Abstract
Cardiovascular disease (CVD) causes high morbidity and mortality worldwide. Accumulating research has indicated the possible roles played by circular RNAs (circRNAs) in the pathogenesis of CVD. CircRNAs are non-coding RNAs with covalently closed loop structures. CircRNAs can function by acting as miRNA sponges, RNA binding protein sponges, mRNA transcriptional regulators and templates for protein translation. The specific characteristics of circRNAs such as high stability, abundant distribution, and tissue- and developmental stage-specific expression make them potential biomarkers for the diagnosis and prognosis of CVD. In this paper, we systematically summarized the current knowledge regarding the biogenesis, biological properties and the action mechanisms of circRNAs, elucidated the roles played by circRNAs in the pathogenesis of CVD, and explored the diagnostic potential of circRNAs in CVD. With in-depth studies, an increasing number of molecular mechanisms underlying the participation of circRNAs in CVD may be elucidated, and the application of circRNAs in the clinical diagnosis and prevention of CVD may eventually be realized.
Keywords: cardiovascular disease, circular RNAs, pathogenesis, diagnosis, clinical application
Introduction
Cardiovascular disease (CVD) is one of the leading causes of morbidity and mortality worldwide. In recent decades, scientists have made considerable progress in the diagnosis and treatment of CVD. However, the increasing tendency of the mortality rate of CVD has not been stopped to date. Therefore, novel and effective strategies for the diagnostic and therapeutic interventions of CVD are strongly warranted. A growing number of studies have determined that non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), participate in the pathological processes of CVD and can serve as biological markers in diagnosis, prognosis and clinical treatment (Yang et al., 2016; Zhang L. et al., 2018, 2020). In the last several years, circular RNAs (circRNAs) have also been reported to be associated with CVD.
CircRNAs are covalently closed loop structures with no 5′ cap and 3′ polyadenylated tail. CircRNAs were first identified in plant viruses (Kolakofsky, 1976) and were thought to have no function or very limited function (Nigro et al., 1991; Cocquerelle et al., 1992; Capel et al., 1993). Subsequently, the existence of circRNAs has also been reported in many organisms, such as yeast (Schroeder et al., 1983) and humans (Cocquerelle et al., 1993). The rapid development of prediction, detection and screening technologies for circRNAs facilitates the discovery of different types of circRNAs. Studies have reported that circRNAs might participate in the regulation of physiological and pathological processes of different kinds of CVD (Fan et al., 2017; Lim et al., 2020), such as myocardial infarction (MI) (Geng et al., 2016; Cai et al., 2019), cardiac senescence (Du et al., 2016; Chen et al., 2018) and coronary artery disease (CAD) (Holdt et al., 2016; Dang et al., 2017; Shan et al., 2017). CircRNAs have a variety of characteristics, including high stability, tissue-and developmental-specific expression, and the altered expression in the pathological and normal conditions of various diseases (Werfel et al., 2016; Siede et al., 2017; Gupta et al., 2018). Due to these characteristics, circRNAs exhibit considerable potential as biomarkers for the detection of CVD from human blood samples (Vausort et al., 2016; Zhao et al., 2017). In this paper, we will summarize the available knowledge on the biogenesis of circRNAs, the functions of circRNAs, the roles of circRNAs in CVD and the diagnostic potential of circRNAs in CVD.
Biogenesis of CircRNAs
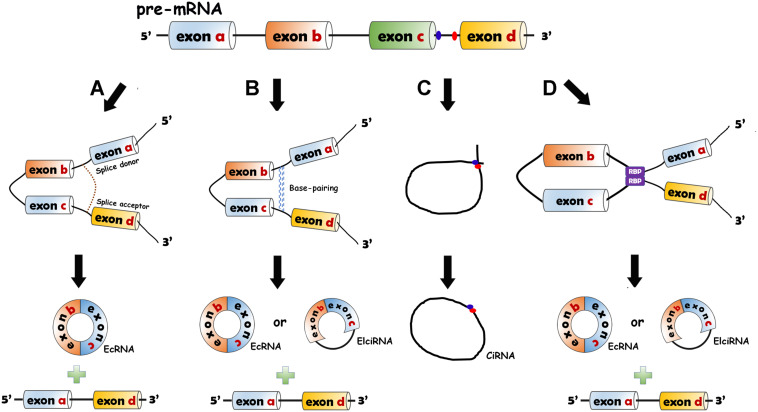
CircRNAs are divided into three categories: exonic circRNAs (ecircRNAs or ecRNAs) (Zhang et al., 2014), circular intronic RNAs (ciRNAs) (Zhang et al., 2013) and exon–intron circRNAs (EIciRNAs) (Li et al., 2015). CircRNAs are generated from pre-mRNAs through backsplicing. Four mechanisms of circRNA formation have been revealed. The 5′ end of the intron (splice donor site, GU) and the 3′ end of the intron (splice acceptor site, AG) can be covalently bound to generate an exon-containing lariat, which will be internally spliced thereafter to form an exonic circle (Jeck et al., 2013; Jeck and Sharpless, 2014) (Figure 1A). The RNA base motifs (e.g., Alu repeats) in the introns of pre-mRNA can pair with the complementary sequences (Jeck et al., 2013; Zhang et al., 2014), and direct cyclization subsequently occurs to generate ecRNAs (introns removed) or EIciRNAs (introns retained) (Jeck et al., 2013) (Figure 1B). In the introns, the C-rich element close to the branch and the GU-rich element close to the 5′ splice site can bind together, and then the other exons and introns are removed by the spliceosome to form ciRNAs (Zhang et al., 2013) (Figure 1C). The bridging of RNA binding proteins (RBPs) with pre-mRNAs has also been elaborated to facilitate the production of circRNAs (ecRNAs or EIciRNAs) (Ashwal-Fluss et al., 2014; Conn et al., 2015) (Figure 1D).
FIGURE 1.
Biogenesis models of circRNAs. (A) Lariat-driven circularization model. The 5′ end of the intron (splice donor site, GU) and the 3′ end of the intron (splice acceptor site, AG) are covalently bound to generate an exon-containing lariat (one or more exons) which is internally spliced to form EcRNA. (B) Intron-pairing-driven circularization model. The RNA base motifs (e.g., Alu repeats) in the introns of pre-mRNA pair with the complementary sequences and the direct cyclization happens to generate ecRNAs or exon–intron circRNAs (EIciRNAs). (C) Circular intronic RNA (ciRNA) formation. The intron lariat is produced by backsplicing process. C-rich element close to the branch and the GU-rich element near the 5′ splice site bind together. The other exons and introns are removed by the spliceosome. (D) RNA binding protein (RBP)-driven model. The bridging of RBPs with pre-mRNAs can give rise to the production of circRNAs (ecRNAs or EIciRNAs).
Multiple factors participate in the biogenesis of circRNAs. Quaking (QKI) and Muscleblind (MBL) proteins function as regulatory activators of the biogenesis of circRNAs (Ashwal-Fluss et al., 2014; Conn et al., 2015). In contrast, an adenosine deaminase acting on RNA-1 inhibits the circRNA formation by destroying the stem structures (Rybak-Wolf et al., 2015). Serine–arginine and heterogeneous nuclear ribonucleoprotein can regulate the generation of circRNAs in Drosophila (Kramer et al., 2015).
Biological Properties of CircRNAs
CircRNAs have some common characteristics and the most important ones are elaborated as follows.
-
(1)
Wide distribution and diversity. CircRNAs are found in many eukaryotic organisms ranging from plants to animals and in all tissues (Jeck et al., 2013). In humans, over 30,000 circRNAs have been found and the number will increase in the future (Xu et al., 2017; Zeng X. X. et al., 2017).
-
(2)
High stability. Because of the covalently closed structures, circRNAs are resistant to the degradation by ribonuclease (RNase) and are more stable than linear RNAs (Suzuki et al., 2006).
-
(3)
Specific expression. CircRNAs are specifically expressed in different tissues, cells and developmental stages (Jakobi et al., 2016; Li Y. S. et al., 2017; Xu et al., 2017). circRNAs have different profiles at four stages of heart differentiation (Li Y. S. et al., 2017). Significant alterations have been detected at different developmental stages in cardiomyocytes derived from induced pluripotent stem cells (Siede et al., 2017).
-
(4)
Evolutionary conservation. Many circRNAs seem to be evolutionarily conserved among species (Jeck et al., 2013; AbouHaidar et al., 2014). Jeck et al. (2013) found the homology of 2121 circRNAs between human fibroblasts and mouse genome. Werfel et al. (2016) reported high homology of 1288 circRNAs across human, mouse and rat. However, Werfel et al. (2016) also revealed that only a small number of circRNAs were conserved. Other studies have also illustrated that many circRNAs are specific to species (Aufiero et al., 2019; Lim et al., 2020).
-
(5)
Dynamic expression profiles between normal and pathological conditions. A lot of circRNAs have altered expression related to diseases. Zheng et al. (2016) revealed altered expression of many circRNAs between normal tissues and cancerous tissues. In many other diseases, the expression differences of circRNAs have also been verified (Werfel et al., 2016; Siede et al., 2017; Gupta et al., 2018). Some studies reported lack of dynamic expression of circRNAs in specific diseases (Werfel et al., 2016; Tan et al., 2017).
The Underlying Mechanisms of CircRNA Functions
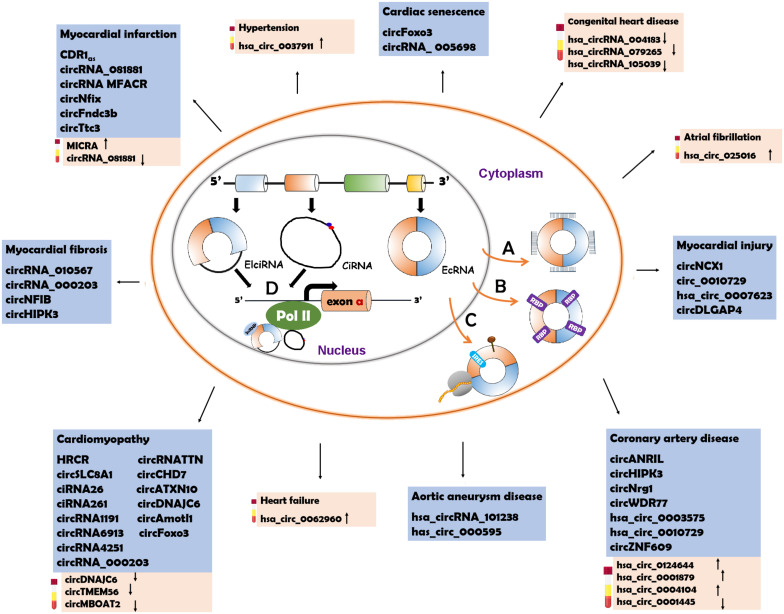
The characteristics of circRNAs, such as wide distribution, high stability, expression specificity and localization, indicate that circRNAs have various biological functions. Recent studies have illustrated that circRNAs can function through different mechanisms (Figure 2).
FIGURE 2.
Underlying action mechanisms of circRNAs and the circRNAs relevant to CVD pathogenesis. (A) CircRNAs can serve as miRNA sponges to regulate the functions of miRNAs. (B) CircRNAs can bind to RNA binding proteins (RBPs ) to influence their functions. (C) CircRNAs can be the templates to encode proteins with the help of internal ribosome entry site (IRES
) to influence their functions. (C) CircRNAs can be the templates to encode proteins with the help of internal ribosome entry site (IRES ) elements or m6A-modification (brown pin
) elements or m6A-modification (brown pin ). (D) circRNAs with intronic sequences can regulate the expression of parental genes through binding to RNA polymerase II (Pol II). The circRNAs involved in CVD are listed in the blue box. The possible diagnostic biomarkers are displayed in the pink box. EcRNA, exonic circRNA; EIciRNA, exon–intron circRNA; CiRNA, circular intronic RNA.
). (D) circRNAs with intronic sequences can regulate the expression of parental genes through binding to RNA polymerase II (Pol II). The circRNAs involved in CVD are listed in the blue box. The possible diagnostic biomarkers are displayed in the pink box. EcRNA, exonic circRNA; EIciRNA, exon–intron circRNA; CiRNA, circular intronic RNA.
Some CircRNAs Act as miRNA Sponges
CircRNAs contain miRNA response elements (MREs) that facilitate the binding of circRNAs and miRNAs. This binding decreases the level of functional miRNAs and thus increases the expression of miRNA targets (Hansen et al., 2013a; Tay et al., 2014). This process is known as the “sponge effect,” as circRNAs can absorb miRNAs similar to sponges (Figure 2A). Many circRNAs can function as miRNA sponges. CiRS-7/CDR1as contains more than 70 binding sites for miR-7 that are involved in the pathogenesis of various diseases (Hansen et al., 2013b; Pan et al., 2018; Liu et al., 2019). The binding of ciRS-7/CDR1as to miR-7 causes downregulation of miR-7 and elevated levels of miR-7 target genes. Sry, a testis-specific circRNA, has 16 conserved MREs for miR-138 (Hansen et al., 2013a). The activity of miR-138 is inhibited due to the sponge effect and the miR-138 target genes are upregulated (Hansen et al., 2013a). CircHIPK3 have different MREs (total number: 18) which allow it to bind to nine different miRNAs (Zheng et al., 2016). HRCR can bind to miR-223 which promotes the pathogenesis of cardiac hypertrophy and heart failure (HF) (Wang et al., 2016). CircNFIB can competitively absorb miR-433 to enhance cardiac fibroblast proliferation induced by the stimulation of TGF-β (Zhu et al., 2019). CircRNA_000203 can directly sponge miR-26b-5p and miR-140-3p to regulate the occurrence of cardiac hypertrophy (Li et al., 2019).
Some CircRNAs Serve as RBP Sponges
CircRNAs can interact with RBPs and inhibit RBP activity (Figure 2B). CircMbl is produced from the same pre-mRNA as the MBL protein and can absorb MBL proteins to regulate the subsequent physiological processes (Ashwal-Fluss et al., 2014). CircPABPN1 can bind to HuR, which is a well-known RBP, to prevent the interaction between HuR and PABPN1 mRNA, suppressing the translation of PABPN1 mRNA (Abdelmohsen et al., 2017). CircANRIL competitively recruits PES1 (an essential 60S-preribosomal assembly factor), leading to inhibition of ribosome biogenesis (Holdt et al., 2016). CircFoxo3 participates in the processes of cardiomyocyte senescence and cell cycle progression through interacting with different RBPs, such as anti-senescent protein ID-1, transcription factor E2F1, anti-stress proteins FAK, p21 (cyclin-dependent kinase inhibitor 1) and CDK2 (cyclin-dependent kinase 2) (Du et al., 2017). CircAmotl1 can protect cardiomyocytes and promote cell proliferation and wound repair by binding to the cardioprotective molecules (PDK1 and AKT1) and to STAT3 (signal transducer and activator of transcription 3) (Yang Z. G. et al., 2017; Zeng Y. et al., 2017).
Some CircRNAs Encode Peptides
Studies conducted in recent years have demonstrated that circRNAs can serve as templates for protein translation (Chen and Sarnow, 1995; Wesselhoeft et al., 2018) (Figure 2C). It was first observed in prokaryotes that circRNAs were able to encode proteins. In 1986, studies in hepatitis D virus showed that circRNAs could be translated into functional proteins (Kos et al., 1986). In Escherichia coli, a circRNA was also reported to act as a translation template (Perriman and Ares, 1998). CircRNAs lack a 5′ cap and a 3′ polyadenylated tail which are typical translation initiation structures of linear RNAs. Nevertheless, instead of recruiting ribosomes, circRNA translation is initiated with the help of specific elements, such as IRES (internal ribosome entry site) and N-methyladenosine (m6A) (Chen and Sarnow, 1995; Wesselhoeft et al., 2018). When IRESs are introduced into a circRNA, the synthetic circRNA initiates translation (Chen and Sarnow, 1995). CircZNF609 has an IRES element and can be translated into a protein that functions in myoblast proliferation (Legnini et al., 2017). CircFBXW7 can be translated into a functional protein that plays a role in the inhibition of glioma tumorigenesis with the assistance of the IRES element (Yang et al., 2018). m6A modification has been illustrated to facilitate the translation of linear mRNAs in a cap-independent manner (Meyer et al., 2015). Some circRNAs also employ m6A to initiate translation (Yang Y. et al., 2017). The m6A reader YTHDF3 can interact with circRNAs and subsequently recruit translation initiation factors to drive the initiation of circRNA translation (Yang Y. et al., 2017). In a study based on ribosome profiling of Drosophila heads, circMbl3 was demonstrated to be the template for splicing-dependent translation (Pamudurti et al., 2017). Another recent study found that circβ-catenin could be translated into the β-CATENIN isoform, a functional protein that activates the Wnt/β-catenin pathway and promotes tumor development in liver cancer (Liang et al., 2019).
Some CircRNAs Regulate the Transcription of Parental Genes
Of the three types of circRNAs, ecRNAs account for the majority and are predominantly located in the cytosol (Hansen et al., 2013a; Memczak et al., 2013). The cytoplasmic localization of ecRNAs facilitates their functions as miRNA sponges, RBP sponges and translational templates. CiRNAs and EIciRNAs are confined to the nucleus due to their intronic sequences (Zhang et al., 2013). Recent studies have verified the roles of nucleus-localized ciRNA and EIciRNA in the transcriptional regulation of their parental genes (Zhang et al., 2013; Li et al., 2015) (Figure 2D). CiRNAs can directly interact with RNA polymerase II (Pol II) and enhance parental gene transcription (Zhang et al., 2013), whereas EIciRNAs bind to the U1 small nuclear ribonucleoproteins (snRNPs) at first, and the complex promotes the interaction with Pol II (Li et al., 2015). CiRNA-ankrd52 can bind to Pol II of the pre-mRNA of the ANKRD52 gene to promote transcription, while the knockdown of ciankrd52 may cause a significant transcriptional decrease in the parental gene (Zhang et al., 2013). CircEIF3J and circPAIP2 are EIciRNAs that can form a complex with U1 snRNP and Pol II (Li et al., 2015). In general, ciRNAs and EIciRNAs can serve as transcription regulators of their parental genes.
CircRNA Identification and Research Databases
Various methods have been developed to identify and study the functions of circRNAs. RNA-seq, microarray, Northern Blot and RT-PCR analyses are the most widely used. RNA-seq can identify new circRNA species and quantify circRNA expression. Next-generation sequencing and depletion of ribosomal RNA methods may be cooperatively utilized for circRNA-seq. The microarray method can only detect and quantify known circRNAs approximately and the results require further confirmation. For high-throughout sequencing, reliable and precise algorithms are of vital importance. Many algorithms have been developed, such as circRNA-finder, MapSplice, CIRCexplorer, and CIRI (Hansen et al., 2016). The combined use of these algorithms may improve the accuracy and sensitivity of circRNA identification. Northern Blot and RT-PCR methods only validate known circRNAs dependent on an exoribonuclease-RNase R, which cleaves linear RNAs from the 3′ end (Asha et al., 1983). With circular structures, circRNAs can avoid the cleavage of RNase R. Thus, in the extraction process, total RNA is digested by RNase R to discard the linear RNAs (Szabo and Salzman, 2016). The collected RNAs are then combined with specific probes (Northern Blot method) or reverse-transcribed to obtain cDNAs of circRNAs (RT-PCR method). Aside from these methods for circRNA identification, some other methods are employed to analyze the subcellular localization (fluorescence in situ hybridization) or the interaction between circRNAs and miRNAs/RBPs (e.g., RNA immunoprecipitation and dual luciferase reporter assay) (Li Y. et al., 2018; Zirkel and Papantonis, 2018). In addition to these known methods, simpler and more efficient methods are still urgently needed for the identification and characterization of circRNAs.
To elucidate the roles of circRNAs, many online databases have been developed (Table 1). Different databases emphasize on different aspects of circRNA research including prediction, identification, protein-coding annotation, circRNA-miRNA interactions and circRNA-RBP interactions. The combined use of these databases may help to establish a foundation for the further studies of circRNAs.
TABLE 1.
Database for circRNA research.
| Name | URL | Function |
| circBase | http://www.circbase.org/ | Collects the circular RNA information of many species, such as human, mouse, C. elegans, etc. |
| CircInteractome | https://circinteractome.nia.nih.gov/ | Maps RBP- and miRNA-binding sites on human circRNAs; Designs primers and siRNA sequence of circRNAs. |
| CircNet | http://circnet.mbc.nctu.edu.tw/ | Integrates the following information: identification of new circRNAs; genome annotation of circRNAs; the expression profile of circRNAs; the network of circRNA-miRNA -mRNA. |
| circRNABase | http://starbase.sysu.edu.cn/mirCircRNA.php | Integrates the published data to construct the interaction network of miRNA and circRNA, or circRNA and RBP. |
| circRNADb | http://reprod.njmu.edu.cn/circrnadb | Summarizes circRNAs that encode proteins; contains 32,914 human circRNAs. |
| Circ2Traits | http://gyanxet-beta.com/circdb/ | Collects circRNAs potentially associated with human diseases or traits. |
| CIRCpedia | http://www.picb.ac.cn/rnomics/circpedia/ | Provides annotations on circRNAs and variable shear events of different cell lines/tissues |
| Deepbase | http://deepbase.sysu.edu.cn/ | Emphasizes the interaction of ceRNA molecular network; integrates the information related to circRNAs. |
| TSCD | http://gb.whu.edu.cn/TSCD/ | Provides information of tissue-specific circRNAs in main tissues of human and mouse. |
| circRNA disease | http://cgga.org.cn:9091/circRNADisease/ | Includes the data of the association between circRNAs and disease, the expression of circRNAs in the patients, the detection methods of circRNA |
| CSCD | http://gb.whu.edu.cn/CSCD | Collects cancer-specific circRNAs; elaborates the interaction between miRNA and circRNA, or circRNA and RBP; predicts variable splicing of related genes. |
TSCD, tissue specific circRNA database; CSCD, cancer-specific circRNA database; RBP, RNA binding protein.
CircRNAs and Cardiovascular Diseases
With the use of advanced technologies in sequencing and data analysis, a lot of circRNAs have been detected in human hearts and been reported to be associated with CVD (Jakobi et al., 2016; Werfel et al., 2016; Fan et al., 2017; Tan et al., 2017) (Table 2 and Figure 2). Moreover, circRNAs have been observed to have great potential as biomarkers for the diagnosis and prognosis of CVD (Vausort et al., 2016; Sonnenschein et al., 2019; Wu et al., 2019; Vilades et al., 2020) (Table 3 and Figure 2).
TABLE 2.
CircRNAs with CVD.
| CVD type | CircRNAs | Source | Action mechanism | Regulation | References |
| Myocardial infarction | CDR1as | Mouse myocardial tissue | Sponge miR-7 | Up | Geng et al. (2016) |
| CircRNA_081881 | Blood | Sponge miR-548 | Down | Deng et al. (2016) | |
| CircRNA MFACR | A/R and I/R mouse models | Sponge miR-652-3p | Up | Wang et al. (2017) | |
| CircNfix | Mouse heart section | Dual function as miR-214 sponge and inducing YBX1 degradation | Down | Huang et al. (2019) | |
| CircFndc3b | Mouse hearts | Interact with RBP-FUS | Down | Garikipati et al. (2019) | |
| CircTtc3 | Rat myocardium | Sponge miR-15b-5p | Up | Cai et al. (2019) | |
| Myocardial fibrosis | CircRNA_010567 | Mouse myocardium, cardiac fibroblasts | Sponge miR-141 | Up | Zhou and Yu (2017) |
| CircRNA_000203 | Mouse myocardium, cardiac fibroblasts | Sponge miR-26b-5p | Up | Tang et al. (2017) | |
| CircNFIB | Mouse heart tissue | Sponge miR-433 | Down | Zhu et al. (2019) | |
| CircHIPK3 | Mouse cardiac fibroblasts | Sponge miR-29b-3p | UP | Ni et al. (2019) | |
| Myocardial injury | CircNCX1 | Mouse cardiomyocytes | Sponge miR-133a-3p | Up | Li M. et al. (2018) |
| Circ_0010729 | Human cardiomyocytes | Sponge miR-145-5p | Down | Jin and Chen (2019) | |
| Hsa_circ_0007623 | Acute ischemia mice | Sponge miR-297 | Up | Zhang Q. et al. (2020) | |
| CircDLGAP4 | Myocardial ischemia-reperfusion injury | Sponge miR-143 | Up | Wang S. et al. (2019) | |
| Cardiomyopathy | HRCR | Mouse cardiomyocytes | Sponge miR-223-5p | Down | Wang et al. (2016) |
| CircSLC8A1 | Mouse cardiomyocytes | Sponge miR-133a | — | Lim et al. (2019) | |
| CircRNA_000203 | NMVCs | Sponge miR-26b-5p and miR-140-3p | Up | Li et al. (2019) | |
| CiRNA26 | Mouse cardiomyocytes | Sponge several miRNAs | Down | Meng et al. (2019) | |
| CiRNA261 | Mouse cardiomyocytes | Sponge several miRNAs | Up | Meng et al. (2019) | |
| CircRNA1191 | Mouse cardiomyocytes | Sponge several miRNAs | Down | Meng et al. (2019) | |
| CircRNA6913 | Mouse cardiomyocytes | Sponge several miRNAs | Up | Meng et al. (2019) | |
| CircRNA4251 | Mouse cardiomyocytes | Sponge several miRNAs | Down | Meng et al. (2019) | |
| CircRNATTN 1/2/4/5 | DCM patient with a heterozygous mutation in RBM20 (E913K). | Unknown | Down | Khan et al. (2016) | |
| CircSLC8A1 | Human dilated cardiomyopathy | Unknown | Up | Siede et al. (2017) | |
| CircCHD7 | Human dilated cardiomyopathy | Unknown | Up | Siede et al. (2017) | |
| CircATXN10 | Human dilated cardiomyopathy | Unknown | Up | Siede et al. (2017) | |
| CircDNAJC6 | Human dilated cardiomyopathy | Unknown | Down | Siede et al. (2017) | |
| CircSLC8A1 | Heart tissues from DCM patient | Unknown | Up | Lei et al. (2018) | |
| CircAmotl1 | Human neonatal cardiac tissue | Interact with AKT and PDK1 | Up | Zeng Y. et al. (2017) | |
| CircFoxo3 | Mouse heart tissue | Interact with ID-1, E2F1, FAK, and HIF1a | Up | Du et al. (2017) | |
| Aortic aneurysm disease | Hsa_circ_000595 | Aortic smooth muscle cells | Sponge miR-19a | Up | Zheng et al. (2015) |
| Hsa_circRNA_101238 | Human aortic segments | Sponge miR-320a | Up | Zou et al. (2017) | |
| Cardiac senescence | CircRNA005698 | Sow cardiac muscle | Sponge seven miRNAs | Up | Chen et al. (2018) |
| CircFoxo3 | Mouse heart tissue | Interact with ID-1, E2F1, FAK, and HIF1a | Up | Du et al. (2017) | |
| Mouse cardiac fibroblast | p21 and CDK2 | Up | Du et al. (2016) | ||
| Coronary artery disease | CircANRIL | Human peripheral blood | Interact with PES1 | Down | Holdt et al. (2016) |
| Different cell lines | interact with INK4/ARF | Down | Burd et al. (2010) | ||
| CircHIPK3 | Diabetic retinopathy | Sponge miR-30a-3p | Up | Shan et al. (2017) | |
| CircNrg1 | MASMCs | Sponge miR-193b-5p | Down | Sun et al. (2019) | |
| CircWDR77 | VSMCs | Sponge miR-124 | Up | Chen et al. (2017) | |
| Nine circRNAs | VSMCs | Sponge miR-130a-3p | — | Pan et al. (2017) | |
| Hsa_circ_0003575 | HUVECs | Sponge miR-9-5p and miR-199-3p | Up | Li C. Y. et al. (2017) | |
| Hsa_circ_0010729 | HUVECs | Sponge miR-186 | Up | Dang et al. (2017) | |
| CircZNF609 | HUVECs | Sponge miR-615-5p | Up | Liu et al. (2017) |
VSMC, vascular smooth mother cell; SMC, smooth mother cell; PBMC, peripheral blood mononuclear cell; HUVEC, human umbilical vein endothelial cells.
TABLE 3.
Circulating circRNAs as diagnostic biomarkers of CVD.
| CVD type | CircRNAs | Source | Regulation | References |
| Myocardial infarction | MICRA | Peripheral blood | Up | Vausort et al. (2016) |
| CircRNA_081881 | Plasma | Down | Deng et al. (2016) | |
| Congenital heart diseases | Hsa_circRNA_004183 | Plasma | Down | Wu et al. (2019) |
| Hsa_circRNA_079265 | Plasma | Down | Wu et al. (2019) | |
| Hsa_circRNA_105039 | Plasma | Down | Wu et al. (2019) | |
| Hypertension | Hsa_circ_0037911 | Whole blood | Up | Bao et al. (2018) |
| Cardiomyopathy | CircDNAJC6 | Serum | Down | Sonnenschein et al. (2019) |
| CircTMEM56 | Serum | Down | Sonnenschein et al. (2019) | |
| CircMBOAT2 | Serum | Down | Sonnenschein et al. (2019) | |
| Heart failure | Hsa_circ_0062960 | Plasma | Up | Sun et al. (2020) |
| Coronary artery disease | Hsa_circ_0124644 | Peripheral blood | Up | Zhao et al. (2017) |
| Hsa_circ_0001879 | PBMCs | Up | Wang L. et al. (2019) | |
| Hsa_circ_0004104 | PBMCs | Up | Wang S. et al. (2019) | |
| Hsa_circ_0001445 | Plasma | Down | Vilades et al. (2020) | |
| Atrial fibrillation | Hsa_circ_025016 | Plasma | Up | Zhang J. et al. (2018) |
PBMCs, peripheral blood mononuclear cells.
Myocardial Infarction
Myocardial infarction provokes cardiac remodeling and is often complicated by arrhythmia, shock or HF. CDR1as has been determined to sponge miR-7 and to have elevated levels in in mice with MI (Geng et al., 2016). The increased levels of CDR1as can upregulate PARP and SP1, the targets of miR-7, which may subsequently enlarge the size of MI (Geng et al., 2016). CircRNA_081881 has been found to be associated with acute MI. CircRNA_081881 has seven binding sites for miR-548, and the competitive binding upregulates the expression of PPARγ which can protect the heart from acute MI (Deng et al., 2016). MTP18 participates in the development of MI. CircRNA MFACR could absorb miR-652-3p to increase the expression of MTP18 and subsequently promote the progression of MI (Wang et al., 2017). CircNfix has been revealed to promote the degradation of Ybx1 and sponge miR-214. The downregulation of circNfix inhibited cardiomyocyte apoptosis after MI and promoted cardiac regeneration and repair (Huang et al., 2019). CircFndc3b exhibited significantly decreased expression levels in post-MI mouse hearts. CircFndc3b could function through interacting with RBP-FUS which regulates the expression and the signaling pathway of vascular endothelial growth factor-A (VEGF-A). The overexpression of circFndc3b in post-MI mouse hearts could decrease cardiomyocyte apoptosis and fibrosis, enhance neovascularization, reduce infarct size and attenuate left ventricular dysfunction after MI (Garikipati et al., 2019). CircTtc3 was reported to be significantly upregulated in rats with MI (Cai et al., 2019). CircTtc3 could recruit miR-15b-5p to repress its inhibitory effect on Arl2, an ADP-ribosylation factor relevant to cardiomyocyte viability (Cai et al., 2019). The knockdown of circTtc3 exacerbated the symptoms of cardiac dysfunction post-MI, suggesting that circTtc3 plays a role in the cardiac protection in MI (Cai et al., 2019).
Myocardial Fibrosis
Myocardial fibrosis is a disease condition in which normal myocardium is replaced by non-beating cardiac fibroblasts, resulting in diastole difficulty. CircRNA_010567 exhibited significantly increased levels in both diabetic mouse myocardium and Angiotensin-II (Ang-II)-induced cardiac fibroblasts (CFs) (Zhou and Yu, 2017). CircRNA_010567 was found to recruit miR-141. The competitive binding of circRNA_010567 and miR-141 released the inhibitory effect of TGF-β1, a pro-fibrotic factor, thereby promoting myocardial fibrosis (Zhou and Yu, 2017). CircRNA_000203 was also revealed to be elevated in diabetic mouse myocardium and Ang-II-induced CFs (Tang et al., 2017). CircRNA_000203 could sponge miR-26b-5p to attenuate the inhibition of its targets, Col1a2 and CTGF, which are fibrosis-associated proteins. Thus, the upregulation of circRNA_000203 may promote the proliferation of CFs (Tang et al., 2017). CircNFIB was downregulated in primary adult CFs treated with TGF-β (Zhu et al., 2019). Overexpression of circNFIB attenuates CF proliferation while inhibition of circNFIB promotes CF proliferation, indicating the cardioprotective role of circNFIB (Zhu et al., 2019). CircHIPK3 was upregulated in CFs treated with Ang-II (Ni et al., 2019). CircHIPK3 was revealed to sponge miR-29b-3p which can target fibrosis-associated proteins such as Col1a2, Col3a1 and a-SMA. The elevated level of circHIPK3 ultimately enhanced the function of Col1a2, Col3a1 and a-SMA, thereby promoting myocardial fibrosis (Ni et al., 2019). In general, circRNA_010567, circRNA_000203 and circHIPK3 are profibrotic while circNFIB is antifibrotic.
Myocardial Injury
Myocardial injury as well as apoptosis is usually correlated to HF, MI, and ischemia–reperfusion (I/R) injury. CircNCX1 was found to have elevated levels during oxidative stress (Li M. et al., 2018). CircNCX1 could bind to miR-133a-3p and subsequently increase the activity of cell death-inducing protein 1 (CDIP1), inducing apoptosis and I/R injury (Li M. et al., 2018). Circ_0010729 was elucidated to play a role in the injury of human cardiomyocytes induced by oxygen-glucose-deprivation (OGD) (Jin and Chen, 2019). The downregulation of circ_0010729 attenuated the OGD-induced cell injury by activating the mTOR and MEK/ERK pathways (Jin and Chen, 2019). Hsa_circ_0007623 was confirmed to have cardioprotective effects in isoproterenol-induced acute ischemia mice. Hsa_circ_0007623 was able to bind to miR-297 and repress the inhibitory effect of miR-297 on VEGF-A, thereby promoting cell proliferation, migration and angiogenesis (Zhang Q. et al., 2020). Wang S. et al. (2019) speculated that circDLGAP4 may regulate cardiomyocyte apoptosis in myocardial I/R injury through targeting miR-143. However, no additional experimental evidence has been reported to date.
Cardiomyopathy
Cardiomyopathy is a disease with abnormal heart muscles. The muscles are stretched, weakened, or have other structural changes, causing pump difficulties of heart. Most patients with cardiomyopathy will have HF (Molkentin et al., 1998; Aaronson and Sackner-Bernstein, 2006; Rajabi et al., 2007; van Rooij et al., 2008; Authors/Task Force et al., 2014; Wang et al., 2016; Lim et al., 2019). Hypertrophic cardiomyopathy (HCM, cardiac hypertrophy) and dilated cardiomyopathy (DCM, cardiac dilatation) are two common subtypes of cardiomyopathy. When HCM happens, the heart muscles are stretched and become thick, thereby decreasing or blocking the blood flow. In DCM, the heart muscles are weakened, leading to the loss of pumping power of the heart. HRCR could bind to miR-223-5p to decrease its activity and could subsequently upregulate its target, ARC (apoptosis inhibitor with CARD domain), resulting in the inhibition of HCM and HF (Wang et al., 2016). CircSLC8A1 has recently been demonstrated to be the sponge of miR-133a (Lim et al., 2019). The knockdown of circSLC8A1 in cardiomyocytes lessened the hypertrophy induced by pressure overload, whereas the overexpression of circSLC8A1 caused HF (Lim et al., 2019). CircRNA_000203 can bind to miR-26b-5p and miR-140-3p to increase the expression level of their target gene-GATA4 (Li et al., 2019). The elevated level of GATA4 promotes the occurrence of cardiac hypertrophy (Li et al., 2019). Several circRNAs (ciRNA26, ciRNA261, circRNA1191, circRNA4251, and circRNA6913) were reported to exhibit altered expression in cardiac cells with HCM when they were cultured in high levels and normal levels of D-glucose, respectively (Meng et al., 2019). These circRNAs might be sponges of more than 60 miRNAs, suggesting that they have vital functions in HCM.
RBM20 plays a critical role in the splicing of many cardiac genes, whose mutation will cause aggressive DCM (Brauch et al., 2009). RBM20 can regulate the generation of TTN circRNA which might be involved in DCM (Khan et al., 2016). CircSLC8A1, circCHD7, and circATXN10 were found to have elevated expression levels in DCM patients compared with control patients, while circDNAJC6 expression levels were reduced (Siede et al., 2017). The increased expression level of circSLC8A1 was also observed in another study in dilated heart tissue compared with control tissues (Lei et al., 2018). CircAmotl1 has been reported to bind to PDK1 and AKT1, two cardioprotective molecules (Zeng Y. et al., 2017). This interaction activated AKT1 through phosphorylation and facilitated the nuclear translocation of AKT1 to protect cardiomyocytes in doxorubicin-induced cardiomyopathy (Zeng Y. et al., 2017). CircFoxo3 has been determined to promote doxorubicin-induced ventricular dilatation (Du et al., 2017).
Aortic Aneurysm Disease
Aortic dissection is the most serious aneurysm disease. Through screening of aortic tissues from patients with aortic dissection aneurysms, Zheng et al. (2015) found an obviously upregulated circRNA, hsa_circ_000595. Hsa_circ_000595 was found to promote apoptosis in vascular smooth mother cells (VSMCs) under hypoxic conditions through upregulating miR-19a expression. Zou et al. (2017) found 162 circRNAs with abnormal expression by microarray analysis of three thoracic aortic dissection (TAD) patients and three control subjects, in which hsa_circRNA_101238 was notably increased. Hsa_circRNA_101238 could sponge miR-320a to inhibit its activity, thereby increasing the levels of its target, MMP9 protein (a TAD related protein) (Zou et al., 2017).
Cardiac Senescence
Cardiac senescence greatly depresses heart function. Through high throughput RNA-seq, Chen et al. (2018) identified 22 circRNAs with dynamic expression in cardiac muscle during aging. Some of them might regulate the pro-coagulation process. CircRNA005698 was found to be a sponge for seven miRNAs and might be a biomarker for cardiac senescence (Chen et al., 2018). CircFoxo3 was reported to bind to several RBPs (ID-1, E2F1, FAK and HIF1ɑ) and inhibit their activities, thereby promoting cardiomyocyte senescence (Du et al., 2017). CircFoxo3 could also absorb two G1 to S phase transition-related proteins (p21 and CDK2) and suppress their functions in the cell cycle, leading to cell cycle repression (Du et al., 2016).
Hypertension
Hypertension is a common chronic disease and a major risk factor for CVD. Through profiling of plasma, Wu et al. (2017) identified 59 circRNAs that exhibited altered expression between hypertensive patients and healthy controls. Additionally, a profiling with blood found 351 circRNAs that had different levels from patients with chronic thromboembolic pulmonary hypertension and healthy people (Miao et al., 2017). However, due to the small cohort (five patients and five controls for both studies), all the results in these studies require further validation. Overall, the number of studies on circRNAs in hypertension remains small. More studies should be conducted to elucidate the mechanisms.
Coronary Artery Disease
Coronary artery disease is a chronic disease mainly caused by atherosclerosis. miRNAs have been shown to function in all pathogenesis processes of CAD (Zhang L. et al., 2018, 2020), such as endothelial dysfunction, lipid metabolism disorder, proliferation and differentiation of smooth muscle cells (SMCs). Recently, circRNAs have also been found to participate in the development of CAD. CircANRIL was testified to interact with PES1 to suppress pre-rRNA maturation and subsequently restrain the biogenesis of ribosomes, which consequently enhances the stability of anti-atherogenic cells (Holdt et al., 2016). The high level of circANRIL might reduce the severity of CAD (Holdt et al., 2016). Thus, circANRIL plays an atheroprotective role. In addition, circANRIL was also illustrated to play a role in the formation of atherosclerosis by regulating the expression of INK4/ARF (Burd et al., 2010). CircHIPK3 was found to have elevated level in diabetic retinopathy. CircHIPK3 could promote endothelial proliferation and vascular dysfunction through binding to miR-30a-3p which can target VEGFC and WNT2 (Shan et al., 2017). Neuregulin-1 (NRG1) participates in vascular physiopathology (Odiete et al., 2012). CircNrg1 was revealed to sponge miR-193b-5p which could target its host mRNA, Nrg1. The overexpression of circNrg1 led to an elevated level of NRG1, whereas the silencing of circNrg1 decreased the level of NRG1 (Sun et al., 2019). CircWDR77 was determined to be increased in VSMCs treated with high glucose (Chen et al., 2017). CircWDR77 could sponge miR-124 to increase the activity of its target, FGF-2 (fibroblast growth factor 2), thereby promoting VSMC proliferation and migration (Chen et al., 2017). Pan et al. (2017) identified 24 circRNAs which were differentially expressed by circRNA microarray. Among these circRNAs, nine circRNAs were found to sponge hsa-miR-130a-3p and then increase the level of TRPM3 which regulates the proliferation and contractility of VSMCs in cooperation with cholesterol (Pan et al., 2017). OxLDL treatment can be employed to induce endothelial cells injury to simulate the pathogenesis of atherosclerosis or CAD. Hsa_circ_0003575 was found to have elevated expression in oxLDL-induced HUVECs (human umbilical vein endothelial cells) (Li C. Y. et al., 2017). The study elaborated that hsa_circ_0003575 could regulate endothelial cells proliferation and angiogenesis probably through interacting with miR-9-5p and miR-199-3p (Li C. Y. et al., 2017). Dang et al. (2017) performed a circRNA microarray in hypoxia-induced HUVECs to identify 36 circRNAs with abnormal expression, and they reported that hsa_circ_0010729 was upregulated. Hsa_circ_0010729 could sponge miR-186 to regulate vascular endothelial cell proliferation and apoptosis via targeting HIF-1α. Circular RNA-ZNF609 was reported to have increased expression in HUVECs under high glucose and hypoxia stress, both in vivo and in vitro. CircZNF609 could regulate endothelial cell function by binding to miR-615-5p which targets the transcription factor MEF2A. The knockdown of circZNF609 promoted endothelial cell migration and inhibited endothelial cell apoptosis (Liu et al., 2017).
CircRNAs as Biomarkers for CVD
Currently, a variety of circulating molecules, such as proteins and miRNAs, have been illustrated to have diagnostic potential for CVD. Such proteins as troponins, creatine kinase-MB and myoglobin have been widely used in the clinic. However, these proteins are not specific and are not applicable for the early diagnosis. Additionally, these proteins are easily influenced by such factors as the heart-associated diseases, medication, patient genetic background, and age (Chen et al., 2008; Lawrie et al., 2008). Therefore, protein biomarkers have limited diagnostic value. Circulating miRNAs have been elaborated to have high specificity and strong potential for early diagnosis. However, circulating miRNAs have not been applied in the clinic due to their low content and time-consuming detection (Zhang L. et al., 2018). Circulating circRNAs have many features resembling circulating miRNAs including high stability, sensitivity and specificity, which are essential for biomarkers. Meanwhile, the circulating levels of circRNAs are not low, and some circRNAs even have high content, making detection easier. Many studies have revealed the considerable potential of circulating circRNAs as novel and promising biomarkers for the early diagnosis of CVD (Table 3 and Figure 2).
CircZNF609 (MICRA) had lower levels in the peripheral blood of MI patients than in healthy controls (Vausort et al., 2016). Circulating MICRA was demonstrated to have a high value of predicting left ventricular dysfunction (Vausort et al., 2016). CircRNA_081881 was downregulated in the plasma of AMI patients and might be a promising target for AMI diagnosis and therapy (Deng et al., 2016). The level of hsa_circ_0124644 was increased in the peripheral blood of CAD patients and was found to have a significant association with CAD. Receiver operating characteristic (ROC) analysis revealed that circulating hsa_circ_0124644 might be a potential diagnostic biomarker for CAD (Zhao et al., 2017). Hsa_circ_0001879 and hsa_circ_0004104 were found to have increased levels in the peripheral blood mononuclear cells (PBMCs) of CAD patients (Wang L. et al., 2019). ROC analysis revealed their high accuracy in the diagnosis of CAD. Furthermore, the combination of hsa_circ_0001879, hsa_circ_0004104 and CAD risk factors had the highest value to discriminate CAD patients from healthy controls (Wang L. et al., 2019). Atrial fibrillation (AF) is a common complication after coronary artery bypass grafting (CABG) (Maesen et al., 2012). Hsa_circ_025016 was upregulated in the plasma of patients with new-onset AF after isolated off-pump CABG. ROC analysis revealed a high diagnostic value (Zhang J. et al., 2018). The analysis with a large validation cohort confirmed the diagnostic power of hsa_circ_025016 (Zhang J. et al., 2018). All results indicated that hsa_circRNA_025016 might be a promising biomarker for the prediction of new-onset AF after isolated off-pump CABG (Zhang J. et al., 2018). Sun et al. (2020) performed circRNA microarrays and found significantly upregulated plasma levels of hsa_circ_0112085, hsa_circ_0062960, hsa_circ_0053919 and hsa_circ_0014010 in HF patients. ROC analysis revealed that hsa_circ_0062960 had great potential to be a diagnostic biomarker of HF (Sun et al., 2020). A study using whole blood revealed that the hsa_circ_0037911 level was significantly increased in hypertensive patients in contrast to the control group (Bao et al., 2018). Another study revealed reduced expression levels of circRNAs (DNAJC6, TMEM56 and MBOAT2) in the serum of patients with HCM (Sonnenschein et al., 2019). All three circRNAs had high discrimination power between HCM patients and the control cohort. Moreover, circTMEM56 and circDNAJC6 could be indicators of disease severity in patients with HCM (Sonnenschein et al., 2019). Wu et al. (2019) reported three notably downregulated circRNAs (hsa_circRNA_004183, hsa_circRNA_079265 and hsa_circRNA_105039) in the plasma of children with congenital heart diseases (CHD) and employed ROC analyses to determine their potential to be biomarkers. They found the great potential of three circRNAs as novel non-invasive diagnostic biomarkers for CHD (Wu et al., 2019). Hsa_circ_0001445 was shown to have lower plasma levels in CAD patients than in the control group (Vilades et al., 2020). Hsa_circ_0001445 is secreted into circulation through being packaged in extracellular vesicles of coronary SMCs (Vilades et al., 2020). The coronary atherosclerotic condition abolished the association of hsa_circ_0001445 and vesicles, leading to the downregulation of plasma hsa_circ_0001445 (Vilades et al., 2020). Therefore, hsa_circ_0001445 might be considered an effective and novel predictor of CAD (Vilades et al., 2020). In general, these studies have illuminated the potential role of circulating circRNAs as biomarkers for the diagnosis and prognosis of CVD.
Conclusion and Future Perspectives
Base on our exploration, the results from a variety of studies have confirmed that circRNAs can participate in the pathogenesis of CVD mainly through acting as miRNA sponges and interacting with RBPs. CircRNAs are widely distributed in different tissues and have tissue- and developmental stage-specific expression. In addition, circRNAs are stable and abundantly present in the circulatory system. Therefore, circRNAs might be promising biomarkers for the diagnosis of CVD, and accumulating research has confirmed this possibility. The clinical use of circRNAs as diagnostic biomarkers will greatly facilitate the prevention and treatment of CVD. However, there are some problems that should be solved before clinical application of circRNAs.
First, there is no generally accepted methodology on the measurement procedures of circulating circRNAs, which might result in the lack of consistency in various studies. Hence, a standardized methodology should be formulated before clinical use. Second, the sample sizes are small in most studies. The insufficient samples might lead to deviation in the test results. A larger cohort is necessary for correct conclusions. Finally, despite these findings, the underlying mechanisms of the functions of many circulating circRNAs have not been elucidated, and our knowledge is still insufficient, which represents a considerable obstacle to clinical application. More and deeper studies should be performed to explore the potential molecular mechanisms.
In summary, studies have confirmed that circRNAs are closely involved in the progression of CVD and might be promising biomarkers for CVD. These findings may provide a new avenue for the prevention, diagnosis and therapeutic intervention of CVD in the future.
Author Contributions
LZ and YZhang drafted the manuscript. YZhao and HD edited the manuscript. YW revised the manuscript. PL and LZ conceived the idea and framework of the review and made the final proofreading. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Liang Xu for the help in the manuscript revision.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Grant Numbers: 91849209 and 31430041) and Innovative Talent Program of Qingdao City, China (18-1-2-6-zhc).
References
- Aaronson K. D., Sackner-Bernstein J. (2006). Risk of death associated with nesiritide in patients with acutely decompensated heart failure. JAMA 296 1465–1466. 10.1001/jama.296.12.1465 [DOI] [PubMed] [Google Scholar]
- Abdelmohsen K., Panda A. C., Munk R., Grammatikakis I., Dudekula D. B., De S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14 361–369. 10.1080/15476286.2017.1279788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbouHaidar M. G., Venkataraman S., Golshani A., Liu B. L., Ahmad T. (2014). Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl. Acad. Sci. U.S.A. 111 14542–14547. 10.1073/pnas.1402814111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha P. K., Blouin R. T., Zaniewski R., Deutscher M. P. (1983). Ribonuclease BN: identification and partial characterization of a new tRNA processing enzyme. Proc. Natl. Acad. Sci. U.S.A. 80 3301–3304. 10.1073/pnas.80.11.3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Aufiero S., Reckman Y. J., Pinto Y. M., Creemers E. E. (2019). Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 16 503–514. [DOI] [PubMed] [Google Scholar]
- Authors/Task Force M., Elliott P. M., Anastasakis A., Borger M. A., Borggrefe M., Cecchi F., et al. (2014). 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur. Heart J. 35 2733–2779. 10.1093/eurheartj/ehu284 [DOI] [PubMed] [Google Scholar]
- Bao X., Zheng S., Mao S., Gu T., Liu S., Sun J., et al. (2018). A potential risk factor of essential hypertension in case-control study: Circular RNA hsa_circ_0037911. Biochem. Biophys. Res. Commun. 498 789–794. 10.1016/j.bbrc.2018.03.059 [DOI] [PubMed] [Google Scholar]
- Brauch K. M., Karst M. L., Herron K. J., de Andrade M., Pellikka P. A., Rodeheffer R. J., et al. (2009). Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 54 930–941. 10.1016/j.jacc.2009.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. E., Jeck W. R., Liu Y., Sanoff H. K., Wang Z., Sharpless N. E. (2010). Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with Atherosclerosis risk. PLoS Genet. 6:e1001233. 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Qi B., Wu X., Peng S., Zhou G., Wei Y., et al. (2019). Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J. Mol. Cell Cardiol. 130 10–22. 10.1016/j.yjmcc.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73 1019–1030. 10.1016/0092-8674(93)90279-y [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Sarnow P. (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268 415–417. 10.1126/science.7536344 [DOI] [PubMed] [Google Scholar]
- Chen J., Zou Q., Lv D., Wei Y., Raza M. A., Chen Y., et al. (2018). Comprehensive transcriptional landscape of porcine cardiac and skeletal muscles reveals differences of aging. Oncotarget 9 1524–1541. 10.18632/oncotarget.23290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. J., Cui L. Q., Yuan J. L., Zhang Y. Q., Sang H. J. (2017). Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophys. Res. Commun. 494 126–132. 10.1016/j.bbrc.2017.10.068 [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18 997–1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- Cocquerelle C., Daubersies P., Majerus M. A., Kerckaert J. P., Bailleul B. (1992). Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 11 1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. (1993). Mis-splicing yields circular RNA molecules. FASEB J. 7 155–160. 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- Conn S. J., Pillman K. A., Toubia J., Conn V. M., Salmanidis M., Phillips C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Dang R. Y., Liu F. L., Li Y. (2017). Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem. Biophys. Res. Commun. 490 104–110. 10.1016/j.bbrc.2017.05.164 [DOI] [PubMed] [Google Scholar]
- Deng Y. Y., Zhang W. P., She J. Q., Zhang L. S., Chen T., Zhou J., et al. (2016). Circular RNA related to PPAR gamma function as ceRNA of microRNA in human acute myocardial infarction. J Am. Coll Cardiol. 68 C51–C52. 10.1016/j.jacc.2016.07.189 [DOI] [Google Scholar]
- Du W. W., Yang W., Chen Y., Wu Z. K., Foster F. S., Yang Z., et al. (2017). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 38 1402–1412. 10.1093/eurheartj/ehw001 [DOI] [PubMed] [Google Scholar]
- Du W. W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucl. Acids Res. 44 2846–2858. 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Weng X., Zhao Y., Chen W., Gan T., Xu D. (2017). Circular RNAs in Cardiovascular Disease: An Overview. Biomed. Res. Int. 2017:5135781. 10.1155/2017/5135781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garikipati V. N. S., Verma S. K., Cheng Z. J., Liang D. M., Truongcao M. M., Cimini M., et al. (2019). Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 10:4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H. H., Li R., Su Y. M., Xiao J., Pan M., Cai X. X., et al. (2016). The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 11:e0151753. 10.1371/journal.pone.0151753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K., Garg A., Bar C., Chatterjee S., Foinquinos A., Milting H., et al. (2018). Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 122 246–254. 10.1161/Circresaha.117.311335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013a). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Kjems J., Damgaard C. K. (2013b). Circular RNA and miR-7 in cancer. Cancer Res. 73 5609–5612. [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Veno M. T., Damgaard C. K., Kjems J. (2016). Comparison of circular RNA prediction tools. Nucl. Acids Res. 44:e58. 10.1093/nar/gkv1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt L. M., Stahringer A., Sass K., Pichler G., Kulak N. A., Wilfert W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7:12429. 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. L., Li X. Z., Zheng H., Si X. Y., Li B., Wei G. Q., et al. (2019). Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 139 2857–2876. 10.1161/Circulationaha.118.038361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobi T., Czaja-Hasse L. F., Reinhardt R., Dieterich C. (2016). Profiling and validation of the circular RNA repertoire in adult murine hearts. Genom. Proteom. Bioinf. 14 216–223. 10.1016/j.gpb.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sharpless N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J. Z., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q. F., Chen Y. Y. (2019). Silencing circular RNA circ_0010729 protects human cardiomyocytes from oxygen-glucose deprivation-induced injury by up-regulating microRNA-145-5p. Mol. Cell Biochem. 462 185–194. [DOI] [PubMed] [Google Scholar]
- Khan M. A., Reckman Y. J., Aufiero S., van den Hoogenhof M. M., van der Made I., Beqqali A., et al. (2016). RBM20 regulates circular RNA production from the titin gene. Circ. Res. 119 996–1003. 10.1161/CIRCRESAHA.116.309568 [DOI] [PubMed] [Google Scholar]
- Kolakofsky D. (1976). Isolation and characterization of Sendai virus DI-RNAs. Cell 8 547–555. [DOI] [PubMed] [Google Scholar]
- Kos A., Dijkema R., Arnberg A. C., van der Meide P. H., Schellekens H. (1986). The hepatitis delta (delta) virus possesses a circular RNA. Nature 323 558–560. 10.1038/323558a0 [DOI] [PubMed] [Google Scholar]
- Kramer M. C., Liang D., Tatomer D. C., Gold B., March Z. M., Cherry S., et al. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29 2168–2182. 10.1101/gad.270421.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie C. H., Gal S., Dunlop H. M., Pushkaran B., Liggins A. P., Pulford K., et al. (2008). Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 141 672–675. 10.1111/j.1365-2141.2008.07077.x [DOI] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66 22–37. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W., Feng T., Fang X., Yu Y., Yang J., Zhao Z. A., et al. (2018). Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem. Cell Res. Ther. 9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Y., Ma L., Yu B. (2017). Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed. Pharmacother. 95 1514–1519. 10.1016/j.biopha.2017.09.064 [DOI] [PubMed] [Google Scholar]
- Li H., Xu J. D., Fang X. H., Zhu J. N., Yang J., Pan R., et al. (2019). Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR26b-5p and miR-140-3p binding to Gata4. Cardiovasc. Res. 116 1323–1334. 10.1093/cvr/cvz215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Ding W., Tariq M. A., Chang W., Zhang X., Xu W., et al. (2018). A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 8 5855–5869. 10.7150/thno.27285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen B., Huang S. (2018). Identification of circRNAs for miRNA Targets by Argonaute2 RNA immunoprecipitation and luciferase screening assays. Methods Mol. Biol. 1724 209–218. 10.1007/978-1-4939-7562-4_17 [DOI] [PubMed] [Google Scholar]
- Li Y. S., Zhang J. W., Huo C. Q., Ding N., Li J. Y., Xiao J., et al. (2017). Dynamic organization of lncrna and circular rna regulators collectively controlled cardiac differentiation in humans. Ebiomedicine 24 137–146. 10.1016/j.ebiom.2017.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Y., Huang C., Bao C., Chen L., Lin M., Wang X. L., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Liang W. C., Wong C. W., Liang P. P., Shi M., Cao Y., Rao S. T., et al. (2019). Translation of the circular RNA circ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 20:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T. B., Aliwarga E., Luu T. D. A., Li Y. P., Ng S. L., Annadoray L., et al. (2019). Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res 115 1998–2007. 10.1093/cvr/cvz130 [DOI] [PubMed] [Google Scholar]
- Lim T. B., Lavenniah A., Foo R. S. (2020). Circles in the heart and cardiovascular system. Cardiovasc. Res. 116 269–278. 10.1093/cvr/cvz227 [DOI] [PubMed] [Google Scholar]
- Liu C., Yao M. D., Li C. P., Shan K., Yang H., Wang J. J., et al. (2017). Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics 7 2863–2877. 10.7150/thno.19353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu F. B., Huang M., Xie K., Xie Q. S., Liu C. H., et al. (2019). Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatob. Pancreat. Dis. Int. 18 580–586. 10.1016/j.hbpd.2019.03.003 [DOI] [PubMed] [Google Scholar]
- Maesen B., Nijs J., Maessen J., Allessie M., Schotten U. (2012). Post-operative atrial fibrillation: a maze of mechanisms. Europace 14 159–174. 10.1093/europace/eur208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Meng Z. Y., Chen C., Cao H. L., Wang J. Y., Shen E. (2019). Whole transcriptome sequencing reveals biologically significant RNA markers and related regulating biological pathways in cardiomyocyte hypertrophy induced by high glucose. J. Cell Biochem. 120 1018–1027. 10.1002/jcb.27546 [DOI] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5′. UTR m(6)A promotes cap-independent translation. Cell 163 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao R., Wang Y., Wan J., Leng D., Gong J., Li J., et al. (2017). Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine 96:e7354. 10.1097/MD.0000000000007354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J. D., Lu J. R., Antos C. L., Markham B., Richardson J., Robbins J., et al. (1998). A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni H., Li W., Zhuge Y., Xu S., Wang Y., Chen Y., et al. (2019). Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int. J. Cardiol. 292 188–196. 10.1016/j.ijcard.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Nigro J. M., Cho K. R., Fearon E. R., Kern S. E., Ruppert J. M., Oliner J. D., et al. (1991). Scrambled exons. Cell 64 607–613. 10.1016/0092-8674(91)90244-s [DOI] [PubMed] [Google Scholar]
- Odiete O., Hill M. F., Sawyer D. B. (2012). Neuregulin in cardiovascular development and disease. Circ. Res. 111 1376–1385. 10.1161/CIRCRESAHA.112.267286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., et al. (2017). Translation of CircRNAs. Mol. Cell 66 9–21e27. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Li T., Jiang Y., Pan C., Ding Y., Huang Z., et al. (2018). Overexpression of circular RNA ciRS-7 Abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J. Cell Biochem. 119 440–446. 10.1002/jcb.26201 [DOI] [PubMed] [Google Scholar]
- Pan R. Y., Liu P., Zhou H. T., Sun W. X., Song J., Shu J., et al. (2017). Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. Oncotarget 8 60280–60290. 10.18632/oncotarget.19941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R., Ares M. (1998). Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 4 1047–1054. 10.1017/S135583829898061x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabi M., Kassiotis C., Razeghi P., Taegtmeyer H. (2007). Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 12 331–343. [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Schroeder R., Breitenbach M., Schweyen R. J. (1983). Mitochondrial circular RNAs are absent in sporulating cells of Saccharomyces cerevisiae. Nucl. Acids Res 11 1735–1746. 10.1093/nar/11.6.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan K., Liu C., Liu B. H., Chen X., Dong R., Liu X., et al. (2017). Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136 1629–1642. 10.1161/CIRCULATIONAHA.117.029004 [DOI] [PubMed] [Google Scholar]
- Siede D., Rapti K., Gorska A. A., Katus H. A., Altmuller J., Boeckel J. N., et al. (2017). Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J. Mol. Cell Cardiol. 109 48–56. 10.1016/j.yjmcc.2017.06.015 [DOI] [PubMed] [Google Scholar]
- Sonnenschein K., Wilczek A. L., de Gonzalo-Calvo D., Pfanne A., Derda A. A., Zwadlo C., et al. (2019). Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci. Rep. 9:20350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jiang X., Lv Y., Liang X., Zhao B., Bian W., et al. (2020). Circular rna expression profiles in plasma from patients with heart failure related to platelet activity. Biomolecules 10:187. 10.3390/biom10020187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhang S. L., Yue M. M., Li Y., Bi J., Liu H. R. (2019). Angiotensin II inhibits apoptosis of mouse aortic smooth muscle cells through regulating the circNRG-1/miR-193b-5p/NRG-1 axis. Cell. Death Dis. 10:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Zuo Y. H., Wang J. H., Zhang M. Q., Malhotra A., Mayeda A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucl. Acids Res. 34:e63. 10.1093/nar/gkl151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L., Salzman J. (2016). Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17 679–692. 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. L., Lim B. T., Anene-Nzelu C. G., Ackers-Johnson M., Dashi A., See K., et al. (2017). A landscape of circular RNA expression in the human heart. Cardiov. Res. 113 298–309. 10.1093/cvr/cvw250 [DOI] [PubMed] [Google Scholar]
- Tang C. M., Zhang M., Huang L., Hu Z. Q., Zhu J. N., Xiao Z., et al. (2017). CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 7:40342. 10.1038/srep40342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y., Rinn J., Pandolfi P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505 344–352. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Marshall W. S., Olson E. N. (2008). Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ. Res. 103 919–928. 10.1161/CIRCRESAHA.108.183426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vausort M., Salgado-Somoza A., Zhang L., Leszek P., Scholz M., Teren A., et al. (2016). Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J. Am. Coll. Cardiol. 68 1247–1248. 10.1016/j.jacc.2016.06.040 [DOI] [PubMed] [Google Scholar]
- Vilades D., Martinez-Camblor P., Ferrero-Gregori A., Bar C., Lu D. C., Xiao K., et al. (2020). Plasma circular RNA hsa_circ_0001445 and coronary artery disease: performance as a biomarker. Faseb J. 34 4403–4414. 10.1096/fj.201902507R [DOI] [PubMed] [Google Scholar]
- Wang K., Gan T. Y., Li N., Liu C. Y., Zhou L. Y., Gao J. N., et al. (2017). Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 24 1111–1120. 10.1038/cdd.2017.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Long B., Liu F., Wang J. X., Liu C. Y., Zhao B., et al. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J 37 2602–2611. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- Wang L., Shen C., Wang Y., Zou T., Zhu H., Lu X., et al. (2019). Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis 286 88–96. 10.1016/j.atherosclerosis.2019.05.006 [DOI] [PubMed] [Google Scholar]
- Wang S., Chen J. Y., Yu W. Q., Deng F. (2019). Circular RNA DLGAP4 ameliorates cardiomyocyte apoptosis through regulating BCL2 via targeting miR-143 in myocardial ischemia-reperfusion injury. Int. J. Cardiol. 279 147–147. 10.1016/j.ijcard.2018.09.023 [DOI] [PubMed] [Google Scholar]
- Werfel S., Nothjunge S., Schwarzmayr T., Strom T. M., Meitinger T., Engelhardt S. (2016). Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell Cardiol. 98 103–107. 10.1016/j.yjmcc.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Wesselhoeft R. A., Kowalski P. S., Anderson D. G. (2018). Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9:2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. H., Li J. Q., Liu H., Yin J. W., Zhang M. J., Yu Z. B., et al. (2019). Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J. Clin. Lab. Anal. 33:e22998. 10.1002/jcla.22998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Jin L., Cai J. (2017). Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin. Exp. Hypertens 39 454–459. 10.1080/10641963.2016.1273944 [DOI] [PubMed] [Google Scholar]
- Xu T. Y., Wu J., Han P., Zhao Z. M., Song X. F. (2017). Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. Bmc Genom. 18:680 10.1186/s12864-017-4029-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 27 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110:435. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. G., Awan F. M., Du W. W., Zeng Y., Lyu J., Wu, et al. (2017). The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 25 2062–2074. 10.1016/j.ymthe.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. G., Guo X. B., Li G. M., Shi Y. L., Li L. P. (2016). Long noncoding RNAs as potential biomarkers in gastric cancer: opportunities and challenges. Cancer Lett. 371 62–70. 10.1016/j.canlet.2015.11.011 [DOI] [PubMed] [Google Scholar]
- Zeng X. X., Lin W., Guo M. Z., Zou Q. (2017). A comprehensive overview and evaluation of circular RNA detection tools. PLos Comp. Biol. 13:e1005420. 10.1371/journal.pcbi.1005420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Du W. W., Wu Y., Yang Z., Awan F. M., Li X., et al. (2017). A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7 3842–3855. 10.7150/thno.19764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xu Y. L., Xu S., Liu Y., Yu L. M., Li Z., et al. (2018). Plasma circular RNAs, Hsa_circRNA_025016, predict postoperative atrial fibrillation after isolated off-pump coronary artery bypass grafting. J. Am. Heart Assoc. 7:e006642 10.1161/JAHA.117.006642 [DOI] [Google Scholar]
- Zhang L., Zhang Y., Xue S., Ding H., Wang Y., Qi H. Z., et al. (2020). Clinical significance of circulating microRNAs as diagnostic biomarkers for coronary artery disease. J. Cell Mol. Med. 24 1146–1150. 10.1111/jcmm.14802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang Y., Zhao Y. F., Wang Y., Ding H., Xue S., et al. (2018). Circulating miRNAs as biomarkers for early diagnosis of coronary artery disease. Expert Opin Ther Pat 28 591–601. 10.1080/13543776.2018.1503650 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Sun W. X., Han J., Cheng S. Y., Yu P., Shen L., et al. (2020). The circular RNA hsa_circ_0007623 acts as a sponge of microRNA-297 and promotes cardiac repair. Biochem. Biophys. Res. Co. 523 993–1000. 10.1016/j.bbrc.2019.12.116 [DOI] [PubMed] [Google Scholar]
- Zhang X. O., Wang H. B., Zhang Y., Lu X. H., Chen L. L., Yang L. (2014). Complementary sequence-mediated exon circularization. Cell 159 134–147. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X. O., Chen T., Xiang J. F., Yin Q. F., Xing Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhao Z. Z., Li X. J., Gao C. Y., Jian D. D., Hao P. Y., Rao L. X., et al. (2017). Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 7:39918. 10.1038/srep39918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Niu H., Li M., Zhang H., Yang Z., Tian L., et al. (2015). Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol. Med. Rep. 12 6656–6662. 10.3892/mmr.2015.4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. P., Bao C. Y., Guo W. J., Li S. Y., Chen J., Chen B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Yu J. W. (2017). A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta 1. Biochem. Biophys. Res. Commun. 487 769–775. 10.1016/j.bbrc.2017.04.044 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Pan W., Yang T., Meng X., Jiang Z., Tao L., et al. (2019). Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR-433. Front. Genet. 10:564. 10.3389/fgene.2019.00564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel A., Papantonis A. (2018). Detecting circular RNAs by RNA fluorescence in situ hybridization. Methods Mol. Biol. 1724 69–75. 10.1007/978-1-4939-7562-4_6 [DOI] [PubMed] [Google Scholar]
- Zou M., Huang C., Li X., He X., Chen Y., Liao W., et al. (2017). Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget 8 81825–81837. 10.18632/oncotarget.18998 [DOI] [PMC free article] [PubMed] [Google Scholar]