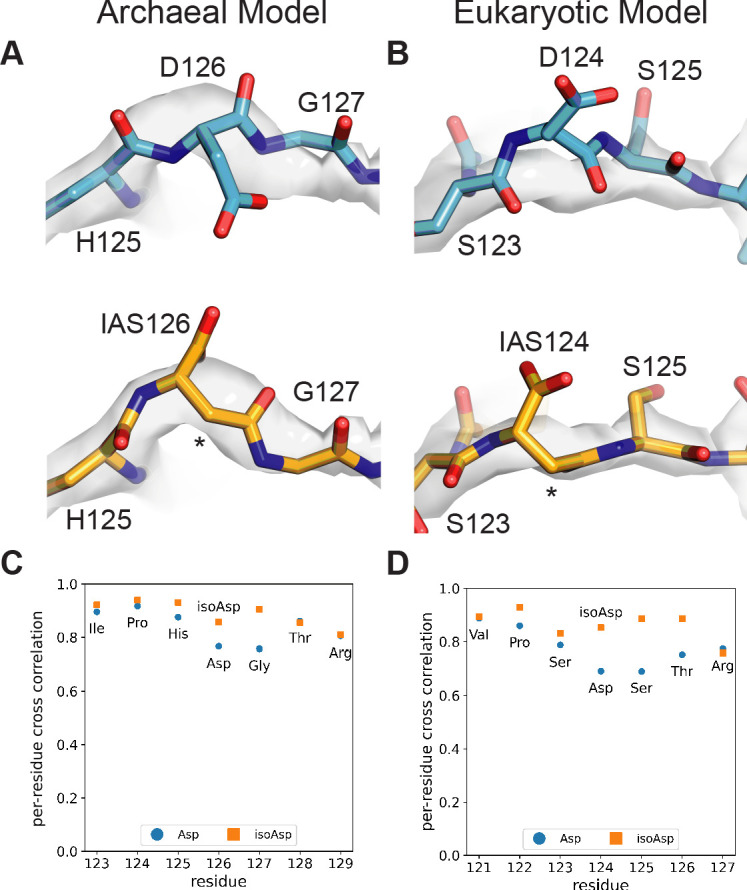
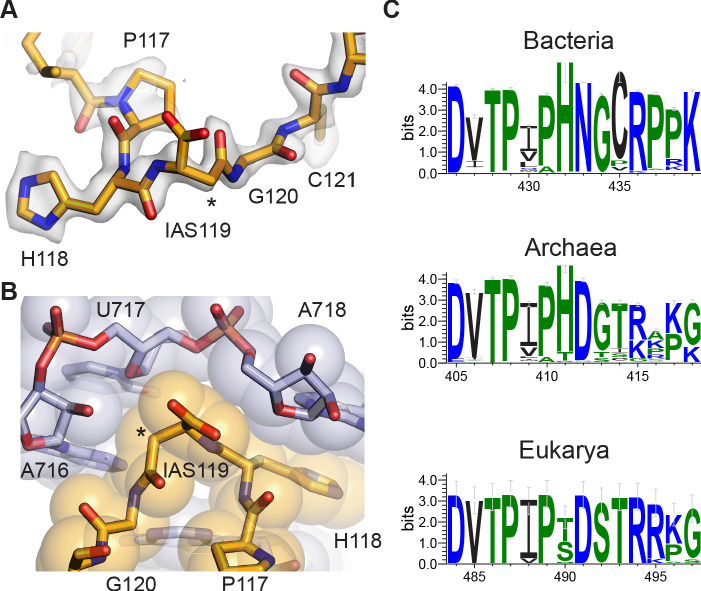
Figure 4. Isoaspartyl residue in protein uS11.
(A) Model of isoAsp at residue position 119 in uS11, with nearby residues and cryo-EM density from the 30S subunit platform-focused refinement. Weak density for the carboxylate is consistent with the effects of damage from the electron beam. The asterisk indicates the position of the additional backbone methylene group. (B) Shape complementarity between uS11 and 16S rRNA nucleotides surrounding IsoAsp119. 16S rRNA is shown in light purple and uS11 in orange, with atomistic coloring for the stick model. (C) Sequence logos of conserved amino acids spanning the putative isoAsp residue in all three domains of life.
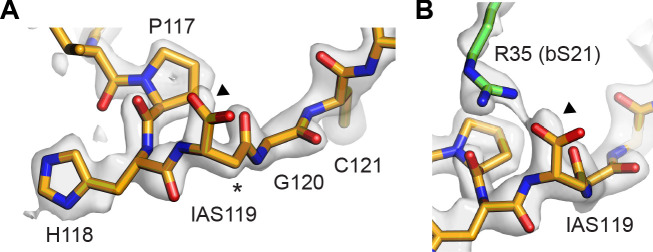
Figure 4—figure supplement 1. Cryo-EM density for uS11 based on early movie frames.
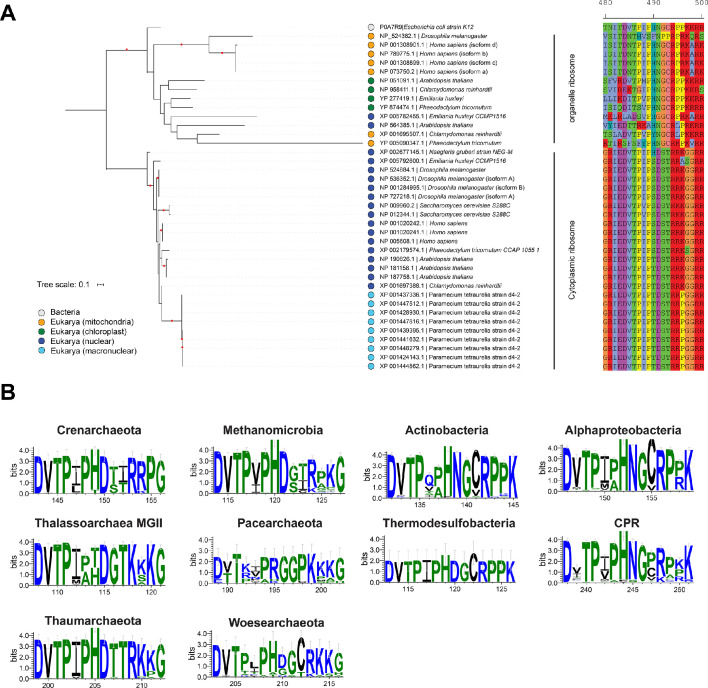
Figure 4—figure supplement 2. Conservation of residues near the isoAsp residue in uS11 homologs.
Figure 4—figure supplement 3. Structural models for isoaspartate in archaeal and eukaryotic ribosomes.