Abstract
Purpose
The aim of the present study was to identify key microRNAs (miRNAs) in porcine follicular fluid (FF) that regulate oocyte growth.
Methods
miRNAs contained in FF were determined by small RNA-seq of exosome RNA. Upstream regulator miRNA was determined by ingenuity pathway analysis using differentially expressed genes in granulosa cells (GCs) between small follicles (1–2 mm in diameter) and large follicles (3–5 mm), and between follicles containing oocytes of high developmental ability and follicles containing oocytes of low developmental ability. The candidate miRNAs overlapping among the three miRNAs group were determined. Lastly, the effect of supplementation with FF, exosome-depleted FFs, or each miRNA on in vitro oocyte growth was examined.
Results
The miRNAs determined were miR-17, -27, -92a, and -145. These miRNAs were found in the spent culture medium of oocytes and granulosa cells complexes and serum by small RNA sequencing. Culturing of oocytes and granulosa cells complexes collected from porcine early antral follicles (0.5–0.7 mm in diameter) with FF for 14 days improved oocyte growth; depletion of exosomes from the FFs neutralized the beneficial effect observed. miR-92a mimic increased the antrum formation and diameter, together with acetylated levels of H4K12 in oocytes. In addition, supplementation of miRNA mimics miR-17b, -92a, and -145b improved the rate of chromatin configuration, and miR-17b and -92a mimics improved the developmental ability of oocytes to the blastocyst stage.
Conclusion
miR-17, -92a, and -145 are major miRNA candidates in follicular fluids regulating oocyte growth.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01909-0) contains supplementary material, which is available to authorized users.
Keywords: In vitro grown oocyte, Follicular fluid, Exosome, miRNA, NGS, miR-92a
Introduction
Oocyte growth is mediated via well-orchestrated communication between granulosa cells (GCs) and oocytes. The developmental competence of oocytes, including the ability to complete meiotic maturation and to develop to the blastocyst stages, increases during follicle development from small to large antral follicles. In addition, along with follicle development, GCs demonstrate major differential gene expression changes between the small and large antral follicles [1].
Follicular fluid (FF) is the exclusive environment that surrounds oocytes and GC complexes (OGCs) in early antral follicles [2, 3]. The components of FF consist of hormones, growth factors, cytokines, and fatty acids; these are secreted from GCs and also moved from the circulatory system and are essential for proper follicular development [4]. Therefore, supplementation of culture medium with FF leads to high oocyte maturation and fertilization outcomes and an overall improvement in oocyte quality [5]. Furthermore, adding FF to the culture media of oocytes derived from early antral follicles (EAFs) enhances the size and developmental abilities of oocytes grown in vitro [6]. Therefore, FF contains specific factors that regulate oocyte growth.
FFs contain small extracellular vesicles, termed exosomes [7]. Exosomes are small vesicles enclosed in a double lipid membrane with diameters ranging from 30 to 100 nm, and contain proteins, mRNA, DNA, and micro RNAs (miRNAs) [7]; GCs can uptake exosomes from exosome-supplemented culture media [8]. These small extracellular vesicles are involved in intracellular communication [9], such as in the induction of cumulus cell expansion in cows [10]. Therefore, many studies have investigated miRNAs in FF [11–13]. In general, studies addressing the effect of miRNA on cellular functions focus on a certain specific signaling pathway, ignoring other complex interactions between target mRNAs and miRNA of interest [14–18]. However, identifying major miRNAs from the myriad of miRNAs present in FF is difficult as one miRNA can regulate hundreds of mRNAs. Here, we predict the major miRNAs in FF based on the following: (1) miRNAs in FF of small and large antral follicles were determined by small RNA-seq; (2) upstream regulator miRNAs in GCs between small antral follicles (SAF; 1–2 mm in diameter) and large antral follicles (LAF; 3–5 mm in diameter) were determined from differentially expressed genes using RNA-seq and ingenuity pathway analysis (IPA); (3) upstream regulator miRNAs of good- or poor-quality OGCs were determined by differentially expressed genes using IPA. Together, this data defines overlapped miRNAs as the major miRNAs that have an impact on follicle development. Furthermore, the effect on oocyte growth after supplementing OGCs culture medium with these miRNAs was examined.
Materials and methods
Reagents and media
Unless otherwise stated, all drugs were purchased from Nacalai Tesque (Kyoto, Japan). The medium used for in vitro growth (IVG) of OGCs was αMEM (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% v/v exosome depleted porcine FF, 2% polyvinylpyrrolidone (Sigma-Aldrich), 26 mM NaHCO3, 2 mM hypoxanthine (Sigma-Aldrich), 10 mM taurine, 0.3% BSA (Fraction-V), 1 μg/mL 17 β-oestradiol (E2), 0.1 mAU/mL FSH (Kyoritsu Seiyaku Corporation, Tokyo, Japan), and Insulin-Transferrin-Selenium × 100 (Gibco BRL, Paisley, UK). The medium used for in vitro maturation (IVM) of oocytes was TCM199 (Gibco BRL) supplemented with 10% v/v FF, 0.5 mM L-cysteine, 0.9 mM sodium pyruvate, 1 mM L-glutamine, 10 ng/mL epidermal growth factor, 5% fetal calf serum, 10 IU/mL equine chorionic gonadotropin (Aska Pharma CO. Ltd, Tokyo, Japan), and 10 IU/mL human chorionic gonadotropin (Fuji Pharma Co. Ltd, Tokyo, Japan). The medium used for in vitro culturing (IVC) of embryos was porcine zygote medium 3 (PZM3) [19]. IVG and IVM were performed at 5% CO2 and 95% air at 38.5 °C, and IVC was performed at 5% O2, 5% CO2, and 90% N2 at 38.5 °C.
Experimental design
In the present study, the miRNAs important for oocyte growth from early antral follicles to antral follicles were determined. Furthermore, we addressed the origin of these miRNAs in FFs. In next, we addressed whether exosomes in FFs would affect oocyte growth in vitro. Finally, the effect of the predicted important miRNAs in FFs on oocyte growth was examined (Fig. 1).
Fig. 1.
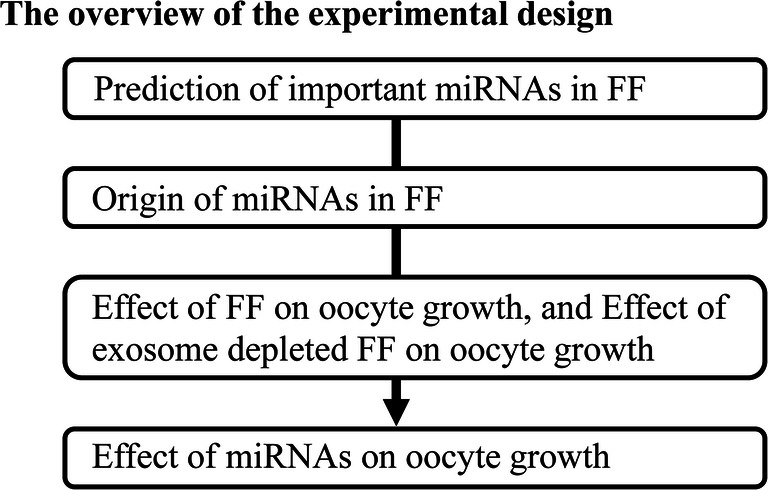
Experimental design overview. The present study addressed the following questions: Which miRNAs in FF are important for oocyte growth? Do miRNAs in FFs come from the in serum, or are they secreted from GCs? Do miRNAs in FF exosomes affect oocyte growth in vitro? Do the predicted miRNAs indeed improve in vitro oocyte growth?
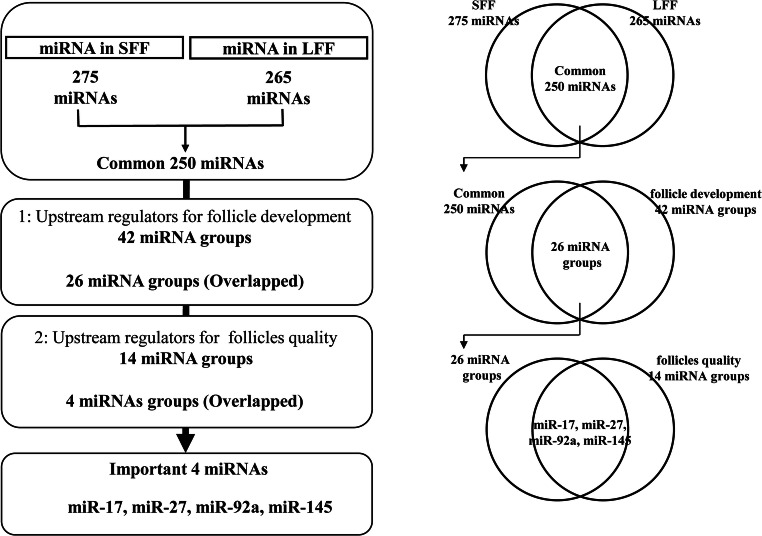
miRNAs important to oocyte growth were predicted using a combination of small RNA-seq and IPA using differentially expressed genes related to follicle development and oocyte quality. miRNA prediction was conducted as described in Fig. 2. First, miRNAs commonly present in FF from SAFs and LAFs were determined by small RNA-seq. From the hundreds of miRNAs found in FF, important miRNAs were elicited by comparison with miRNA groups predicted as upstream regulators that could regulate follicle development and oocyte quality. Two miRNA groups of upstream regulators were predicted by IPA using differentially expressed genes. These differentially expressed genes were determined by previous RNA-seq (DRA004449) [1], which compared GCs of SAFs and LAFs or GCs of follicles containing good oocytes having high developmental ability and that of follicles containing poor oocytes with low developmental ability. Overlapping miRNAs among the three miRNA groups were determined to be important miRNAs for follicle development.
Fig. 2.
Schematic design for identification of important miRNAs for follicle development. miRNA in follicular fluid derived from small antral follicles (SFFs) and large antral follicles (LFF) were determined by small RNA-seq. Overlapped 250 miRNAs between SFF and LFF were filtered down to 26 miRNA groups by comparison with the upstream regulator miRNA (42 miRNA groups) for follicle development. The upstream regulators were predicted from differentially expressed genes (1751 genes; DDJB, DRA004449) in granulosa cells between small and large follicles. The 26 miRNAs were also filtered down to four miRNAs by comparison with upstream regulator miRNAs (14 miRNA groups) for follicle qualities. The upstream regulators were predicted from differentially expressed genes (174 genes; DDJB, DRA006323) in granulosa cells between good and poor follicles. A detailed explanation (left) and the corresponding Venn diagram (right) are provided
In addition, to address the origins of miRNAs found in FF, miRNAs in FF were compared with the miRNAs found in spent culture medium of OGCs (via small RNA-seq) and those reportedly found in serum (DDJB:SRA985010).
To verify the effect of the overlapping miRNAs predicted to be important to follicle development, OGCs collected from EAFs (500–700 μm) were cultured with the predicted miRNA mimic or control mimic for 14 days; in vitro oocytes were examined for their developmental rate to the blastocyst stage, together with markers of oocyte quality, including chromatin configuration (SN ratio), histone acetylation (H4K12), and oocyte diameters.
Preparation of FFs for small RNA-seq
Follicular fluid was aspirated from SAFs and LAFs of 60 gilts; FF was centrifuged (10,000×g for 5 min), filtered (0.22 μm), and then used for exosome extraction.
Extraction of exosome RNA from FF and spent culture medium
FF derived from small antral follicles (SFFs), large antral follicles (LFFs), and spent culture medium was used for exosome collection. Thirty OGCs were cultured individually for 14 days. At the end of the culture period, culture medium in which OGCs formed antra were collected and mixed. Exosomes were collected from SFFs, LFFs, and the spent culture medium using ExoQuick (System Biosciences, LLC, Palo Alto, CA, USA). Exosome RNA was extracted from exosomes using a SeraMir Exosome RNA Amplification Kit (System Biosciences). Three batches of SFF, LFF, and a spent culture medium were used for small RNA-seq.
Small RNA sequences of miRNAs in exosomes
RNA quality and concentration were examined using a Bioanalyzer (Agilent technologies, Palo Alto, CA, USA); a cDNA library was constructed using an Illumina TruSeq Small RNA Library Preparation Kit (Illumina, San Diego, CA, USA) for RNA from FFs, and the SMARTer® smRNA-Seq Kit for Illumina®(Takara, Tokyo, Japan) for RNA of spent culture medium. The average length of all derived libraries was confirmed using an Agilent Bioanalyzer with a High Sensitivity DNA Kit (Agilent Technologies, Palo Alto, CA, USA), and the concentration of each library was adjusted to 10 nM based on the results of qPCR (KAPA Biosystems, Boston, MA, USA). Sequencing was conducted using NextSeq 500 (Illumina) to sequence 75 bp (single read).
Raw reads derived from RNA of FFs were trimmed by removing ligated adapters and filtered using Cutadapt [20]. Data quality was re-checked, following trimming. Small RNA reads obtained from FF were mapped to miRbase 21.0 (http://www.mirbase.org/) to identify known pig miRNAs using miRDeep2 [21]. Raw reads derived from small RNA of spent culture medium were checked for quality control, trimmed, and mapped using CLC Genomics Workbench 12. At trim reads process, 3′ poly A added by SMARTer® was removed. Small RNA reads were mapped to miRbase 21.0 (http://www.mirbase.org/) to identify known porcine miRNAs. Data related to small RNA-seq of FF and medium were registered in DDJB (DRA006647 and DRA009489, respectively).
Prediction of miRNAs as upstream regulators of follicle development
miRNA groups regulating follicle development and follicle quality were predicted from differentially expressed genes determined by two previous RNA-seq methods. First, RNA-seq was conducted using granulosa cells collected from SAF and LAF on the surface of ovaries of 60 gilt (registered in DDJB database; DRA004449) [1]. The second RNA-seq was conducted using granulosa cells collected from follicles containing oocytes with high developmental ability and follicles containing poor oocytes (DRA006323) [22].
Filtering, mapping, and subsequent analysis were performed using the CLC genomics workbench (Qiagen, Redwood City, CA, USA). For prediction of upstream transcriptional regulators, differentially expressed genes that were significantly differentially expressed were analyzed using the upstream regulator function of ingenuity pathway analysis (IPA, Qiagen).
For the first RNA-seq, we predicted upstream regulator miRNA groups (42 miRNAs) determined from 1751 differentially expressed genes (cut-off for expression P < 0.05, Q value < 0.05, fold change > 2). In the second RNA-seq, the upstream regulator miRNA group (14 miRNAs) was predicted from 670 differentially expressed genes (cut-off for expression P value < 0.05, Q value < 0.2, fold change > 1.5).
Preparation of exosome depleted FF for in vitro oocyte growth culture
FF was aspirated from SAF of ovaries of 60 gilts and was centrifuged (10,000×g for 5 min). After that, it was filtered (0.22 μm). Exosomes in FF were depleted using the FBS Exosome Depletion Kit (Norgen Biotek Corp, Thorold, Canada) according to the manufacturer’s protocol. The exosome depleted SFF was stored at − 80 °C until use and added to IVG medium at a concentration of 10%.
Ovaries and collection of OGCs from early antral follicles
Porcine ovaries were collected from prepubescent gilts at a local slaughterhouse and transported to the laboratory within 1 h in phosphate-buffered saline solution (PBS) containing antibiotics and maintained at 37 °C. The slice was dissected from the ovaries and OGCs were retrieved from EAFs using an 18-G needle connected to a 1-mL syringe, under a stereomicroscope (Olympus, Tokyo, Japan).
In vitro growth of oocytes derived from EAFs
OGCs collected from EAFs were individually transferred to wells containing 200 μL of IVG medium (96-well plates, Becton Dickinson) and cultured for 14 days. Half of the medium was replaced with fresh medium, and antrum formation was examined at 4-day intervals. The culture medium and methods were described in previous reports [23]. Using these methods, oocytes developed in vitro have developmental abilities as high as those of in vivo grown oocytes.
Transfection of miRNA mimics
Each of the four miRNAs was diluted in Opti-MEM (Invitrogen) and co-incubated with Lipofectamine®2000 (Invitrogen) for 15 min to form miRNA-lipid complexes, which were then added to the IVG medium. The final concentration of miRNA was 20 nM. The miRNAs were purchased from Ambion (Grand Island, NY, USA). The miRNA concentration was determined in a preliminary experiment: BLOCK-iT™ Alexa Fluor® Red Fluorescent siRNA (50 nM; Thermo Fisher Scientific) was added to IVG medium and the fluorescent signals in GCs were detected under a fluorescent microscope (Keyence, Tokyo, Japan). Sequences of miR-17, -27b, -92a, and -145 were retrieved from the miRNA database miRbase 22.1. OGCs were collected from EAFs and cultured with mimic or control mimic for 14 days. The IVG medium used in this experiment was supplemented with 10% exosome free FF.
Evaluation of antrum formation and oocyte diameter in vitro
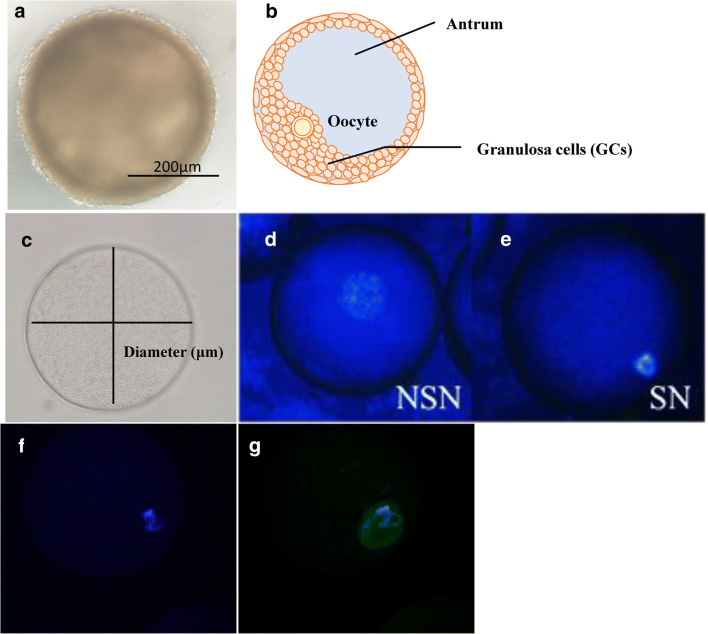
After IVG, the presence of antrum was morphologically evaluated (Fig. 3a, b), and oocytes were picked up from the OGCs having antrum. The oocytes were completely removed from the surrounding GCs, and their vertical and horizontal diameters were measured under a digital microscope (BZ-8000; Keyence, Tokyo, Japan). The average of the vertical and horizontal diameters was calculated as the oocyte diameter (Fig. 3c).
Fig. 3.
Representative images of OGCs and oocytes. OGCs derived from early antral follicles were cultured for 14 days. Follicle development and the quality of oocytes were determined using several markers. a, b OGCs forming an antrum (a, representative picture and b image of antrum formation). c Diameter of oocytes grown in vitro. d, e Images of oocytes with non-surrounded nucleolus (d, NSN) or surrounding nucleoli (e, SN). f, g Oocytes immuno-stained against H4K12 (f, representative pictures of nuclei stained with DAPI and g, representative pictures of oocytes immuno-stained against H4K12ace)
Observation of chromatin configuration
After IVG, oocytes denuded from OGCs were fixed using 4 % paraformaldehyde. Oocytes were washed several times and stained in PBS containing 1 μg/mL Hoechst 33342 (Sigma) for 15 min. Non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) oocytes (Fig. 3d, e) were categorized according to a previous report [24] using a digital microscope (Keyence) following staining with Hoechst 33342. SN represents the oocyte of an advanced stage, while an NSN is considered as a stage prior to SN [25].
Detection of acetylated H4K12 by fluorescence immunostaining
After IVG, oocytes denuded from OGCs were fixed using 4% paraformaldehyde. Oocytes were permeabilized with PBS containing 0.25% TritonX-100 (Sigma-Aldrich) for 30 min at approximately 25 °C. Next, oocytes were blocked (PBS containing 5% BSA, 1% Tween20, 5% goat serum) for 1 h, followed by overnight incubation with rabbit polyclonal anti-acetylated H4K12 primary antibody (H4K12ace, 1:200; Novus International Saint Charles, MO, USA) diluted in blocking solution. Oocytes were then incubated for 1 h with anti-rabbit IgG secondary antibody (1:500; Cell Signalling Technology Japan). Oocytes were mounted on microscope slides with Pro-long gold antifade reagent and DAPI (Invitrogen); fluorescence images were acquired using a fluorescence digital microscope (Keyence) and fluorescence intensity was measured using Image-J software (NIH). Representative images of oocytes stained with H4K12ace are shown in Fig. 3f, g.
Preparation of FF for in vitro oocyte maturation
FF was collected from antral follicles (3–5 mm in diameter) of ovaries of 60 gilt. FF was centrifuged (10,000×g for 5 min), filtered (0.2 μm), and stored at − 80 °C until use. The FF was added to the IVM medium at a concentration of 10%.
IVM, activation, and IVC
Following IVG, OGCs with an antrum cavity were subjected to IVM for 48 h. After IVM, oocytes were denuded from surrounding GCs, parthenogenetically activated by incubation in IVC medium containing 10 μg/mL ionomycin for 5 min, followed by incubation with PZM3 containing 10 μg/mL cytochalasin B and 10 μg/mL cycloheximide for 5 h. Following activation, oocytes were cultured in PZM3 for 8 days to determine the rate of blastulation and blastomere number of the blastocysts.
Statistical analysis
Data were compared using one-way ANOVA followed by Tukey’s post hoc test. The rates of antrum formation and blastocyst stage were arcsine-transformed prior to analysis. Statistical significance was set at P < 0.05.
Results
Identification of miRNAs predicted to have a substantial impact on follicle development
As described in the “Materials and Methods” section, we identified miRNAs in FFs. Small RNA-seq revealed 275 and 265 miRNAs in SFF and LFFs, respectively, and 250 miRNAs were present (overlapped) in both populations (Supplementary Table 1). These miRNAs were filtered down with two miRNA groups predicted as upstream regulators by IPA. Comparison of the 250 overlapping miRNA with the first miRNA group (42 miRNAs, upstream regulators of SAFs and LAFs, Table 1) identified 26 miRNA that overlapped in both groups. The 26 miRNAs were compared with the second miRNA group of upstream regulators (14 miRNAs, Table 1, upstream regulators of good and poor follicles). Finally, miR-17, -27, -92a, and 145 were found in each of the three miRNA groups.
Table 1.
miRNA of upstream regulator between SAF and LAF and between good and poor
| SAF—LAF | Good—poor | ||||
|---|---|---|---|---|---|
| Upstream regulator | Activation z-score | p value of overlap | Upstream regulator | Activation z-score | p value of overlap |
| let-7 | − 0.607 | 1.26E-02 | miR-33-5p | 8.05E-03 | |
| miR-1 | − 0.249 | 1.25E-02 | miR-27 | 8.49E-03 | |
| miR-124-3p | 1.296 | 2.87E-02 | miR-145-5p | 1.09E-02 | |
| miR-126 | 1.949 | 4.98E-02 | miR-19 | 1.22E-02 | |
| miR-130 | 0.391 | 3.33E-03 | miR-379 | 1.60E-02 | |
| miR-133 | 0 | 8.57E-05 | miR-370 | 1.60E-02 | |
| miR-135 | 1.528 | 4.10E-03 | miR-342 | 1.60E-02 | |
| miR-143 | − 0.293 | 4.85E-02 | miR-17 | 1.62E-02 | |
| miR-145-5p | 3.579 | 4.02E-04 | miR-199a-5p | 1.89E-02 | |
| miR-146 | − 0.895 | 3.97E-02 | miR-92a-3p | 2.12E-02 | |
| miR-15 | − 1.156 | 4.08E-02 | miR-541 | 2.40E-02 | |
| miR-150 | − 0.036 | 2.36E-02 | miR-370-3p | 2.40E-02 | |
| miR-154 | 3.87E-02 | miR-291a-3p | 3.42E-02 | ||
| miR-17-5p | − 1.661 | 7.62E-03 | miR-665 | 3.96E-02 | |
| miR-181 | 0.493 | 2.69E-03 | |||
| miR-183 | − 0.139 | 4.85E-02 | |||
| miR-18a-5p | 0.283 | 2.61E-02 | |||
| miR-193 | − 1.026 | 5.10E-03 | |||
| miR-194 | 2.87E-02 | ||||
| miR-199a-5p | 1.947 | 2.75E-07 | |||
| miR-200b-3p | − 0.456 | 4.98E-02 | |||
| miR-203a-3p | 0.647 | 5.94E-03 | |||
| miR-204 | 1.35E-02 | ||||
| miR-208a-3p | 1.982 | 8.13E-03 | |||
| miR-21 | − 0.91 | 1.31E-04 | |||
| miR-210 | 1.97 | 4.13E-02 | |||
| miR-217 | 3.35E-02 | ||||
| miR-221 | − 1.675 | 9.60E-04 | |||
| miR-223 | − 0.318 | 1.16E-03 | |||
| miR-224-5p | 1.067 | 8.13E-03 | |||
| miR-23 | 0.856 | 1.13E-06 | |||
| miR-24 | 0.845 | 6.77E-05 | |||
| miR-26a-5p | − 0.807 | 1.68E-02 | |||
| miR-27 | − 1.757 | 4.98E-02 | |||
| miR-29 | − 0.995 | 4.57E-04 | |||
| miR-293-5p | 1.387 | 2.13E-02 | |||
| miR-30 | 1.937 | 1.53E-02 | |||
| miR-34 | 0.941 | 8.90E-03 | |||
| miR-382-5p | 4.35E-03 | ||||
| miR-486 | − 1.982 | 5.77E-02 | |||
| miR-494-3p | 1.35E-02 | ||||
| miR-92a-3p | − 1.395 | 2.00E-02 | |||
Comparison of miRNAs in FFs, serum, and culture medium
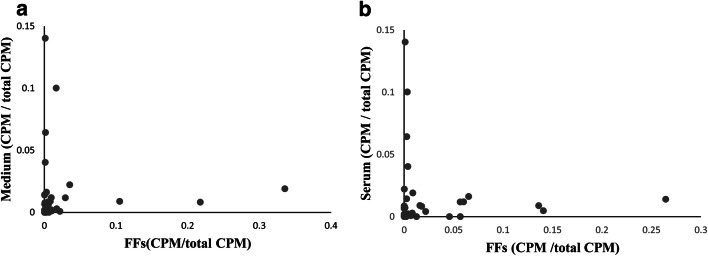
Small RNA-seq of the spent culture medium of OGCs revealed 108 miRNAs. After comparison with the 294 miRNA found in FFs (total miRNAs in both SFF and LFF) 94 overlapping miRNAs were found; the database registered as DDJB:SRA985010 shows 280 miRNAs in pig serum. Comparison of the miRNAs in serum with miRNAs found in FFs revealed that 235 miRNAs overlapped; 81 miRNAs overlapped among FFs, medium, and serum (Supplementary table 2).
Comparison of miRNAs found in FFs and those found in serum showed that miRNAs found at high frequency in FFs were not necessarily found at a high frequency in serum and vice versa. Similar trends were also found between miRNAs in FFs and miRNA in spent culture medium (Fig. 4a, b).
Fig. 4.
Comparison of miRNA frequency between medium and FFs or between serum and FFs. a Relation of count per million (CPM) between spent culture medium and FFs. b Relation of count per million (CPM) between serum and FFs
Effect of supplementation of IVG medium with small-FF and exosome-depleted FFs on oocyte development
This experiment was designed to address whether exosomes in SFF play a role in oocyte growth. Remarkably, SFF enhanced several markers associated with oocyte growth (oocyte diameter, H4K12 acetylation levels, and the developmental rate to the blastocyst stage), whereas chromatin condensation, determined by the SN rate was not affected by SFF supplementation (Table 2). Next, OGCs were cultured in a medium containing control-SFF or exosomes’ depleted SFF (SFF-free) for 14 days. Exosome depletion reduced oocyte diameter, the levels of H4K12 acetylation, and the rate of SN, while the reduction of the development to the blastocyst stage was not significantly impacted compared with the control-SFF condition (Table 2). Of note, the number of trials and the total number of oocytes examined are presented (Supplementary table 3).
Table 2.
Oocytes grown with FFs, exosome-free FF, control mimic, or miRNA-mimics
| Groups | Antral formation (%) | Oocyte diameter (μm)** | Rate of SN (%) | H4K12 acetylation | Rate of blastulation (%) |
|---|---|---|---|---|---|
| Control* | 90.0 ± 3.7 | 123.4 ± 0.6a | 82.2 ± 5.1 | 1.0 ± 0.1a | 5.6 ± 2.6a |
| Small-FF | 90.0 ± 2.6 | 132.3 ± 0.7b | 90.5 ± 1.9 | 2.5 ± 0.2b | 23.6 ± 3.6b |
| Small-FF | 75.6 ± 0.1 | 126.6 ± 0.7a | 92.5 ± 2.5a | 1.0 ± 0.1a | 23.6 ± 3.6 |
| Exo-free FF | 84.4 ± 0.1 | 116.9 ± 1.0b | 59.3 ± 6.9b | 0.7 ± 0.0b | 15.5 ± 4.6 |
| Control | 91.2 ± 2.2 | 123.7 ± 1.4 | 69.4 ± 2.0a | 1.0 ± 0.1 | 6.9 ± 2.8a |
| miR-17 mimic | 95.8 ± 1.6 | 124.1 ± 1.1 | 85.8 ± 2.9b | 1.2 ± 0.1 | 14.7 ± 1.8b |
| Control | 88.9 ± 4.1 | 123.5 ± 1.1 | 79.6 ± 6.9 | 1.0 ± 0.1 | 9.1 ± 2.4 |
| miR-27b mimic | 88.9 ± 2.8 | 125.6 ± 0.9 | 78.8 ± 7.7 | 1.2 ± 0.1 | 20.4 ± 6.5 |
| Control | 86.1 ± 1.9a | 121.6 ± 1.7a | 72.0 ± 6.1a | 1.0 ± 0.1a | 9.1 ± 2.4a |
| miR-92a mimic | 93.2 ± 1.9b | 129.0 ± 0.9b | 85.9 ± 2.9b | 1.3 ± 0.1b | 22.0 ± 5.5b |
| Control | 91.7 ± 2.2 | 123.7 ± 1.4 | 69.4 ± 2.0a | 1.0 ± 0.1 | 6.9 ± 2.8 |
| miR-145 mimic | 91.7 ± 2.2 | 125.8 ± 0.8 | 81.7 ± 0.7b | 0.9 ± 0.1 | 12.3 ± 3.9 |
| Small-FF | × | ○ | × | ○ | ○ |
| Exo-free FF | × | ○ | ○ | ○ | × |
| miR-17 mimic | × | × | ○ | × | ○ |
| miR-27b mimic | × | × | × | × | × |
| miR-92a mimic | ○ | ○ | ○ | ○ | ○ |
| miR-145 mimic | × | × | ○ | × | × |
Small-FF, small follicular fluids; Exo-free FF, exosome-depleted follicular fluids. Data are presented means ± standard error of the mean (SEM). OGCs were cultured in IVG medium containing 3 mg/ml BSA (control*) or 10% small-FF, 10% small-FF or 10% exosome depleted FF, negative control mimic (control) or miRNA mimics. **, oocyte diameter is excluding zona pellucida; a–b, P < 0.05; ○, means significant difference; ×, means comparable with control
Effect of supplementation of IVG medium with mimic-miRNAs on oocyte development
As shown in Table 2, the diameter of oocytes grown increased after supplementation with miR-92a. Chromatin condensation (SN ratio) was enhanced by miR-17b, -92a, and -145, and the acetylation levels of H4K12 were enhanced by miR-92a. The rate of development to the blastocyst stage following in vitro maturation and activation was improved by miR-17b and -92a. These data are summarized in Table 2. The number of trials and total number of oocytes examined are presented (supplementary table 3).
Discussion
In this study, important miRNA candidates that regulate follicle growth and oocyte development were determined. Supplementation of the IVG medium with the predicted miRNAs revealed that the miRNAs have the ability to improve oocyte quality and developmental ability to the blastocyst stage.
FF contains myriads of molecules that support follicle development. First, we addressed whether FF would improve oocyte development since the effects of FFs on in vitro oocyte growth have rarely been explored; of note, several studies report the beneficial effect of FFs on oocyte nucleic maturation [26–28], Importantly, here, we found that FF improves oocyte’s developmental markers (oocyte diameters, H4K12, and acetylation levels) and developmental ability. Among the factors contained in FFs, miRNAs are believed to play a role in regulating oocyte development [29]. Next, we have confirmed that the depletion of exosomes from FFs neutralized its beneficial effects of FF on oocyte development. FF contains exosome vesicles derived from GCs; the exosomes are known to carry miRNAs [29]. However, FFs are expected to contain hundreds of miRNAs, each of which may target hundreds of mRNAs. These complex networks underlie the difficulty of specifying the most important miRNAs in FF for oocyte development. The present study determined miRNAs in SFF and LFF and found that the majority of miRNAs overlapped between the two follicle stages. On extraction of RNA in FF, the amount of a small size RNA from 250 μL of SFFs was 2.07 μg ± 1.14, which was higher than that of LFF (1.37 μg ± 1.36, P = 0.06). Since the same volume of cDNA was used for small RNA-seq, a direct comparison of miRNA concentrations between SFF and LFF is impossible. Therefore, we used miRNA overlapping between SFF and LFF as candidates for further analysis. It has been reported that miRNAs in FF correlate with those observed in serum in humans [30]. Compared with miRNA found by small RNA-seq of FF with a serum-miRNA database in pigs (DDJB:SRA985010), 83.9% of serum miRNA overlapped with those found in small RNA-seq of FF. However, miRNAs found in FF at a high concentration are found at very low levels in serum (DDJB:SRA985010). Furthermore, the present study found 108 miRNAs in the spent culture medium of OGCs, 87% of which also overlapped with those found in FF. miRNA present in spent culture medium with high frequency was not necessarily found in FF at high concentrations. From these results, we cannot determine the origin of miRNAs in FF as the physiological condition of pigs and culture conditions of OGCs could affect each miRNA concentration; however, it is possible that some miRNAs are secreted from GCs and accumulate in FFs.
The previous two RNA-seq studies identified differentially expressed genes (DEGs) in GCs either between small antral follicles and large antral follicles [1], registered as (DDJB database; DRA004449) or between follicles containing good oocytes and those containing poor oocytes [22] registered as DDJB database: DRA006323). Upstream regulators of these DEG, as predicted by IPA, are key regulators for follicle development and good-quality oocytes on the premise that miRNAs regulate follicle development. Comparing the miRNAs predicted as upstream regulators with miRNAs determined by small RNA-seq of FF identified four overlapped miRNAs (miR-17, -27, -92a, and -145), which were determined as important miRNAs regulating follicle development. The presence of these four miRNAs in FFs and follicle cells has been reported in cows, mice, and humans [29, 31–34]. To the best of our knowledge, this is the first report of important miRNAs determined by bidirectional predictions that consider both the presence of miRNA in FF and crosstalk between miRNAs and GCs.
Prior studies on the effect of miRNA on GCs considered GCs cultured in vitro without oocytes and under high oxygen tension, conditions which differ significantly from those in vivo, and no report has addressed the effect of miRNAs on oocyte growth in large animals. We developed an in vitro culture system of porcine oocytes derived from early antral follicles in pigs, in which oocytes developed in vitro have developmental competence as high as those developed in vivo [23], and the gene expression profiles of the OGCs are closer to those in vivo. Using this culture system, we examined the effect of miRNA on in vitro oocyte growth and developmental ability. The quality of oocytes grown in vitro has been evaluated using several markers such as oocyte diameters, chromatin configurations, H4K12 acetylation, and the ability to develop into the blastocyst stage. Oocyte diameter and developmental rate to the blastocyst stage are commonly used markers. The SN rate has been reported to reflect the extent of oocyte growth because the rate of SN increases as an oocyte develops [35]. Acetylation levels of H4K12 increase with oocyte development [36], and it has been reported that the acetylation level of lysine as well as H4K12 is good for oocytes grown in vitro with high developmental competence [37, 38].
The miR-17-92 cluster (miR-17, miR-18, miR-19a, miR-20a, miR-19b-1, and miR-92a-1) are located on chromosome 12 in pigs, and of these, miR-18 and miR-20 were absent in FF, medium, and serum in the present small RNA-seq (DDJB:SRA985010). Germ cell ablation of the miRNA 17-92 cluster causes subfertility in female mice [39]. In addition, expression of miR-92a in FF was associated with the fertilization ability of human oocytes [40]. Liu et al. [15] cultured GCs with miR-92 mimic and found that miR-92a repressed porcine GC apoptosis by targeting SMAD7, while Andreas et al. [33] cultured bovine GC and found that miR-92 and -17b targeted PTEN and BMRP2 and enhanced CCND and PCNA expression as well as GC proliferation [33]. Furthermore, miR-17b enhanced the proliferation of porcine GCs and suppressed apoptosis via E2F1 suppression [41]. The present study shows the beneficial effect of miR-17 and -92a on the SN ratio and developmental ability to the blastocyst stage; miR-92a additionally improved oocyte diameter and acetylation levels of H4K12. Thus, these miRNAs may improve oocyte growth and quality; however, the molecular background is still unclear. In this study, the number of GCs consisting of OGCs and survival rate of GCs did not differ between mimic control and miR-17b and -92a. Furthermore, when OGCs were cultured in vitro, miR-92a did not affect the expression levels of SMAD7 in GCs (supplementary experiment).
miR-27b is contained in a cluster with miR-23 and miR-24 on chromosome 10 in pigs. These miRNAs were predicted as upstream regulators of the DEG of GCs between small and large antral follicles and poor- and good-quality follicles. These miRNAs were also found in FF (Table 1) and spent culture medium of OGCs. miR-23 and -27b are reported to regulate human GC apoptosis through SMAD5 [42]. The present study has not found a significant effect of miR-27b on oocyte growth; only a trend was observed on oocyte developmental ability to the blastocyst stages (P = 0.13) suggesting the effect of miR-27b on oocyte growth needs to further addressed.
miR-143 and -145 are clustered on chromosome 2 in pigs, and both miRNAs were found in serum, spent culture medium, and FF. These miRNAs were also predicted as upstream regulators for DEG between GCs of small and large follicles and between and poor- and good-quality follicles (Table 1). miR-145 levels in GCs were low in polycystic ovary syndrome (PCOS) patients [30]. miR-145 attenuates hydrogen peroxide induced apoptosis in GCs in mice by targeting KLF4 [43]. For oocyte maturation, supplementation of in vitro maturation medium with miR-145 reduced human oocyte maturation ability through inhibition of EGF and GDNF receptors [44]. We observed a beneficial effect of miR-145 only in the SN ratio of oocytes grown in vitro. This is the first report that confirms that miRNAs have an effect on in vitro oocyte growth and quality. However, the fundamental unsolved question is whether miRNAs in the exosomes of FF are taken up by GCs and show functions in them. What are the major targets of miRNAs among hundreds of possible target mRNAs? The previous studies on the effect of miRNA on GCs used culture conditions that differ substantially from in vivo conditions; for instance, GCs were cultured in vitro without oocytes. Furthermore, during oocyte growth and culture of OGCs, gene expression profiles, including those of housekeeping and metabolic genes differ extensively [23, 37]. Therefore, comprehensive gene expression analysis using an oocyte growth system that resembles the in vivo environment is needed to determine the main targets of each miRNA.
In conclusion, miR-17-5p, -92a, and -145 are potential candidates in regulation of follicle and oocyte growth in FFs based on the presence in FFs, the secretion from OGCs, and the association with differentially expressed genes between small and large follicles and poor- and good-quality follicles.
Electronic supplementary materials
(DOCX 31 kb)
Acknowledgments
We thank Rumi Ohtake and Naoko Kosuge (Tokyo University of Agriculture) for technical support.
Author’s contributions
I.Y, and M.Y conducted culture experiment; S.A, M.R, and M.Y conducted RNA-seq and small RNA-seq; H.I designed this experiment; H.I, K.S, M.Y, and I.Y wrote this paper.
Funding information
This study was supported by the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan.
Data availability
All data sets about RNA-seq and small RNA-seq data are registered in DDJB as described in text.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Based on the policy of the animal ethics committee of Tokyo University of Agriculture, the use of oocytes collected from porcine ovaries was approved, because all ovary samples were collected from a slaughterhouse where ovaries were discarded due to a lack of edible use.
Consent to participate and for publications
All authors have approved the manuscript and agree to its resubmission.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Munakata Y, Kawahara-Miki R, Shiratsuki S, Tasaki H, Itami N, Shirasuna K, Kuwayama T, Iwata H. Gene expression patterns in granulosa cells and oocytes at various stages of follicle development as well as in in vitro grown oocyte-and-granulosa cell complexes. J Reprod Dev. 2016;62:359–366. doi: 10.1262/jrd.2016-022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122:829–838. doi: 10.1530/rep.0.1220829. [DOI] [PubMed] [Google Scholar]
- 3.Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296:2170–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 4.Rodgers RJ, Irving-Rodgers HF. Formation of the ovarian follicular antrum and follicular fluid. Biol Reprod. 2010;82:1021–1029. doi: 10.1095/biolreprod.109.082941. [DOI] [PubMed] [Google Scholar]
- 5.Mao J, Whitworth KM, Spate LD, Walters EM, Zhao J, Prather RS. Regulation of oocyte mitochondrial DNA copy number by follicular fluid, EGF, and neuregulin 1 during in vitro maturation affects embryo development in pigs. Theriogenology. 2012;78:887–897. doi: 10.1016/j.theriogenology.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibahara H, Ishiguro A, Shirasuna K, Kuwayama T, Iwata H. Follicular factors determining the developmental competence of porcine oocyte. Reprod Med Biol. 2019; In press. [DOI] [PMC free article] [PubMed]
- 7.Di Pietro C. Exosome-mediated communication in the ovarian follicle. J Assist Reprod Genet. 2016;33:303–311. doi: 10.1007/s10815-016-0657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol Reprod. 2012;86:71. doi: 10.1095/biolreprod.111.093252. [DOI] [PubMed] [Google Scholar]
- 9.da Silveira JC, de Ávila ACFCM, Garrett HL, Bruemmer JE, Winger QA, Bouma GJ. Cell-secreted vesicles containing microRNAs as regulators of gamete maturation. J Endocrinol. 2018;236:R15–R27. doi: 10.1530/JOE-17-0200. [DOI] [PubMed] [Google Scholar]
- 10.Hung WT, Hong X, Christenson LK, McGinnis LK. Extracellular vesicles from bovine follicular fluid support cumulus expansion. Biol Reprod. 2015;93:117. doi: 10.1095/biolreprod.115.132977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navakanitworakul R, Hung W-T, Gunewardena S, Davis JS, Chotigeat W, Christenson LK. Characterization and small RNA content of extracellular vesicles in follicular fluid of developing bovine antral follicles. Sci Rep. 2016;6:25486. doi: 10.1038/srep25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasquariello R, Manzoni EFM, Fiandanese N, Viglino A, Pocar P, Brevini TAL, Williams JL, Gandolfi F. Implications of miRNA expression pattern in bovine oocytes and follicular fluids for developmental competence. Theriogenology. 2020;145:77–85. doi: 10.1016/j.theriogenology.2020.01.027. [DOI] [PubMed] [Google Scholar]
- 13.Sohel MM, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft C, Rings F, Uddin MJ, Spencer TE, Schellander K, Tesfaye D. Exosomal and Non-exosomal transport of extra-cellular microRNAs in follicular fluid: implications for bovine oocyte developmental competence. PLoS One. 2013;8(11):e78505. doi: 10.1371/journal.pone.0078505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diez-Fraile A, Lammens T, Tilleman K, Witkowski W, Verhasselt B, De Sutter P, Benoit Y, Espeel M, D’Herde K. Age-associated differential microRNA levels in human follicular fluid reveal pathways potentially determining fertility and success of in vitro fertilization. Hum Fertill (Camb) 2014;17:90–98. doi: 10.3109/14647273.2014.897006. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Yao W, Yao Y, Du X, Zhou J, Ma B, Liu H, Li Q, Pan Z. MiR-92a inhibits porcine ovarian granulosa cell apoptosis by targeting Smad7 gene. FEBS Lett. 2014;588:4497–4503. doi: 10.1016/j.febslet.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Roth LW, McCallie B, Alvero R, Schoolcraft WB, Minjarez D, Katz-Jaffe MG. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. J Assist Reprod Genet. 2014;31:355–362. doi: 10.1007/s10815-013-0161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng JY, An XP, Fang F, Gao KX, Xin HY, Han P, Bao LJ, Ma HD, Cao BY. MicroRNA-10b suppresses goat granulosa cell proliferation by targeting brain-derived neurotropic factor. Domest Anim Endocrinol. 2016;54:60–67. doi: 10.1016/j.domaniend.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Machtinger R, Rodosthenous RS, Adir M, Mansour A, Racowsky C, Baccarelli AA, Hauser R. Extracellular microRNAs in follicular fluid and their potential association with oocyte fertilization and embryo quality: an exploratory study. J Assist Reprod Genet. 2017;34:525–533. doi: 10.1007/s10815-017-0876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshioka K, Suzuki C, Tanaka A, Anas IM, Iwamura S. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod. 2002;66:112–119. doi: 10.1095/biolreprod66.1.112. [DOI] [PubMed] [Google Scholar]
- 20.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011;17:10–12. [Google Scholar]
- 21.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munakata Y, Ueda M, Kawahara-Miki R, Kansaku K, Itami N, Shirasuna K, Kuwayama T, Iwata H. Follicular factors determining granulosa cell number and developmental competence of porcine oocytes. J Assist Reprod Genet. 2018;35:1809–1819. doi: 10.1007/s10815-018-1247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munakata Y, Kawahara-Miki R, Shirasuna K, Kuwayama T, Iwata H. Polyacrylamide gel as a culture substrate improves in vitro oocyte growth from porcine early antral follicles. Mol Reprod Dev. 2017;84:44–54. doi: 10.1002/mrd.22758. [DOI] [PubMed] [Google Scholar]
- 24.Sun MJ, Zhu S, Li YW, Lin J, Gong S, Jiao GZ, Chen F, Tan JH. An essential role for the intra-oocyte MAPK activity in the NSN-to-SN transition of germinal vesicle chromatin configuration in porcine oocytes. Sci Rep. 2016;6:23555. doi: 10.1038/srep23555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292:1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Bijttebier J, Van Soom A, Meyer E, Mateusen B, Maes D. Preovulatory follicular fluid during in vitro maturation decreases polyspermic fertilization of cumulus-intact porcine oocytes in vitro maturation of porcine oocytes. Theriogenology. 2008;70:715–724. doi: 10.1016/j.theriogenology.2008.04.046. [DOI] [PubMed] [Google Scholar]
- 27.Somfai T, Inaba Y, Watanabe S, Geshi M, Nagai T. Follicular fluid supplementation during in vitro maturation promotes sperm penetration in bovine oocytes by enhancing cumulus expansion and increasing mitochondrial activity in oocytes. Reprod Fertil Dev. 2012;24:743–752. doi: 10.1071/RD11251. [DOI] [PubMed] [Google Scholar]
- 28.Lopes JS, Canha-Gouveia A, París-Oller E, Coy P. Supplementation of bovine follicular fluid during in vitro maturation increases oocyte cumulus expansion, blastocyst developmental kinetics, and blastocyst cell number. Theriogenology. 2019;126:222–229. doi: 10.1016/j.theriogenology.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Machtinger R, Laurent LC, Baccarelli AA. Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum Reprod Update. 2016;22:182–193. doi: 10.1093/humupd/dmv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naji M, Nekoonam S, Aleyasin A, Arefian E, Mahdian R, Azizi E, et al. Expression of miR-15a, miR-145, and miR-182 in granulosa-lutein cells, follicular fluid, and serum of women with polycystic ovary syndrome (PCOS). 2018;297:221–31. [DOI] [PubMed]
- 31.Salilew-Wondim D, Ahmad I, Gebremedhn S, Sahadevan S, Hossain MD, Rings F, Hoelker M, Tholen E, Neuhoff C, Looft C, et al. The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PLoS One. 2014;9:e106795. doi: 10.1371/journal.pone.0106795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan G, Zhang L, Fang T, Zhang Q, Wu S, Jiang Y, Sun H, Hu Y. MicroRNA-145 suppresses mouse granulosa cell proliferation by targeting activin receptor IB. FEBS Lett. 2012;586:3263–3270. doi: 10.1016/j.febslet.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 33.Andreas E, Hoelker M, Neuhoff C, Tholen E, Schellander K, Tesfaye D, Salilew-Wondim D. MicroRNA 17-92 cluster regulates proliferation and differentiation of bovine granulosa cells by targeting PTEN and BMPR2 genes. Cell Tissue Res. 2016;366:219–230. doi: 10.1007/s00441-016-2425-7. [DOI] [PubMed] [Google Scholar]
- 34.Cuomo D, Porreca I, Ceccarelli M, Threadgill DW, Barrington WT, Petriella A, D'Angelo F, Cobellis G, De Stefano F, D'Agostino MN, et al. Transcriptional landscape of mouse-aged ovaries reveals a unique set of non-coding RNAs associated with physiological and environmental ovarian dysfunctions. Cell Death Dis. 2018;4:112. doi: 10.1038/s41420-018-0121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun XS, Liu Y, Yue KZ, Ma SF, Tan JH. Changes in germinal vesicle (GV) chromatin configurations during growth and maturation of porcine oocytes. Mol Reprod Dev. 2004;69:228–234. doi: 10.1002/mrd.20123. [DOI] [PubMed] [Google Scholar]
- 36.Kageyama S, Liu H, Kaneko N, Ooga M, Nagata M, Aoki F. Alterations in epigenetic modifications during oocyte growth in mice. Reproduction. 2007;133:85–94. doi: 10.1530/REP-06-0025. [DOI] [PubMed] [Google Scholar]
- 37.Munakata Y, Ichinose T, Ogawa K, Itami N, Tasaki H, Shirasuna K, Kuwayama T, Iwata H. Relationship between the number of cells surrounding oocytes and energy states of oocytes. Theriogenology. 2016;86:1789–1798. doi: 10.1016/j.theriogenology.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 38.Sugiyama M, Sumiya M, Shirasuna K, Kuwayama T, Iwata H. Addition of granulosa cell mass to the culture medium of oocytes derived from early antral follicles increases oocyte growth, ATP content, and acetylation of H4K12. Zygote. 2016;24:848–856. doi: 10.1017/S0967199416000198. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Xu B, Tian GG, Sun T, Wu J. Ablation of the MiR-17-92 microrna cluster in germ cells causes subfertility in female mice. Cell Physiol Biochem. 2018;45:491–504. doi: 10.1159/000487028. [DOI] [PubMed] [Google Scholar]
- 40.Martinez RM, Liang L, Racowsky C, Dioni L, Mansur A, Adir M, Bollati V, Baccarelli AA, Hauser R, Machtinger R. Extracellular microRNAs profile in human follicular fluid and IVF outcomes. Sci Rep. 2018;8:17036. doi: 10.1038/s41598-018-35379-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, Wang L, Wang L, Chen Y, Li F. miR-17-5p affects porcine granulosa cell growth and oestradiol synthesis by targeting E2F1 gene. Reprod Domest Anim. 2019:1459–69. [DOI] [PubMed]
- 42.Nie M, Yu S, Peng S, Fang Y, Wang H, Yang X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol Reprod. 2015;93(4):98. doi: 10.1095/biolreprod.115.130690. [DOI] [PubMed] [Google Scholar]
- 43.Xu L, Sun H, Zhang M, Jiang Y, Zhang C, Zhou J, Ding L, Hu Y, Yan G. MicroRNA-145 protects follicular granulosa cells against oxidative stress-induced apoptosis by targeting Krüppel-like factor 4. Mol Cell Endocrinol. 2017;452:138–147. doi: 10.1016/j.mce.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 44.Cui L, Fang L, Mao X, Chang HM, Leung PCK, Ye Y. GDNF-Induced downregulation of miR-145-5p enhances human oocyte maturation and cumulus cell viability. J Clin Endocrinol Metab. 2018;103(7):2510–2521. doi: 10.1210/jc.2017-02742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 31 kb)
Data Availability Statement
All data sets about RNA-seq and small RNA-seq data are registered in DDJB as described in text.