one in the lung. The differentially expressed genes would more likely explain the variation in tissue vulnerability than ubiquitously expressed genes. Tissue-distinctive alternative splicing adds another layer of regulation and variation that may contribute to tissue vulnerability and interactions with SARS-CoV-2 virus.
The wide-spread eQTLs affecting SARS-CoV-2 host gene expression in the normal population show the extent to which genetic variants contribute to expression variation (and hence to variations in disease manifestation). Our results indicate a potential value of eQTL analysis of COVID-19. Large scale genotypes and/or transcriptome data of COVID-19 patients, when made available in the future, will further refine the analyses and better identify genetic underlying of the susceptibility to COVID-19. For example, Ellinghaus et al. [54] performed a genome wide association study for severe COVID-19 with respiratory failure and identified two genetic markers: rs11385942 and rs657152. We checked the GTEx resource and found that rs11385942 is associated with the splicing variation of the host gene FYCO1. This may help to interpret the reported disease association for SNP rs11385942.
Some SARS-CoV-2 host genes show sex or age dependency. Interestingly, ACE2 exhibits the strongest age effect, however, when excluding the PEER factors in the regression, the age effect is insignificant (P = 0.62). PEER factors represent hidden determinants of expression variability including batch effects, experimental confounders, and systemic gene expression effect (e.g., simultaneous activation of many genes by a major pathway or transcription factor). Removal of these confounding factors derives a more accurate assessment of the age effect.
Beyond age and sex effects, people with underlying medical conditions constitute a higher-risk population to COVID-19. It would be interesting to examine the effect of common comorbidities on the expression of SARS-CoV-2 host genes in the same age and sex group, and to cross reference disease-dependent gene expression markers, if any, with the age- and sex-dependent COVID-19-associated gene expression.
Our analysis is based on the provisional list of human genes interacting with SARS-Cov-2 proteins. We caution that further investigation is needed to validate biological functions of these interactions.
4. Methods
4.1. Assembly of SARS-CoV-2 host genes
The list of provisional SARS-CoV-2 host genes were downloaded from the Human Protein Atlas website (https://www.proteinatlas.org/humanproteome/sars-cov-2, as of April 10, 2020, based on [11]) as well as those inferred by [12] (http://korkinlab.org/wuhan). A total of 420 genes were assembled and all of them are protein coding. They were related to the GENCODE annotation (v26) [19]. A total of 944 cassette exons were derived from gene annotation. They are internal exons included in some but not all transcripts of a gene.
4.2. GTEx data analysis
RNA-seq data which were profiled in normal tissue samples were downloaded from the GTEx portal (V8) (https://www.gtexportal.org/). Specifically, we downloaded gene TPM values and converted them to log2 (TPM+1). Average gene expression values were calculated based on samples from the same tissue.
We also downloaded exon-exon junction read counts to calculate inclusion ratios for cassette exons. We required at least 20 inclusive junction reads (sum of upstream and downstream inclusive junction reads) or 10 exclusive junction reads to obtain a valid inclusion ratio. The PSI (percent-spliced-in) index was calculated as 0.5×inclusive junction reads/(0.5×inclusive junction reads + exclusive junction reads) ×100 to represent the inclusion ratio in percent. To calculate the average PSI of a cassette exon for a tissue, we required at least five samples passing the coverage threshold mentioned above, otherwise we assigned a ‘NA’ for that tissue. We then focused on 298 cassette exons which exhibited a PSI range of at least 10 (maxPSI−minPSI ≥ 10) across all tissues (kidney.medulla was excluded due to insufficient read coverage in the splicing analysis) and less than five ‘NA’ values. For t-SNE visualization, ‘NA’ values (0.6%) were imputed as the average inclusion ratio of a cassette exon across different tissues. In addition to the approach with qualified samples, we also obtained the aggregated inclusion ratio for a cassette exon in a tissue by pooling all junction reads from samples of the same tissue. A total of 371 cassette exons exhibited ranges of at least 10 in aggregated PSI across tissues (kidney.medulla excluded) and less than five ‘NA’ values due to zero mapped junction reads (0.1% missing values). The t-SNE visualization results were similar for these two approaches.
The lists of eGenes with qval ≤0.05 for cis-eQTL for were downloaded from GTEx. The downloaded covariates for lung expression analysis include the top 5 genotyping principal components, 60 expression confounding factors identified using the Probabilistic Estimation of Expression Residuals (PEER) method [41], sequencing platform, sequencing protocol, sex, and age. There are 395 lung samples from males and 183 lung samples from females. The age distribution is: 33 samples for age 20–29, 40 samples for age 30–39, 93 samples for age 40–49, 200 samples for age 50–59, 190 samples for age 60–69, and 22 samples for age 70–79. Among the 578 samples, 515 have the complete covariate information available (i.e., with genotypes). For those 515 samples fitted in regression models, there are 349 males and 166 females; 33, 36, 82, 187, 162, 15 samples for each of the six age groups, respectively.
The 52 tissues considered in GTEx include: Adipose.Subcutaneous (Adipose-1), Adipose.Visceral.Omentum (Adipose-2), AdrenalGland (Adrenal), Artery.Aorta (Artery-1), Artery.Coronary (Artery-2), Artery.Tibial (Atery-3), Bladder, Brain.Amygdala (Brain-1), Brain.Anteriorcingulatecortex.BA24 (Brain-2), Brain.Caudate.basalganglia (Brain-3), Brain.CerebellarHemisphere (Brain-4), Brain.Cerebellum (Brain-5),Brain.Cortex (Brain-6),Brain.FrontalCortex.BA9 (Brain-7), Brain.Hippocampus (Brain-8), Brain.Hypothalamus (Brain-9), Brain.Nucleusaccumbens.basalganglia (Brain-10), Brain.Putamen.basalganglia (Brain-11), Brain.Spinalcord.cervical.c1 (Brain-12), Brain.Substantianigra (Brain-13),Breast.MammaryTissue (Breast),Cervix.Ectocervix (Cervix-1),Cervix.Endocervix (Cervix-2),Colon.Sigmoid (Colon-1),Colon.Transverse (Colon-2), Esophagus.GastroesophagealJunction (Esophagus-1), Esophagus.Mucosa (Esophagus-2), Esophagus.Muscularis (Esophagus-3), FallopianTube, Heart.AtrialAppendage (Heart-1), Heart.LeftVentricle (Heart-2), Kidney.Cortex (Kidney-1), Kidney.Medulla (Kidney-2),Liver, Lung, MinorSalivaryGland (M.salivary.G), Muscle.Skeletal (Muscle), Nerve.Tibial, Ovary, Pancreas, Pituitary, Prostate, Skin.NotSunExposed.Suprapubic (Skin-1), Skin.SunExposed.Lowerleg (Skin-2), SmallIntestine.TerminalIleum (S.intestine), Spleen, Stomach, Testis, Thyroid, Uterus, Vagina, WholeBlood (W.Blood).
4.3. Other data analysis
Gene-disease associations were downloaded from the DisGeNet database (v6) [52] https://www.disgenet.org/. The DisGeNet database integrates human gene-disease associations from multiple source databases including curated and inferred associations.
All statistical analyses were performed in R (3.6.1). P-values are two-sided unless specified.
Author contributions
L.C. and S.Z. conceived of the work and designed the study; L.C. performed the data analysis; L.C. and S.Z. interpreted the results and wrote the manuscript.
Declaration of competing interest
None declared.
Acknowledgments
This work has been supported by the National Institutes of Health [grant number R01GM137428 to L.C., R01MH116220 and R01NS104041 to S.Z.]. The founding source has no role in the conduct of the research and in the preparation of the article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100443.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Expression profiles of important caspases related to host cell death.
Lists of tissue-distinctive SARS-CoV-2 host genes and tissue-distinctive cassette exons belonging to SARS-CoV-2 host genes.
List of tissues in which SARS-CoV-2 host genes are eGenes and their related genetic variants. Only the most significant genetic variants are listed, and others can be found in the GTEx portal.
References
- 1.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. J Am Med Assoc. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colaneri M., Sacchi P., Zuccaro V., Biscarini S., Sachs M., Roda S., Pieri T.C., Valsecchi P., Piralla A., Seminari E., Di Matteo A., Novati S., Maiocchi L., Pagnucco L., Tirani M., Baldanti F., Mojoli F., Perlini S., Bruno R., The Covid Irccs San Matteo Pavia Task F. Clinical characteristics of coronavirus disease (COVID-19) early findings from a teaching hospital in Pavia, North Italy, 21 to 28 February 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.16.2000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siordia J.A., Jr. Epidemiology and clinical features of COVID-19: a review of current literature. J Clin Virol. 2020;127:104357. doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in wuhan, China. JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X., Ma H., Chen W., Lin Y., Zheng Y., Wang J., Yi Y., Shen H. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Liu Y., Liu L., Wang X., Luo N., Ling L. Clinical outcome of 55 asymptomatic cases at the time of hospital admission infected with SARS-Coronavirus-2 in Shenzhen, China. J Infect Dis. 2020;221(11):1770–1774. doi: 10.1093/infdis/jiaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls; Treasure Island (FL): 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- 10.Rodel F., Arenas M., Ott O.J., Fournier C., Georgakilas A.G., Tapio S., Trott K.R., Gaipl U.S. Low-dose radiation therapy for COVID-19 pneumopathy: what is the evidence? Strahlenther Onkol. 2020;196:679–682. doi: 10.1007/s00066-020-01635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O'meara M., Guo J., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Al E. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. 2020. Preprint at bioRxiv. [DOI] [PMC free article] [PubMed]
- 12.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12 doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., Mcveigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C.M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V.S., Kodali V.K., Li W., Maglott D., Masterson P., Mcgarvey K.M., Murphy M.R., O'neill K., Pujar S., Rangwala S.H., Rausch D., Riddick L.D., Schoch C., Shkeda A., Storz S.S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R.E., Vatsan A.R., Wallin C., Webb D., Wu W., Landrum M.J., Kimchi A., Tatusova T., Dicuccio M., Kitts P., Murphy T.D., Pruitt K.D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., Von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., Von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 16.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. J Am Med Assoc. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L., Xie W., Li D., Shi L., Mao Y., Xiong Y., Zhang Y., Zhang M. Effect of SARS-CoV-2 infection upon male gonadal function: a single center-based study. 2020. Preprint at medRxiv. [DOI]
- 18.Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., Barnes I., Berry A., Bignell A., Carbonell Sala S., Chrast J., Cunningham F., Di Domenico T., Donaldson S., Fiddes I.T., Garcia Giron C., Gonzalez J.M., Grego T., Hardy M., Hourlier T., Hunt T., Izuogu O.G., Lagarde J., Martin F.J., Martinez L., Mohanan S., Muir P., Navarro F.C.P., Parker A., Pei B., Pozo F., Ruffier M., Schmitt B.M., Stapleton E., Suner M.M., Sycheva I., Uszczynska-Ratajczak B., Xu J., Yates A., Zerbino D., Zhang Y., Aken B., Choudhary J.S., Gerstein M., Guigo R., Hubbard T.J.P., Kellis M., Paten B., Reymond A., Tress M.L., Flicek P. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashirova A.A., Geijtenbeek T.B., Van Duijnhoven G.C., Van Vliet S.J., Eilering J.B., Martin M.P., Wu L., Martin T.D., Viebig N., Knolle P.A., Kewalramani V.N., Van Kooyk Y., Carrington M. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gramberg T., Hofmann H., Moller P., Lalor P.F., Marzi A., Geier M., Krumbiegel M., Winkler T., Kirchhoff F., Adams D.H., Becker S., Munch J., Pohlmann S. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340:224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan K.Y., Xu M.S., Ching J.C., So T.M., Lai S.T., Chu C.M., Yam L.Y., Wong A.T., Chung P.H., Chan V.S., Lin C.L., Sham P.C., Leung G.M., Peiris J.S., Khoo U.S. CD209 (DC-SIGN) -336A>G promoter polymorphism and severe acute respiratory syndrome in Hong Kong Chinese. Hum Immunol. 2010;71:702–707. doi: 10.1016/j.humimm.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito H., Kubota M., Roberts R.W., Chi Q., Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Behrens M., Bartelt J., Reichling C., Winnig M., Kuhn C., Meyerhof W. Members of RTP and REEP gene families influence functional bitter taste receptor expression. J Biol Chem. 2006;281:20650–20659. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- 26.Arno G., Agrawal S.A., Eblimit A., Bellingham J., Xu M., Wang F., Chakarova C., Parfitt D.A., Lane A., Burgoyne T., Hull S., Carss K.J., Fiorentino A., Hayes M.J., Munro P.M., Nicols R., Pontikos N., Holder G.E., Asomugha C., Raymond F.L., Moore A.T., Plagnol V., Michaelides M., Hardcastle A.J., Li Y., Cukras C., Webster A.R., Cheetham M.E., Chen R. Mutations in REEP6 cause autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2016;99:1305–1315. doi: 10.1016/j.ajhg.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fusco D.N., Pratt H., Kandilas S., Cheon S.S., Lin W., Cronkite D.A., Basavappa M., Jeffrey K.L., Anselmo A., Sadreyev R., Yapp C., Shi X., O'sullivan J.F., Gerszten R.E., Tomaru T., Yoshino S., Satoh T., Chung R.T. HELZ2 is an IFN effector mediating suppression of Dengue virus. Front Microbiol. 2017;8:240. doi: 10.3389/fmicb.2017.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L., Fu B., Li W., Patil G., Liu L., Dorf M.E., Li S. Comparative influenza protein interactomes identify the role of plakophilin 2 in virus restriction. Nat Commun. 2017;8:13876. doi: 10.1038/ncomms13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morham S.G., Langenbach R., Loftin C.D., Tiano H.F., Vouloumanos N., Jennette J.C., Mahler J.F., Kluckman K.D., Ledford A., Lee C.A., Smithies O. Prostaglandin synthase 2 gene disruption causes severe renal pathology in the mouse. Cell. 1995;83:473–482. doi: 10.1016/0092-8674(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 30.Ricciotti E., Fitzgerald G.A. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov A.I., Romanovsky A.A. Prostaglandin E2 as a mediator of fever: synthesis and catabolism. Front Biosci. 2004;9:1977–1993. doi: 10.2741/1383. [DOI] [PubMed] [Google Scholar]
- 32.Feldberg W., Saxena P.N. Fever produced by prostaglandin E1. J Physiol. 1971;217:547–556. doi: 10.1113/jphysiol.1971.sp009585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams T.J., Peck M.J. Role of prostaglandin-mediated vasodilatation in inflammation. Nature. 1977;270:530–532. doi: 10.1038/270530a0. [DOI] [PubMed] [Google Scholar]
- 34.Klok F.A., Kruip M., Van Der Meer N.J.M., Arbous M.S., Gommers D., Kant K.M. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poissy J., Goutay J., Caplan M., Parmentier E., Duburcq T., Lassalle F. Pulmonary embolism in COVID-19 patients: awareness of an increased prevalence. Circulation. 2020;142(2):184–186. doi: 10.1161/CIRCULATIONAHA.120.047430. [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Zhu R., Liu Y., Song J., Xu J., Yang Y. Medium-chain acyl-coenzyme A dehydrogenase deficiency: six cases in the Chinese population. Pediatr Int. 2019;61:551–557. doi: 10.1111/ped.13872. [DOI] [PubMed] [Google Scholar]
- 37.Lu X., Zhang L., Du H., Zhang J., Li Y.Y., Qu J., Zhang W., Wang Y., Bao S., Li Y., Wu C., Liu H., Liu D., Shao J., Peng X., Yang Y., Liu Z., Xiang Y., Zhang F., Silva R.M., Pinkerton K.E., Shen K., Xiao H., Xu S., Wong G.W.K. SARS-CoV-2 infection in children. N Engl J Med. 2020;382:1663–1665. doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.2020 b. Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, february 12-march 16, 2020. MMWR Morb Mortal Wkly Rep, 69, 343-346. [DOI] [PMC free article] [PubMed]
- 39.2020 a. Characteristics of Health care personnel with COVID-19 - United States, february 12-april 9, 2020. MMWR Morb Mortal Wkly Rep, 69, 477-481. [DOI] [PMC free article] [PubMed]
- 40.Jin J.-M., Bai P., He W., Wu F., Liu X.-F., Han D.-M., Liu S., Yang J.-K. Gender differences in patients with COVID-19: focus on severity and mortality. Frontiers in Public Health. 2020;8 doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stegle O., Parts L., Durbin R., Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koehler C.M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R.J., Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 43.Breuza L., Halbeisen R., Jeno P., Otte S., Barlowe C., Hong W., Hauri H.P. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem. 2004;279:47242–47253. doi: 10.1074/jbc.M406644200. [DOI] [PubMed] [Google Scholar]
- 44.Yokoyama-Kobayashi M., Saeki M., Sekine S., Kato S. Human cDNA encoding a novel TGF-beta superfamily protein highly expressed in placenta. J Biochem. 1997;122:622–626. doi: 10.1093/oxfordjournals.jbchem.a021798. [DOI] [PubMed] [Google Scholar]
- 45.Fujita Y., Taniguchi Y., Shinkai S., Tanaka M., Ito M. Secreted growth differentiation factor 15 as a potential biomarker for mitochondrial dysfunctions in aging and age-related disorders. Geriatr Gerontol Int. 2016;16(Suppl 1):17–29. doi: 10.1111/ggi.12724. [DOI] [PubMed] [Google Scholar]
- 46.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riphagen I.J., Keyzer C.A., Drummen N.E.A., De Borst M.H., Beulens J.W.J., Gansevoort R.T., Geleijnse J.M., Muskiet F.a.J., Navis G., Visser S.T., Vermeer C., Kema I.P., Bakker S.J.L. Prevalence and effects of functional vitamin K insufficiency: the PREVEND study. Nutrients. 2017;9 doi: 10.3390/nu9121334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dofferhoff A., Piscaer I., Schurgers L., Walk J., Ouweland J., T H., Lux P., Maassen C., Karssemeijer E., Wouters E., Jassen R. Reduced vitamin K status as a potentially modifiable prognostic risk factor in COVID-19. 2020. https://www.preprints.org/manuscript/202004.0457/v1 Preprint at.
- 50.Alperin J.B. Coagulopathy caused by vitamin K deficiency in critically ill, hospitalized patients. J Am Med Assoc. 1987;258:1916–1919. [PubMed] [Google Scholar]
- 51.Arachchillage D.R., Laffan M. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemostasis. 2020;18(5):1233–1234. doi: 10.1111/jth.14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinero J., Ramirez-Anguita J.M., Sauch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C. Neurologic features in severe SARS-CoV-2 infection. N Engl J Med. 2020;382(23):2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P. Genomewide association study of severe covid-19 with respiratory failure. N Engl J Med. 2020 doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiles of important caspases related to host cell death.
Lists of tissue-distinctive SARS-CoV-2 host genes and tissue-distinctive cassette exons belonging to SARS-CoV-2 host genes.
List of tissues in which SARS-CoV-2 host genes are eGenes and their related genetic variants. Only the most significant genetic variants are listed, and others can be found in the GTEx portal.