Abstract
Purpose
Housekeeping genes (HKGs), reference or endogenous control genes, are vital to normalize mRNA levels between different samples. Since using inappropriate HKGs can lead to unreliable results, selecting the proper ones is critical for gene expression studies. To this end, normal human ovaries, as well as those from patients diagnosed with ovarian endometrioid adenocarcinoma (OEA), ovarian mucinous adenocarcinoma (OMA), ovarian serous papillary carcinoma (OSPC), and polycystic ovary syndrome (PCOS), were used to identify the most suitable housekeeping genes.
Methods
RNA was isolated from 5 normal human ovaries (52–79 years of age), 9 cancerous ovaries (3 OEA, 3 OMA, 3 OSPC; 49–75 years of age), and 4 PCOS ovaries (18–35 years of age) in women undergoing hysterectomy. cDNA was synthesized using a whole transcriptome kit, and quantitative real-time PCR was performed using TaqMan array 96-well plates containing 32 human endogenous controls in triplicate.
Results
Among 32 HKGs studied, RPS17, RPL37A, PPIA, 18srRNA, B2M, RPLP0, RPLP30, HPRT1, POP4, CDKN1B, and ELF1 were selected as the best reference genes.
Conclusions
This study confirms recent investigations demonstrating that conventional HKGs, such as GAPDH and beta-actin, are not suitable reference genes for specific pathological conditions, emphasizing the importance of determining the best HKGs on a case-by-case basis and according to tissue type. Our results have identified reliable HKGs for studies of normal human ovaries and those affected by OEA, OMA, OSPC, or PCOS, as well as combined studies of control subjects vs. each cancer or PCOS group.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01901-8) contains supplementary material, which is available to authorized users.
Keywords: Housekeeping/reference/endogenous control genes, Gene expression, Ovarian endometrioid adenocarcinoma (OEA), Ovarian mucinous adenocarcinoma (OMA), Ovarian serous papillary carcinoma (OSPC), Polycystic ovary syndrome (PCOS)
Introduction
Transcriptional regulation is a key factor in cellular responses, so analysis of gene expression profiles has become an essential part of biomedical research [1]. Real-time quantitative polymerase chain reaction (qPCR) is the current method of choice for specific and highly sensitive expression profiling of selected groups of target genes. However, despite being a powerful technique, data normalization remains problematic. At present, use of the so-called housekeeping genes (HKGs), endogenous control genes or reference genes, is the gold standard to normalize mRNA fractions of interest. HKGs are often used to correct sample-to-sample variations, including the amount and integrity of materials, enzymatic efficiency, cDNA sample loading, and differences between tissues or cells in overall transcription activity [2, 3].
By definition, HKGs selected for normalization should not be influenced by experimental or biological conditions, and their own expression levels should be identical in tumors and adjacent healthy sites. A growing number of studies show that expression levels of commonly used HKGs are affected by experimental conditions or vary in pathological states, particularly cancer, highlighting the risk of blind use of classic HKGs [4–7]. Although GAPDH and beta-actin are widely used, they are not suitable HKGs in many cases involving cell proliferation, differentiation, metabolic changes, hypoxia, and cancer [8–13]. Since no universal reference gene has yet been detected, the choice of HKG(s) should be validated in each investigated condition. To this end, bioinformatic tools like NormFinder, BestKeeper, and geNorm have been developed [3, 14, 15].
Regarding the incidence of polycystic ovary syndrome (PCOS) and ovarian cancers, it is assumed that PCOS elevates ovarian cancer risk through increased androgen exposure [16, 17], so gene analyses for early diagnosis are highly recommended [17]. In order to obtain reliable results from qPCR, it is essential to use the correct reference gene(s). With this in mind, our study set out to assess 32 HKGs in healthy ovarian tissue as a control group, ovarian endometrioid adenocarcinoma (OEA), ovarian mucinous adenocarcinoma (OMA), ovarian serous papillary carcinoma (OSPC), and PCOS groups individually, all 5 groups together, and healthy ovaries vs. pathological cases.
Materials and methods
Source of ovaries and ethical approval
Biopsies of ovarian tissue were removed from five healthy women to investigate the presence of ovarian metastasis. However, all of them proved to be free of any pathology. Moreover, samples of OEA (n = 3), OMA (n = 3), and OSPC (n = 3) were obtained from the anatomic pathology laboratory of Saint-Luc’s Hospital. Use of human ovarian tissue was approved by the Institutional Review Board of the Université Catholique de Louvain on November 28, 2016 (IRB reference 2012/23MAR/125, registration number B403201213872). The tissue was frozen in Tissue-Tek® O.C.T.™ compound (Sakura, Netherlands) and cut with the Microm HM 560 microtome (Thermo Fisher, USA) into 5-μm sections (Table 1). Four PCOS samples (Table 1) were procured from anovulatory patients resistant to clomiphene, who had undergone laparoscopy to remove benign ovarian cysts and assess previous ovarian resection. At the moment of surgery, biopsies of the ovarian tissue (far from the cysts) were collected for this study. Samples were acquired as snap-frozen tissue in cryotubes filled with RNAlater™-ICE frozen tissue transition solution (Invitrogen, USA) from the assisted reproduction technology center of the Careggi University Hospital in Florence, Italy (ethical approval n. 11314_bio).
Table 1.
Description of study groups, ovarian tissue origin, and biopsy storage conditions
| Groups | Tissue type | Patient age | Code | Tissue size | Tissue condition |
|---|---|---|---|---|---|
| G1 | Healthy ovarian tissue (controls) | 59 | 1 | 10 fragments of 5 μm each | Frozen in OCT* |
| 61 | 2 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| 68 | 3 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| 52 | 4 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| 79 | 5 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| G2 | Endometrioid adenocarcinoma | 73 | 6 | 10 fragments of 5 μm each | Frozen in OCT* |
| 49 | 7 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| 73 | 8 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| G3 | Mucinous adenocarcinoma | 42 | 9 | 10 fragments of 5 μm each | Frozen in OCT* |
| 53 | 10 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| 56 | 11 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| G4 | Serous papillary carcinoma | 73 | 12 | 10 fragments of 5 μm each | Frozen in OCT* |
| 75 | 13 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| 61 | 14 | 10 fragments of 5 μm each | Frozen in OCT* | ||
| G5 | Polycystic ovary syndrome | 35 | 15 | 0.2 cm × 0.2 cm × 0.2 cm | Snap-frozen** |
| 18 | 16 | 0.2 cm × 0.2 cm × 0.3 cm | Snap-frozen** | ||
| 26 | 17 | 0.3 cm × 0.3 cm × 0.5 cm | Snap-frozen** | ||
| 35 | 18 | 0.3 cm × 0.3 cm × 0.3 cm | Snap-frozen** |
*OCT: optimal cutting temperature compound
**Snap-frozen at − 80 °C
RNA extraction and cDNA synthesis
Tissue fragments were placed in lysing matrix Z tubes (MP Biomedicals, USA) containing 2 mm yttria-stabilized zirconium beads with 600 μl RLT Plus buffer from the AllPrep DNA/RNA micro kit (QIAGEN, Germany) with 1% 2-mercaptoethanol (Sigma-Aldrich, Germany). Samples were lysed by bead beating for two 30-s cycles at 6 m/s in FastPrep lysing matrix Z tubes. A cooling step on ice (40-s cycles) was performed between the two bead beatings. Tubes containing the lysates were then centrifuged for 3 min at 14,000g. Supernatant was collected and RNA was extracted according to the manufacturer’s instructions. Briefly, the supernatant with RLT Plus and 2-mercaptoethanol was first passed through the DNA column to selectively bind genomic DNA. After centrifugation at 8000g for 30 s, the eluate containing RNA was mixed with 70% ethanol, before placing inside a RNeasy MinElute spin column and centrifuging at 8000g for 30 s. At this point, the eluate was discarded and the column was then washed once each with RW1 and RPE1 buffers, respectively, provided by the kit, with centrifugation after each washing step (8000g for 30 s). Eighty percent ethanol was added to the column, followed by centrifugation (8000g for 2 min), before finally eluting the RNA in 14 μl RNase-free water.
Isolated RNA concentrations were measured using the Qubit 4 fluorometer (Thermo Fisher Scientific, USA) and the Qubit RNA broad range assay kit. Since part of our samples were cryostat slices of OCT-embedded frozen tissue [18], the QuantiTect whole transcriptome kit (QIAGEN, Germany) was used to synthesize and amplify cDNA from 20 ng of isolated RNA. This kit does not amplify small RNA molecules, such as tRNAs or miRNAs, or degraded RNA. In fact, RNA transcripts should be longer than 500 nucleotides. Synthesized cDNA concentrations were measured using the Qubit 4 fluorometer (Thermo Fisher Scientific) and Qubit dsDNA broad range assay kit (Thermo Fisher Scientific) to be sure of loading 25 ng of cDNA in each well.
Quantitative real-time PCR
Expression levels of 32 commonly used candidates for HKGs (Table 2) [19, 20] were studied using predesigned TaqMan array 96-well fast plates for human endogenous controls (Applied Biosystems, CA, USA). Each TaqMan® gene expression assay consisted of a fluorogenic carboxyfluorescein (FAM™) dye-labeled minor groove binder (MGB) probe and two amplification primers (forward and reverse) provided in a pre-formulated 20X mix; 1X final concentrations were 250 nM for the probe and 900 nM for each primer. Each assay is estimated to have an amplification efficiency of 100 ± 10% (Thermo Fisher Scientific) [20]. A single 96-well plate was used for each individual sample. Ten microliters of pyrogen together with DNAse- and RNAse-free diethyl pyrocarbonate (DEPC) water (Invitrogen, USA) containing 25 ng cDNA was added to each well, followed by 10 μl TaqMan gene expression Master Mix (Thermo Fisher, Lithuania) in a total volume of 20 μl. For every study group, all the reactions for reference genes were performed at least in triplicate both biologically and technically. Standard cycling conditions were as follows: 2 min at 50 °C, 10 min at 95 °C, 50 cycles of 15 s at 95 °C, and 1 min at 60 °C. Thermal cycling and fluorescence detection were performed using the StepOnePlus Real-Time PCR system (Applied Biosystems).
Table 2.
Assay ID, gene symbol, and name of 32 investigated predesigned HKGs in 96 plates in triplicate
| Target | Assay ID | Gene symbol | Gene name |
|---|---|---|---|
| 1 | Hs99999901_s1 | 18S rRNA | Eukaryotic 18S rRNA |
| 2 | Hs99999903_m1 | ACTB | Actin; beta |
| 3 | Hs99999906_m1 | PGK1 | Phosphoglycerate kinase 1 |
| 4 | Hs00824723_m1 | UBC | Ubiquitin C |
| 5 | Hs00201226_m1 | CASC3 | Cancer susceptibility candidate 3 |
| 6 | Hs00206469_m1 | PUM1 | Pumilio homolog 1 (drosophila) |
| 7 | Hs00245445_m1 | ABL1 | C-Abl oncogene 1; non-receptor tyrosine kinase |
| 8 | Hs00198357_m1 | POP4 | Ribonuclease P protein subunit p29 |
| 9 | Hs99999905_m1 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| 10 | Hs99999907_m1 | B2M | Beta-2-microglobulin |
| 11 | Hs99999902_m1 | RPLP0 | Ribosomal protein; large; P0 |
| 12 | Hs00237047_m1 | YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein |
| 13 | Hs00355782_m1 | CDKN1A | Cyclin-dependent kinase inhibitor 1A |
| 14 | Hs00197826_m1 | PSMC4 | Proteasome (prosome; macropain) 26S subunit |
| 15 | Hs00152844_m1 | ELF1 | E74-like factor 1 (ETS domain transcription factor) |
| 16 | Hs01102345_m1 | RPL37A | Ribosomal protein L37a |
| 17 | Hs99999909_m1 | HPRT1 | Hypoxanthine phosphoribosyltransferase 1 |
| 18 | Hs00609297_m1 | HMBS | Hydroxymethylbilane synthase |
| 19 | Hs99999910_m1 | TBP | TATA box-binding protein |
| 20 | Hs99999904_m1 | PPIA | Peptidylprolyl isomerase A (cyclophilin A) |
| 21 | Hs00153277_m1 | CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27; Kip1) |
| 22 | Hs00426752_m1 | EIF2B1 | Eukaryotic translation inhibition factor 2B |
| 23 | Hs02596862_g1 | MT-ATP6 | Mitochondrially encoded ATP synthese 6 |
| 24 | Hs00265497_m1 | RPL30 | Ribosomal protein L30 |
| 25 | Hs99999908_m1 | GUSB | Glucuronidase; beta |
| 26 | Hs00183533_m1 | IPO8 | Importin 8 |
| 27 | Hs99999911_m1 | TFRC | Transferrin receptor (p90; CD71) |
| 28 | Hs00172187_m1 | POLR2A | Polymerase (RNA) II (DNA directed) polypeptide A |
| 29 | Hs00169255_m1 | GADD45A | Growth arrest and DNA damage-inducible |
| 30 | Hs00362795_g1 | PES1 | Pescadillo ribosomal biogenesis factor 1 |
| 31 | Hs00608519_m1 | MRPL19 | Mitochondrial ribosomal protein L19 |
| 32 | Hs00734303_g1 | RPS17 | Ribosomal protein S17 |
Data analysis
To analyze our data, the bioinformatic tool geNorm was used. The geNorm algorithm was developed by Vandesompele et al. [3] and is considered the gold standard for determining the most stable set and number of HKGs to use for accurate relative quantification. An advantage of the geNorm algorithm is that it is only slightly affected by the expression intensity of candidate genes [2, 3]. In a first step, the average of qPCR technical triplicates was computed for each target gene. If a single technical replicate out of three was undetermined, it was removed from downstream analysis, as previously recommended [21]. Because the variability of the technical replicates sharply increased when the Ct value exceeded 35 (Supplementary Fig. 1), samples with Ct > 35 were withdrawn before HKG selection. The “selectHKs” function of the NormqPCR program was then used to pick out the most stable HKGs by applying the geNorm algorithm. GeNorm uses mean pairwise variation for a given reference gene candidate compared with other candidates (M-value) and excludes the least stable genes before repeating the analysis, until only the most stable genes are left [3]. Since an appropriate reference gene should have an M-value below 0.5 or 1.0 in homogeneous and heterogeneous cell/tissue sample sets respectively, and we were working with cortex and medulla adjacent to the cortex, the threshold for the M-value in this study was set at < 1 [22]. As all samples in this study belong to 5 distinct groups (G1: normal human ovary; G2: OEA; G3: OMA; G4: OSPC; and G5: PCOS), selection of the best HKGs was done on samples from each group individually and all groups together, before being applied to samples from G1 vs. G2, G1 vs. G3, G1 vs. G4, and G1 vs. G5 (Figs. 1, 2, and 3).
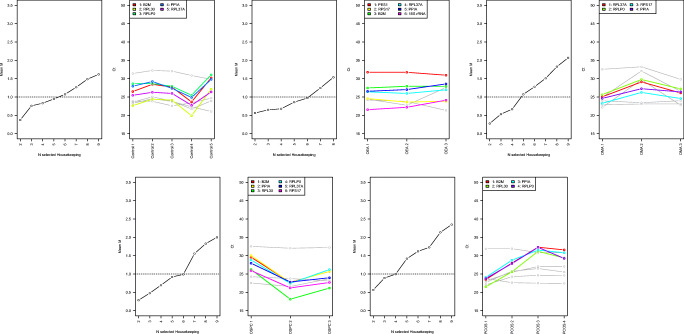
Fig. 1.
geNorm analyses in each studied group. G1 is healthy control ovarian tissue, G2 is ovarian endometrioid adenocarcinoma (OEA), G3 is ovarian mucinous adenocarcinoma (OMA), G4 is ovarian serous papillary carcinoma (OSPC), and G5 is polycystic ovary syndrome (PCOS). In each group, in the graph on the left, the X-axis represents the codes of the studied genes and the Y-axis the M-value. In the graph on the right, the X-axis represents the studied samples and the Y-axis the Ct value of the genes. The legend reports only HKGs selected by geNorm. The lower the M-value, the more stable the HKG, as shown below. G1: B2M > RPL30 > RPLP0 > PPIA > RPL37AG2: PES1 > RPS17 > B2M > RPL37A > PPIA > 18S rRNAG3: RPL37A > RPLP0 > RPS17 > PPIAG4: B2M > PPIA > RPL30 > RPLP0 > RPL37A > RPS17G5: B2M > RPL30 > PPIA > RPLP0
Fig. 2.
geNorm analyses in all groups together (18 samples), including control samples, ovarian endometrioid adenocarcinoma (OEA), ovarian mucinous adenocarcinoma (OMA), ovarian serous papillary carcinoma (OSPC), and polycystic ovary syndrome (PCOS). The graph on the left represents the codes of the studied genes (X-axis) and their mean value (Y-axis). The graph on the right represents the studied groups (X-axis) and Ct values (Y-axis). The legend reports only the HKGs selected by geNorm. The most stable HKGs in all groups were RPL37A > RPS17
Fig. 3.
geNorm analyses in control samples (G1) vs. G2, G3, G4, and G5. a G1 vs. OEA: RPL37A > RPS17 > PPIA. b G1 vs. OMA: RPL37A > RPS17. c G1 vs. OSPC: B2m > RPL30 > PPIA > RPLP0. d G1 vs. PCOS: RPL37A > RPS17. In each comparison, the graph on the left represents the codes of the studied genes (X-axis) and their mean value (Y-axis). The graph on the right represents the studied groups (X-axis) and Ct values (Y-axis). The legend reports only the HKGs selected by geNorm
Results
We analyzed a predesigned human endogenous control panel consisting of 32 primers, one in each well in triplicate, top identify the most stable HKGs.
The most stable HKGs per group
In five samples of normal human ovary (G1), 5 HKGs were suggested, namely beta 2-microglobulin (B2M), ribosomal protein L30 (RPL30), ribosomal protein P0 (RPLP0), peptidylprolyl isomerase A (cyclophilin A) (PPIA), and ribosomal protein L37a (RPL37A) (Fig. 1G1). In three OEA samples, 6 HKGs, namely pescadillo ribosomal biogenesis factor 1 (PES1), ribosomal protein S17-like (RPS17), B2M, RPL37A, PPIA, and eukaryotic 18s rRNA (18S rRNA), were endorsed (Fig. 1G2). In three OMA samples, RPL37A, RPLP0, and RPS17 were the most stable HKGs respectively (Fig. 1G3). In three OSPC samples, 6 HKGs emerged, namely B2M, PPIA, RPL30, RPLP0, RPL37A, and RPS17 (Fig. 1G4). Finally, in four PCOS samples, B2M, RPL30, PPIA, and RPLP0 ranged from the most to the least stable HKGs in the set threshold (Fig. 1G5).
-
2.
The most stable HKGs in all groups
Among 18 investigated samples, RPL37A and RPS17 were shown to be the most stable HKGs (Fig. 2).
-
3.
The most stable HKGs in the control group vs. OEA, OMA, OSPC, and PCOS subjects
In the study of five healthy control ovarian tissue fragments vs. three OEA samples, RPL37A, RPS17, and PPIA showed the least variation among all investigated HKGs. On the other hand, in an analysis of the five controls vs. three OMA samples, RPL37A and RPS17 showed the best stability to be chosen as HKGs. Controls vs. three OSPC samples identified B2M, RPL30, PPIA, and RPLP0 as having the least variation. Finally, in a comparison of the five controls vs. four PCOS samples, RPL37A and RPS17 were the most stable HKGs out of all 32 studied genes (Fig. 3).
Discussion
Although qPCR is recommended as an accurate method to investigate precise gene expression levels with high sensitivity [23, 24] in basic research, molecular medicine, and biotechnology [25], it is crucial to use the right HKGs to normalize data [26, 27]. In this study, we conducted extensive analysis of human ovarian tissue in an attempt to recommend specific use of optimal HKGs for this tissue. Due to tissue specificity for HKGs [4–7], we evaluated a panel of 32 HKGs in 18 samples from normal human ovaries as controls, PCOS ovaries, and three types of ovarian cancer, namely OEA, OMA, and OSPC. In each experiment, we introduced HKGs with an M-value less than 1 [22], and as it decreased, there was less variation in chosen gene expression in the different analyses. Our results suggest RPL37A, PPIA, and RPS17 to be the most stable HKGs in expression levels in the human ovary. RPL37A was also provided by Siemens Healthcare Diagnostics (Cologne, Germany) for use by Sinn et al. [28] in their studies on patients with high-stage, high-grade serous adenocarcinomas, consistent with the general population of ovarian cancer patients in their study. Looking into the literature and HKG studies in human ovaries, our findings on PPIA appear to corroborate those of Yan-Li et al. [29], who suggested PPIA as a reference gene in postmenopausal ovaries and serous ovarian cancer subtypes. PPIA was also proposed as a reference gene in native and vitrified/thawed human ovarian tissue by Nikishin et al. [30]. Both geNorm and NormFinder found PPIA to be a reliable reference gene in studies by Li et al. [29] on serous ovarian cancer. Sharan et al. [31] recommended that a combination of PPIA and either GAPDH, ACTB, HPRT, or TBP, or appropriate combinations of two or three of these genes, be used to verify results from different studies on various human cancers. RPS17 along with 18S rRNA was suggested as a reliable HKG in Wharton’s jelly–derived mesenchymal stem cells (WJ-MSCs) that were undergoing transduction and differentiation by Borkowska et al. [32]. Our results also indicate that RPLP0, B2M, RPL30, 18S rRNA, and PES1 may be suitable HKGs in human ovary studies. RPLP0 encodes ribosomal protein components of the 60S subunit, and Nikishin et al. [30] considered RPLP0 to be the most stable reference gene in human native and vitrified/thawed ovarian tissue. In another study, Shen et al. [33] demonstrated that the RPLP0 gene may prove reliable to normalize gene expression in human cervical tissue. RPLP0 together with RPL4 and HSPCB are also recommended for normalization in all ovarian-related research [34]. B2M was reported by Ofinran et al. [35] as an appropriate reference gene in human ovarian cancer, borderline ovarian cancer, and normal ovarian tissue. RPL30 was recommended as a suitable HKG in a study of the Scandinavian population, including borderline ovarian tumors [36]. Our findings on 18S rRNA as an applicable HKG in ovarian cancer are consistent with other studies on human ovarian cancer [34, 37]. PES1, to our knowledge, has not been stablished as a reliable HKG in human ovary, but has been used as an HKG in human lung cancer [38]. Li et al. [39] showed that PES1 differentially regulates the expression balance of ERα and ERβ and may play a crucial role in ovarian cancer.
While our data could have been reinforced by more extensive sampling, human ovarian tissue remains a precious material to be used in research, particularly when it comes from reproductive-age women. Similar to our study, Ofinran et al. [35] also used ovarian tissue fragments to represent the whole ovary. Such practice was also performed by Aithal and Rajeswari [40], who collected brain samples from paraffin-embedded blocks. Furthermore, it is important to bear in mind that sample sizes comparable or smaller than ours have been described in studies in different human tissues [2, 20, 40–43].
It is also important to stress that there are other factors that could play a role in our findings, such as age difference between control and PCOS groups, and menstrual cycle stage at the moment of ovarian biopsy collection. Moreover, we did not evaluate genome-wide transcript expression. However, it is important to bear in mind that, among the 32 most popular HKGs, we obtained quite similar gene results in different types of investigations.
In conclusion, our study showed RPL37A, PPIA, RPS17, RPLP0, B2M, RPL30, 18S rRNA, and PES1 to be the most suitable HKGs out of the 32 investigated reference genes in 18 samples from individual studies in normal human ovaries, OEA, OMA, OSPC, and PCOS samples, all groups together and control tissue vs. patient groups (Table 3). On the other hand, as there is also disease specificity for HKGs in the same tissue, we propose particular HKGs for some common ovarian pathologies, like the 3 types of ovarian cancer and PCOS. Our results can be used to provide a good starting point for developing primers to use or be selected in gene array studies in order to ensure generation of accurate and robust data in the same tissue conditions in different gene expression studies.
Table 3.
Summary of the selected HKGs with an M-value < 1 in all studied groups
| B2M | RPL30 | RPLP0 | PPIA | RPL37A | RPS17 | 18S rRNA | PES1 | |
|---|---|---|---|---|---|---|---|---|
| Control | * | * | * | * | * | |||
| OEA | * | * | * | * | * | * | ||
| OMA | * | * | * | * | ||||
| OSPC | * | * | * | * | * | * | ||
| PCOS | * | * | * | * | ||||
| All | * | * | ||||||
| Control vs. OEA | * | * | * | |||||
| Control vs. OMA | * | * | ||||||
| Control vs. OSPC | * | * | * | * | ||||
| Control vs. PCOS | * | * | ||||||
| Frequency out of 10 | 5 | 4 | 5 | 7 | 8 | 7 | 1 | 1 |
Control normal human ovary, OEA ovarian endometrioid adenocarcinoma, OMA ovarian mucinous adenocarcinoma, OSPC ovarian serous papillary carcinoma, PCOS polycystic ovary syndrome. Frequency out of ten referring to the repetition of each gene in a total of ten experiments. Beta-2-microglobulin (B2M), ribosomal protein L30 (RPL30), ribosomal protein, large, P0 (RPLP0), peptidylprolyl isomerase A (cyclophilin A) (PPIA), ribosomal protein L37a (RPL37A), ribosomal protein S17-like (RPS17), eukaryotic 18s rRNA (18S rRNA), and pescadillo ribosomal biogenesis factor 1 (PES1)
Electronic supplementary material
Standard deviation for qPCR technical triplicates as a function of the average value of qPCR technical triplicates. A sharp increase in variability was observed for Ct > 35. (PNG 752 kb)
Acknowledgments
We are grateful to Mira Hryniuk for reviewing the English language of the manuscript and Dolores Gonzalez and Olivier Van Kerk for their technical assistance. We are also thankful to Professor Etienne Marbaix for his kind collaboration and providing ovarian control and cancerous samples.
Authors’ contributions
P. A.: study design, experimental procedures, analysis, interpretation of data, and manuscript preparation. J. A.: statistical analysis. C. G. and M. E. C.: PCOS tissue supply. B. B.: manuscript revision. M. C. C.: PCOS tissue supply and manuscript revision. M. M. D.: manuscript preparation and revision. C. A. A.: experimental design, experimental procedures, interpretation of results, and manuscript revision.
Funding information
This study was supported by grants from the Fonds National de la Recherche Scientifique de Belgique (FNRS) (C. A. Amorim is an FRS-FNRS Research Associate; grant MIS #F4535 16 awarded to C. A. Amorim; grant 5/4/150/5 awarded to M. M. Dolmans; grant ASP-RE314 awarded to P. Asiabi; FNRS-PDR Convention T.0077.14 and EOS grant 30443682).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Use of human ovarian tissue was approved by the Institutional Review Board of the Université Catholique de Louvain on November 28, 2016 (IRB reference 2012/23MAR/125, registration number B403201213872).
Four PCOS samples came from the assisted reproduction technology center of the Careggi University Hospital in Florence, Italy (Ethical approval n. 11314_bio).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC®: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida TA, Quispe-Ricalde A, Montes de Oca F, Foronda P, Hernández MM. A high-throughput open-array qPCR gene panel to identify housekeeping genes suitable for myometrium and leiomyoma expression analysis. Gynecol Oncol. 2014;134(1):138–143. doi: 10.1016/j.ygyno.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034. 1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendriks-Balk MC, Michel MC, Alewijnse AE. Pitfalls in the normalization of real-time polymerase chain reaction data. Basic Res Cardiol. 2007;102(3):195–197. doi: 10.1007/s00395-007-0649-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocha-Martins M, Njaine B, Silveira MS. Avoiding pitfalls of internal controls: validation of reference genes for analysis by qRT-PCR and Western blot throughout rat retinal development. PLoS One. 2012;7(8):e43028. doi: 10.1371/journal.pone.0043028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thellin O, Zorzi W, Lakaye B, de Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75(2–3):291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 7.Guénin S, et al. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot. 2009;60(2):487–493. doi: 10.1093/jxb/ern305. [DOI] [PubMed] [Google Scholar]
- 8.Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. β-Actin—an unsuitable internal control for RT-PCR. Mol Cell Probes. 2001;15(5):307–311. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
- 9.Ruan W, Lai M. Actin, a reliable marker of internal control? Clin Chim Acta. 2007;385(1–2):1–5. doi: 10.1016/j.cca.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Caradec J, et al. ‘Desperate house genes’: the dramatic example of hypoxia. Br J Cancer. 2010;102(6):1037. doi: 10.1038/sj.bjc.6605573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caradec J, Sirab N, Revaud D, Keumeugni C, Loric S. Is GAPDH a relevant housekeeping gene for normalisation in colorectal cancer experiments? Br J Cancer. 2010;103(9):1475–1476. doi: 10.1038/sj.bjc.6605851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansur NR, Meyer-Siegler K, Wurzer JC, Sirover MA. Cell cycle regulation of the glyceraldehyde3phosphate dehydrogenaseluracil DNA glycosylase gene in normal human cells. Nucleic Acids Res. 1993;21(4):993–998. doi: 10.1093/nar/21.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilà MR, et al. Increased glyceraldehyde-3-phosphate dehydrogenase expression in renal cell carcinoma identified by RNA-based, arbitrarily primed polymerase chain reaction. Cancer. 2000;89(1):152–164. [PubMed] [Google Scholar]
- 14.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 16.Risch HA. Hormonal etiology of epithelial ovarian cancer, with a hypothesis concerning the role of androgens and progesterone. J Natl Cancer Inst (JNCI) 1998;90(23):1774–1786. doi: 10.1093/jnci/90.23.1774. [DOI] [PubMed] [Google Scholar]
- 17.Jiao J, Sagnelli M, Shi B, Fang Y, Shen Z, Tang T, Dong B, Li D, Wang X. Genetic and epigenetic characteristics in ovarian tissues from polycystic ovary syndrome patients with irregular menstruation resemble those of ovarian cancer. BMC Endocr Disord. 2019;19(1):30. doi: 10.1186/s12902-019-0356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupasquier S, Delmarcelle AS, Marbaix E, Cosyns JP, Courtoy PJ, Pierreux CE. Validation of housekeeping gene and impact on normalized gene expression in clear cell renal cell carcinoma: critical reassessment of YBX3/ZONAB/CSDA expression. BMC Mol Biol. 2014;15(1):9. doi: 10.1186/1471-2199-15-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 2009;55(4):611–22. [DOI] [PubMed]
- 20.Ayakannu T, Taylor AH, Willets JM, Brown L, Lambert DG, McDonald J, Davies Q, Moss EL, Konje JC. Validation of endogenous control reference genes for normalizing gene expression studies in endometrial carcinoma. Mol Hum Reprod. 2015;21(9):723–735. doi: 10.1093/molehr/gav033. [DOI] [PubMed] [Google Scholar]
- 21.Pabinger S, Rödiger S, Kriegner A, Vierlinger K, Weinhäusel A. A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol Detect Quantif. 2014;1(1):23–33. doi: 10.1016/j.bdq.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellemans J, Mortier G, de Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gachon C, Mingam A, Charrier B. Real-time PCR: what relevance to plant studies? J Exp Bot. 2004;55(402):1445–1454. doi: 10.1093/jxb/erh181. [DOI] [PubMed] [Google Scholar]
- 24.Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54(4):391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rebouças EL, et al. Real time PCR and importance of housekeepings genes for normalization and quantification of mRNA expression in different tissues. Braz Arch Biol Technol. 2013;56(1):143–154. [Google Scholar]
- 26.Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29(2):332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 27.Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313(4):856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 28.Sinn BV, Darb-Esfahani S, Wirtz RM, Faggad A, Weichert W, Buckendahl AC, Noske A, Müller BM, Budczies J, Sehouli J, Braicu EI, Dietel M, Denkert C. Vascular endothelial growth factor C mRNA expression is a prognostic factor in epithelial ovarian cancer as detected by kinetic RT-PCR in formalin-fixed paraffin-embedded tissue. Virchows Arch. 2009;455(6):461–467. doi: 10.1007/s00428-009-0851-6. [DOI] [PubMed] [Google Scholar]
- 29.Li Y-L, Ye F, Hu Y, Lu WG, Xie X. Identification of suitable reference genes for gene expression studies of human serous ovarian cancer by real-time polymerase chain reaction. Anal Biochem. 2009;394(1):110–116. doi: 10.1016/j.ab.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 30.Nikishin DA, Filatov MA, Kiseleva MV, Bagaeva TS, Konduktorova VV, Khramova YV, et al. Selection of stable expressed reference genes in native and vitrified/thawed human ovarian tissue for analysis by qRT-PCR and Western blot. J Assist Reprod Genet. 2018;35(10):1851–60. [DOI] [PMC free article] [PubMed]
- 31.Sharan R, et al. Consensus reference gene (s) for gene expression studies in human cancers: end of the tunnel visible? Cell Oncol. 2015;38(6):419–431. doi: 10.1007/s13402-015-0244-6. [DOI] [PubMed] [Google Scholar]
- 32.Borkowska P, Zielińska A, Paul-Samojedny M, Stojko R, Kowalski J. Evaluation of reference genes for quantitative real-time PCR in Wharton’s jelly-derived mesenchymal stem cells after lentiviral transduction and differentiation. Mol Biol Rep. 2020;47(2):1107–1115. doi: 10.1007/s11033-019-05207-6. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Li Y, Ye F, Wang F, Lu W, Xie X. Identification of suitable reference genes for measurement of gene expression in human cervical tissues. Anal Biochem. 2010;405(2):224–229. doi: 10.1016/j.ab.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 34.Fu J, Bian L, Zhao L, Dong Z, Gao X, Luan H, Sun Y, Song H. Identification of genes for normalization of quantitative real-time PCR data in ovarian tissues. Acta Biochim Biophys Sin. 2010;42(8):568–574. doi: 10.1093/abbs/gmq062. [DOI] [PubMed] [Google Scholar]
- 35.Ofinran O, Bose U, Hay D, Abdul S, Tufatelli C, Khan R. Selection of suitable reference genes for gene expression studies in normal human ovarian tissues, borderline ovarian tumours and ovarian cancer. Mol Med Rep. 2016;14(6):5725–5731. doi: 10.3892/mmr.2016.5933. [DOI] [PubMed] [Google Scholar]
- 36.Kolkova Z, Arakelyan A, Casslén B, Hansson S, Kriegova E. Normalizing to GADPH jeopardises correct quantification of gene expression in ovarian tumours–IPO8 and RPL4 are reliable reference genes. J Ovarian Res. 2013;6(1):60. doi: 10.1186/1757-2215-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, He J, Huang S, Zhang X, Bian Y, He N, Zhang H, Xie J. Activation of hedgehog signaling is not a frequent event in ovarian cancers. Mol Cancer. 2009;8(1):112. doi: 10.1186/1476-4598-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharungbam GD, Schwager C, Chiblak S, Brons S, Hlatky L, Haberer T, Debus J, Abdollahi A. Identification of stable endogenous control genes for transcriptional profiling of photon, proton and carbon-ion irradiated cells. Radiat Oncol. 2012;7(1):70. doi: 10.1186/1748-717X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Zhuang Q, Lan X, Zeng G, Jiang X, Huang Z. PES1 differentially regulates the expression of ERα and ERβ in ovarian cancer. IUBMB Life. 2013;65(12):1017–1025. doi: 10.1002/iub.1228. [DOI] [PubMed] [Google Scholar]
- 40.Aithal MG, Rajeswari N. Validation of housekeeping genes for gene expression analysis in glioblastoma using quantitative real-time polymerase chain reaction. Brain Tumor Res Treat. 2015;3(1):24–29. doi: 10.14791/btrt.2015.3.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nazet U, Schröder A, Grässel S, Muschter D, Proff P, Kirschneck C. Housekeeping gene validation for RT-qPCR studies on synovial fibroblasts derived from healthy and osteoarthritic patients with focus on mechanical loading. PLoS One. 2019;14(12):e0225790. doi: 10.1371/journal.pone.0225790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biade S, Marinucci M, Schick J, Roberts D, Workman G, Sage EH, O’Dwyer PJ, LiVolsi VA, Johnson SW. Gene expression profiling of human ovarian tumours. Br J Cancer. 2006;95(8):1092–1100. doi: 10.1038/sj.bjc.6603346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caracausi M, Piovesan A, Antonaros F, Strippoli P, Vitale L, Pelleri MC. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol Med Rep. 2017;16(3):2397–2410. doi: 10.3892/mmr.2017.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Standard deviation for qPCR technical triplicates as a function of the average value of qPCR technical triplicates. A sharp increase in variability was observed for Ct > 35. (PNG 752 kb)