Abstract
Primary cilia are microtubule-based protrusions from the cell surface that are approximately 0.3 µm in diameter and 3 µm in length. Because size approximates the optical diffraction limit, ciliary structures at the subdiffraction level can be observed only by using a superresolution microscope or electron microscope. Expansion microscopy (ExM) is an alternative superresolution imaging technique that uses a swellable hydrogel that enables the physical expansion of specimens. However, the efficacy of ExM has not been fully verified, and further improvements in the method are anticipated. In this study, we applied ExM to the observation of primary cilia and centrioles and compared the acquired images with those obtained using conventional superresolution microscopy. Furthermore, we developed a new tool, called the amplibody, for fluorescence signal amplification, to compensate for the substantial decrease in fluorescence signal per unit volume inherent to physical expansion and for the partial proteolytic digestion of cellular proteins before expansion. We also demonstrate that the combinatorial use of the ExM protocol optimized for amplibodies and Airyscan superresolution microscopy enables the practical observation of cilia and centrioles with high brightness and resolution.
INTRODUCTION
The primary cilium is a small antenna-like organelle protruding from the surface of eukaryotic cells. It is composed of a microtubule-based scaffold called the axoneme, which extends from the mother centriole (MC)-derived basal body and is surrounded by the ciliary membrane. The primary cilium has a diameter of 0.2–0.3 µm and a length of 1–10 μm (Ishikawa and Marshall, 2011). Electron microscopy has been used to observe the fine structures of cilia, because conventional fluorescence microscopy cannot distinguish the detailed structures owing to the diffraction limit (Fisch and Dupuis-Williams, 2011). However, in recent years, various types of superresolution (SR) microscopy that can observe structures below the diffraction limit have been developed and applied to cilia research (Sonnen et al., 2012; Yang et al., 2015, 2018; Shi et al., 2017; Bowler et al., 2019; Kashihara et al., 2019).
Expansion microscopy (ExM) is an innovative SR microscopy technique developed by Boyden and colleagues (Chen et al., 2015). ExM improves resolution by the physical expansion of samples using water-absorbent polymers (see Figure 1, A and B). The same group then reported an improved method, named protein-retention expansion microscopy (proExM), which uses fluorescent proteins and commercially available fluorescent antibodies (Tillberg et al., 2016). In the same year, two other groups independently developed different protocols for ExM (Chozinski et al., 2016; Ku et al., 2016). Although there have been reports on the application of ExM for the observation of samples of various organisms, such as humans (Zhao et al., 2017), zebrafish (Freifeld et al., 2017), Drosophila (Cahoon et al., 2017; Jiang et al., 2018), planaria (Wang et al., 2016), and Escherichia coli (Zhang et al., 2016), further validation and improvements on the effectiveness of ExM are anticipated.
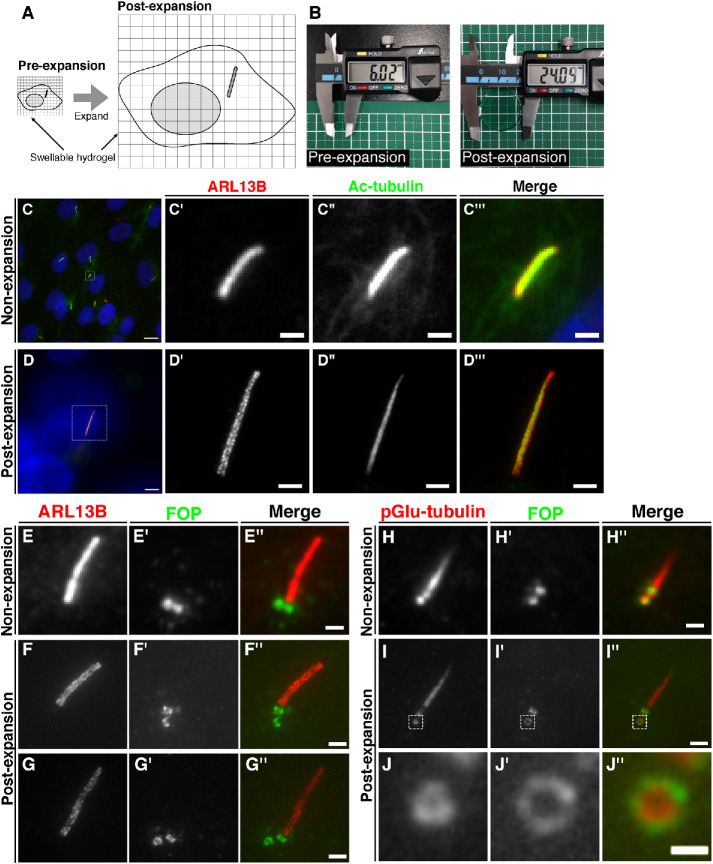
FIGURE 1:
Comparison of cilia and centrioles in nonexpanded and expanded cells observed by conventional fluorescence microscopy. (A) Schematic diagram of the process of cell expansion. The immunostained cells are cross-linked to a polymer in a swellable hydrogel and treated with proteinase K. By immersing the gel in water, the cells physically expand about four times. (B) Comparison of the diameters of gel pieces before and after expansion. A circular piece of gel with a diameter of approximately 6 mm was expanded to approximately 24 mm. (C–J) RPE1 cells were serum-starved for 24 h to induce ciliogenesis, fixed, and immunostained with or without expansion. (C, D) Representative images of nonexpanded cells (C) and expanded cells (D) doubly immunostained for ARL13B and Ac-tubulin. Enlarged images of the boxed regions in C and D are shown in C′–C′′′ and D′–D′′′, respectively. Scale bars, 10 µm (C, D), 1 µm (C′–C′′′), and 4 µm (D′–D′′′). (E–G) Representative images of nonexpanded cells (E) and expanded cells (F and G) doubly immunostained for ARL13B and FOP. Scale bars, 1 µm (E) and 4 µm (F and G). (H–J) Representative images of nonexpanded cells (H) and expanded cells (I) doubly immunostained with an anti-FOP antibody and the GT335 antibody, which recognizes pGlu-tubulin. Enlarged images of the boxed regions in I–I′′ are shown in J–J′′. Scale bars, 1 µm (H), 4 µm (I), and 1 µm (J).
In this study, we show that SR imaging of primary cilia and centrioles is possible using proExM. We also demonstrate that observation of expanded samples by two types of SR microscopy, that is, structured illumination microscopy (SIM) and the Zeiss Airyscan, further improve the resolution. In addition, we developed a new tool, called the amplibody, to overcome the problem of the greatly reduced fluorescence signal per unit volume inherent to physical sample expansion. We demonstrate that the combinatorial use of the optimized proExM protocol and Airyscan microscopy enables practical subdiffraction imaging of primary cilia and centrioles.
RESULTS AND DISCUSSION
Observation of primary cilia and centrioles by ExM
We first set out to validate whether proExM enables the observation of the structures of primary cilia that are indistinguishable using conventional fluorescence microscopy. Primary cilia in human telomerase reverse transcriptase–immortalized retinal pigmented epithelial (hTERT-RPE1) cells were serum-starved to induce ciliogenesis and immunostained using antibodies against acetylated α-tubulin (Ac-tubulin), which constitutes the axonemal microtubules, and ARL13B, which is a lipid-anchored protein localized on the ciliary membrane. By conventional fluorescence microscopy, uniform staining patterns of Ac-tubulin and ARL13B overlapped and were indistinguishable on the ciliary structures (Figure 1C). In striking contrast, when the cells were expanded by the proExM protocol, ciliary staining for ARL13B demonstrated a broader distribution than that for Ac-tubulin (Figure 1D). Furthermore, ARL13B staining was not uniform but showed a patched distribution on the ciliary membrane. This patched ARL13B distribution is unlikely to be an artifact resulting from the expansion process, as similar patch-like structures were observed using SIM and stimulated emission depletion (STED) microscopy (Kohli et al., 2017; Thorpe et al., 2017).
We then validated whether ExM enables the observation of centriolar structures at the subdiffraction level. FOP (recently renamed as CEP43) is localized to both the mother and daughter centrioles and participates in the import of the intraflagellar transport (IFT) complex into cilia by sequentially recruiting CEP19 and the small GTPase RABL2 (Kanie et al., 2017; Nishijima et al., 2017). A previous study using SIM showed that FOP demonstrates a ring-like localization around the centrioles, with a diameter of 458.5 ± 22.9 nm (Mojarad et al., 2017). By conventional microscopy, FOP was observed as two dots located at the ciliary base (Figure 1E). By contrast, using ExM, we observed FOP staining on both sides of vertically oriented cylindrical centrioles (Figure 1F) and were able to distinguish between their vertical and horizontal orientations (Figure 1G), in the latter of which FOP had a ring-like distribution (Figure 1G′). We then validated whether proExM can distinguish the distribution of two centriolar proteins with ring-like structures of different diameters. A previous SIM study reported that staining for polyglutamylated-tubulin (pGlu-tubulin) using the GT335 antibody is observed as ring-like structures with a diameter of 166.3 ± 20.5 nm (Sonnen et al., 2012). By conventional microscopy, the daughter centrioles immunostained for pGlu-tubulin and FOP were observed as indistinguishable dots (Figure 1H). By contrast, ExM demonstrated ring-like structures of pGlu-tubulin and FOP with different diameters (Figure 1, I and J). These results demonstrate that structures of primary cilia and centrioles below the diffraction limit can be distinguished by proExM.
Observation of primary cilia and centrioles by ExM combined with SR microscopy
We next investigated whether the combination of proExM and SR microscopy would further improve the resolution. SR microscopy techniques include photoactivated localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM), STED, SIM, and Airyscan (Sahl et al., 2017). We considered that the combinatorial use of single-molecule localization microscopy, such as PALM and STORM, with ExM is difficult, although not impossible, as both SR microscopy techniques require blinking fluorescent proteins or dyes and are unsuitable for thick samples. Although the combinatorial use of ExM and STED has already been reported (Gao et al., 2018), we considered that this combination is not practical, particularly for the imaging of primary cilia, because of the limitation of fluorescent proteins and dyes applicable to STED, and the photobleaching caused by the intense STED beam. On the other hand, we considered SIM and Airyscan to be suitable for combinatorial use with ExM, as both SR microscopy techniques are applicable to the observation of thick samples, and neither has limitations regarding the fluorescent proteins and dyes that can be used.
We first compared images of primary cilia acquired by SIM and Airyscan. As shown in Figure 2, A and E, the immunostaining signals for ARL13B and Ac-tubulin in serum-starved RPE1 cells without expansion overlapped and were difficult to be distinguished by SIM and Airyscan. By contrast, when expanded cells were analyzed by SIM and Airyscan, ARL13B and Ac-tubulin demonstrated distinct distributions (Figure 2, B and F).
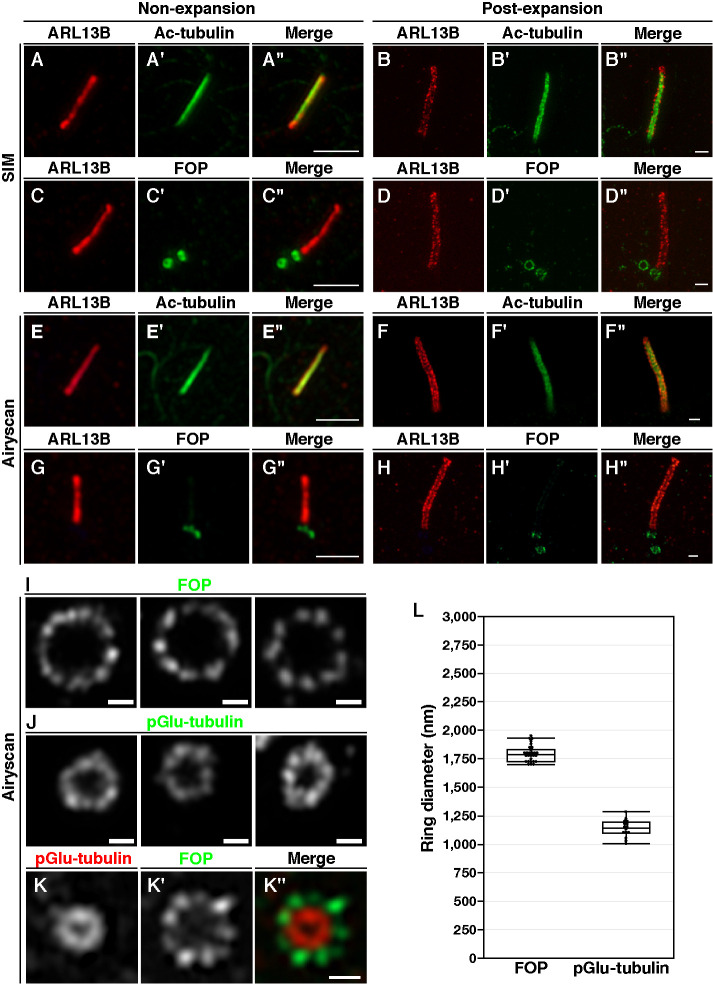
FIGURE 2:
Comparison of cilia and centrioles in nonexpanded and expanded cells observed by SR microscopy. (A–K) RPE1 cells serum-starved for 24 h were fixed and immunostained with or without expansion. (A–H) Nonexpanded cells (A, C, E, G) or expanded cells (B, D, F, H) were doubly immunostained for ARL13B and either Ac-tubulin (A, B, E, F) or FOP (C, D, G, H) and imaged by SIM (A–D) or Airyscan microscopy (E–H). Scale bar, 2 µm. (I–K) Expanded cells were immunostained with either an anti-FOP (I) or the GT335 (J) antibody alone or both anti-FOP and GT335 antibodies (K) and imaged by Airyscan microscopy. Scale bars, 2 µm (A–H) and 500 nm (I–K). (L) Average diameters of the ring-like structures of FOP (n = 28) and pGlu-tubulin (n = 16).
Next we compared the images of FOP staining with or without expansion by SIM and Airyscan. As shown in Figure 2, C and G, the ring-like structures of FOP were not clearly observed by SIM or Airyscan in nonexpanded cells. By contrast, observation of expanded cells by SIM and Airyscan demonstrated distinct ring-like structures of FOP (Figure 2, D and H). SIM and Airyscan have approximately 2- and 1.7-fold higher resolutions, respectively, than conventional microscopy; thus, theoretically, SIM enables the acquisition of higher-resolution images than Airyscan. However, in combination with proExM, the difference in resolution between the images obtained by the two SR microscopy techniques appeared negligible. As it is easier for us to use Airyscan than SIM with respect to changing the scan area and zoom magnification, we used Airyscan in the following experiments.
When expanded cells were observed with Airyscan, distinct ring-like structures surrounding the daughter centrioles that were stained with antibodies against FOP and pGlu-tubulin were apparent (Figure 2, I and J). Notably, FOP was found as nearly ninefold symmetry structures. The average diameters of these rings were 1793.2 ± 68.2 nm for FOP and 1137.8 ± 72.7 nm for pGlu-tubulin (Figure 2L). Assuming that the expansion factor is 4, the diameters of the FOP ring and the pGlu-tubulin ring are estimated to be 448.3 and 284.45 nm, respectively. The diameter of the FOP ring reported in a previous SIM study was 458.5 nm (Mojarad et al., 2017), which is in good agreement with our results. On the other hand, the diameter of pGlu-tubulin–positive rings as determined in another SIM study was 166.3 nm (Sonnen et al., 2012), which is smaller than the diameter estimated in this study (Figure 2L). Although the exact reason for the difference in the diameter of the pGlu-tubulin–positive rings determined in the two studies is unknown, our estimation might involve an error resulting from anisotropic macromolecular expansion. Finally, we performed double immunostaining of the expanded cells with antibodies against FOP and pGlu-tubulin and observed the nearly ninefold symmetric structures of FOP outside of the pGlu-tubulin–positive ring (Figure 2K). Thus, the combinatorial use of proExM and Airyscan microscopy was demonstrated to enable practical high-resolution imaging of primary cilia and centrioles.
Design and validation of amplibodies for fluorescence signal amplification
During the course of the ExM experiments, we encountered difficulties in observing dim fluorescence signals in the expanded samples. As the gel expands by a factor of 4, the fluorescence signal per unit volume decreases to 1/64 (=1/43). Partial digestion of cellular proteins with proteinase K in the gel before expansion also causes a signal reduction, approximately by half (Tillberg et al., 2016). Thus, in theory, the expansion process results in the fluorescence signal becoming less than 1/100. Therefore, to overcome the practical problem inherent to ExM, we aimed to develop a novel method for fluorescence signal amplification.
In our novel method, we use a fluorescent protein fused with anti-immunoglobulin (Ig) nanobodies (Nbs) (Pleiner et al., 2018) (Figure 3A, inset); we hereafter refer to these fusion proteins as amplibodies, which stands for Nb for amplification of fluorescence signals. We fused mClover3 or mCherry with either of the two Nbs that recognize the mouse Ig kappa (κ) light chain or the rabbit IgG Fc region. The principle of amplification of the fluorescence signals using amplibodies is as follows (Figure 3A): 1) A primary antibody recognizes the protein of interest; 2) an amplibody binds the primary antibody; 3) an anti–fluorescent protein antibody binds the fluorescent protein portion of the amplibody; 4) an anti–IgG antibody conjugated with a fluorescent dye binds both the primary antibody and the anti–fluorescent protein antibody.
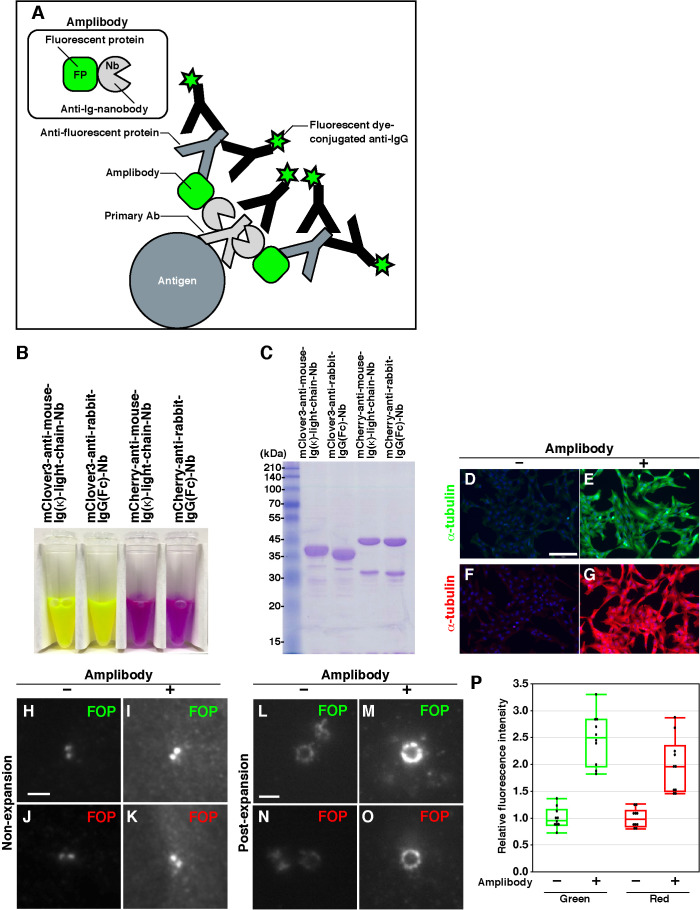
FIGURE 3:
Design of amplibodies and validation of their use in combination with proExM. (A) Schematic diagram of a method for amplifying fluorescence signals using amplibodies, which are anti-Ig Nbs fused to a fluorescent protein. An antigen is recognized by a primary antibody, to which the amplibody binds. Then, an anti–fluorescent protein antibody binds to the fluorescent protein portion of the amplibody, and finally, a fluorescent dye–conjugated secondary antibody binds to the primary and anti–fluorescent protein antibodies. Staining with the fluorescent dye–conjugated antibody can be performed either before or after gelation. (B) Photograph of purified amplibodies. The fluorescence of amplibodies is visible with the naked eye. (C) SDS–PAGE analysis of the purified amplibodies, followed by staining with CBB. (D–O) Validation of the amplification of fluorescence signals with amplibodies by conventional fluorescence microscopy. (D–G) Fixed RPE1 cells were treated with a polyclonal rabbit anti–α-tubulin antibody followed by Alexa488-conjugated (D, E) or Alexa555-conjugated (F, G) anti-rabbit IgG with (E, G) or without (D, F) intervening amplibody treatment (E, mClover3-fused anti-rabbit IgG(Fc) Nb; G, mCherry-fused anti-rabbit IgG(Fc) Nb). (H–O) Nonexpanded (H–K) and expanded (L–O) RPE1 cells were fixed and treated with a monoclonal mouse anti-FOP antibody followed by Alexa488-conjugated (H, I, L, M) or Alexa555-conjugated (J, K, N, O) anti-mouse IgG with (I, K, M, O) or without (H, J, L, N) intervening amplibody treatment (I and M, mClover3-fused anti-mouse Ig κ light chain Nb; K and O, mCherry-fused anti-mouse Ig κ light chain Nb). Scale bars, 150 µm (D–G) and 2 µm (H–O). (P) Comparison of fluorescence signal intensities of FOP-positive rings in expanded cells with or without amplibody treatment (n = 10).
The reason why we chose amplibodies (i.e., Nbs fused to a fluorescent protein) instead of Nbs conjugated with a fluorescent dye is that the fluorescent protein portion of an amplibody is expected to be retained even after digestion with protease K, because of its rigid structure, whereas the Nb portion is degraded (Gao et al., 2018). In addition, the fluorescence of the fluorescent protein part itself can also contribute to signal enhancement, and high-performance antibodies that recognize fluorescent proteins are available from various manufacturers. Thus, the final step of the signal amplification involves secondary antibodies conjugated with fluorescent dyes used for conventional immunostaining and, in conjunction with the availability of the amplibody expression vectors (see below), makes the whole amplification process practical for typical cell biology laboratories.
Amplibodies as glutathione S-transferase (GST)-fusion proteins can be easily produced in E. coli transformed with their expression vectors, which we have deposited to Addgene, and can be purified from cell lysates using glutathione-Sepharose beads. After removal of the GST tag by digestion with PreScission protease (Figure 3, B and C), the purified amplibodies are practically applicable to ExM studies (see below).
We first compared the signal amplification protocol involving amplibodies with a standard immunostaining protocol. When RPE1 cells incubated with an anti–α-tubulin antibody were subsequently incubated with a dye-conjugated secondary antibody with or without interposed amplibody treatment, the fluorescence signals were found to be considerably amplified by the amplibody treatment (Figure 3, compare E with D, and G with F), demonstrating the usefulness of amplibodies in conventional immunostaining. We next investigated whether amplibodies can be applied to ExM. When RPE1 cells were incubated with an anti-FOP antibody, subjected to the modified proExM protocol (see Materials and Methods), and observed under a conventional microscope, the amplification protocol was found to enhance the green- and red-fluorescence signals of the ring-like structures by averages of approximately 2.5- and 2-fold, respectively (Figure 3, L–P), demonstrating that amplibodies are useful tools for signal amplification, which are applicable to proExM.
Airyscan imaging of amplibody-mediated signal-amplified centriolar and ciliary proteins by proExM
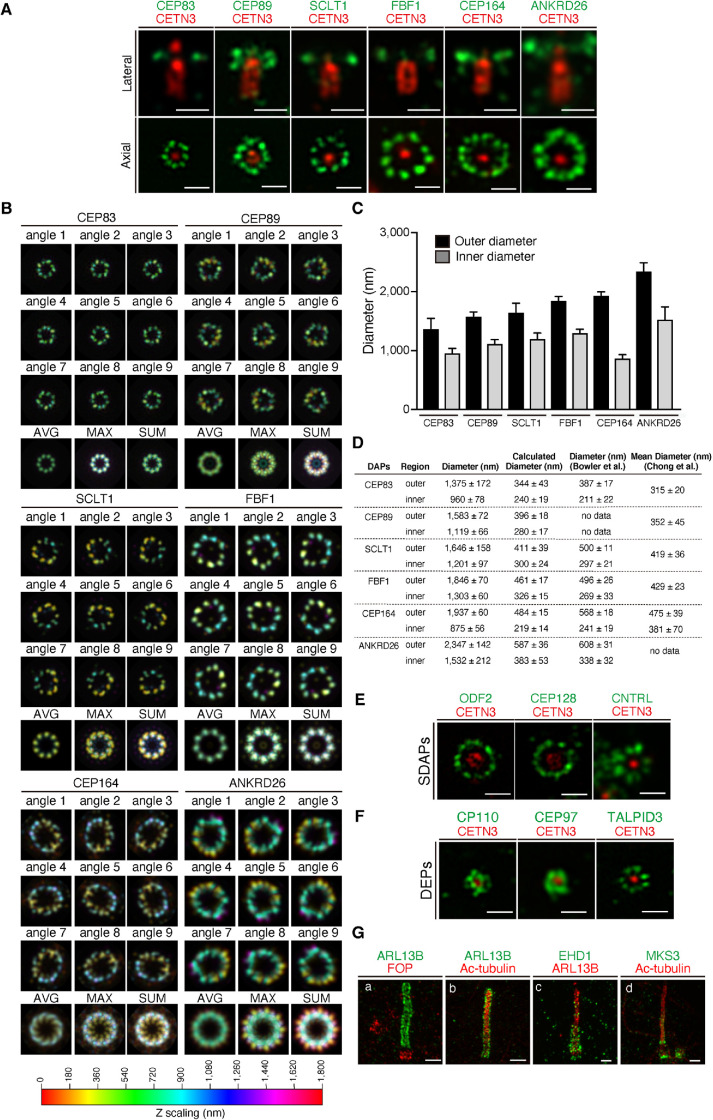
To assess the performance of the combinatorial method of amplibody and proExM (amplibody-proExM), we analyzed the localizations of various centriolar proteins by Airyscan microscopy (Figure 4). We first analyzed the spatial localizations of distal appendage proteins (DAPs) in association with centrin 3 (CETN3), a protein localized at the distal end of the centriole (Figure 4A). The relative localization of DAPs and CETN3 in the lateral view demonstrated the distinct arrangement of individual DAPs in the proximal-distal axis of the MC, which is positive for CETN3. In the axial view, CEP83, SCLT1, and FBF1 were found on discrete ninefold symmetric clusters around the MC. On the other hand, CEP89, CEP164, and ANKRD26 were localized as irregularly shaped, ambiguous ninefold symmetric densities. To emphasize the symmetrical distributions of DAPs, we processed a series of three-dimensional Airyscan images to obtain averaged signals from all nine appendages (Figure 4B; see Materials and Methods). The averaged maximum projection images demonstrated that DAPs have ninefold symmetric densities with characteristic shapes of clusters and with distinct ring sizes (Figure 4B). The estimated ring diameters of individual DAPs were approximately four times larger than those reported in the previous STORM studies (Figure 4, C and D) (Bowler et al., 2019; Chong et al., 2020).
FIGURE 4:
Airyscan analysis of the localization of centriolar and ciliary proteins in cells subjected to amplibody-proExM. (A) Fixed RPE1 cells were incubated with antibodies against CETN3 and the indicated DAP before amplibody-proExM treatment and analyzed by Airyscan microscopy. Images of laterally (top panels) or axially (bottom panels) oriented MCs are shown. Scale bars, 1 µm. (B) Maximum-intensity projections with color code in the Airyscan images were rotated nine times at 40° intervals around the CETN3 signal. The image set was then merged into one stack, and the average (AVG), maximum (MAX), and sum (SUM) projections were generated. The Z-scaling color coding is illustrated at the bottom. (C) Outer and inner diameters of rings positive for individual DAPs were measured and are expressed as bar graphs. Data are shown as means ± SD; n = 10. (D) Summary of the average outer and inner diameters of rings positive for individual DAPs determined in this study (C) and in the STORM study by Bowler et al. (2019) and Chong et al. (2020). Calculated diameters are shown as values divided by an expansion factor of 4. Diameters from Chong et al. represent mean diameters. (E, F) Fixed RPE1 cells were incubated with antibodies against CETN3 and the indicated SDAP (E) or DEP (F) before amplibody-proExM treatment, and analyzed by Airyscan microscopy. Scale bars, 1 µm. (G) RPE1 cells serum-starved for 24 h were fixed, incubated with antibodies against ARL13B (a, b, c), FOP (a), Ac-tubulin (b, d), EHD1 (c), and MKS3 (d) before amplibody-proExM treatment, and analyzed by Airyscan microscopy. Scale bars, 2 µm
We further used amplibody-proExM followed by Airyscan imaging to observe subdistal appendage proteins (SDAPs) (Figure 4E). Previous studies reported that, unlike the distal appendages, the number of subdistal appendages is variable depending on the cell types, their functional state, and external factors (Anderson, 1972; Kong and Loncarek, 2015; Uzbekov and Alieva, 2018). As expected, clusters positive for ODF2, CEP128, and centriolin (CNTRL) were variable in their number per MC (7–10) (Figure 4E). The distal end proteins (DEPs), CP110, CEP97, and TALPID3, were more closely adjacent to CETN3 than DAPs and SDAPs (Figure 4F).
We also imaged ciliary proteins and found that localizations of ARL13B and Ac-tubulin, as well as that of FOP, were essentially the same as those detected without amplibody treatment (compare Figure 4G with Figure 2, F and H). The signals of EHD1, a membrane-reshaping protein found on the ciliary pocket membrane, and those of MKS3, a transition zone (TZ) protein, had distributions as expected from previous studies (Williams et al., 2011; Lu et al., 2015). Overall, imaging of amplibody-proExM samples by Airyscan microscopy was found to be useful for analyzing the distribution of centriolar and ciliary proteins.
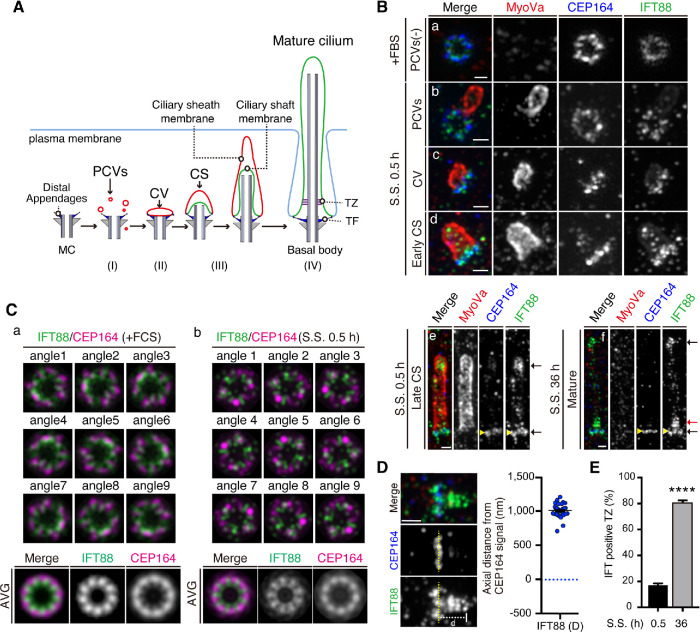
Amplibody-combined three-color proExM followed by Airyscan imaging reveals dynamic changes in the localization of IFT88 during ciliogenesis
Multicolor imaging can be crucial for analyzing molecular processes in their proper context. We next set out to develop a new proExM that enables SR imaging in three colors using two different amplibodies together with a fluorescent secondary antibody (see Materials and Methods). To clarify whether amplibody-combined three-color proExM is applicable for investigating the ultrastructural organization of molecules that act during ciliogenesis, we analyzed the spatial and temporal distributions of IFT88, CEP164, and myosin Va (MyoVa); IFT88 is a subunit of the IFT-B complex, which mediates entry into cilia and the anterograde trafficking of ciliary proteins, including the αβ-tubulin dimer required for axonemal extension (Bhogaraju et al., 2013; Nakayama and Katoh, 2018), and MyoVa is a marker for ciliary membrane structures, such as preciliary vesicles (PCVs), the ciliary vesicle (CV), and the ciliary sheath (CS) (Wu et al., 2018) (Figure 5A and Supplemental Figure S2). RPE1 cells were cultured under serum-fed conditions (fetal bovine serum [+FBS]) or serum-starved for 0.5 or 36 h and analyzed by Airyscan microscopy after amplibody-proExM treatment. Under serum-fed conditions, PCVs positive for MyoVa were not detected around the MC (Figure 5Ba). During the 0.5 h of serum starvation, MyoVa-positive structures gradually accumulated around the MC (Figure 5B, b–e). IFT88 was shown to be closely adjacent to CEP164 in both serum-fed and serum-starved cells (Figure 5B, a–f). In the axial view, IFT88 signals were broadly distributed near the ninefold symmetric densities of CEP164 (Figure 5C, a and b). The averaged images of IFT88 and CEP164 demonstrated that IFT88 is localized near the distal appendages in a pattern occupying gaps between the CEP164 blades (Figure 5C, a and b, bottom row). Intriguingly, the radial localization of IFT88 relative to CEP164 supports the recent findings that a new structural element, designated the distal appendage matrix (DAM), where IFT proteins are concentrated, is present between the blades of transition fibers (TFs)/distal appendages (Yang et al., 2018). In the early stage of CS extension (early CS stage), a small number of IFT88 particles appeared to enter the cilium, probably concurrently with axoneme extension (Figure 5Bd). With further CS extension (late CS stage), IFT88 was concentrated at the tip of the elongating cilium, as well as at the TFs (Figure 5Be). We further imaged IFT88 in MyoVa-negative, mature primary cilia, in which the membrane is directly exposed to the extracellular space (Figure 5Bf and Supplemental Figure 2F). Unlike in the late CS stage, IFT88 signals were found above the TFs as well as at the distal tip and TFs in the mature cilium (Figure 5Bf). These IFT88 puncta occupied the area in the range of 1007 ± 14.4 nm from the distal end of TFs positive for CEP164 (Figure 5D). Notably, IFT88 puncta in the TZ were found in 80.7% ± 1.8% of mature cilia in cells serum-starved for 36 h, whereas they were found in 16.7% ± 1.8% of MyoVa-associated growing cilia in the cells serum-starved for 0.5 h (Figure 5E). These observations are consistent with the previous proposal that the TZ acts as a potential waiting place for IFT particles to enter and function in the cilium (Yang et al., 2015) and demonstrate that rapid axonemal extension is likely to be supported by the IFT-B complex, which delivers the αβ-tubulin dimer toward the growing tip.
FIGURE 5:
Three-color Airyscan analysis of changes in IFT88 localization during ciliogenesis in cells subjected to amplibody-proExM. (A) Schematic representation of stages of the ciliogenesis process: (I) After serum starvation, PCVs positive for MyoVa (red) start to accumulate around and dock to the distal appendages of the MC (blue) beneath the plasma membrane; (II) PCVs are fused to form a large membrane structure, CV; (III) coupled with axonemal growth (formation of a ciliary shaft), PCVs are further docked to the CV, resulting in elongation of the CV to form the CS; and (IV) the CS membrane is finally fused with the plasma membrane, resulting in direct exposure of the ciliary membrane to the extracellular environment. During the ciliogenesis process, TFs and the TZ are formed at the ciliary base and together function as a gate that controls the entry and exit of soluble and ciliary membrane proteins. (B) Three-color Airyscan analysis, coupled with amplibody-proExM, of the localization of IFT88 (green), MyoVa (red), and CEP164 (blue) during ciliogenesis. RPE1 cells were cultured under serum-fed conditions (a) or under serum-starved conditions for 0.5 h (b–e) or 36 h (f), fixed, and incubated with antibodies against IFT88, MyoVa, and CEP164 before the amplibody-proExM treatment, and analyzed by Airyscan microscopy. Representative images of cells without PCVs (a), with PCVs (b), CV (c), or CS (d, early CS; e, late CS), or with mature cilia (f) are shown. Black arrows indicate the IFT88 signals at the axoneme tip and TFs (e and f), and the red arrow indicates IFT88 signals above the TFs (f). Scale bars, 1 µm. (C) Maximum-intensity projection images of IFT88 (green) and CEP164 (magenta) in cells cultured under serum-fed conditions (a) or serum-starved (S.S.) for 0.5 h (b) were created by rotating the image nine times at 40° intervals around the physical center. The set of nine images was then merged into one stack, and average projections were generated. (D) The axial distance of the distal edge of IFT88 signals from the distal end of CEP164-positive TFs in mature cilia. The yellow dashed line represents the position of CEP164. The axial distance (d) from IFT88 to CEP164 was measured (n = 40). (E) After serum starvation for 0.5 or 36 h, cells with IFT88 signals in the TZ were counted, and the percentages are shown as bar graphs. Values are means ± SEM from three independent experiments, and 100 cells were analyzed in each set of experiments. ****p < 0.0001 (unpaired t test).
We here show that amplibodies can be easily incorporated into the existing procedure of immunofluorescence microscopy to increase the signal intensity and that the combinatorial use of amplibodies and proExM is a practical SR imaging strategy that compensates for the reduction in fluorescence intensity inherent to the expansion of samples. For example, we demonstrated that three-color immunofluorescence analysis in combination with the amplibody-proExM method is useful for ultrastructural analyses of biological processes, such as ciliogenesis. Although the amplibody-proExM method proposed here is based on Boyden’s protocol (Tillberg et al., 2016; Asano et al., 2018), in principle, amplibodies can be incorporated into other ExM protocols, such as U-ExM (Gambarotto et al., 2018) and centriole-MAP (Sahabandu et al., 2019).
Finally, we compare ExM with existing SR microscopy techniques in terms of advantages and disadvantages. The major advantage of ExM is that it allows us to distinguish subcellular structures below the diffraction limit using a conventional microscope. The resolution of ExM (approximately 70 nm) is higher than that of SIM and Airyscan and comparable to that of STED microscopy. As shown in this study, the resolution of ExM can be further improved in combination with SIM or Airyscan, comparable to that of PALM/STORM. Moreover, compared with STED microscopy and PALM/STORM, the combinatorial use of ExM and either SIM or Airyscan is much more practical with respect to multicolor SR imaging and observation of thick samples.
On the other hand, a disadvantage inherent to ExM is that it is not applicable, in principle, to the observation of living cells; therefore, other microscopy techniques are required for live cell SR imaging, although the application of some techniques is challenging. Another disadvantage of ExM is the variation in the sample expansion, which makes it difficult to determine the actual sizes of objects. By comparing the gel sizes before and after expansion, we estimated the expansion factor of approximately 4 (see Figure 1B). However, the actual expansion factors can vary from sample to sample or even locally within the same sample (e.g., depending on organelles): previous studies calculated the expansion factors between 3.96 and 4.42 (Tillberg et al., 2016) and between 4.2 and 4.7 (Zhao et al., 2017). Several methods have been proposed to overcome the problem of anisotropic or differential expansion: 1) a method using DNA origami nanorulers as traceable distance measurement standards (Scheible and Tinnefeld, 2018); 2) a method creating an intrinsic ruler in a sample by fluorescence photobleaching (Vanheusden et al., 2020); and (3) a computational image analysis by machine learning (Gatti et al., 2019; Pernal et al., 2020). Using these methods, it is possible to estimate local expansion factors and correct variations from sample to sample. In the near future, with the addition of various technological innovations, ExM is expected to become a more reliable microscopy technique, bridging the gap between electron microscopy and conventional fluorescence microscopy.
MATERIALS AND METHODS
Antibodies, reagents, and instruments
Antibodies used in this study are listed in Supplemental Table S1. The following reagents were purchased from the manufacturers as noted: sodium acrylate (SA) (#408220) from Sigma-Aldrich; acrylamide (AA) (#00809-85), N,N′-methylenebisacrylamide (#22402-02), ammonium persulfate (APS) (#02627-34), N,N,N′,N′-tetramethylethylenediamine (TEMED) (#33401-72), guanidine hydrochloride (#17318-95), EDTA (#15105-35), Triton X-100 (#12967-45), and proteinase K (#29442-14) from Nacalai Tesque; 6-((acryloyl)amino)hexanoic acid, succinimidyl ester (Acryloyl-X, SE) (#A20770), and UltraPure Tris (#15504-020) from Invitrogen; and SeaPlaque Agarose (#50100) from Lonza.
The monomer solution containing 8.6 g/100 ml of SA, 2.5 g/100 ml of AA, 0.15 g/100 ml of N,N′-methylenebisacrylamide, 11.7 g/100 ml of sodium chloride, and 1× phosphate-buffered saline (PBS) was prepared and stored at 4°C until use. Digestion buffer containing 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.5% Triton X-100, and 0.8 M guanidine hydrochloride was prepared and stored at 4°C until use.
The following materials were purchased from the manufacturers: Grace Bio-Labs CultureWell removable chambered cover glass (Sigma-Aldrich; #GBL112358), six-well glass-bottom plates with high-performance #1.5 cover glass (Cellvis; P06-1.5H-N), and coverslips (9-mm diameter) (Matsunami Glass; #CS01029).
Design and preparation of amplibodies
Plasmids encoding four kinds of amplibodies were constructed in two steps. First, cDNA of the fluorescent proteins mClover3 (Addgene #74253) (Bajar et al., 2016) or mCherry (a kind gift from Roger Tsien, University of California, San Diego) (Shaner et al., 2004) was inserted into the GST tag vector, pGEX-6P-1 (GE Healthcare). Next, the cDNA of a Nb that recognizes the mouse Ig κ light chain or the rabbit IgG Fc region (Addgene #104161 and #104164, respectively) (Pleiner et al., 2018) was then inserted into the C-terminus of the fluorescent protein cDNA. The amino acid sequences of the amplibodies are shown in Supplemental Figure S1. The plasmids have been deposited to Addgene (#138128–#138131).
E. coli (BL21-CodonPlus(DE3)-RIPL strain; Agilent Technologies) transformed with the amplibody vector were incubated for 2–4 h at 37°C with vigorous shaking until the culture reached OD600 = 0.5. To induce expression of the GST-tagged amplibody, E. coli cells were treated with 0.1 mM isopropyl β-D-1-thiogalactopyranoside and cultured overnight at 20°C with vigorous shaking; amplibody expression was easily confirmed by the change in color of the E. coli culture. The cells were harvested by centrifugation at 5,550 × g for 15 min at 4°C. The cell pellet was resuspended in 9 ml of binding buffer (PBS containing 5 mM dithiothreitol [DTT] and protease inhibitors), and the cells were then disrupted by sonication. The cells were lysed by adding Triton X-100 to 1% and incubated on ice for 20 min. The cell lysates were centrifuged at 20,700 × g for 20 min at 4°C, and the supernatants were transferred to new tubes. The supernatant was mixed with 500 µl (bed vol) of glutathione-Sepharose 4B beads (GE Healthcare) and binding of the GST-tagged amplibody to the beads was allowed by rotating the tube overnight (12–24 h) at 4°C. The beads were centrifuged at 780 × g for 2 min at 4°C and then washed eight times with 10 ml of washing buffer (PBS containing 5 mM DTT and 0.1% Triton X-100). Finally, the beads mixed with cleavage buffer (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.01% Triton X-100) to a 50% slurry were incubated with PreScission protease (GE Healthcare) for 24 h at 4°C, with constant rotation to cleave the GST tag. The colored supernatants were collected by sedimentation of the beads by spinning down for 30 s. After protein concentrations were determined by measuring the absorbance at 280 nm, the amplibodies were subjected to SDS–PAGE followed by staining with Coomassie Brilliant Blue (CBB) to confirm the purity. After the protein concentration was adjusted to 5 µg/µl with cleavage buffer, the purified proteins were aliquoted to a microcentrifuge tube, rapidly frozen, and stored at –20°C. A working solution was stored at 4°C.
Cell culture
hTERT-RPE1 cells (CRL-4000; American Type Culture Collection) were cultured in DMEM/F-12 (Nacalai Tesque) supplemented with 10% FBS and 0.348% sodium bicarbonate at 37°C in 5% CO2. To induce ciliogenesis, cells were serum-starved for 30 min, 24 h, or 36 h in Opti-MEM (Invitrogen) containing 0.2% bovine serum albumin, as described previously (Katoh et al., 2017; Nozaki et al., 2017).
Conventional immunostaining
hTERT-RPE1 cells were fixed with 3% paraformaldehyde (PFA) in PBS for 15 min at 37°C, permeabilized with 0.1% Triton-X 100 in PBS for 5 min at room temperature (RT), and incubated in PBS containing 10% FBS at RT for 1 h. For the detection of pGlu-tubulin, cells were fixed and permeabilized with methanol for 5 min at –20°C and washed three times with PBS. The fixed/permeabilized cells were blocked with 10% FBS. The cells were then incubated with primary antibodies diluted with 5% FBS for 2 h at RT or overnight at 4°C. Cells were washed three times with PBS and incubated with secondary antibodies for 1 h at RT. After being washed three times with PBS, the cells on a coverslip were mounted on a glass slide with Mowiol (Calbiochem).
Original proExM protocol
Samples were prepared according to Boyden’s proExM protocol (Tillberg et al., 2016; Asano et al., 2018). Briefly, cells were cultured on a removal chambered cover glass and serum-starved to induce ciliogenesis and immunostained as described above. The cells were incubated with 0.1 mg/ml Acryloyl-X, SE in PBS for 6–12 h at RT. The gelling solution containing 188 µl of monomer solution, 4 µl of 10% TEMED, 4 µl of 10% APS, and 4 µl of H2O was prepared on ice. Forty microliters of the gelling solution was added into each well. After a glass slide wrapped with Parafilm was placed on top of the silicone gasket to seal the gelling solution within the wells, the gelling solution was incubated for 1 h at 37°C. After the glass slide wrapped with Parafilm and the silicone gasket were removed, pieces of the gel on the cover glass were soaked in digestion buffer containing 8 U/ml proteinase K overnight at RT. The gel was transferred into a six-well glass-bottom plate, and milli-Q water containing 1 µg/ml Hoechst 33342 was added. The water was exchanged three or four times every hour until the gel was fully expanded. After the water was removed, the gel was immobilized with 2% low-melting-point agarose solution to prevent sample drift during the imaging.
Modified proExM protocol using amplibodies
hTERT-RPE1 cells were cultured on a coverslip (9-mm diameter) in a 24-well plate. For detection of EHD1, SDAPs, and DEPs, cells were fixed and permeabilized with methanol for 5 min at –20°C. For the detection of DAPs, cells were permeabilized with 0.1% Triton X-100 in PHEM buffer (60 mM PIPES [pH 6.8], 25 mM HEPES [pH 6.8], 2 mM MgCl2, 10 mM ethylene glycol tetraacetic acid) for 1 min at RT and fixed in ice-cold methanol for 7 min at –20°C. For the detection of MyoVa and IFT88, cells were fixed with 3% PFA in PBS for 15 min at 37°C, washed three times with PBS, and permeabilized with ice-cold methanol for 7 min at –20°C. The fixed/permeabilized cells were blocked with 10% FBS. The amplibody was diluted to 1 µg/ml (for PFA and methanol fixation) or 0.25 µg/ml (for PHEM/methanol fixation) in a primary antibody solution. A drop (30 µl) of amplibody and primary antibody solution was placed on Parafilm in a moist chamber, and the coverslip was put on the drop and then incubated overnight at 4°C. Optionally, the cells were washed three times with PBS and incubated with CF633-conjugated anti-goat IgG (H+L) for 2 h at RT in a moist chamber. Then, the cells were washed three times with PBS and incubated with anti–fluorescent protein antibodies for 2 h at RT. The cells were again washed three times with PBS and incubated with 0.1 mg/ml Acryloyl-X, SE in PBS for 3 h at RT. The gelling solution was prepared on ice as in the original proExM protocol. A drop of 15 µl of the gelling solution was placed on a slide glass wrapped with Parafilm, and the coverslip was put on the drop and incubated for 20 min at 37°C. Then, the coverslips with the gel attached were peeled off from the slide using forceps and transferred into a six-well glass-bottom plate. The gels were incubated in the digestion buffer containing 8 U/ml proteinase K for 30–60 min at 37°C until the gel swelled slightly and came off the coverslip. After removal of the digestion buffer, the gel was washed three times with PBS and incubated with fluorescent dye–conjugated secondary antibodies diluted in 5% FBS overnight at RT with gentle shaking. The gel was washed three times with PBS containing 0.1% Triton X-100 for 20 min at RT. Finally, the gel was soaked in milli-Q water containing 1 µg/ml Hoechst 33342, which was exchanged three or four times every 30 min until the gel was fully expanded. After removal of the water, the gel was immobilized with 2% low-melting agarose solution to prevent sample drift during imaging.
Microscopy and image analysis
Conventional wide-field microscopy was performed using the Axiovert 200M microscope equipped with a 63×/1.40 NA oil-immersion objective and AxioCam MRm CCD camera (Carl Zeiss). SR-Airyscan imaging was performed using the LSM800 or LSM880 microscope equipped with a 63×/1.40 NA or 100×/1.46 NA oil-immersion objective (Carl Zeiss). SR-SIM imaging was performed using an ELYRA S.1 microscope (Carl Zeiss) equipped with an Andor iXon 885 EMCCD camera, a 100×/1.40 NA oil-immersion objective, and four laser beams (405, 488, 561, and 642 nm).
The images taken in superresolution Airyscan mode were Airy-processed in three dimensions with a strength value of 7.0–7.5. Image stacks were collected with a z-step size of 0.18 µm and processed with Zen software (Carl Zeiss) to build SR and maximum projection images. In the SR-SIM imaging, serial z-stack sectioning was carried out at 101-nm intervals. Z stacks were recorded with three phase changes and five grating rotations for each section. The microscope was calibrated with 100 nm fluorescent beads to calculate both lateral and axial limits of image resolution. Images were reconstituted with Zen software.
The diameters of the ring-like structures of the centriolar proteins were measured using Zen software. Box-and-whisker diagrams and scatter plots were generated using JMP Pro13 software (SAS Institute). Maximum-intensity projection of the z-series was rotated nine times at 40° intervals around the physical center of the MC to average the signals from the nine appendages. The image set was then merged into one stack, and the AVG, MAX, and SUM projections were generated by FIJI software.
Supplementary Material
Acknowledgments
We thank Dirk Görlich, Michael Lin, and Roger Tsien for kindly providing plasmids of anti-IgG nanobodies, mClover3, and mCherry, respectively, Fumiyoshi Ishidate for technical support for SR imaging, and Helena Akiko Popiel for critical reading of the manuscript. We used SR-microscopes in the iCeMS Analysis Center of Kyoto University, in the Center for Medical Research and Education of Osaka University, and in the Research Support Platform of Osaka City University. This work was supported in part by grants from the Japan Society for the Promotion of Science (grant numbers 18H02403 to Y.K., 15H05596 and 19K06644 to S.C., and 19H00980 to K.N.) and OCU “Think globally, act locally” Research Grant for Young Scientist 2019 through the hometown donation fund of Osaka City to S.C.
Abbreviations used:
- Ac-tubulin
acetylated α-tubulin
- CS
ciliary sheath
- CV
ciliary vesicle
- DAP
distal appendage protein
- DEP
distal end protein
- ExM
expansion microscopy
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- hTERT-RPE1
human telomerase reverse transcriptase–immortalized retinal pigment epithelial 1
- IFT
intraflagellar transport
- Ig
immunoglobulin
- MC
mother centriole
- MyoVa
myosin Va
- Nb
nanobody
- PALM
photoactivated localization microscopy
- PBS
phosphate-buffered saline
- PCV
preciliary vesicle
- pGlu-tubulin
polyglutamylated-tubulin
- proExM
protein-retention ExM
- SDAP
subdistal appendage protein
- SIM
structured illumination microscopy
- SR
superresolution
- STED
stimulated emission depletion
- STORM
stochastic optical reconstruction microscopy
- TF
transition fiber
- TZ
transition zone.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-04-0250) on July 29, 2020.
REFERENCES
- Anderson RG. (1972). The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol , 246–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano SM, Gao R, Wassie AT, Tillberg PW, Chen F, Boyden ES. (2018). Expansion microscopy: protocols for imaging proteins and RNA in cells and tissues. Curr Protoc Cell Biol 80, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajar BT, Wang ES, Lam AJ, Kim BB, Jacobs CL, Howe ES, Davidson MW, Lin MZ, Chu J. (2016). Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep , 20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. (2013). Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science , 1009–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler M, Kong D, Sun S, Nanjundappa R, Evans L, Farmer V, Holland A, Mahjoub MR, Sui H, Loncarek J. (2019). High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat Commun , 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon CK, Yu Z, Wang Y, Guo F, Unruh JR, Slaughter BD, Hawley RS. (2017). Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc Natl Acad Sci USA , E6857–E6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. (2015). Optical imaging. Expansion microscopy. Science , 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong WM, Wang W-J, Lo C-H, Chiu T-Y, Chang T-J, Liu Y-P, Tanos B, Mazo G, Tsou M-FB, Jane W-N, et al. (2020). Super-resolution microscopy reveals coupling between mammalian centriole subdistal appendages and distal appendages. eLife , 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chozinski TJ, Halpern AR, Okawa H, Kim H-J, Tremel GJ, Wong ROL, Vaughan JC. (2016). Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods , 485–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch C, Dupuis-Williams P. (2011). Ultrastructure of cilia and flagella—back to the future! Biol Cell , 249–270. [DOI] [PubMed] [Google Scholar]
- Freifeld L, Odstrcil I, Förster D, Ramirez A, Gagnon JA, Randlett O, Costa EK, Asano S, Celiker OT, Gao R, et al. (2017). Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proc Natl Acad Sci USA , E10799–E10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotto D, Zwettler FU, Le Guennec M, Schmidt-Cernohorska M, Fortun D, Borgers S, Heine J, Schloetel J-G, Reuss M, Unser M, et al. (2018). Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat Methods , 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Maraspini R, Beutel O, Zehtabian A, Eickholt B, Honigmann A, Ewers H. (2018). Expansion stimulated emission depletion microscopy (ExSTED). ACS Nano , 4178–4185. [DOI] [PubMed] [Google Scholar]
- Gatti DL, Arslanturk S, Lal S, Jena BP. (2019). Deep learning strategies for differential expansion microscopy. bioRxiv. 10.1101/743682 [DOI] [Google Scholar]
- Ishikawa H, Marshall WF. (2011). Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol , 222–234. [DOI] [PubMed] [Google Scholar]
- Jiang N, Kim H-J, Chozinski TJ, Azpurua JE, Eaton BA, Vaughan JC, Parrish JZ. (2018). Superresolution imaging of Drosophila tissues using expansion microscopy. Mol Biol Cell , 1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie T, Abbott KL, Mooney NA, Plowey ED, Demeter J, Jackson PK. (2017). The CEP19-RABL2 GTPase complex binds IFT-B to initiate intraflagellar transport at the ciliary base. Dev Cell , 22–36.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashihara H, Chiba S, Kanno S-I, Suzuki K, Yano T, Tsukita S. (2019). Cep128 associates with Odf2 to form the subdistal appendage of the centriole. Genes Cells , 231–243. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Michisaka S, Nozaki S, Funabashi T, Hirano T, Takei R, Nakayama K. (2017). Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated homology-independent knock-in system. Mol Biol Cell , 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli P, Höhne M, Jüngst C, Bertsch S, Ebert LK, Schauss AC, Benzing T, Rinschen MM, Schermer B. (2017). The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep , 1521–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Loncarek J. (2015). Correlative light and electron microscopy analysis of the centrosome: a step-by-step protocol. Methods Cell Biol , 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T, Swaney J, Park J-Y, Albanese A, Murray E, Cho JH, Park Y-G, Mangena V, Chen J, Chung K. (2016). Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol , 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang Y-S, et al. (2015). Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol , 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojarad BA, Gupta GD, Hasegan M, Goudiam O, Basto R, Gingras A-C, Pelletier L. (2017). CEP19 cooperates with FOP and CEP350 to drive early steps in the ciliogenesis programme. Open Biol , 170114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y. (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J Biochem , 155–164. [DOI] [PubMed] [Google Scholar]
- Nishijima Y, Hagiya Y, Kubo T, Takei R, Katoh Y, Nakayama K. (2017). RABL2 interacts with the IFT-B complex and CEP19, and participates in ciliary assembly. Mol Biol Cell , 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K. (2017). Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci , 563–576. [DOI] [PubMed] [Google Scholar]
- Pernal SP, Liyanaarachchi A, Gatti DL, Formosa B, Pulvender R, Kuhn ER, Ramos R, Naik AR, George K, Arslanturk S, et al. (2020). Nanoscale imaging using differential expansion microscopy. Histochem Cell Biol , 469–480. [DOI] [PubMed] [Google Scholar]
- Pleiner T, Bates M, Görlich D. (2018). A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J Cell Biol , 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahabandu N, Kong D, Magidson V, Nanjundappa R, Sullenberger C, Mahjoub MR, Loncarek J. (2019). Expansion microscopy for the analysis of centrioles and cilia. J Microsc , 145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl SJ, Hell SW, Jakobs S. (2017). Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol , 685–701. [DOI] [PubMed] [Google Scholar]
- Scheible MB, Tinnefeld P. (2018). Quantifying expansion microscopy with DNA origami expansion nanorulers. bioRxiv. 10.1101/265405 [DOI] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol , 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shi X, Garcia G, Van De Weghe JC, McGorty R, Pazour GJ, Doherty D, Huang B, Reiter JF. (2017). Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat Cell Biol , 1178–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. (2012). 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open , 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe SD, Gambassi S, Thompson CL, Chandrakumar C, Santucci A, Knight MM. (2017). Reduced primary cilia length and altered Arl13b expression are associated with deregulated chondrocyte Hedgehog signaling in alkaptonuria. J Cell Physiol , 2407–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu C-CJ, English BP, Gao L, Martorell A, Suk H-J, Yoshida F, et al. (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol , 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R, Alieva I. (2018). Who are you, subdistal appendages of centriole? Open Biol , 180062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanheusden M, Vitale R, Camacho R, Janssen KPF, Acke A, Rocha S, Hofkens J. (2020). Fluorescence photobleaching as an intrinsic tool to quantify the 3D expansion factor of biological samples in expansion microscopy. ACS Omega , 6792–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IE, Lapan SW, Scimone ML, Clandinin TR, Reddien PW. (2016). Hedgehog signaling regulates gene expression in planarian glia. eLife , 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, Bialas NJ, Stupay RM, Chen N, Blacque OE, et al. (2011). MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol , 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-T, Chen H-Y, Tang TK. (2018). Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat Cell Biol , 175–185. [DOI] [PubMed] [Google Scholar]
- Yang TT, Chong WM, Wang W-J, Mazo G, Tanos B, Chen Z, Tran TMN, Chen Y-D, Weng RR, Huang C-E, et al. (2018). Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun , 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang W-J, Craige B, Witman GB, Tsou M-FB, Liao J-C. (2015). Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep , 14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YS, Chang J-B, Alvarez MM, Trujillo-de Santiago G, Aleman J, Batzaya B, Krishnadoss V, Ramanujam AA, Kazemzadeh-Narbat M, Chen F, et al. (2016). Hybrid microscopy: enabling inexpensive high-performance imaging through combined physical and optical magnifications. Sci Rep , 22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Bucur O, Irshad H, Chen F, Weins A, Stancu AL, Oh E-Y, DiStasio M, Torous V, Glass B, et al. (2017). Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat Biotechnol , 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.