Abstract
Background
Strong evidence has suggested an important role of telomeres in meiosis, fertilization, and embryo development.
Purpose
To determine if sperm telomere length (STL) in sperm purified by differential gradient centrifugation followed by swim-up (selected STL) is correlated with sperm quality and clinical outcomes.
Methods
Relative selected STL was assessed by quantitative polymerase chain reaction (Q-PCR) in 78 consecutive assisted reproductive technology (ART) treatments during 2017. Statistical analyses were performed in the totality of patients, and in normozoospermic and non-normozoospermic patients. These included correlations between selected STL and sperm quality parameters, embryological parameters (multivariable linear regression), and clinical parameters (multivariable logistic regression).
Results
No significant correlations were found between selected STL and sperm quality in the total population. However, selected STL was significantly correlated with total sperm count (r = 0.361; P = 0.039) and sperm DNA fragmentation-post-acrosomal region pattern (r = − 0.464; P = 0.030) in normozoospermic patients. No relation was observed between selected STL and clinical outcomes in any clinical group.
Conclusions
As the correlations observed in normozoospermic patients were not representative of the whole heterogeneous population, differences in the sperm characteristics of the study population may lead to discrepant results when evaluating the association of STL with sperm quality. Since the total population selected STL was not related with sperm quality and with clinical outcomes, results do not support the use of selected STL measurement to evaluate the reproductive potential of the male patient or to predict the success rates of ART treatments.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01897-1) contains supplementary material, which is available to authorized users.
Keywords: Assisted reproduction, Male infertility, Sperm telomere length, Infertility biomarkers
Introduction
Globally, infertility is estimated to affect 10–15% of couples. In this epidemiologic context, it is generally assumed that 50% of the cases are due to female factors, 20–30% to male factors, and 20–30% to a combination of mixed factors [1]. Male factor infertility is a current global health concern with a multifactorial etiology, including a genetic, environmental, and lifestyle influence. However, infertility of unknown origin still accounts for a large percentage of men [2]. It is thus important to investigate new pathways that may influence clinical outcomes, in order to assist the diagnosis of infertility and develop new lines of treatment.
Telomeres are specialized nucleoprotein structures present at the ends of eukaryotic chromosomes, with vital functions in the maintenance of genomic stability and integrity [3]. Telomeres are also thought to play a role in reproduction, by promoting the alignment and pairing of homologous chromosomes during meiosis, as well as the establishment of synapsis and chiasma, and the formation of the meiotic spindle [4, 5]. In addition, telomeres of sperm are particularly decondensed and characteristically organized in specific positions of the sperm nuclei, which may contribute to facilitating the access of ooplasm factors to male chromosomes after fertilization, and the sequential activation of the male genome [6, 7].
Human telomeres contain tandem repeats of the 5′-TTAGGG-3′ sequence and they are very variable in size [8]. Telomeres are progressively subjected to shortening due to incomplete replication during cell division, processing by nucleases or reactive oxidative species (ROS), or as a result of recombination events. On the other hand, telomeres lengthen by the activity of telomerase, a reverse transcriptase that specifically binds telomeres [9]. However, telomerase is exclusively expressed in certain types of cells, such as germ and stem cells [10]. In proliferating cells lacking telomerase, telomeres eventually become critically short, leading to cell senescence or apoptosis [11].
The lengthening of telomeres by telomerase in germ cells is thought to ensure that a maximum telomere length (TL) is transmitted through gametes to the zygote, from where TL will continue to rise, being exponential at the inner cell mass of the blastocyst [12–14]. At this stage, the TL is reset for a particular organism and for the species [15–17]. However, variations in sperm TL (STL) have been reported, indicating the possible existence of critically short telomeres in sperm that may affect the development of the zygote [18]. In line with this hypothesis, several studies have emerged attempting to understand the impact of STL in male infertility. However, the published results have not been consensual [19]. Some studies have reported that STL is positively associated with semen parameters [20–22] and pregnancy outcomes [20–23], whereas other studies did not confirm these observations [14, 24, 25]. It is thus not clear whether STL is a suitable new biomarker of male infertility.
The heterogeneity of the samples used in these studies may be a limitation for their comparison. In assisted reproductive technology (ART) treatments, sperm samples are submitted to purification methods, such as the sequential method of differential gradient centrifugation (DGC) followed by swim-up [26], which isolates a higher quality sperm population, known to have higher STL in comparison with raw semen [22, 27, 28]. Therefore, STL in raw semen does not represent STL of the actual sperm used in ART treatments. In addition, raw semen may contain cells other than sperm that might influence the results. Differences in the spermiogram of patients may also be a relevant limitation in these studies since altered semen parameters, independently of STL, may mask its association with sperm quality.
In the present study, we assessed the STL of DGC-swim-up-selected sperm to clarify whether it was related with sperm quality, and with embryological and clinical outcomes, using different clinical groups according to the spermiogram of patients.
Materials and methods
Ethical approval
This study followed all the rules of ethical conduct regarding originality, data processing and analysis, duplicate publication, and patient material. Ethical guidelines were also followed in the conduction of the research, with patient written informed consent obtained before experiments. This study did not involve experiments in humans or animals, as only donated samples of surplus sperm were used. The approval of the Ethics Committee and the Declaration of Helsinki, revised in Tokyo 2004, on human experimentation does not apply to this study. The procedures at the infertility clinic are under the determinations of the National Law on Medically Assisted Procreation (Law of 2017) and supervised by the National Council on Medically Assisted Procreation (CNPMA-2018). According to these rules and guidelines, the use of clinic databases and patient biological material for diagnosis and research can be used without further ethical approval, under strict individual anonymity, and after patient written informed consent. Regarding the use of semen samples for laboratorial experimentation, the Ethics Committee authorization number is Project 2019/CE/P017 (266/CETI/ICBAS).
Statement of meaning
In the present manuscript, STL refers to the TL measured in sperm selected after DGC-swim-up.
Patients and sample size determination
The sample size was determined using the online calculation software (https://select-statistics.co.uk/calculators/sample-size-calculator-population-proportion/) with a 95% confidence level, and with the following reasoning: from the established 20–30% rate of pure male causes, we considered the minimum value of 20%, and from the established 20–30% rate of mixed causes, we considered the maximum value of 30%, with half (15%) corresponding to male causes [1]. This gives a total male factor of 35%, which corresponds to a likely sample proportion of 5.25% of male causes from the 15% global infertility rate. Using this likely sample proportion, the recommended sample size was 77 patients.
This study was performed on sperm samples retrieved after DGC-swim-up from 78 consecutive treatment cycles (therefore meeting the minimum recommended sample size), happening between October and December of 2017. From the 78 patients, 33 were normozoospermic (NZ) and 45 were non-normozoospermic (non-NZ). The treatment cycles used freshly ejaculated sperm for in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) procedures. All ART treatments were performed at the infertility clinic. During the period of the study, there were 181 IVF/ICSI cycles. Cycles that were not included in the study corresponded to cases with testicular sperm or donor sperm with unauthorized sample use for research by patients, or in which all swim-up sperm was used in ART treatments.
Karyotype analysis
Karyotypes were evaluated using G-banding with an analysis of at least 30 metaphases from peripheral blood lymphocytes, according to general protocols [29].
Sperm preparation
Semen analysis was performed before the beginning of each treatment cycle. For this, semen samples were collected by masturbation into sterile cups (Deltalab, Barcelona, Spain) following 2–4 days of sexual abstinence. Samples were left to liquefy, for 30 min over a thermal plate, at 37 °C (Minitube, Tiefenbach, Germany), and subsequently used for assessment of semen parameters according to the World Health Organization guidelines [30].
For IVF/ICSI treatments, fresh semen samples were left to liquefy at 33 °C, 5% CO2 in humidified air (Sanyo, Osaka, Japan), and then submitted to DGC using PureSperm 100 (Nidacon Int AB, Mölndal, Sweden) and SpermRinse (Vitrolife, Frölunda, Sweden) in Falcon tubes (Falkon, Corning, NY, USA). After centrifugation at 380g (Gyrosen, Daejeon, South Korea), for 20 min, at room temperature (RT), the bottom gradient layer was recovered. The pellet was washed two times (380 g, 10 min, RT) in sperm preparation medium (SPM) containing HEPES buffer (Origio, Malöv, Denmark). Then, it was layered with 100–1000 μL of Sequential Fert medium (Origio) and incubated (1 h, 33 °C, 6% CO2) to collect the swim-up fraction. After ART treatments, the remaining swim-up samples were used for research.
Using appropriate concentrations from each retrieved swim-up sample, aliquots were separated for DNA and protein extraction. Aliquots for chromosomic alterations analysis were fixed with 3:1 methanol/acetic acid (VWR International, Stockholm, Sweden/Panreac, Barcelona, Spain). Smears were also performed using 10 μL of the retrieved swim-up samples in adhesion microscope slides (Waldemar Knittel, Braunschweig, Germany), for DNA fragmentation and chromatin maturity assessment, and left to air-dry at RT. All aliquots and slides were stored at − 20 °C until use.
Ovarian controlled hyperstimulation
Women underwent controlled ovarian hyperstimulation with a gonadotropin-releasing hormone (GnRH) antagonist protocol (0.25 mg cetrorelix, Cetrotide; Merck-Serono, London, UK; or 0.25 mg ganirelix, Orgalutran; MSD, Hertfordshire, UK) in the large majority of the cases, and with a long agonist protocol (0.1 mg triptorelin, Decapeptyl; Ipsen Pharma Biotech, Signes, France) in the remaining cycles. For stimulation, recombinant follicle-stimulating hormone (rFSH, follitropin beta, Puregon; MSD, Haarlem, Netherlands; or rFSH, follitropin alfa, Gonal-F; Merck-Serono) was used alone in most of the cases. In some cases, rFSH was combined with human menopausal gonadotropin (HMG, Menopur; Ferring, Kiel, Germany), or with recombinant luteinizing hormone (follitropin alfa + lutropin alfa rFSH + rLH, Pergoveris, Merck-Serono; or rLH, Luveris, Merck-Serono). In other cases, HMG was also used alone. Ovulation trigger was performed with recombinant choriogonadotropin alfa (rHCG, 250 μg, Ovitrelle; Merck-Serono) in the majority of the cases. In other cases, ovulation was triggered with a GnRH agonist (0.2 mg triptorelin), or with a dual trigger, using triptorelin (0.2 mg) and rHCG (250 μg). Estradiol serum levels were assayed at the day of rHCG or 1 day before [31, 32].
Gamete and embryo handling
All procedures were performed on a K-Systems laminar flow with a thermal base (Cooper Surgical, Malöv, Denmark). All media were devoid of phenol red. For IVF, cumulus-oocyte complexes (COC) were collected in SynVitro Flush medium (without heparin, Origio). After this step, all procedures were performed under paraffin oil (Ovoil-100, Vitrolife). COC were washed with SynVitro Flush in 1-well culture dishes (Falcon). They were then transferred to an ESCO incubator (MRI-6A10, EscoMedical, Singapore, Singapore) (37 °C, 5% O2, 6% CO2, 89% N2) in 5-well culture dishes (Vitrolife) with Sequential Fert medium (Origio), 500 μL/well (up to 4 COC/well), and inseminated (4–5 h after oocyte pick-up) with 50,000–100,000 sperm/mL. About 16–19 h after insemination, COC were mechanically denuded in wells, to observe oocyte maturity and fertilization. Oocytes were then transferred individually to 12-well microdroplet embryo slide culture dishes (Vitrolife) with a Sequential Cleavage medium (Origio), 30 μL/droplet, up to day 3. Thereafter, embryos were transferred to new dishes with Sequential Blast medium (Origio), up to day 5.
For ICSI, COC allocated in 1-well culture dishes with a Sequential Fert medium were incubated in the ESCO for 2 h. Thereafter, they were denuded, for 30 s, with recombinant hyaluronidase (ICSI Cumulase, Origio), washed with SPM, and then mechanically dissociated from granulosa cells in SPM with oocyte denudation micropipettes (Vitrolife). Denudation was performed at 37 °C (thermic laminar flow base). Denuded oocytes were then incubated in 5-well culture dishes up to microinjection (1–2 h) in a Sequential Cleavage medium. Microinjection was performed under a described technique [33, 34] in an inverted microscope (Nikon, Tokyo, Japan) equipped with Hoffman optics and Narishige micromanipulators (Narishige, Tokyo, Japan), using microinjection and holding micropipettes (Origio, Charlottesville, VA, USA), polyvinylpyrrolidone (PVP Clinical Grade, Origio) and ICSI dishes (Falcon). After ICSI, oocytes were transferred individually to 12-well microdroplet culture dishes with a Sequential Cleavage medium, up to day 3, and embryos then transferred to new dishes with a Sequential Blast medium, up to day 5. When incubated in EmbryoScopes (Vitrolife, FertiliTech, Viby, Denmark), microinjected oocytes were transferred to time-lapse dishes (Vitrolife, Viby, Denmark) with a Sequential Cleavage medium (25 μL/well), up to day 3, and embryos then in Sequential Blast. In this case, the medium was replaced (20 μL), but not the culture dish.
Normal fertilization was assessed 14–18 h after microinjection (2 pronuclei, 2 polar bodies). At day 3, embryo quality was evaluated according to the number, size, and regularity of the blastomeres, and percentage of fragments, with high-quality embryos being those with the correct number of cells, of similar size and regularity, and with less than 25% of fragments (grade A/B) [35]. Blastocysts were scored at day 5, with high-quality blastocysts being those developed to grades BL1 and BL2 with good morphology and to BL3–BL5 if the inner cell mass and trophectoderm were of grade A/B [36].
Ultrasound-guided embryo transfer used a Sure View Wallace Embryo Replacement Catheter (Cooper Surgical, Trumbull, CT, USA) or a Wallace malleable stylet (Cooper Surgical).
Luteal supplementation
All patients had luteal supplementation with intravaginal administration of natural-micronized progesterone (Projeffik; Effik, Meudon-la-Forêt, France), 200 mg (t.i.d.), from the day of oocyte retrieval. Implantation was confirmed by a rise in β-HCG serum, 12 days after embryo transfer. Clinical pregnancy was established by ultrasound at 7 weeks of gestation, with sac visualization and presence of fetal heartbeat. When a GnRH agonist was used for triggering final oocyte maturation, progesterone luteal support was associated with oral estradiol (Estrofen, Isdin, Lisbon, Portugal) and a bolus of rHCG (Ovitrelle, 250 μg) given at the day of oocyte pick-up [37].
Sperm DNA fragmentation assessment
The incidence of morphological normal spermatozoa displaying nuclear DNA strand breaks was identified by the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay using the In Situ Cell Death Detection kit, Fluorescein (Roche Diagnostics, Mannheim, Germany), according to manufacturer’s instructions for in situ technique, as previously described [38, 39]. For this, 10 μL of the swim-up fraction was spread onto adhesion microscope slides (Waldemar Knittel). After air-drying at RT, slides were stored at − 20 °C until use. Cells were fixed for 1 h at RT with 4% paraformaldehyde (Merck, Darmstadt, Germany) in 0.2 M phosphate-buffered saline (PBS; Sigma, Steinheim, Germany). Slides were then washed in PBS and permeabilized with 0.1% Triton X-100 (Sigma) in 0.1% sodium citrate (Sigma) for 2 min at 4 °C. After washing twice with PBS, sperm were incubated in 50 μL of labeling solution containing the terminal deoxynucleotidyl transferase (TdT) enzyme for 1 h at 37 °C in a dark moist chamber. After incubation, slides were washed and counterstained with VECTASHIELD Antifade medium containing 4′,6-diamino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). On each slide, a minimum of 200 sperm were double-blindly scored under the × 100 objective of a Nikon Eclipse E400 fluorescence microscope fitted with a CCD camera and NIS-Elements BR V.4.60 software (Nikon). Of the total patients, 39 had sufficient sperm for sperm DNA fragmentation (sDNAfrag) analysis. A number of 8022 sperm were evaluated, with a mean of 206 sperm per patient. For each experiment, a negative (TdT enzyme omitted and replaced by distilled water) and positive (deoxyribonuclease treatment) controls were performed to ensure the reproducibility of the assay. Each spermatozoon was assigned as with the presence of DNA fragmentation (if displaying intense green fluorescence) or normal (DAPI staining only). The percentage of TUNEL-positive sperm was expressed as the percentage of cells exhibiting sDNAfrag. The TUNEL test presents low inter- and intra-variability, giving consistent results in different countries. This test is considered a highly valuable indicator for sperm quality, with increased sDNAfrag being related to poor embryo development and clinical outcomes [40–42]. A single experienced researcher (R.S.) performed these experiments.
Sperm chromatin maturity assessment
Sperm chromatin maturity, which refers to the levels of chromatin condensation due to the replacement of somatic cell histones by sperm-specific protamines in sperm during spermatogenesis, was evaluated by acidic aniline blue (AB) staining. Sperm smears (10 μL of the swim-up fraction) were fixed in 3% glutaraldehyde (Merck) in PBS (Sigma) for 30 min at RT. Microscope slides (ground edges, double frosted area; Süsse; Gudensberg, Germany) were then stained for 5 min at RT with 5% aqueous aniline blue (Merck) and acidified with 4% acetic acid (Merck) to pH 3.5. Slides were then washed with tap water and allowed to completely air-dry at RT. At least two hundred spermatozoa were blindly evaluated per slide. Of the total patients, 41 had sufficient sperm for AB analysis. A number of 9259 sperm were evaluated, with a mean of 226 sperm per patient. The percentages of sperm heads stained dark blue (indicates histone-rich immature chromatin) and those remaining unstained (indicates protamine-rich mature chromatin) were calculated. Controls are not used in this test. The discrimination between lysine-rich histones (AB+) and arginine-rich protamines (AB−) reveals the level of sperm chromatin maturity, which is an important factor for the vulnerability to sDNAfrag and abnormal embryo development [41, 43, 44]. A single experienced researcher (R.S.) performed these experiments.
Sperm ploidy assessment
The percentage of sperm with euploidy, diploidy, and overall aneuploidy was obtained by fluorescent in situ hybridization (FISH). Of the total patients, 44 had sufficient sperm for FISH analysis. A number of 22,265 sperm were evaluated, with a mean of 504 sperm per patient. Approximately 500 sperm per patient were blindly evaluated. Aneuploid sperm included disomy 18, disomy X, disomy Y, disomy XY, or diploidies. Diploid sperm cases were considered those with disomy 18 simultaneously with disomy X, or disomy Y, or disomy XY [45, 46]. Swim-up samples were washed in PBS (2 × 10 min, 504 g) and then fixed with methanol/acetic acid (fixative solution; 3:1; VWR/Panreac). After the washes with the fixative solution (2 × 10 min, 504 g), samples were kept at − 20 °C until the FISH procedure. FISH was performed using a probe mixture containing CEPX (Spectrum Green), CEPY (Spectrum Orange), and CEP18 (Spectrum Aqua) from AneuVysion Multicolour DNA Probe Kit (Abbott Molecular, IL, USA). Briefly, 10 μL of the fixed semen sample was spread on a slide (ground edges, double frosted area; Süsse), washed in 1:10 saline-sodium citrate buffer (SSC; Invitrogen, Scotland, UK), dehydrated in ethanol series and sperm heads decondensed with 1,4-dithiothreitol (DTT; Roche Applied Systems, Penzberg, Germany). Sperm and probe mixture were denatured separately and hybridization occurred overnight at 37 °C in a humidified chamber. In order to counterstain the DNA (blue), 5 μL of DAPI (Abbott Molecular) was applied. Sperm were observed and analyzed in a fluorescence microscope (Axio Imager Z1, Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA) fitted with a CCD camera (AxioCam MRm, Zeiss) and an automated image software (FISH Imaging System, version 5.1, MetaSystems GmbH, Altlussheim, Germany). Controls are not used in this test. A single experienced researcher (C.A.) performed these experiments.
STL measurement
Sperm DNA was extracted from all samples using the NZY Tissue gDNA Isolation Kit (NZYtech, Lisbon, Portugal), according to manufacturer instructions. STL was measured by the quantitative polymerase chain reaction (Q-PCR) described method, in which the average TL per cell is given relatively to a single-copy control gene in a given sample [47]. As a single-copy gene, we used β-2-microglobulin (B2M), which encodes for a serum protein that associates with the major histocompatibility complex class I and has been tested for gene-dosage studies [48]. For this, in a CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, California, CA, USA), with the Bio-Rad CFX Manager 3.1 software (Bio-Rad), two PCRs were performed in separate plates for each sample (which maintained the same position in each plate). One PCR included the telomere forward and reverse primer pair [49], for telomere (T) amplification, and the other PCR included the B2M forward and reverse primer pair [50], for single-copy (S) control gene amplification. Each sample was run in triplicate with 20 ng of sample DNA, 1× Green Master Mix NZY q-PCR (NZYtech), and 0.4 μM of each primer. Each plate included a no template control (negative control) and serial dilutions of a positive control DNA from the HT-1376 cell line (ATCC CRL-1472, Manassas, VA, USA) to generate a reference curve. A melting curve was obtained for each sample to access primer specificity. A single experienced researcher (A.C.L.) performed these experiments.
The coefficient of variation (CoV) of cycle threshold (Ct) values was ≤ 2%. Mean Ct values of each sample were used to calculate their relative STL (T/S ratio), according to the following formulas: ΔCt(sample) = ΔCt(telomere) − ΔCt(control); ΔCt(reference curve) = ΔCt(telomere) − ΔCt(control); ΔΔCt = ΔCt(sample) − ΔCt(reference curve); T/S = 2−ΔΔCt.
Sperm carbonyl and nitro groups evaluation
To characterize the oxidative profile of sperm, protein carbonylation and protein nitration, commonly used oxidation biomarkers, were evaluated by quantification of their resulting products: 2,4-dinitrophenyl (DNP) and nitro-tyrosine, respectively. The content of these products was evaluated using the slot blot technique, as previously described [51]. Of the total patients, 47 and 45 patients had sufficient sperm for protein carbonylation and protein nitration analysis, respectively. Total proteins were extracted from sperm using the Mammalian Protein Extraction Reagent (M-PER; Thermo Fisher Scientific, Waltham, MA, USA), according to manufacturer’s instructions. After protein transfer to nitrocellulose blotting membranes (GE Healthcare Life Sciences, Chicago, IL, USA), these were incubated overnight at 4 °C with a rabbit anti-DNP (1:1000; D9656, Sigma-Aldrich), or with a rabbit anti-nitro-tyrosine (1:1000; #9691, Cell Signaling Technology, Leiden, Netherlands). In each membrane, there was a negative control. Posteriorly, all membranes were conjugated with a goat anti-rabbit IgG-alkaline phosphatase antibody (1:5000; sc-2007, Santa Cruz Biotechnology, Heidelberg, Germany). The membranes were then revealed with ECF substrate (GE Healthcare) using a ChemiDoc XRS+ System (Bio-Rad). The densities of each band were quantified using the Image Lab software, version 5.2 (Bio-Rad). A single experienced researcher (A.C.L.) performed these experiments.
Statistical analysis
Shapiro-Wilk test was used to evaluate the normality of the data. Different normalization transformations were tested according to data distribution, including negative reciprocal, logarithm, square root, cube root, fourth root, and arcsine. For correlation analysis between relative STL and the different sperm parameters analyzed, Pearson (rp) or Spearman rank (rs) correlation coefficients were used, depending on the distribution of the variables. Correlation coefficients were interpreted according to the rule of thumb [52]. Comparisons of relative STL among different groups were addressed with t test, Mann-Whitney U test, chi-square test, Kruskal-Wallis test, or one-way ANOVA, as appropriate. Multiple comparison Dunn’s post hoc test, adjusted for sequential Bonferroni significance, was performed in several-sample tests. Multivariable linear regressions were performed to analyze the association between relative STL and fertilization, embryo cleavage, high-quality embryos, embryo fragmentation, blastocyst, and implantation rates, which were considered dependent variables. Relative STL, as well as TSC, male and female ages, anti-Müllerian hormone (AMH) levels, and number of mature oocytes (MII) were included as independent variables to all models, except for implantation rate that was adjusted for relative STL, TSC, male and female ages, number of MII, number of embryos transferred, and day of embryo transfer. Multivariable logistic regressions were used to investigate the predictability of relative STL on biochemical, clinical, and ongoing pregnancy rates (dependent variables). For normalization in logistic regressions, we also used relative STL, TSC, male and female ages, number of MII, number of embryos transferred, and day of embryo transfer as covariables. All statistical analysis was performed in the totality of patients, and in NZ and non-NZ patients. Statistical tests were carried out using the Past 3 software, version 3.20 [53], except for chi-square test, multivariable linear regression, and multivariable logistic regression, which were performed using the SPSS Statistics software, version 25 (IBM Corp, Foster City, CA, USA). Graphics were obtained using the GraphPad Prism software, version 6.01 (San Diego, CA, USA). P value (P) < 0.05 was considered statistically significant.
Results
Characterization of patients and samples
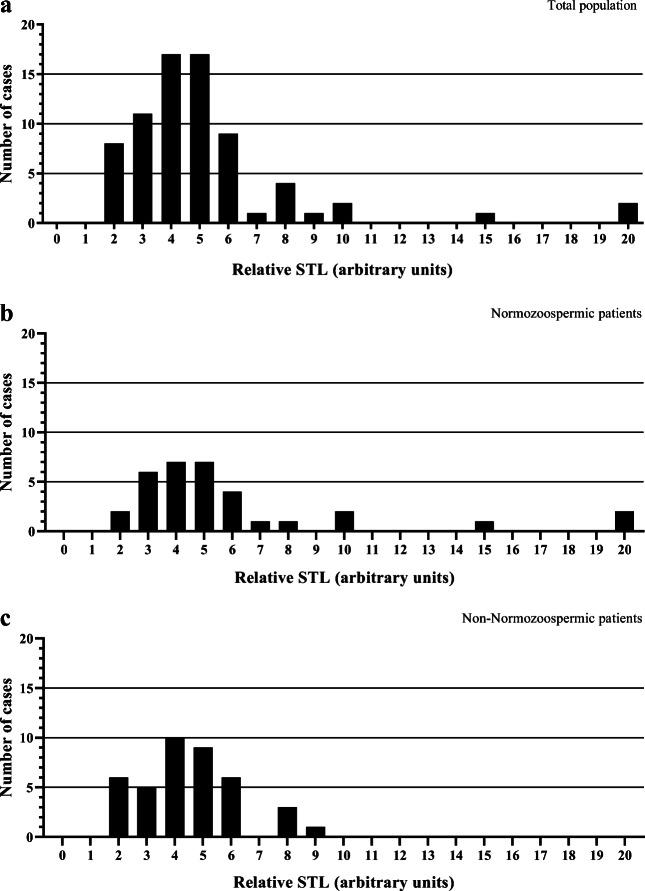
The measurement of relative STL was performed in 73 of the total 78 patients, as five samples provided insufficient DNA for analysis. Relative STL in the total population presented a logarithmic distribution (Shapiro-Wilk, P < 0.001) and revealed high inter-individual variations, with the distribution of 5.22 ± 3.26 (1.65–19.67), as shown in Fig. 1a. Three STL values were identified as outliers (14.69, 19.60, and 19.67), without which the distribution of the population was 4.68 ± 1.87 (1.65–10.46). No biological reason would justify excluding these samples from the statistical analysis. Nonetheless, to address their influence on the statistical results, all statistical tests were repeated with these three samples excluded. All figures and tables contain data with outliers. Only the cases in which the analysis without outliers gave a result of different statistical significance are going to be referred.
Fig. 1.
Frequency distribution histograms of relative sperm telomere length (STL) in the study population. a Total population (n = 73), with a relative STL of 5.22 ± 3.26 (1.65–19.67). b Normozoospermic patients (n = 33), with relative STL of 6.10 ± 4.36 (1.80–19.67). c Non-normozoospermic patients (n = 40), with relative STL of 4.54 ± 1.74 (1.65–8.87)
The demographic and stimulation characteristics of the patients studied are described in Table 1. The clinical characteristics and sperm quality parameters of the totality of patients, and of NZ and non-NZ patients are listed in Supplementary Table 1. The semen parameters analyzed included the following: total sperm count (TSC), sperm concentration (Conc), total motility (TM), progressive motility (PM) and normal morphology (NM). According to WHO guidelines [30], patients were classified in the following clinical groups: NZ (Conc: ≥ 15 × 106/mL + PM: ≥ 32% + NM: ≥ 4%); oligozoospermic (Conc: < 15 × 106/mL); asthenozoospermic (PM: < 32%); and teratozoospermic (NM: < 4%). We considered the oligozoospermic, asthenozoospermic, and teratozoospermic patients together with the non-NZ group. Figure 1b and c represent the distribution of relative STL in NZ and non-NZ patients, respectively. The comparison of the relative STL of NZ patients with the relative STL of the remaining clinical groups did not reveal statistically significant differences (Supplementary Table 2).
Table 1.
Demographic and stimulation characteristics of the 78 couples undergoing infertility treatments
| Parameter | Values |
|---|---|
| Age | |
| Female age (n = 78) (years) | 38.3 ± 4.3 (27–49) |
| Male age (n = 78) (years) | 39.3 ± 4.1 (29–48) |
| Karyotype | |
| Normal male karyotype rate (%) | 68/71 (95.8) |
| Abnormal male karyotype rate (%)a | 3/71 (4.2) |
| Normal female karyotype rate (%) | 69/72 (95.8) |
| Abnormal female karyotype rate (%)b | 3/72 (4.2) |
| Infertility | |
| Time of infertility (n = 77) (years) | 2.8 ± 2.1 (1–13) |
| Male factor only rate (%) | 31/78 (39.7) |
| Female factor only rate (%) | 32/78 (41.0) |
| Mixed factors-male + female-rate (%) | 6/78 (7.7) |
| Mixed factors-female + female-rate (%) | 9/78 (11.5) |
| Stimulation | |
| bFSH (n = 73) (mIU/mL) | 9.2 ± 4.9 (2.4–31.5) |
| AMH (n = 72) (ng/mL) | 2.4 ± 2.2 (0.1–13.5) |
| Follicle number (n = 77) | 9.5 ± 4.5 (1–20) |
| GnRH antagonist rate (%) | 75/78 (96.2) |
| GnRH agonist rate (%) | 3/78 (3.8) |
| Total GnRH dose (n = 78) (IU/mL) | 2647.1 ± 1239.2 (11–7250) |
| rFSH rate (%) | 31/78 (39.7) |
| rFSH + HMG rate (%) | 33/78 (42.3) |
| rFSH + rLH rate (%) | 12/78 (15.4) |
| HMG rate (%) | 2/78 (2.6) |
| Time of stimulation (n = 78) (days) | 9.0 ± 1.9 (1–13) |
| Estradiol (n = 74) (pg/mL) | 1596.8 ± 930.6 (229.3–4725.8) |
| Ovulation trigger | |
| rHCG rate (%) | 30/78 (38.5) |
| GnRH agonist rate (%) | 23/78 (29.5) |
| Dual trigger rate (rHCG + agonist) (%) | 25/78 (32.1) |
bFSH, baseline follicle-stimulating hormone; rFSH, recombinant FSH; AMH, anti-Müllerian hormone; GnRH, gonadotropin-releasing hormone; HMG, human menopausal gonadotropin; rLH, recombinant luteinizing hormone; rHCG, recombinant human choriogonadotropin
Values in n (rate); mean ± standard deviation (range)
aAbnormal male karyotypes 46Xinv(Y)(p11,2q11,22); 46XY,inv(3)(p26.1q13.1); 47XYY
bAbnormal female karyotypes 45X(3)/47XXX(2)/46XX(53); 46XX,t(10;12); 45X(3)46XX(55)
Relation of STL with demographic and sperm quality parameters
Supplementary Figure 1 exemplifies the observations obtained in the TUNEL, AB, and FISH techniques. The correlations obtained between relative STL and the demographic characteristics of the patients and their sperm quality parameters, for the total population and within the different clinical groups, are listed in Table 2. No correlation was found between relative STL and TM, NM, time of infertility, sperm chromosomic alterations, and sperm oxidative profile.
Table 2.
Correlation of relative sperm telomere length with time of infertility, male age, semen parameters, sperm DNA fragmentation, chromatin maturity, chromosomic alterations, and oxidative profile, within different clinical groups according to the spermiogram of patients
| Parameter | Population | n | rp | rs | P value |
|---|---|---|---|---|---|
| Time of infertility (years) | |||||
| Total | 72 | 0.071 | 0.556 | ||
| Normozoospermic | 33 | 0.329 | 0.061 | ||
| Non-normozoospermic | 39 | − 0.141 | 0.391 | ||
| Oligozoospermic | 11 | − 0.065 | 0.850 | ||
| Asthenozoospermic | 9 | − 0.055 | 0.909 | ||
| Teratozoospermic | 31 | − 0.152 | 0.413 | ||
| Male age (years) | |||||
| Total | 73 | 0.030 | 0.821 | ||
| Normozoospermic | 33 | 0.114 | 0.526 | ||
| Non-normozoospermic | 40 | − 0.085 | 0.600 | ||
| Oligozoospermic | 11 | − 0.328 | 0.326 | ||
| Asthenozoospermic | 9 | − 0.939 | 0.0002 | ||
| Teratozoospermic | 32 | − 0.265 | 0.143 | ||
| Semen parameters | |||||
| Total count (× 106/ejaculate) | |||||
| Total | 73 | 0.064 | 0.590 | ||
| Normozoospermic | 33 | 0.361 | 0.039 | ||
| Non-normozoospermic | 40 | − 0.329 | 0.038 | ||
| Oligozoospermic | 11 | − 0.42 | 0.199 | ||
| Asthenozoospermic | 9 | − 0.409 | 0.275 | ||
| Teratozoospermic | 32 | − 0.217 | 0.234 | ||
| Concentration (× 106/mL) | |||||
| Total | 73 | 0.006 | 0.957 | ||
| Normozoospermic | 33 | 0.188 | 0.294 | ||
| Non-normozoospermic | 40 | − 0.324 | 0.041 | ||
| Oligozoospermic | 11 | − 0.574 | 0.065 | ||
| Asthenozoospermic | 9 | − 0.428 | 0.251 | ||
| Teratozoospermic | 32 | − 0.288 | 0.110 | ||
| Total motility (%) | |||||
| Total | 73 | − 0.110 | 0.354 | ||
| Normozoospermic | 33 | − 0.176 | 0.328 | ||
| Non-normozoospermic | 40 | − 0.169 | 0.297 | ||
| Oligozoospermic | 11 | − 0.252 | 0.454 | ||
| Asthenozoospermic | 9 | − 0.265 | 0.491 | ||
| Teratozoospermic | 32 | − 0.299 | 0.097 | ||
| Progressive motility (%) | |||||
| Total | 73 | − 0.040 | 0.736 | ||
| Normozoospermic | 33 | − 0.081 | 0.653 | ||
| Non-normozoospermic | 40 | − 0.303 | 0.057 | ||
| Oligozoospermic | 11 | − 0.38 | 0.249 | ||
| Asthenozoospermic | 9 | − 0.894 | 0.003 | ||
| Teratozoospermic | 32 | − 0.393 | 0.026 | ||
| Normal morphology (%) | |||||
| Total | 71 | 0.165 | 0.169 | ||
| Normozoospermic | 33 | − 0.053 | 0.768 | ||
| Non-normozoospermic | 38 | 0.271 | 0.100 | ||
| Oligozoospermic | 11 | − 0.092 | 0.788 | ||
| Asthenozoospermic | 9 | − 0.038 | 0.924 | ||
| Teratozoospermic | 32 | 0.189 | 0.300 | ||
| Sperm DNA fragmentation (%) | |||||
| Total | 38 | − 0.045 | 0.788 | ||
| Normozoospermic | 22 | 0.025 | 0.913 | ||
| Non-normozoospermica | 16 | 0.052 | 0.850 | ||
| Staining patterns | |||||
| H (%) | |||||
| Total | 38 | 0.348 | 0.032 | ||
| Normozoospermic | 22 | 0.412 | 0.057 | ||
| Non-normozoospermica | 16 | 0.300 | 0.260 | ||
| AVR (%) | |||||
| Total | 38 | 0.05 | 0.763 | ||
| Normozoospermic | 22 | 0.054 | 0.811 | ||
| Non-normozoospermica | 16 | 0.160 | 0.553 | ||
| ER (%) | |||||
| Total | 38 | − 0.25 | 0.137 | ||
| Normozoospermic | 22 | − 0.251 | 0.260 | ||
| Non-normozoospermica | 16 | − 0.208 | 0.440 | ||
| PAR (%) | |||||
| Total | 38 | − 0.371 | 0.022 | ||
| Normozoospermic | 22 | − 0.464 | 0.030 | ||
| Non-normozoospermica | 16 | − 0.270 | 0.312 | ||
| Sperm chromatin maturity (%) | |||||
| Total | 40 | − 0.197 | 0.223 | ||
| Normozoospermic | 22 | − 0.027 | 0.905 | ||
| Non-normozoospermica | 18 | − 0.683 | 0.002 | ||
| Sperm chromosomic alterations | |||||
| Euploidy rate (%) | Total | 43 | − 0.108 | 0.490 | |
| Diploidy rate (%) | Total | 6 | 0.015 | 0.924 | |
| Aneuploidy rate (%) | |||||
| Total | 43 | 0.507 | 0.333 | ||
| Normozoospermic | 25 | 0.115 | 0.584 | ||
| Non-normozoospermica | 18 | − 0.046 | 0.855 | ||
| Sperm oxidative profile | |||||
| Protein carbonylation (protein expression) | |||||
| Total | 46 | − 0.003 | 0.983 | ||
| Normozoospermic | 25 | − 0.352 | 0.084 | ||
| Non-normozoospermica | 21 | 0.118 | 0.611 | ||
| Protein nitration (protein expression) | |||||
| Total | 45 | 0.040 | 0.794 | ||
| Normozoospermic | 25 | − 0.066 | 0.753 | ||
| Non-normozoospermica | 20 | 0.225 | 0.340 | ||
H, head; AVR, acrosomal vesicle region; ER, equatorial region; PAR, post-acrosomal region
rs, Spearman’s rank correlation coefficient; rp, Pearson’s correlation coefficient
P < 0.05 is presented in italics
aInsufficient n for further differentiation in oligozoospermic, asthenozoospermic and teratozoospermic patients
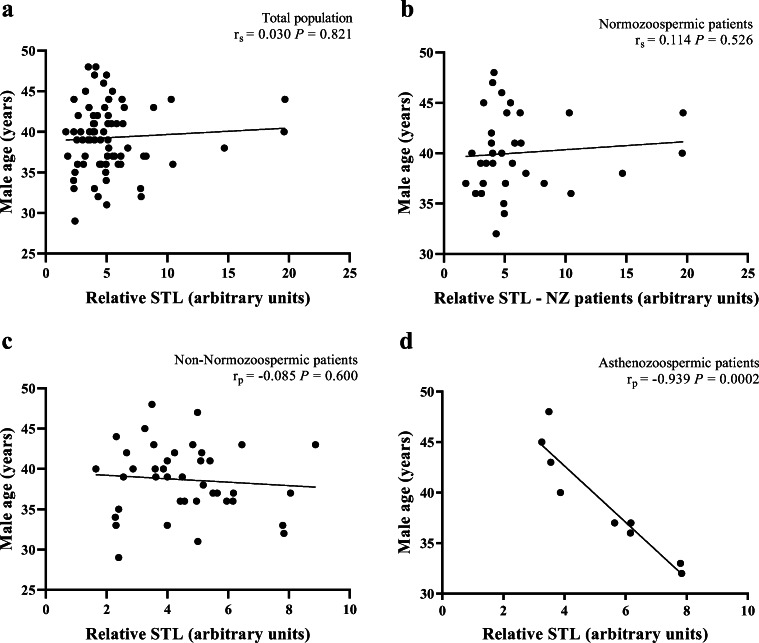
To investigate a possible effect of age on STL we compared, in the total population, the relative STL between males aged < 35 and ≥ 35 years, a cut-off determined in previous studies [54], but no statistically significant difference was found (Mann-Whitney test; P = 0.857). No significant correlations were also obtained between relative STL and male age in the totality of patients (r = 0.030; P = 0.821) (Fig. 2a), in NZ patients (r = 0.114; P = 0.526) (Fig. 2b) or in non-NZ patients (r = − 0.085; P = 0.600) (Fig. 2c). However, when dividing non-NZ patients into subgroups, we observed a significant negative correlation between relative STL and male age only in patients with asthenozoospermia (r = − 0.939; P = 0.0002) (Fig. 2d).
Fig. 2.
Correlations between relative sperm telomere length (STL) and male age. a Absence of a significant correlation in the total population (n = 73). b Absence of a significant correlation in normozoospermic patients (n = 33). c Absence of a significant correlation in non-normozoospermic patients (n = 40). d A significant correlation in asthenozoospermic patients (n = 9). rp, Pearson’s correlation coefficient; rs, Spearman’s rank correlation coefficient; P, P value
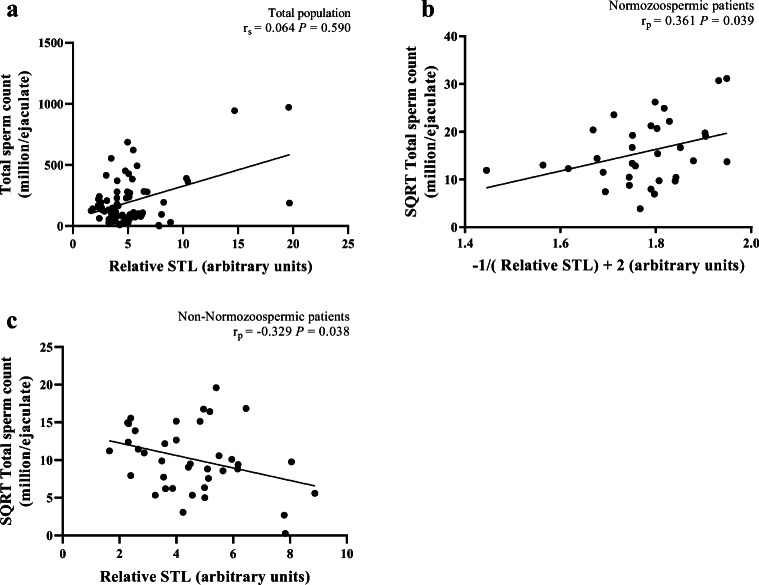
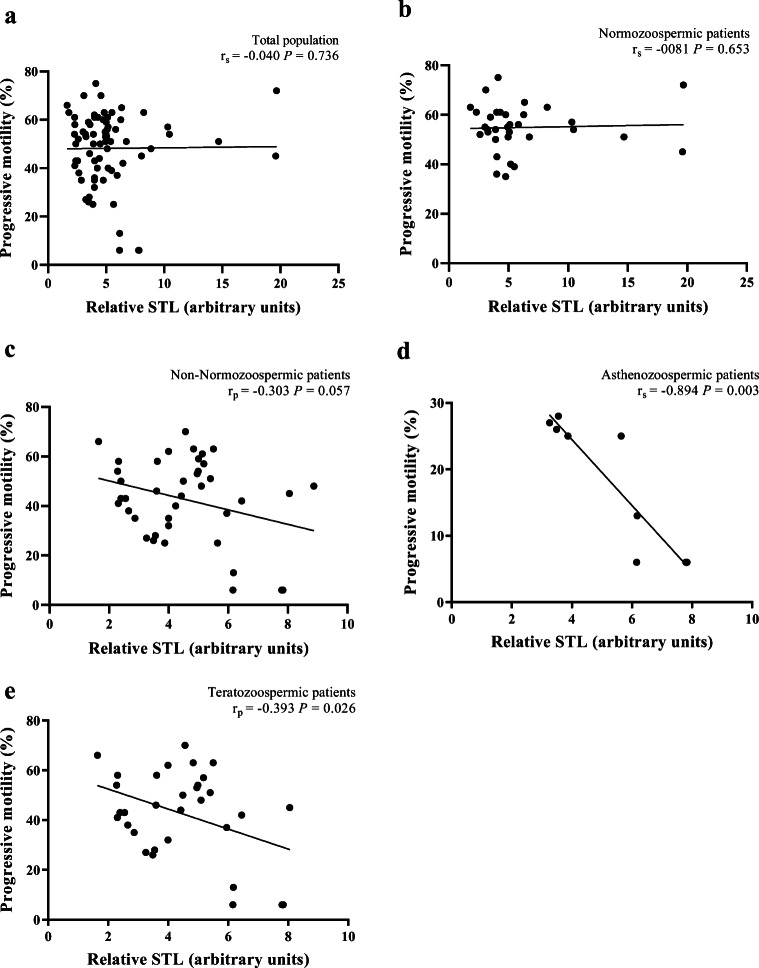
There was no significant correlation between relative STL and TSC in the totality of patients (r = 0.064; P = 0.590) (Fig. 3a), but significant correlations were found in both NZ (r = 0.361, P = 0.039) (Fig. 3b) and non-NZ patients (r = − 0.329, P = 0.038) (Fig. 3c). However, the removal of the outliers from the statistical analysis abolished the significance of the correlation found in NZ patients (r = 0.192, P = 0.309). Relative STL was not also significantly correlated with PM in the totality of patients (r = − 0.040; P = 0.736) (Fig. 4a), in NZ patients (r = − 0.081; P = 0.653) (Fig. 4b) or in non-NZ patients (r = − 0.303; P = 0.057) (Fig. 4c). However, significant correlations were found in patients with asthenozoospermia (r = − 0.894, P = 0.003) (Fig. 4d) and teratozoospermia (r = − 0.393, P = 0.026) (Fig. 4e).
Fig. 3.
Correlations between relative sperm telomere length (STL) and total sperm count. a Absence of a significant correlation in the total population (n = 73). b A significant correlation in normozoospermic patients (n = 33). c A significant correlation in non-normozoospermic patients (n = 40). SQRT, square root.; rp, Pearson’s correlation coefficient; rs, Spearman’s rank correlation coefficient; P, P value
Fig. 4.
Correlations between relative sperm telomere length (STL) and progressive motility. a Absence of a significant correlation in the total population (n = 73). b Absence of a significant correlation in normozoospermic patients (n = 33). c Absence of a significant correlation in non-normozoospermic patients (n = 40). d A significant correlation in asthenozoospermic patients (n = 9). e A significant correlation in teratozoospermic patients (n = 32). rp, Pearson’s correlation coefficient; rs, Spearman’s rank correlation coefficient; P, P value
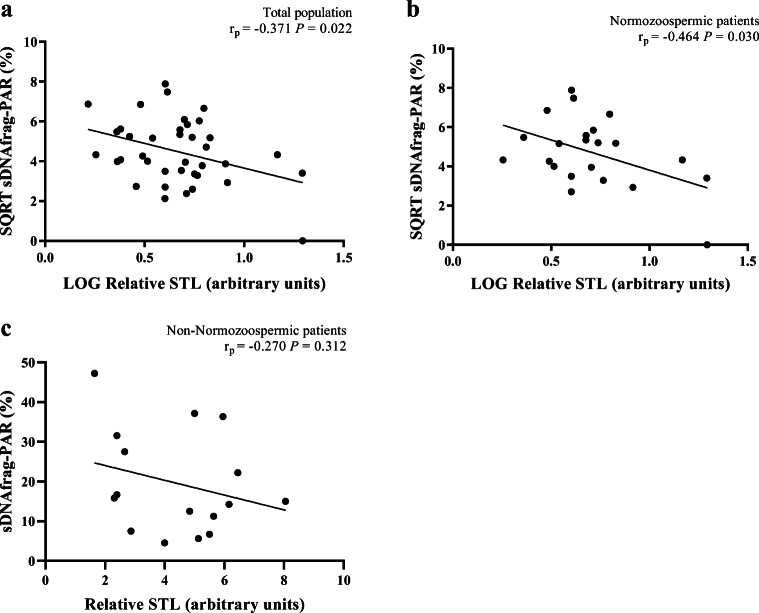
In the matter of sDNAfrag, relative STL was significantly negatively correlated with sDNAfrag in the post-acrosomal region (PAR) pattern, in the totality of patients (r = − 0.371, P = 0.022) (Fig. 5a), in NZ patients (r = − 0.464, P = 0.030) (Fig. 5b), but not in non-NZ patients (r = − 0.270, P = 0.312) (Fig. 5c). Relative STL was also significantly positively correlated with sDNAfrag in the head (H) pattern, for the totality of patients (r = 0.348, P = 0.032). However, since the positive correlation between relative STL and sDNAfrag in the H pattern was not observed either in the NZ or in the non-NZ group, the positive correlation observed in the total population may not be clinically relevant. No correlations were found in the remaining sDNAfrag patterns. The removal of the outliers from the statistical analysis abolished the significance from all three sDNAfrag correlations with relative STL, in the totality of patients: H pattern (r = 0.164, P = 0.347) and PAR pattern (r = − 0.186, P = 0.286); and in NZ patients: PAR pattern (r = − 0.130, P = 0.596).
Fig. 5.
Correlations between relative sperm telomere length (STL) and sperm DNA fragmentation (sDNAfrag) in the post-acrosomal region (PAR). a A significant correlation in the total population (n = 38). b A significant correlation in normozoospermic patients (n = 22). c Absence of a significant correlation in non-normozoospermic patients (n = 16). SQRT, square root; LOG, logarithm; rp, Pearson’s correlation coefficient; P, P value
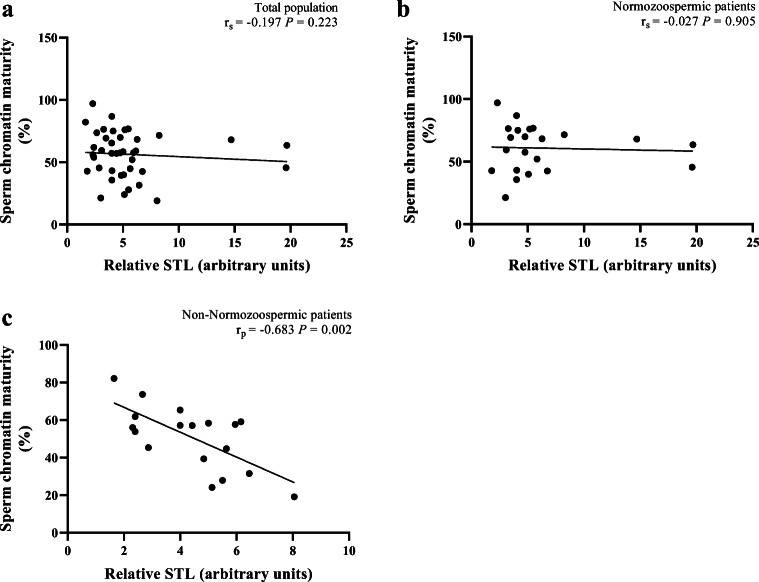
For sperm chromatin maturity, there were no significant correlations with relative STL in the totality of patients (r = − 0.197; P = 0.223) (Fig. 6a), or in NZ patients (r = − 0.027; P = 0.905) (Fig. 6b), but a significant negative correlation was detected in the non-NZ group (r = − 0.683, P = 0.002) (Fig. 6c).
Fig. 6.
Correlations between relative sperm telomere length (STL) and sperm chromatin maturity. a Absence of a significant correlation in total population (n = 40). b Absence of a significant correlation in normozoospermic patients (n = 22). c A significant correlation in non-normozoospermic patients (n = 18). rp, Pearson’s correlation coefficient; rs, Spearman’s rank correlation coefficient; P, P value
Relation of STL with clinical and embryological outcomes
The clinical and embryological outcomes of the treatment cycles, in the totality of patients, and in NZ and non-NZ patients, are described and compared in Table 3. From the 65 patients with embryo transfer cycles (ETC), 61 had relative STL measurement. These 61 ETC originated 23 clinical pregnancies, 18 ongoing pregnancies, and 17 live-birth deliveries, with 19 newborns (two twins) (Supplementary Fig. 2).
Table 3.
Embryological and clinical outcomes in the total study population, and in normozoospermic and non-normozoospermic patients
| Parameter | Total | Normozoospermic | Non-normozoospermic | P value |
|---|---|---|---|---|
| Patients (n) | 78 | 33 | 45 | - |
| Treatment cycles (n) | 78 | 33 | 45 | |
| IVF cycles (n, rate) | 26/78 (33.3) | 20/33 (60.6) | 6/45 (13.3) | 0.000 |
| ICSI cycles (n, rate) | 52/78 (66.7) | 13/33 (39.4) | 39/45 (86.7) | 0.000 |
| ETC (n) | 65 | 29 | 36 | - |
| ETC (rate) | 65/78 (83.3) | 29/33 (87.9) | 36/45 (80) | 0.356 |
| Canceled cycles (n, rate) | 13/78 (16.7) | 4/33 (12.1) | 9/45 (20) | - |
| COC (n, mean, range) | (621) 8.0 ± 4.6 (1–20) | (277) 8.4 ± 4.6 (1–20) | (344) 7.6 ± 4.7 (1–17) | 0.484 |
| MII (n, mean, range) | (470) 6.0 ± 3.5 (0–15) | (220) 6.6 ± 3.3 (1–12) | (250) 5.6 ± 3.6 (0–15) | 0.168 |
| MR (MII/COC) (n, rate) | 470/621 (75.7) | 220/277 (79.4) | 250/344 (72.7) | 0.051 |
| 2PN/2PB (n, mean, range) | (388) 5.0 ± 3.3 (0–15) | (189) 5.5 ± 3.1 (1–12) | (199) 4.5 ± 3.3 (0–15) | 0.108 |
| FR (2PN/MII) (n, rate) | 388/470 (82.6) | 189/220 (85.9) | 199/250 (79.6) | 0.072 |
| Day 2 embryos (n, mean, range) | (378) 5.0 ± 3.0 (1–12) | (187) 5.7 ± 3.1 (1–12) | (191) 4.5 ± 3.0 (1–12) | 0.114 |
| ECR (D2/2PN) (n, rate) | 378/388 (97.4) | 187/189 (98.9) | 191/199 (96) | 0.066 |
| Day 3 embryos (n, mean, range) | (338) 5.4 ± 2.7 (1–12) | (164) 5.7 ± 2.7 (1–12) | (174) 5.1 ± 2.7 (1–12) | 0.439 |
| AB embryos (n, mean, range) | (332) 5.3 ± 2.7 (1–42) | (160) 5.5 ± 2.6 (1–12) | (172) 5.1 ± 2.8 (1–12) | 0.505 |
| AB rate (AB/D3) (n, rate) | 332/338 (98.2) | 160/164 (97.6) | 172/174 (98.9) | 0.370 |
| Day 4 embryos (n, mean, range) | (268) 5.2 ± 2.4 (1–11) | (126) 5.3 ± 2.1 (2–11) | (142) 5.1 ± 2.7 (1–11) | 0.796 |
| Day 5 embryos (n, mean, range) | (209) 4.3 ± 2.1 (1–11) | (105) 4.6 ± 2 (2–10) | (104) 4.2 ± 2.3 (1–11) | 0.516 |
| BL rate (D5/D2-**) (n, rate) | 157/250 (62.8) | 79/122 (64.8) | 78/128 (60.9) | 0.533 |
| No. TE (n, mean, range) | (97) 1.5 ± 0.5 (1–2) | (47) 1.6 ± 0.5 (1–2) | (50) 1.4 ± 0.5 (1–2) | 0.065 |
| Day of embryo transfer | - | |||
| Day 2 (n, rate) | 12/65 (18.5) | 4/29 (13.8) | 8/36 (22.2) | 0.384 |
| Day 3 (n, rate) | 9/65 (13.8) | 5/29 (17.2) | 4/36 (11.1) | 0.477 |
| Day 4 (n, rate) | 4/65 (6.2) | 1/29 (3.4) | 3/36 (8.3) | 0.415 |
| Day 5 (n, rate) | 40/65 (61.5) | 19/29 (65.5) | 21/36 (58.3) | 0.554 |
| BP (n) | 30 | 16 | 14 | - |
| BP rate (/ETC) (n, rate) | 30/65 (46.2) | 16/29 (55.2) | 14/36 (38.9) | 0.191 |
| Sacs (n) | 27 | 15 | 12 | - |
| IR (sacs/n° TE) (n, rate) | 27/97 (27.8) | 15/47 (31.9) | 12/50 (24) | 0.385 |
| CP (n) | 25 | 13 | 12 | - |
| CP rate (/ETC) (n, rate) | 25/65 (38.5) | 13/29 (44.8) | 12/36 (33.3) | 0.344 |
| Singletons (/CP) (n, rate) | 23/25 (92) | 11/13 (84.6) | 12/12 (100) | 0.157 |
| Twins (/CP) (n, rate) | 2/25 (8) | 2/13 (15.4) | - | 0.157 |
| Abortion (n) | 6 | 2 | 4 | - |
| Abortion rate (/CP) (n, rate) | 6/25 (24) | 2/13 (15.4) | 4/12 (33.3) | 0.294 |
| OP (CP-Abortion) (n) | 19 | 11 | 8 | – |
| OP rate (/ETC) (n, rate)a | 19/65 (29.2) | 11/29 (37.9) | 8/36 (22.2) | 0.166 |
| Singletons (/OP) (n, rate) | 17/19 (89.5) | 9/11 (81.8) | 8/8 (100) | 0.202 |
| Twins (/OP) (n, rate) | 2/19 (10.5) | 2/11 (18.2) | - | 0.202 |
| LBD (n) | 18 | 10 | 8 | - |
| LBDR (/ETC) (n, rate) | 18/65 (27.7) | 10/29 (34.5) | 8/36 (22.2) | 0.272 |
| Singletons (LBD) (n, rate) | 16/18 (88.9) | 8/10 (80) | 8/8 | 0.180 |
| Twins (LBD) (n, rate) | 2/18 (11.1) | 2/10 (20) | - | 0.180 |
IVF, in vitro fertilization; ICSI, intracytoplasmic sperm injection; ETC, embryo transfer cycles; COC, aspirated cumulus-oocyte complexes; MII, mature oocytes, at metaphase II of meiosis; MR, oocyte maturation rate; 2PN/2PB, normally fertilized oocytes, with two pronuclei (PN) and two polar bodies (PB); FR, fertilization rate; ECR, embryo cleavage rate; AB, top embryos at day 3; BL, blastocyst; No. TE, number of transferred embryos; BP, biochemical pregnancy; IR, implantation rate; CP, clinical pregnancy; OP, ongoing pregnancy; LBD, live-birth delivery
aThere were no triplets or ectopic pregnancies
Values in (n), (n, rate), (n, mean ± standard deviation, range)
P values refer to the comparison between the normozoospermic and non-normozoospermic groups
Comparisons of the relative STL between the different pregnancy stages did not reveal statistically significant differences, in the total population (Kruskal-Wallis test, P = 0.257) (Supplementary Fig. 3a), in NZ patients (Kruskal-Wallis test, P = 0.082) (Supplementary Fig. 3b), or in non-NZ patients (One-way ANOVA, P = 0.208) (Supplementary Fig. 3c). However, a crescent ongoing pregnancy rate was observed when dividing the population into three intervals, according to their relative STL: pregnancy rates increased non-significantly from 0.00 to 29.63 and 40.00%, respectively, from the lowest to the highest relative STL interval. This was no longer observed when the population was homogeneously divided into quartiles, in which no pregnancy rate stood out from neither of the quartiles (Supplementary Table 3).
Multivariable regression analysis revealed no influence of relative STL in fertilization, embryo cleavage, AB embryo grade, embryo fragmentation, blastocyst, or implantation rates (Table 4), as it did not for biochemical, clinical, and ongoing pregnancy rates (Table 5) in the totality of patients, or in the NZ or non-NZ groups. As there was only one case with late abortion (singleton pregnancy) from the ongoing pregnancies, no additional statistical test was performed to analyze the influence of relative STL in the live-birth delivery rate.
Table 4.
Multivariable linear regression analysis of fertilization, embryo cleavage, AB embryo, embryo fragmentation, blastocyst and implantation rates on relative sperm telomere length, and independent variables in the total population and in normozoospermic and non-normozoospermic patients
| Population | Dependent variables | Independent variables | Coefficient | 95% confidence interval | P value |
|---|---|---|---|---|---|
| Total | |||||
| Fertilization rate | |||||
| Constant | 0.542 | − 0.184 to 1.269 | 0.141 | ||
| Relative STL | 0.008 | − 0.012 to 0.029 | 0.411 | ||
| TSC | < − 0.000 | 0.000 to 0.000 | 0.965 | ||
| Female age | − 0.002 | − 0.020 to 0.016 | 0.834 | ||
| Male age | 0.004 | − 0.013 to 0.020 | 0.672 | ||
| AMH | 0.005 | − 0.025 to 0.036 | 0.724 | ||
| Number of MII | 0.019 | 0.001 to 0.037 | 0.040 | ||
| Embryo cleavage rate | |||||
| Constant | 0.957 | 0.679 to 1.236 | 0.000 | ||
| Relative STL | 0.000 | − 0.008 to 0.007 | 0.900 | ||
| TSC | < − 0.000 | 0.000 to 0.000 | 0.804 | ||
| Female age | 0.002 | − 0.005 to 0.009 | 0.570 | ||
| Male age | − 0.001 | − 0.007 to 0.006 | 0.838 | ||
| AMH | 0.002 | − 0.010 to 0.013 | 0.789 | ||
| Number of MII | − 0.004 | − 0.011 to 0.003 | 0.252 | ||
| AB embryo rate | |||||
| Constant | 0.946 | 0.767 to 1.125 | 0.000 | ||
| Relative STL | − 0.004 | − 0.009 to 0.001 | 0.123 | ||
| TSC | < 0.000 | 0.000 to 0.000 | 0.033 | ||
| Female age | 0.001 | − 0.003 to 0.006 | 0.621 | ||
| Male age | < 0.000 | − 0.004 to 0.004 | 0.987 | ||
| AMH | − 0.010 | − 0.17 to − 0.002 | 0.011 | ||
| Number of MII | 0.003 | − 0.002 to 0.009 | 0.195 | ||
| Embryo fragmentation rate | |||||
| Constant | 0.069 | − 0.041 to 0.180 | 0.213 | ||
| Relative STL | 0.002 | − 0.001 to 0.005 | 0.136 | ||
| TSC | < − 0.000 | 0.000 to 0.000 | 0.078 | ||
| Female age | 0.000 | − 0.003 to 0.002 | 0.843 | ||
| Male age | − 0.001 | − 0.004 to 0.001 | 0.285 | ||
| AMH | 0.001 | − 0.003 to 0.006 | 0.618 | ||
| Number of MII | − 0.001 | − 0.004 to 0.002 | 0.595 | ||
| Blastocyst rate | |||||
| Constant | 1.758 | 0.758 to 2.758 | 0.001 | ||
| Relative STL | − 0.003 | − 0.037 to 0.030 | 0.836 | ||
| TSC | < − 0.000 | − 0.001 to 0.000 | 0.874 | ||
| Female age | − 0.005 | − 0.028 to 0.018 | 0.647 | ||
| Male age | − 0.012 | − 0.034 to 0.011 | 0.296 | ||
| AMH | − 0.044 | − 0.061 to 0.010 | 0.157 | ||
| Number of MII | − 0.025 | − 0.075 to 0.012 | 0.008 | ||
| Implantation rate | |||||
| Constant | 1.496 | 0.276 to 2.716 | 0.017 | ||
| Relative STL | − 0.011 | − 0.044 to 0.022 | 0.508 | ||
| TSC | 0.000 | 0.000 to 0.001 | 0.146 | ||
| Female age | 0.004 | − 0.021 to 0.030 | 0.735 | ||
| Male age | − 0.035 | − 0.062 to − 0.008 | 0.013 | ||
| Number of MII | − 0.025 | − 0.066 to 0.015 | 0.213 | ||
| Number of embryos transferred | − 0.176 | − 0.399 to 0.047 | 0.120 | ||
| Day of embryo transfer | 0.096 | − 0.011 to 0.203 | 0.077 | ||
| Normozoospermic | |||||
| Fertilization rate | |||||
| Constant | 1.269 | 0.284 to 2.255 | 0.014 | ||
| Relative STL | 0.003 | − 0.017 to 0.022 | 0.782 | ||
| TSC | < 0.000 | 0.000 to 0.000 | 0.887 | ||
| Female age | − 0.002 | − 0.023 to 0.019 | 0.862 | ||
| Male age | − 0.010 | − 0.029 to 0.009 | 0.281 | ||
| AMH | 0.013 | − 0.024 to 0.025 | 0.599 | ||
| Number of MII | 0.000 | − 0.036 to 0.061 | 0.973 | ||
| Embryo cleavage rate | |||||
| Constant | 1.039 | 0.904 to 1.173 | 0.000 | ||
| Relative STL | − 0.001 | − 0.004 to 0.002 | 0.403 | ||
| TSC | < − 0.000 | 0.000 to 0.000 | 0.117 | ||
| Female age | − 0.001 | − 0.004 to 0.002 | 0.581 | ||
| Male age | 0.000 | − 0.002 to 0.003 | 0.844 | ||
| AMH | − 0.003 | − 0.010 to 0.003 | 0.309 | ||
| Number of MII | 0.000 | − 0.003 to 0.004 | 0.901 | ||
| AB embryo rate | |||||
| Constant | 0.848 | 0.577 to 1.119 | 0.000 | ||
| Relative STL | − 0.001 | − 0.006 to 0.004 | 0.609 | ||
| TSC | < 0.000 | 0.000 to 0.000 | 0.224 | ||
| Female age | 0.002 | − 0.004 to 0.007 | 0.511 | ||
| Male age | 0.001 | − 0.004 to 0.007 | 0.587 | ||
| AMH | 0.002 | − 0.011 to 0.015 | 0.704 | ||
| Number of MII | 0.001 | − 0.007 to 0.009 | 0.858 | ||
| Embryo fragmentation rate | |||||
| Constant | 0.126 | − 0.102 to 0.354 | 0.265 | ||
| Relative STL | 0.001 | − 0.003 to 0.006 | 0.500 | ||
| TSC | < − 0.000 | 0.000 to 0.000 | 0.231 | ||
| Female age | − 0.002 | − 0.007 to 0.003 | 0.451 | ||
| Male age | − 0.001 | − 0.005 to 0.003 | 0.638 | ||
| AMH | − 0.003 | − 0.014 to 0.008 | 0.565 | ||
| Number of MII | 0.001 | − 0.005 to 0.007 | 0.745 | ||
| Blastocyst rate | |||||
| Constant | 1.006 | − 0.784 to 2.795 | 0.235 | ||
| Relative STL | − 0.023 | − 0.090 to 0.045 | 0.465 | ||
| TSC | 0.000 | − 0.001 to 0.001 | 0.704 | ||
| Female age | 0.012 | − 0.020 to 0.043 | 0.432 | ||
| Male age | − 0.008 | − 0.037 to 0.022 | 0.558 | ||
| AMH | − 0.017 | − 0.113 to 0.080 | 0.706 | ||
| Number of MII | − 0.042 | − 0.126 to 0.041 | 0.282 | ||
| Implantation rate | |||||
| Constant | 2.298 | 0.206 to 4.390 | 0.033 | ||
| Relative STL | − 0.003 | − 0.050 to 0.044 | 0.904 | ||
| TSC | < 0.000 | − 0.001 to 0.001 | 0.858 | ||
| Female age | − 0.016 | − 0.055 to 0.023 | 0.394 | ||
| Male age | − 0.039 | − 0.084 to 0.006 | 0.084 | ||
| Number of MII | − 0.032 | − 0.105 to 0.040 | 0.363 | ||
| Number of embryos transferred | 0.007 | − 0.408 to 0.422 | 0.973 | ||
| Day of embryo transfer | 0.103 | − 0.092 to 0.299 | 0.284 | ||
| Non-normozoospermic | |||||
| Fertilization rate | |||||
| Constant | 0.170 | − 1.009 to 1.348 | 0.771 | ||
| Relative STL | − 0.012 | − 0.065 to 0.042 | 0.660 | ||
| TSC | − 0.001 | − 0.002 to 0.000 | 0.011 | ||
| Female age | 0.007 | − 0.025 to 0.040 | 0.644 | ||
| Male age | 0.008 | − 0.018 to 0.033 | 0.534 | ||
| AMH | 0.004 | − 0.036 to 0.044 | 0.829 | ||
| Number of MII | 0.031 | 0.004 to 0.057 | 0.024 | ||
| Embryo cleavage rate | |||||
| Constant | 1.032 | 0.423 to 1.641 | 0.002 | ||
| Relative STL | 0.002 | − 0.026 to 0.031 | 0.867 | ||
| TSC | 0.000 | − 0.001 to 0.000 | 0.692 | ||
| Female age | 0.002 | − 0.014 to 0.019 | 0.774 | ||
| Male age | − 0.003 | − 0.016 to 0.010 | 0.644 | ||
| AMH | 0.005 | − 0.016 to 0.025 | 0.652 | ||
| Number of MII | − 0.008 | − 0.022 to 0.006 | 0.245 | ||
| AB embryo rate | |||||
| Constant | 0.983 | 0.661 to 1.305 | 0.000 | ||
| Relative STL | − 0.013 | − 0.028 to 0.003 | 0.097 | ||
| TSC | 0.000 | 0.000 to 0.000 | 0.494 | ||
| Female age | 0.000 | − 0.009 to 0.009 | 0.937 | ||
| Male age | 0.001 | − 0.007 to 0.009 | 0.724 | ||
| AMH | − 0.016 | − 0.27 to − 0.004 | 0.012 | ||
| Number of MII | 0.004 | − 0.004 to 0.012 | 0.341 | ||
| Embryo fragmentation rate | |||||
| Constant | 0.050 | − 0.101 to 0.200 | 0.506 | ||
| Relative STL | 0.007 | 0.000 to 0.014 | 0.049 | ||
| TSC | < − 0.000 | 0.000 to 0.000 | 0.399 | ||
| Female age | 0.001 | − 0.003 to 0.005 | 0.682 | ||
| Male age | − 0.003 | − 0.006 to 0.001 | 0.120 | ||
| AMH | 0.002 | − 0.001 to 0.009 | 0.209 | ||
| Number of MII | 0.004 | − 0.005 to 0.001 | 0.140 | ||
| Blastocyst rate | |||||
| Constant | 3.254 | 1.745 to 4.764 | 0.001 | ||
| Relative STL | − 0.052 | − 0.129 to 0.025 | 0.164 | ||
| TSC | − 0.002 | − 0.004 to 0.000 | 0.027 | ||
| Female age | − 0.032 | − 0.074 to 0.011 | 0.125 | ||
| Male age | − 0.020 | − 0.059 to 0.019 | 0.280 | ||
| AMH | − 0.025 | − 0.073 to 0.022 | 0.157 | ||
| Number of MII | − 0.025 | − 0.061 to 0.011 | 0.264 | ||
| Implantation rate | |||||
| Constant | 0.732 | − 1.426 to 2.890 | 0.490 | ||
| Relative STL | 0.024 | − 0.078 to 0.127 | 0.627 | ||
| TSC | 0.000 | − 0.002 to 0.002 | 0.679 | ||
| Female age | 0.032 | − 0.011 to 0.075 | 0.142 | ||
| Male age | − 0.043 | − 0.086 to 0.000 | 0.048 | ||
| Number of MII | − 0.024 | − 0.081 to 0.034 | 0.405 | ||
| Number of embryos transferred | − 0.366 | − 0.700 to − 0.033 | 0.033 | ||
| Day of embryo transfer | 0.133 | − 0.025 to 0.290 | 0.095 | ||
AB embryo, high-quality embryos; STL, sperm telomere length; TSC, total sperm count; AMH, anti-Müllerian hormone; MII, mature oocytes, at metaphase II of meiosis
Table 5.
Multivariable logistic regression analysis of biochemical, clinical, and ongoing pregnancy rates on relative sperm telomere length and covariables, in the total population, and in normozoospermic and non-normozoospermic patients
| Population | Dependent variables | Covariables | Odds ratio | 95% confidence interval | P value |
|---|---|---|---|---|---|
| Total | |||||
| Biochemical pregnancy rate | |||||
| Constant | 10.385 | 0.517 | |||
| Relative STL | 0.918 | 0.743 to 1.133 | 0.424 | ||
| TSC | 1.003 | 0.999 to 1.006 | 0.142 | ||
| Female age | 1.036 | 0.897 to 1.197 | 0.629 | ||
| Male age | 0.860 | 0.728 to 1.016 | 0.076 | ||
| Number of MII | 0.924 | 0.737 to 1.158 | 0.492 | ||
| Number of embryos transferred | 1.410 | 0.401 to 4.967 | 0.592 | ||
| Day of embryo transfer | 1.631 | 0.853 to 3.117 | 0.139 | ||
| Clinical pregnancy rate | |||||
| Constant | 51.967 | 0.283 | |||
| Relative STL | 0.979 | 0.798 to 1.201 | 0.838 | ||
| TSC | 1.003 | 0.999 to 1.007 | 0.100 | ||
| Female age | 1.019 | 0.895 to 1.174 | 0.789 | ||
| Male age | 0.837 | 0.704 to 0.996 | 0.045 | ||
| Number of MII | 0.881 | 0.699 to 1.109 | 0.281 | ||
| Number of embryos transferred | 0.869 | 0.234 to 3.223 | 0.834 | ||
| Day of embryo transfer | 1.711 | 0.881 to 3.323 | 0.113 | ||
| Ongoing pregnancy rate | |||||
| Constant | 3.878 | 0.706 | |||
| Relative STL | 1.054 | 0.871 to 1.275 | 0.588 | ||
| TSC | 1.001 | 0.998 to 1.004 | 0.554 | ||
| Female age | 0.970 | 0.839 to 1.121 | 0.679 | ||
| Male age | 0.923 | 0.782 to 1.089 | 0.342 | ||
| Number of MII | 0.989 | 0.787 to 1.243 | 0.925 | ||
| Number of embryos transferred | 1.415 | 0.380 to 5.261 | 0.605 | ||
| Day of embryo transfer | 1.309 | 0.675 to 2.538 | 0.425 | ||
| Normozoospermic | |||||
| Biochemical pregnancy rate | |||||
| Constant | 2380.012 | 0.248 | |||
| Relative STL | 0.858 | 0.606 to 1.216 | 0.390 | ||
| TSC | 1.0004 | 0.998 to 1.010 | 0.182 | ||
| Female age | 0.873 | 0.694 to 1.098 | 0.246 | ||
| Male age | 0.904 | 0.704 to 1.160 | 0.427 | ||
| Number of MII | 1.124 | 0.743 to 1.699 | 0.581 | ||
| Number of embryos transferred | 1.541 | 0.167 to 14.246 | 0.703 | ||
| Day of embryo transfer | 1.056 | 0.367 to 3.038 | 0.919 | ||
| Clinical pregnancy rate | |||||
| Constant | 1171.566 | 0.237 | |||
| Relative STL | 1.017 | 0.764 to 1.355 | 0.907 | ||
| TSC | 1.001 | 0.996 to 1.007 | 0.613 | ||
| Female age | 0.914 | 0.742 to 1.125 | 0.394 | ||
| Male age | 0.841 | 0.649 to 1.089 | 0.188 | ||
| Number of MII | 0.843 | 0.562 to 1.265 | 0.410 | ||
| Number of embryos transferred | 2.181 | 0.217 to 21.915 | 0.508 | ||
| Day of embryo transfer | 1.817 | 0.589 to 5.603 | 0.299 | ||
| Ongoing pregnancy rate | |||||
| Constant | 153,275.158 | 0.082 | |||
| Relative STL | 1.071 | 0.788 to 1.445 | 0.663 | ||
| TSC | 0.999 | 0.993 to 1.005 | 0.841 | ||
| Female age | 0.804 | 0.612 to 1.055 | 0.115 | ||
| Male age | 0.830 | 0.624 to 1.105 | 0.202 | ||
| Number of MII | 0.879 | 0.578 to 1.337 | 0.547 | ||
| Number of embryos transferred | 5.267 | 0.454 to 61.073 | 0.184 | ||
| Day of embryo transfer | 1.394 | 0.437 to 4.448 | 0.574 | ||
| Non-normozoospermic | |||||
| Biochemical pregnancy rate | |||||
| Constant | 2.515 | 0.897 | |||
| Relative STL | 0.946 | 0.525 to 1.703 | 0.853 | ||
| TSC | 1.512 | 0.983 to 1.007 | 0.402 | ||
| Female age | 0.634 | 0.989 to 2.313 | 0.056 | ||
| Male age | 0.877 | 0.417 to 0.962 | 0.032 | ||
| Number of MII | 0.723 | 0.630 to 1.221 | 0.437 | ||
| Number of embryos transferred | 2.120 | 0.093 to 5.646 | 0.757 | ||
| Day of embryo transfer | 2.515 | 0.722 to 6.224 | 0.172 | ||
| Clinical pregnancy rate | |||||
| Constant | 13.049 | 0.699 | |||
| Relative STL | 1.099 | 0.607 to 1.990 | 0.755 | ||
| TSC | 1.002 | 0.990 to 1.013 | 0.791 | ||
| Female age | 1.243 | 0.927 to 1.666 | 0.147 | ||
| Male age | 0.721 | 0.517 to 1.006 | 0.054 | ||
| Number of MII | 0.908 | 0.661 to 1.249 | 0.555 | ||
| Number of embryos transferred | 0.349 | 0.047 to 2.559 | 0.300 | ||
| Day of embryo transfer | 1.819 | 0.677 to 4.886 | 0.235 | ||
| Ongoing pregnancy rate | |||||
| Constant | 0.000 | 0.290 | |||
| Relative STL | 1.518 | 0.671 to 3.436 | 0.317 | ||
| TSC | 0.988 | 0.969 to 1.007 | 0.219 | ||
| Female age | 1.291 | 0.915 to 1.820 | 0.146 | ||
| Male age | 0.941 | 0.677 to 1.308 | 0.718 | ||
| Number of MII | 1.193 | 0.791 to 1.799 | 0.401 | ||
| Number of embryos transferred | 0.194 | 0.014 to 2.717 | 0.223 | ||
| Day of embryo transfer | 1.484 | 0.462 to 4.770 | 0.507 | ||
STL, sperm telomere length; TSC, total sperm count; AMH, anti-Müllerian hormone; MII, mature oocytes, at metaphase II of meiosis
Discussion
It is widely known that sperm quality plays an essential role in determining the success rate of ART treatments, which has encouraged research studies concerning the efficiency of sperm selection methods. The DGC-swim-up procedure selects a high-quality sperm population, being routinely used in infertility clinics for insemination. In this study, we deliberately selected sperm from DGC-swim-up, in order to ascertain if STL is related with sperm quality and treatment outcomes in this high-quality pre-selected sperm population.
The originality of this study also resided on the separate analysis of different clinical groups according to the spermiogram of patients, to understand their influence on STL associations with sperm quality and treatment outcomes. This study involved a broad panoply of parameters analyzed and robust statistics. We provide a complete analysis of sperm quality, including standard semen parameters, sDNAfrag, chromatin maturity, chromosomic alterations, and oxidative profile. In addition, we included sDNAfrag staining patterns to better characterize the relation of STL with sDNAfrag. A full characterization of the embryological and clinical outcomes is also provided, including the rates of fertilization; embryo cleavage; embryo quality; blastocyst formation; implantation; abortion; biochemical, clinical, and ongoing pregnancies; and live-birth deliveries. The results only intend to demonstrate the population study, since the total number of cycles is too low for comprehensive studies.
The technique used for STL measurements, Q-PCR, reflects the differences in average TL of different cell samples and it has been adopted by several research studies to measure relative STL. Its main advantage is the requirement of small amounts of DNA, an aspect particularly important in the analysis of sperm, which has low DNA yield. Q-PCR is faster and cheaper and has higher-throughput in comparison with other technologies that measure TL. However, TL measured by Q-PCR is not comparable between studies, due to inter-assay variation and deviations introduced by differential DNA extraction methods or amplification efficiency. In addition, Q-PCR only provides one measurement of TL in a given population of cells, not allowing the differentiation of critically short or long telomeres in the same sample. Techniques such as Q-FISH or flow cytometry FISH may be considered more appropriate methods to determine TL.
We obtained a logarithmic distribution of relative STL, indicating that sperm samples with longer telomeres are under-represented in the population, which has been previously demonstrated [55]. We also showed that the longest STL values belonged to the NZ patients (Fig. 1).
In the totality of patients, we found no significant correlations between relative STL and semen parameters, global sDNAfrag, sperm chromatin maturity, aneuploidy rate, or oxidative profile, which emphasizes the efficacy of the sperm purification method used in selecting a high-quality sperm population to be used in ART treatments. However, the analysis of non-NZ subgroups revealed significant negative correlations between relative STL and some semen parameters (TSC, Conc, PM). Yet, as these subgroups presented a low number of cases, results cannot be taken into deep consideration until confirmed by additional experiments. Nevertheless, if confirmed, the contradictory results found in the literature about the relationships between STL and semen parameters could be explained by our findings, as they evidence that results can be dependent on the heterogeneity of characteristics in the population under analysis, emphasizing the need for a detailed description of the studied population.
Analysis of testicular spreads revealed that altered telomerase function in primary spermatocytes of azoospermic patients could lead to meiotic recombination errors and germ cell apoptosis, causing severe decrease in sperm count and thus non-obstructive azoospermia [56]. Analysis of the ejaculate also showed a significant decrease in sperm concentration in association with an increase in the apoptotic index [57]. However, this was only observed in NZ samples, and the authors suggested that the absence of correlations in low-quality sperm samples could be justified by the presence of a disrupted regulation of apoptosis [57]. This could explain why we only found a significant positive correlation between STL and TSC in NZ patients. However, this correlation was abolished with the removal of the outliers, indicating the need for further confirmation by increasing the sample size.
A previous study demonstrated a significant negative correlation between sDNAfrag and relative STL in sperm of NZ patients after Percoll gradient centrifugation [21]. Although we did not find this correlation in the totality of the population or within the clinical groups, we found for the first time a negative significant correlation of relative STL with the sDNAfrag-PAR pattern. The majority of sperm with DNA fragmentation showed a predominance of the H pattern, followed by PAR, which is in line with a previous study demonstrating that PAR is the only pattern that increased from raw semen to swim-up sperm, in which it was the second most frequent sDNAfrag region [38]. It was suggested that since PAR is a gene-poor region, with lower content of protamines, it has a higher susceptibility to oxidative damage than the other regions of the sperm head, which have more compact chromatin. Although we did not find a correlation between relative STL and sperm oxidative profile, one study evaluating the superoxide content in sperm purified by gradient centrifugation or swim-up (n = 150) reported a negative significant correlation between STL and ROS production [22]. Furthermore, when subdividing sDNAfrag-PAR into clinical groups, we detected a significant negative correlation in the NZ group, but not in the non-NZ group, which suggests that in pathological groups the high levels of sDNAfrag may reflect apoptotic events that are independent of STL. This result emphasizes the importance of quantifying the different staining patterns when performing sDNAfrag analysis. Nonetheless, relative STL correlations with sDNAfrag-PAR were lost when removing the outliers from the analysis, meaning that further studies are necessary to confirm these observations.
In the non-NZ group of patients, unexpected results were obtained. We observed significant negative correlations of relative STL with TSC and sperm Conc. Significant negative correlations of relative STL were also observed with sperm PM in asthenozoospermic and teratozoospermic patients. Since STL has been shown to increase from raw semen to selected sperm by the swim-up procedure [22, 27], suggesting a natural selection of highly motile sperm with longer telomeres, it would also be expected in our results a positive correlation between STL and PM. Nevertheless, our surprising results are in line with a recent study that also reported negative correlations of STL from swim-up sperm with sperm Conc and PM [23]. In addition, we did not find a positive correlation between relative STL and chromatin maturity as expected. In a previous study, chromatin maturity was shown to be positively related with STL in sperm of NZ patients after Percoll gradient centrifugation, suggesting that a lower chromatin condensation increases STL susceptibility to damage and shortening [21]. Instead, we found a significant negative correlation between relative STL and chromatin maturity, in non-NZ patients. It seems that sperm from non-NZ patients have an alteration of the telomere length regulation mechanisms described to date in sperm. Thus, additional research is necessary to clarify this phenomenon.
Although telomeres are vital for the maintenance of genomic stability, no correlation was found between relative STL average values and percentage of chromosomic abnormalities in sperm samples, in agreement with a previous report [20]. This may be explained by the regulatory mechanism in spermatocytes believed to promote the elimination of aneuploid spermatocytes [58, 59].
Few studies have addressed the association between STL and embryological and clinical outcomes. Our results showed no relation between these parameters, which is in line with previous studies performed in sperm obtained after swim-up from 60 donor samples [25], and in frozen-thaw sperm obtained from raw semen of 50 patients [14]. Other study performed in sperm obtained after gradient-swim-up from 418 patients did not also find a relation between STL and clinical outcomes but revealed a positive association between STL and good quality embryo rate [60]. Nonetheless, it seems plausible that the relation of STL with embryo quality does not reflect in clinical outcomes because only the high-quality embryos are deliberately selected for transfer. In a different study, using sperm obtained after gradient centrifugation from 54 patients, the authors determined a relative STL interval in which the OP rate was 35.7%, whereas outside the pregnancy rate was null [20]. From the analysis of relative STL intervals, we observed that, although not significantly, OP rates increased from lower to higher relative STL intervals. However, the population was not evenly distributed between these intervals. When dividing the population homogeneously in intervals, such as in quartiles, we could no longer observe an increase of OP rates with the increase of relative STL.
Because male fertility and sperm quality are known to decrease with age [61, 62], the male age was also a parameter evaluated in our study. We did not observe a significant correlation between relative STL and male age in the totality of patients, or in the NZ and non-NZ patients. This could be explained by our limited age range, having only one patient bellow 30 years and none above 50 years. However, the absence of correlation between STL and male age has been confirmed in previous studies [21, 22, 24]. Other studies have reported a positive correlation between STL and male age; however, even with high population size and large age range, the correlations (r value) obtained were weak or negligible (DGC purified sperm, r = 0.34, P < 0.02, n = 47 [18]; DGC purified sperm, r = 0.1, P = 0.04, n = 418 [60]; raw semen, r = 0.32, P = 0.0002, n = 135 [63]; raw semen, r = 0.32, P = 0.01, n = 63 [64]), which calls into question their biological relevance.
In the present study, we found a significant negative correlation between relative STL and male age only in patients with asthenozoospermia. It appears that in asthenozoospermic patients, relative STL decreases with age, perhaps because the factor negatively affecting PM is also negatively affecting STL and accumulating with age. As DNA damage in the male germline was shown to increase with age, possibly as a result of ROS accumulation [65, 66], and ROS are known to impair semen parameters [67] and sperm motility in particular [68], it is possible that the accumulation of ROS with age could promote both STL shortening and impairment of PM. We could not confirm this hypothesis, as the asthenozoospermic subgroup of patients did not provide enough sperm for the characterization of their oxidative profile.
In conclusion, this is the largest and deepest study on STL evaluated in sperm samples prepared sequentially by gradient centrifugation followed by swim-up, in which a large panoply of molecular techniques and robust statistics were used. Although we could not relate STL with sperm quality in the total study population, we observed significant correlations in different clinical groups of patients. Our study clarifies that different sperm sample characteristics may lead to discrepant results when evaluating the importance of STL in male infertility, as cases with altered spermiogram values may have interfering factors that disassociate STL from sperm quality. This indicates that STL may not be considered a suitable biomarker for sperm quality. As the presented results showed no relation of STL with embryological and clinical outcomes, data also suggest that STL may not be a reliable prognostic biomarker for ART treatments.
The present study, although representative, includes a relatively limited sample size. This sample size may significantly limit the ability to find significance, being a strong limitation of the present study. Thus, as a pilot work, in a future study, the expansion of the sample size for consolidation of results is necessary to validate the statistical results.
Electronic supplementary material
Representative observations of the analysis of sperm samples. (a) TUNEL assay for sperm DNA fragmentation evaluation, considering the different staining patterns. Evaluation was performed in morphologically normal spermatozoa. Upper panel: (1) sperm head; (2) acrosome vesicle region; (3) post-acrosome region; (4) equatorial region. Lower panel: schematic representation of the same images of the upper panel, with the same corresponding letters, to facilitate interpretation. Scale bar = 10 μm. (b) Aniline blue (AB) assay for sperm chromatin maturity evaluation: (1): AB-negative spermatozoon, revealing sperm chromatin fully condensed; (2): AB-positive spermatozoon, revealing sperm immature chromatin condensation. Scale bar = 20 μm. (c) FISH assay for sperm chromosomic numeric alterations evaluation: (1) haploid (X,18); (2) haploid (Y,18); (3) sex chromosome nullisomy (18+); (4) chromosome 18 nullisomy (Y+); (5) chromosome 18 nullisomy (X+); (6) sex chromosome disomy (XX/18); (7) sex chromosome disomy (YY/18); (8) sex chromosome disomy (XY/18); (9) chromosome 18 disomy (X/18,18); (10) chromosome 18 disomy (Y/18,18); (11) sexual and autosomal disomy (XX/18,18); (12) sexual and autosomal disomy (XY/18,18). Color legends: X chromosome (green); Y chromosome (orange); chromosome 18 (aqua-blue); nucleus (DAPI-blue). Scale bar = 1 μm. (DOCX 931 kb)
Frequency distribution histogram of relative STL in the total population (Total), and in cases with embryo transfer cycles (ETC), biochemical pregnancy (BP), clinical pregnancy (CP), ongoing pregnancy (OP) and live-birth delivery (LBD). (DOCX 19 kb)
Comparison of relative sperm telomere length (STL) within clinical outcomes: negative, for no pregnancy; biochemical pregnancy only (BP); clinical pregnancy only (CP) and ongoing pregnancy (OP). Relative STL did not significantly differ among the different pregnancy status. a Total population: Kruskal-Wallis test, P = 0.257. Medians are represented. b Normozoospermic patients: Kruskal-Wallis test, P = 0.082. Medians are represented. c Non-normozoospermic patients: One-way ANOVA, P = 0.208. Means are represented. (DOCX 53 kb)
(DOCX 23 kb)
(DOCX 16 kb)
(DOCX 14 kb)
Acknowledgments
We would like to acknowledge to the team of the IVF clinic: Jorge Beires, MD, and José Manuel Teixeira da Silva, MD, Gynecology, and Obstetrics (for oocyte retrieval); José Correia, MD, Anesthesiology (for anesthesia); Cristiano Oliveira, MD, José Teixeira da Silva, MD, Pedro Xavier, MD, António Couceiro MD, and Sandra Soares, MD, Gynecology and Obstetrics, subspecialty in Reproductive Medicine (for patient evaluation, controlled ovarian hyperstimulation, embryo transfer, and patient follow-up); Joaquina Silva, MD, Mariana Cunha, MSc, and Paulo Viana, MSc, Senior Clinical Embryologists (ESHRE), and Nuno Barros, MSc, Clinical Embryologist (for embryology laboratorial work); Ana Gonçalves, MSc, and Cláudia Osório, MSc (for andrology laboratorial work); Carolina Lemos, PhD (for additional statistical assistance).
Authors’ contributions
A.C.L. was involved in performing molecular biology experimental procedures, acquisition of data, analysis and interpretation of data, and writing of the article. P.F.O. was involved in the supervision of molecular biology experimental procedures, analysis and interpretation of data, and critical approval of the final article. S.P. was involved in performing sperm preparation, semen analysis and embryological work, and critical approval of the final article. C.A. and M.J.P. were involved in performing the determination of sperm aneuploidies and critical approval of the final article. R.S. was involved in performing the determination of sperm chromatin maturity and sperm DNA fragmentation and critical approval of the final article. E.R was involved in the supervision of the statistical work and critical approval of the final article. A.B. was involved in patient recruitment and in the supervision of the IVF laboratory and critical approval of the final article. M.S. was involved in the conception and design of the study, analysis and interpretation of data, and final writing of the article.
Funding information
UMIB (Pest-OE/SAU/UI0215/2014) is funded by the National Funds through FCT-Foundation for Science and Technology.
Compliance with ethical standards
The procedures of the infertility clinic CGR A.Barros are under the determinations of the National Law on Medically Assisted Procreation (Law of 2017) and supervised by the National Council on Medically Assisted Procreation (CNPMA-2018). According to these rules and guidelines, the use of clinic databases and patient biological material for diagnosis and research can be used without further ethical approval, under strict individual anonymity, and after patient written informed consent. Regarding the use of semen samples for laboratorial experimentation at ICBAS-UP, the Ethics Committee authorization number is Project 2019/CE/P017 (266/CETI/ICBAS).
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
A draft of the present manuscript was awarded with the first Prize for the Best Basic Sciences Project by the Portuguese Society of Reproductive Medicine.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ana Catarina Lopes, Email: acv.lopes@campus.fct.unl.pt.
Pedro Fontes Oliveira, Email: pfobox@gmail.com.
Soraia Pinto, Email: soraiapinto.pt@gmail.com.
Carolina Almeida, Email: carola@med.up.pt.
Maria João Pinho, Email: maria.pinho@gmail.com.
Rosália Sá, Email: rmsa@icbas.up.pt.
Eduardo Rocha, Email: erocha@icbas.up.pt.
Alberto Barros, Email: geral@cgrabarros.pt.
Mário Sousa, Email: msousa@icbas.up.pt.
References
- 1.Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;1337. [DOI] [PMC free article] [PubMed]
- 2.Hamada A, Esteves SC, Agarwal A. Unexplained male infertility: potential causes and management. Hum Andrology. 2011;1(1):2–16. [Google Scholar]
- 3.Blackburn EH. Structure and function of telomeres. Nature. 1991;350(6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 4.Scherthan H. Telomeres and meiosis in health and disease. Cell Mol Life Sci. 2007;64(2):117–124. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klutstein M, Fennell A, Fernández-Álvarez A, Cooper JP. The telomere bouquet regulates meiotic centromere assembly. Nat Cell Biol. 2015;17(4):458–469. doi: 10.1038/ncb3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zalensky A, Zalenskaya I. Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans. 2007;35(Pt 3):609–611. doi: 10.1042/BST0350609. [DOI] [PubMed] [Google Scholar]
- 7.Ioannou D, Millan NM, Jordan E, Tempest HG. A new model of sperm nuclear architecture following assessment of the organization of centromeres and telomeres in three-dimensions. Sci Rep. 2017;7:41585. doi: 10.1038/srep41585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samassekou O, Gadji M, Drouin R, Yan J. Sizing the ends: normal length of human telomeres. Ann Anat. 2010;192(5):284–291. doi: 10.1016/j.aanat.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 9.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 10.Forsyth NR, Wright WE, Shay JW. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69(4–5):188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- 11.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326(5955):948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozik A, Bradbury M, Zalensky A. Increased telomere size in sperm cells of mammals with long terminal (TTAGGG)n arrays. Mol Reprod Dev. 1998;51(1):98–104. doi: 10.1002/(SICI)1098-2795(199809)51:1<98::AID-MRD12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 13.Brenner CA, Wolny YM, Adler RR, Cohen J. Alternative splicing of the telomerase catalytic subunit in human oocytes and embryos. Mol Hum Reprod. 1999;5(9):845–850. doi: 10.1093/molehr/5.9.845. [DOI] [PubMed] [Google Scholar]
- 14.Turner S, Hartshorne GM. Telomere lengths in human pronuclei, oocytes and spermatozoa. Mol Hum Reprod. 2013;19(8):510–518. doi: 10.1093/molehr/gat021. [DOI] [PubMed] [Google Scholar]
- 15.Schaetzlein S, Lucas-Hahn A, Lemme E, Kues WA, Dorsch M, Manns MP, Niemann H, Rudolph KL. Telomere length is reset during early mammalian embryogenesis. Proc Natl Acad Sci U S A. 2004;101(21):8034–8038. doi: 10.1073/pnas.0402400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner S, Wong HP, Rai J, Hartshorne GM. Telomere lengths in human oocytes, cleavage stage embryos and blastocysts. Mol Hum Reprod. 2010;16(9):685–694. doi: 10.1093/molehr/gaq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varela E, Schneider RP, Ortega S, Blasco MA. Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc Natl Acad Sci U S A. 2011;108(37):15207–15212. doi: 10.1073/pnas.1105414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird DM, Britt-Compton B, Rowson J, Amso NN, Gregory L, Kipling D. Telomere instability in the male germline. Hum Mol Genet. 2006;15(1):45–51. doi: 10.1093/hmg/ddi424. [DOI] [PubMed] [Google Scholar]
- 19.Lopes AC, Oliveira PF, Sousa M. Shedding light into the relevance of telomeres in human reproduction and male factor infertility. Biol Reprod. 2019;100(2):318–330. doi: 10.1093/biolre/ioy215. [DOI] [PubMed] [Google Scholar]
- 20.Cariati F, Jaroudi S, Alfarawati S, Raberi A, Alviggi C, Pivonello R, Wells D. Investigation of sperm telomere length as a potential marker of paternal genome integrity and semen quality. Reprod BioMed Online. 2016;33(3):404–411. doi: 10.1016/j.rbmo.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Rocca MS, Speltra E, Menegazzo M, Garolla A, Foresta C, Ferlin A. Sperm telomere length as a parameter of sperm quality in normozoospermic men. Hum Reprod. 2016;31(6):1158–1163. doi: 10.1093/humrep/dew061. [DOI] [PubMed] [Google Scholar]
- 22.Zhao F, Yang Q, Shi S, Luo X, Sun Y. Semen preparation methods and sperm telomere length: density gradient centrifugation versus the swim up procedure. Sci Rep. 2016;639051. [DOI] [PMC free article] [PubMed]
- 23.Lafuente R, Bosch-Rue E, Ribas-Maynou J, Alvarez J, Brassesco C, Amengual MJ, et al. Sperm telomere length in motile sperm selection techniques: a qFISH approach. Andrologia. 2018;50(2). [DOI] [PubMed]
- 24.Thilagavathi J, Kumar M, Mishra SS, Venkatesh S, Kumar R, Dada R. Analysis of sperm telomere length in men with idiopathic infertility. Arch Gynecol Obstet. 2013;287(4):803–807. doi: 10.1007/s00404-012-2632-8. [DOI] [PubMed] [Google Scholar]
- 25.Torra-Massana M, Barragan M, Bellu E, Oliva R, Rodriguez A, Vassena R. Sperm telomere length in donor samples is not related to ICSI outcome. J Assist Reprod Genet. 2018;35(4):649–657. doi: 10.1007/s10815-017-1104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bucar S, Goncalves A, Rocha E, Barros A, Sousa M, Sa R. DNA fragmentation in human sperm after magnetic-activated cell sorting. J Assist Reprod Genet. 2015;32(1):147–154. doi: 10.1007/s10815-014-0370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santiso R, Tamayo M, Gosalvez J, Meseguer M, Garrido N, Fernandez JL. Swim-up procedure selects spermatozoa with longer telomere length. Mutat Res. 2010;688(1–2):88–90. doi: 10.1016/j.mrfmmm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Yang Q, Zhang N, Zhao F, Zhao W, Dai S, Liu J, Bukhari I, Xin H, Niu W, Sun Y. Processing of semen by density gradient centrifugation selects spermatozoa with longer telomeres for assisted reproduction techniques. Reprod BioMed Online. 2015;31(1):44–50. doi: 10.1016/j.rbmo.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Rooney DE, Czepulkowski BH. Human chromosome preparation: essential techniques. New York: Wiley; 1997. [Google Scholar]
- 30.World Health Organization . WHO laboratory manual for the examination and processing of human semen. 5. Geneva: WHO Press; 2010. [Google Scholar]
- 31.Huirne JA, Homburg R, Lambalk CB. Are GnRH antagonists comparable to agonists for use in IVF? Hum Reprod. 2007;22(11):2805–2813. doi: 10.1093/humrep/dem270. [DOI] [PubMed] [Google Scholar]
- 32.Pinto F, Oliveira C, Cardoso MF, Teixeira-da-Silva J, Silva J, Sousa M, et al. Impact of GnRH ovarian stimulation protocols on intracytoplasmic sperm injection outcomes. Reprod Biol Endocrinol. 2009;75. [DOI] [PMC free article] [PubMed]
- 33.Tesarik J, Sousa M, Testart J. Human oocyte activation after intracytoplasmic sperm injection. Hum Reprod. 1994;9(3):511–518. doi: 10.1093/oxfordjournals.humrep.a138537. [DOI] [PubMed] [Google Scholar]
- 34.Tesarik J, Sousa M. Key elements of a highly efficient intracytoplasmic sperm injection technique: Ca2+ fluxes and oocyte cytoplasmic dislocation. Fertil Steril. 1995;64(4):770–776. doi: 10.1016/s0015-0282(16)57853-4. [DOI] [PubMed] [Google Scholar]
- 35.Vandervorst M, Liebaers I, Sermon K, Staessen C, De Vos A, Van de Velde H, et al. Successful preimplantation genetic diagnosis is related to the number of available cumulus–oocyte complexes. Hum Reprod. 1998;13(11):3169–3176. doi: 10.1093/humrep/13.11.3169. [DOI] [PubMed] [Google Scholar]
- 36.Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73(6):1155–1158. doi: 10.1016/s0015-0282(00)00518-5. [DOI] [PubMed] [Google Scholar]
- 37.Radesic B, Tremellen K. Oocyte maturation employing a GnRH agonist in combination with low-dose hcg luteal rescue minimizes the severity of ovarian hyperstimulation syndrome while maintaining excellent pregnancy rates. Hum Reprod. 2011;26(12):3437–3442. doi: 10.1093/humrep/der333. [DOI] [PubMed] [Google Scholar]
- 38.Sá R, Cunha M, Rocha E, Barros A, Sousa M. Sperm DNA fragmentation is related to sperm morphological staining patterns. Reprod BioMed Online. 2015;31(4):506–515. doi: 10.1016/j.rbmo.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Gomes M, Goncalves A, Rocha E, Sa R, Alves A, Silva J, et al. Effect of in vitro exposure to lead chloride on semen quality and sperm DNA fragmentation. Zygote. 2015;23(3):384–393. doi: 10.1017/S0967199413000671. [DOI] [PubMed] [Google Scholar]
- 40.Sergerie M, Laforest G, Bujan L, Bissonnette F, Bleau G. Sperm DNA fragmentation: threshold value in male fertility. Hum Reprod. 2005;20(12):3446–3451. doi: 10.1093/humrep/dei231. [DOI] [PubMed] [Google Scholar]
- 41.Zidi-Jrah I, Hajlaoui A, Mougou-Zerelli S, Kammoun M, Meniaoui I, Sallem A, Brahem S, Fekih M, Bibi M, Saad A, Ibala-Romdhane S. Relationship between sperm aneuploidy, sperm DNA integrity, chromatin packaging, traditional semen parameters, and recurrent pregnancy loss. Fertil Steril. 2016;105(1):58–64. doi: 10.1016/j.fertnstert.2015.09.041. [DOI] [PubMed] [Google Scholar]
- 42.Majzoub A, Agarwal A, Cho CL, Esteves SC. Sperm DNA fragmentation testing: a cross sectional survey on current practices of fertility specialists. Transl Androl Urol. 2017;6(Suppl 4):S710–S719. doi: 10.21037/tau.2017.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofmann N, Hilscher B. Use of aniline blue to assess chromatin condensation in morphologically normal spermatozoa in normal and infertile men. Hum Reprod. 1991;6(7):979–982. doi: 10.1093/oxfordjournals.humrep.a137472. [DOI] [PubMed] [Google Scholar]
- 44.Sellami A, Chakroun N, Ben Zarrouk S, Sellami H, Kebaili S, Rebai T, et al. Assessment of chromatin maturity in human spermatozoa: useful aniline blue assay for routine diagnosis of male infertility. Adv Urol. 2013;2013578631. [DOI] [PMC free article] [PubMed]
- 45.Vegetti W, Van Assche E, Frias A, Verheyen G, Bianchi M, Bonduelle M, et al. Correlation between semen parameters and sperm aneuploidy rates investigated by fluorescence in-situ hybridization in infertile men. Hum Reprod. 2000;15(2):351–365. doi: 10.1093/humrep/15.2.351. [DOI] [PubMed] [Google Scholar]
- 46.Turnpenny P, Ellard S. Emery’s elements of medical genetics. 14. Philadelphia: Elsevier Churchil Livingstone; 2012. [Google Scholar]
- 47.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Covault J, Abreu C, Kranzler H, Oncken C. Quantitative real-time PCR for gene dosage determinations in microdeletion genotypes. Biotechniques. 2003;35(3):594–598. doi: 10.2144/03353dd02. [DOI] [PubMed] [Google Scholar]
- 49.O’Callaghan N, Dhillon V, Thomas P, Fenech M. A quantitative real-time PCR method for absolute telomere length. Biotechniques. 2008;44(6):807–809. doi: 10.2144/000112761. [DOI] [PubMed] [Google Scholar]
- 50.Bernardino RL, Dias TR, Moreira BP, Cunha M, Barros A, Oliveira E, Sousa M, Alves MG, Oliveira PF. Carbonic anhydrases are involved in mitochondrial biogenesis and control the production of lactate by human sertoli cells. FEBS J. 2019;286(7):1393–1406. doi: 10.1111/febs.14779. [DOI] [PubMed] [Google Scholar]
- 51.Dias TR, Alves MG, Bernardino RL, Martins AD, Moreira AC, Silva J, et al. Dose-dependent effects of caffeine in human sertoli cells metabolism and oxidative profile: relevance for male fertility. Toxicology. 2015:32812–20. [DOI] [PubMed]
- 52.Mukaka M. Statistics corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24(3):69–71. [PMC free article] [PubMed] [Google Scholar]
- 53.Hammer Ø, Harper D, Ryan P. Past: paleontological statistics software package for education and data analysis. Palaeontol Electronica. 2001;4(1):1–9. [Google Scholar]
- 54.Bray I, Gunnell D, Davey SG. Advanced paternal age: how old is too old? J Epidemiol Community Health. 2006;60(10):851–853. doi: 10.1136/jech.2005.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura M, Cherkas LF, Kato BS, Demissie S, Hjelmborg JB, Brimacombe M, Cupples A, Hunkin JL, Gardner JP, Lu X, Cao X, Sastrasinh M, Province MA, Hunt SC, Christensen K, Levy D, Spector TD, Aviv A. Offspring’s leukocyte telomere length, paternal age, and telomere elongation in sperm. PLoS Genet. 2008;4(2):e37. doi: 10.1371/journal.pgen.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reig-Viader R, Capilla L, Vila-Cejudo M, Garcia F, Anguita B, Garcia-Caldes M, et al. Telomere homeostasis is compromised in spermatocytes from patients with idiopathic infertility. Fertil Steril. 2014;102(3):728–738. doi: 10.1016/j.fertnstert.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Ricci G, Perticarari S, Fragonas E, Giolo E, Canova S, Pozzobon C, Guaschino S, Presani G. Apoptosis in human sperm: its correlation with semen quality and the presence of leukocytes. Hum Reprod. 2002;17(10):2665–2672. doi: 10.1093/humrep/17.10.2665. [DOI] [PubMed] [Google Scholar]
- 58.Hemann MT, Rudolph KL, Strong MA, DePinho RA, Chin L, Greider CW. Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol Biol Cell. 2001;12(7):2023–2030. doi: 10.1091/mbc.12.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Franco S, Spyropoulos B, Moens PB, Blasco MA, Keefe DL. Irregular telomeres impair meiotic synapsis and recombination in mice. Proc Natl Acad Sci U S A. 2004;101(17):6496–6501. doi: 10.1073/pnas.0400755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Q, Zhao F, Dai S, Zhang N, Zhao W, Bai R, Sun Y. Sperm telomere length is positively associated with the quality of early embryonic development. Hum Reprod. 2015;30(8):1876–1881. doi: 10.1093/humrep/dev144. [DOI] [PubMed] [Google Scholar]
- 61.Kidd SA, Eskenazi B, Wyrobek AJ. Effects of male age on semen quality and fertility: a review of the literature. Fertil Steril. 2001;75(2):237–248. doi: 10.1016/s0015-0282(00)01679-4. [DOI] [PubMed] [Google Scholar]
- 62.Hassan MAM, Killick SR. Effect of male age on fertility: evidence for the decline in male fertility with increasing age. Fertil Steril. 2003:791520–7. [DOI] [PubMed]
- 63.Aston KI, Hunt SC, Susser E, Kimura M, Factor-Litvak P, Carrell D, Aviv A. Divergence of sperm and leukocyte age-dependent telomere dynamics: implications for male-driven evolution of telomere length in humans. Mol Hum Reprod. 2012;18(11):517–522. doi: 10.1093/molehr/gas028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allsopp R, Vaziri H, Patterson C, Goldstein S, Younglai E, Futcher A, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A. 1992;89(21):10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh NP, Muller CH, Berger RE. Effects of age on DNA double-strand breaks and apoptosis in human sperm. Fertil Steril. 2003;80(6):1420–1430. doi: 10.1016/j.fertnstert.2003.04.002. [DOI] [PubMed] [Google Scholar]
- 66.Schmid TE, Eskenazi B, Baumgartner A, Marchetti F, Young S, Weldon R, Anderson D, Wyrobek AJ. The effects of male age on sperm DNA damage in healthy non-smokers. Hum Reprod. 2007;22(1):180–187. doi: 10.1093/humrep/del338. [DOI] [PubMed] [Google Scholar]
- 67.Agarwal A, Ikemoto I, Loughlin KR. Relationship of sperm parameters with levels of reactive oxygen species in semen specimens. J Urol. 1994;152(1):107–110. doi: 10.1016/s0022-5347(17)32829-x. [DOI] [PubMed] [Google Scholar]
- 68.Pereira R, Sá R, Barros A, Sousa M. Major regulatory mechanisms involved in sperm motility. Asian J Androl. 2017;19(1):5–14. doi: 10.4103/1008-682X.167716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative observations of the analysis of sperm samples. (a) TUNEL assay for sperm DNA fragmentation evaluation, considering the different staining patterns. Evaluation was performed in morphologically normal spermatozoa. Upper panel: (1) sperm head; (2) acrosome vesicle region; (3) post-acrosome region; (4) equatorial region. Lower panel: schematic representation of the same images of the upper panel, with the same corresponding letters, to facilitate interpretation. Scale bar = 10 μm. (b) Aniline blue (AB) assay for sperm chromatin maturity evaluation: (1): AB-negative spermatozoon, revealing sperm chromatin fully condensed; (2): AB-positive spermatozoon, revealing sperm immature chromatin condensation. Scale bar = 20 μm. (c) FISH assay for sperm chromosomic numeric alterations evaluation: (1) haploid (X,18); (2) haploid (Y,18); (3) sex chromosome nullisomy (18+); (4) chromosome 18 nullisomy (Y+); (5) chromosome 18 nullisomy (X+); (6) sex chromosome disomy (XX/18); (7) sex chromosome disomy (YY/18); (8) sex chromosome disomy (XY/18); (9) chromosome 18 disomy (X/18,18); (10) chromosome 18 disomy (Y/18,18); (11) sexual and autosomal disomy (XX/18,18); (12) sexual and autosomal disomy (XY/18,18). Color legends: X chromosome (green); Y chromosome (orange); chromosome 18 (aqua-blue); nucleus (DAPI-blue). Scale bar = 1 μm. (DOCX 931 kb)
Frequency distribution histogram of relative STL in the total population (Total), and in cases with embryo transfer cycles (ETC), biochemical pregnancy (BP), clinical pregnancy (CP), ongoing pregnancy (OP) and live-birth delivery (LBD). (DOCX 19 kb)
Comparison of relative sperm telomere length (STL) within clinical outcomes: negative, for no pregnancy; biochemical pregnancy only (BP); clinical pregnancy only (CP) and ongoing pregnancy (OP). Relative STL did not significantly differ among the different pregnancy status. a Total population: Kruskal-Wallis test, P = 0.257. Medians are represented. b Normozoospermic patients: Kruskal-Wallis test, P = 0.082. Medians are represented. c Non-normozoospermic patients: One-way ANOVA, P = 0.208. Means are represented. (DOCX 53 kb)
(DOCX 23 kb)
(DOCX 16 kb)
(DOCX 14 kb)