Abstract.
Purpose: Prostate cancer primarily arises from the glandular epithelium. Histomophometric techniques have been used to assess the glandular epithelium in automated detection and classification pipelines; however, they are often rigid in their implementation, and their performance suffers on large datasets where variation in staining, imaging, and preparation is difficult to control. The purpose of this study is to quantify performance of a pixelwise segmentation algorithm that was trained using different combinations of weak and strong stroma, epithelium, and lumen labels in a prostate histology dataset.
Approach: We have combined weakly labeled datasets generated using simple morphometric techniques and high-quality labeled datasets from human observers in prostate biopsy cores to train a convolutional neural network for use in whole mount prostate labeling pipelines. With trained networks, we characterize pixelwise segmentation of stromal, epithelium, and lumen (SEL) regions on both biopsy core and whole-mount H&E-stained tissue.
Results: We provide evidence that by simply training a deep learning algorithm on weakly labeled data generated from rigid morphometric methods, we can improve the robustness of classification over the morphometric methods used to train the classifier.
Conclusions: We show that not only does our approach of combining weak and strong labels for training the CNN improve qualitative SEL labeling within tissue but also the deep learning generated labels are superior for cancer classification in a higher-order algorithm over the morphometrically derived labels it was trained on.
Keywords: prostate cancer, deep learning, machine learning, histology, epithelium, segmentation
1. Introduction
It is estimated that one in seven men will develop prostate cancer (PCa) in their lifetime and that PCa itself accounts for one in five new cancer diagnoses.1 Invasive removal of prostate tissue is currently required to confirm cancer diagnosis and drive treatment. From these samples, pathologists use the well-characterized Gleason criteria2,3 to interpret histomorphometric features and grade the tissues,4 which can then be used to assign a grade group (GG).5 While these grading criteria have been shown to hold great prognostic value, they are inherently subjective, relying on a pathologist’s interpretation.6
In the histological analysis of prostates, it can be useful to divide the tissue into three major components: stroma (connective muscular tissue), epithelium, and lumen (SEL). Identifying these features using quantitative histomorphometry (QH) requires either manual or automatic segmentation. SEL segmentation itself, which if not a requisite, is a stepping-stone for the more complex problem of automated cancer classification and grading using QH. In PCa, cancerous growth commonly occurs within glands, whose structure is delineated and characterized by an epithelial border that normally surrounds a luminal space. Substantial effort has already been invested in segmenting epithelium and stroma in a host of tissues (e.g., breast, colorectal, and prostate) using both hand-engineered features7,8 and approaches using deep learning.9–11
In the interest of cost, time, and resources, methods that segment stroma and epithelium using common general stains (such as H&E) are of particular utility. Recently, Bulten et al. reported the use of both a fully convolutional network (FCN)12 and U-net13 to address this problem. They report impressive accuracies of 0.89 and 0.90 for the U-Net and FCN methods, respectively, on the two-class problem (stroma and epithelium). However, they lament “We suspect that most of the errors are, first of all, caused by a lack of training examples and not due to a limitation of the models.”
The performance of machine learning methods is highly dependent on the training data provided14,15 and, subsequently, the ground-truth annotations. Methods have been developed to address these issues that both expand the dataset through data augmentation16 (e.g., image translation, rotation, flipping), as well as expanding the number of examples of ground-truth annotations with weak supervision.17,18 Weak supervision is a broad category of methods that may rely on heuristics but ultimately assumes noisy ground truth labels.
It is the goal of this study to compare the segmentation of stroma, epithelium, and lumen when using different combinations and sources of strong and weak labels. Further, we assess the segmentation of whole-mount prostate samples using a deep learning framework trained on biopsy cores from a separate institution. First, this study compares the accuracy of SEL segmentation when a similar training dataset, with both manually (strong) and computationally (weak) derived annotations, is provided to the deep convolution encoder–decoder network, SegNet.19 Second, we demonstrate that in our dataset, labels generated from a deep learning framework trained using weak morphological segmentation are more accurate than the labels used to train the network. Third, we demonstrate the utility of this biopsy-trained algorithm by applying it to whole-mount prostate histology processed and digitized at a separate institution. Finally, we demonstrate an improvement in the discrimination of benign regions (atrophy and HGPIN) and cancerous (Gleason pattern 3+) regions using the proposed training methods for a convolutional neural network when compared to the rigid morphological methods used, in part, to train the deep learning method.
2. Materials and Methods
2.1. Patient Population
The histology from two patient groups was digitally analyzed for this interinstitutional study. Patients () from the University of Wisconsin (UW, group 1) underwent biopsy for suspected PCa, although all samples included in this study showed no presence of PCa. Each patient had cores acquired as part of the standard biopsy protocol. Patients from the Medical College of Wisconsin (MCW, group 2) undergoing a radical prostatectomy were prospectively recruited to participate in this study (). A summary of patient demographics and diagnoses is shown in Table 1. Data collected from group 1 were approved under the University of Wisconsin Madison’s Institutional Review Board (IRB) and data collected for group 2 were approved under the MCW’s IRB.
Table 1.
Demographic information for training and testing datasets.
| Subject demographics | ||
|---|---|---|
| Metric | Group 1 | Group 2 |
| Patients (#) | 145 | 26 |
| Samples (#) | 146 | 32 |
| Age (years) | ||
| PSA (ng/mL) | ||
| Grade | — | — |
| Atrophy + HGPIN | — | 4 |
| Gleason 3+ | — | 19 |
2.2. Histological Preparation and Digitization
2.2.1. Biopsy cores
Tissues obtained from biopsy procedures in patients from group 1 were paraffin embedded, sliced at thickness, and hematoxylin and eosin (H&E) stained at UW as part of standard of care. Each slide was digitized, and images were transferred electronically to MCW for further analysis.
2.2.2. Whole mount prostate histology
In addition to the prostate biopsy cores, 32 previously reported20,21 whole mount prostate slides (H&E stained—sectioned at ) (group 2) were digitized at per pixel using an Olympus VS120 automated microscope. Each digital slide was then annotated by a urological fellowship trained pathologist (KAI) using the Gleason pattern classification system. This resulted in the manual annotation of regions containing the benign abnormalities of atrophy and HGPIN () and cancer (Gleason 3+, ). For purposes of this study, the definition of annotated region describes all pixels that the pathologist labeled in a single tissue slide. These annotated regions may include connected and nonconnected pixels. Segmentation in this paper refers to computationally derived labels.
2.2.3. Ground truth segmentation
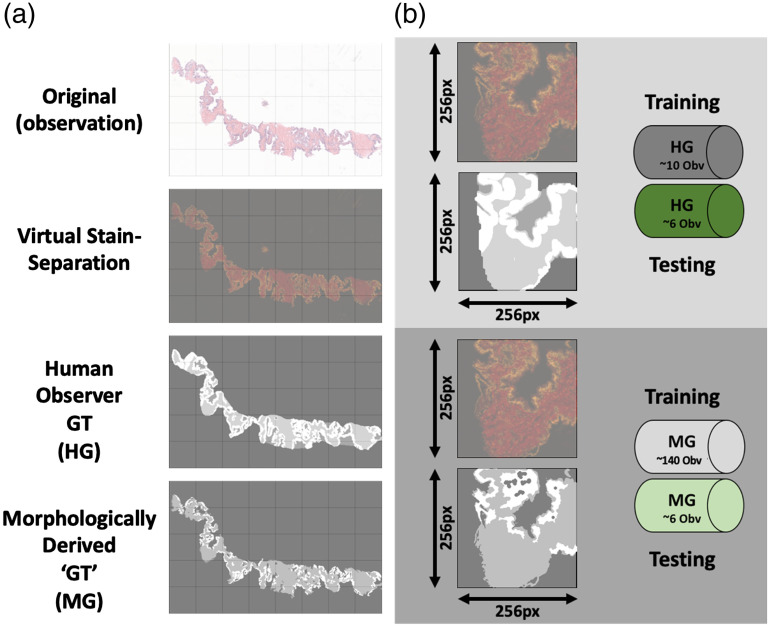
Pixelwise image segmentation was performed on the biopsy core images to label SEL associated foreground pixels in two ways (Fig. 1). The whole mount prostate slides did not have SEL ground truth annotations.
Fig. 1.
Preparation of training and testing dataset from prostate needle biopsies. (a) Examples are given of a representative sample from the prostate biopsy dataset. Grid pattern denotes blocks that the images would later be divided up into. Top row gives the original true color (RGB) image from the original scan. Second row shows virtual stain separation of image with the red and green color channels representing separate stain intensities (eosin and hematoxylin, respectively). Third row displays labels from the human observer ground truth dataset (white, epithelium; mid-gray, stroma; dark-gray, lumen). Fourth row displays labels from the computer generated, morphologically derived ground truth dataset (white, epithelium; mid-gray, stroma; dark-gray, lumen). (b) Top: Magnified example of a training image with paired virtual stain separation and HG-labeled ground truth. HG dataset included 16 total images, split into training (10 images) and testing (6 images). Bottom: Magnified example of a training image with paired virtual stain separation and morphologically generated (MG) labeled ground truth. MG dataset included 146 total images, split into training (140 images) and testing (6 images).
2.2.4. Biopsy core group assignment
The segmented biopsy cores were then separated into training and testing group subsets. This resulted in computer-generated (MG) and human-generated (HG) labeled datasets each containing 140 and 10 training images, respectively. The test set used for all trained classifiers consisted of the same six images that were randomly selected from the dataset.
2.2.5. Human-generated annotation
The HG ground truth annotation was performed on a subset of 16 randomly selected images (32 total cores) from the full 146 image dataset. Each of the 16 core images was SEL segmented by a trained human observer (H.F.) using a Microsoft Surface tablet computer and a stylus (Microsoft Corp., Seattle, Washington).
2.2.6. Computer/morphologically generated segmentation
The MG ground truth segmentation was created using a custom intensity-based morphological algorithm written in MATLAB, using the Image Processing Toolbox (Mathworks Inc. Natick, Massachusetts) as previously reported.21 In short, following contrast enhancement each biopsy core was located and masked. Intensity thresholds were then applied to the images to separate SEL into three separate masks. To correct potential noise, spurious small regions surrounded by pixels of another segmentation were removed from the lumen and epithelium masks. This MG segmentation was applied to 146 biopsy images ( cores total). The algorithm performed segmentations in less than a second per sample.
2.2.7. Class label summary
With ground truth labels assigned for each image in training and testing dataset, image areas containing background were excluded. The class percent breakdown for pixels in each split is given in Table 2. All trained classifiers were tested against the HG strong labels.
Table 2.
Class makeup of training/testing dataset splits. This table describes the percent makeup of each of the three classes (stroma, epithelium, and lumen) used to train and test the classifiers.
| Stroma (%) | Epithelium (%) | Lumen (%) | |
|---|---|---|---|
| MG training set | |||
| HG training set | |||
| MG test set | |||
| HG test set |
2.2.8. Histology tiling
The deep learning algorithms required images for training and testing. Custom Matlab code was therefore developed to divide the resulting images into tiles constrained to include SEL segmentations. This resulted in the MG training dataset containing 6042 unique, nonoverlapping, tiles that included at least one pixel with a labeled class. Data augmentation of 90-deg rotations and mirroring was performed to increase the training dataset to 42,294 images. The HG dataset likewise contained 531 unique, nonoverlapping tiles that included at least one pixel with a labeled class. Using data augmentation, this dataset was expanded to 3717 images. The HG dataset consisted entirely of a subset of the MG dataset, with the only difference being the source of the ground truth labels.
2.3. Digital Histology Preprocessing
2.3.1. Biopsy cores
For training purposes, all images in the training and testing datasets were color deconvolved and virtually stain separated using an automated method described by Macenko et al.22 Using this method, color basis vectors were solved for each individual image, and the Eosin and hematoxylin stain intensities were separated into different channels. The combined training dataset was constructed with three channel images corresponding to eosin, hematoxylin, and residual.
2.3.2. Whole mount prostate histology
To further improve robustness and decrease variation between slides stained and digitized at separate institutions, the whole mount samples from MCW were color normalized to a reference biopsy core from UW using the automated method describe by Khan et al.23 implemented in MATLAB (MathWorks Inc., Natick, Massachusetts). Resulting color normalized images were then color deconvolved as described above Ref. 22 to be consistent with the training dataset.
2.3.3. Convolutional neural network design and training: Arm1, Arm2, and Arm3
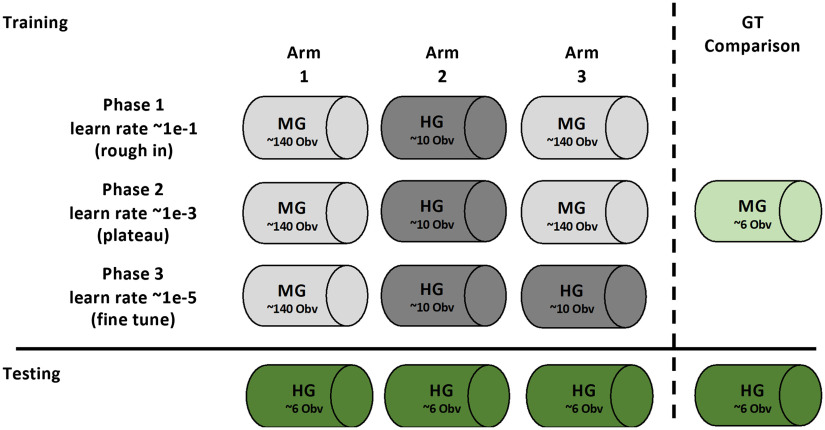
The deep learning encoder–decoder SegNet19 was used to perform pixelwise segmentation of the images. To initialize SegNet, a transfer learning approach was employed using pretrained weights and design associated with the MATLAB implementation of VGG16-trained SegNet.19,24 Implementation and training of SegNet were split into three separate Arms (Fig. 2). Three separate training phases comprised each Arm. Phase 1 was considered a “rough-in” phase characterized by a high learning rate (0.1) and low number of epochs (30). Phase 2 was considered a “plateau” phase, which was comprised by lower learning rate (1e-3) and higher number of epochs (1000+). Phase 3 was considered the “fine-tune” phase, where learning rate was dropped further (1e-5) and a small number of epochs were performed (). The number of epochs was chosen based on plateauing of training loss and accuracy. The three different arms, or trained classifiers, were distinguished by the training dataset used in each phase. This training and dataset schedule are provided in Fig. 2. Regardless of arm, phase, or label source, the six-image test dataset was held out for all training. A fully trained network was able to generate probability masks per class, followed by the final layer, which performed the formal pixelwise segmentation.
Fig. 2.
Training and test schedule for SEL segmentation algorithm. Using the datasets identified in Fig. 1, training and testing scheduling are outlines above. The three arms represent the training of the same segmentation architecture with differing training data provided. Arm 1 represents the segmentation algorithm trained only with morphologically generated (MG) ground truth. Arm 2 represents the segmentation algorithm trained only with HG ground truth. Arm 3 represents segmentation algorithm trained initially with MG and fine-tuned with HG. Training of the three arms is split into three different “phases” using associated training data. Performance of all three Arms is compared using only the HG dataset as it represents the “stronger” ground truth. Comparisons were also made between the MG ground truth and HG ground truth.
2.3.4. Combination of trained classifiers: mArm
To incorporate benefits observed from each individually trained classifier, a combination of the classifiers (mArm) was used to analyze regional SEL segmentation in whole mount prostate samples. Pixels classified by any arm as epithelium were labeled as such in the mArm segmentation. Remaining pixels, if labeled as lumen in any arm, were labeled as lumen in mArm. Any pixels marked as tissue, yet not labeled as epithelium or lumen, were then classified as stroma. This strategy was used to weight epithelium most important due to its relevance in PCa Gleason pattern classification. While this may have introduced bias for SEL classification into the resulting mArm classifier, the mArm classifier’s intended use is for benign/cancerous detection pipeline, not strictly SEL classification.
2.4. Experimental Endpoints
2.4.1. Experiment 1: pixelwise probability maps
Probability maps generated for each class within each arm and compared classwise to the human SEL-labeled test dataset. Receiver operating characteristic (ROC) curves were generated for each class and arm for each test image in the dataset. The area-under-the-curve (AUC) of each ROC curve was averaged per condition and used to compare arms.
2.4.2. Experiment 2: dice and BF-score comparisons for trained models
The classification layer of the trained network was used to generate SEL labels for each of the test images. The dice coefficient and BF score were then calculated for each arm’s classification on each of the test images in comparison to the human-labeled ground truth. Larger dice coefficients indicate greater overlap, and BF scores are larger when boundaries of class annotations are similar.
2.4.3. Experiment 3: comparison of SEL segmentation-based Gleason pattern recognition
It has previously been shown that the density of SEL differs between Gleason patterns. To determine whether mArm SEL classification constituted a clinically relevant and meaningful improvement over conventional methods and to assess our method in a generalized test case, we compared the accuracy of benign versus cancerous pattern classification using SEL features derived from mArm and MG, using pathologist annotations as a ground-truth. Specifically, we implemented a commonly used machine learning algorithm, support vector machines (SVM), trained to differentiate the pathologist annotations in a region based on its SEL signature (percentage make-up). To test the clinical relevance, we used a repeated k-fold cross validation method in a “paired” fashion. Within these datasets, each observation was a slide averaged SEL signature derived from mArm or MG, within pattern regions of interest. The accuracy was then calculated for each SVM training/test instance and compared between MG and mArm.
3. Results
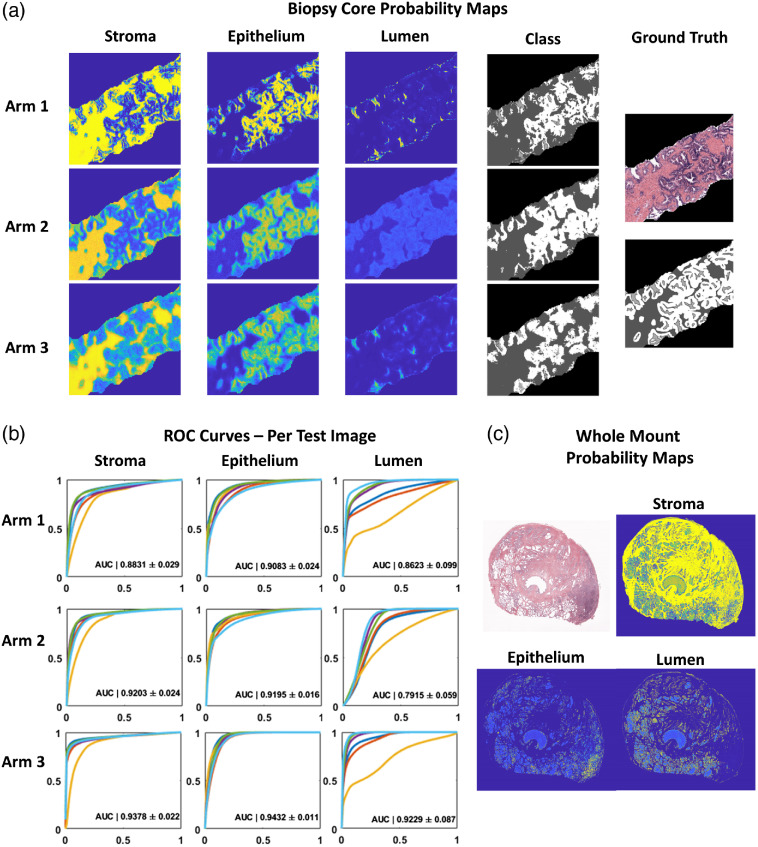
3.1. Experiment 1: Pixelwise Probability Maps
Probability masks pertaining to each of the three classes (SEL) were assessed after being generated by each of the three trained classifiers (Arm 1, Arm 2, and Arm 3). A representative sample of a biopsy core in full color (top left) and associated HG ground truth labels are shown in Fig. 3 (top). Probability maps for each class from the three trained classifiers are presented to the right. Qualitatively, the probability masks pertaining to Arm 1 (MG-generated ground truth labels) show the greatest confidence in all classes, evident in the hard boundaries present, whereas the probability maps generated by Arms 2 and 3 remain softer. This results in a heavy overlap between probability maps generated for lumen and epithelium.
Fig. 3.
Application of morphological operations and combined SEL segmentation algorithms on pathologist annotated whole mount prostate. (a) Original color example of prostate core from test set (right, top) with (right bottom) stroma (dark gray), epithelium (white), and lumen labels (light gray). Accompanying probability maps generated for each class (column) and from each arm of CNN training (row). Note that with increased number of training samples: Arm1, the probabilities are stronger, whereas Arms 2 and 3 show softer probabilities. In addition, fewer training examples (Arm 2) lead to lower overall probabilities for lumen segmentation with large overlap of epithelium probabilities. This contributes to lack of lumen classification seen in segmentation (Fig. 4). (b) Individual ROC curves generated for each class using probability maps solved for the test set. Each plot shows the ROC curve generated from each of the six test set images. The AUCs presented represent the mean from the six test images and the corresponding standard deviation. Significant differences were found between both arms and classes by two-way ANOVA (). Significant differences were found by post-hoc Holm Sidak method when comparing both Arms 1 and 2 to Arm 3 (). (c) With training performed solely on prostate biopsy cores collected, stained, and imaged at a different institution, probability maps generated for whole mount prostate slides show qualitatively good performance. Note increased probability of epithelium in bottom right corner associated with cancerous tissue.
The ROC curves for each of the six test images were plotted by arm and class and shown in Fig. 3(b). Progressive improvement is generally seen for each subsequent Arm (stroma: Arm 1 , Arm 2 , Arm 3 ; epithelium: Arm 1 , Arm 2 , Arm 3 ; lumen: Arm 1 , Arm 2 , Arm 3 ). Significant differences were found between both arms and classes by two-way ANOVA (). Significant differences were found by post-hoc Holm Sidak method when comparing both Arms 1 and 2 to Arm 3 (). This suggests that a viable strategy is to first learn coarse features using noisier samples, then subsequently fine tune with high quality labels. As demonstration of classifier robustness, an example of the biopsy trained Arm 3 network applied to a whole mount prostatectomy sample is shown in Fig. 3 (bottom). A region of high-grade cancer was identified by a pathologist in the lower right quadrant. This is clearly delineated by the patch of increased epithelium probability and decreased lumen probability.
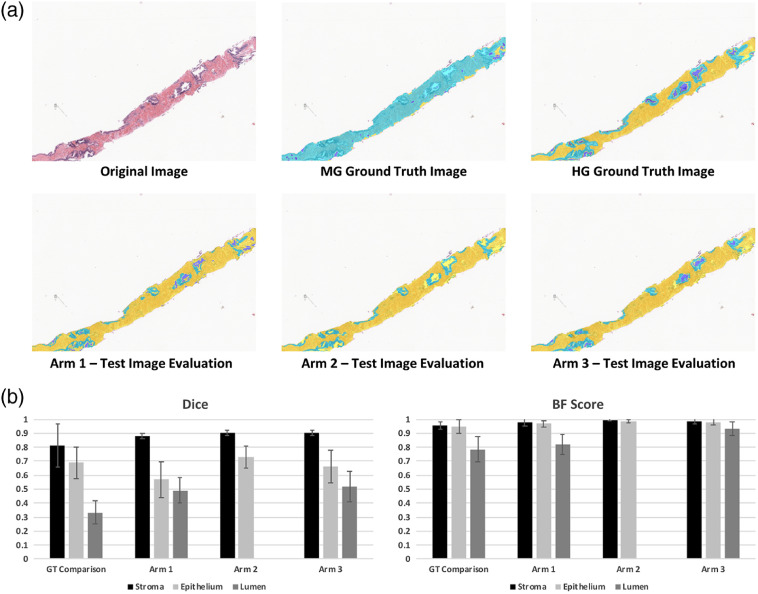
3.2. Experiment 2: Dice and BF-Score Comparisons for Trained Models
The final three-class segmented output for each Arm was compared via Dice coefficient25 and BF score26 to the HG ground-truth. Significant differences were found between both region and classification by a two-way ANOVA (). An example image from the test set is presented in Figs. 4(a)–4(f), chosen to illustrate the potential inaccuracy of the MG segmentation. The top of the figure indicates that a more robust segmentation is reached with a DL algorithm compared to the conventional MG approach. Dice coefficients associated with the stroma class did not differ between any of the models or the standard comparison. Significant differences were found by post-hoc Holm Sidak method between lumen classification for each comparison (each ) except for Arm 1 to Arm 3 (). Significant differences were found for epithelium classification in Arm 1 versus Arm 2 () and standard versus arm 1 (). Notably, Arm 2 failed to classify lumen, reflected in the overlap of the epithelium and lumen probabilities shown in Fig. 3(a).
Fig. 4.
Evaluation of test set using individually trained SEL classification algorithms. (a) Using a biopsy sample that showed poor performance with the morphologically generated (MG) labeling, the fully trained segmentation algorithms (Arms 1 to 3) are compared to the MG ground truth and the HG ground truth. Background and foreground image regions were premasked for comparison. In all labeled images, stroma is labeled yellow, epithelium is labeled light blue, lumen is labeled dark blue. Note that the MG ground truth example shows noisy labeling compared to HG ground truth. Arm 1 shows a good balance between lumen and epithelium labeled, although many glands are left with incomplete epithelium label. Arm 2 shows good epithelium labeling; however, lumen label is almost completely missing. Arm 3 shows good balance between both models (Arm 1 and Arm 2). (B) Dice and BF Score comparison between the ground truth datasets (MG and HG).
The bottom bar charts further illustrate this point with the decrease in variability seen when comparing the Arms to the original MG method. No significant difference was found between the Dice mean for class “stroma” between the standard and Arm 1, variance within the Arm 1 stroma class was found to be significantly decreased compared to standard (Bartlett test: ). This suggests that while the MG class labels may have been more noisy, deep learning may have distilled features pertinent to the HG ground truth thereby improving robustness of the classifier.
3.3. Experiment 3: Comparison of SEL Segmentation-Based Gleason Pattern Recognition
To further demonstrate the benefit gained from trained SEL classification, the three experimental arms were combined (mArm) and applied to whole mount prostates and compared against the morphologically generated labels. To compensate for staining differences and provide the best method comparison, the whole mount prostates were first normalized as per the Khan method.23
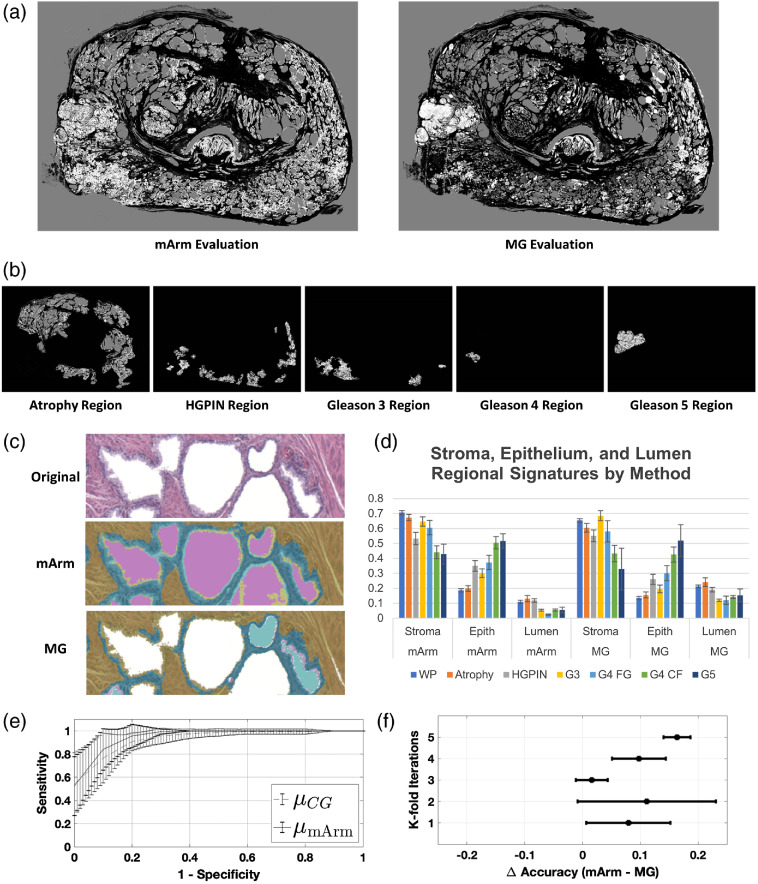
Figures 5(a) and 5(b) show a comparison of the mArm labeling and MG labeling for a prostate that was determined by pathologist to contain examples of atrophy, HGPIN, and Gleason 3 to 5 regions. Figure 5(c) show pathologist’s annotated regions with mArm-generated SEL labels. These regions are a visual depiction of the regional SEL “signatures” derived from mArm and MG methods [Fig. 5(d)]. Similar to results found in experiment 1, when regional standard deviations were compared between the two methods, mArm labeling was found to be less variable than MG labeling (; paired student’s -test).
Fig. 5.
Application of morphological operations and combined SEL segmentation algorithms on pathologist annotated whole mount prostate. (a) By combining all three classifiers (arms 1, 2, and 3)—mArm—pixels from whole mount prostate images were labeled as either SEL. A comparison of the mArm labeling and MG labeling is shown for a prostate that was determined to contain examples of atrophy, HGPIN, and Gleason 3 to 5 regions. (b) Pathologist annotated regions corresponding to each of the regional labels are shown with associated SEL mArm labeling. (c) An example classification of a magnified region within a whole mount slide. Stroma, yellow; epithelium, blue; lumen, pink. Notice improved epithelial segmentation with mArm method over MG method. (d) Using 32 pixel wise annotated whole mount prostate sections; SEL regional signatures were generated for both mArm and MG labeling methods. Each signature is defined by the regional percent makeup of SEL, which all sum to one. Mean and standard error are plotted. Using a paired Student’s t-test, the set of standard deviations from assessed mArm regions was found to be significantly less than () the set of standard deviations from the same regions assessed by MG. (e) An SVM was trained to separate benign (atrophy and HGPIN lesions) from cancerous (G3+ lesions) using the three-feature SEL signatures generated from the mArm and MG classifier. Three-fold cross validation with five repetitions was performed, the accuracy of mArm SEL labels was found to be for the mArm features versus () for the MG features, supporting use of SEL labels as features for future classification methods (). (f) Individual train/test cohort data showing difference in accuracy plotted between each paired SVM.
In order to translate the impact of our observed improvement in SEL classification into the clinical application of cancer detection, we next used a supervised learning technique for the classification of benign (atrophy and HGPIN) versus cancerous (Gleason scores 3+) on predefined regions of whole-mount prostates. We then compared the supervised learning method using two datasets: the SEL classification from the new method presented here and the morphological SEL classification. Region signatures, or observations, were generated using all pixels of a given label (e.g., G3) in a single whole-mount prostate. These signatures described the percent contribution of SEL for a given region.
In order to control for potentially confounding effects of the training dataset, we used a paired repeated -fold cross validation approach using three folds and five iterations. This was paired because each of the classifiers was trained on the same set of observations with the only difference being the origin of the labels (mArm or MG). The 62 observations of benign and Gleason 3+ pattern regions were subclassified as benign (atrophy and HGPIN | ) versus cancer (Gleason ) and repeated for five iterations of randomly sampled cohorts for training/testing (66/33 split). Two separate SVMs were trained for each pair of labels (mArm and MG) within the randomly sampled datasets. The resultant comparative groups, therefore, consisted of identical images each with two sets of SEL scores, one from the new method and one from the morphological SEL classification. The accuracy resulting from these groups was then compared. Average ROCs were generated for all train/test cohorts of the SVMs and plotted in Fig. 5(e). Using a nonparametric Mann–Whitney U-test across all paired train/test splits, mArm accuracy was shown to be significantly higher at versus the MG accuracy of () (). Corresponding delta accuracies between mArm and MG within the paired datasets are shown in Fig. 5(f).
4. Discussion
Using histological preparations of prostate tissue from multiple institutions, we have described a practical method for using transfer learning combined with both high-quality (human annotation) and low-quality (heuristically generated) ground truth labels to train a semantic segmentation algorithm. In addition, we have presented a well characterized and robust pixelwise classification method for labeling H&E-stained prostate tissue into SEL classes. Finally, we have shown demonstrable improvement in the classification of benign versus cancerous regions in the context of whole-mount prostate tissue using region of interest signatures generated from our improved methods, against the morphological model used in training.
Segmentation algorithms based on morphological heuristics have a long history in image processing pipelines.27 They have been used to varying degrees of success, with the simplest of algorithms often designed around hard coded intensity values for one-off applications. While this may have limited their application to the niche cases they were designed for, our study provides evidence that implementation of deep learning frameworks using one of these previously described operations may add robustness for segmentation tasks.
Within this study, we have demonstrated two forms of this increased robustness. First is separating the lumen into a separate class. While it is a trivial task to classify nontissue regions of a slide as lumen, this method is not robust against tissue artifacts such as tearing. However, we see improved segmentation performance of the lumen in the combined method (Arm3) when comparing all methods to human annotations. In addition, using training data obtained solely through the morphological operations that generated the clearly mislabeled image in Fig. 4(b), a deep learning architecture distills the salient features and returns an algorithm that much more closely matches the human observer annotations. This is further demonstrated in our final experiment, which shows improved benign versus cancer discrimination using the refined features compared to the original morphological features.
Observation-hungry machine learning methods show tremendous promise in image analysis and interpretation in rad-path applications.21 These algorithms are in large part hampered only by limitations in available training data. We sought practical ways to bridge this gap of annotated data by examining the use of weakly supervised data in a histological dataset. The benefit of this method is that heuristic algorithms may be used to generate larger training datasets. A small dataset from a classically trained observer can then serve as a fine-tuning step in training and final test dataset. This study provides evidence, or at a minimum impetus, for applying “naïve observer” heuristic algorithms alongside the valuable subject matter expert when training deep learning methods.
While our proposed training approach is targeted at mitigating the decreased availability of high-quality human labels, we recognize that a shortcoming of this study is our modest dataset. Fittingly, in the third experiment of our study we encountered the same problem that our study was designed to address – scarcity of strongly labeled data. Our dataset lacked ground truth SEL labels for our whole-mount slides. As a surrogate for direct SEL labels, we applied our trained algorithm to the more clinically relevant question of discriminating between benign and cancerous regions in whole mount tissue. We do not claim that the 62 regions presented in experiment three of this paper form a near-perfect representation of the true distribution of PCa histology or that unguided use of tissue signatures will solve PCa segmentation. However, the demonstrated improvement in cancer classification using features from mArm does suggest that our proposed labeling enhances a signal that is relevant to cancer classification over the previous morphological method. In addition, we believe this further emphasizes the need for approaches that circumvent limited amounts of labeled data.
We envision several use cases and future studies for our characterized algorithm. Most directly, we could see the algorithm being incorporated in-line with a region proposal algorithm, or as a “second opinion” in a computer-aided diagnostic workflow where a trained observer annotates suspicious regions. In addition, this algorithm could be used in the generation of higher level, human interpretable, metrics such as epithelial thickness and tortuosity that may more closely capture the patterns of Gleason’s original criteria. While these future studies may still rely on a pathologist’s annotated ground truth, we see it as an important piece in the way forward to fully automated cancer detection algorithms.
In conclusion, this study provides a robust algorithm for SEL segmentation in bright field H&E-stained prostate histology. We demonstrated a practical application of weak supervision to bolster a smaller dataset of high-quality domain expert annotation for repurposing a pretrained deep learning network. The performance of this network improved when fine-tuned with fewer, and more precious, high quality expert annotated samples. This ultimately demonstrates that using a small set of human annotated histology, when combined with a much larger dataset of heuristically derived segmented histology, can improve classification above the same network trained with either dataset alone. This prompts a revisitation of the field’s bespoke segmentation algorithms and their adaption to deep learning pipelines.
Acknowledgments
We would like to thank the patients for participating in this study and Mellissa Hollister for clinical coordination efforts. This research was completed in part with computational resources and technical support provided by the Research Computing Center at the Medical College of Wisconsin. Funding was provided by the State of Wisconsin Tax Check-off Program for Prostate Cancer Research (RO1CA218144 and R01CA113580) and the National Center for Advancing Translational Sciences (NIH UL1TR001436 and TL1TR001437).
Biography
Biographies of the authors are not available.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
Contributor Information
John D. Bukowy, Email: bukowy@msoe.edu.
Halle Foss, Email: hfoss@mcw.edu.
Sean D. McGarry, Email: smcgarry@mcw.edu.
Allison K. Lowman, Email: alowman@mcw.edu.
Sarah L. Hurrell, Email: shurrell15@jcu.edu.
Kenneth A. Iczkowski, Email: kaiczkowski@mcw.edu.
Anjishnu Banerjee, Email: abanerjee@mcw.edu.
Samuel A. Bobholz, Email: sbobholz@mcw.edu.
Alexander Barrington, Email: abarrington@mcw.edu.
Alex Dayton, Email: alexander.r.dayton@gmail.com.
Jackson Unteriner, Email: junteriner@mcw.edu.
Kenneth Jacobsohn, Email: kjacobsohn@mcw.edu.
William A. See, Email: wsee@mcw.edu.
Marja T. Nevalainen, Email: mnevalainen@mcw.edu.
Andrew S. Nencka, Email: anencka@mcw.edu.
Tyler Ethridge, Email: tetheridge@wisc.edu.
David F. Jarrard, Email: jarrard@urology.wisc.edu.
Peter S. LaViolette, Email: plaviole@mcw.edu.
References
- 1.Siegel R. L., Miller K. D., Jemal A., “Cancer statistics, 2016,” CA Cancer J. Clin. 66, 7–30 (2016). 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Delahunt B., Srigley J. R., Lamb D. S., “Gleason grading: consensus and controversy,” Pathology 41(7), 613–614 (2009). 10.3109/00313020903308823 [DOI] [PubMed] [Google Scholar]
- 3.Gleason D., “Classification of prostatic carcinomas,” Cancer Chemother. Rep. 50, 125 (1966). [PubMed] [Google Scholar]
- 4.Epstein J. I., “An update of the Gleason grading system,” J. Urol. 183, 433–440 (2010). 10.1016/j.juro.2009.10.046 [DOI] [PubMed] [Google Scholar]
- 5.Epstein J. I., et al. , “A contemporary prostate cancer grading system: a validated alternative to the Gleason score,” Eur. Urol. 69, 428–435 (2016). 10.1016/j.eururo.2015.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nir G., et al. , “Automatic grading of prostate cancer in digitized histopathology images: learning from multiple experts,” Med. Image Anal. 50, 167–180 (2018). 10.1016/j.media.2018.09.005 [DOI] [PubMed] [Google Scholar]
- 7.Gertych A., et al. , “Machine learning approaches to analyze histological images of tissues from radical prostatectomies,” Comput. Med. Imaging Graphics 46, 197–208 (2015). 10.1016/j.compmedimag.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naik S., et al. , “Gland segmentation and computerized Gleason grading of prostate histology by integrating low-, high-level and domain specific information,” in MIAAB Workshop, Citeseer, pp. 1–8 (2007). [Google Scholar]
- 9.Bulten W., et al. , “Automated segmentation of epithelial tissue in prostatectomy slides using deep learning,” Proc. SPIE 10581, 105810S (2018). 10.1117/12.2292872 [DOI] [Google Scholar]
- 10.Serag A., et al. , “A multi-level deep learning algorithm to estimate tumor content and cellularity of prostate cancer” (2018).
- 11.Xu J., et al. , “A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images,” Neurocomputing 191, 214–223 (2016). 10.1016/j.neucom.2016.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long J., Shelhamer E., Darrell T., “Fully convolutional networks for semantic segmentation,” in Proc. IEEE Conf. Comput. Vision and Pattern Recognit., pp. 3431–3440 (2015). 10.1109/CVPR.2015.7298965 [DOI] [PubMed] [Google Scholar]
- 13.Ronneberger O., Fischer P., Brox T., “U-net: convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. 9351, 234–241 (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 14.Halevy A., Norvig P., Pereira F., “The unreasonable effectiveness of data,” IEEE Intell. Syst. 24, 8–12 (2009). 10.1109/MIS.2009.36 [DOI] [Google Scholar]
- 15.Sun C., et al. , “Revisiting unreasonable effectiveness of data in deep learning era,” in IEEE Int. Conf. Comput. Vision, IEEE, pp. 843–852 (2017). 10.1109/ICCV.2017.97 [DOI] [Google Scholar]
- 16.Guo Y., et al. , “Deep learning for visual understanding: a review,” Neurocomputing 187, 27–48 (2016). 10.1016/j.neucom.2015.09.116 [DOI] [Google Scholar]
- 17.Ratner A., et al. , “Snorkel: rapid training data creation with weak supervision,” Proc. VLDB Endowment 11, 269–282 (2017). 10.14778/3157794.3157797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campanella G., et al. , “Clinical-grade computational pathology using weakly supervised deep learning on whole slide images,” Nat. Med. 25, 1301–1309 (2019). 10.1038/s41591-019-0508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badrinarayanan V., Kendall A., Cipolla R., “SegNet: a deep convolutional encoder-decoder architecture for image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017). 10.1109/TPAMI.2016.2644615 [DOI] [PubMed] [Google Scholar]
- 20.Hurrell S. L., et al. , “Optimized b-value selection for the discrimination of prostate cancer grades, including the cribriform pattern, using diffusion weighted imaging,” J. Med. Imaging 5, 011004 (2018). 10.1117/1.JMI.5.1.011004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGarry S. D., et al. , “Radio-pathomic maps of epithelium and lumen density predict the location of high-grade prostate cancer,” Int. J. Radiat. Oncol. Biol. Phys. 101(5), 1179–1187 (2018). 10.1016/j.ijrobp.2018.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macenko M., et al. , “A method for normalizing histology slides for quantitative analysis,” IEEE Int. Symp. Biomed. Imaging: From Nano to Macro, IEEE, pp. 1107–1110 (2009). 10.1109/ISBI.2009.5193250 [DOI] [Google Scholar]
- 23.Khan A. M., et al. , “A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution,” IEEE Trans. Biomed. Eng. 61, 1729–1738 (2014). 10.1109/TBME.2014.2303294 [DOI] [PubMed] [Google Scholar]
- 24.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” arXiv:14091556 (2014).
- 25.Dice L. R., “Measures of the amount of ecologic association between species,” Ecology 26, 297–302 (1945). 10.2307/1932409 [DOI] [Google Scholar]
- 26.Csurka G., et al. , “What is a good evaluation measure for semantic segmentation?” in Proc. Br. Mach. Vision Conf., pp. 32.1–32.11 (2013). [Google Scholar]
- 27.Gurcan M. N., et al. , “Histopathological image analysis: a review,” IEEE Rev. Biomed. Eng. 2, 147 (2009). 10.1109/RBME.2009.2034865 [DOI] [PMC free article] [PubMed] [Google Scholar]