Abstract
Background
The degree of protective immunity conferred by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently unknown. As such, the possibility of reinfection with SARS-CoV-2 is not well understood. We describe an investigation of two instances of SARS-CoV-2 infection in the same individual.
Methods
A 25-year-old man who was a resident of Washoe County in the US state of Nevada presented to health authorities on two occasions with symptoms of viral infection, once at a community testing event in April, 2020, and a second time to primary care then hospital at the end of May and beginning of June, 2020. Nasopharyngeal swabs were obtained from the patient at each presentation and twice during follow-up. Nucleic acid amplification testing was done to confirm SARS-CoV-2 infection. We did next-generation sequencing of SARS-CoV-2 extracted from nasopharyngeal swabs. Sequence data were assessed by two different bioinformatic methodologies. A short tandem repeat marker was used for fragment analysis to confirm that samples from both infections came from the same individual.
Findings
The patient had two positive tests for SARS-CoV-2, the first on April 18, 2020, and the second on June 5, 2020, separated by two negative tests done during follow-up in May, 2020. Genomic analysis of SARS-CoV-2 showed genetically significant differences between each variant associated with each instance of infection. The second infection was symptomatically more severe than the first.
Interpretation
Genetic discordance of the two SARS-CoV-2 specimens was greater than could be accounted for by short-term in vivo evolution. These findings suggest that the patient was infected by SARS-CoV-2 on two separate occasions by a genetically distinct virus. Thus, previous exposure to SARS-CoV-2 might not guarantee total immunity in all cases. All individuals, whether previously diagnosed with COVID-19 or not, should take identical precautions to avoid infection with SARS-CoV-2. The implications of reinfections could be relevant for vaccine development and application.
Funding
Nevada IDEA Network of Biomedical Research, and the National Institute of General Medical Sciences (National Institutes of Health).
Introduction
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) leads to a detectable immune response, but the susceptibility of previously infected individuals to reinfection with SARS-CoV-2 is not well understood. SARS-CoV-2 infection results in generation of neutralising antibodies in patients.1 However, the degree to which this immune response indicates a protective immunity to subsequent infection with SARS-CoV-2 has not yet been elucidated. In studies of immunity to other coronaviruses,2, 3, 4, 5, 6, 7, 8, 9 loss of immunity can occur within 1–3 years. Cases of primary illness due to infection followed by a discrete secondary infection or illness with the same biological agent can best be ascertained as distinct infection events by genetic analysis of the agents associated with each illness event. Reports of secondary infection events with SARS-CoV-2 have been published from Hong Kong,10 the Netherlands and Belgium,11 and Ecuador.12 We present a case report of an individual who had two distinct COVID-19 illnesses from genetically distinct SARS-CoV-2 agents.
Methods
Case history
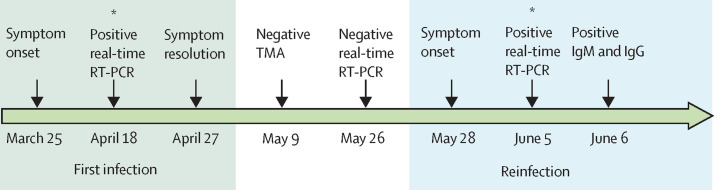
We present a case report of a 25-year-old male patient who was a resident of Washoe County in the US state of Nevada. The patient presented to a community testing event held by the Washoe County Health District on April 18, 2020. He had symptoms consistent with viral infection (sore throat, cough, headache, nausea, and diarrhoea), which had started on March 25, 2020 (figure 1 ). The patient had no history of clinically significant underlying conditions, and no indications of compromised immunity were identified. During isolation, the patient's symptoms resolved (reported on April 27, 2020) and he continued to feel well until May 28, 2020. On May 31, 2020, the patient sought care at an urgent care centre with self-reported fever, headache, dizziness, cough, nausea, and diarrhoea, at which time chest radiography was done and he was discharged home. 5 days later (on June 5, 2020), the patient presented to a primary care doctor and was found to be hypoxic with shortness of breath. He was instructed to go to the emergency department after provision of oxygen.
Figure 1.
Timeline of symptom onset, molecular diagnosis, and sequencing of specimens
TMA=transcription-mediated amplification. *Sequenced specimens.
Research in context.
Evidence before this study
We searched PubMed, preprint servers (MedRxiv, BioRxiv, and SSRN), and general news channels (via Google search) from June 30 to Sept 9, 2020, for reports of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection, using keywords including “reinfection”, “SARS-CoV-2”, and “secondary infection”. We restricted our search to publications in English. Three reports of reinfection, with variable symptom severity on reinfection, have been published worldwide to date, supporting the possibly for SARS-CoV-2 reinfection.
Added value of this study
We present, to our knowledge, the first North American case of reinfection with SARS-CoV-2. A 25-year-old man, who was a resident of Washoe County in the US state of Nevada, had laboratory-confirmed SARS-CoV-2 infection in April, 2020, followed by secondary infection within a period of around 6 weeks, in June, 2020. The second infection was symptomatically more severe than the first. Genomic analysis showed the two viral agents were genetically distinct. The patient's immune reaction in vitro was not assessed and, thus, conclusions cannot be made about the duration or degree of immunity.
Implications of all the available evidence
Reinfection with SARS-CoV-2 has been reported in at least four individuals worldwide. Thus, previous exposure to SARS-CoV-2 does not necessarily translate to guaranteed total immunity. The implications of reinfections could be relevant for vaccine development and application. From a public health perspective, all individuals—whether previously diagnosed or not—must take identical precautions to prevent infection with SARS-CoV-2. Further work is needed to assess immune reactions in vitro after reinfection.
This work was done under an emergency order by the Chief Medical Officer of the Division of Public and Behavioral Health for the State of Nevada. Ethics approval was waived by the University of Nevada, Reno Institutional Review Board. The patient provided written consent to publish this report.
Procedures
Specimens were obtained from the patient by nasopharyngeal swab at the community testing event, during the period of isolation and recovery, and on presentation to hospital. Swabs were transported to the Nevada State Public Health Laboratory (Reno, NV, USA) in either viral transport medium or Aptima Multiswab Transport Media (Hologic, San Diego, CA, USA). Specimens were transported on cold packs and stored by refrigeration (4–8°C) for no longer than 72 h before nucleic acid extraction and subsequent real-time RT-PCR.
Nucleic acid extraction was done using Omega Biotek MagBind Viral DNA/RNA 96 Kit (Omega Bio-tek, Norcross, GA, USA), per manufacturer's instructions and with an elution volume of 100 μL. Aliquots of eluted RNA underwent real-time RT-PCR with either the Taqpath COVID-19 Emergency Use Authorized (EUA) Multiplex Assay (ThermoScientific, Waltham, MA, USA; 10 μL aliquots) or the US Centers for Disease Control and Prevention (CDC) 2019-nCoV Real-Time RT-qPCR Diagnostic Panel (CDC, Atlanta, GA, USA; 5 μL aliquots). Specimens transported on Aptima Multiswab Transport Media were tested by transcription-mediated amplification using the Aptima SARS-CoV-2 (Panther System) assay (Hologic, Marlborough, MA, USA). Assays were done according to their respective EUA procedures, unless otherwise indicated. For the Taqpath real-time RT-PCR test, the threshold for calling a specimen positive was reactivity of two of three target sequences, each with reactivity at a cycle threshold of less than 40·00. A positive or negative result on the Hologic Aptima assay was based on proprietary processes. Antibody testing was done with the Roche Elecsys Anti SARS-CoV-2 test (Roche Diagnostics, Indianapolis, IN, USA).
For viral genomic sequencing, total RNA was extracted from nasopharyngeal swabs as described. 70 μL of extracted RNA was treated for 30 min at room temperature with Qiagen DNase I (Qiagen, Germantown, MD, USA) and then cleaned and concentrated with silica spin columns (Qiagen RNeasy MinElute; Qiagen) with a 12 μL water elution. A portion (7 μL) of this RNA was annealed to an rRNA inhibitor (Qiagen FastSelect rRNA HMR; Qiagen) and then reverse-transcribed (cDNA) using random hexamers. The synthesised DNA was strand-ligated and isothermally amplified into micrograms of DNA (Qiagen FX Single Cell RNA Library Kit; Qiagen). A portion (1 μg) of this amplified DNA was sheared and ligated to Illumina-compatible sequencing adapters, followed by six cycles of PCR amplification (KAPA HiFi HotStart Library Amplification Kit; Roche Sequencing and Life Science, Kapa Biosystems, Wilmington, MA, USA) to enrich for library molecules with adapters at both ends. Next, these sequencing libraries were enriched for a sequence specific to SARS-CoV-2 using biotinylated oligonucleotide baits (myBaits Expert Virus, Arbor Biosciences, Ann Arbor, MI, USA). A further eight to 16 cycles of PCR were done after enrichment (98°C for 45 s, 98°C for 15 s, 60°C for 30 s, repeat for eight to 16 cycles, then 72°C for 60 s and 4°C to complete), and these SARS-CoV-2-enriched sequencing libraries were pooled and sequenced with an Illumina NextSeq 500 (Illumina, San Diego, CA USA) as paired-end 2 × 75 base pair reads using the NextSeq version 2.5 mid-output 150 cycle kit (Illumina).
For bioinformatics analysis of the two SARS-CoV-2 agents (referred to herein as specimen A and specimen B), after sequencing of each library, FASTQ files were imported into CLC Genomics Workbench version 20.0.4 (Qiagen A/S, Vedbæk, Denmark) with the CLC Microbial Genomics Module, CLC Genome Finishing Module, and Biomedical Genomics Analysis. Briefly, reads were imported, trimmed, and mapped to National Center for Biotechnology Information SARS-CoV-2 reference sequence MN908947.3. The alignment was refined using the InDels and Structural Variants module, followed by the Local Realignment module. Variants were identified by a minimum coverage of five reads, minimum count of five, and minimum frequency of 70·0%.
To ascertain repeatability of results, a second bioinformatics analysis was done using an independent process and open source tools. Potential reinfection sequence libraries were trimmed using Trimmomatic version 0.39, with the ILLUMINACLIP adapter-clipping setting 2:30:10:2:keepBothReads. Sequence pairs were aligned to the SARS-CoV-2 reference genome (MN908947.3) using Bowtie 2 version 2.3.13 PCR optical duplicates were flagged using Picard MarkDuplicates in picard-slim version 2.22.5. Variants were called for both samples in concert using Freebayes version 1.0.2, with ploidy settings of 1, a minimum allele frequency of 0·70, and a minimum depth of five reads for any variant call. The genome sequence of each sample was constructed using coverage statistics from BBtools pileup.sh and applyvariants.sh version 38.86, whereby only variants supported by coverage of five or more reads were written to bcftools consensus version 1.10.2, and all positions supported by fewer than five reads, whether reference or alternative, were replaced with Ns.14
For phylogenetic analysis, the whole genome sequences of the isolates (specimen A and specimen B) were compared with those of 171 contemporaneous sequences from Nevada,15 the SARS-CoV-2 reference strain (MN908947.3), and one sequence derived from isolate USA-WA1/2020 (Bei Resources, Manassas, VA, USA). After trimming six 5′ uncalled bases (Ns) from specimen A and 98 Ns from specimen B, genomic sequences were aligned and related using NGPhylogeny.fr PhyML+SMS.16 Sequences were then first-aligned using MAFFT with automatic flavour selection.17 Informative regions were selected using Block Mapping and Gathering with Entropy, a sliding window size of 3, and maximum entropy of 0·5.18 Unrooted trees were constructed by PhyML with Smart Model Selection, the Akaike information criterion, and Subtree Pruning and Regrafting.19 Newick trees were visualised using Interactive Tree Of Live version 4 and rooted at the Wuhan reference strain.20 Major SARS-CoV-2 clade memberships were predicted using Nextclade.
To confirm specimens A and B were from the same individual, the original swab specimens, transport media, and residual samples of extracted RNA supplied to the sequencing core facility underwent short tandem repeat (STR) analysis for identity comparison, by the Washoe County Sheriff's Forensic Science Division (Reno, NV, USA). 2 μL of extracted DNA was quantified using the Quantifiler Trio DNA Quantification Kit (Applied Biosystems, Foster City, CA, USA) on the 7500 Real-Time PCR System and analysed with 7500 HID software version 1.3 (Applied Biosystems). Amplification of 24 GlobalFiler STR markers (Thermo Fisher Scientific, Waltham, MA, USA) was accomplished on the ProFlex PCR Instrument (Thermo Fisher Scientific) for 29 cycles. The 3500xL Genetic Analyzer (Applied Biosystems) was used for fragment analysis of the amplified STR marker regions in conjunction with HID Data Collection Software version 4.0.1 (Applied Biosystems) and Genemapper ID-X software version 1.6 (Thermo Fisher Scientific). Statistical interpretation of STR data was achieved using allele frequencies maintained in the National Institute of Standards and Technology population database.21
Role of the funding source
The funder had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Results
The first nasopharygeal swab, obtained at the community screening event on April 18, 2020, was positive for SARS-CoV-2 on real-time RT-PCR testing. Two subsequent nucleic acid amplification tests obtained after resolution of symptoms were negative for SARS-CoV-2 RNA (table 1 ). The patient's symptoms returned on May 28, 2020, and he was admitted to hospital on June 5, 2020, at which time a second nasopharyngeal swab was obtained and was positive for SARS-CoV-2 infection by real-time RT-PCR testing. The patient required ongoing oxygen support in hospital and reported symptoms that included myalgia, cough, and shortness of breath. Chest radiography showed development of patchy, bilateral, interstitial opacities suggestive of viral or atypical pneumonia. On June 6, 2020, the patient was tested for IgG and IgM against SARS-CoV-2 and positive results were obtained (figure 1).
Table 1.
Summary of laboratory results
|
Specimen A |
Specimen B |
||||
|---|---|---|---|---|---|
| April 18, 2020 | May 9, 2020 | May 26, 2020 | June 5, 2020 | June 6, 2020 | |
| Test methodology | Real-time RT-PCR | TMA | Real-time RT-PCR | Real-time RT-PCR | Immunoassay (IgG and IgM antibody detection) |
| Test result | Positive | Negative | Negative | Positive | Positive |
| Quantitative result | Ct 35·24 | RLU 299 | .. | Ct 35·31 | .. |
TMA=transcription-mediated amplification. Ct=cycle threshold. RLU=relative light units.
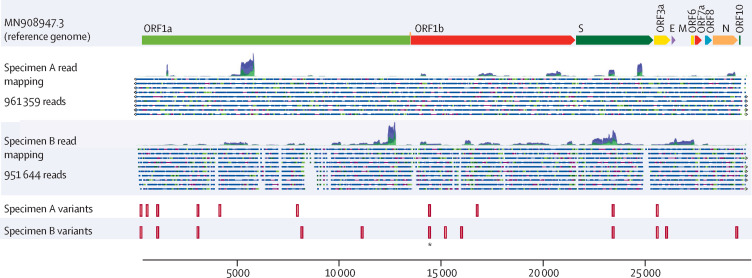
With two episodes of symptoms consistent with COVID-19, and two specimens positive for SARS-CoV-2 separated by a period of 48 days, in addition to resolution of symptoms and two non-reactive (negative) SARS-CoV-2 test results in between positive test results, nucleic acid sequencing was done of the viruses associated with the two positive tests. Illumina sequencing yielded 738 617 read pairs for the specimen obtained in April, 2020 (specimen A), and 1 410 885 read pairs for the specimen obtained in June, 2020 (specimen B). Sequence data indicated that specimen A was a member of clade 20C, because genomic sequence analysis identified five mutations (single nucleotide variants [SNVs]) that were hallmarks of the 20C clade (3037C→T, 14408C→T, 23403A→G, 1059C→T, and 25563G→T). Specimen B was also a member of clade 20C and presented the same five hallmark SNVs. Specimen A had five further SNVs compared with the reference genome. Specimen B showed six additional SNVs and a mutation at position 14 407, adjacent to the SNV 14408C→T and recorded as a dinucleotide multinucleotide variant (MNV) at positions 14 407 and 14 408 of the genome. Six SNVs were shared between specimen A and specimen B (table 2 ). Specimen A had four additional SNVs not seen in specimen B, whereas specimen B had seven SNVs that were absent in specimen A. A visualisation of the relation of sequence data sets between specimens A and B is shown in figure 2 . An additional three deletions and one insertion were noted in the sequence of specimen B relative to the reference genome (appendix p 2). These findings were confirmed by additional analyses of FASTQ files generated from specimens A and B (only the SNV at locus 4113 in specimen A was not verified). Predictions of insertions and deletions were less stable, with only the deletion at loci 2084 and the insertion at 6018 confirmed. The Freebayes analysis detected a deletion at 22 832 in specimen B that was not identified by the first sequence analysis (appendix p 3), but insertion and deletion predictions from short-read alignments are less reliable than are SNV predictions22 and are merely presented for completeness.
Table 2.
Variants noted in specimens A and B compared with the reference genome
| Coverage (reads) | Allele frequency (%) | Forward/reverse balance* | Average quality† | ||
|---|---|---|---|---|---|
| Shared variants of specimens A and B versus reference genome | |||||
| 241C→T | |||||
| Specimen A | 67 | 100% | 0·37 | 35·6 | |
| Specimen B | 6 | 100% | 0·38 | 36·0 | |
| 1059C→T | |||||
| Specimen A | 144 | 100% | 0·48 | 35·6 | |
| Specimen B | 55 | 92·7% | 0·26 | 35·4 | |
| 3037C→T | |||||
| Specimen A | 89 | 100% | 0·42 | 35·6 | |
| Specimen B | 425 | 99·8% | 0·19 | 35·5 | |
| 14408C→T‡ | |||||
| Specimen A | 73 | 100% | 0·40 | 35·7 | |
| Specimen B | 1145 | 99·6% | 0·43 | 35·6 | |
| 23403A→G | |||||
| Specimen A | 6859 | 99·9% | 0·19 | 35·7 | |
| Specimen B | 10 484 | 99·9% | 0·46 | 35·6 | |
| 25563G→T | |||||
| Specimen A | 421 | 100% | 0·45 | 35·2 | |
| Specimen B | 757 | 99·1% | 0·48 | 35·4 | |
| Specimen A-specific variants versus reference genome | |||||
| 539C→T | 141 | 99·3% | 0·45 | 35·6 | |
| 4113C→T | 159 | 70·4% | 0·38 | 35·6 | |
| 7921A→G | 182 | 98·9% | 0·49 | 35·7 | |
| 16741G→T | 173 | 99·4% | 0·47 | 35·6 | |
| Specimen B-specific variants versus reference genome | |||||
| 8140C→T | 1046 | 85·0% | 0·43 | 35·6 | |
| 11102C→T | 1713 | 99·9% | 0·44 | 35·5 | |
| 14407C→T‡ | 1145 | 99·7% | 0·43 | 35·6 | |
| 15190G→C | 139 | 90·6% | 0·33 | 35·7 | |
| 15981C→T | 224 | 100% | 0·38 | 35·5 | |
| 26013C→T | 1415 | 99·2% | 0·38 | 35·5 | |
| 29466C→T | 86 | 98·8% | 0·07 | 35·8 | |
Reference genome was Wuhan Hu 1 (GenBank MN908947.3).
Ratio of forward to reverse reads covering the locus.
Phred score. Phred is a measure of base calling accuracy, a higher score indicates higher quality. A Phred score of 30 indicates a base-calling accuracy of 99·9%.
CLC Genomics classified this variant as a dinucleotide multinucleotide variant. The two variants have been split in this table for clarity.
Figure 2.
Variant mapping of specimens A and B against the reference genome
ORF1a and ORF1b encode replicase proteins. The other ORFs encode assembly proteins. ORF=open reading frame. S=spike. E=envelope. M=membrane. N=nucleocapsid. *Identifies variant 14 407 in specimen A and variants 14 407 and 14 408 in specimen B.
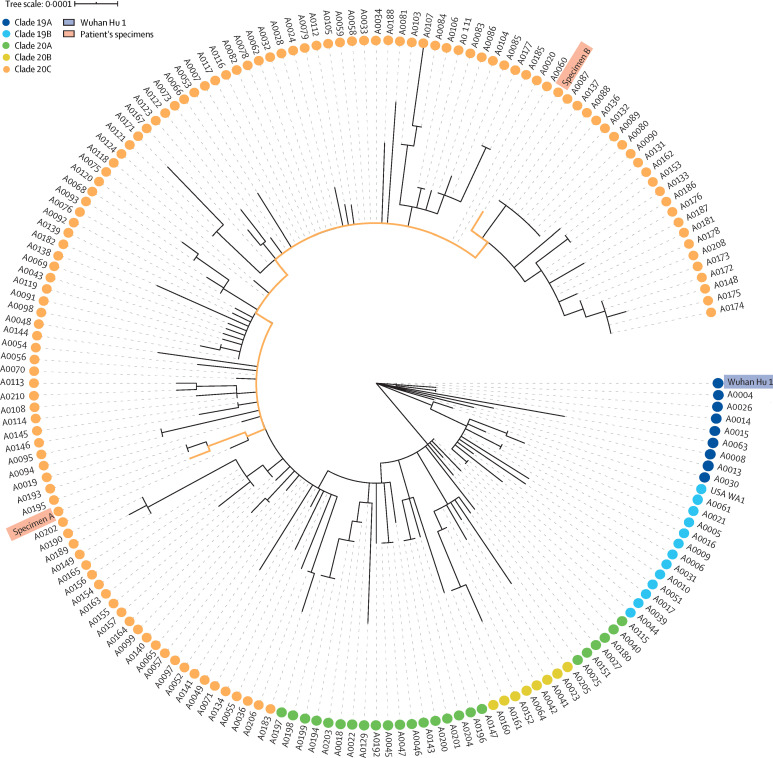
Specimens A and B were among 171 samples obtained in the US state of Nevada between March 5 and June 5, 2020, and sequenced. Phylogenetic analysis showed the relatedness of specimens A and B to each other and their comparative distance among additional positive samples (figure 3 ). To rule out the possibility of specimen mishandling, or mislabelling errors during RNA extractions, forensic identity testing was done to investigate the source and intermediate materials of specimens A and B. Analysis of each of the specimens, residual extractions, and aliquot residuals showed that that specimens A and B were derived from the same individual, with a one in 53·48 × 1024 chance of the specimens being from different people.
Figure 3.
Phylogenetic placement of specimens A and B within Nevada isolates, reference genomes, and global clades
171 sequences were from Nevada. Wuhan Hu 1 was the reference genome (GenBank MN908947.3). USA WA1 was the isolate USA-WA1/2020 (Bei Resources, Manassas, VA, USA).
Discussion
Our case report presents details of the first individual in North America to have symptomatic reinfection with SARS-CoV-2. Similar to observations with the reinfection case in Ecuador,12 our patient showed increased symptom severity in their second infection, whereas the cases from Belgium and the Netherlands11 and Hong Kong10 did not show a difference in severity of symptoms. The mechanisms that could account for a more severe secondary infection can only be speculated. First, a very high dose of virus might have led to the second instance of infection and induced more severe disease.23 Second, it is possible that reinfection was caused by a version of the virus that was more virulent, or more virulent in this patient's context. Third, a mechanism of antibody-dependent enhancement might be the cause, a means by which specific Fc-bearing immune cells become infected with virus by binding to specific antibodies. This mechanism has been seen previously with the betacoronavirus causing severe acute respiratory syndrome.24 In that case, the patient recovered and was discharged from hospital.
The individual associated with these two SARS-CoV-2 infections had no immunological disorders that would imply facilitation of reinfection. They were not taking any immunosuppressive drugs. The individual was negative for HIV by antibody and RNA testing (data not shown) and had no obvious cell count abnormalities. The secondary positive case (reinfection) occurred simultaneously to a positive case in a cohabitant (parent), who also provided a specimen on June 5, 2020, that was positive by nucleic acid amplification testing (transcription-mediated amplification). Sequencing is underway on the co-habitant specimen to ascertain its potential role in reinfection. However, the positive specimen from the co-habitant was obtained and tested in the Hologic Aptima format, which did not align with the procedures established at our sequencing laboratory. Nevertheless, the co-habitant positive case provides a possible source for secondary exposure and reinfection of our patient.
It is possible that we have reported a case of continuous infection entailing deactivation and reactivation. However, for such a hypothesis to be true, a mutational rate of SARS-CoV-2 would be required that has not yet been recorded.25, 26, 27, 28 Specimens A and B showed an extrapolated rate of SNV and MNV accumulation of 83·64 substitutions per year, a rate that greatly exceeds the currently observed rate of 23·12.28 However, even more importantly, the four substitutions noted in specimen A would have to revert to the ancestral genotype, and the odds of this reversion occurring are remote. Of course, if such an amount of base change did occur in that timeframe, the remarkable nature of specimens A and B would shift from a case of possible reinfection to one of high-rate evolution within an infected individual. Another alternative explanation for the observed differences in specimens A and B would be that of co-infection. In a co-infection hypothesis, the patient would have been infected with viruses of both genotypes at the time of sample collection. Such a hypothesis would then further require that the specimen B type virus be present, yet undetected in April, 2020, and then conversely, specimen A type virions become depleted before the June, 2020, sample collection date. Specimens A and B were both in clade 20C, which was the predominant major clade seen in northern Nevada at the time samples were obtained. Our survey of viruses in Nevada identified samples resembling each of the case genotypes.15 Although evidence exists that SARS-CoV-2 quasispecies exist at low and fluctuating frequencies in infected samples,29 whereby low-frequency (eg, 1%) SNVs could be seen in various samples from the same patient, this possible situation would not itself account for the genotype switch observed between the first infection and reinfection.
Our findings have implications for the role of vaccination in response to COVID-19. If we have truly reported a case of reinfection, initial exposure to SARS-CoV-2 might not result in a level of immunity that is 100% protective for all individuals. With respect to vaccination, this understanding is established, with influenza regularly showing the challenges of effective vaccine design.30 A major limitation of our case study is that we were unable to undertake any assessment of the immune response to the first episode of SARS-CoV-2 infection. We also could not assess fully the effectiveness of the immune responses (eg, neutralising antibody titres) during the second episode, when the individual was antibody-positive for total antibody assay to the SARS-CoV-2 nucleocapsid protein. If our patient is a case of natural viral evolution in vivo (although highly unlikely in view of the requirement of four reversions to reference genotypes) then the implications of these data are that SARS-CoV-2 can adapt with enough genetic dexterity to avoid a natural immune response in a manner to re-establish detectable levels of infection in an individual. If our patient is a case of reinfection, it is crucial to note that the frequency of such an occurrence is not defined by one case study: this event could be rare. The absence of comprehensive genomic sequencing of positive cases in the USA and worldwide limits the advances in public health surveillance needed to find these cases. Certainly, limitations in screening and testing availability for SARS-CoV-2 exacerbate the poor surveillance efforts being undertaken not only to diagnose COVID-19 but also to obtain actionable genetic tracking of this agent.
Data sharing
The CLC workflow, combined mapping report, parameters for CLC modules, FASTA-format sequences, the open-source workflow, BAM alignments, and VCF-format files are available online.
Acknowledgments
Acknowledgments
We thank Nevada IDEA Network of Biomedical Research for funding this work. We acknowledge grants from the National Institute of General Medical Sciences (National Institutes of Health), which enabled publication of these findings (GM103440 and GM104944). We thank the Washoe County Health District and Washoe County Sheriff's Department for helping to identify and confirm these findings.
Contributors
RLT contributed to writing of the report and data analysis. JRS contributed to review and editing of the report and data analysis. PDH contributed to sequencing and analysis. HK contributed to public health intelligence and case identification. NC contributed to clinical data and clinical care. AG and CL contributed to diagnostics analysis. SCV and CCR contributed to writing and editing of the report and figure generation. DJ and MJF contributed to identity testing and confirmation. SVH contributed to diagnostics and laboratory management. MP had the idea for the study and contributed to diagnostics, formal analysis, and writing and editing of the report.
Declaration of interests
JRS reports personal fees from Qiagen Digital Insights, outside of the submitted work. All other authors declare no competing interests.
Supplementary Material
References
- 1.Ju B, Zhang Q, Ge J. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 2.Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S-C, Wang J-T, Huang L-M. Longitudinal analysis of severe acute respiratory syndrome (SARS) coronavirus-specific antibody in SARS patients. Clin Diagn Lab Immunol. 2005;12:1455–1457. doi: 10.1128/CDLI.12.12.1455-1457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang AT, Garcia-Carreras B, Hitchings MDT. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. 2020 doi: 10.1101/2020.04.14.20065771. published online April 17. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu W, Fontanet A, Zhang P-H. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis. 2006;193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mo H, Zeng G, Ren X. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11:49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed SE. The behaviour of recent isolates of human respiratory coronavirus in vitro and in volunteers: evidence of heterogeneity among 229E-related strains. J Med Virol. 1984;13:179–192. doi: 10.1002/jmv.1890130208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo PCY, Lau SKP, Wong BHL. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin Diagn Lab Immunol. 2004;11:665–668. doi: 10.1128/CDLI.11.4.665-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L-P, Wang N-C, Chang Y-H. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To KK-W, Hung IF-N, Ip JD. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. published online Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Elslande J, Vermeersch P, Vandervoort K. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1330. published online Sept 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prado-Vivar B, Becerra-Wong M, Guadalupe JJ. COVID-19 re-infection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in South America. SSRN. 2020 doi: 10.2139/ssrn.3686174. published online Sept 8. (preprint) [DOI] [Google Scholar]
- 13.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartley P, Tillett RL, Xu Y. Genomic surveillance revealed prevalence of unique SARS-CoV-2 variants bearing mutation in the RdRp gene among Nevada patients. medRxiv. 2020 doi: 10.1101/2020.08.21.20178863. published online Sept 11. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemoine F, Correia D, Lefort V. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47:W260–W265. doi: 10.1093/nar/gkz303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefort V, Longueville J-E, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Institute of Standards and Technology Population studies conducted by the NIST Forensics/Human Identity Project Team. Sept 23, 2014. https://strbase.nist.gov/NISTpop.htm
- 22.Abnizova I, te Boekhorst R, Orlov YL. Computational errors and biases in short read next generation sequencing. J Proteomics Bioinform. 2017 doi: 10.4172/jpb.1000420. published online Jan 26. [DOI] [Google Scholar]
- 23.Guallar MP, Meiriño R, Donat-Vargas C, Corral O, Jouvé N, Soriano V. Inoculum at the time of SARS-CoV-2 exposure and risk of disease severity. Int J Infect Dis. 2020;97:290–292. doi: 10.1016/j.ijid.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yip MS, Leung NH, Cheung CY. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill V, Rambaut A. Phylogenetic analysis of SARS-CoV-2 genomes. March 6, 2020. https://virological.org/t/phylodynamic-analysis-of-sars-cov-2-update-2020-03-06/420
- 26.Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pachetti M, Marini B, Benedetti F. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadfield J, Megill C, Bell SM. Genomic epidemiology of novel coronavirus: global subsampling. Sept 16, 2020. https://nextstrain.org/ncov/global?c=region&l=clock
- 29.Jary A, Leducq V, Malet I. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.07.032. published online July 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CLC workflow, combined mapping report, parameters for CLC modules, FASTA-format sequences, the open-source workflow, BAM alignments, and VCF-format files are available online.