Abstract
This study characterizes the transcriptome and the cellular tumor microenvironment (TME) of conjunctival melanoma (CM) and identifies prognostically relevant biomarkers. 12 formalin-fixed and paraffin-embedded CM were analyzed by MACE RNA sequencing, including six cases each with good or poor clinical outcome, the latter being defined by local recurrence and/or systemic metastases. Eight healthy conjunctival specimens served as controls. The TME of CM, as determined by bioinformatic cell type enrichment analysis, was characterized by the enrichment of melanocytes, pericytes and especially various immune cell types, such as plasmacytoid dendritic cells, natural killer T cells, B cells and mast cells. Differentially expressed genes between CM and control were mainly involved in inhibition of apoptosis, proteolysis and response to growth factors. POU3F3, BIRC5 and 7 were among the top expressed genes associated with inhibition of apoptosis. 20 genes, among them CENPK, INHA, USP33, CASP3, SNORA73B, AAR2, SNRNP48 and GPN1, were identified as prognostically relevant factors reaching high classification accuracy (area under the curve: 1.0). The present study provides new insights into the TME and the transcriptional profile of CM and additionally identifies new prognostic biomarkers. These results add new diagnostic tools and may lead to new options of targeted therapy for CM.
Subject terms: Prognostic markers, Cancer genomics, Eye cancer, Tumour biomarkers, Cancer microenvironment
Introduction
Conjunctival melanoma (CM) is a rare but potentially life-threatening tumor of the ocular surface associated with systemic metastasis within 10 years in 18–26% and a 10 year survival rate of 41–78%1. Age-adjusted incidence is estimated at 0.55 per million per year in the United States with a tendency to increase from 0.26 in 1973 to 0.55 per million per year in 19992 accounting for 0.24% of all melanoma3. The majority of CM originates from primary acquired melanosis (57–76%), whereas a smaller proportion of CM develops de novo (16–25%) or from a pre-existing nevus (1–6%)1,4. Confirmed risk factors are the localization, with worse prognosis being reported for fornices, caruncle, plica semilunaris, palpebral conjunctiva and lid margins5, the tumor thickness5, the TNM stage6 and de novo CM1. There is evidence that UV radiation contributes to the development of CM7, although this factor is still debated8. It has also been shown that melanization stimulates HIF-1α expression of melanoma cells9, affects clinical outcome of patients with cutaneous and uveal melanoma10,11 and modifies the sensitivity to anticancer treatment, including radio- and immunotherapy12–14. The expression of progesterone and estrogen receptors on conjunctival melanomas could explain hormone-dependent changes in tumor morphology15. Based on the genetic profile, CM seems to be a distinct subset of melanoma, which is more similar to cutaneous melanoma than mucosal melanoma and is significantly different from uveal melanoma16. It is well known that CM is often associated with mutations in BRAF, representing a potential therapeutic target17. Nevertheless, little is known about the transcriptome and the cellular tumor microenvironment (TME) of CM, which could provide additional information about the pathways, molecular mechanisms and cell types involved in the pathogenesis of CM and thus define potentially new diagnostic and therapeutic targets.
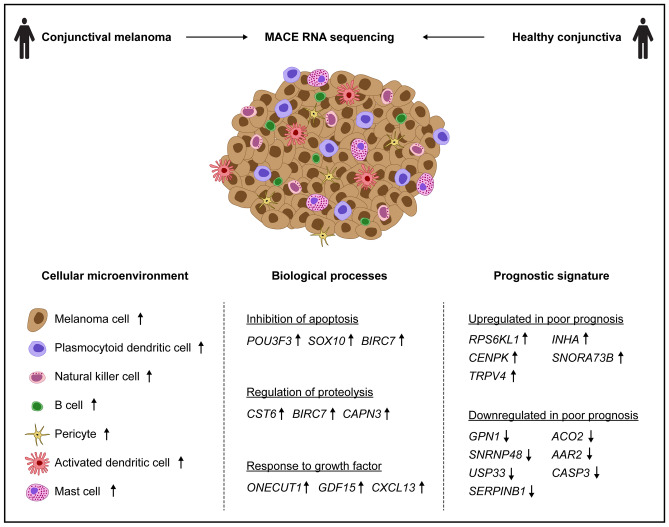
This study characterizes the cellular tumor microenvironment and provides a transcriptional profile of CM compared to healthy conjunctiva and analyzes the transcriptional differences between CM with poor and good clinical outcome (Fig. 1). The results of this study add new diagnostic and prognostic tools and may lead to new options of targeted therapy for CM.
Figure 1.
Study overview. The study characterizes the transcriptional profile of conjunctival melanoma compared to healthy conjunctiva, provides new insights into the cellular tumor microenvironment and identifies a prognostic signature classifying clinical outcome of conjunctival melanoma.
Results
Patient characteristics
A total of 20 conjunctival samples from 20 patients were included in this study. Histological analysis confirmed CM in 12 patients and healthy conjunctiva in eight patients. Patient characteristics are summarized in Table 1. Mean age in the melanoma and the control group was 58.9 (range: 27.3–85.3) and 55.7 years (range: 43.0—69.0), respectively (p = 0.692). There were three male and nine female patients in the melanoma group and six male and two female patients in the control group, respectively (p = 0.081). Pathologic T (pT) categories according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual (eighth edition)18 were pT1a in 9 (75.0%), pT1b in 1 (8.3%) and pT3b in 2 (16.6%) cases. Mean tumor thickness was 944.1 µm (range: 199.0 µm—2720.0 µm). Nine (75.0%) cases were located at the bulbar conjunctiva and one (8.3%) each at the fornix, tarsal conjunctiva and limbus. There were four patients with local recurrence (three cases with different localization between primary tumor and recurrence), one patient with local recurrence and systemic metastasis and one patient with systemic metastasis, which were included in the poor prognosis group. The mean follow-up time in the melanoma group was 7.1 years (min: 2.2, max: 16.7) (see Table 1).
Table 1.
Patient characteristics.
| Group | Conjunctival melanoma | Healthy conjunctiva | p |
|---|---|---|---|
| n | 12 | 8 | – |
| Age at surgery (years) | 58.9 (27.3–85.3) | 55.7 (43.0–69.0) | 0.692 |
| Sex | 0.081 | ||
| Male | 3 (25.0%) | 6 (75.0%) | |
| Female | 9 (75.0%) | 2 (25.0%) | |
| pT-category | – | – | |
| pT1a | 9 (75.0%) | ||
| pT1b | 1 (8.3%) | ||
| pT3b | 2 (16.6%) | ||
| Tumor-thickness (µm) | 944.1 (199.0–2720.0) | – | – |
| Localization | – | – | |
| Bulbar conjunctiva | 9 (75.0%) | ||
| Fornix | 1 (8.3%) | ||
| Tarsal conjunctiva | 1 (8.3%) | ||
| Limbus | 1 (8.3%) | ||
| Local recurrence | – | – | |
| Different localization | 3 | ||
| Similar localization | 1 | ||
| Systemic metastasis | 1 | – | – |
| Local recurrence and systemic metastasis | 1 | – | – |
| Follow-up (years) | 7.1 (2.2–16.7) | – | – |
pT-classification according to the eighth edition of the AJCC conjunctival melanoma staging system18. Data is shown as mean (range) or as absolute (relative) numbers.
Unsupervised transcriptomic analysis
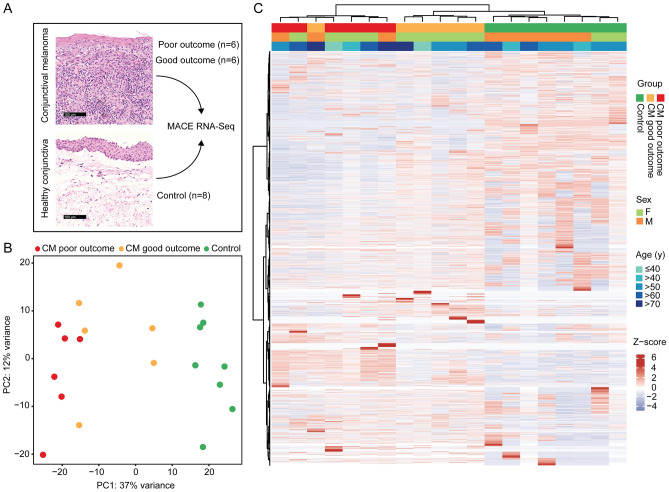
Unsupervised analysis revealed distinct differences in the transcriptome of CM when compared to normal conjunctiva. In addition, melanoma with poor and good clinical outcome also differed significantly with regard to their transcriptional profile (see PCA and unsupervised heatmap, Fig. 2).
Figure 2.
Unsupervised cluster analysis of twelve conjunctival melanoma (CM, six each with good or poor outcome) and eight normal conjunctival samples. Clinical outcome was classified based on the presence or absence of local recurrence and/or systemic metastases, respectively, with a follow up of at least 24 months. (A) Representative histological specimens of melanoma and normal conjunctiva as well as an overview of the study design. (B) Principle component analysis. (C) Unsupervised heatmap: basic demographic and clinical data are shown above. Each column represents one sample (red: melanoma with poor outcome, orange: melanoma with good outcome and green: healthy conjunctiva) and each row one expressed gene. There were 20.189 genes with at least one raw read. Unsupervised clustering was performed for both the rows and the columns (see dendrogram). The z-score represents a gene’s expression in relation to its mean expression by standard deviation units (red: upregulation, blue: downregulation). CM: melanoma, PC: principle component.
Cellular tumor microenvironment of conjunctival melanoma
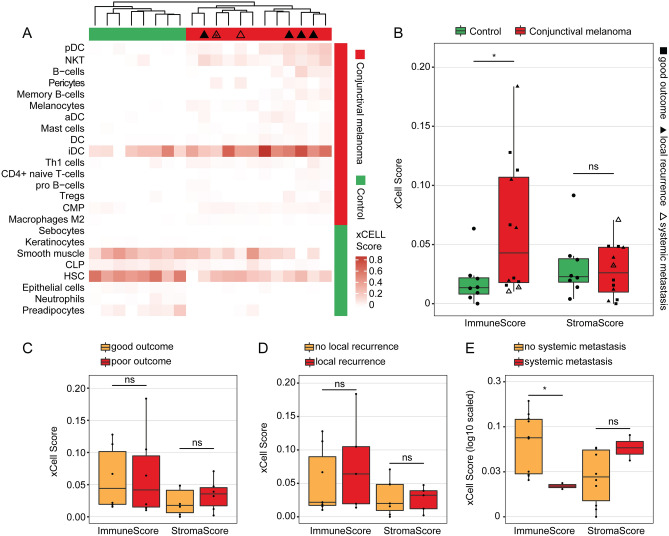
Cell type enrichment analysis using xCell19 revealed that numerous immune cell types were enriched in the tumor microenvironment (TME) of CM. Among them, plasmacytoid dendritic cells (pDC), natural killer T cells (NKT), B cells, activated dendritic cells (aDC) and mast cells were most significantly increased in CM compared to healthy conjunctiva (log2FC of enrichment scores between CM and control: 2.5, 2.3, 2.1, 1.4 and 1.3, respectively, p < 0.05, Fig. 3A). Furthermore, melanocytes and pericytes were significantly enriched in CM compared to control conjunctiva (log2FC: 1.6 and 1.8, respectively, p < 0.05). An overview of the enrichment scores of all 64 analyzed immune and stroma cell types is shown in Supplementary Fig. 2. The cell type analysis also demonstrated that melanoma and control samples clustered according to their histological diagnosis based on their cell type enrichment scores, indicating the significance of the TME in CM (Fig. 3A). This finding was unaffected when all 64 cell types were included in the analysis (data not shown). Immune enrichment scores (composite score of all analyzed immune cell types) were significantly higher in CM (mean: 0.063, SD: 0.057) when compared to healthy conjunctiva (mean: 0.019, SD: 0.020, p = 0.034), whereas stroma scores (composite score of all analyzed stroma cell types) did not significantly differ (mean: 0.028, SD: 0.023 and mean: 0.032, SD: 0.027, p = 0.847) (Fig. 3B). Next, we investigated whether immune and stroma scores differed between melanoma with poor and good clinical outcome and found that neither immune or stroma scores, nor the cell composition differed significantly when taking both clinical endpoints—metastasis and local recurrence—into account (Fig. 3A, C). While the immune scores between melanoma with and without local recurrence did not differ significantly (mean: 0.077, SD: 0.070 and mean: 0.053, SD: 0.050, p = 0.745), the immune scores in the two melanoma samples with systemic metastasis (mean: 0.012, SD: 0.002) were significantly lower compared to all other melanoma (mean: 0.074 SD 0.057, p = 0.041, (Fig. 3D, E). Stroma scores, in contrast, were not significantly different in metastasized or recurrent tumors (Fig. 3 D-E).
Figure 3.
Cellular tumor microenvironment of conjunctival melanoma characterized by cell type enrichment analysis using xCell. The tool uses gene expression profiles of 64 immune and stromal cell types to calculate cell type enrichment scores. (A) Heatmap illustrating xCell enrichment scores of 24 of the 64 cell types which were significantly enriched in melanoma compared to normal conjunctiva (p < 0.05, Mann–Whitney U test). 16 cell types were up- and 8 cell types were downregulated in melanoma (see annotation on the right of the heatmap). Each row represents one cell type, each column represents one sample. Rows are ordered according to the fold change of mean enrichment scores for melanoma and normal conjunctiva, respectively. Columns are clustered according to similarities in xCell enrichment scores (see dendrogram). Samples with local recurrence and/or systemic metastasis are labeled (see legend in B, *: local recurrence and systemic metastasis). pDC: plasmacytoid dendritic cell, NKT: natural killer T cell, aDC: activated dendritic cell, DC: dendritic cell, iDC: immature dendritic cells, Th1: type 1 T-helper cells, Tregs: regulatory T cells, CMP: common myeloid progenitor, CLP: common lymphoid progenitor, HSC: hematopoietic stem cell. (B) Boxplots of the xCell immune and stroma score between melanoma and healthy conjunctiva. Each symbol represents one sample and the shape represents the prognostic groups also shown in (C–E). (C–E): boxplots of the xCell immune and stroma score between melanoma with poor and good outcome (C), melanoma with and without local recurrence (D) and with or without systemic metastasis (E), respectively. *p < 0.05, ns: not significant (Mann–Whitney U test).
Transcriptional characterization of conjunctival melanoma
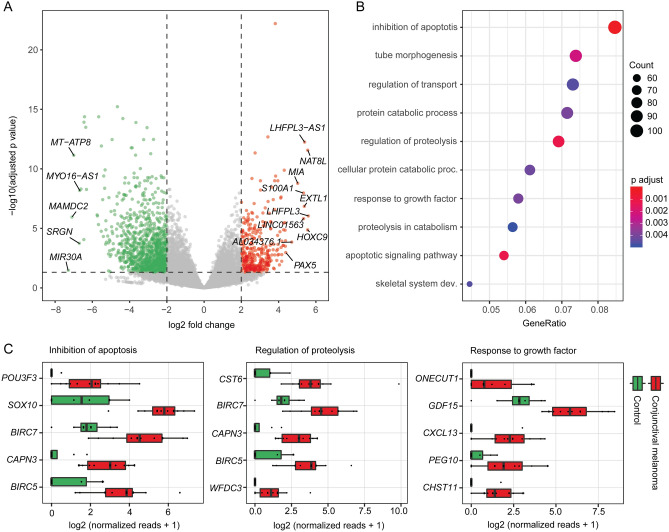
Differential gene expression analysis revealed 363 up- and 1096 downregulated genes in CM compared to healthy conjunctiva (Fig. 4A). Among them, LHFPL3 (LHFPL tetraspan subfamily member 3), NAT8L (N-acetyltransferase 8 like), HOXC9 (homeobox C9), LHFPL3-AS1 (LHFPL3 antisense RNA 1) and EXTL1 (exostosin like glycosyltransferase 1) were the five most significantly upregulated factors in CM (Fig. 4A). MIR30A (microRNA 30a), MAMDC2 (MAM domain containing 2), MT-ATP8 (mitochondrially encoded ATP synthase membrane subunit 8), SRGN (serglycin) and MYO16-AS1 (MYO16 antisense RNA 1) were the top five downregulated genes in CM (Fig. 4A). Gene ontology (GO) analysis revealed, that the DEG contributed to biological processes such as inhibition of apoptosis (GO:0043066, GO:0043069), apoptotic signaling pathway (GO:0097190), proteolysis (GO:0030162, GO:0051603), tube morphogenesis (GO:0035239), protein catabolism (GO:0030163, GO:0044257), response to growth factor (GO:0071363) and transport of substances (GO:0051050) (Fig. 4B). A total of 106 DEG were associated to inhibition of apoptosis, among which 20 where up- and 86 were downregulated in CM (Supplementary Fig. 2). The top five upregulated genes in CM with regard to inhibition of apoptosis were POU3F3 (POU class 3 homeobox 3), SOX10 (SRY-box transcription factor 10), BIRC7 (baculoviral IAP repeat containing 7), CAPN3 (calpain 3) and BIRC5 (baculoviral IAP repeat containing 5) (Fig. 4C). CAPN3, BIRC5 and 7, as well as CST6 (cystatin E/M) and WFDC3 (WAP four-disulfide core domain 3), also appeared among the top five genes playing a role in proteolysis (Fig. 4C). ONECUT1 (one cut homeobox 1), GDF15 (growth differentiation factor 15), CXCL13 (C-X-C motif chemokine ligand 13), PEG10 (paternally expressed 10) and CHST11 (carbohydrate sulfotransferase 11) were the top five DEG in response to growth factor (Fig. 4C).
Figure 4.
Characterization of the transcriptome of conjunctival melanoma (CM). (A) Volcano plot showing the up- and downregulated genes (red and green, respectively) in CM in comparison to normal conjunctiva. Genes not differentially expressed are shown in grey. The top ten upregulated as well as the top five downregulated genes in melanoma are labeled. (B) Gene ontology (GO) analysis of the differentially expressed genes in conjunctival melanoma. The top ten biological processes, which the DEG were involved in, are shown in the dot plot. “Inhibition of apoptosis” summarizes the two GO terms “negative regulation of apoptotic process” and “negative regulation of programmed cell death”, which overlapped with all 106 associated DEG. The size of the dots represents the number of associated genes (count). The adjusted p value of each GO term is shown by color. The gene ratio describes the ratio of the count to the number of all DEG. (C) Box plots illustrating normalized reads of the top five upregulated factors of three disease-relevant GO terms arranged by log2 fold change.
Prognostic transcriptome signature of conjunctival melanoma
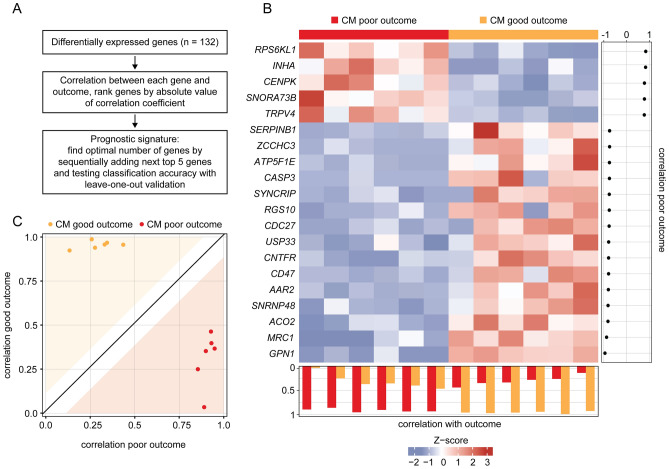
Genes with prognostic relevance for CM were identified by determining the DEG between melanoma with poor and good clinical outcome in a first step (Fig. 5A). Poor clinical outcome was defined based on the presence or absence of local recurrence and/or systemic metastases, respectively, with a follow up of at least 24 months (mean follow-up: 7.1 years, min: 2.2, max: 16.7). In a second step, the correlation coefficient between each gene and outcome was calculated and the top genes were selected according to the absolute value of their correlation coefficient. By stepwise increasing the number of genes and testing the classification accuracy using ROC and leave-one-out validation (see methods for details), it became evident that a gene signature of twenty genes had the optimal accuracy for classifying clinical outcome in CM (AUC = 1.0). The expression profile of these twenty genes in all 12 melanoma samples is shown in the heatmap in Fig. 5B, with the genes and samples arranged according to their correlation with clinical outcome (Fig. 5B, description and right and bottom panel). Five prognostic genes were identified, which were upregulated in CM with poor outcome: RPS6KL1 (ribosomal protein S6 kinase like 1), INHA (inhibin subunit alpha), CENPK (centromere protein K), SNORA73B (small nucleolar RNA, H/ACA box 73B) and TRPV4 (transient receptor potential cation channel subfamily V member 4). The top five of 15 downregulated genes in CM with poor outcome were GPN1 (GPN-loop GTPase 1), MRC1 (mannose receptor C-type 1), ACO2 (aconitase 2), SNRNP48 (small nuclear ribonucleoprotein U11/U12 subunit 48) and AAR2 (AAR2 splicing factor) (Fig. 5B). The classification of poor and good outcome based on the twenty signature genes is shown in Fig. 5C (see methods for details). GO analysis revealed, that the twenty signature genes were mainly involved in biological processes such as cell proliferation, apoptosis and several immune processes as illustrated in the cnetplot in Supplementary Fig. 3.
Figure 5.
Prognostic transcriptome signature of conjunctival melanoma (CM). (A) Workflow of analysis. DEG between melanoma with poor and good clinical outcome were used to define a prognostic signature. Clinical outcome was defined based on the presence or absence of local recurrence and/or systemic metastases with a follow up of at least 24 months. (B) Heatmap of expression data of 20 prognostic marker genes of CM with good (orange) and poor outcome (red). Each row represents one gene and each column one tumor. The z-score represents a gene’s expression in relation to its mean expression by standard deviation units (red: upregulation, blue: downregulation). The genes are ordered according to their correlation with poor outcome, placing the gene with the highest correlation coefficient at the top (correlation coefficients are shown beside the heatmap). Tumors in the columns are ordered according to the correlation between the expression values of all 20 signature genes of one sample to the mean expression values of the other samples of both outcomes using leave-one-out validation (see methods). The correlation coefficients for both outcomes are shown below the heatmap and in (C): The white space around the diagonal is defined by the standard deviation of the distances of each sample from the diagonal. A sample that lies within the colored area can be assigned to an outcome with high probability.
Finally, we investigated whether clinical or histopathological parameters, such as age, gender, pT category, tumor thickness, localization of the CM or tumor pigmentation, were associated with clinical outcome (Table 2). Uni- and multivariate logistic regression analysis revealed that the clinical and histopathological parameters assessed were not significantly associated with outcome, whereas the transcriptome signature remained significant in multivariate analysis (p < 0.001).
Table 2.
Multivariate analysis of clinical and histopathological factors regarding the clinical outcome of conjunctival melanoma.
| Poor prognosis | Good prognosis | p | |
|---|---|---|---|
| n | 6 | 6 | – |
| Age at surgery (years) | 57.7 (37.1–85.3) | 60.1 (27.3–81.0) | 0.818 |
| Sex | > 0.999 | ||
| Male | 2 (33.3%) | 1 (16.6%) | |
| Female | 4 (66.6%) | 5 (83.3%) | |
| Skin color | > 0.999 | ||
| White | 6 (100.0%) | 6 (100.0%) | |
| pT-category | 0.135 | ||
| pT1a | 3 (50%) | 6 (100%) | |
| pT1b | 1 (16.7%) | 0 (0%) | |
| pT3b | 2 (33.3%) | 0 (0%) | |
| Tumor-thickness (µm) | 1201.0 (376.0–2720.0) | 687.2 (199.0–1002.0) | 0.630 |
| Localization | 0.262 | ||
| Bulbar conjunctiva | 3 (50%) | 6 (100%) | |
| Fornix | 1 (16.7%) | 0 (0%) | |
| Tarsal conjunctiva | 1 (16.7%) | 0 (0%) | |
| Limbus | 1 (16.7%) | 0 (0%) | |
| Pigmentation | 0.439 | ||
| Melanotic | 6 (100.0%) | 4 (66.7%) | |
| Amelanotic | 0 (0.0%) | 2 (33.3%) | |
| Transcriptome signature | < 0.001 | ||
| Classified: poor | 6 (100.0%) | 0 (0.0%) | |
| Classified: good | 0 (0.0%) | 6 (100.0%) |
pT-classification according to the eighth edition of the AJCC conjunctival melanoma staging system18. Data is shown as mean (range) or as absolute (relative) numbers.
Discussion
Gene expression analysis provides important insights into the molecular mechanisms of tumorigenesis and -progression and has helped to define new therapeutic targets in numerous malignancies. However, the transcriptional analysis of rare tumors, such as CM, has so far been hampered by their low incidence, challenging a prospective analysis of fresh tissue. 3′-RNA sequencing methods, such as the Massive Analysis of cDNA Ends (MACE), allow transcriptome analysis of FFPE samples20, in which RNA degradation predominantly occurs at the 5′ end21. The present study uses MACE RNA sequencing of FFPE samples to characterize the cellular tumor microenvironment (TME) and the transcriptional profile of CM compared to healthy conjunctiva and identifies a prognostic transcriptome signature that allows the classification of poor and good clinical outcome for CM.
The cellular tumor microenvironment is known to modulate tumorigenesis, tumor progression, therapeutic response and clinical outcome of various malignancies22. Transcriptome-based cell type enrichment analysis using xCell19 revealed that the tumor microenvironment of CM was predominantly characterized by an enrichment of numerous immune cell types, including plasmacytoid dendritic cells (pDC), natural killer T cells (NKT), B cells and mast cells. pDCs and NKTs are known to be important components of the TME in various malignancies and are essentially involved in the regulation of anti-tumor immunity23,24. There is also evidence of tumor-resident B-cells in skin melanoma, which are associated with a favorable prognosis, suggesting an involvement in anti-tumor immunity and highlighting their potential as a therapeutic target25. Finally, mast cell infiltration has been observed in various tumors, including malignant cutaneous melanoma, breast and colorectal cancer26. Mast cells are able to modulate the TME by releasing different mediators, including a variety of proangiogenic factors and several matrix metalloproteinases that can increase the invasiveness of the tumor26. Taken together, our results suggest that the enriched immune cells might play a crucial role in the development of CM and thus may represent potential therapeutic targets for a specific anti-tumor immune response. Additionally, the present study revealed that CM with systemic metastases had significantly lower immune scores than non-metastatic tumors, a finding that has previously been reported for different cancer types27,28. In a study of Mlecnik et al.27, a low immune score was associated with an increased risk of systematic metastases in patients with colorectal cancer. The authors conclude that patients in early stages could benefit most from checkpoint T-cell therapies to prevent the development of distant metastases because these tumors have higher immune scores27. In CM, the immune analysis could therefore be a promising diagnostic approach to predict the development of systemic metastases and might also be used to predict the response to immune therapy, for which, in contrast to other tumor localizations, a local application could also be considered. However, it is important to note that of the twelve CM included, only two were associated with metastasis, which requires further investigation to validate their diagnostic and therapeutic potential.
The transcriptional signature of CM provided in this study differed significantly from healthy conjunctival samples and revealed 1459 differentially expressed genes (DEG). Gene ontology (GO) analysis demonstrated that these DEG were mainly involved in biological processes such as inhibition of apoptosis, apoptotic signaling pathway, proteolysis and response to growth factor. Among the apoptosis inhibitors, POU3F3 was the top upregulated DEG in CM. This factor and especially the long non coding (lnc) RNA linc-POU3F3, which was also among the upregulated DEG, are known to play a role in tumor cell proliferation, inhibition of apoptosis, as well as in angiogenesis in several malignancies29–35. siRNA-mediated knockdown of linc-POU3F3 reduces tumor cell proliferation and invasion and increase apoptosis in colorectal cancer cells33 and may therefore represent a therapeutic approach for the treatment of CM. Another top expressed factor in inhibition of apoptosis was SOX10, a nuclear transcription factor that is involved in the differentiation of neural crest progenitor cells to melanocytes and is known as a sensitive and specific marker for skin melanoma36, that has not yet been described in CM. In skin melanoma cells, SOX10 mediates the invasion through MIA (Melanoma Inhibitory Activity)37, which was also among the top upregulated DEG in CM. In addition, SOX10 was recently identified as an oncogene in skin melanoma that could be inhibited by the miRNA miR-3138. BIRC5 and BIRC7 were among the top 5 DEG associated with inhibition of apoptosis and regulation of proteolysis. Both factors belong to the "inhibitors of apoptosis proteins" family and are upregulated in uveal melanoma as well39. The most significantly upregulated DEG in CM was LHFPL3, which is also highly expressed in malignant glioma and can be inhibited by miRNA-218-5p thus reducing the invasiveness of glioma cells40. Further studies will be necessary to investigate the presented factors and signaling pathways in the development of CM in more detail and to validate them as potential therapeutic targets for the treatment of CM.
In search of a prognostic tool that provides information on the aggressiveness of CM in relation to local recurrence or systemic metastases, this study identified a prognostic transcriptional signature comprised of twenty genes allowing a classification of the clinical outcome of CM with high accuracy. The clinical and histopathological standard predictors, such as TNM stage, tumor thickness and tumor localization6 were not significantly associated with the clinical outcome in our study, although some tendency was observed which may be explained by the low sample size of our study. These results underline the potential of a transcriptional analysis to categorize clinical outcome. While most of the genes predicting outcome of CM have already been described as prognostic factors in other malignancies, such as CENPK41–46, INHA47–50, RPS6KL151, CASP3 (Caspase 3)52, SERPINB1 (Serpin Family B Member 1)53, USP33 (Ubiquitin Specific Peptidase 33)54–59, TRPV460 and ACO261, some of the predicting factors have not yet been associated to cancer prognosis, such as GPN1, SNRNP48, AAR2 and SNORA73B. Among the mentioned factors, GPN1 was the gene with the highest correlation coefficient with good clinical outcome in CM. GPN1 is involved in the nuclear translocation of XPA (XPA, DNA Damage Recognition And Repair Factor), an important factor that controls nucleotide excision repair62, and has been linked to the development of oral cancer in a genome-wide association study63. RNA splicing regulators such as AAR2 and SNRNP48, on the other hand, are emerging as a new class of oncoproteins and tumor suppressors64, and small nucleolar RNAs such as SNORA73B seem to play important roles in tumorigenesis65. However, none of the aforementioned factors has been linked to cancer prognosis and further studies are warranted to validate their prognostic value in CM and other malignancies.
We acknowledge that this study is limited by its retrospective single center design and its relatively small sample size. However, a prospective single center study does not appear feasible due to the very low incidence of CM. A prospective multicenter study would be necessary to overcome these limitations. Another limitation is the lack of external validation of the prognostic transcription signature, since no sequencing data of CM are available so far. Instead, we used the concept of leave-one-out validation, as previously described66,67. Furthermore, in contrast to single cell RNA sequencing (scRNA), bulk RNA sequencing cannot provide insights into cell heterogeneity and thus cannot reveal cell-specific transcriptional profiles to discern possible subtypes of tumor cells. However, scRNA sequencing is not feasible on FFPE samples. Therefore, we employed a bulk RNA sequencing-based cell type enrichment analysis using xCell19 which is one of the most accurate tools available68 to characterize the cell types involved in the tumor microenvironment of CM. It is important to emphasize that the cell type investigations are based on in silico analysis and have not been validated histologically due to the limited number of specimens. This needs to be considered in future studies.
In summary, the present study provides new insights into the cellular tumor microenvironment and the transcriptional profile of CM. It adds new prognostic biomarkers and diagnostic tools which can help to improve identification of high-risk patients and may lead to new options of targeted therapy for CM.
Methods
Patients and clinical outcome
A total of 12 CM samples from 12 patients who underwent tumor resection at the Eye Center of the University of Freiburg between 1996 and 2017 were retrospectively included in this study. Six cases each with good or poor clinical outcome with a follow-up of at least 24 months were examined, the latter being defined by local recurrence and/or systemic metastases. Eight healthy conjunctival samples from eight patients who underwent retinal detachment surgery between 2013 and 2016 served as controls. All methods were carried out in accordance with relevant guidelines and regulations and informed consent was obtained from all subjects. Ethics approval was granted from Ethics Committee of the Albert-Ludwigs-University Freiburg (approval number 481/19).
Formalin fixation and paraffin embedding
Formalin fixation and paraffin embedding (FFPE) of tissue samples was performed immediately after surgery according to routine protocols, as previously described69,70. Briefly, samples were fixed immediately after surgery in 4% formalin for 12 h, dehydrated in alcohol and processed for paraffin embedding. Histological diagnoses were made by an experienced ophthalmic pathologist (CAH). Hematoxylin and eosin stained slides were imaged using a Hamamatsu NanoZoomer S60 (Hamamatsu Photonics, Herrsching, Germany).
RNA isolation
After melting the block, tumor-free tissue areas were removed and the tumor, as well as the control FFPE samples were stored in tubes until RNA isolation, which was performed as previously described20,70. Briefly, total RNA was isolated from FFPE samples using the Quick-RNA FFPE Kit (Zymo Research). Following a DNAse I digestion using the Baseline-ZERO kit (Epicentre), the RNA concentration was measured with the Qubit RNA HS Assay Kit on a Qubit Fluorometer (Life Technologies). The RNA quality was determined with the RNA Pico Sensitivity Assay on a LabChip GXII Touch (PerkinElmer).
RNA sequencing
RNA sequencing was performed using massive analysis of cDNA ends (MACE), a 3′ RNA sequencing method, as previously described20,70. We recently demonstrated that MACE allows sequencing of FFPE samples with high accuracy71. Briefly, 20 barcoded libraries comprising unique molecule identifiers were sequenced on the NextSeq 500 (Illumina) with 1 × 75 bp. PCR bias was removed using unique molecular identifiers.
Bioinformatics
Sequencing data (fastq files) were uploaded to and analyzed on the Galaxy web platform (usegalaxy.eu)72, as previously described73. Quality control was performed with FastQC Galaxy Version 0.72 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ last access on 11/19/2019). Reads were mapped to the human reference genome (Gencode, release 32, hg38) with RNA STAR Galaxy Version 2.7.2b74 with default parameters using the Gencode annotation file (Gencode, release 32, https://www.gencodegenes.org/human/releases.html). Reads mapped to the human reference genome were counted using featureCounts Galaxy Version 1.6.475 with default parameters using the aforementioned annotation file. The output of featureCounts was imported to RStudio (Version 1.2.1335, R Version 3.5.3). Gene symbols and gene types were determined based on ENSEMBL release 98 (Human genes, GRCh38.p12, download on 11/19/2019)76. Genes without EntrezID and with zero mean reads were removed from analysis. Principal component analysis (PCA)77 was applied to check for potential batch effects, which were removed by the limma function removeBatchEffect78 and by consideration within the linear model of DESeq277. Differential gene expression was analyzed using the R package DESeq2 Version 1.22.277 with default parameters (Benjamini–Hochberg adjusted p values). Transcripts with log2 fold change (log2 FC) > 2 or < -2 and adjusted p value < 0.05 were considered as differentially expressed genes (DEG). Heatmaps were created with the R package ComplexHeatmap 1.20.079. Other data visualization was performed using the ggplot2 package80. Gene enrichment analysis and its visualization were done using the R package clusterProfiler 3.10.181. Cell type enrichment analysis was performed using xCell19. The tool uses sequencing-derived transcriptomic signatures of 64 distinct immune and stroma cell types to estimate the relative contributions of these cells to a bulk RNA transcriptome. Transcripts per million were calculated as an input for the analysis based on the output of featureCounts (assigned reads and feature length), as previously described82. xCell enrichment scores were compared between different groups using the Mann–Whitney U test. The log2 fold change of enrichment scores between different groups was defined as the log2 of the quotient of mean enrichment scores of each group. To define a prognostic transcriptome signature of CM, a three-step method, as described by van´t Veer et al.66, was applied. In a first step, DEG between CM with poor and good outcome were determined, as described above. Subsequently, the Pearson correlation between each gene and outcome was calculated. All genes were then arranged by the absolute value of their correlation coefficient. The top five genes were selected and their expression profile in one sample was correlated to each gene’s mean expression of the remaining samples of the poor and good prognosis group, respectively. These steps were repeated until each sample was left out once (leave-one-out validation), as described previously66,67. Classification accuracy was determined by calculating the area under the curve (AUC) of receiver operating curves (ROC). To find the optimal number of prognostic genes, the next top five genes were added to the signature and its classification accuracy was evaluated, as described above. This process was repeated until the AUC stopped improving.
Supplementary information
Acknowledgement
The authors thank Sylvia Zeitler for excellent technical assistance.
Author contributions
J.W., C.L., C.A.H. and G.S. designed the study, collected, analysed and interpreted the data. J.W. and C.L. carried out literature research, generated figures and wrote the original draft of the manuscript. All authors were involved in reviewing and editing the paper and had final approval of the submitted and published version.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The sequencing data are available in the Gene Expression Omnibus Database under the accession number GSE148387.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72864-0.
References
- 1.Wong JR, Nanji AA, Galor A, Karp CL. Management of conjunctival malignant melanoma: a review and update. Expert Rev Ophthalmol. 2014;9:185–204. doi: 10.1586/17469899.2014.921119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu GP, Hu DN, McCormick S, Finger PT. Conjunctival melanoma: is it increasing in the United States? Am. J. Ophthalmol. 2003;135:800–806. doi: 10.1016/s0002-9394(02)02288-2. [DOI] [PubMed] [Google Scholar]
- 3.McLaughlin CC, et al. Incidence of noncutaneous melanomas in the U.S. Cancer. 2005;103:1000–1007. doi: 10.1002/cncr.20866. [DOI] [PubMed] [Google Scholar]
- 4.Atzrodt L, Lapp T, Reinhard T, Auw-Haedrich C. Transformation of a naevus into a mixed pyogenic granuloma-naevus mimicking conjunctival melanoma. Pathology. 2020;52:269–271. doi: 10.1016/j.pathol.2019.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Paridaens AD, Minassian DC, McCartney AC, Hungerford JL. Prognostic factors in primary malignant melanoma of the conjunctiva: a clinicopathological study of 256 cases. Br. J. Ophthalmol. 1994;78:252–259. doi: 10.1136/bjo.78.4.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain P, et al. Multicenter, international assessment of the eighth edition of the American Joint Committee on cancer cancer staging manual for conjunctival melanoma. JAMA Ophthalmol. 2019;137:905–911. doi: 10.1001/jamaophthalmol.2019.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaminathan SS, et al. Molecular characteristics of conjunctival melanoma using whole-exome sequencing. JAMA Ophthalmol. 2017;135:1434–1437. doi: 10.1001/jamaophthalmol.2017.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pane AR, Hirst LW. Ultraviolet light exposure as a risk factor for ocular melanoma in Queensland, Australia. Ophthalmic Epidemiol. 2000;7:159–167. [PubMed] [Google Scholar]
- 9.Slominski A, et al. The role of melanogenesis in regulation of melanoma behavior: melanogenesis leads to stimulation of HIF-1alpha expression and HIF-dependent attendant pathways. Arch. Biochem. Biophys. 2014;563:79–93. doi: 10.1016/j.abb.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brozyna AA, Jozwicki W, Carlson JA, Slominski AT. Melanogenesis affects overall and disease-free survival in patients with stage III and IV melanoma. Hum. Pathol. 2013;44:2071–2074. doi: 10.1016/j.humpath.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brozyna AA, Jozwicki W, Roszkowski K, Filipiak J, Slominski AT. Melanin content in melanoma metastases affects the outcome of radiotherapy. Oncotarget. 2016;7:17844–17853. doi: 10.18632/oncotarget.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brozyna AA, VanMiddlesworth L, Slominski AT. Inhibition of melanogenesis as a radiation sensitizer for melanoma therapy. Int. J. Cancer. 2008;123:1448–1456. doi: 10.1002/ijc.23664. [DOI] [PubMed] [Google Scholar]
- 13.Pawlikowska M, et al. Coriolus versicolor-derived protein-bound polysaccharides trigger the caspase-independent cell death pathway in amelanotic but not melanotic melanoma cells. Phytother. Res. 2020;34:173–183. doi: 10.1002/ptr.6513. [DOI] [PubMed] [Google Scholar]
- 14.Slominski A, Zbytek B, Slominski R. Inhibitors of melanogenesis increase toxicity of cyclophosphamide and lymphocytes against melanoma cells. Int. J. Cancer. 2009;124:1470–1477. doi: 10.1002/ijc.24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bredow L, et al. Progesterone and estrogen receptors in conjunctival melanoma and nevi. Graefes Arch. Clin. Exp. Ophthalmol. 2014;252:359–365. doi: 10.1007/s00417-013-2523-0. [DOI] [PubMed] [Google Scholar]
- 16.Rossi E, et al. Conjunctival melanoma: genetic and epigenetic insights of a distinct type of melanoma. Int. J. Mol. Sci. 2019;20:5447. doi: 10.3390/ijms20215447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mor JM, Heindl LM. Systemic BRAF/MEK inhibitors as a potential treatment option in metastatic conjunctival melanoma. Ocul. Oncol. Pathol. 2017;3:133–141. doi: 10.1159/000452473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coupland, S.E., Barnhill, R. & Conway, R. Conjunctival melanoma, Cancer Staging Manual, 8th edition. 8, 803–812 (2017).
- 19.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange CAK, et al. Increased expression of hypoxia-inducible factor-1 alpha and its impact on transcriptional changes and prognosis in malignant tumours of the ocular adnexa. Eye (Lond.) 2018;32:1772–1782. doi: 10.1038/s41433-018-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdueva D, Wing M, Schaub B, Triche T, Davicioni E. Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J. Mol. Diagn. 2010;12:409–417. doi: 10.2353/jmoldx.2010.090155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell D, Chintala S, Dey M. Plasmacytoid dendritic cell in immunity and cancer. J. Neuroimmunol. 2018;322:63–73. doi: 10.1016/j.jneuroim.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Nair S, Dhodapkar MV. Natural killer T cells in cancer immunotherapy. Front Immunol. 2017;8:1178. doi: 10.3389/fimmu.2017.01178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiaruttini G, et al. B cells and the humoral response in melanoma: the overlooked players of the tumor microenvironment. Oncoimmunology. 2017;6:e1294296. doi: 10.1080/2162402X.2017.1294296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komi DEA, Redegeld FA. Role of mast cells in shaping the tumor microenvironment. Clin. Rev. Allergy Immunol. 2020;58:313–325. doi: 10.1007/s12016-019-08753-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mlecnik B, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci. Transl. Med. 2016;8:327ra326. doi: 10.1126/scitranslmed.aad6352. [DOI] [PubMed] [Google Scholar]
- 28.Li X, et al. Identification of prognostic genes in adrenocortical carcinoma microenvironment based on bioinformatic methods. Cancer Med. 2020;9:1161–1172. doi: 10.1002/cam4.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, et al. LncRNA POU3F3 promotes proliferation and inhibits apoptosis of cancer cells in triple-negative breast cancer by inactivating caspase 9. Biosci. Biotechnol. Biochem. 2019;83:1117–1123. doi: 10.1080/09168451.2019.1588097. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Wu X, She W. LncRNA POU3F3 promotes cancer cell migration and invasion in nasopharyngeal carcinoma by up-regulating TGF-beta1. Biosci. Rep. 2019 doi: 10.1042/BSR20181632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Wu L, Yang Q, Ye M, Zhu X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene. 2015;554:114–119. doi: 10.1016/j.gene.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Lang HL, et al. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 2017;21:959–972. [PubMed] [Google Scholar]
- 33.Shan TD, et al. Knockdown of linc-POU3F3 suppresses the proliferation, apoptosis, and migration resistance of colorectal cancer. Oncotarget. 2016;7:961–975. doi: 10.18632/oncotarget.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishii J, et al. Class III/IV POU transcription factors expressed in small cell lung cancer cells are involved in proneural/neuroendocrine differentiation. Pathol. Int. 2014;64:415–422. doi: 10.1111/pin.12198. [DOI] [PubMed] [Google Scholar]
- 35.Imaoka T, et al. Gene expression profiling distinguishes between spontaneous and radiation-induced rat mammary carcinomas. J. Radiat. Res. 2008;49:349–360. doi: 10.1269/jrr.07126. [DOI] [PubMed] [Google Scholar]
- 36.Mohamed A, Gonzalez RS, Lawson D, Wang J, Cohen C. SOX10 expression in malignant melanoma, carcinoma, and normal tissues. Appl. Immunohistochem. Mol. Morphol. 2013;21:506–510. doi: 10.1097/PAI.0b013e318279bc0a. [DOI] [PubMed] [Google Scholar]
- 37.Graf SA, Busch C, Bosserhoff AK, Besch R, Berking C. SOX10 promotes melanoma cell invasion by regulating melanoma inhibitory activity. J. Invest. Dermatol. 2014;134:2212–2220. doi: 10.1038/jid.2014.128. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Y, et al. The miR-31-SOX10 axis regulates tumor growth and chemotherapy resistance of melanoma via PI3K/AKT pathway. Biochem. Biophys. Res. Commun. 2018;503:2451–2458. doi: 10.1016/j.bbrc.2018.06.175. [DOI] [PubMed] [Google Scholar]
- 39.Lederman M, Meir T, Zeschnigk M, Pe'er J, Chowers I. Inhibitor of apoptosis proteins gene expression and its correlation with prognostic factors in primary and metastatic uveal melanoma. Curr. Eye Res. 2008;33:876–884. doi: 10.1080/02713680802382989. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, Qian R, Zhang J, Shi X. MiR-218-5p targets LHFPL3 to regulate proliferation, migration, and epithelial-mesenchymal transitions of human glioma cells. Biosci. Rep. 2019 doi: 10.1042/BSR20180879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YC, Huang CC, Lin DY, Chang WC, Lee KH. Overexpression of centromere protein K (CENPK) in ovarian cancer is correlated with poor patient survival and associated with predictive and prognostic relevance. PeerJ. 2015;3:e1386. doi: 10.7717/peerj.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, et al. Co-expression of mitosis-regulating genes contributes to malignant progression and prognosis in oligodendrogliomas. Oncotarget. 2015;6:38257–38269. doi: 10.18632/oncotarget.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, et al. Overexpression of centromere protein K (CENP-K) gene in hepatocellular carcinoma promote cell proliferation by activating AKT/TP53 signal pathway. Oncotarget. 2017;8:73994–74005. doi: 10.18632/oncotarget.18172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komatsu M, et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int. J. Oncol. 2013;42:478–506. doi: 10.3892/ijo.2012.1744. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, et al. Downregulation of CENPK suppresses hepatocellular carcinoma malignant progression through regulating YAP1. Oncol. Targets Ther. 2019;12:869–882. doi: 10.2147/OTT.S190061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, et al. Analysis of gene expression in bladder cancer: possible involvement of mitosis and complement and coagulation cascades signaling pathway. J. Comput. Biol. 2019 doi: 10.1089/cmb.2019.0237. [DOI] [PubMed] [Google Scholar]
- 47.Balanathan P, et al. Elevated level of inhibin-alpha subunit is pro-tumourigenic and pro-metastatic and associated with extracapsular spread in advanced prostate cancer. Br. J. Cancer. 2009;100:1784–1793. doi: 10.1038/sj.bjc.6605089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mylonas I, et al. Inhibin/activin subunits (inhibin-alpha, -betaA and -betaB) are differentially expressed in human breast cancer and their metastasis. Oncol. Rep. 2005;13:81–88. [PubMed] [Google Scholar]
- 49.Mylonas I. Inhibin-alpha subunit expression in uterine endometrioid adenocarcinomas and endometrial cancer cell lines: a potential prognostic factor. Int. J. Mol. Med. 2011;27:309–318. doi: 10.3892/ijmm.2010.586. [DOI] [PubMed] [Google Scholar]
- 50.Yoon W, Yoo Y, Chae YS, Kee SH, Kim BM. Therapeutic advantage of genetically engineered Salmonella typhimurium carrying short hairpin RNA against inhibin alpha subunit in cancer treatment. Ann. Oncol. 2018;29:2010–2017. doi: 10.1093/annonc/mdy240. [DOI] [PubMed] [Google Scholar]
- 51.Xie H, Xie C. A six-gene signature predicts survival of adenocarcinoma type of non-small-cell lung cancer patients: a comprehensive study based on integrated analysis and weighted gene coexpression network. Biomed. Res. Int. 2019;2019:4250613. doi: 10.1155/2019/4250613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang KH, et al. Caspase-3, a key apoptotic protein, as a prognostic marker in gastric cancer after curative surgery. Int. J. Surg. 2018;52:258–263. doi: 10.1016/j.ijsu.2018.02.055. [DOI] [PubMed] [Google Scholar]
- 53.Willmes C, et al. SERPINB1 expression is predictive for sensitivity and outcome of cisplatin-based chemotherapy in melanoma. Oncotarget. 2016;7:10117–10132. doi: 10.18632/oncotarget.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia Y, et al. Reduced USP33 expression in gastric cancer decreases inhibitory effects of Slit2-Robo1 signalling on cell migration and EMT. Cell Prolif. 2019;52:e12606. doi: 10.1111/cpr.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Z, et al. USP33 mediates Slit-Robo signaling in inhibiting colorectal cancer cell migration. Int. J. Cancer. 2015;136:1792–1802. doi: 10.1002/ijc.29226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, Pang X, Ji L, Sun Y, Ji Y. Reduced expression of deubiquitinase USP33 is associated with tumor progression and poor prognosis of gastric adenocarcinoma. Med. Sci. Monit. 2018;24:3496–3505. doi: 10.12659/MSM.908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia M, Guo Y, Lu X. USP33 is a biomarker of disease recurrence in papillary thyroid carcinoma. Cell Physiol. Biochem. 2018;45:2044–2053. doi: 10.1159/000488041. [DOI] [PubMed] [Google Scholar]
- 58.Wen P, et al. USP33, a new player in lung cancer, mediates Slit-Robo signaling. Protein Cell. 2014;5:704–713. doi: 10.1007/s13238-014-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuasa-Kawada J, Kinoshita-Kawada M, Rao Y, Wu JY. Deubiquitinating enzyme USP33/VDU1 is required for Slit signaling in inhibiting breast cancer cell migration. Proc. Natl. Acad. Sci USA. 2009;106:14530–14535. doi: 10.1073/pnas.0801262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee WH, et al. TRPV4 regulates breast cancer cell extravasation, stiffness and actin cortex. Sci. Rep. 2016;6:27903. doi: 10.1038/srep27903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang P, et al. Decreased expression of the mitochondrial metabolic enzyme aconitase (ACO2) is associated with poor prognosis in gastric cancer. Med. Oncol. 2013;30:552. doi: 10.1007/s12032-013-0552-5. [DOI] [PubMed] [Google Scholar]
- 62.Dong L, et al. Melanocyte-stimulating hormone directly enhances UV-Induced DNA repair in keratinocytes by a xeroderma pigmentosum group A-dependent mechanism. Cancer Res. 2010;70:3547–3556. doi: 10.1158/0008-5472.CAN-09-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lesseur C, et al. Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat. Genet. 2016;48:1544–1550. doi: 10.1038/ng.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urbanski LM, Leclair N, Anczukow O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA. 2018;9:e1476. doi: 10.1002/wrna.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan X, et al. Analysis of expression pattern of snoRNAs in different cancer types with machine learning algorithms. Int. J. Mol. Sci. 2019;20:2185. doi: 10.3390/ijms20092185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 67.Lee JS, Ruppin E. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol. 2019;5:1614–1618. doi: 10.1001/jamaoncol.2019.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sturm G, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lange CA, et al. Enhanced TKTL1 expression in malignant tumors of the ocular adnexa predicts clinical outcome. Ophthalmology. 2012;119:1924–1929. doi: 10.1016/j.ophtha.2012.03.037. [DOI] [PubMed] [Google Scholar]
- 70.Schlecht A, et al. Transcriptomic characterization of human choroidal neovascular membranes identifies calprotectin as a novel biomarker for patients with age-related macular degeneration. Am. J. Pathol. 2020;190:1632–1642. doi: 10.1016/j.ajpath.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 71.Boneva, S., et al. 3′ MACE RNA-sequencing allows for transcriptome profiling in human tissue samples after long-term storage. Laboratory Investigation, Online ahead of print (2020). [DOI] [PMC free article] [PubMed]
- 72.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boeck M, et al. Temporospatial distribution and transcriptional profile of retinal microglia in the oxygen-induced retinopathy mouse model. Glia. 2020;68:1859–1873. doi: 10.1002/glia.23810. [DOI] [PubMed] [Google Scholar]
- 74.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 76.Cunningham F, et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 80.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2016. [Google Scholar]
- 81.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data are available in the Gene Expression Omnibus Database under the accession number GSE148387.