Abstract
Microtubule nucleation is spatiotemporally regulated in cells by several known molecules, including the template γ-tubulin and the polymerase XMAP215. The role of XMAP215 in nucleation is under debate, specifically whether it acts independently as a polymerase or acts dependently with γ-tubulin. We first confirm XMAP215 as a classically defined nucleator that reduces the nucleation lag seen in bulk tubulin assembly. Secondly, using deletion constructs, we probe the domain requirements for XMAP215 to promote microtubule nucleation. We show that its ability to nucleate microtubules in purified solutions correlates with its ability to elongate existing microtubules and does not depend on the number of tumor overexpressed gene (TOG) domains. Finally, we show that XMAP215 and γ-tubulin promote αβ-tubulin assembly in an additive, not synergistic, manner. Thus, their modes of action during microtubule nucleation are distinct. These findings suggest there are at least two independent processes in nucleation, one promoted by γ-tubulin and one promoted by XMAP215. We propose that XMAP215 accelerates the addition of subunits to existing nucleation intermediates formed either spontaneously or by oligomers of γ-tubulin.
INTRODUCTION
Spontaneous nucleation of a new microtubule from nanometer-sized soluble αβ-tubulin dimers is an event that is currently impossible to observe directly. Tubulin dimers must associate longitudinally and laterally in order to form the final product: a hollow tube of side-by-side protofilaments. Models of the spontaneous microtubule nucleation pathway have disagreed on the number of rate-determining steps and key intermediates in the nucleation pathway (Voter and Erickson, 1984; Kuchnir Fygenson et al., 1995; Flyvbjerg et al., 1996).
More recently, studies of nucleation have centered on the γ-tubulin ring complex (γ-TuRC), which is essential for microtubule formation across organisms (Oakley et al., 1990; Horio et al., 1991; Stearns et al., 1991; Zheng et al., 1991; Joshi et al., 1992; Sobel and Snyder, 1995). The γ-TuRC stably binds the microtubule minus end and is thought to act as a template onto which αβ-tubulin dimers associate (Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000), theoretically promoting lateral interactions between adjacent αβ-tubulin dimers (Aldaz et al., 2005). However, increasingly higher resolution structures of purified γ-TuRC show that the template is not perfect—the 13-member γ-tubulin ring presented by the complex does not perfectly match the geometry of the microtubule (Kollman et al., 2015; Consolati et al., 2019; Wieczorek et al., 2019). Further, the fraction of purified γ-TuRC that nucleates in vitro is consistently low (Choi et al., 2010; Kollman et al., 2010; Consolati et al., 2019; Thawani et al., 2019). It is hypothesized that, in order to nucleate well, γ-TuRC must be activated in vivo through interactions with other proteins (Klotz et al., 1990; Buendia et al., 1992; Moritz et al., 1998).
There is a rapidly growing body of evidence that at least one of the crucial cofactors for the γ-TuRC is the XMAP215 family of microtubule polymerases. The XMAP215 family promotes microtubule growth from various templates, including the γ-TuRC, isolated centrosomes, and GMPCPP-stabilized seeds (Popov et al., 2002; Wieczorek et al., 2015; Flor-Parra et al., 2018; Thawani et al., 2018, 2019; Consolati et al., 2019), and it also promotes microtubule nucleation in reactions without any template (Popov et al., 2002; Slep and Vale, 2007; Roostalu et al., 2015; Hussmann et al., 2016; Woodruff et al., 2017). Across these studies, the role of XMAP215 in nucleation was generally thought to be due to its polymerase activity. Recently, however, it was reported that XMAP215 and the γ-TuRC promote microtubule nucleation in a manner that is greater than the sum of their individual effects (Flor-Parra et al., 2018; Thawani et al., 2018; Consolati et al., 2019). The mechanism underlying this reported synergy is unknown, although Thawani et al. hypothesized it is due to a direct binding interaction between the unstructured C-terminal tail of XMAP215 and γ-tubulin.
Here, we sought to characterize the mode of action of XMAP215 in a simplified system, using recombinantly expressed and purified protein in the classical turbidity assay. We first show that γ-tubulin alone forms laterally associated arrays that reduce the nucleation lag associated with αβ-tubulin assembly. We also confirm that XMAP215 alone reduces the nucleation lag and probe its mechanism of action using domain analysis, finding that nucleation activity strongly correlates with polymerase activity. We then explore the nucleation activity of the γ-tubulin arrays together with XMAP215. Across a range of concentrations of XMAP215 and across XMAP215 deletion constructs, we find additive action with γ-tubulin. These results provide evidence for two distinct rate-limiting processes during microtubule nucleation that are independently promoted by XMAP215 and γ-tubulin.
RESULTS AND DISCUSSION
γ-Tubulin and XMAP215 each promote microtubule nucleation in the classical turbidity assay
Nucleation of a new microtubule is impossible to observe directly. Fluorescence microscopy can visualize dynamics of individual microtubule formation events, but it is complicated by the requirement of fluorophore-labeled αβ-tubulin heterodimers. In order for αβ-tubulin to assemble, only a small fraction of αβ-tubulin may carry a fluorophore; this not only makes early nucleation intermediates impossible to visualize, but it also indicates that unlabeled and labeled αβ-tubulin assemble differently.
Light scattering, on the other hand, uses unlabeled αβ-tubulin heterodimers to monitor nucleation. This assay, performed in bulk, is classically used in the study of polymerization dynamics (Gaskin et al., 1974; Wegner and Engel, 1975). The initial delay or lag between the start of the reaction and when the turbidity reaches one-tenth of its maximum provides a sensitive measure of polymer nucleation efficiency (Voter and Erickson, 1984; Flyvbjerg et al., 1996).
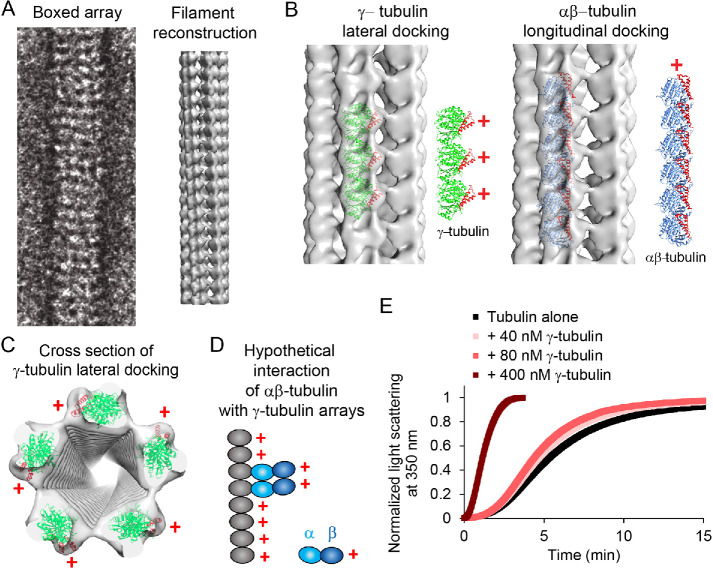
To establish the light scattering assay in our hands, we first tested an accepted αβ-tubulin nucleator, γ-tubulin. γ-Tubulin is a potent microtubule nucleator in the absence of other γ-TuRC components (see below), most likely due to its ability to oligomerize with itself in a manner distinct from αβ-tubulin, providing a binding surface that promotes and stabilizes αβ-tubulin polymerization (Figure 1). At sufficiently high concentrations, γ-tubulin forms laterally associated arrays, as revealed by negative stain electron microscopy and reconstruction (Supplemental Figure 1A), and as seen previously (Aldaz et al., 2005; Thawani et al., 2020). These lateral arrays form hollow tubes with γ-tubulin plus ends oriented outward, available for interaction with αβ-tubulin.
FIGURE 1:
Negative-stain electron microscopy reveals that γ-tubulin forms arrays that are distinct from microtubules. (A) γ-Tubulin arrays were boxed and averaged for helical reconstruction. Helical reconstruction reveals fivefold symmetry in the array, with a hollow center. (B) Left, docking of the human γ-tubulin crystal structure (1Z5W) into the 3D reconstruction revealed that γ-tubulins are laterally associated along the long axis of the arrays. The lateral packing of γ-tubulins is similar to that observed in crystals (Aldaz et al., 2005). γ-Tubulins in the arrays are oriented with their plus ends outward (red +). Helices 3 and 4 are red in the models to illustrate the orientation of the plus end of the γ- or αβ-tubulins. Right, docking of αβ-tubulins (1JFF) as a proxy for a hypothetical longitudinal assembly of γ-tubulins. The periodicity of the longitudinal model does not match that of the EM map. (C) Docking of the human γ-tubulin crystal structure (1Z5W) into the cross-section of the 3D reconstruction. The filaments are approximately 150 Å in diameter and show fivefold symmetry. (D) Cartoon representation of how αβ-tubulin dimers may interact with each other and the γ-tubulin arrays, based on the location of the plus ends of γ-tubulin. (E) Characterization of 40, 80, or 400 nM γ-tubulin in the turbidity assay with 12 μM αβ-tubulin.
Further characterization revealed that the arrays form at 250 nM γ-tubulin and above but not at 80 nM (Supplemental Figure 1A). They alone do not scatter light (Supplemental Figure 1B), evidently due to the relatively low concentration of γ-tubulin required for array formation. Also, the arrays promoted formation of microtubules rather than other noncanonical tubulin polymers (Supplemental Figure 1C). Finally, nucleation capacity correlated with array formation because concentrations of γ-tubulin that do not allow array formation (e.g., 40 and 80 nM) promote little nucleation (Figure 1E).
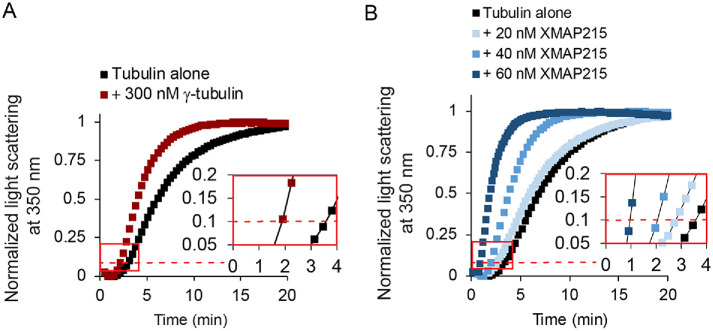
To quantify the nucleation activity of γ-tubulin arrays for our comparisons with XMAP215, we used 300 nM γ-tubulin. At this concentration, the γ-tubulin arrays decreased the time to reach 10% polymer formation, hereafter known as the nucleation lag, by approximately 50% (Figure 2A).
FIGURE 2:
Laterally associated γ-tubulin arrays and XMAP215 are classically defined nucleators. (A) Representative results of a turbidity assay for tubulin assembly at 15 μM free αβ-tubulin, with γ-tubulin added at 300 nM. The red dashed line represents 10% of the maximum light scattering. The inset shows the same data from the red box plotted on a larger scale with the data plotted as points connected by lines for clarity. The raw data from all replicates are given in Supplemental Figure 1. (B) Representative results of turbidity assay for tubulin assembly for 20, 40, or 60 nM XMAP215 with 15 μM free tubulin. XMAP215 promotes spontaneous tubulin assembly in a concentration-dependent manner. The red dashed line represents 10% of the normalized maximum light scattering. The inset shows the same data from the red box plotted on a larger scale with the data plotted as points connected by lines for clarity. SDS–PAGE of XMAP215 and the raw data for all replicates of the turbidity assay are given in Supplemental Figure 2.
We then tested the nucleation activity of XMAP215, finding significant effects. The nucleation lag decreased monotonically with increasing concentrations of XMAP215, with a decrease of 75% at the highest concentration (60 nM; Figure 2B). This provided independent confirmation that XMAP215 is a nucleator, with similar potency to γ-tubulin arrays.
XMAP215 functions as a microtubule polymerase during microtubule nucleation
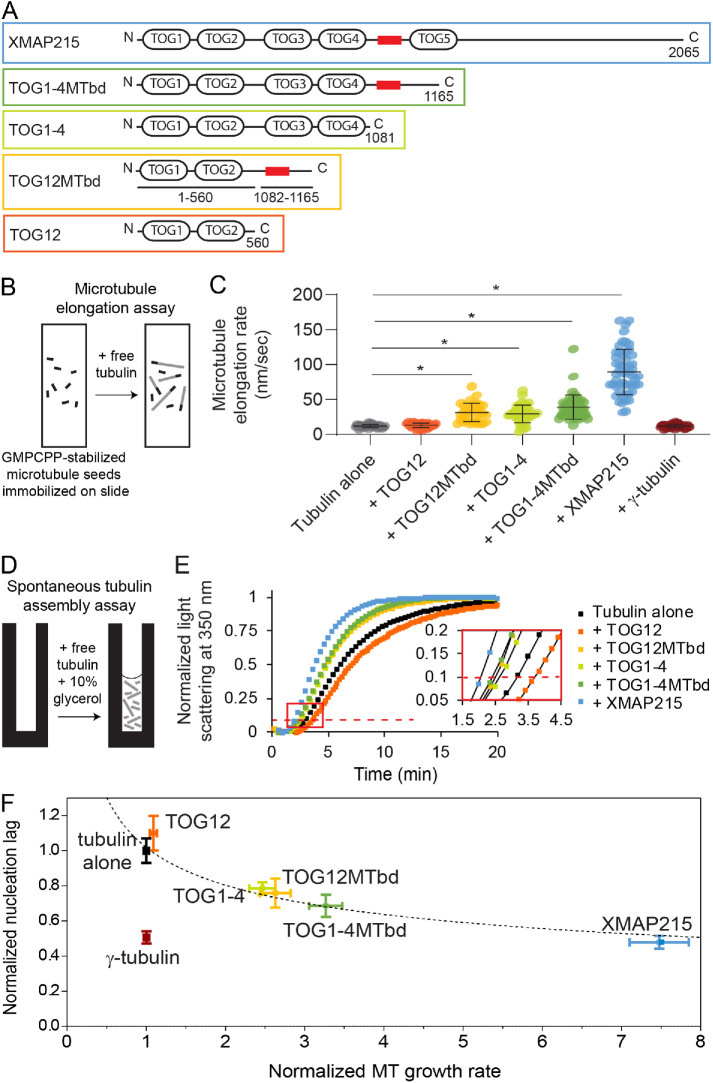
XMAP215 has five tumor overexpressed gene (TOG) domains as well as a basic region referred to as the microtubule binding domain (MTbd). Previous domain analyses of XMAP215 have shown that two separate types of domains are required for polymerase activity: 1) TOGs 1 and 2, which bind free αβ-tubulin dimers with high affinity, and 2) a microtubule-lattice binding region, which can be provided by either the MTbd or by TOGs 3 and 4 (Slep and Vale, 2007; Widlund et al., 2011; Fox et al., 2014; Flor-Parra et al., 2018). To test if the XMAP215 domain requirements for nucleation activity are the same as those for polymerase activity, we created an array of constructs with varying polymerase activities. In our constructs, we maintained the presence of TOGs 1 and 2, which are essential for polymerase activity, and varied the domains required for localization to the microtubule lattice (Figure 3A).
FIGURE 3:
The microtubule nucleation activity of XMAP215 correlates strongly with its polymerase activity. (A) XMAP215 and XMAP215 deletion constructs used in this work. The red box represents the basic microtubule binding domain. An SDS–polyacrylamide gel showing the deletion constructs is given in Supplemental Figure 2. (B) Schematic of the assay used to measure tip growth rates from immobilized GMPCPP-stabilized microtubule seeds by microscopy. (C) Microtubule elongation rates at 12 μM free αβ-tubulin, with full-length XMAP215 or the indicated deletion constructs added at 40 nM, or with γ-tubulin added at 300 nM. Error bars represent SD (N = 41, 35, 33, 41, 49, 53, 41 microtubules measured for each respective condition). The asterisk represents p < 0.0001, two-sided t test. (D) Schematic of the assay used to measure nontemplated tubulin assembly by turbidity. (E) Representative turbidity assay at 15 μM free αβ-tubulin, with XMAP215 or the indicated deletion constructs added at 40 nM. The red dashed line represents 10% of the maximum light scattering. The inset shows the data indicated by the red box replotted on a larger scale with the data plotted as points connected by lines for clarity. (F) Nucleation activity plotted against polymerase activity. Data for XMAP215 and its deletion constructs, each added at 40 nM, are fit by a power curve with the equation 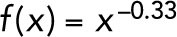 (see Materials and Methods). Normalized microtubule growth rate means ± SEM represent the data from Figure 3C normalized to the growth rate of tubulin alone. Relative nucleation activity for each construct is represented by a delay between the start of the polymerization reaction and when the turbidity reached 10% of its maximum, divided by the delay measured in controls with αβ-tubulin alone. Normalized nucleation rate means ± SEM represent the continuous turbidity reaction data of experimental conditions normalized to the nucleation lag of tubulin alone (N = 12, 7, 6, 4, 6, 5, and 7 for tubulin alone, γ-tubulin, TOG12, TOG12MTbd, TOG1-4, TOG1-4MTbd, and XMAP215, respectively).
(see Materials and Methods). Normalized microtubule growth rate means ± SEM represent the data from Figure 3C normalized to the growth rate of tubulin alone. Relative nucleation activity for each construct is represented by a delay between the start of the polymerization reaction and when the turbidity reached 10% of its maximum, divided by the delay measured in controls with αβ-tubulin alone. Normalized nucleation rate means ± SEM represent the continuous turbidity reaction data of experimental conditions normalized to the nucleation lag of tubulin alone (N = 12, 7, 6, 4, 6, 5, and 7 for tubulin alone, γ-tubulin, TOG12, TOG12MTbd, TOG1-4, TOG1-4MTbd, and XMAP215, respectively).
To examine the polymerase activities of the XMAP215 constructs, we measured microtubule elongation rates for individual microtubule plus-end tips assembling continuously from GMPCPP-stabilized seeds (Figure 3, B and C). As seen in previous studies, the addition of full-length XMAP215 strongly accelerated microtubule elongation, increasing the plus-end growth rate by up to 10-fold over αβ-tubulin alone. The addition of TOG1-4MTbd, TOG12MTbd, or TOG1-4 at identical concentrations also accelerated growth, but increased the rate more modestly, by 2.5- to 3.3-fold. The addition of TOG12 had no significant effect. The necessity of TOGs 1 and 2 plus either the MTbd or TOGs 3 and 4 for polymerase activity confirms previous findings (Widlund et al., 2011; Fox et al., 2014; Flor-Parra et al., 2018).
We then tested the activity of our XMAP215 constructs in microtubule nucleation. Three of the deletion constructs added at identical concentrations had moderate nucleation activity compared with full-length XMAP215: TOG1-4MTbd, TOG12MTbd, and TOG1-4. Of these, TOG1-4MTbd had the largest effect, reducing the nucleation lag by 31% compared with αβ-tubulin alone (Figure 3E). TOG12MTbd and TOG1–4 promoted nucleation similarly; each reduced the nucleation lag by ∼25%. Only one of the deletion constructs we tested, TOG12, showed no effect on nucleation. Notably, the activity of the constructs in nucleation was not dependent on the number of TOG domains.
Similarities in the domain requirements for nucleation versus elongation activity were striking. Deletion of TOG5 and the C-terminus of XMAP215 significantly reduced both elongation and nucleation activity. Further reduction of microtubule-lattice affinity, by deleting either TOGs 3 or 4 (TOG12MTbd construct) or the basic region following TOG4 (TOG1–4 construct), predictably reduced elongation activity and likewise reduced nucleation activity. Elimination of microtubule-lattice affinity altogether (TOG12) eliminated both activities. Taken together, these results show that elongation activity and nucleation activity both require only TOGs1 and 2 and a lattice-binding region. Therefore, the nucleation activity of XMAP215 does not depend on any particular lattice-binding domain or on the number of TOG domains, but rather it correlates directly with microtubule elongation activity.
Plotting the turbidity and tip growth data against one another illustrates their correlation (Figure 3F). To fit the data, a power-law function was chosen because it fits the theoretical relationship between microtubule elongation and nucleation. As the microtubule growth rate approaches zero, the nucleation lag should increase and approach infinity. In contrast, as the microtubule growth rate approaches infinity, the nucleation lag should decrease and approach zero. The curve fit was constrained to go through (1,1) because elongation and nucleation are defined as 1 in the absence of XMAP215 constructs. The resulting power curve fit is 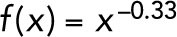 . The exponent, −0.33, is negative. Therefore, as the elongation rate increases, the nucleation lag decreases. The magnitude of the exponent is less than 1, which indicates that XMAP215 constructs increase elongation rates at growing microtubule tips more potently than they increase the addition of tubulin dimers to nucleation intermediates.
. The exponent, −0.33, is negative. Therefore, as the elongation rate increases, the nucleation lag decreases. The magnitude of the exponent is less than 1, which indicates that XMAP215 constructs increase elongation rates at growing microtubule tips more potently than they increase the addition of tubulin dimers to nucleation intermediates.
γ-Tubulin promotes microtubule nucleation without affecting elongation rate
For comparison with XMAP215, it was instructive to also compare the effects of γ-tubulin in the tip growth and turbidity assays. The addition of purified γ-tubulin alone had no effect on plus-end growth rates (Figure 3C), which is not surprising because γ-tubulin caps minus ends and does not associate with microtubule plus ends. However, the same concentration of γ-tubulin dramatically reduced the nucleation lag in the turbidity assay by 50% (Figures 2A and 3F). These observations confirm that γ-tubulin promotes microtubule nucleation without affecting microtubule plus-end elongation, consistent with its expected role as a minus-end-binding template.
XMAP215 and γ-tubulin function additively during microtubule nucleation
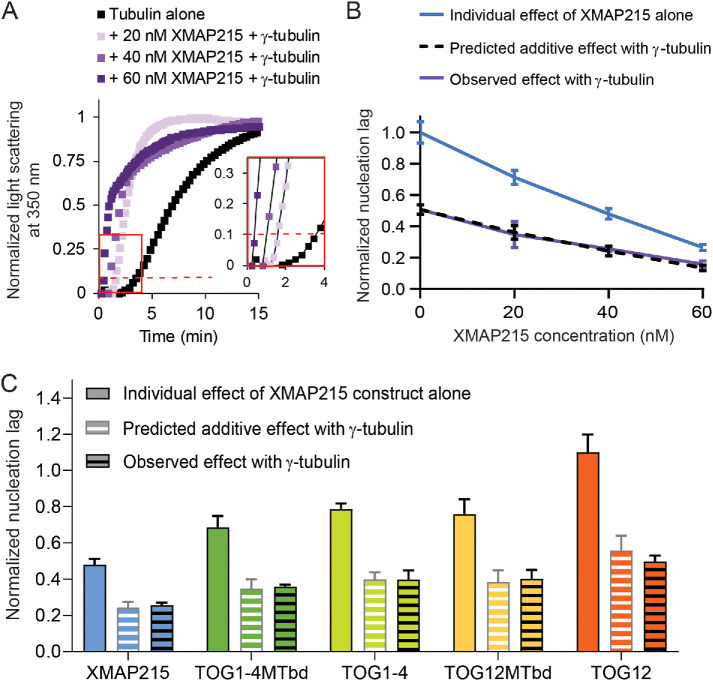
If XMAP215 functions strictly to elongate nucleation intermediates during microtubule formation, then its role should not overlap with γ-tubulin, and thus the effects of XMAP215 and γ-tubulin arrays in the turbidity assay should be additive. In contrast, if XMAP215 and γ-tubulin arrays function synergistically (or redundantly), they should produce a larger (or smaller) effect when combined than the sum of their independent contributions.
To distinguish the type of relationship XMAP215 and γ-tubulin arrays display, we calculated the effect on nucleation lag that would occur in a purely additive scenario (Supplemental Figure 3 shows example calculations). Then, over the previously tested concentrations of XMAP215, we measured the combined effects of XMAP215 and γ-tubulin arrays (Figure 4A). We compared the observed nucleation lags with the predicted nucleation lags, and found they closely matched across all XMAP215 concentrations (Figure 4B).
FIGURE 4:
XMAP215 and γ-tubulin promote nucleation additively in pure tubulin solutions. (A) Representative tubulin assembly assay for 20, 40, or 60 nM XMAP215 in combination with 300 nM γ-tubulin with 15 μM free tubulin. The red dashed line represents 10% of the normalized maximum light scattering. The inset shows the same data from the red box plotted on a larger scale with the data plotted as points connected by lines for clarity. For 40 and 60 nM XMAP215 in combination with 300 nM γ-tubulin, microtubule bundling occurs at later time points, which causes a difference in the shape of the curve of tubulin assembly. See Supplemental Figure 4. (B) XMAP215 functions additively with γ-tubulin at all concentrations measured (see Supplemental Figure 3). The data plotted are mean values ± SEM (N = 12, 4, 6, 4 for 0, 20, 40, and 60 nM XMAP215, respectively; N = 7, 4, 4, and 4 for 0, 20, 40, and 60 nM XMAP215 + γ-tubulin, respectively). (C) XMAP215 deletion constructs all show additive action with γ-tubulin (see Supplemental Figure 3). The data plotted for each XMAP215 deletion construct are mean values ± SEM replotted from Figure 3F. The data plotted for each XMAP215 deletion construct + γ-tubulin are mean values ± SEM (N = 4 for each condition).
To further test if XMAP215 and γ-tubulin act in an additive manner, we measured the combined effect of each of our XMAP215 deletion constructs with γ-tubulin. In every case, the nucleation lag observed when the two were combined was not significantly different from the predicted additive value (Figure 4C). Thus, the removal of various domains from XMAP215 does not allow for either synergy or redundancy with γ-tubulin. Overall, the close correspondence of the measured nucleation lags with the predicted additive values, across different XMAP215 concentrations and across the different deletion constructs, shows that XMAP215 and γ-tubulin function additively in the absence of other ring complex proteins.
During polymerization, XMAP215 increases the on-rate for tubulin dimers associating with the microtubule tip (Ayaz et al., 2014). Our results suggest that this is its primary contribution during microtubule nucleation as well. XMAP215’s effect on microtubule nucleation correlates with its polymerase activity, independent of the domains present. This provides support for a model of independent action by XMAP215 during microtubule nucleation, as suggested previously (Roostalu et al., 2015; Reid et al., 2016; Thawani et al., 2019).
Our results also show that γ-tubulin and XMAP215 do not show the synergistic interaction that was previously reported for the full γ-TuRC and XMAP215. Several possibilities could explain this difference. It is thought that the γ-TuRC is activated through a conformational change, and the reported interaction between the unstructured C-terminal tail of XMAP215 and γ-tubulin might be responsible for this. It is also possible that the full γ-TuRC offers higher affinity binding sites for this interaction with XMAP215 than arrays of γ-tubulin alone. Extra molecules of XMAP215 would increase the local concentration of αβ-tubulin near the template and thereby increase the rate of addition of αβ-tubulin. Alternatively, the arrangement of a γ-tubulin template into a ring might activate it for synergistic action by forming better substrates for XMAP215. Laterally associated γ-tubulin arrays and γ-TuRCs both provide templates that should promote lateral interactions between αβ-tubulin dimers. However, the templates are not identical and each might promote a distinct ensemble of nucleation intermediates. According to this model, XMAP215 would act preferentially on nucleation intermediates that form on the 13-member γ-tubulin template provided by the ring complex.
We show here that XMAP215 functions additively with laterally associated arrays of γ-tubulin. That a microtubule polymerase, which promotes elongation, and a template, which promotes lateral interactions, act independently during the early stages of microtubule formation supports early nucleation models that include several rate-limiting steps (Voter and Erickson, 1984; Flyvbjerg et al., 1996) and a more recent model that proposes microtubules form by progressively faster tubulin accretion (Rice et al., 2019). Our results suggest that there must be at least two distinct types of rate-limiting processes in the nucleation pathway. Given the theoretical activities of the template and the polymerase, we propose that these are 1) the formation of initial lateral αβ-tubulin bonds, which is specifically accelerated by γ-tubulin, and 2) the formation of subsequent αβ-tubulin bonds, which is specifically accelerated by XMAP215.
MATERIALS AND METHODS
Purification of tubulin
αβ-Tubulin was purified from fresh bovine brain using a high-concentration PIPES buffer as previously described (Castoldi and Popov, 2003).
γ-Tubulin and XMAP215 expression plasmids
A human γ-tubulin construct with a C-terminal myc-His6 tag was previously described (Murphy et al., 1998). A construct with XMAP215 with a C-terminal GFP-His7 tag was previously described (Brouhard et al., 2008). The fragment containing GFP was removed using SalI digestion (pRK009) and QuikChange was performed to produce TOG1-4MTbd (pRK087) and TOG1-4 (pRK031). pPW263, a bacterial expression plasmid containing TOG12 and three K loops with a C-terminal myc-His6 tag was previously described (Widlund et al., 2011). QuikChange was performed to produce TOG12MTbd (pRK074) and TOG12 (pRK056).
Purification of γ-tubulin and XMAP215 constructs
γ-Tubulin, XMAP215, TOG1-4MTbd, and TOG1-4 were expressed in Sf9 cells using the Bac-to-Bac system from Invitrogen (Thermo Fisher Scientific, Waltham, MA). Sf9 cells at a density (1–2) × 106 cells/ml were infected with baculovirus. Cell pellets were harvested 48–72 h postinfection. Infections were done both in-house, at the National Cell Culture Center (Minneapolis, MN), or at the Tissue Culture Core Facility, University of Colorado Cancer Center, UCHSC at Fitzsimons (Aurora, CO). TOG12MTbd and TOG12 were expressed in Escherichia coli Rosetta 2 cells with plasmid pRARE. All XMAP215 and TOG constructs were purified as previously described (Brouhard et al., 2008), and γ-tubulin was purified as previously described (Aldaz et al., 2005).
Spontaneous tubulin assembly assay
The spontaneous assembly of purified bovine brain tubulin was measured by light scattering at 350 nm as described previously (Voter and Erickson, 1984). Tubulin was thawed on ice slurry (0°C) and pelleted in a TLA100 rotor at 90,000 rpm for 10 min to remove microtubule polymers and/or inactive tubulin. The supernatant was removed, kept at 0°C, and used for the assembly reactions. Experimental reactions were prepared without tubulin in 1.7 ml Eppendorf tubes at 0°C, with 30% glycerol, 1 mM dithiothreitol (DTT; Millipore Sigma, St. Louis, MO), 1 mM GTP (Millipore Sigma) in BRB80 buffer (80 mM potassium PIPES buffer, pH 6.8 [EMD Chemicals, Gibbstown, NJ], 1 mM MgCl2 [Millipore Sigma], and 1 mM EGTA [Millipore Sigma]). Immediately before measuring light scattering, tubulin was added at either 12 or 15 μM to experimental reactions at 0°C for a total volume of 150 µl, and then the entire reaction was moved to a prewarmed cuvette at 37°C in a DU 800 spectrophotometer (Beckman Coulter Life Sciences, Indianapolis, IN). The A350 nm was recorded every 6–20 s on two to six samples at a time for greater than 40 min. Recording on the spectrophotometer was started before tubulin being added to the experimental reaction mix. For XMAP215 and γ-tubulin, at least seven experimental samples per condition were tested, for two independent purifications of each protein. For XMAP215 deletion constructs, at least four experimental samples were tested. Experiments were performed using two different tubulin preps.
For data quantification, a tubulin alone control was run simultaneously with each reaction, using the same freshly thawed and cleared tubulin aliquot and the same buffers. Nucleation lag was quantified by comparing time to reach 0.1 maximum polymer, or 10% maximum polymer, between experimental reactions and their corresponding tubulin alone control reaction. The effect of each protein was reproducible.
Microtubule growth rate assay
Small flow chambers were constructed by adhering a KOH etched coverslip onto a glass slide with two strips of double-sided tape. Flow chambers were incubated with prewarmed 1 mg/ml biotin-BSA (bovine serum albumin; Vector Laboratories, Burlingame, CA) for 15 min at room temperature, then washed with prewarmed BRB80 buffer (80 mM potassium PIPES [EMD Chemicals], 1 mM MgCl2 [Millipore Sigma], and 1 mM EGTA [Millipore Sigma], pH 6.8). GMPCPP-stabilized microtubule seeds assembled from a mixture of biotinylated porcine tubulin, 7 or 14% of total (Cytoskeleton, Denver, CO) and purified bovine tubulin were secured to the glass coverslip surface using 0.25 mg/ml avidin (Vector Laboratories; Franck et al., 2007, 2010; Powers et al., 2009; Akiyoshi et al., 2010). After 5 min, microtubule seeds were washed with BRB80 containing 1 mM GTP (Millipore Sigma), 8 mg/ml BSA (Millipore Sigma), and 1 mg/ml Κ-casein (Millipore Sigma), until reaction mixture was added.
Dynamic microtubule extensions were grown in the absence, or presence, of 40 nM XMAP215 constructs. Reaction mixture consisted of freshly thawed 12 µM purified bovine brain tubulin in microtubule growth buffer (BRB80 buffer supplemented with 1 mM GTP [Millipore Sigma], 5 mM DTT [Millipore Sigma], 25 mM glucose [Millipore Sigma], 200 µg/ml glucose oxidase [Millipore Sigma], 35 µg/ml catalase [Millipore Sigma]). Microtubule extensions were imaged by recording video-enhanced differential interference contrast, which allowed us to visualize the dynamic microtubule tips. A field of view (FOV) was imaged for 15–20 min. Four to eight FOVs per flow chamber were analyzed per experiment.
Growing microtubule tips were manually tracked using MTrackJ. X-Y coordinates of the microtubule tips were projected onto a line corresponding to the microtubule long axis. Projected tip positions were then plotted against time, and during episodes of steady growth were fitted to a line. The slopes of these line fits were recorded as the growth velocities.
Nucleation lag and elongation rate correlational analysis
Normalized nucleation lag was plotted against normalized microtubule (MT) growth rate using Igor Pro software (WaveMetrics, Portland, OR). Data points used in the power curve fit included tubulin alone, TOG12, TOG12MTbd, TOG1-4, TOG1-4MTbd, and XMAP215. The line was constrained to pass through (1,1), and the curve was weighted in relation to the SEM for the normalized nucleation lag data.
Electron microscopy and helical reconstructions
γ-Tubulin arrays were prepared by diluting pure γ-tubulin to 0.5–1 μM out of 0.5 M KCl-containing buffer to ≤100 mM KCl, no GTP. Notably, arrays form at 4 and 37°C, with or without GTP. For negative stained samples, carbon-coated grids were glow-discharged or plasma-cleaned before use, followed by application of 3–5 μl of protein solution for 60 s. Grids were then washed in ddH2O and stained with filtered 1% uranyl acetate for 30 s. Images were obtained on a Tecnai20 transmission electron microscope (FEI, Hillsboro, OR) at 62,000 magnification and 200 kV in low-dose mode with the use of either a 1024 × 1024 pixel CCD camera or a 4096 × 4096 pixel CCD camera (Gatan, Pleasanton, CA) at −1.3–1.5 μm defocus. For γ-tubulin array reconstructions, individual arrays 300 pixels wide were extracted from the original micrographs using EMAN2 Helixboxer (Tang et al., 2007), after rotation to vertical along the long array axis. SPIDER (Frank et al., 1996; Sachse et al., 2007) was then used to window the arrays along the long axis, shifting by 270 pixels for 90% overlap between adjacent images. The image stacks were then padded by 90 pixels and binned twice for a final square image size of 195 pixels. Image stacks corresponding to individually picked arrays were aligned in 2D to initially characterize array symmetry. Using a simple rectangular initial reference image based on an unaligned average of the image stacks, 10 rounds of reference-based alignment and averaging were run to calculate a final average image. Three similar, long and well-ordered arrays consisting of 560 overlapping segments were chosen as an initial dataset for 3D reconstruction. A repeat distance of ∼52 Å was measured from the 2D averages and used as a starting parameter for 3D reconstruction. The IHRSR program was used as the 3D helical alignment algorithm (Egelman, 2010). Fifty rounds of refinement were run for each experiment using a simple cylinder with a diameter similar to the width of the 2D averages of the images. Multiple runs of IHRSR were performed using different initial azimuthal angles. Several different point-group symmetries were also applied. The best results were obtained using a fivefold point-group symmetry with a final refined azimuthal angle of 6.5° and a rise of 52.9 Å. The final volume was contoured to an estimated 8.4 MDa and x-ray structures of γ-tubulin trimers (Aldaz et al., 2005) were manually fit to the corresponding density. Molecular graphics and analyses of EM reconstructions were performed with UCSF chimera (Pettersen et al., 2004).
Microtubule pelleting assay
Experimental reactions were prepared as described in the spontaneous tubulin assembly, and incubated at 37°C for 30 min. The reactions were fixed by 1:4 dilution in 2% glutaraldehyde (Electron Microscopy Services, Hatfield, PA), and 2 min incubation at room temperature. Dilution of samples (1:1200) before pelleting was required to differentiate individual microtubules. Pelleting onto coverslips was performed as previously described (Graczyk and Davis, 2011).
To visualize microtubules on a coverslip, immunostaining was performed using anti–α-tubulin—FITC antibody (Millipore Sigma, St. Louis, MO). The slides were imaged using an AxioObserver Z1 microscope (Zeiss, Jena, Germany) equipped with an ORCA-Flash4.0 camera (Hamamatsu Photonics K.K., Bridgewater, NJ) and an α Plan-Apochromat 63× objective (1.46 NA; Zeiss). Immunostained microtubules were imaged with 0.4 s exposures and were not binned with a final resolution of 2048 × 2048.
Supplementary Material
Acknowledgments
Regarding the γ-tubulin arrays, we thank Albion Baucom and Koji Yonekura for help with reconstructions, we thank Joel Quispe for help with data acquisition, and we thank Justin Kollman for helpful discussion. We thank Per Widlund for XMAP215 expression plasmids and helpful discussion. This study was supported by the following grants from the National Institutes of Health: Grant no. P01 GM-105537 to M.W., T.D., D.A., and C.A.; Grant no. R35 GM-130293 to T.D.; Grant no. R35 GM-134842 to C.A.; Grant no. R35 GM-118099 to D.A.; and Grant no. T32 GM-007270 to B.R.K.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-02-0160) on July 29, 2020.
REFERENCES
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. (2010). Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature , 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaz H, Rice LM, Stearns T, Agard DA. (2005). Insights into microtubule nucleation from the crystal structure of human γ-tubulin. Nature , 523–527. [DOI] [PubMed] [Google Scholar]
- Ayaz P, Munyoki S, Geyer EA, Piedra F-A, Vu ES, Bromberg R, Otwinowski Z, Grishin NV, Brautigam CA, Rice LM. (2014). A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. ELife , e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. (2008). XMAP215 is a processive microtubule polymerase. Cell , 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia B, Draetta G, Karsenti E. (1992). Regulation of the microtubule nucleating activity of centrosomes in Xenopus egg extracts: role of cyclin A–associated protein kinase. J Cell Biol , 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov Av. (2003). Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif , 83–88. [DOI] [PubMed] [Google Scholar]
- Choi YK, Liu P, Sze SK, Dai C, Qi RZ. (2010). CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J Cell Biol , 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consolati T, Locke J, Roostalu J, Asthana J, Lim WM, Gannon J, Martino F, Costa A, Surrey T. (2019). Microtubule nucleation by single human γTuRC in a partly open asymmetric conformation. BioRxiv 853218. [Google Scholar]
- Egelman EH. (2010). Reconstruction of helical filaments and tubes. In: Methods in Enzymology, Vol. , Issue C, New York: Academic Press, pp. 167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor-Parra I, Iglesias-Romero AB, Chang F. (2018). The XMAP215 ortholog Alp14 promotes microtubule nucleation in fission yeast. Curr Biol , 1681–1691.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyvbjerg H, Jobs E, Leibler S. (1996). Kinetics of self-assembling microtubules: An “inverse problem” in biochemistry. Proc Natl Acad Sci USA , 5975–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JC, Howard AE, Currie JD, Rogers SL, Slep KC. (2014). The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol Biol Cell , 2375–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Davis TN, Asbury CL. (2010). Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods , 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. (2007). Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol , 832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. (1996). SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol , 190–199. [DOI] [PubMed] [Google Scholar]
- Gaskin F, Cantor CR, Shelanski ML. (1974). Turbidimetric studies of the in vitro assembly and disassembly of porcine neurotubules. J Mol Biol , 737–740. [DOI] [PubMed] [Google Scholar]
- Graczyk B, Davis TN. (2011). An assay to measure the affinity of proteins for microtubules by quantitative fluorescent microscopy. Anal Biochem , 313–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. (1991). The fission yeast γ-tubulin is essential for mitosis and is localized at microtubule organizing centres. J Cell Sci , 693–700. [DOI] [PubMed] [Google Scholar]
- Hussmann F, Drummond DR, Peet DR, Martin DS, Cross RA. (2016). Alp7/TACC-Alp14/TOG generates long-lived, fast-growing MTs by an unconventional mechanism. Sci Rep , 20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. (1992). γ-Tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature , 80–83. [DOI] [PubMed] [Google Scholar]
- Keating TJ, Borisy GG. (2000). Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol , 352–357. [DOI] [PubMed] [Google Scholar]
- Klotz C, Dabauvalle M-C, Paintrand M, Weber T, Bornens M, Karsenti E. (1990). Parthenogenesis in Xenopus eggs requires entrosomal integrity. J Cell Biol , 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong KK, Fernandez J-J, Sali A, Kilmartin J, Davis TN, Agard DA. (2015). Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Nat Struct Mol Biol , 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. (2010). Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature , 879–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchnir Fygenson D, Flyvbjerg H, Sneppen K, Libchaber A, Leibler S. (1995). Spontaneous nucleation of microtubules. Phys Rev E , 5058–5063. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guénebaut V, Heuser J, Agard DA. (2000). Structure of the γ-tubulin ring complex: A template for microtubule nucleation. Nat Cell Biol , 365–370. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts BM, Oegema K. (1998). Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol , 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. (1998). The mammalian γ-tubulin complex contains homologues of the yeast spindle pole body components Spc97p and Spc98p. J Cell Biol , 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. (1990). γ-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell , 1289–1301. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004). UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem , 1605–1612. [DOI] [PubMed] [Google Scholar]
- Popov AV, Severin F, Karsenti E. (2002). XMAP215 is required for the microtubule-nucleating activity of centrosomes. Curr Biol , 1326–1330. [DOI] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. (2009). The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell , 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid TA, Schuster BM, Mann BJ, Balchand SK, Plooster M, McClellan M, Coombes CE, Wadsworth P, Gardner MK. (2016). Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J Cell Sci , 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L, Moritz M, Agard DA. (2019). Microtubules form by progressively faster tubulin accretion, not by nucleation-elongation. BioRxiv 545236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Cade NI, Surrey T. (2015). Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol , 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse C, Chen JZ, Coureux PD, Stroupe ME, Fändrich M, Grigorieff N. (2007). High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol , 812–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Vale RD. (2007). Structural basis of microtubule pus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell , 976–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. (1995). A highly divergent γ-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomycescerevisiae. J Cell Biol , 1775–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. (1991). γ-Tubulin is a highly conserved component of the centrosome. Cell , 825–836. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. (2007). EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol , 38–46. [DOI] [PubMed] [Google Scholar]
- Thawani A, Kadzik RS, Petry S. (2018). XMAP215 is a microtubule nucleation factor that functions synergistically with the γ-tubulin ring complex. Nat Cell Biol , 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A, Rale MJ, Coudray N, Bhabha G, Stone HA, Shaevitz JW, Petry S. (2020). The transition state and regulation of γ-TuRC-mediated microtubule nucleation revealed by single molecule microscopy. ELife , 54253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A, Stone HA, Shaevitz JW, Petry S. (2019). Molecular mechanism of microtubule nucleation from gamma-tubulin ring complex. BioRxiv 853010. [Google Scholar]
- Voter W, Erickson H. (1984). The kinetics of microtubule assembly. Evidence for a two-stage mechanism. J Biol Chem , 10430–10443. [PubMed] [Google Scholar]
- Wegner A, Engel J. (1975). Kinetics of the cooperative association of actin to actin filament. Biophys Chem , 215–225. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA , 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. (2015). Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat Cell Biol , 907–918. [DOI] [PubMed] [Google Scholar]
- Wieczorek M, Urnavicius L, Ti S-C, Molloy KR, Chait BT, Kapoor TM. (2019). Asymmetric molecular architecture of the human γ-tubulin ring complex. BioRxiv 820142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. (2000). A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol , 358–364. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. (2017). The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell , 1066–1077. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. (1991). γ-Tubulin is present in Drosophilamelanogaster and Homo sapiens and is associated with the centrosome. Cell , 817–823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.