Dear Editor,
JAK2V617F is the most common driver mutation identified in primary myelofibrosis (PMF), followed by CALR and MPL mutations. In addition to the International Prognostic Scoring System (IPSS), Dynamic IPSS (DIPSS), and DIPSS plus, three widely adopted prognostic systems for PMF patients1–3, the prognostic implication of JAK2V617F allele burden on the survival of PMF patients has also been evaluated4–6. Tefferi et al. first reported that PMF patients with lower JAK2V617F allele burden had a poorer prognosis compared to those with higher allele burden, and the observation was verified by Guglielmelli et al.5.
Besides, ASXL1, EZH2, SRSF2, and IDH mutations were reported to contribute to the disease progression and acute leukemia transformation of PMF, and thus classified as high-molecular risk (HMR) mutations7,8. Among them, ASXL1 mutation is the most frequently harbored mutation.
While the JAK2V617F allele burden predicts the prognosis of PMF patients, its clinical association with ASXL1 mutation remains unexplored. In this study, we found that ASXL1 mutation confers poor prognosis in PMF patients with low JAK2V617F allele burden but not in those with high allele burden, irrespective of DIPSS-plus/DIPSS/IPSS risk classification and other HMR mutations.
We retrospectively enrolled 122 adult patients with PMF diagnosed at the National Taiwan University Hospital (NTUH) from 2005 to 2019. The pathological diagnoses based on the 2016 World Health Organization classifications9,10 and fibrosis grading were reviewed by two hematopathologists. A TruSight myeloid sequencing panel and the HiSeq platform were adopted to analyze the gene alterations and mutant allele burden of 54 myeloid-neoplasm relevant genes (Supplementary Table 1) on bone marrow or whole blood cells obtained at the time of PMF diagnosis or referral. This study was approved by the Research Ethics Committee of NTUH (Project number: 201709072RINC).
The median age of the 122 PMF patients was 61 years. Thirteen patients had pre-PMF, whereas 109 patients had overt PMF. The clinical and laboratory characteristics of these patients at diagnosis or first referral are shown in Supplemental Table 2. JAK2V617F was the most common driver mutation (64.8%), followed by CALR (18%) and MPL mutations (9%).
The mutational landscape of 122 PMF patients is illustrated in Supplemental Fig. 1. ASXL1 mutation was the most common (37%) mutation other than driver mutations, followed by TET2 (16%) and EZH2 mutations (12%). Overall, 88 (72%) patients harbored at least one additional mutation; 33, 16, 10, and 13% of the patients harbored 1, 2, 3, or 4 or more mutations other than JAK2/CALR/MPL, respectively. Fifty-two patients (42.6%) had at least one HMR mutation (ASXL1, EZH2, SRSF2, and IDH mutations). Details of mutation status and allele burden are shown in Supplemental Table 3.
With a median follow-up time of 28.2 months, the median overall survival (OS) of all cohort was not reached (NR) and was not different among patients with different driver mutations (p = 0.342, Supplementary Fig. 2a). Meanwhile, patients with type 1/like CALR mutation tended to have a better OS than those with type 2/like CALR mutations (p = 0.051, Supplementary Fig. 2b). Patients with pre-PMF and overt-PMF had significantly different leukemia-free survival (LFS) and OS (Supplementary Fig. 3a and b, respectively). Moreover, patients with at least one HMR mutation had significantly shorter LFS and OS than those without (Supplementary Fig. 3c and d, respectively). Conceivably, patients harboring more HMR mutations had worse LFS and OS (Supplementary Fig. 3e and f, respectively). The patients’ OS was well risk-stratified by prognostic scoring systems, including IPSS, DIPSS plus, MIPSS70, and GIPSS (Supplementary Fig. 4 and Supplemental Table 4).
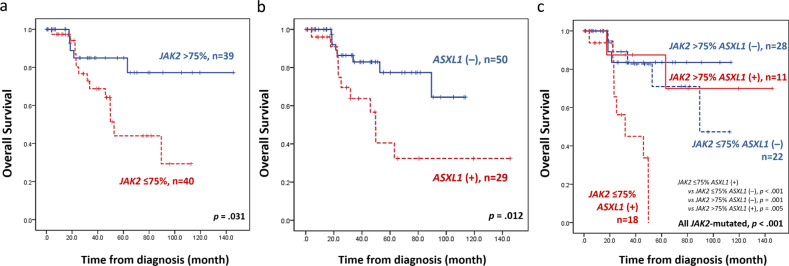
We divided the 79 patients who had JAK2V617F and available data of allele burden into low- and high-allele burden groups using 75%, the median, as the cutoff. Concurring with previous studies, patients with lower JAK2V617F allele burden had significantly inferior OS than those with higher burden (p = 0.031, Fig. 1a). Among the JAK2-mutated patients, those with ASXL1 mutation had a significantly shorter OS than those without (p = 0.012, Fig. 1b). Taken ASXL1 mutation and allele burden of JAK2V617F together, the mutation of ASXL1 conferred worse OS in those with lower JAK2V617F allele burden but not in those with higher allele burden (p < 0.001 and p = 0.703, Supplemental Fig. 5). By and large, patients with concurrent ASXL1 mutation and low JAK2V617F allele burden had a distinctively worse survival than those with other three mutation combinations (p < 0.001, Fig. 1c). Besides, this group of patients also had the shortest survival among the total cohort with JAK2-wild patients included (p < 0.001, Supplementary Fig. 6). We further levelled off the cutoff value of allele burden to 50%, approximately the lower quadrant of this cohort. Consistently, ASXL1 mutation conferred poor prognosis in those with lower JAK2V617F allele burden but not in those with higher allele burden (Supplementary Fig. 7).
Fig. 1. Kaplan–Meier survival curves of 79 JAK2-mutated PMF patients.
a OS stratified by JAK2 allele burden. Patients with lower JAK2 allele burden had inferior survival. b OS stratified by mutation status of ASXL1 among JAK2-mutated patients. Patients with concurrent ASXL1 mutation had inferior survival. c OS stratified by combined JAK2 allele burden and ASXL1 mutation status. Patients with concurrent ASXL1 mutation and low JAK2 allele burden had distinctively inferior survival.
In light of this interesting finding, we compared the clinical and laboratory characteristics between patients with higher and lower JAK2V617F allele burden. Nevertheless, there was no significant difference between groups, except that patients with lower JAK2 allele burden more frequently harbored SRSF2 mutation (Supplementary Table 5). The mutation details of ASXL1 in JAK2-mutated patients are displayed in Supplementary Table 6.
In multivariable analysis, we used DIPSS-plus, ASXL1 mutation/JAK2V617F allele burden, and mutation statuses of EZH2, SRSF2, and IDH as variables. SETBP1, ETV6, and KDM6A mutations were not included due to low prevalence in this cohort despite of their prognostic impact in univariate analysis (Supplemental Table 7). Mutation of ASXL1 along with low allele burden of JAK2V617F appeared as an independent adverse risk factor for both LFS and OS (hazard ratio, HR: 2.061, p = 0.045; and HR: 2.361, p = 0.012, respectively, Table 1), irrespective of DIPSS-plus risk groups and mutation statuses of other HMR genes. The prognostic significance of combined ASXL1 mutation and low JAK2V617F allele burden remained valid for OS in the analysis applying IPSS and DIPSS, respectively (Supplementary Tables 8 and 9).
Table 1.
Multivariable analysis for LFS and OS in the 79 JAK2-mutated PMF patients, adopting DIPSS-plus, ASXL1 mutation and JAK2 allele burden, and other HMR gene mutations as variables.
| LFS 95% CI |
OS 95% CI |
|||||||
|---|---|---|---|---|---|---|---|---|
| Variable | HR | Lower | Upper | P | HR | Lower | Upper | P |
| DIPSS-plusa | 1.984 | 0.981 | 4.012 | 0.057 | 2.034 | 0.983 | 4.212 | 0.056 |
| ASXL1/JAK2 allele burdenb | 2.061 | 1.015 | 4.186 | 0.045 | 2.361 | 1.206 | 4.624 | 0.012 |
| EZH2 | 1.932 | 0.456 | 8.181 | 0.371 | 1.567 | 0.384 | 6.388 | 0.531 |
| SRSF2 | 2.074 | 0.414 | 10.394 | 0.375 | 1.504 | 0.324 | 6.979 | 0.602 |
| IDH | 2.319 | 0.079 | 68.062 | 0.626 | 4.134 | 0.152 | 112.1 | 0.399 |
P values < 0.05 are considered statistically significant.
HMR high-molecular risk, HR hazard ratios, CI confidence interval.
aDIPSS-plus: low vs. intermediate-1 vs. intermediate-2 vs. high-risk groups.
bASXL1 mutation with low JAK2 allele burden (<75%) versus others.
Since the survival of PMF patients may vary from months to more than a decade, the identification of prognostic factors has been of great interest to physicians and scientists. Tefferi et al. first described the impact of JAK2V617F allele burden on PMF patients’ survival: patients with JAK2V617F allele burden in the lower quartile had significantly reduced OS, compared with those with allele burden in middle or upper quartile, or those without a mutant JAK24. This finding was supported by the study of Guglielmelli et al.5, within which a low JAK2V617F allele burden was confirmed as an independent adverse risk factor5.
Although the pathophysiologic mechanism underlying this correlation and the optimal cutoff value remain unknown, the contention that a lower JAK2V617F allele burden at diagnosis is associated with inferior survival in PMF patients is widely accepted. On the other hand, the role of a higher JAK2V617F allele burden in the MF phenotype is yet to be established. In this cohort, patients with higher and lower allele burden did not differ in clinical and laboratory features, except for a higher incidence of SRSF2 mutation in the latter group.
The ASXL1 mutations are correlated with adverse prognosis in PMF5. In a study of Guglielmelli et al.7, mutant ASXL1 and other three mutations (EZH2, SRSF2, and IDH1/2 mutations) were categorized as high-molecular risk mutations due to their detrimental effects on PMF patients’ survival. Recently, Tefferi et al identified seven adverse mutations by targeted deep sequencing and multivariable analysis8. Among them, ASXL1 mutation was the most frequently detected and significantly associated with inferior survival.
In this study, we analyzed the combined effect of JAK2V617F allele burden and ASXL1 mutation on the survival of PMF patients. We identified a distinct patient population, characterized by having concurrent ASXL1 mutation and low JAK2V617F allele burden, who had a significantly shorter OS than others not only in patients with JAK2V617F, but also in the total cohort. The prognostic impact of combined ASXL1 mutation and low allele burden of JAK2V617F remained significant in multivariable analysis for OS. As hematopoietic stem cell transplantation (HSCT) is currently the only curative treatment for PMF, the optimal timing and selection of candidates for transplant remain to be defined11–13. The International Working Group-Myeloproliferative Neoplasms Research and Treatment and European LeukemiaNet suggest that patients with intermediate-1 risk may consider HSCT if they harbor ASXL1 mutation14. However, in clinical practice, physicians may not optimally adhere to the guideline due to the risks HSCT introduces. Incorporation of the JAK2V617F allele burden to current risk stratification might provide complementary information to treatment decisions in the future11.
This study was limited by its retrospective nature; thereby diverse confounding factors might become difficult to assess. Additionally, while most specimens were obtained at the time of diagnosis, a small proportion of patients’ samples were drawn at their referral to NTUH. As the allele burden of JAK2V617F is known to evolve throughout disease and treatment, the allele burden assessment could be, therefore, confounded. Despite of these, we have demonstrated that ASXL1 mutation accompanied by a low JAK2V617F allele burden dictated a distinct patient population with significantly reduced survival, and its independent prognostic relevance was validated in multivariable analysis. Prospective and experimental studies are warranted to support these observations and ascertain the underlying mechanisms.
Supplementary information
Acknowledgements
We thank all the patients who participated in this study. The study was partially supported by grants from Ministry of Health and Welfare, Taiwan (MOHW 107-TDU-B-211-114009) and Ministry of Science and Technology, Taiwan (MOST 107-2314-B-002-013).
Data availability
The data reported in this article could be accessed through 10.6084/m9.figshare.12952022 once the manuscript is published, or request with the corresponding author.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yu-Hung Wang, Chien-Chin Lin
These authors jointly supervised this work: Jih-Luh Tang, Hwei-Fang Tien
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41408-020-00364-5).
References
- 1.Cervantes F, et al. New prognostic scoring system for primary myelofibrosis based on a study of the International Working Group for Myelofibrosis Research and Treatment. Blood. 2009;113:2895–2901. doi: 10.1182/blood-2008-07-170449. [DOI] [PubMed] [Google Scholar]
- 2.Passamonti F, et al. A dynamic prognostic model to predict survival in primary myelofibrosis: a study by the IWG-MRT (International Working Group for Myeloproliferative Neoplasms Research and Treatment) Blood. 2010;115:1703–1708. doi: 10.1182/blood-2009-09-245837. [DOI] [PubMed] [Google Scholar]
- 3.Gangat N, et al. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J. Clin. Oncol. 2011;29:392–397. doi: 10.1200/JCO.2010.32.2446. [DOI] [PubMed] [Google Scholar]
- 4.Tefferi A, et al. Low JAK2V617F allele burden in primary myelofibrosis, compared to either a higher allele burden or unmutated status, is associated with inferior overall and leukemia-free survival. Leukemia. 2008;22:756–761. doi: 10.1038/sj.leu.2405097. [DOI] [PubMed] [Google Scholar]
- 5.Guglielmelli P, et al. Identification of patients with poorer survival in primary myelofibrosis based on the burden of JAK2V617F mutated allele. Blood. 2009;114:1477–1483. doi: 10.1182/blood-2009-04-216044. [DOI] [PubMed] [Google Scholar]
- 6.Vannucchi AM, Pieri L, Guglielmelli P. JAK2 Allele burden in the myeloproliferative neoplasms: effects on phenotype, prognosis and change with treatment. Ther. Adv. Hematol. 2011;2:21–32. doi: 10.1177/2040620710394474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guglielmelli P, et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia. 2014;28:1804–1810. doi: 10.1038/leu.2014.76. [DOI] [PubMed] [Google Scholar]
- 8.Tefferi A, et al. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016;1:105–111. doi: 10.1182/bloodadvances.2016000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 10.Barbui T, et al. The 2016 WHO classification and diagnostic criteria for myeloproliferative neoplasms: document summary and in-depth discussion. Blood Cancer J. 2018;8:15. doi: 10.1038/s41408-018-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tefferi A, et al. Allogeneic hematopoietic stem cell transplant overcomes the adverse survival effect of very high risk and unfavorable karyotype in myelofibrosis. Am. J. Hematol. 2018;93:649–654. doi: 10.1002/ajh.25053. [DOI] [PubMed] [Google Scholar]
- 12.Kröger N. Stem cell transplantation in myelofibrosis. HemaSphere. 2018;2:146–148. doi: 10.1097/HS9.0000000000000146. [DOI] [Google Scholar]
- 13.Tiribelli M, et al. The role of allogeneic stem-cell transplant in myelofibrosis in the era of JAK inhibitors: a case-based review. Bone Marrow Transplant. 2020;55:708–716. doi: 10.1038/s41409-019-0683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tefferi A, et al. Revised response criteria for myelofibrosis: International Working Group-Myeloproliferative Neoplasms Research and Treatment (IWG-MRT) and European LeukemiaNet (ELN) consensus report. Blood. 2013;122:1395–1398. doi: 10.1182/blood-2013-03-488098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this article could be accessed through 10.6084/m9.figshare.12952022 once the manuscript is published, or request with the corresponding author.