Abstract
SCAR/WAVE proteins and Arp2/3 complex assemble branched actin networks at the leading edge. Two isoforms of SCAR/WAVE, WAVE1 and WAVE2, reside at the leading edge, yet it has remained unclear whether they perform similar or distinct roles. Further, there have been conflicting reports about the Arp2/3-independent biochemical activities of WAVE1 on actin filament elongation. To investigate this in vivo, we knocked out WAVE1 and WAVE2 genes, individually and together, in B16-F1 melanoma cells. We demonstrate that WAVE1 and WAVE2 are redundant for lamellipodia formation and motility. However, there is a significant decrease in the rate of leading edge actin extension in WAVE2 KO cells, and an increase in WAVE1 KO cells. The faster rates of actin extension in WAVE1 KO cells are offset by faster retrograde flow, and therefore do not translate into faster lamellipodium protrusion. Thus, WAVE1 restricts the rate of actin extension at the leading edge, and appears to couple actin networks to the membrane to drive protrusion. Overall, these results suggest that WAVE1 and WAVE2 have redundant roles in promoting Arp2/3-dependent actin nucleation and lamellipodia formation, but distinct roles in controlling actin network extension and harnessing network growth to cell protrusion.
INTRODUCTION
Rapid cell migration depends on Arp2/3 complex-dependent branched actin network assembly at the leading edge, which drives the extension of lamellipodia (Svitkina et al., 1997, 1999; Small et al. 2002; Welch and Mullins, 2002). Branched actin nucleation by Arp2/3 complex is stimulated by members of the SCAR/WAVE family of nucleation-promoting factors (NPFs), and depends on their C-terminal VCA domains (Campellone and Welch, 2010; Padrick and Rosen, 2010; Krause and Gautreau, 2014). Mammals express three different SCAR/WAVE proteins (herein referred to as WAVE), from different genes: WAVE1, WAVE2, and WAVE3 (Suetsugu et al., 1999; Stovold et al., 2005). WAVE1 and WAVE2 are widely expressed in diverse cell types and tissues, whereas WAVE3 is neural specific (Suetsugu et al., 1999; Sossey-Alaoui et al., 2003). WAVE1 and WAVE2 both localize to the leading edge of cells (Nozumi et al., 2003; Yamazaki et al., 2005), but their respective roles there are still not well understood. Genetic disruptions of WAVE2 have been reported to cause severe defects in lamellipodia formation and cell motility (Yamazaki et al., 2003; Yan, 2003; Kawamura et al., 2004; Kurisu et al., 2004; Yamazaki et al., 2005; Stuart et al., 2006; Danson et al., 2007; Mendoza et al., 2011; Yamashita et al., 2011; Leithner et al., 2016). However, the roles of WAVE1 have been more elusive. In nonneuronal cells, WAVE1 transcripts are often less abundant than WAVE2 (Block et al., 2008; e.g., in B16 cells the WAVE1 transcript is approximately threefold lower than WAVE2), but the significance of these differences in expression levels is unknown. Overall, it has remained an open question why so many different cell types express both WAVE1 and WAVE2 (Block et al., 2008), and to what degree these two WAVE proteins contribute, respectively, to cell protrusion and motility.
The primary function assigned to WAVE proteins is as NPFs for the Arp2/3 complex (Pollard, 2007). NPF activity is mediated by the C-terminal VCA (verproline/WH2, central and acidic) region, in which the CA portion binds Arp2/3 complex and the V (or WH2) domain binds actin monomers (Robinson et al., 2001; Panchal et al., 2003; Boczkowska et al., 2008; Padrick et al., 2011; Ti et al., 2011; Espinoza-Sanchez et al., 2018). More specifically, the WH2 domain binds to actin in the hydrophobic cleft located between subdomains 1 and 3 (Chereau et al., 2005). Importantly, this WH2-binding site is also exposed at the barbed end of actin filaments (Co et al., 2007), and independent of its interactions with Arp2/3 complex VCA can dynamically link filament ends to membranes (Co et al., 2007; Khanduja and Kuhn, 2014; Bieling et al., 2017). Furthermore, when VCA activates Arp2/3 complex on the side of a mother filament, the new daughter filament does not begin elongating until WH2 dissociates from the barbed end of the first actin subunit (Smith et al., 2013). Thus, WH2 domains in NPFs not only facilitate Arp2/3-dependent actin nucleation of daughter branches, but also interact with the barbed ends of actin filaments independently of Arp2/3 complex to influence rates of filament elongation.
Previously, we showed that the VCA domains of WAVE1 and WAVE2 each have robust NPF effects on Arp2/3 complex (Sweeney et al., 2015). We also made the surprising discovery that the VCA domain of WAVE1, but not WAVE2, retards barbed end elongation of actin filaments in both the presence and absence of Arp2/3 complex. We demonstrated that these inhibitory effects on filament elongation were specified by the unique sequence of the WH2 domain in WAVE1. More recently, the inhibitory effects of soluble WAVE1 VCA domain on filament elongation were confirmed by another group (Bieling et al., 2017). However, the same study reported that a longer fragment of WAVE1 (PVCA, which includes adjacent polyproline stretches), when concentrated onto 2D patches and in the presence of profilin, accelerated rather than slowed elongation. These in vitro observations, in which WAVE1 has opposite effects on filament elongation depending on the particular in vitro setup and conditions, have led to uncertainty about whether WAVE1 serves to promote or restrict barbed end growth in vivo. Further, as mentioned above, most genetic studies to date have focused on only a single WAVE isoform, or disrupted all WAVE isoforms together. Few studies have compared the genetic contributions of WAVE1 and WAVE2 to leading edge actin assembly.
Here we have addressed these open questions by knocking out WAVE1 and WAVE2, alone and together, in B16-F1 cells and assessing their respective contributions to lamellipodia extension and cell motility. Our data show that WAVE1 and WAVE2 are redundant for lamellipodia formation (likely due to their similar, robust NPF activities), yet they have distinct mechanistic roles in controlling the rate of actin network extension at the leading edge, with WAVE2 promoting extension and WAVE1 restricting it. These observations suggest that WAVE1 and WAVE2 have both overlapping and distinct roles as actin regulators, and that their unique functions may provide balancing effects at the leading edge that help tune actin network growth.
RESULTS AND DISCUSSION
WAVE1 and WAVE2 have redundant roles in lamellipodia formation despite their strikingly different abundances
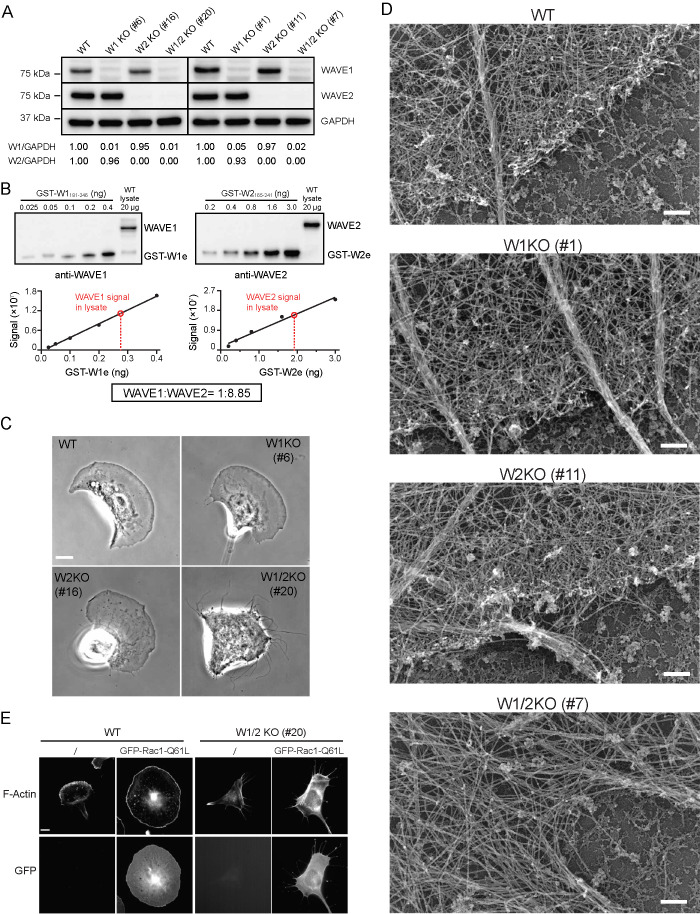
To investigate the respective contributions of WAVE1 and WAVE2 to lamellipodia dynamics and cell motility, we used CRISPR/Cas9-mediated genome editing to generate WAVE1 KO (W1 KO), WAVE2 KO (W2 KO), and WAVE1/2 double KO (W1/2 KO) lines in B16-F1 melanoma cells (Ran et al., 2013). The presence and/or absence of WAVE1 and WAVE2 isoforms was confirmed by genomic sequencing at the targeted loci (Supplemental Figure 1A) and Western blotting of cell lysates probed with isoform-specific antibodies (Figure 1A and Supplemental Figure 1, B and C). Importantly, expression of WAVE1 was unaltered by WAVE2 knockout (KO), and expression of WAVE2 was unaltered by WAVE1 KO (Figure 1A). Thus, the phenotypes arising from each KO are not due to compensatory changes in isoform expression. Two independent clones for each single and double KO line (W1KO #1 and #6; W2KO #11 and #16; W1/2KO #7 and #20) were used in most of the experiments below, as indicated. WAVE3 transcript is undetectable in B16-F1 cells (Block et al., 2008), and we could not detect WAVE3 protein in B16-F1 cells by Western blotting (Supplemental Figure 1D).
FIGURE 1:
WAVE1 and WAVE2 have redundant roles in lamellipodia formation. (A) Representative Western blots showing levels of WAVE1 and WAVE2 proteins in two independent clones of each B16-F1 cell line. Below each lane is the WAVE to GAPDH intensity ratio normalized against WT cells (set to 1.00). (B) Top panels, representative blots showing signal of endogenous WAVE1 or WAVE2 in 20 µg of cell lysate compared with known amounts of purified GST-WAVE1(aa 181–246) (GST-W1e) or GST-WAVE2(aa 165–241) (GST-W2e). Bands were quantified by densitometry and compared with the standard curves (see graphs). Black filled circles, intensity of GST-W1e or GST-W2e. Black lines, linear fits for GST-W1e or GST-W2e. Red open circles, interpolated values of endogenous WAVE1 and WAVE2 in cell lysates. The concentration of WAVE1 in cell extracts was determined in three independent experiments (22.0, 30.8, and 39.4 nM), and WAVE2 in two independent experiments (285.8 and 263.4 nM), and the means were used to estimate their molar ratio in cells (boxed). (C) Representative phase-contrast images of B16-F1 WT and KO cell lines highlighting the morphological defects in W1/2KO cells. Scale bar, 10 µm. (D) Representative platinum replica electron micrographs of actin organization at the leading edge in the same cell lines. Scale bars, 200 nm. (E) Representative images of WT and W1/2 KO cells with or without ectopic expression of GFP-Rac1-Q61L, showing F-actin stained with Alexa568-phalloidin and GFP-Rac1-Q61L. Scale bar, 10 µm.
Previous microarray analysis showed that WAVE2 transcript levels are more than threefold higher than WAVE1 in B16-F1 cells (Block et al., 2008), but to our knowledge WAVE1 and WAVE2 protein levels have never been directly compared in any cell type. To define the levels of endogenous WAVE1 and WAVE2 proteins in B16-F1 cells, we performed quantitative Western blotting using a commercial WAVE2 antibody that recognizes a purified GST-tagged fragment of WAVE2 (residues 165–241), and a homemade WAVE1 antibody that recognizes a purified GST-tagged fragment of WAVE1 (residues 181–246; Figure 1B). Using these GST-tagged fragments as quantitative standards on blots, we determined the levels of endogenous WAVE1 and WAVE2 in B16-F1 lysates, which revealed that WAVE2 protein is approximately nine times more abundant than WAVE1 (Figure 1B). This suggests that WAVE1 makes up ∼10%, and WAVE2 the remaining ∼90% of total WAVE protein. This is also consistent with our observation of a modest decrease in specific subunits of WAVE regulatory complex (WRC) in WAVE1 KO cells, and a much more dramatic decrease in WAVE2 KO cells (Supplemental Figure 1, E–G). Based on estimates of total cytosolic protein (100 mg/ml; Zeskind et al., 2007), the concentrations of WAVE1 and WAVE2 are ∼31 and ∼274.5 nM, respectively (see Materials and Methods); however, these levels may be higher at the leading edge where both proteins are enriched (Nozumi et al., 2003; Yamazaki et al., 2005). Importantly, the relative abundance of WAVE1 and WAVE2 specifically at the leading edge is still unknown, and may be distinct from the difference we observe in their total cellular abundance.
Morphologically, WAVE1 and WAVE2 single KO lines were similar to wild-type (WT) cells; however, WAVE1/2 double KO cells were strikingly different (Figure 1C). WAVE1/2 double KO cells lacked obvious lamellipodia, and instead were dominated by long filopodia-like protrusions, similar to fibroblasts lacking Arp2/3 complex (Steffen et al., 2006; Suraneni et al., 2012, 2015; Wu et al., 2012). Live imaging further revealed that in the WAVE1/2 double KO cells, the thin protrusions dynamically extend and retract, and cells advance forward by slowly filling in the gaps between the extending protrusions (Supplemental Movie S1). This was in stark contrast to WT and single KO cells, which moved by extending lamellipodia.
Movie S1.
Protrusion dynamics at the leading edge. Representative phase contrast time-lapse imaging for 5 min of WT, WAVE1 KO, WAVE2 KO, and WAVE1/2 KO B16-F1 cells, shown at 25 frames per second. Zoom shows highlighted region in right most panel at higher magnification (yellow box), with green arrowhead tracking one of the extending filopodia-like structures, while the yellow arrowhead tracking one of the retracting structures. Scale bars, 10 μm.
Consistent with these results, an examination of actin ultrastructure at the leading edge using platinum replica electron microscopy revealed densely branched networks in WT and single KO cells, but not in WAVE1/2 KO cells (Figure 1D and Supplemental Figure 2A). In addition, we analyzed motility by tracking individual cells over a 10-h period, and found that WAVE1 KO and WAVE2 KO cells were similar to WT cells in their migration speed, distance migrated (between start and end points), frequency of pausing during migration, and mean square displacement (Supplemental Figure 2, B–F). However, WAVE1/2 KO cells showed reduced migration speed and directedness, distance migrated, and increased frequency of pausing. Reexpression of EGFP-WAVE1 or EGFP-WAVE2 was sufficient to rescue lamellipodia formation (Supplemental Figure 3A), and partially rescue migration speed defects in WAVE1/2 KO cells (Supplemental Figure 3D). These results further support a redundancy in the roles of WAVE1 and WAVE2 in forming lamellipodia.
The observation that WAVE2 KO cells still form lamellipodia and move normally was somewhat unexpected, given that these cells are missing ∼90% of their total WAVE protein. This observation suggests that relatively low levels of WAVE1 are sufficient to support lamellipodia formation in the absence of WAVE2. They also raise the possibility that only a fraction of the total cellular pool of WAVE2 protein localizes to the leading edge.
Our results in B16-F1 cells differ somewhat from reported results in mouse embryonic fibroblasts (MEFs), where WAVE2 KO virtually abolished lamellipodia formation (Yamazaki et al., 2005). However, the relative levels of WAVE1 and WAVE2 have not been determined in MEFs, and thus MEFs may not contain enough WAVE1 to support lamellipodia in the absence of WAVE2. As mentioned above, B16-F1 cells lack detectable WAVE3 transcript or protein, but ectopic expression of EGFP-WAVE3 was sufficient to rescue lamellipodia formation in WAVE1/2 KO cells (46% of transfected cells formed lamellipodia, n = 95 cells; Supplemental Figure 3A), which was at least as efficient as EGFP-WAVE1 (28% of transfected cells formed lamellipodia, n = 96 cells) and EGFP-WAVE2 (34% transfected cells, n = 92 cells). These results suggest that even though WAVE3 is not normally expressed in B16-F1 cells, when expression is forced, it can serve as an effective NPF for Arp2/3 complex in building a lamellipodial actin network. We also expressed constitutively active Rac1 (Q61L), because Rac1 activity is normally required for WRC activation (Steffen et al., 2013; Schaks et al., 2018). In WT cells expressing Rac1 (Q61L), we observed the formation of enlarged lamellipodia spreading in all directions. In contrast, WAVE1/2 KO cells expressing Rac1 (Q61L) completely failed to form lamellipodia (Figure 1E), providing further evidence that the WAVE1/2 KO cells lack any appreciable amount of WAVE protein.
WAVE1 and WAVE2 have distinct roles in controlling F-actin density at the leading edge
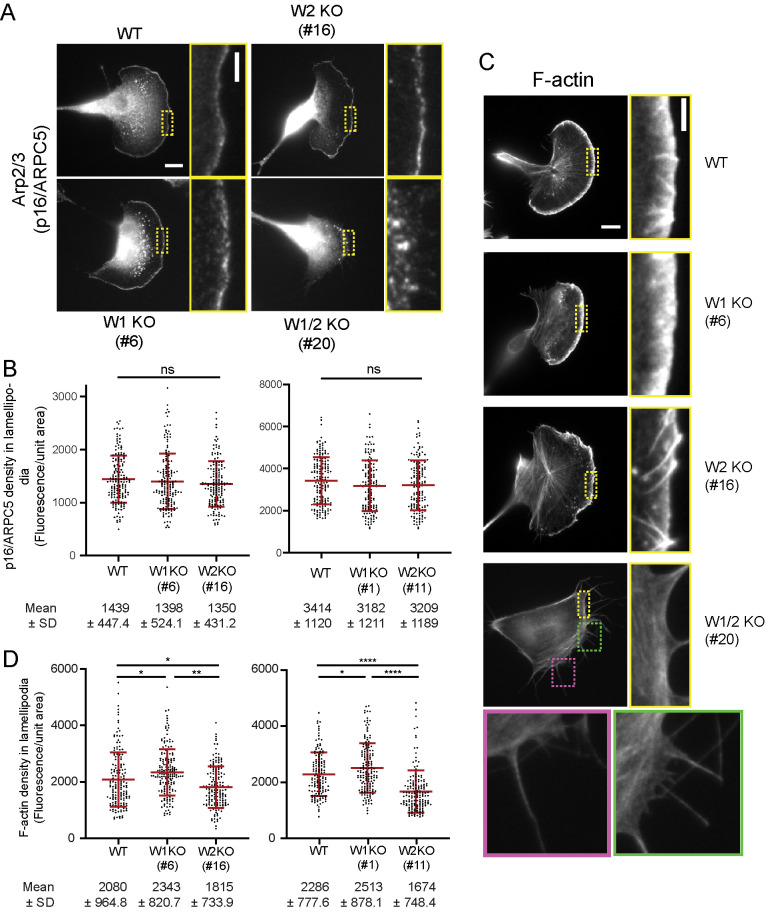
We next asked how WAVE1 and WAVE2 contribute to the densities of Arp2/3 complex and F-actin at the leading edge. Given that Arp2/3 complex is both a nucleator and a structural component of branched networks, levels of Arp2/3 complex incorporation at the leading edge reflect levels of dendritic branching in these networks. As expected, Arp2/3 complex immunostaining was largely absent from the leading edge of WAVE1/2 KO cells, and instead we observed bright puncta that possibly represent invadopodia-like adhesive structures (Figure 2A and Supplemental Figure 3C; Linder et al., 2011). However, Arp2/3 complex staining was clearly visible at the leading edge of WT and single KO cell lines (Figure 2A and Supplemental Figure 3C), consistent with the ability of these cells to form lamellipodia. Somewhat surprisingly, the levels of Arp2/3 complex were unchanged in single KO cells compared with WT cells (Figure 2B), despite substantial differences in the total levels of WAVE protein (Figure 1B). Thus, WAVE1 and WAVE2 appear to be redundant in supporting lamellipodia formation, likely because they are both strong NPFs for Arp2/3 complex. Given that the total cellular levels of WAVE1 and WAVE2 are so strikingly different, it suggests that only a fraction of the total WAVE in WT cells may be required for Arp2/3 activation at the leading edge, and that cells may maintain a large reservoir of WAVE protein in the cytosol, either to rapidly respond to other signals to form additional protrusions, or to serve additional, hitherto uncharacterized activities of WAVE proteins.
FIGURE 2:
WAVE1 and WAVE2 have distinct effects on F-actin density at the leading edge. (A) Representative cell images showing distribution of Arp2/3 complex (p16/ARPC5 subunit) at the leading edge determined by immunofluorescence. Scale bar, 10 µm. Highlighted regions (dotted boxes) shown at higher magnification. Scale bar, 3 µm. (B) Density of Arp2/3 complex (mean ± SD) in lamellipodia, quantified from cell images as in A. Data pooled from three independent experiments (left to right: n = 162, 159, 151, 165, 146, 156 cells). (C) Representative cell images showing F-actin stained with Alexa568-phalloidin. Scale bar, 10 µm. Highlighted regions (dotted boxes) shown at higher magnification. Scale bar, 3 µm. (D) Density of F-actin at the leading edge, quantified from cell images as in C. Mean ± SD shown. Data pooled from three independent experiments (left to right: n = 168, 175, 161, 139, 157, 193 cells). One-way ANOVA and Tukey’s multiple comparison tests were performed in B and D. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns (p > 0.05), not significant.
Although WT and single KO cells showed no differences in Arp2/3 density in lamellipodia, they did show differences in F-actin densities (Figure 2 and Supplemental Figure 3, B and C). WAVE2 KO cells had reduced F-actin density compared with WT cells, and WAVE1 KO cells instead had increased F-actin density (Figure 2D). As discussed earlier, WAVE1 has been hypothesized to have Arp2/3-independent functions in restricting actin filament elongation at the leading edge based on interactions of its WH2 domain with barbed ends (Sweeney et al., 2015). As a test of this model, we expressed EGFP-WAVE1 in WAVE2 KO cells and examined F-actin density. Cells expressing EGFP-WAVE1 exhibited a decrease in F-actin density compared with control WAVE2 KO cells (Supplemental Figure 3, E and F), which is consistent with the view that WAVE1 restricts barbed end growth at the leading edge. However, this result should be interpreted with some caution, as ectopic overexpression of WAVE could lead to unregulated VCA activity, in turn activating Arp2/3-dependent nucleation in the cytosol, and diminishing the actin monomer pool available for incorporation at the leading edge (Machesky and Insall, 1998; Koestler et al., 2013).
WAVE1 and WAVE2 have opposite effects on actin growth rate at the leading edge
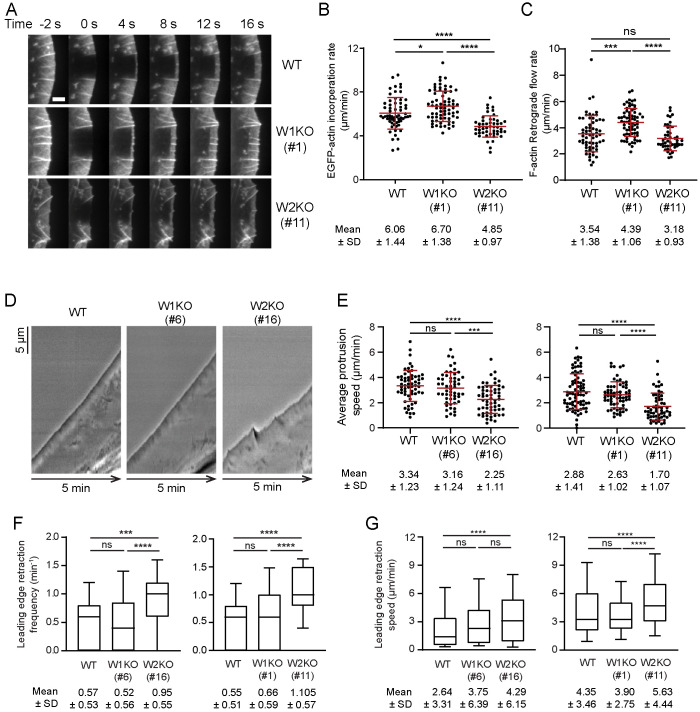
To directly determine how WAVE1 and WAVE2 KOs affect the rate of actin assembly at the leading edge, we expressed EGFP-actin in these cell lines. Then we photobleached rectangular regions of their lamellipodia and measured rates of EGFP-actin recovery in these regions of interest (Figure 3A). When actin polymerization is coupled to the plasma membrane it drives lamellipodial protrusion in 2D motility, but the growing actin network also undergoes retrograde flow (Symons and Mitchison, 1991; Waterman-Storer et al., 1998; Watanabe, 2002; Lai et al., 2008). Thus, monitoring incorporation of EGFP-actin after photobleaching allows measurement of both the rate of actin network growth/extension at the leading edge (Figure 3B) and the rate of actin network retrograde flow (Figure 3C).
FIGURE 3:
WAVE1 and WAVE2 oppositely affect actin network growth rate. (A) Representative time-lapse images of lamellipodia in WT, W1KO (#1), and W2KO (#11) cells expressing EGFP-actin, showing fluorescence recovery after photobleaching (FRAP). Scale bar, 3 µm. (B) Rates of actin assembly (EGFP-actin incorporation, mean ± SD) at the leading edge determined from FRAP analysis as in A. Data pooled from two independent experiments (left to right: n = 62, 69, 49 cells). (C) Rates of actin retrograde flow (mean ± SD) for the same cells as in B. (D) Representative kymographs of lamellipodial dynamics. (E) Lamellipodia protrusion speeds (mean ± SD). Data pooled from three independent experiments (left to right: n = 60, 54, 59, 78, 65, 58 cells). (F) Box and whisker plot (10–90 percentile) showing frequency of leading edge retraction during the 5-min window for the same cells as in E. Mean ± SD shown below the plot. (G) Box and whisker plot (10th–90th percentile) showing leading edge retraction speed for the same cells as in E. Data pooled from three independent experiments (left to right: n = 168, 141, 280, 213, 214, 314 retraction events). Mean ± SD shown below the plot. One-way ANOVA and Tukey’s multiple comparison tests were performed in B, C, and E. Kruskal-Wallis tests were performed in F and G. *, p < 0.05; **, p < 0.01;***, p < 0.01, **** p < 0.0001; ns (p > 0.05), not significant.
We observed a statistically significant increase in the rate of actin extension in WAVE1 KO cells compared with WT cells, and a marked decrease in WAVE2 KO cells (Figure 3B). These observations are in good agreement with our F-actin density measurements, and demonstrate that WAVE1 and WAVE2 have opposing roles in controlling the rate of actin incorporation at the leading edge. In WAVE1 KO cells, but not WAVE2 KO cells, we also observed a significant increase in the retrograde flow rate of actin networks (Figure 3C). This suggests that the actin networks in WAVE1 KO cells may not be coupled properly to the membrane or substrate, which is required for the increase in actin extension rate to be translated into faster membrane protrusion.
To investigate this further, we used live imaging to monitor lamellipodial dynamics (Figure 3D) and quantify protrusion rates (Figure 3E). We observed a significant decrease in the rate of lamellipodial protrusion in WAVE2 KO cells compared with WT cells, and no change in WAVE1 KO cells. These results are consistent with our predictions for WAVE1 KO cells based on their increased rate of actin extension balanced by an increased rate of retrograde flow. The reduced rate of protrusion in WAVE2 KO cells agrees with their slower rate of actin extension and unchanged rate of retrograde flow. Additionally, in WAVE2 KO cells (but not WAVE1 KO cells) there were more frequent transitions from lamellipodial protrusion to retraction, and faster retraction speeds (Figure 3, F and G). These effects may stem from slower actin assembly and protrusion rates (Yamazaki et al., 2005; Ryan et al., 2017). Based on these results, we cannot exclude the possibility that our WAVE1 KO and/or WAVE2 KO alter focal adhesions to influence actin network dynamics (Gardel et al., 2008; Yamashiro et al., 2014). Indeed, genetic disruptions of WRC and WAVE can alter focal adhesion levels and activity, possibly through an indirect mechanism (Yamazaki et al., 2005; Silva et al., 2009; Tang et al., 2013). However, for a change in focal adhesions to account for the changes we observe in retrograde actin flow in WAVE1 KO cells, we would also expect to see a change in motility rate, because motility depends strongly on adhesion, yet we do not. Instead, we primarily see changes at the leading edge in WAVE1 KO cells, including an increase in F-actin density and an increase in actin incorporation rate. These observations are most readily explained by loss of WAVE1 functions at the leading edge.
Overall, these observations suggest that even though lamellipodial actin networks undergo faster growth or extension in WAVE1 KO cells, this does not result in an increased rate of lamellipodial protrusion. This points to a role for WAVE1 in coupling actin networks to membranes. WH2 domains bind to surfaces exposed at the barbed end of the actin filament, and thus WH2 domains can influence barbed end growth rate and transiently link barbed ends to membranes (Co et al., 2007; Khanduja and Kuhn, 2014; Sweeney et al., 2015; Bieling et al., 2017). Our observations suggest that WAVE1 may perform such roles at the leading edge, consistent with our previous in vitro results showing that the VCA domain of WAVE1 binds to actin with higher affinity than the VCA domain of WAVE2 and slows barbed end growth independent of its NPF effects on Arp2/3 complex (Sweeney et al., 2015).
These results also have broad implications for actin regulation in other cell types, including neurons where WAVE1 is highly expressed (Etienne-Manneville, 2013). The enrichment of WAVE1 in neurons may reflect an increased need for restricting actin assembly and/or coupling actin assembly to membrane remodeling in synaptic function. Indeed, WAVE1 KO mice show severe defects in dendritic spine morphogenesis, synapse architecture and plasticity, central nervous system myelination, and memory and cognition (Soderling et al., 2003; Kim et al., 2006a,b; Soderling et al., 2007; Sung et al., 2008; Hazai et al., 2013). Given the distinct roles of WAVE1 and WAVE2 described here, it is possible that the expression of WAVE1 is carefully tuned in different cell types to produce peripheral actin protrusion dynamics tailored to their distinct physiological roles.
Finally, our results in B16-F1 cells support the predictions of our earlier in vitro work on WAVE1 (Sweeney et al., 2015), but reach a different conclusion from another in vitro study, which showed that immobilized, concentrated patches of WAVE1 PVCA accelerate rather than slow barbed end actin filament elongation (Bieling et al., 2017). One explanation for this discrepancy is that WAVE1 molecules at the leading edge may not be organized in this manner. Another possibility is that WAVE1 could slow versus accelerate the growth rate of actin arrays at different sites in cells depending on how WAVE1 molecules are spatially organized.
MATERIALS AND METHODS
Cell culture
B16-F1 cells (ATCC CRL-6323), and derived WAVE1 KO, WAVE2 KO, and WAVE1/2 KO cell lines generated by CRISPR/Cas9 (below) were cultured in DMEM (4.5 g/l glucose; Life Technologies, Thermofisher Scientific, Waltham, MA) supplemented with 2 mM l-glutamine (Genclone), 10% fetal calf serum (FCS; PAA Laboratory, Austria), 50 units/ml penicillin and 50 µg/ml streptomycin (Life Technologies, Thermofisher), and 10 mM HEPES (Life Technologies, Thermofisher). Unless otherwise specified, cells were transfected in six-well plates or 35-mm dishes at 50% confluency using 2 µl per well of Jetprime transfection reagent (Genesee Scientific, El Cajon, CA) and 0.5 µg of plasmid DNA.
CRISPR/Cas9-mediated genome editing
Wasf1 (WAVE1) and Wasf2 (WAVE2) genes were knocked out individually and sequentially in B16-F1 cells by CRISPR/Cas9-mediated genome editing (Cong et al., 2013). Specifically, WAVE1/2 KO cell lines were generated by disrupting Wasf1 (WAVE1) in WAVE2 KO (#11). Targeting/guide sequences GGCTGAGCTCAAGATGCCGT (for WAVE1) and GTGCCTTGGCTCGATGTTCC (for WAVE2) were cloned into plasmid pSpCas9(BB)-2A-Puro (Addgene; ID 48139), plasmids were transfected into B16-F1 cells, and transfected cells were enriched by selection for 3 d in medium containing 2.5 µg/ml puromycin (Sigma-Aldrich, Taufkirchen, Germany). Puromycin-resistant cells were diluted to grow single cell–derived colonies. After 5–7 d of growth, without agitation, individual colonies were isolated and expanded for screening by Western blotting with anti-WAVE1 and/or anti-WAVE2 antibodies, then confirmed by genomic sequencing of the target loci (for more details, see Supplemental Figure 1A).
Plasmid construction
The coding sequences of mouse WAVE1 and WAVE2 were synthesized as DNA inserts (Eurofins Scientific, Luxembourg, Belgium) and cloned into pEGFP-C2 plasmids (Clontech, Mountain View, CA) using BglII/SacII sites. The cDNA of murine WAVE3 was amplified with primers 5′-CGAATTCATGCCATTAGTCAAGAGAAACATC-3′ and 5′-CGTCGACCTCAGTCAGACCAGTCATTC-3′ and cloned into pEGFP-C2 using EcoRI/SalI. DNA sequences corresponding to amino acid residues 181–246 of human WAVE1, and 165–241 of mouse WAVE2 were cloned separately into pGEX-6P-1 plasmid using EcoRI/XhoI sites. pEGFP-β-actin plasmid was acquired from Clontech.
Protein purification
GST-WAVE1(aa181–246) and GST-WAVE2 (aa165–241) proteins were expressed in Rossetta 2 Escherichia coli cells transformed with the pGEX-6P-1-WAVE plasmids. Cells were grown to OD600 = 2.5 in terrific broth and expression was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside at 18°C for 16 h. Cells were pelleted and lysed by sonication in lysis/wash buffer (10 mM imidazole, pH 7.4, 300 mM NaCl, 1 mM EDTA, 1 mM NaN3) supplemented with 1 mM dithiothreitol (DTT), 1× protease inhibitor cocktail (0.5 μg/ml each of leupeptin, aprotinin, antipain, chymostatin, and pepstatin A), and 1 mM phenylmethylsulphonyl fluoride. The lysate was clarified by centrifugation at 20,000 × g for 25 min at 4°C and incubated for 1–2 h at 4°C with 2 ml packed glutathione agarose resin (ThermoFisher Scientific). The resin was washed with 50 column volumes of lysis/wash buffer and the bound GST-WAVE fusion proteins were eluted with 8–10 column volumes of 10 mM reduced glutathione in lysis/wash buffer, dialyzed into storage buffer (10 mM imidazole, pH 7.4, 300 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM NaN3, 50% [vol/vol] glycerol), and stored at −20°C. For antibody production, the WAVE1 fragment was purified as above, but with an additional wash of three column volumes of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4), then cleaved from GST by digestion at 4°C for 16 h with PreScission protease in PBS buffer.
Antibody generation
To generate polyclonal antibodies, bacterially expressed WAVE1 polypeptide (residues 181–246) was purified as above and injected into rabbits (Cocalico Biologicals, Stevens, PA), and antibodies were affinity purified from terminal bleeds. Briefly, 150 µg of purified GST-WAVE1(aa181–246) (GST-W1e) was run out on an SDS–PAGE gel and transferred to polyvinylidene difluoride (PVDF) membrane. The portion of the blot containing GST-W1e was excised, and incubated overnight at 4°C with the polyclonal antibody serum diluted 1:4 (v:v) in TBST buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% [vol/vol] Tween 20) containing 3% (wt/vol) bovine serum albumin (BSA) and 5 mM NaN3. The blot was washed three times with TBST and three times with high salt TBST (20 mM Tris, pH 7.5, 300 mM NaCl, 0.1% [vol/vol] Tween 20). Bound antibodies were eluted by incubating the blot for 10 min at 23°C with 0.1 M glycine (pH 2.5), with gentle agitation, and then the eluate was immediately neutralized by the addition of 0.15 volume of 1 M Tris-HCl (pH 8.5). Antibodies were concentrated using Amicon Ultra centrifugal filters (MilliporeSigma, Burlington, MA). Then 1 mg/ml BSA was added and antibodies were dialyzed against PBS supplemented with 2 mM NaN3.
Western blotting
To prepare cell lysates, B16-F1 cells were pelleted, washed with PBS, and lysed with ice-cold RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 1× cOmplete protease inhibitor cocktail (Roche, Basel, Switzerland). Lysates were then clarified by centrifugation at 17,000 × g for 15 min at 4°C, and the total soluble Z protein concentration was measured by Bradford assay (Biorad, Hercules, CA). Samples of lysates were mixed with SDS–PAGE sample buffer containing β-mercaptoethanol and boiled for 5 min, then run on SDS–PAGE gels, transferred to PVDF membranes (MilliporeSigma), and blocked with TBST buffer containing 5% (wt/vol) nonfat milk. The blot was incubated overnight at 4°C with primary antibodies in TBST plus 5% milk at the following dilutions: 1:1000 anti-WAVE1, 1:3000 anti-WAVE2 (Cell Signaling; #3659), 1:1000 anti-WAVE3 (Cell Signaling; #2806), 1:2000 anti-GAPDH (Abcam; #9485), 1:10,000 anti-Nap1 (Steffen et al., 2004), 1:5000 anti-Sra-1 (Steffen et al., 2004), and 1:1000 anti-α-tubulin (Santa Cruz; #32293). Horseradish peroxidase (HRP)–conjugated goat anti-rabbit or HRP-conjugated goat anti-mouse IgG secondary antibodies (ThermoFisher Scientific) were used at 1:2000-1:10,000. The chemiluminescence signals were developed using ECL Prime Western blotting detection reagent (GE Healthcare, Chicago, IL). Images of blots were acquired and bands were quantified by densitometry using a Chemidoc MP imaging system (BioRad).
For quantitative Western blotting, variable amounts of purified GST-WAVE1 and GST-WAVE2 polypeptides, corresponding to the regions used as antigens, were included on gels and blots as standards alongside 20 µg of cell lysate. Standard curves were generated and used to determine the amounts of endogenous WAVE1 and WAVE2 in the cell lysates. Quantities were determined from three independent blots (WAVE1) or two independent blots (WAVE2). Using the estimated concentration of total protein in the cytoplasm, 100 mg/ml (Zeskind et al., 2007), and the predicted molecular weights of WAVE1 and WAVE2 (61.5 and 54.1 kDa, respectively), we calculated the molar concentrations of WAVE1 and WAVE2 in the cytoplasm.
Fixed cell imaging
Cells were seeded onto 18-mm-diameter acid-washed glass coverslips coated with 25 µg/ml laminin and grown overnight to allow attachment. Then cells were fixed with 4% paraformaldehyde in PBS (prewarmed to 37°C) for 20 min, washed with PBS, and permeabilized with 0.1% (vol/vol) Triton X-100 in PBS for 1 or 2 min at 23°C. For phalloidin staining, cells were fixed as above, but using 4% (vol/vol) paraformaldehyde and 0.25% (vol/vol) glutaraldehyde. To visualize F-actin, fixed cells were incubated for 1 h at 23°C with 16.5 nM Alexa568-phalloidin (ThermoScientific) or 200 nM Alexa594-phalloidin in PBS. To visualize Arp2/3 complex or the WRC subunit Abl interacting protein (Abi), fixed cells were first incubated for 30 min with 3% (wt/vol) BSA in PBS, then incubated with undiluted anti-ARPC5A antiserum (Olazabal et al., 2002), or a monoclonal anti-Abi1 antibody (1:20, clone W8.3; kindly provided by Giorgio Scita, IFOM Milan, Italy) for 1 h, washed three times in PBS, followed by incubation for 1 h with a 1:200 dilution of Alexa Fluor Plus 488–goat anti-mouse IgG (ThermoScientific) in PBS containing 3% (wt/vol) BSA. Then coverslips were washed three times in PBS and mounted onto glass slides with AQUA-MOUNT anti-fade agent (ThermoScientific). Images were acquired at 0.11 µm/pixel resolution on a Nikon Eclipse Ti inverted microscope (Nikon instruments, Melville, NY) equipped with a Nikon Plan Apo λ 60× /1.4 NA oil objective and Zyla sCMOS camera (Andor Technology, Belfast, UK), driven by NIS elements software. To quantify actin, Arp2/3 complex or Abi1 fluorescence signal per area, lamellipodial regions (for F-actin) or the more distal edge of the lamellipodia (for Arp2/3 complex) were outlined and measured using the manual measurement tool from NIS element analysis software package (version 4.20; Nikon), and the background signal intensity was subtracted using the intensity of regions outside of cells. To score lamellipodia formation in W1/2KO cells carrying plasmids (expressing EGFP-WAVE1, EGFP-WAVE2, or EGFP-WAVE3), cells with visible EGFP signal were assessed for the presence of an F-actin–rich leading edge after fixation and staining with Alexa594-phalloidin.
Cell motility and lamellipodial dynamics
For random cell migration assays, 35-mm glass-bottom imaging dishes (MatTek, Ashland, MA) were precoated with laminin (25 µg/ml) for 1 h at 23°C, and then seeded with cells to ∼2.5% confluency. Cells were allowed to attach to dishes and spread for at least 6 h at 37°C, and then imaged on a Nikon Eclipse Ti inverted microscope (Nikon instruments) enclosed in an environmental chamber maintained at 37°C and 90% humidity supplied with 5% CO2. Phase-contrast images were acquired every 2 min (Supplemental Figure 2, B–F) or 5 min (Supplemental Figure 3D) for 10 h at multiple positions on the dish using a Nikon Plan Fluor 10×/0.3 NA air objective at a resolution of 0.65 µm per pixel, and a Zyla sCMOS camera (Andor Technology). The migration of all cell lines (Supplemental Figure 2, B–F) were tracked by MetaVi Lab tracking service (https://www.metavilabs.com/) using the Chemotaxis PC3050 algorithm using a 50-µm total pathlength threshold. Only those cells that remained in the field of view for the duration of imaging were included in the analysis. The migration of cells in Supplemental Figure 3D was tracked using a manual tracking plugin in ImageJ (National Institutes of Health [NIH]). For WAVE1/2 KO cells ectopically expressing EGFP-WAVE1 or EGFP-WAVE2, only those cells that displayed a lamellipodia were tracked, as only those WAVE1/2 KO cells that expressed EGFP-WAVE could form lamellipodia. Migration trajectory plots along with migration speeds and distances were determined using Chemotaxis and Migration Tool version 2.0 software (Ibidi, Martinsried, Planegg, Germany). Pausing was defined as a zero displacement between two or more continuous time frames in the movies, and pause frequency was defined as the number of pauses that occur (for an individual cell) over the 10-h duration of its migration. Mean square displacement was calculated using a previously published macro for Excel (Gorelik and Gautreau, 2014).
To monitor lamellipodial dynamics, cells were seeded on dishes and imaged as above except images were acquired every 2 s for 5 min using a heated Nikon Plan Apo λ 60×/1.4 NA oil objective (Nikon). The manual measurement tool in NIS element analysis software (version 4.20; Nikon) was used to generate kymographs of lamellipodial protrusion dynamics and calculate average protrusion velocity (from total protrusion distance over 5 min). For each cell, the protrusion velocity was averaged from measurements taken at three separate regions of the leading edge. Data were combined from at least three independent experiments (analyzing a total of at least 54 cells per condition). Retraction frequencies and velocities were also calculated from kymographs, from at least three independent experiments (analyzing a total of at least 54 cells per condition).
Actin assembly and retrograde flow at the leading edge
Cells were seeded in six-well plates or on 35-mm dishes and transfected with 0.5 µg of EGFP-actin plasmid for 16–24 h, then seeded at ∼2.5% confluency on glass chamber slides (Ibidi) coated with laminin as above, and allowed to grow overnight. During live imaging, cells were maintained in F12 HAM HEPES-buffered medium (MilliporeSigma) supplemented with 10% FCS, 2 mM l-glutamine, and penicillin (50 units/ml)/streptomycin (50 mg/ml). For fluorescence recovery after photobleaching (FRAP) experiments, cells were imaged on an inverted Axio Observer microscope (Zeiss, Oberkochen, Germany) equipped with a DG4 light source (Shutter instrument), a VIS-LED for phase-contrast optics, and a Coolsnap-HQ2 camera (Photometrics) driven by VisiView software (Visitron Systems, Puchheim, Germany). Photobleaching was achieved using a 405-nm wavelength diode laser at 70–80 mW output power, controlled by the 2D-VisiFRAP Realtime Scanner (Visitron System). Images were acquired every 2 s using a 100×/1.4 NA apochromatic oil objective before and after photobleaching of selected rectangular regions. Actin assembly rates at the leading edge were determined using ImageJ (NIH), calculated as the distance between the front and the back of the fluorescent region that is recovering divided by time (µm/min). The rate of actin assembly for each individual cell was determined from either one rectangular region, or two (in which case the rates were averaged). Data were combined from two independent experiments (analyzing a total of at least 49 cells per condition). Rates of actin retrograde flow were determined from the same live imaging data, by tracking the distance of rearward movement of the trailing edge of the bleached zone over time.
Platinum replica and electron microscopy
WT, WAVE1 KO, WAVE2 KO, and WAVE1/2 KO B16-F1 cells were plated on coverslips coated with 25 μg/ml laminin and cultured overnight. Cells were extracted with 1% Triton X-100 in PEM buffer (100 mM Pipes-KOH, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) containing 2 µM phalloidin and 10 µM taxol for 3 min at room temperature. After three quick rinses with PEM buffer containing 2 µM phalloidin and 10 µM taxol, the extracted cells were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3) for 20 min. Sample processing for platinum replica and electron microscopy (PREM) was performed as described previously (Svitkina, 2016; Yang and Svitkina, 2019). Briefly, glutaraldehyde-fixed cells were postfixed sequentially with 0.1% tannic acid and 0.2% uranyl acetate in water, dehydrated in a graded ethanol series (10%, 20%, 40%, 60%, and 80% for 5 min each, twice with 100% ethanol for 5 min each), treated with 0.2% uranyl acetate in 100% ethanol for 20 min, and washed with 100% ethanol four times for 5 min each. Samples were then critical-point dried, coated with platinum and carbon, and then “replicas” were transferred to EM grids after separation from glass coverslips with hydrofluoric acid. The PREM samples were examined on a JEM 1011 transmission electron microscope (JEOL USA, Peabody, MA) operated at 100 kV. Images were acquired by an ORIUS 832.10 W CCD camera (Gatan, Pleasanton, CA) and presented in inverted contrast.
Graphing and statistical analysis
Data were graphed and statistical analysis performed using GraphPad Prism version 8.1.2 (GraphPad Software, San Diego, CA). For statistical analysis comparing three or more groups, one-way ANOVA test (or nonparametric Kruskal-Wallis test) and Tukey’s (or Dunn’s) multiple comparison tests were performed as indicated in figure legends. p value is indicated as not significant or “ns” for p > 0.05, and as statistically significant with “*” for p ≤ 0.05, “**” for p ≤ 0.01, “***” for p ≤ 0.001, and “****” for p ≤ 0.0001.
Supplementary Material
Acknowledgments
We thank M. Angeles Juanes for providing advice throughout this project, and M. Angeles Juanes and C. P. Fees for critically evaluating the manuscript. We also thank Giorgio Scita for generously providing Abi1 antibodies, and Julia Ehinger for EGFP-WAVE3. This work was supported by an NIH R35 award (GM134895) to B.L.G., an NIH postdoctoral fellowship (F32 GM131541) to L.W.P., an NIH award (R01 GM095977) to T.S., by the Deutsche Forschunsgemeinschaft (GRK2223/1), and by intramural funding from the Helmholtz Society to K.R.
Abbreviations used:
- EGFP
enhanced green fluorescent protein
- GST
glutathione S-transferase
- KO
knockout
- MEF
mouse embryonic fibroblast
- NPF
nucleation promoting factor
- SCAR
suppressor of CAR
- VCA
verproline/WH2, central and acidic
- WAVE
WASP family Verprolin-homologous protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-12-0705) on July 22, 2020.
REFERENCES
- Bieling P, Hansen SD, Akin O, Li TD, Hayden CC, Fletcher DA, Mullins RD. (2017). WH2 and proline-rich domains of WASP-family proteins collaborate to accelerate actin filament elongation. EMBO J , 102–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, Stradal TEB, Hänisch J, Geffers R, Köstler SA, Urban E, Small JV, Rottner K, Faix J. (2008). Filopodia formation induced by active mDia2/Drf3 J Microsc , 506–517. [DOI] [PubMed] [Google Scholar]
- Boczkowska M, Rebowski G, Petoukhov MV, Hayes DB, Svergun DI, Dominguez R. (2008). X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure , 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. (2010). A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol , 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. (2005). Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA , 16644–16649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J. (2007). Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell , 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science , 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson CM, Pocha SM, Bloomberg GB, Cory GO. (2007). Phosphorylation of WAVE2 by MAP kinases regulates persistent cell migration and polarity. J Cell Sci , 4144–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Sanchez S, Metskas LA, Chou SZ, Rhoades E, Pollard TD. (2018). Conformational changes in Arp2/3 complex induced by ATP, WASp-VCA, and actin filaments. Proc Natl Acad Sci USA , E8642–E8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. (2013). Microtubules in cell migration. Annu Rev Cell Dev Biol , 471–499. [DOI] [PubMed] [Google Scholar]
- Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. (2008). Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol , 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik R, Gautreau A. (2014). Quantitative and unbiased analysis of directional persistence in cell migration. Nat Protoc , 1931–1943. [DOI] [PubMed] [Google Scholar]
- Hazai D, Szudoczki R, Ding J, Soderling SH, Weinberg RJ, Sótonyi P, Rácz B. (2013). Ultrastructural abnormalities in CA1 hippocampus caused by deletion of the actin regulator WAVE-1. PLoS One , e75248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Takano K, Suetsugu S, Kurisu S, Yamazaki D, Miki H, Takenawa T, Endo T. (2004). N-WASP and WAVE2 acting downstream of phosphatidylinositol 3-kinase are required for myogenic cell migration induced by hepatocyte growth factor. J Biol Chem , 54862–54871. [DOI] [PubMed] [Google Scholar]
- Khanduja N, Kuhn JR. (2014). Processive acceleration of actin barbed-end assembly by N-WASP. Mol Biol Cell , 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-J, DiBernardo AB, Sloane JA, Rasband MN, Solomon D, Kosaras B, Kwak SP, Vartanian TK. (2006a). WAVE1 is required for oligodendrocyte morphogenesis and normal CNS myelination. J Neurosci , 5849–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sung JY, Ceglia I, Lee K-W, Ahn J-H, Halford JM, Kim AM, Kwak SP, Park JB, Ho Ryu S, et al. (2006b). Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature , 814. [DOI] [PubMed] [Google Scholar]
- Koestler SA, Steffen A, Nemethova M, Winterhoff M, Luo N, Holleboom JM, Krupp J, Jacob S, Vinzenz M, Schur F, et al. (2013). Arp2/3 complex is essential for actin network treadmilling as well as for targeting of capping protein and cofilin. Mol Biol Cell , 2861–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Gautreau A. (2014). Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol , 577–590. [DOI] [PubMed] [Google Scholar]
- Kurisu S, Suetsugu S, Yamazaki D, Yamaguchi H, Takenawa T. (2004). Rac-WAVE2 signaling is involved in the invasive and metastatic phenotypes of murine melanoma cells. Oncogene , 1309–1319. [DOI] [PubMed] [Google Scholar]
- Lai FP, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TE, Dunn GA, Small JV, Rottner K. (2008). Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J , 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leithner A, Eichner A, Muller J, Reversat A, Brown M, Schwarz J, Merrin J, de Gorter DJ, Schur F, Bayerl J, et al. (2016). Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat Cell Biol , 1253–1259. [DOI] [PubMed] [Google Scholar]
- Linder S, Wiesner C, Himmel M. (2011). Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol , 185–211. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. (1998). Scar1 and the related Wiskott–Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex Curr Biol , 1347–1356. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Zhang W, Ballif BA, Elliott HL, Danuser G, Blenis J. (2011). ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol Cell , 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozumi M, Nakagawa H, Miki H, Takenawa T, Miyamoto S. (2003). Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J Cell Sci , 239–246. [DOI] [PubMed] [Google Scholar]
- Olazabal IM, Caron E, May RC, Schilling K, Knecht DA, Machesky LM. (2002). Rho-kinase and myosin-II control phagocytic cup formation during CR, but not FcγR, Phagocytosis , 1413–1418. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. (2011). Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA , E472–E479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK. (2010). Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem , 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. (2003). A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol , 591. [DOI] [PubMed] [Google Scholar]
- Pollard TD. (2007). Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct , 451–477. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc , 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand J-B, Higgs HN, Choe S, Pollard TD. (2001). Crystal structure of Arp2/3 complex. Science , 1679–1684. [DOI] [PubMed] [Google Scholar]
- Ryan GL, Holz D, Yamashiro S, Taniguchi D, Watanabe N, Vavylonis D. (2017). Cell protrusion and retraction driven by fluctuations in actin polymerization: a two-dimensional model. Cytoskeleton , 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaks M, Singh SP, Kage F, Thomason P, Klunemann T, Steffen A, Blankenfeldt W, Stradal TE, Insall RH, Rottner K. (2018). Distinct interaction sites of Rac GTPase with WAVE regulatory cmplex have non-redundant functions in vivo. Curr Biol , 3674–3684.e3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Ezhkova E, Silva J, Heart S, Castillo M, Campos Y, Castro V, Bonilla F, Cordon-Cardo C, Muthuswamy SK, et al. (2009). Cyfip1 is a putative invasion suppressor in epithelial cancers Cell , 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol , 112–120. [DOI] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Correa IR, Jr, Xu MQ, Goode BL, Rosen MK, Gelles J. (2013). Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife , e01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, Langeberg LK, Banker G, Raber J, Scott JD. (2007). A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci , 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD. (2003). Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci USA , 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Head K, Nowak N, Cowell JK. (2003). Genomic organization and expression profile of the human and mouse WAVE gene family. Mamm Genome , 314–322. [DOI] [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TEB. (2006). Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell , 2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Ladwein M, Dimchev GA, Hein A, Schwenkmezger L, Arens S, Ladwein KI, Margit Holleboom J, Schur F, Victor Small J, et al. (2013). Rac function is crucial for cell migration but is not required for spreading and focal adhesion formation. J Cell Sci , 4572–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TE. (2004). Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J , 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovold CF, Millard TH, Machesky LM. (2005). Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC Cell Biol , 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JR, Gonzalez FH, Kawai H, Yuan ZM. (2006). c-Abl interacts with the WAVE2 signaling complex to induce membrane ruffling and cell spreading. J Biol Chem , 31290–31297. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T. (1999). Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem Biophys Res Commun , 296–302. [DOI] [PubMed] [Google Scholar]
- Sung JY, Engmann O, Teylan MA, Nairn AC, Greengard P, Kim Y. (2008). WAVE1 controls neuronal activity-induced mitochondrial distribution in dendritic spines , 3112–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Fogelson B, Rubinstein B, Noguera P, Volkmann N, Hanein D, Mogilner A, Li R. (2015). A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol Biol Cell , 901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. (2012). The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol , 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. (2016). Imaging Cytoskeleton Components by Electron Microscopy, New York: Springer, 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. (1999). Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol , 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. (1997). Analysis of the actin–myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol , 397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MO, Collins A, Padrick SB, Goode BL. (2015). A novel role for WAVE1 in controlling actin network growth rate and architecture. Mol Biol Cell , 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons MH, Mitchison TJ. (1991). Control of actin polymerization in live and permeabilized fibroblasts. J Cell Biol , 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Li A, Bi J, Veltmen DM, Zech T, Spence HJ, Yu X, Timpson P, Insall RH, Frame MC, Machesky LM. (2013). Loss of scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Curr Biol , 107–117. [DOI] [PubMed] [Google Scholar]
- Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. (2011). Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA , E463–E471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N. (2002). Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science , 1083–1086. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Chloe Bulinski J, Salmon ED. (1998). Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol , 1227–1230. [DOI] [PubMed] [Google Scholar]
- Welch MD, Mullins RD. (2002). Cellular control of actin nucleation. Annu Rev Cell Dev Biol , 247–288. [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. (2012). Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell , 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Mizuno H, Smith MB, Ryan GL, Kiuchi T, Vavylonis D, Watanabe N. (2014). New single-molecule speckle microscopy reveals modification of the retrograde actin flow by focal adhesions at nanometer scales Mol Biol Cell , 1010–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Ueda K, Kioka N. (2011). WAVE2 forms a complex with PKA and is involved in PKA enhancement of membrane protrusions. J Biol Chem , 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Fujiwara T, Suetsugu S, Takenawa T. (2005). A novel function of WAVE in lamellipodia: WAVE1 is required for stabilization of lamellipodial protrusions during cell spreading. Genes Cells , 381–392. [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S-I, Fujiwara T, Yoshida N, Takenawa T. (2003). WAVE2 is required for directed cell migration and cardiovascular development. Nature , 452–456. [DOI] [PubMed] [Google Scholar]
- Yan C. (2003). WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility EMBOJ , 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Svitkina TM. (2019). Ultrastructure and dynamics of the actin−myosin II cytoskeleton during mitochondrial fission. Nat Cell Biol , 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeskind BJ, Jordan CD, Timp W, Trapani L, Waller G, Horodincu V, Ehrlich DJ, Matsudaira P. (2007). Nucleic acid and protein mass mapping by live-cell deep-ultraviolet microscopy Nat Methods , 567–569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.