Abstract
Hybrid strains of Escherichia coli combine virulence traits of diarrheagenic (DEC) and extraintestinal pathogenic E. coli (ExPEC), but it is poorly understood whether these combined features improve the virulence potential of such strains. We have previously identified a uropathogenic E. coli (UPEC) strain (UPEC 252) harboring the eae gene that encodes the adhesin intimin and is located in the locus of enterocyte effacement (LEE) pathogenicity island. The LEE-encoded proteins allow enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC) to form attaching and effacing (A/E) lesions in enterocytes. We sought to characterize UPEC 252 through whole-genome sequencing and phenotypic virulence assays. Genome analysis unveiled that this strain harbors a complete LEE region, with more than 97% of identity comparing to E2348/69 (EPEC) and O157:H7 Sakai (EHEC) prototype strains, which was functional, since UPEC 252 expressed the LEE-encoded proteins EspB and intimin and induced actin accumulation foci in HeLa cells. Phylogenetic analysis performed comparing 1,000 single-copy shared genes clustered UPEC 252 with atypical EPEC strains that belong to the sequence type 10, phylogroup A. Additionally, UPEC 252 was resistant to the bactericidal power of human serum and colonized cells of the urinary (T24 and HEK293-T) and intestinal (Caco-2 and LS174T) tracts. Our findings suggest that UPEC 252 is an atypical EPEC strain that emerges as a hybrid strain (aEPEC/UPEC), which could colonize new niches and potentially cause intestinal and extraintestinal infections.
Keywords: Escherichia coli, UPEC, hybrid pathogenic strain, virulence factor, ExPEC, urinary tract infection, atypical EPEC
Introduction
Urinary tract infection (UTI) is one of the most prevalent diseases worldwide, affecting both community and hospitalized subjects, resulting in impairment of the patients' life quality (Terpstra and Geerlings, 2016; Wurpel et al., 2016). Several factors, such as surgical manipulation, diabetes, previous hospitalizations, and catheterization, are related to a higher risk for developing this disease (Saltoglu et al., 2015; Redder et al., 2016). UTI most often results from bacteria transitorily present in the gut microbiota that colonize the perineum and ascend through the urethra. Alternatively, UTI may occur after the introduction of bacteria directly into the urinary tract, during the use of a bladder catheter or certain sexual practices (Donnenberg and Welch, 1996; Finer and Landau, 2004). Characteristics of the host, especially those associated with immunity, also contribute to the occurrence and severity of infections in the urinary tract.
The main agents of UTI are Gram-negative bacilli of the family Enterobacteriaceae. Among them, uropathogenic Escherichia coli (UPEC) are the most frequent (Donnenberg and Welch, 1996; Heilberg and Schor, 2003), responding for about 80% of all cases worldwide (Ronald, 2003; Hooton, 2012; Korbel et al., 2017). UPEC strains belong to a group of E. coli clones that are collectively referred to as extraintestinal pathogenic E. coli (ExPEC), which are adapted to colonize and cause disease in different extraintestinal host sites (Russo and Johnson, 2000). The ability of ExPEC strains to colonize and develop extraintestinal diseases is due to the presence of a variety of virulence factors, including adhesins, toxins, capsules, invasins, and iron uptake systems that are encoded in Pathogenicity islands (PAI) present in chromosomes or plasmids (Dale and Woodford, 2015). Currently, more than 50 virulence factors have their role in ExPEC pathogenicity established (Johnson and Russo, 2018), but the ability of ExPEC to colonize a variety of host sites varies among strains reflecting their genetic diversity.
Despite the number of virulence factors related to ExPEC pathogenicity, its classification is primarily performed by the isolation site, and their pathotypes defined by the infection source, with uropathogenic E. coli (UPEC) being the most prevalent ExPEC pathotype. Many studies focus on UPEC virulence and epidemiology, mainly because of the disease's burden. Johnson et al. (2003) described a set of virulence factors that could identify ExPEC strains by their intrinsic virulence, and are thus considered more virulent strains (presence of at least two among five markers: pap, sfa/foc, afa/dra, iuc/iut, kpsMTII). Later, Spurbeck et al. (2012) described another set of virulence markers that are epidemiologically linked with the uropathogenic potential (simultaneous presence of chuA, fyuA, vat, and yfcV). Together these methodologies enable the screening of the ExPEC strains that either harbor intrinsic virulence or uropathogenic potential, in diverse sources, like water, food, and gut, favoring the identification of their reservoirs. However, these criteria fail to classify all strains isolated from extraintestinal infections (Spurbeck et al., 2012; Santos et al., 2013, 2020b; Freire et al., 2020) and some clinical lineages with epidemiological importance also do not meet these criteria (Bert et al., 2010; Olesen et al., 2012; Manges and Johnson, 2015; Manges et al., 2017, 2019; Campos et al., 2018; Yamaji et al., 2018; Santos et al., 2020a,b).
Other clones of E. coli can cause intestinal infections by different mechanisms and are collectively referred to as diarrheagenic E. coli (DEC). DEC strains are classified into six pathotypes named enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), Shiga toxin-producing E. coli (STEC), diffusely adherent E. coli (DAEC), and enteropathogenic E. coli (EPEC) (Kaper and Nataro, 1998; Croxen et al., 2013; Gomes et al., 2016). EPEC strains are divided into typical (tEPEC) and atypical (aEPEC) based on the presence of the EAF (EPEC adherence factor) plasmid (pEAF) in tEPEC and its absence in aEPEC (Trabulsi et al., 2002). pEAF encodes a bundle forming pilus (BFP) that promotes bacterial aggregation and, consequently, the formation of compact microcolonies that characterize a localized adhesion (LA) pattern on the surface of HeLa and HEp-2 epithelial cells (Scaletsky et al., 1984; Girón et al., 1991).
Reports of E. coli strains that caused intestinal and extraintestinal infections in the same host are uncommon. However, recent studies have identified ExPEC strains promoting intestinal infections followed by bloodstream infections (Mariani-Kurkdjian et al., 2014; Kessler et al., 2015). These strains that contain virulence factors of intestinal and extraintestinal pathotypes are considered hybrid and potentially more virulent pathogens. In these studies, the horizontal gene transfer (HGT) of virulence factors occurred from ExPEC to DEC and DEC to ExPEC.
Despite the recognized potential of HGT between pathogenic E. coli strains, only a few studies have described this phenomenon among DEC and UPEC in strains isolated from UTI and in these studies the genetic background of strains involved was not evaluated (Olesen et al., 1994; Garcia et al., 2000; Keller et al., 2002; Matar et al., 2005; Ogura et al., 2007; Wallace-Gadsden et al., 2007; Abe et al., 2008; Lara et al., 2017; Santos et al., 2020a). A study evaluating fecal EAEC isolates identified that some strains produced alpha-hemolysin (hlyA) and P fimbria (pap) (Suzart et al., 1999), characteristics often found in UPEC strains, suggesting that certain EAEC strains with specific virulence genes would be potentially uropathogenic. Assessing the virulence factors of strains isolated from UTI, we have previously (Falsetti, 1998; Abe et al., 2008) demonstrated in some UPEC isolates the presence of the aggR and aat genes, as well as the aggregative adhesion (AA) pattern on HeLa cells, which are properties that define the EAEC pathotype. In addition to the UPEC isolates bearing EAEC virulence genes, in the referred study, we identified a UPEC strain (UPEC 252) carrying the eae gene (Abe et al., 2008), which encodes intimin, an outer membrane adhesive protein that is fundamental for the intimate adhesion of EPEC and EHEC strains to intestinal cells and subsequent formation of attaching/effacing (A/E) lesions (Kaper et al., 2004). A/E lesions are characterized by intimate bacterial adhesion to the host cells, which undergoes the eradication of microvilli and accumulation of actin filaments underneath the adhered bacteria, thus promoting the formation of pedestal-like structures on the cell surface (Moon et al., 1983). This type of lesion can be identified in non-intestinal (HeLa and HEp-2 cells) and intestinal epithelial (Caco-2) cell lineages in vitro, as well as in animal and human enterocytes in vivo (Kaper et al., 2004). The genes involved in A/E lesion formation are contained in a chromosomal PAI called the locus of enterocyte effacement (LEE). Besides eae, LEE encodes a type III secretion system (T3SS) and several effectors and regulatory proteins, whose activities culminate in the production of the lesion (Mcdaniel et al., 1995). Regardless of the detection of the eae gene in UPEC 252, the genetic background of the strain has not been properly evaluated, and its relationship with other pathogenic E. coli is unknown. Furthermore, it is not known whether the DEC virulence genes found in this strain are complete, nor if they are expressed and play a role in the bacterial interaction with intestinal and urinary tract cells. Therefore, this study aimed to provide information about the genetic and virulence properties of UPEC 252, an E. coli that was isolated from UTI and carries the eae gene characteristic of EPEC and EHEC.
Materials and Methods
Bacterial Strain
UPEC 252 was isolated, in 1998, from a 1-month-old female patient hospitalized at Hospital São Paulo, São Paulo, Brazil, with urinary tract infection (Falsetti, 1998; Abe et al., 2008). The strain was stored in Lysogeny broth (LB) supplemented with 15% glycerol and kept at −80°C. For routine use, it was grown in LB at 37°C for ~18 h.
Genome Sequencing, Assembly, Annotation, and Data Analysis
The genome of UPEC 252 was sequenced as previously published (Valiatti et al., 2019). Briefly, DNA was extracted using the Wizard® Genomic DNA Purification Kit and sequenced on an Illumina Hiseq1500 platform. The paired-end reads were assembled de novo using SPAdes (version 3.12.0) (Bankevich et al., 2012) and the annotation was performed using Prokka version 1.13.3 (Seemann, 2014). The reads used for UPEC 252 genome assembly were deposited in the Sequence Read Archive (SRA) at NCBI under the accession number SRR9317828, and the whole-genome sequences (WGS) were deposited in the GenBank database under the accession number VFST00000000 (Valiatti et al., 2019).
The WGS of UPEC 252 was submitted to the Center for Genomic Epidemiology (CGE) to determine the strain sequencing type (Multi-Locus Sequence Typing - MLST version 2.0) (Larsen et al., 2012), virulence profile (VirulenceFinder 2.0) (Joensen et al., 2014), resistance profile (ResFinder 3.1) (Zankari et al., 2012), and serotype (SerotypeFinder version 1.1) (Joensen et al., 2015). To analyze the presence of virulence and resistance genes, we also used ecoli_VF collection v0.1 (Leimbach, 2016) and the ABRicate program1, which employes multiple databases: VFDB (Chen et al., 2016), ARG-ANNOT (Gupta et al., 2014), CARD (Jia et al., 2017), PlasmidFinder (Carattoli et al., 2014), EcOH (Ingle et al., 2016), MEGARes 2.0 (Doster et al., 2020), and NCBI AMRFinderPlus (Feldgarden et al., 2019). The phylogenetic typing of the strain was determined using the ClermonTyping tool (Beghain et al., 2018). Confirmation of the identified genes of the LEE region and the non-LEE encoded genes was carried out by aligning the DNA sequences of UPEC 252 with the DNA sequences deposited at the National Center for Biotechnological Information (NCBI) using the BLAST program.
Comparative Genome Analyses of UPEC 252
To identify the UPEC 252 genome rearrangement, gain and loss, different strategies were used. First, the annotated genome of UPEC 252 was aligned using progressiveMAUVE (Darling et al., 2010). In another approach, pangenome analysis was performed using the Roary pipeline (Page et al., 2015). In both methods, the genome of UPEC 252 was compared with the genome of the following strains: prototype EPEC E2348/69 (GCF_000026545.1), aEPEC 4581-2 (QYYF00000000.1), prototype EHEC O157:H7 Sakai (GCF_000008865.2), EHEC O111:H- 11128 (GCF_000010765.1), prototype UPEC CFT073 (GCF_000007445.1), prototype UPEC 536 (GCF_000013305.1), prototype UPEC UMN026 (GCF_000026325.1), an isolate from asymptomatic bacteriuria VR50 (GCF_000968515.1), and K12 derived MG1655 (GCF_000005845.2).
UPEC 252 Phylogenetic Analysis
The phylogenetic tree that includes UPEC 252 was built using the Phylogenetic tree tool of the Pathosystems Resource Integration Center (PATRIC) (Wattam et al., 2017). A total of 140 E. coli public genomes were used to build the tree, 95 genomes of which were randomly chosen among strains belonging to ST10 (Supplementary Table 1), while 45 others were selected by similarity using the Mash algorithm in the PATRIC similarity genome service (Supplementary Table 1). The tree was built applying the codon tree methodology employing the alignment of 1,000 single-copy coding-genes in the RAxML matrix. The E. coli strains UMN026 (ST597-D), CFT073 (ST73-B2), 536 (ST127-B2), S88 (ST95-B2), E2348/69 (ST15-B2), O157:H7 Sakai (ST11-E), and O111:H- 11128 (ST16-B1) were added to the phylogenetic tree as outgroups as well as the Escherichia fergusonni strain ATCC35469. The final layout and annotation were accomplished using iTOL v.4. (Letunic and Bork, 2019).
Analysis of Intimin Production and EspB Secretion by Immunoblotting
For the immunoblotting assays, the bacterial inoculum was standardized sub-culturing the strains at a dilution of 1/50 into DMEM until they reached an OD600 ≅1.
Intimin production was analyzed using total protein extract, followed by immunoblotting, as previously described by Towbin et al. (1979) using a monospecific polyclonal antibody against the conserved region of this protein (Int388-667) (Menezes et al., 2010).
Secreted proteins were extracted as described by Zarivach et al. (2007), and an immunoblotting assay using an anti-EspB primary antibody (Guirro et al., 2013) and anti-rabbit secondary IgG antibody (whole molecule)-Peroxidase (Sigma, Saint Louis, EUA) was employed to evaluate the ability of UPEC 252 to produce T3SS. A protein extract from the typical EPEC prototype strain E2348/69 was used as a positive control (Santos et al., 2019).
Cell Culture and Maintenance
HeLa (ATCC® CCL-2), HEK293T (ATCC® CRL-1573), Caco-2 (ATCC® HTB-37TM), and LS174T (ATCC® CL-188) cell lineages were cultivated using Dulbecco's Modified Eagle Medium (DMEM), high glucose, GlutaMax™ (Gibco-ThermoFisher Scientific, USA) (2 g/L sodium bicarbonate, 4.5 g/L glucose) containing 15 mM HEPES (Sigma, Saint Louis, USA), 1x Penicillin-Streptomycin-Neomycin (PSN) antibiotic mixture (GiBCO, USA), and 10% bovine fetal serum (BFS) (GIBCO, USA). T24 cells (ATCC® HTB-4) were grown using McCoy 5A (modified) medium (Gibco, USA) supplemented with 10% BFS and 1x PSN. All cell lineages were kept in 5% CO2 atmosphere at 37°C.
Suspensions of each cell type were seeded into 24-well plates (1 × 105 cells/ mL per well) and kept in culture according to the assay type. For qualitative assays, all cell lineages were cultivated for 2–5 days until they reached 80% confluency, except for Caco-2 cells, which were cultivated for 10 days to reach confluency, polarization, and differentiation. For quantitative assays, all cells were cultivated until they reached full confluency.
Determination of the Bacterial Adherence Pattern
The adherence pattern was determined as described (Santos et al., 2019) using HeLa cells in 6 h of interaction in the presence of 2% D-mannose (Sigma - USA) to prevent Type 1 fimbria-mediated adhesion. Bacteria previously grown in LB for 18 h at 37°C were inoculated in 1 mL of cell media, at a 1:50 dilution, and incubated at 37°C for 3 h. The preparation was then washed with phosphate-buffered saline (PBS), fresh medium was added, and incubation proceeded for an additional 3 h. After the 6 h period, the cells were washed, fixed with methanol, stained with May Grünwald and Giemsa (Merck, New Jersey, USA) and evaluated by light microscopy (Hernandes et al., 2013). As controls, the E. coli prototype strains producing the LA (tEPEC E2348/69), AA (EAEC 042), and diffuse adhesion (DA) (DAEC C1845) patterns were used, as well as a non-adherent laboratory strain (E. coli HB101) (Hernandes et al., 2013).
Fluorescence Actin Staining (FAS) Assay
The FAS test was used as an indirect approach to verify the ability of UPEC 252 to cause A/E lesions (Knutton et al., 1989). The test was performed in HeLa cells precisely as described for the determination of adhesion patterns. At the end of the assay, cells were fixed in 3% formaldehyde in PBS, permeabilized with 1% Triton X-100, and incubated with phalloidin for 20 min. Subsequently, coverslips were washed in PBS, mounted on glycerol, and observed under fluorescence microscopy (OLYMPUS BX60) (Olympus, Tokyo, Japan) with immersion lenses. As positive and negative controls, the E. coli E2348/69 (Jerse et al., 1990) and C1845 strains, respectively, were used.
The percentage of cells with pedestal produced by the UPEC 252 and EPEC E2348/69 strains was quantified in three different fields of view in a triplicated FAS assay. The results were shown by the means of the number of infected cells with F-actin accumulation foci (pedestals) ± SD. The unpaired bi-directional Student's t-test was used to compare means of the two strains, in which P ≤ 0.05 was considered statistically significant.
Transmission Electron Microscopy
For Transmission Electron Microscopy (TEM), HeLa cells were grown onto ACLAR® film inserted into 24-wells plates, and subsequently subjected to an adhesion assay for 3 h as described above. In the end, the preparation was washed twice with 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.2 and kept overnight in the same buffer. The material used was gradually infiltrated and embedded in gelatin capsules and allowed to polymerize for 48 h in an oven at 60°C; semi-thin sections (300–500 μm) were obtained and hot-stained with 1% toluidine blue. Finally, ultrathin sections (70 nm) were contrasted with uranium and lead citrate and visualized on a Jeol JEM 1200EX II transmission electron microscope. The typical EPEC E2348/69 and DAEC C1845 prototype strains were used as a positive and negative controls, respectively.
Bacterial Interaction With Intestinal and Urinary Tract Cell Lineages
The ability of UPEC 252 to interact with intestinal (Caco-2 and LS174T) and urinary tract (T24 and HEK293) cell lineages was evaluated.
Adherence assays were performed as described above but in 3 h and the absence of D-mannose. For the quantitative adhesion assays, after the 3 h incubation period, cells were incubated for ~30 min with 1 mL sterile distilled water for cell lysis (Santos et al., 2019). Then, the suspension containing lysed cells and bacteria was subjected to serial dilutions and seeding onto MacConkey Agar to determine the number of colony-forming units (CFU) associated with the cells. The UPEC CFT073, aEPEC 4581-2 and non-adherent E. coli HB101 were included as controls. The One-way ANOVA followed by post hoc Tukey HSD test was used to compare the results obtained in this experiment.
The quantitative invasion assay was performed in duplicates, however, at the end of 3 h of incubation, one of the duplicates was washed with PBS, lysed by incubation with sterile distilled water for about 30 min, serially diluted, and plated onto MacConkey Agar to obtain the total number of bacteria interacting with the cells. The second duplicate was washed with PBS, incubated for an additional hour with DMEM containing 100 μg/mL gentamicin to kill the extracellular bacteria; after the incubation period, cells were washed, lysed, and the resulting preparations, diluted and seeded onto MacConkey agar to obtain the number of internalized bacteria (Luck et al., 2005). The invasion index was defined by the ratio of the total number of internalized CFUs by the total number of CFUs associated with the cells (externally and internally), expressed as a percentage. The invasive Shigella flexneri strain M90T was used as positive control while the non-invasive strain Escherichia albertii 1551-2::eae, mutated in the eae gene (Hernandes et al., 2008), was employed as a non-invasive control in all cellular lineages tested, except for the HEK293T cell lineage, where the DAEC C1845 strain was used as a negative control. All quantitative assays were performed in biological and experimental triplicates. The One-way ANOVA followed by post hoc Tukey HSD test was used to compare the results obtained in this experiment.
Bacterial Resistance to the Human Serum Complement
To determine the resistance of UPEC 252 to the human serum complement, lyophilized human complement (Sigma, USA) reconstituted in PBS was used. Bacterial cells grown overnight in LB were washed twice and resuspended in PBS. The bacterial suspension was seeded onto MacConkey agar for CFU determination. The bacterial culture concentration was then adjusted to ~1 × 107 CFU / mL in PBS. Then, 200 μl of the adjusted inoculum and 200 μl of human serum pool were mixed and incubated at 37°C for 4 h. Aliquots of the preparation were collected at three different times (30, 60, and 240 min), serially diluted, and plated onto MacConkey agar to obtain the survived bacterial counts. Simultaneously, another assay using serum inactivated by heating for 30 min at 56°C was performed. The experiments were carried out in biological triplicates, and the E. coli strain C600 and the Enterobacter spp. strain EB046 were used as serum sensitive and resistant controls, respectively (Keller et al., 1998). Additionally, the UPEC CFT073 and aEPEC 4581-2 strains were used as controls.
Biofilm Formation
Analysis of biofilm formation was carried out as described previously (Lima et al., 2019). UPEC 252 was grown in LB at 37°C for 18 h. Then, 20 μl of the culture were added to 24-well polystyrene plates containing 1 mL of DMEM GlutaMax™ and incubated at 37°C for 24 or 48 h. After incubation, bacteria were washed with PBS, fixed overnight with 1 mL of formaldehyde (3%), and stained with 1% violet crystal. After successive washes, the dye impregnated in the cells was solubilized with 95% ethanol (Merck), supernatants were transferred to 96-well plates, and the optical density of the solution was measured in a spectrophotometer at 620 nm (Hernandes et al., 2013). The experiments were carried out in biological and experimental triplicates. The EAEC 042, UPEC CFT073, aEPEC 4581-2, and E. coli HB101 strains were used as controls. The One-way ANOVA followed by post hoc Tukey HSD test was used to compare the results obtained in this experiment.
Antimicrobial Sensitivity Test
The diffusion disc technique was used as proposed by Bauer et al. (1966) to evaluate the antimicrobial susceptibility profile. The diameters of growth inhibition halos were measured and interpreted following the standards of the Brazilian Committee on Antimicrobial Susceptibility Testing - BrCAST breakpoints (EUCAST, 2020).
The antibiotics employed in this test were: amoxicillin-clavulanic acid (20 + 10 μg), aztreonam (30 μg), cefepime (30 μg), ceftazidime (10 μg), ceftriaxone (30 μg), meropenem (10 μg), imipenem (10 μg), ertapenem (10 μg), gentamicin (10 μg), amikacin (30 μg), tigecycline (15 μg), cefoxetin (30 μg), and ciprofloxacin (5 μg).
Results
Genetic Characterization of the LEE Region and Non-LEE Effectors in UPEC 252
UPEC 252 was originally reported to harbor the eae gene, which is a property characteristic of the EPEC and EHEC pathotypes (Abe et al., 2008). This fact raised the question of whether this strain would carry an intact LEE region, where the eae gene is located, and, therefore, could be potentially more virulent. Based on manual verification of the recently published genome sequence of UPEC 252 (Valiatti et al., 2019), we observed that the strain carried a complete LEE region (Table 1). Moreover, of all the 55 non-LEE effectors screened, only the cif, nleB, nleB1, nleB2, nleC, nleE, nleF, nleG, nleH1, nleH2, espL, and espJ genes were found.
Table 1.
Genes that comprise the LEE region of UPEC252a.
| Gene | Coverage | %Coverage | %Identity | Accession |
|---|---|---|---|---|
| espG | 1–1,197/1,197 | 100.00 | 100.00 | gi:260869665 |
| escE | 1–219/219 | 100.00 | 99.54 | NP_290287 |
| escK | 1–199/199 | 100.00 | 99.50 | AF022236.1 |
| escR | 1–654/654 | 100.00 | 100.00 | gi:215488994 |
| escT | 1–777/777 | 100.00 | 99.87 | gi:260869699 |
| etgA | 1–459/459 | 100.00 | 99.35 | NP_290279 |
| grlA | 1–414/414 | 100.00 | 99.76 | gi:260869695 |
| escC | 1–1,539/1,539 | 100.00 | 99.94 | gi:215488986 |
| escJ | 1–573/573 | 100.00 | 99.83 | gi:260869691 |
| espZ | 1–297/297 | 100.00 | 99.66 | gi:260869689 |
| escV | 1–2,028/2,028 | 100.00 | 100.00 | gi:260869687 |
| escO | 1–378/378 | 100.00 | 98.41 | NP_290267 |
| sepQ/escQ | 1–918/918 | 100.00 | 93.14 | NP_290265 |
| cesF | 1–363/363 | 100.00 | 99.45 | gi:260869681 |
| tir | 1–1,656/1,656 | 100.00 | 99.94 | gi:260869679 |
| eae | 1-2808/2808 | 100.00 | 100.00 | gi:260869677 |
| sepL | 1–1,056/1,056 | 100.00 | 100.00 | gi:260869675 |
| espD | 1–1,143/1,143 | 100.00 | 100.00 | gi:260869673 |
| cesD2 | 135–408/408 | 67.16 | 99.27 | gi:260869671 |
| escG | 1–279/279 | 100.00 | 96.77 | NP_290251 |
| espF | 1–504/789 | 63.88 | 90.71 | gi:260869668 |
| escF | 1–222/222 | 100.00 | 100.00 | gi:260869670 |
| espB | 1–963/963 | 100.00 | 100.00 | gi:260869672 |
| espA | 1–579/579 | 100.00 | 100.00 | gi:260869674 |
| escD | 1–1,221/1,221 | 100.00 | 99.84 | gi:260869676 |
| cesT | 1–471/471 | 100.00 | 100.00 | gi:260869678 |
| map | 1–612/612 | 100.00 | 99.84 | gi:260869680 |
| espH | 1–510/510 | 100.00 | 100.00 | gi:260869682 |
| escP | 1–276/276 | 100.00 | 98.19 | NP_290266 |
| escN | 1–1,341/1,341 | 100.00 | 99.92 | gi:260869686 |
| cesL | 1–354/354 | 100.00 | 99.44 | NP_290270 |
| escI | 1–429/429 | 100.00 | 93.24 | NP_290272 |
| sepD | 1–456/456 | 100.00 | 100.00 | gi:260869692 |
| cesD | 1–456/456 | 100.00 | 100.00 | gi:260869694 |
| grlR | 1–372/372 | 100.00 | 100.00 | gi:260869696 |
| escU | 1–1,038/1,038 | 100.00 | 100.00 | gi:260869698 |
| escS | 1–273/273 | 100.00 | 99.63 | gi:291285070 |
| escL | 1–615/615 | 100.00 | 99.67 | NP_290284 |
| cesAB | 1–324/324 | 100.00 | 99.69 | gi:15804247 |
| ler | 1–390/390 | 100.00 | 99.74 | gi:215488999 |
| rorf1 | 1–272/272 | 100.00 | 90.81 | AF022236.1 |
Summary of LEE genes identified by the ABRicate program. The percentage of coverage and identity displayed were the highest identified, comparing UPEC 252 with O157: H7 Sakai (EHEC), O127: H6 E2348/69 (EPEC), and O111: H- 11128 (EHEC). Most proteins presented belong to the E. coli O111:H- str 11128.
By examining the sequence of the LEE region using the BLAST program, UPEC 252 showed identities ranging from 93 to 99% with EPEC, EHEC, and E. albertii strains isolated from different sources and countries (Table 2). The highest identity was found between UPEC 252 and aEPEC strain 13E0767 isolated from cattle in Germany, and eight EHEC strains, five of them belonging to the O111 serogroup, including the EHEC O111:H- strain 11128 (Table 2).
Table 2.
Comparison of the LEE region of UPEC 252 with strains available at the NCBI.
| Strain | Isolation year | Place | Pathotype | Serogroup | Source | Identity | Access number |
|---|---|---|---|---|---|---|---|
| Escherichia coli 13E0767 | 1998 | Germany | EPEC | O156 | Cattle | 99.97% | CP020107.1 |
| Escherichia coli FORC_042 | 2013 | South Korea | EPEC | NAa | Pork meat | 99.93% | CP025318.1 |
| Escherichia coli 2013C-3304 | 2013 | USA | EHEC | O71 | Stool human | 99.92% | CP027593.1 |
| Escherichia coli O111 RM9322 | 2009 | USA | EHEC | O111 | Stream | 99.92% | CP028117.1 |
| Escherichia coli 2015C-3101 | 2014 | USA | EHEC | O111 | Stool human | 99.92% | CP027221.1 |
| Escherichia coli O111:H- 11128 | 2001 | Japan | EHEC | O111 | Stool human | 99.92% | AP010960.1 |
| Escherichia coli RM9975 | 2009 | USA | EHEC | O111 | Stool human | 99.91% | CP028432.1 |
| Escherichia coli 95JB1 | 1995 | Australia | EHEC | O111 | Stool human | 99.91% | CP021335.1 |
| Escherichia coli O157:H7 Sakai | 1996 | Japan | EHEC | O157 | Stool human | 97.46% | BA000007.3 |
| Escherichia coli O127:H6 E2348/69 | 1969 | United kingdom | EPEC | O127 | Stool human | 97.19% | FM180568.1 |
| Escherichia coli O121 RM8352 | 2009 | USA | EHEC | O121 | Sedimentb | 93.17% | CP028110.1 |
| Escherichia coli O121:H19 16-9255 | 2016 | Canada | EHEC | O121 | Flour | 93.17% | CP022407.1 |
| Escherichia albertii _EC06_170 | 2006 | Japan | NA | NA | Human | 93.20% | AP014857.1 |
NA – Information not available or not applicable.
the strain was isolated from sediments from a stream.
Assessment of the LEE Region Functionality
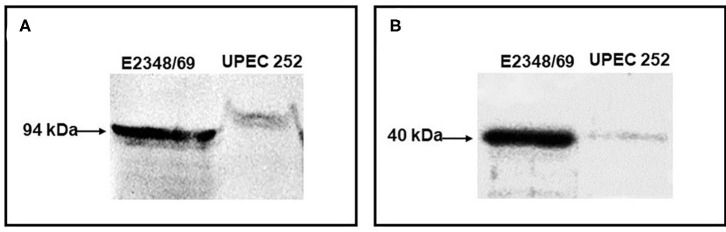
The functionality of the LEE was verified phenotypically. Initially, Intimin and EspB expression was confirmed by immunoblotting, in which both proteins exhibited their predicted molecular sizes (Figure 1).
Figure 1.
Immunodetection of Intimin and EspB. The production of the adhesin intimin and the secreted protein EspB were detected by immunoblotting. (A) Immunoblotting assay of UPEC 252 using an anti-intimin antibody (1:100) against the conserved region of the protein (Int388-667); (B) Immunoblotting assay of UPEC 252 using an anti-EspB antibody (1:3000). tEPEC strain E2348/69, positive control of production of intimin (94 kDa) and EspB (~ 40 kDa).
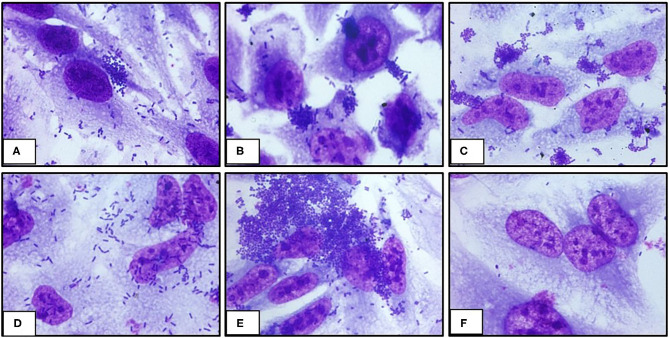
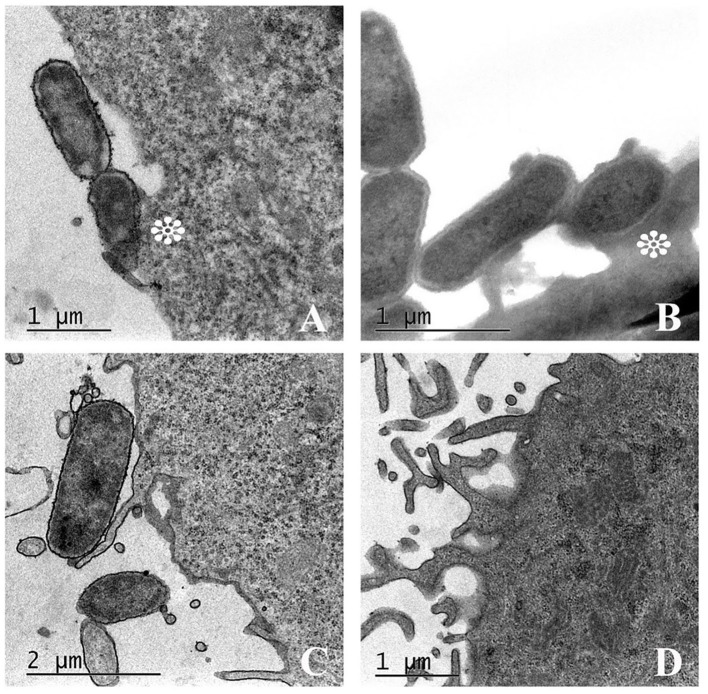
The ability of UPEC 252 to promote A/E lesion was then assessed in HeLa cells, where it presented a mixed adherence pattern either in the presence or absence of D-mannose, with occasional loose clusters resembling the Localized Adherence-Like (LAL) pattern (Figure 2), as well as bacteria diffusely attached to the cell surface, after 6 h of interaction. The ability of UPEC 252 to produce A/E lesion was then assessed by the FAS test, which revealed the capacity of the strain to mobilize actin. However, as observed by optical microscopy, actin mobilization occurred only occasionally, ~30% of the infected cells, 3-fold less of the mobilization caused by E2348/69 (Supplementary Figures 1, 2). Pedestal formation due to actin mobilization was found on the cells surface by TEM (Figure 3), showing that UPEC 252 bears a functional LEE capable of producing A/E lesion in vitro.
Figure 2.
Adherence pattern of UPEC 252 to HeLa cells. The adherence pattern of the UPEC 252 (A) was obtained in a 6-h interaction assay in the presence of 2% D-mannose. Light microscopy images (microscopic magnification 1,000 ×) shows the production of loose bacterial microcolonies similar to the LAL (localized adherence-like) pattern of atypical EPEC. Controls: Localized adhesion: typical EPEC E2348/69 (B); LAL: aEPEC 3991-1 (C); Diffuse adhesion: DAEC C1845 (D), Aggregative adhesion: EAEC 042 (E); Non-adherent: E. coli HB101 (F).
Figure 3.
Attaching-effacing lesion promoted by UPEC 252. Transmission electron microscopy (TEM) of UPEC 252 evidencing cytoskeleton rearrangement and pedestal formation under the intimate bacterial adherence to HeLa cell. (A) A/E lesion produced by UPEC 252; (B) A/E lesion produced by positive control typical EPEC E2348/69; (C) Absence of A/E lesion in cells infected with the DAEC strain C1845, used as a negative control; (D) Non-infected cells. Asterisks indicate the pedestal-like structure formed under the adhered bacterium.
UPEC 252 Genetic Background and Other Virulence-Associated Genes
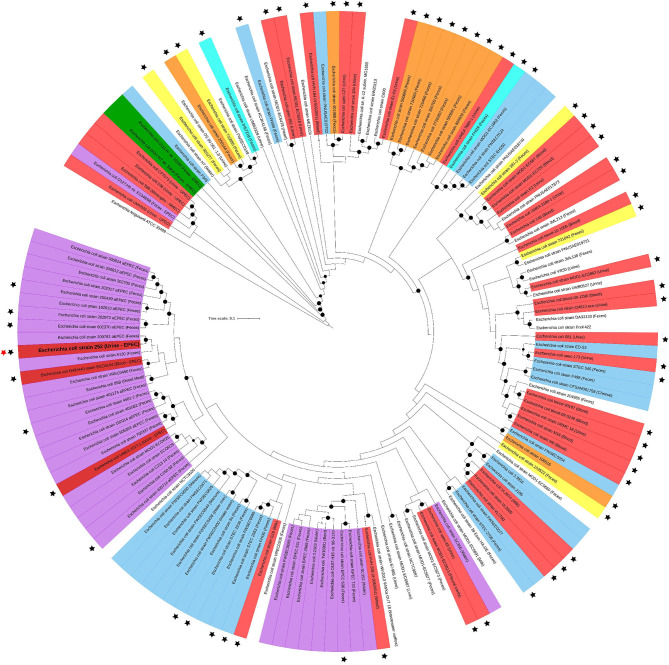
To understand the general genomic relationship of UPEC 252 with other E. coli isolates, phylogenetic analyses were performed, showing that it belongs to serotype O71:H40 and to phylogroup A. Multilocus sequence analysis conducted according to the Warwick scheme identified the strain as belonging to ST10. The phylogenetic tree built with strains of the ST10 showed that this ST is composed of strains from various DEC pathotypes, including strains presenting virulence encoding-genes of different DEC pathotypes and ExPEC strains isolated from different sites of infection (Figure 4). UPEC 252 was allocated in a cluster composed of aEPEC strains and two E. coli strains isolated from extraintestinal infections (Figure 4). Among the aEPEC strains with higher similarity, most were isolated from humans without intestinal infection (Figure 4 and Supplementary Table 1).
Figure 4.
UPEC 252 phylogenetic tree. The phylogenetic tree was built with 140 public E. coli genomes belonging to ST10 using the codon-tree methodology of 1,000 single-copy proteins in the Maximum Likelihood-based matrix (RAxML). Labels are colored according to the E. coli pathotypes, which were determined by the presence of specific virulence factors or type of infection: red, extraintestinal pathogenic E. coli (strains isolated from extraintestinal infections); orange, enterotoxigenic E. coli (elt or est positive); light blue, Shiga toxin-producing E. coli (stx positive and eae and escV negative); green, enterohemorrhagic E. coli (stx, eae and escV positive) cyan, strains harboring elt and stx (ETEC/STEC); yellow, strains isolated from diarrhea but of unknown DEC pathotype; lilac, enteropathogenic E. coli (eae and escV positive, stx negative); dark red, extraintestinal pathogenic E. coli strains that, like UPEC 252, were isolated from extraintestinal infections but harbored EPEC virulence genes; uncolored, strains with no pathotype assignment or not confirmed as causing disease. Strains from known origins are indicated in parenthesis. Black stars indicate strains isolated from confirmed infections. UPEC 252 is labeled with a bold letter and indicated by a red star aside from the label. Bootstrap values higher than 90 are indicated by a black circle in the branch.
Regarding the virulence-encoding genes investigated, in silico analysis revealed the presence of 17 virulence genes/operons, as shown in Table 3, which are related to several pathogenicity mechanisms. Genes related to iron acquisition, serum resistance, and some adhesins were identified; although none of them are recognized as classically related to extraintestinal infections, except for malX (Table 3), they might contribute to the occurrence of extraintestinal infections. Several genes of different efflux pumps related to resistance to antimicrobials and other organic compounds were found in the UPEC 252 genome (Supplementary Table 2). However, no gene directly related to antimicrobial resistance was identified. Additionally, a phenotypic assay demonstrated that our strain was sensitive to all tested antimicrobials (Supplementary Table 3). The pangenome analyzes performed reinforced the phylogenetic relationship of UPEC 252 and aEPEC (Supplementary Figure 3), but did not evidence any gene that could be related to the establishment of UTI. Moreover, the genome alignment performed using progressiveMAUVE highlighted some insertion regions (composed of genes with unknown function or bacteriophages) in the UPEC 252 genome (Supplementary Figure 4) in comparison with EPEC/EHEC strains.
Table 3.
Virulence-encoding genes detected in UPEC 252.
| Traits | Virulence factor | Gene/operon |
|---|---|---|
| Adhesins/Invasins | E. coli laminin-binding fimbriae (ELF) | elfACDG |
| Hemorrhagic E. coli pilus (HCP) | hcpABC | |
| Type I fimbriae | fimABCDEFGHI | |
| E. coli common pilus (ECP) | ecpRABCDE | |
| Curli adhesin | csgABC-csgEFG | |
| Intimin-like adhesin FdeC | eaeH | |
| IbeB (CusC) invasive protein | cusC | |
| EhaB autotransporter Protein | ehaB | |
| Protectins | Surface exclusion protein | traT |
| Increased serum survival | iss | |
| Colicin V production protein | cvpA | |
| Iron acquisition system | Ferric hydroxamate system | fhuACDB |
| Fe3+ uptake coprogen receptor | fhuE | |
| Ferric enterobactin receptor | fepA | |
| Toxins | Hemolysin/cytolysin A | hlyE |
| Cytoskeleton-binding toxin CbtA | cbtA | |
| Pathogenicity island marker | PTS system maltose-specific | malX |
Virulence genes detected by VFBD and manually confirmed by BLAST.
UPEC 252 Is Resistant to Serum Complement Activity
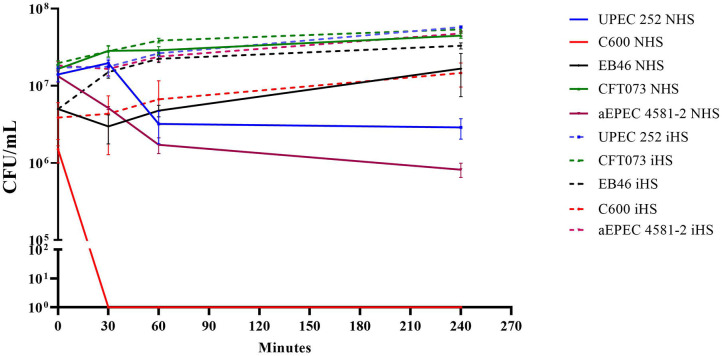
As the analyses of the genome revealed the presence of the traT and iss genes, UPEC 252 was tested regarding its ability to resist the bactericidal power of human serum. Data demonstrated that UPEC 252 was serum resistant in the assayed condition, in which bacteria were incubated with serum complement in the physiologic concentration (50%) for at least 4 h, like the serum-resistant control Enterobacter sp. strain EB046 (Figure 5).
Figure 5.
UPEC 252 serum resistance assay. Resistance to the bactericidal effect of the human serum was evaluated using a pool of human serum in PBS (50% of final serum concentration) in a 4-h assay. NHS, normal human serum; iHS, heat-inactivated human serum. The prototypical K-12 derived laboratory strain, E. coli C600, was used as a negative control, and the Enterobacter sp. strain EB046 was used as a positive control. Strains CFT073 and aEPEC4581 were also included in the experiment. The experiments were performed in biological triplicates.
UPEC 252 Does Not Produce Biofilm on Abiotic Surfaces
The ability to form biofilm on an abiotic surface (polystyrene) was tested at two different periods (24 and 48 h), however, under the conditions tested (cultivation in DMEM at 37°C) and regardless of the incubation period, the UPEC 252 was not able to form biofilm in vitro (Supplementary Figure 5).
Evaluation of the Ability of UPEC 252 to Colonize and Invade Urinary and Intestinal Tract Cells in vitro
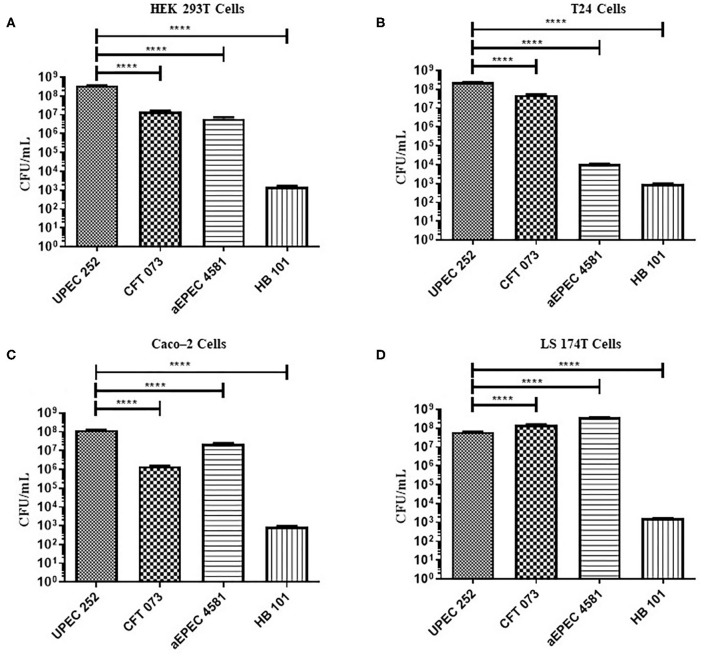
The ability of UPEC 252 to adhere to urinary (T24 and HEK 293T) and intestinal (Caco-2 and LS174T) epithelial cells was assessed using qualitative (Supplementary Figures 6, 7) and quantitative (Figure 6) adherence assays, with incubation periods of 3 h. UPEC 252 was able to adhere efficiently to all cell lines tested, and, when compared to UPEC (CFT073) and aEPEC (4581-2) controls, it showed higher adherence capacity in HEK293T, T24, and Caco-2 cells (p < 0.0001) than in the LS174T cell lineage (p < 0.001).
Figure 6.
Quantitative adherence of UPEC 252 in different eukaryotic cell lineages. Quantitative assays were performed in a 3-h incubation period, in urinary (T24 and HEK 293T), and intestinal (Caco-2 and LS174T) tract cell lineages, in the absence of D-mannose. (A) HEK-293T cells; (B) T24 cells; (C) Caco-2 cells; (D) LS174T cells. The bacterial strains CFT073, aEPEC 4581-2, and E. coli HB101 were used as UPEC, aEPEC and poorly adherent controls, respectively. ****P ≤ 0.0001.
Comparing UPEC 252 adherence capacity among the diverse cell lineages used, the number of bacteria that adhered to the cells of the urinary tract was higher when compared to the cells of the intestinal tract (*** p < 0.01) (Supplementary Figure 8).
Invasion assays were performed to verify the ability of UPEC 252 to invade epithelial cells. To this end, HeLa, HEK293T, and Caco-2 cells were infected for 3 h in the absence of mannose. Despite recovering internalized bacterial counts in the order of 104 CFU/mL, the invasion indexes observed were 0.028% (HeLa cells), 0.024% (HEK293T cells), and 0.05% (Caco-2 cells) (Supplementary Figures 9–11). These indexes were considerably low in comparison with the index of the classical invasive strain, Shigella flexneri M90T, used as a positive control.
Discussion
In this study, we sought to perform a genotypic and phenotypic characterization of UPEC 252, which carried the eae gene (Falsetti, 1998; Abe et al., 2008) that is characteristic of the EPEC and EHEC pathotypes, what suggested that this strain could be capable of producing intestinal and extraintestinal infections in the same patient.
The alignment of the sequence of the LEE of UPEC 252 with the LEE of other strains revealed high similarities (more than 99.9% of identity) with aEPEC and EHEC strains isolated in Europe, Asia, Oceania, and North America. Interestingly, the strain whose genome showed the highest similarity was an aEPEC strain isolated in Germany in the same year as UPEC 252 (1998). In addition, we investigated if UPEC 252 harbored non-LEE genes that encode effector proteins that are secreted by A/E lesion-producing pathogens by a T3SS (Deng et al., 2012). Twelve among 55 known non-LEE genes (cif, nleB, nleB1, nleB2, nleC, nleE, nleF, nleG, nleH1, nleH2, espL, and espJ) were identified. It should be noted that the non-LEE proteins are not directly related to A/E lesion formation, but their activities may contribute to increased bacterial virulence by other pathways (reviewed by Gomes et al., 2016). The non-LEE genes found in UPEC 252 are related to several pathogenic mechanisms, such as inhibition of cell detachment and modulation of cell death (cif and nleF); inhibition of phagocytosis (nleG, espJ, nleH1, and nleH2), modulation of pro-inflammatory signaling (nleB, nleB1, nleB2, nleC, nleE, nleG, nleH1, and nleH2), and cytoskeleton modulation (espL) (Dean and Kenny, 2009; Vieira et al., 2010; Vossenkämper et al., 2011; Wong et al., 2011; Salvador et al., 2014).
Immunoblotting assays revealed that UPEC 252 produced intimin and secreted the EspB protein, thus suggesting that the LEE region was functional in this strain. Given the similarity of UPEC 252 with aEPEC strains (presence of the LEE region, and absence of pEAF and the stx genes), its adhesion pattern was investigated on HeLa cells, using prolonged assays (6-h assay) in the presence of D-mannose. Although UPEC 252 presented a non-characteristic adhesion pattern, it adhered randomly, forming loose microcolonies, which is the same pattern observed in some aEPEC strains. Thus, the presence of the LEE region in the UPEC 252 genome is probably contributing to the formation of the observed loose clusters, but further studies should be performed to confirm this hypothesis.
To corroborate the hypothesis of the ability of the UPEC 252 to produce A/E lesion, we employed the FAS test, which is commonly used as an indirect measure to determine the capacity of EPEC, EHEC, E. albertii, and Citrobacter rodentium strains to remodel the host cell cytoskeleton (Knutton et al., 1991; Lima et al., 2019; Tanabe et al., 2019). Although we have verified that UPEC 252 can promote actin accumulation in HeLa cells at the site of bacterial adhesion at only some fields and 3-fold less than a typical EPEC strain, TEM images revealed the occurrence of pedestal-like structures on the cell surface. Additional experiments are necessary to confirm further the ability of this strain to promote A/E lesions in vivo since it has been shown that not all LEE-positive strains that produce these lesions are FAS positive in vitro (Bai et al., 2008).
Genomic analyses have improved our understanding of how E. coli strains have evolved, revealing their genetic backgrounds, and highlighting the occurrence of horizontal gene transfer. Our phylogenetic analysis evidenced that UPEC 252 belonged to phylogroup A, ST10, and serotype O71:H40. E. coli strains of the ST10 have been frequently associated with UTI in several countries, including Brazil (Gonçalves et al., 2016; Hertz et al., 2016; Campos et al., 2018). Other studies have also reported the isolation of E. coli strains of the ST10 from the environment and food, as well as from bloodstream infections and diarrhea, evidencing the versatility of colonization of this ST (Arais et al., 2018; Liu et al., 2018; Mohsin et al., 2018; Yamaji et al., 2018).
In addition, our analysis showed that ST10 is composed of E. coli strains from various DEC pathotypes (ETEC, STEC, EPEC), as well as commensals and strains isolated from a variety of extraintestinal infections. Moreover, strains harboring combined virulence-encoding-genes used as diagnostic markers of the different DEC pathotypes (e.g., ETEC/STEC) were identified among the ST10 strains. Furthermore, two strains isolated in Europe (Sweden and England) from extraintestinal infections and harboring the LEE region were detected. One of them was isolated in 1995 from the urine of an inpatient that developed urosepsis, while the other was isolated in a surveillance program from a bloodstream infection in 2008 (Kallonen et al., 2017). Altogether, these data corroborate with the hypotheses that some E. coli STs could be considered a “melting pot” that favors the emergence of hybrid strains, i.e., strains presenting concomitantly virulence markers that define different E. coli pathotypes (Gati et al., 2019). The ST10 complex could be a candidate for this emergence because it encompasses all E. coli pathotypes, and a variety of hybrid strains have been identified in this ST (STEC/ETEC, EAEC/UPEC, and EPEC/ExPEC).
In this context, two hypotheses were raised: UPEC 252 could be an ExPEC strain that acquired the LEE island, or an aEPEC strain that, over time, received ExPEC genes that allowed it to establish UTI. Our phylogenetic analyses supported the latter hypothesis because UPEC 252 is inserted into a cluster composed of aEPEC strains, suggesting that it has an EPEC genomic background. The diversity of aEPEC strains and their ability to acquire virulence genes by horizontal transfer were demonstrated by Hazen et al. (2017) through genome analyses. These authors showed that EPEC/ETEC hybrid strains were genetically closer to EPEC, and later acquired ETEC virulence genes. It is worth to mention that most aEPEC strains in the UPEC 252 cluster were isolated from the gut of asymptomatic human carriers, besides the two ExPEC strains that also harbored the LEE and were isolated from extraintestinal infection of non-diarrheic inpatients. These data suggested that this specific cluster could harbor strains with an attenuated diarrheagenic phenotype, which is reinforced by the few FAS positive signals produced by UPEC 252 in HeLa cells mentioned above. However, more studies are necessary to confirm this hypothesis because ST10 bears other EPEC clusters in which several pathogenic EPEC strains that had been isolated from diarrhea were detected (Ferdous et al., 2016; Arais et al., 2018; Santos et al., 2019). Besides the EPEC related genes, other virulence factors were identified in the genome of UPEC 252, among which genes that are involved in bacterial adherence to host cells, like E. coli laminin-binding fimbriae (ELF), E. coli common pilus (ECP), and Type 1 fimbriae. In UPEC strains, Type I fimbriae were demonstrated to be essential for adherence to the bladder epithelium (Paneth cells) at the early stage of UTI establishment (Connell et al., 1996; Sokurenko et al., 1998). Analysis of the genome of UPEC 252 also revealed the presence of the iss and traT genes, which confer to E. coli strains resistance to the bactericidal power of human serum (Binns et al., 1979) that is the first line of host defense (Walport, 2001). The serum resistance ability of UPEC 252 was confirmed in experiments conducted in vitro. This mechanism is essential for the pathogenicity of strains that reach the bloodstream since it allows bacteria to move across the whole human body, evading such a system and, consequently, aggravating the patient's clinical condition.
We also identified the presence of malX in the genome of UPEC 252, which encodes enzyme II of the phosphotransferase system, and is used as the pathogenicity island marker of the UPEC prototype strain CFT073, whose relationship with UTI persistence and recurrence was previously demonstrated (Ejrnæs, 2011; Ejrnæs et al., 2011). It is noteworthy that malX is essential for the persistence of E. coli strains in the intestinal tract of healthy babies (Östblom et al., 2011).
Biofilm production allows bacteria to persist in the host and can be associated with the gravity of the infection (Høiby et al., 2011). UPEC strains have been shown to present varying levels of biofilm production (Watts et al., 2010; Novais et al., 2012; Ponnusamy et al., 2012; Agarwal et al., 2013; Tapiainen et al., 2014; Flament-Simon et al., 2019). Herein, we showed that UPEC 252 is not capable of producing biofilm on abiotic surfaces under the conditions tested.
The ability of bacterial strains to adhere to and invade host cells are important mechanisms of virulence, which directly implicates increased pathogenicity since these processes are associated with persistent infections. We showed that UPEC 252 could adhere to different cell lines of the urinary and intestinal tracts in vitro. A higher affinity with the urinary tract epithelial cells was observed, which was expected, considering that it was isolated from a urinary tract infection. However, whether UPEC 252 can interact with the intestinal epithelium and cause disease in the intestinal tract remains to be investigated. Furthermore, the ability to invade the host cells allows bacteria to overcome the actions of the immune system and certain antibiotics (Lewis et al., 2016). Especially in female patients, UTIs often recur even after antibiotic therapy (Dielubanza and Schaeffer, 2011; Barber et al., 2013). Studies estimate that ~1-3rd of women up to 24 years old will be affected by UTI, and about 25% of these women will have at least one recurrent UTI in the following 6 months (Foxman, 2014). According to Lewis et al. (2016), recurrent UTIs are probably related to the invasiveness, persistence, and multiplication of bacteria within the urinary epithelium. Although the internalization rates of UPEC 252 were considerably lower than those of a classical invasive strain (S. flexneri), our findings evidenced the ability of this strain to invade the three different cell lines tested (HeLa, HEK293T, and Caco-2), suggesting that it may cause recurrent UTI, or even persistent diarrhea, since it was also able to invade differentiated intestinal Caco-2 cells. As in the present study, Owrangi et al. (2018) verified the ability of UPEC strains to adhere to and invade Caco-2 cells, with 104 CFU being recovered from the intracellular compartment.
The occurrence in UPEC 252 of genes characteristic of DEC, the isolation of an symptomatic urinary tract infection, and the presence of ExPEC traits simultaneously suggests that it may cause infections in both the intestinal and extraintestinal tracts, and can be classified as a hybrid aEPEC/UPEC strain (Santos et al., 2020a). Likewise, Lara et al. (2017) identified two UPEC strains carrying EAEC and UPEC genes simultaneously. In the same study, an E. coli strain isolated from bacteremia with the same hybrid characteristics as the UPEC strains was also found. By analyzing the presence of specific genes of the DEC pathotypes, Salmani et al. (2016) identified UPEC strains that carried the st, lt, aggR, invE, and daaD genes; however, none of them was positive for the eae gene. In contrast, Kim et al. (2018) found six UPEC strains of serogroups O2, O15, O18, and O25 that carried eae; however, these authors did not demonstrate whether these strains had complete and functional LEE regions.
In addition to our previous study (Abe et al., 2008), four other studies have reported the occurrence of EPEC/ExPEC hybrid strains (Toval et al., 2014; Kessler et al., 2015; Riveros et al., 2017; Lindstedt et al., 2018). In two of them, the strains were isolated from urinary tract infections and carried EPEC virulence genes but were devoid of the most common ExPEC virulence factors, like P fimbriae (Toval et al., 2014; Riveros et al., 2017). Interestingly, these strains also lacked the virulence genes used to classify the intrinsic virulence of pathogenic ExPEC strains, which is determined by the presence of at least two among the following virulence genes: iuc/iut, pap, sfa, afa, and kpsMTII (Johnson et al., 2003). In this context, these strains were similar to UPEC 252, whose genome carried a few ExPEC virulence-encoding genes. The other two studies reported on the occurrence of E. coli strains from diarrheal patients that harbored the ExPEC intrinsic virulence encoding-genes; in one of these patients, bacteremia and multiorgan dysfunction development were reported (Kessler et al., 2015; Lindstedt et al., 2018). Notably, these reports included strains from three different STs (ST12, ST28, and ST2018) of phylogroups B2 (Toval et al., 2014; Kessler et al., 2015; Lindstedt et al., 2018) and one strain from phylogroup A, whose ST was not determined (Riveros et al., 2017).
Although some E. coli clones of ST10 are recognized as ExPEC strains involved in various human extraintestinal infections (Bert et al., 2010; Manges et al., 2017; Yamaji et al., 2018), they do not fulfill the molecular criteria to be recognized as intrinsic virulent (Johnson et al., 2003, 2015) or uropathogenic (Spurbeck et al., 2012) strains. The absence of the most common traits related to ExPEC virulence does not limit their capacity to cause more than 10% of infections in some countries (Manges and Johnson, 2015; Campos et al., 2018; Manges et al., 2019) nor their importance as a recognized foodborne cause of extraintestinal infections that has been continuously reported worldwide (Campos et al., 2018; Yamaji et al., 2018; Manges et al., 2019). Additionally, we identified two other strains that harbored the LEE and were isolated from extraintestinal infections in the same phylogenetic cluster of UPEC 252.
Despite the frequent reports about E. coli from ST10 harboring multidrug resistance genes, including mcr-1, blaNDM, and blaCTX(Zhang et al., 2017; García et al., 2018; Mohsin et al., 2018; dos Anjos et al., 2019), UPEC 252 was shown to be sensitive to the tested antimicrobials. The detection of some resistance-related genes in UPEC 252 suggests that there is a potential phenotype of resistance for this strain, which perhaps was not detected due to the lack of other essential genes and /or gene expression.
Finally, the data presented here contribute to the discussion about hybrid E. coli strains, more specifically, hybrid strains of UPEC, which have additional virulence mechanisms that are characteristic of other E. coli pathotypes. Therefore, hybrid UPEC strains may emerge as important pathogens in the world scenario, and further studies are necessary to better understand their virulence mechanisms, so that prevention, diagnosis, and treatment can be more appropriately addressed.
Conclusion
UPEC 252 strain exhibits characteristics of ExPEC, EPEC and EHEC strains, along with the ability to adhere to and invade cells of the urinary and intestinal tracts in vitro. Our findings suggest that UPEC 252 is an atypical EPEC strain that emerges as a hybrid strain (aEPEC/UPEC), which could colonize new niches and potentially cause intestinal and extraintestinal infections.
Data Availability Statement
The reads used for UPEC 252 genome assembly were deposited in the Sequence Read Archive (SRA) at NCBI under the accession number SRR9317828, and the whole-genome sequences (WGS) were deposited in the GenBank database under the accession number VFST00000000.
Ethics Statement
This work was evaluated and approved by the local Research Ethics Committee of the Federal University of São Paulo-UNIFESP/São Paulo Hospital, under CEP N 8580081117. Written and informed consent was not required in accordance with local guidelines.
Author Contributions
TG, TV, RMS, AS, and FS conceptualized the study. TV, AS, FS, EC, and TG contributed to the formal analysis. TG and TV were responsible for funding acquisition. TV, FS, AS, JN, and EC carried out the investigation. TV, FS, AS, JN, EC, and RS worked on the methodology and validated the study. TG helped with the project administration. TG and RMS supervised the study. TV, FS, and AS wrote the original draft. TV, FS, AS, JN, EC, and TG reviewed and edited the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank The Electronic Microscopy Center (CEME) of the Federal University of São Paulo for support with electron microscopy experiments.
1Seemann, T. ABRicate. Available online at: https://github.com/tseemann/abricate
Funding. This study was conducted with a scholarship funded by grant 2017/21947-7 to TV and grant 2018/17353-7 to TG from the São Paulo Research Foundation (FAPESP).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00492/full#supplementary-material
References
- Abe C. M., Salvador F. A., Falsetti I. N., Vieira M. A. M., Blanco J., Blanco J. E., et al. (2008). Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol. Med. Microbiol. 52, 397–406. 10.1111/j.1574-695X.2008.00388.x [DOI] [PubMed] [Google Scholar]
- Agarwal J., Mishra B., Srivastava S., Srivastava R. (2013). Genotypic characteristics and biofilm formation among Escherichia coli isolates from Indian women with acute cystitis. Trans. R. Soc. Trop. Med. Hyg. 107, 183–187. 10.1093/trstmh/trs090 [DOI] [PubMed] [Google Scholar]
- Arais L. R., Barbosa A. V., Andrade J. R. C., Gomes T. A. T., Asensi M. D., Aires C. A. M., et al. (2018). Zoonotic potential of atypical enteropathogenic Escherichia coli (aEPEC) isolated from puppies with diarrhoea in Brazil. Vet. Microbiol. 227, 45–51. 10.1016/j.vetmic.2018.10.023 [DOI] [PubMed] [Google Scholar]
- Bai L., Schüller S., Whale A., Mousnier A., Marches O., Wang L., et al. (2008). Enteropathogenic Escherichia coli O125:H6 triggers attaching and effacing lesions on human intestinal biopsy specimens independently of Nck and TccP/TccP2. Infect. Immun. 76, 361–368. 10.1128/IAI.01199-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber A. E., Norton J. P., Spivak A. M., Mulvey M. A. (2013). Urinary tract infections: current and emerging management strategies. Clin. Infect. Dis. 57, 719–724. 10.1093/cid/cit284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. 10.1093/ajcp/45.4_ts.493 [DOI] [PubMed] [Google Scholar]
- Beghain J., Bridier-Nahmias A., Le Nagard H., Denamur E., Clermont O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genomics 4:e000192. 10.1099/mgen.0.000192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert F., Johnson J. R., Ouattara B., Leflon-Guibout V., Johnston B., Marcon E., et al. (2010). Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J. Clin. Microbiol. 48, 2709–2714. 10.1128/JCM.00516-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns M. M., Davies D. L., Hardy K. G. (1979). Cloned fragments of the plasmid ColV,I-K94 specifying virulence and serum resistance. Nature 279, 778–781. 10.1038/279778a0 [DOI] [PubMed] [Google Scholar]
- Campos A. C. C., Andrade N. L., Ferdous M., Chlebowicz M. A., Santos C. C., Correal J. C. D., et al. (2018). Comprehensive molecular characterization of Escherichia coli isolates from urine samples of hospitalized patients in Rio de Janeiro, Brazil. Front. Microbiol. 9:243. 10.3389/fmicb.2018.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zheng D., Liu B., Yang J., Jin Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res. 44, D694–D697. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell H., Agace W., Klemm P., Schembri M., Mårild S., Svanborg C. (1996). Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 93, 9827–9832. 10.1073/pnas.93.18.9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M. A., Law R. J., Scholz R., Keeney K. M., Wlodarska M., Finlay B. B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale A. P., Woodford N. (2015). Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J. Infect. 71, 615–626. 10.1016/j.jinf.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P., Kenny B. (2009). The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12, 101–109. 10.1016/j.mib.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Yu H. B., De Hoog C. L., Stoynov N., Li Y., Foster L. J., et al. (2012). Quantitative proteomic analysis of type III secretome of enteropathogenic Escherichia coli reveals an expanded effector repertoire for attaching/effacing bacterial pathogens. Mol. Cell. Proteomics 11, 692–709. 10.1074/mcp.M111.013672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dielubanza E. J., Schaeffer A. J. (2011). Urinary tract infections in women. Med. Clin. North Am. 95, 27–41. 10.1016/j.mcna.2010.08.023 [DOI] [PubMed] [Google Scholar]
- Donnenberg M. S., Welch R. A. (1996). Virulence determinants of uropathogenic Escherichia coli, in Urinary Tract Infections: Molecular Pathogenesis and Clinical Management, eds Mobley H. L. T., Warren J. W. (Washington, DC: ASM Press; ), 135–174. [Google Scholar]
- dos Anjos C., Sabino C. P., Bueris V., Fernandes M. R., Pogliani F. C., Lincopan N., et al. (2019). Antimicrobial blue light inactivation of international clones of multidrug-resistant Escherichia coli ST10, ST131 and ST648. Photodiagnosis Photodyn. Ther. 27, 51–53. 10.1016/j.pdpdt.2019.05.014 [DOI] [PubMed] [Google Scholar]
- Doster E., Lakin S. M., Dean C. J., Wolfe C., Young J. G., Boucher C., et al. (2020). MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 48, 561–569. 10.1093/nar/gkz1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejrnæs K. (2011). Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli. Dan. Med. Bull. 58:B4187. Available online at: https://ugeskriftet.dk/files/scientific_article_files/2018-11/b4187.pdf [PubMed] [Google Scholar]
- Ejrnæs K., Stegger M., Reisner A., Ferry S., Monsen T., Holm S. E., et al. (2011). Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: phylogenetic groups, virulence factors and biofilm formation. Virulence 2, 528–537. 10.4161/viru.2.6.18189 [DOI] [PubMed] [Google Scholar]
- EUCAST (2020). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Available online at: http://www.eucast.org (accessed March 16, 2020).
- Falsetti I. N. (1998). Identificação de Caracteristicas Fenotipicas e Genotipicas de Escherichia coli Enteroagregativa (EAEC) em Amostras de Escherichia coli Uropatogenica (UPEC). Available online at: http://repositorio.unifesp.br/handle/11600/16065 (acessed January 19, 2020).
- Feldgarden M., Brover V., Haft D. H., Prasad A. B., Slotta D. J., Tolstoy I., et al. (2019). Validating the AMRFINder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63:e00483-19. 10.1128/AAC.00483-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous M., Friedrich A. W., Grundmann H., de Boer R. F., Croughs P. D., Islam M. A., et al. (2016). Molecular characterization and phylogeny of Shiga toxin–producing Escherichia coli isolates obtained from two Dutch regions using whole genome sequencing. Clin. Microbiol. Infect. 22, 642.e1–642.e9. 10.1016/j.cmi.2016.03.028 [DOI] [PubMed] [Google Scholar]
- Finer G., Landau D. (2004). Pathogenesis of urinary tract infections with normal female anatomy. Lancet Infect. Dis. 4, 631–635. 10.1016/S1473-3099(04)01147-8 [DOI] [PubMed] [Google Scholar]
- Flament-Simon S. C., Duprilot M., Mayer N., García V., Alonso M. P., Blanco J., et al. (2019). Association between kinetics of early biofilm formation and clonal lineage in Escherichia coli. Front. Microbiol. 10:1183. 10.3389/fmicb.2019.01183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman B. (2014). Urinary tract infection syndromes. Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. North Am. 28, 1–13. 10.1016/j.idc.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Freire C. A., Santos A. C. M., Pignatari A. C., Silva R. M., Elias W. P. (2020). Serine protease autotransporters of enterobacteriaceae (SPATEs) are largely distributed among Escherichia coli isolated from the bloodstream. Brazilian J. Microbiol. 51, 447–454. 10.1007/s42770-020-00224-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. I., Jouve M., Nataro J. P., Gounon P., Le Bouguénec C. (2000). Characterization of the AfaD-like family of invasins encoded by pathogenic Escherichia coli associated with intestinal and extra-intestinal infections. FEBS Lett. 479, 111–117. 10.1016/S0014-5793(00)01898-6 [DOI] [PubMed] [Google Scholar]
- García V., García-Meniño I., Mora A., Flament-Simon S. C., Díaz-Jiménez D., Blanco J. E., et al. (2018). Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006-2017). Int. J. Antimicrob. Agents 52, 104–108. 10.1016/j.ijantimicag.2018.03.022 [DOI] [PubMed] [Google Scholar]
- Gati N. S., Middendorf-Bauchart B., Bletz S., Dobrindt U., Mellmann A. (2019). Origin and evolution of hybrid Shiga toxin-producing and uropathogenic (STEC/UPEC) Escherichia coli of sequence type 141. J. Clin. Microbiol. 58:01309–19. 10.1128/JCM.01309-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girón J. A., Ho A. S. Y., Schoolnik G. K. (1991). An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254, 710–713. 10.1126/science.1683004 [DOI] [PubMed] [Google Scholar]
- Gomes T. A. T., Elias W. P., Scaletsky I. C. A., Guth B. E. C., Rodrigues J. F., Piazza R. M. F., et al. (2016). Diarrheagenic Escherichia coli. Br. J. Microbiol. 47, 3–30. 10.1016/j.bjm.2016.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves L. F., de Oliveira Martins P., De Melo A. B. F., Da Silva R. C. R. M., de Paulo Martins V., Pitondo-Silva A., et al. (2016). Multidrug resistance dissemination by extended-spectrum β-lactamase-producing Escherichia coli causing community-acquired urinary tract infection in the Central-Western Region, Brazil. J. Glob. Antimicrob. Resist. 6, 1–4. 10.1016/j.jgar.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Guirro M., de Souza R. L., Piazza R. M. F., Guth B. E. C. (2013). Antibodies to intimin and Escherichia coli-secreted proteins EspA and EspB in sera of Brazilian children with hemolytic uremic syndrome and healthy controls. Vet. Immunol. Immunopathol. 152, 121–125. 10.1016/j.vetimm.2012.09.016 [DOI] [PubMed] [Google Scholar]
- Gupta S. K., Padmanabhan B. R., Diene S. M., Lopez-Rojas R., Kempf M., Landraud L., et al. (2014). ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. 10.1128/AAC.01310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen T. H., Michalski J., Luo Q., Shetty A. C., Daugherty S. C., Fleckenstein J. M., et al. (2017). Comparative genomics and transcriptomics of Escherichia coli isolates carrying virulence factors of both enteropathogenic and enterotoxigenic E. coli. Sci. Rep. 7:3513. 10.1038/s41598-017-03489-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilberg I. P., Schor N. (2003). Diagnosis and clinical management of urinary tract infection. Rev. Assoc. Med. Bras. 49, 109–116. 10.1590/S0104-42302003000100043 [DOI] [PubMed] [Google Scholar]
- Hernandes R. T., De la Cruz M. A., Yamamoto D., Girón J. A., Gomes T. A. T. (2013). Dissection of the role of pili and type 2 and 3 secretion systems in adherence and biofilm formation of an atypical enteropathogenic Escherichia coli strain. Infect. Immun. 81, 3793–3802. 10.1128/IAI.00620-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandes R. T., Silva R. M., Carneiro S. M., Salvador F. A., Fernandes M. C. D. C., Padovan A. C. B., et al. (2008). The localized adherence pattern of an atypical enteropathogenic Escherichia coli is mediated by intimin omicron and unexpectedly promotes HeLa cell invasion. Cell. Microbiol. 10, 415–425. 10.1111/j.1462-5822.2007.01054.x [DOI] [PubMed] [Google Scholar]
- Hertz F. B., Nielsen J. B., Schønning K., Littauer P., Knudsen J. D., Løbner-Olesen A., et al. (2016). Population structure of drug-susceptible, -resistant and ESBL-producing Escherichia coli from community-acquired urinary tract infections. BMC Microbiol. 16:63 10.1186/s12866-016-0725-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N., Ciofu O., Johansen H. K., Song Z. J., Moser C., Jensen P. Ø., et al. (2011). The clinical impact of bacterial biofilms. Int. J. Oral Sci. 3, 55–65. 10.4248/IJOS11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton T. M. (2012). Uncomplicated urinary tract infection. N. Engl. J. Med. 366, 1028–1037. 10.1056/NEJMcp1104429 [DOI] [PubMed] [Google Scholar]
- Ingle D. J., Valcanis M., Kuzevski A., Tauschek M., Inouye M., Stinear T., et al. (2016). In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genomics 2:e000064 10.1099/mgen.0.000064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse A. E., Yu J., Tall B. D., Kaper J. B. (1990). A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 87, 7839–7843. 10.1073/pnas.87.20.7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acid Res. 45, D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen K. G., Scheutz F., Lund O., Hasman H., Kaas R. S., Nielsen E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. 10.1128/JCM.03617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen K. G., Tetzschner A. M. M., Iguchi A., Aarestrup F. M., Scheutz F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. 10.1128/JCM.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Murray A. C., Gajewski A., Sullivan M., Snippes P., Kuskowski M. A., et al. (2003). Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47, 2161–2168. 10.1128/AAC.47.7.2161-2168.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Porter S., Johnston B., Kuskowski M. A., Spurbeck R. R., Mobley H. L. T., et al. (2015). Host characteristics and bacterial traits predict experimental virulence for Escherichia coli bloodstream isolates from patients with urosepsis. Open Forum Infect. Dis. 2:eofv083. 10.1093/ofid/ofv083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Russo T. A. (2018). Molecular epidemiology of extraintestinal pathogenic Escherichia coli. EcoSal. Plus 8:ESP-0004-2017. 10.1128/ecosalplus.ESP-0004-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallonen T., Brodrick H. J., Harris S. R., Corander J., Brown N. M., Martin V., et al. (2017). Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 27, 1437–1449. 10.1101/gr.216606.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Nataro J. P. (1998). Diarrheagenic Escherichia coli strains. Clin. Microbiol. Rev. 11, 142–201. 10.1128/CMR.11.1.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Nataro J. P., Mobley H. L. T. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Keller R., Ordoñez J. G., De Oliveira R. R., Trabulsi L. R., Baldwin T. J., Knutton S. (2002). Afa, a diffuse adherence fibrillar adhesin associated with enteropathogenic Escherichia coli. Infect. Immun. 70, 2681–2689. 10.1128/IAI.70.5.2681-2689.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R., Pedroso M. Z., Ritchmann R., Silva R. M. (1998). Occurrence of virulence-associated properties in enterobacter cloacae. Infect. Immun. 66, 645–649. 10.1128/IAI.66.2.645-649.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R., Nisa S., Hazen T. H., Horneman A., Amoroso A., Rasko D. A., et al. (2015). Diarrhea, bacteremia and multiorgan dysfunction due to an extraintestinal pathogenic Escherichia coli strain with enteropathogenic E. coli genes. Pathog. Dis. 73:ftv076. 10.1093/femspd/ftv076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Subhadra B., Kang H. Y., Woo K., Kim J., Son Y. J., et al. (2018). Virulence properties of uropathogenic Escherichia coli isolated from children with urinary tract infection in Korea. Genes Genomics 40, 625–634. 10.1007/s13258-018-0664-6 [DOI] [PubMed] [Google Scholar]
- Knutton S., Baldwin T., Williams P. H., McNeish A. S. (1989). Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57, 1290–1298. 10.1128/IAI.57.4.1290-1298.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S., Phillips A. D., Smith H. R., Gross R. J., Shaw R., Watson P., et al. (1991). Screening for enteropathogenic Escherichia coli in infants with diarrhea by the fluorescent-actin staining test. Infect. Immun. 59, 365–371. 10.1128/IAI.59.1.365-371.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel L., Howell M., Spencer J. D. (2017). The clinical diagnosis and management of urinary tract infections in children and adolescents. Paediatr. Int. Child Health 37, 273–279. 10.1080/20469047.2017.1382046 [DOI] [PubMed] [Google Scholar]
- Lara F. B. M., Nery D. R., de Oliveira P. M., Araujo M. L., Carvalho F. R. Q., Messias-Silva L. C. F., et al. (2017). Virulence markers and phylogenetic analysis of Escherichia coli strains with hybrid EAEC/UPEC genotypes recovered from sporadic cases of extraintestinal infections. Front. Microbiol. 8:146. 10.3389/fmicb.2017.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimbach A. (2016). Database: ecoli_VF_collection: v0.1. Zenodo. 10.5281/zenodo.56686 [DOI] [Google Scholar]
- Letunic I., Bork P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A. J., Richards A. C., Mulvey M. A. (2016). Invasion of host cells and tissues by uropathogenic bacteria. Microbiol. Spectr. 4:UTI-0026-2016. 10.1128/microbiolspec.UTI-0026-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima M. P., Yamamoto D., Santos A. C., de M., Ooka T., Hernandes R. T., et al. (2019). Phenotypic characterization and virulence-related properties of Escherichia albertii strains isolated from children with diarrhea in Brazil. Pathog. Dis. 77:ftz014. 10.1093/femspd/ftz014 [DOI] [PubMed] [Google Scholar]
- Lindstedt B.-A., Finton M. D., Porcellato D., Brandal L. T. (2018). High frequency of hybrid Escherichia coli strains with combined intestinal pathogenic Escherichia coli (IPEC) and extraintestinal pathogenic Escherichia coli (ExPEC) virulence factors isolated from human faecal samples. BMC Infect. Dis. 18:544. 10.1186/s12879-018-3449-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Liu H., Wang L., Peng Q., Li Y., Zhou H., et al. (2018). Molecular characterization of extended-spectrum β-lactamase-producing multidrug resistant Escherichia coli from Swine in Northwest China. Front. Microbiol. 9:1756. 10.3389/fmicb.2018.01756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck S. N., Bennett-Wood V., Poon R., Robins-Browne R. M., Hartland E. L. (2005). Invasion of epithelial cells by locus of enterocyte effacement-negative enterohemorrhagic Escherichia coli. Infect. Immun. 73, 3063–3071. 10.1128/IAI.73.5.3063-3071.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges A. R., Geum H. M., Guo A., Edens T. J., Fibke C. D., Pitout J. D. D. (2019). Global extraintestinal pathogenic Escherichia coli (Expec) lineages. Clin. Microbiol. Rev. 32:e00135-18. 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges A. R., Johnson J. R. (2015). Reservoirs of extraintestinal pathogenic Escherichia coli. Microbiol. Spectr. 3:UTI-0006-2012. 10.1128/microbiolspec.UTI-0006-2012 [DOI] [PubMed] [Google Scholar]
- Manges A. R., Mende K., Murray C. K., Johnston B. D., Sokurenko E. V., Tchesnokova V., et al. (2017). Clonal distribution and associated characteristics of Escherichia coli clinical and surveillance isolates from a military medical center. Diagn. Microbiol. Infect. Dis. 87, 382–385. 10.1016/j.diagmicrobio.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Mariani-Kurkdjian P., Lemaître C., Bidet P., Perez D., Boggini L., Kwon T., et al. (2014). Haemolytic-uraemic syndrome with bacteraemia caused by a new hybrid Escherichia coli pathotype. New Microbes New Infect. 2, 127–131. 10.1002/nmi2.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matar G. M., Daou R., Araj G. F., Uwaydah M. (2005). Expression of DR-II adhesins encoded by the draE2 genes in Escherichia coli strains from urinary tract infections-related bacteremia. J. Appl. Res. 5, 275–281. [Google Scholar]
- Mcdaniel T. K., Jarvis K. G., Donnenberg M. S., Kaper J. B. (1995). A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 92, 1664–1668. 10.1073/pnas.92.5.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes M. A., Rocha L. B., Koga P. C. M., Fernandes I., Nara J. M., Magalhães C. A., et al. (2010). Identification of enteropathogenic and enterohaemorrhagic Escherichia coli strains by immunoserological detection of intimin. J. Appl. Microbiol. 108, 878–887. 10.1111/j.1365-2672.2009.04484.x [DOI] [PubMed] [Google Scholar]
- Mohsin J., Pál T., Petersen J. E., Darwish D., Ghazawi A., Ashraf T., et al. (2018). Plasmid-mediated colistin resistance gene mcr-1 in an Escherichia coli ST10 bloodstream isolate in the sultanate of Oman. Microb. Drug Resist. 24, 278–282. 10.1089/mdr.2017.0131 [DOI] [PubMed] [Google Scholar]
- Moon H. W., Whipp S. C., Argenzio R. A., Levine M. M., Giannella R. A. (1983). Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41, 1340–1351. 10.1128/IAI.41.3.1340-1351.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais Â., Pires J., Ferreira H., Costa L., Montenegro C., Vuotto C., et al. (2012). Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob. Agents Chemother. 56, 3973–3976. 10.1128/AAC.00475-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Ooka T., Asadulghani Terajima J., Nougayrède J. P., Kurokawa K., et al. (2007). Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 8:R138. 10.1186/gb-2007-8-7-r138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen B., Kolmos H. J., Ørskov F., Ørskov I. (1994). Cluster of multiresistant Escherichia coli O78:H10 in greater copenhagen. Scand. J. Infect. Dis. 26, 406–410. 10.3109/00365549409008613 [DOI] [PubMed] [Google Scholar]
- Olesen B., Scheutz F., Andersen R. L., Menard M., Boisen N., Johnston B., et al. (2012). Enteroaggregative Escherichia coli O78:H10, the cause of an outbreak of urinary tract infection. J. Clin. Microbiol. 50, 3703–3711. 10.1128/JCM.01909-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östblom A., Adlerberth I., Wold A. E., Nowrouzian F. L. (2011). Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants' commensal microbiotas. Appl. Environ. Microbiol. 77, 2303–2308. 10.1128/AEM.02405-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owrangi B., Masters N., Kuballa A., O'Dea C., Vollmerhausen T. L., Katouli M. (2018). Invasion and translocation of uropathogenic Escherichia coli isolated from urosepsis and patients with community-acquired urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 37, 833–839. 10.1007/s10096-017-3176-4 [DOI] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T. G., et al. (2015). Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy P., Natarajan V., Sevanan M. (2012). In vitro biofilm formation by uropathogenic Escherichia coli and their antimicrobial susceptibility pattern. Asian Pac. J. Trop. Med. 5, 210–213. 10.1016/S1995-7645(12)60026-1 [DOI] [PubMed] [Google Scholar]
- Redder J. D., Leth R. A., Møller J. K. (2016). Analysing risk factors for urinary tract infection based on automated monitoring of hospital-acquired infection. J. Hosp. Infect. 92, 397–400. 10.1016/j.jhin.2015.12.009 [DOI] [PubMed] [Google Scholar]
- Riveros M., García W., García C., Durand D., Mercado E., Ruiz J., et al. (2017). Molecular and phenotypic characterization of diarrheagenic Escherichia coli strains isolated from bacteremic children. Am. J. Trop. Med. Hyg. 97, 1329–1336. 10.4269/ajtmh.17-0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A. (2003). The etiology of urinary tract infection: traditional and emerging pathogens. Dis. Mon. 49, 71–82. 10.1067/mda.2003.8 [DOI] [PubMed] [Google Scholar]
- Russo T. A., Johnson J. R. (2000). Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181, 1753–1754. 10.1086/315418 [DOI] [PubMed] [Google Scholar]
- Salmani H., Azarnezhad A., Fayazi M. R., Hosseini A. (2016). Pathotypic and phylogenetic study of diarrheagenic Escherichia coli and uropathogenic E. coli using multiplex polymerase chain reaction. Jundishapur J. Microbiol. 9:e28331. 10.5812/jjm.28331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltoglu N., Karali R., Yemisen M., Ozaras R., Balkan I. I., Mete B., et al. (2015). Comparison of community-onset healthcare-associated and hospital-acquired urinary infections caused by extended-spectrum beta-lactamase-producing Escherichia coli and antimicrobial activities. Int. J. Clin. Pract. 69, 766–770. 10.1111/ijcp.12608 [DOI] [PubMed] [Google Scholar]
- Salvador F. A., Hernandes R. T., Vieira M. A. M., Rockstroh A. C., Gomes T. A. T. (2014). Distribution of non-LEE-encoded type 3 secretion system dependent effectors in enteropathogenic Escherichia coli. Brazilian J. Microbiol. 45, 851–855. 10.1590/S1517-83822014000300014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. C. M., Santos F. F., Silva R. M., Gomes T. A. T. (2020a). Diversity of Hybrid- and Hetero-Pathogenic Escherichia coli and their potential implication in more severe diseases. Front. Cell. Infect. Microbiol. 10:339. 10.3389/fcimb.2020.00339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. C. M., Silva R. M., Valiatti T. B., Santos F. F., Santos-Neto J. F., Cayô R., et al. (2020b). Virulence potential of a multidrug-resistant Escherichia coli strain belonging to the emerging clonal group ST101-B1 isolated from bloodstream infection. Microorganisms 8:827. 10.3390/microorganisms8060827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. C. M., Zidko A. C. M., Pignatari A. C., Silva R. M. (2013). Assessing the diversity of the virulence potential of Escherichia coli isolated from bacteremia in São Paulo, Brazil. Br. J. Med. Biol. Res. 46, 968–973. 10.1590/1414-431X20133184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F. F., Yamamoto D., Abe C. M., Bryant J. A., Hernandes R. T., Kitamura F. C., et al. (2019). The Type III Secretion System (T3SS)-translocon of atypical enteropathogenic Escherichia coli (aEPEC) can mediate adherence. Front. Microbiol. 10:1527. 10.3389/fmicb.2019.01527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaletsky I. C. A., Silva M. L. M., Trabulsi L. R. (1984). Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45, 534–536. 10.1128/IAI.45.2.534-536.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Genome analysis prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sokurenko E. V., Chesnokova V., Dykhuizen D. E., Ofek I., Wu X. R., Krogfelt K. A., et al. (1998). Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. U.S.A. 95, 8922–8926. 10.1073/pnas.95.15.8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck R. R., Dinh P. C., Walk S. T., Stapleton A. E., Hooton T. M., Nolan L. K., et al. (2012). Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 80, 4115–4122. 10.1128/IAI.00752-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzart S., Gomes T. A. T., Guth B. E. C. (1999). Characterization of serotypes and outer membrane protein profiles in enteroaggregative Escherichia coli strains. Microbiol. Immunol. 43, 201–205. 10.1111/j.1348-0421.1999.tb02394.x [DOI] [PubMed] [Google Scholar]
- Tanabe R. H. S., Vieira M. A., Mariano N. A. B., Dias R. C. B., da Silva R. V., Castro C. M., et al. (2019). Identification and characterization of atypical enteropathogenic and Shiga toxin-producing Escherichia coli isolated from ground beef and poultry breast purchased in Botucatu, Brazil. Brazilian J. Microbiol. 50, 1099–1103. 10.1007/s42770-019-00101-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapiainen T., Hanni A. M., Salo J., Ikäheimo I., Uhari M. (2014). Escherichia coli biofilm formation and recurrences of urinary tract infections in children. Eur. J. Clin. Microbiol. Infect. Dis. 33, 111–115. 10.1007/s10096-013-1935-4 [DOI] [PubMed] [Google Scholar]
- Terpstra M. L., Geerlings S. E. (2016). Urinary tract infections: how new findings create new research questions. Curr. Opin. Infect. Dis. 29, 70–72. 10.1097/QCO.0000000000000232 [DOI] [PubMed] [Google Scholar]
- Toval F., Köhler C. D., Vogel U., Wagenlehner F., Mellmann A., Fruth A., et al. (2014). Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 52, 407–418. 10.1128/JCM.02069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354. 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabulsi L. R., Keller R., Tardelli Gomes T. A. (2002). Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8, 508–513. 10.3201/eid0805.010385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiatti T. B., Santos F. F., Santos A. C., de M., Silva R. M., Carvalho E., et al. (2019). Draft whole-genome sequence of a uropathogenic Escherichia coli strain carrying the eae gene. Microbiol. Resour. Announc. 8:e00980-19. 10.1128/MRA.00980-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira M. A. M., Salvador F. A., Silva R. M., Irino K., Vaz T. M. I., Rockstroh A. C., et al. (2010). Prevalence and characteristics of the O122 pathogenicity Island in typical and atypical enteropathogenic Escherichia coli strains. J. Clin. Microbiol. 48, 1452–1455. 10.1128/JCM.01944-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossenkämper A., MacDonald T. T., Marchès O. (2011). Always one step ahead: how pathogenic bacteria use the type III secretion system to manipulate the intestinal mucosal immune system. J. Inflamm. 8:11. 10.1186/1476-9255-8-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace-Gadsden F., Johnson J. R., Wain J., Okeke I. N. (2007). Enteroaggregative Escherichia coli related to uropathogenic clonal group A. Emerg. Infect. Dis. 13, 757–760. 10.3201/eid1305.061057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walport M. J. (2001). Advances in immunology: complement (first of two parts). N. Engl. J. Med. 344, 1058–1066. 10.1056/NEJM200104053441406 [DOI] [PubMed] [Google Scholar]
- Wattam A. R., Davis J. J., Assaf R., Boisvert S., Brettin T., Bun C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts R. E., Hancock V., Ong C. L. Y., Vejborg R. M., Mabbett A. N., Totsika M., et al. (2010). Escherichia coli isolates causing asymptomatic bacteriuria in catheterized and noncatheterized individuals possess similar virulence properties. J. Clin. Microbiol. 48, 2449–2458. 10.1128/JCM.01611-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. R. C., Pearson J. S., Bright M. D., Munera D., Robinson K. S., Lee S. F., et al. (2011). Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80, 1420–1438. 10.1111/j.1365-2958.2011.07661.x [DOI] [PubMed] [Google Scholar]
- Wurpel D. J., Totsika M., Allsopp L. P., Webb R. I., Moriel D. G., Schembri M. A. (2016). Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. J. Proteomics 131, 177–189. 10.1016/j.jprot.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Yamaji R., Friedman C. R., Rubin J., Suh J., Thys E., McDermott P., et al. (2018). A Population-based surveillance study of shared genotypes of Escherichia coli isolates from retail meat and suspected cases of urinary tract infections. mSphere 3, e00179–e00118. 10.1128/mSphere.00179-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarivach R., Vuckovic M., Deng W., Finlay B. B., Strynadka N. C. J. (2007). Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14, 131–137. 10.1038/nsmb1196 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liao K., Gao H., Wang Q., Wang X., Li H., et al. (2017). Decreased fitness and virulence in ST10 Escherichia coli harboring blaNDM-5 and mcr-1 against a ST4981 strain with blaNDM-5. Front. Cell. Infect. Microbiol. 7:242. 10.3389/fcimb.2017.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The reads used for UPEC 252 genome assembly were deposited in the Sequence Read Archive (SRA) at NCBI under the accession number SRR9317828, and the whole-genome sequences (WGS) were deposited in the GenBank database under the accession number VFST00000000.