Abstract
We used social network analysis (SNA) to study the novel coronavirus (COVID-19) outbreak in Karnataka, India, and to assess the potential of SNA as a tool for outbreak monitoring and control. We analysed contact tracing data of 1147 COVID-19 positive cases (mean age 34.91 years, 61.99% aged 11–40, 742 males), anonymised and made public by the Karnataka government. Software tools, Cytoscape and Gephi, were used to create SNA graphics and determine network attributes of nodes (cases) and edges (directed links from source to target patients). Outdegree was 1–47 for 199 (17.35%) nodes, and betweenness, 0.5–87 for 89 (7.76%) nodes. Men had higher mean outdegree and women, higher mean betweenness. Delhi was the exogenous source of 17.44% cases. Bangalore city had the highest caseload in the state (229, 20%), but comparatively low cluster formation. Thirty-four (2.96%) ‘super-spreaders’ (outdegree ⩾ 5) caused 60% of the transmissions. Real-time social network visualisation can allow healthcare administrators to flag evolving hotspots and pinpoint key actors in transmission. Prioritising these areas and individuals for rigorous containment could help minimise resource outlay and potentially achieve a significant reduction in COVID-19 transmission.
Key words: Analysis of data, contact tracing, COVID-19, infectious disease epidemiology, social network analysis
Introduction
The novel coronavirus (COVID-19) outbreak has attained the proportions of a global calamity. Not only has the virus sickened millions, it has also affected economic growth worldwide. Researchers have pointed to overpopulation, globalisation, and hyper-connectivity as factors responsible for intensifying the spread of infection, turning the outbreak into a pandemic [1]. The first case of coronavirus disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in India was reported on 30 January 2020, and the first case in Karnataka, a southern state of India, was reported on 9 March 2020 [2]. In the initial phases of the infection's spread through the country, Karnataka reported fewer cases than other Indian states. It was among the earliest states to deploy modern technology tools as part of its logistics and containment measures [3, 4]. As of 17 May 2020, Karnataka had declared 1147 diagnosed cases and 18 648 individuals under observation [5]. Among the 1147, there were 600 active cases, 509 who had recovered and 37 who died due to COVID (a fatality rate of 3.2%); one person died by suicide after being diagnosed.
Researchers from several countries have used mathematical modelling to predict the transmission of COVID-19 [6–8] and to identify predictors of mortality [9, 10]. However, social network analysis (SNA) has not, thus far, been optimally utilised in the endeavour to understand the characteristics of this disease.
SNA is a technique to study the configurations of social relations between individuals or other social units. Social network models can be used to measure variables that shape relationships between social actors and the extent to which they affect health-related outcomes [11, 12]. Researchers are exploring the use of SNA to study various facets of the COVID-19 pandemic, such as the role of public figures in communication [13], and clustering patterns within the broader patient network [14].
Since the pandemic is imposing a considerable burden on healthcare delivery systems, any solution that can potentially aid in controlling its spread deserves serious exploration. One such approach was the Karnataka healthcare task force's extensive use of contact tracing. We expanded on this approach by applying SNA to the corpus of contact tracing data generated by the task force's efforts. We had two main research questions in mind. First, can SNA improve our understanding of the transmission patterns of SARS-CoV-2? Second, can SNA produce actionable findings that can help in timely control of the spread of this disease?
Methods
Study area
Karnataka is a southern state of India, consisting of 30 administrative districts with a population of over 60 million, accounting for approximately 5% of the total Indian population. Bangalore, a densely populated metropolis, is the capital city of Karnataka [15].
Data source
The government of Karnataka initiated measures to control the spread of COVID-19 in early February 2020 [16]. A government appointed task force formulated guidelines for quarantine and contact tracing. Field workers, trained to elicit travel and contact history, carried out telephonic and house-to-house surveys to identify primary and secondary contacts of positive cases. On average, 47.4 contacts were tested for each confirmed case [17]. Data collected at the community level was collated by the State War Room. Daily consolidated bulletins, containing anonymised patient and contact data, were uploaded by the government to the portal it created to share information on COVID-19 [5].
As a part of its effort to contain the outbreak, the Karnataka government implemented a phased lockdown, closing shops and offices, and shutting down interdistrict and interstate travel. Phase 1 of the lockdown, with the most stringent curbs on travel and socialisation, was from 24 March to 14 April. The second phase was from 15 April to 3 May, and the third phase was from 4 May to 17 May.
Study design
For our analysis, we downloaded the daily bulletins containing information for all cases reported positive for COVID-19 from 9 March to 17 May 2020, spanning the period from detection of the first case in the state to the end of phase-3 of the preventive lockdown. We extracted relevant demographic and contact data from the bulletins and created a dataset consisting of anonymised data of 1147 COVID-19 patients. We tabulated and summarised demographic details such as age, district of residence and history of travel, using Microsoft Excel. We created a nodes and edges datasheet in Excel, with each node representing a patient and each edge, a confirmed link or contact between a source and a target patient. We imported this dataset into Gephi version 0.9.2 and applied the following sequence of layout algorithms: Yifan Hu Proportional, Fruchterman Reingold and No Overlap, to achieve a visual representation in which the more connected nodes are placed centrally, and ones with lower connectivity are placed towards the periphery of the network [18].
We wanted to combine the capabilities of two of the leading SNA software tools [19], Gephi and Cytoscape, and utilise the features missing in one but available in the other. The use of Gephi's network analysis tools results in the nodes and edges datasheet being populated with additional attribute variables. These values, such as node betweenness and edge betweenness, can then be displayed as visual features of the network elements in other software tools such as Cytoscape [20]. We reformatted the data exported from Gephi to make it compatible with the data model acceptable to Cytoscape version 3.8.0, which we used to create network graphics highlighting pertinent demographic characteristics of the nodes. Layout algorithms provided in Cytoscape were applied in the following sequence: Compound Spring Embedder (CoSE) and yFiles Remove Overlap, followed by a few manual adjustments to improve clarity.
We analysed the network attributes generated by Gephi using MS Excel to explain relevant aspects of the network and its components. We discussed the characteristics and evolution of the graphs and attempted to explain them in the context of facts and events on the ground.
Ethical considerations
We have used anonymised, secondary data in the public domain, available at the COVID-19 information portal of the Karnataka state government, the copyright policy of which indicates that material featured on the site may be reproduced free of charge in any format or media without requiring specific permission, subject to acknowledgement of the source [5]. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees, and with the Helsinki Declaration of 1975, as revised in 2008.
Important definitions
(Also displayed in Supplementary Table S1):
Degree centrality is a measure of the number of social connections or links that a node has. It is expressed as an integer or count [21].
The indegree of a node is the number of incoming links to it from source nodes and refers to the number of infectious patients who had confirmed contact with a given target patient.
Outdegree is the number of links to target nodes from a source node and is a measure of the number of secondary cases infected by a given patient. The direction of the links is denoted by arrowheads at the target ends, in our visual representations.
Betweenness centrality is a measure of the number of times a node appears on the shortest path between other nodes [22]. It reflects the role a patient plays in creating a bridge of infectious transmission between patients who would not have had direct contact with each other.
Closeness centrality is the average of the shortest path lengths from a node to every other node in the network. It is calculated as the inverted sum of the distances from the node to all other nodes [23].
We used harmonic closeness to measure closeness centrality, due to the presence of unconnected nodes in our network. It is calculated as the sum of the inverted distances from a node to all other nodes, instead of the reciprocal of the sum of all distances [23].
Edge betweenness is the number of the shortest paths that go through an edge in a graph or network, with a high score indicative of a bridge-like connection between two parts of a network, crucial to transmission between many pairs of nodes [24].
Clustering coefficient measures the degree to which nodes in a graph tend to cluster together [25].
Network density is the number of existing ties between nodes, divided by the number of possible ties [26].
Network diameter is the shortest path between the two most distant nodes in a network [25].
Mean path length is the average of the shortest path lengths between all possible node pairs [25].
Network component is an island of interlinked nodes that are disconnected from other nodes of the network. Many networks consist of one large component, sometimes together with several smaller ones and singleton actors [12].
Super-spreader (operational definition): Any node with an outdegree ⩾5 was considered a super-spreader. Individuals represented by these nodes would have infected five or more contacts.
Results
Demography
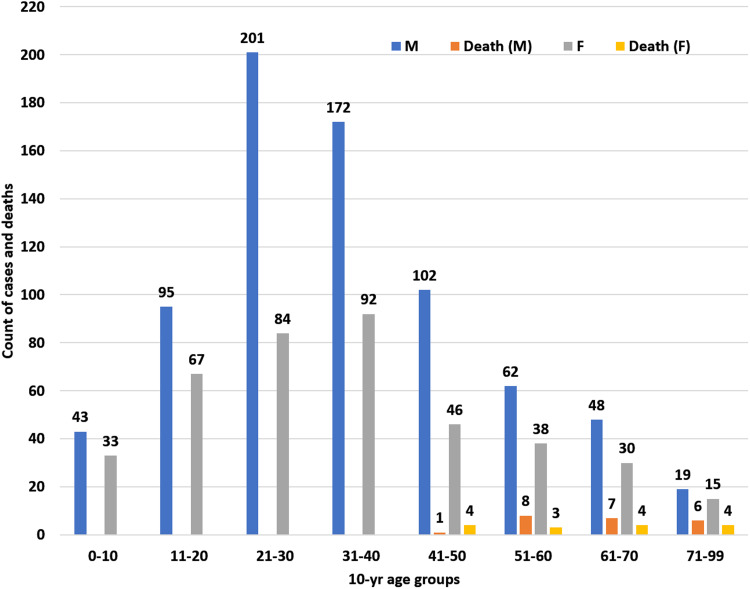
We analysed 1147 patients of whom 742 (64.69%) were males, aged 34.91 years on average (standard deviation 17.34 years) (Fig. 1). Most of these individuals (711/1147; 61.99%) belonged to the 11–40 years age range. Most deaths, however, occurred among older patients. We observed maximum mortality in patients aged over 70 years. There were 34 patients in this age-group, of whom 10 (29.41%) had died. Further socio-demographical details of these patients are available in the public domain [27].
Fig. 1.
Age–sex distribution of cases and deaths. Case counts are inclusive of deaths. Death counts are also shown separately.
Network parameters
We found 948 nodes with zero outdegree. The remaining 199 (17.35%) nodes had an outdegree range of 1–47 and were the source of infection to 657 targets through 706 links (edges). Among the target nodes, 36 had indegree >1 (range 2–5), implying more than one source. We noted equal means, but widely differing standard deviations for outdegree and indegree centralities. This difference is due to the wider range of outdegree compared to indegree (Table 1).
Table 1.
Network parameters
| Network | C1 | C2 | ||
|---|---|---|---|---|
| Nodes | 1147 | 75 | 45 | |
| Edges | 706 | 76 | 50 | |
| Node attributes | Range | Mean (s.d.) | Range (mean) | |
| Outdegree | 0–47 | 0.616 (2.78) | 0–47 (1.013) | 0–15 (1.111) |
| Indegree | 0–5 | 0.616 (0.60) | 0–2 (1.013) | 0–3 (1.111) |
| Degree | 0–47 | 1.231 (2.79) | 1–47 (2.027) | 1–15 (2.222) |
| Betweenness | 0–87 | 0.469 (3.69) | 0–9 (0.426) | 0–12 (1.133) |
| Harmonic closeness | 1 (n = 143) | 0.161 (0.35) | 0–1 (0.231) | 0–1 (0.263) |
| 0 < HC < 1 (n = 56) | ||||
| 0 (n = 949) | ||||
| Eccentricity | 0–4 | 0.242 (0.61) | 0–3 (0.307) | 0–3 (0.4) |
| Network attributes | Value | |||
| Diameter | 4 | 3 | 3 | |
| Radius | 1 | 1 | 1 | |
| Mean path length | 1.623 | 1.321 | 1.314 | |
| Mean no. of neighbours | 1.231 | 2.027 | 2.222 | |
| Density | 0.001 | 0.025 | 0.014 | |
| Clustering coefficient | 0.004 | 0.011 | 0.000 | |
C1, largest component; C2, second-largest component; s.d., standard deviation; HC, harmonic closeness.
There were 490 nodes with zero indegree, of which 383 had zero outdegree. The latter were isolated nodes, with degree centrality value of zero.
The range of betweenness centrality was 0.5–87 for 89 (7.76%) nodes. The network had 143 nodes with a harmonic closeness centrality (HCC) of one, and 56 nodes with HCC between zero and one. Our network density was 0.001, network diameter was 4 and the clustering coefficient was 0.004.
Table 2 shows that men had a higher mean outdegree (0.628, M vs. 0.593, F) and women, higher mean betweenness (0.573, F vs. 0.412, M).
Table 2.
Mean outdegree and betweenness by sex and age group
| Age groups | Mean outdegree | Mean betweenness | ||||
|---|---|---|---|---|---|---|
| Women | Men | Combined | Women | Men | Combined | |
| 0–10 | 0.000 | 0.140 | 0.079 | 0.000 | 0.233 | 0.132 |
| 11–20 | 0.463 | 0.368 | 0.407 | 0.179 | 0.242 | 0.216 |
| 21–30 | 0.655 | 0.289 | 0.396 | 1.561 | 0.095 | 0.527 |
| 31–40 | 0.500 | 0.994 | 0.822 | 0.114 | 1.159 | 0.795 |
| 41–50 | 0.870 | 0.480 | 0.601 | 0.783 | 0.216 | 0.392 |
| 51–60 | 1.158 | 1.145 | 1.150 | 0.474 | 0.210 | 0.310 |
| 61–70 | 0.467 | 1.083 | 0.846 | 0.033 | 0.406 | 0.263 |
| 71–99 | 0.667 | 1.263 | 1.000 | 1.572 | 0.000 | 0.694 |
| Combined | 0.593 | 0.628 | 0.616 | 0.573 | 0.412 | 0.469 |
The 95th percentile cut-off values for outdegree and betweenness were 3 and 2, respectively. There were 77 (6.71% of 1147) nodes with outdegree ⩾3, and combined, they accounted for 556 (78.75% of 706) edges. More than two-thirds of these 77 nodes were men (54, 70.13%). The average HCC for the 77 nodes with outdegree ⩾3 was 0.887, compared to 0.161 for the entire network. Of the 59 nodes with betweenness ⩾2, more than half (33, 55.93%) were men, though women had a higher mean betweenness overall.
We noted 34 super-spreaders with outdegree ⩾5, with a cumulative outdegree of 410, and after deducting 17 duplicate edges for target nodes with indegree >1, they accounted for 393 (59.81%) of the 657 target cases.
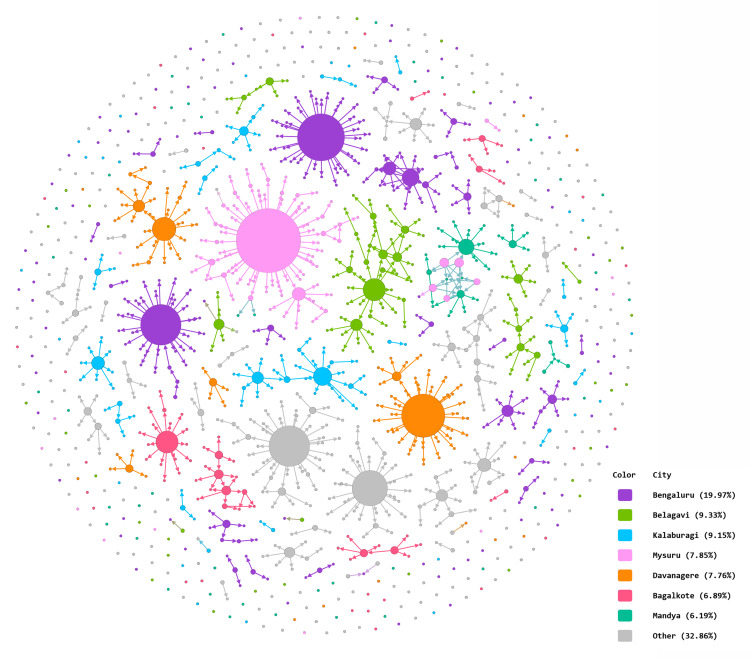
The aggregate network graphic (Fig. 2), created using Gephi, shows nodes representing patients, and components representing case clusters. The nodes are coloured according to district and sized by outdegree, making the larger nodes represent individuals who infected a greater number of targets. The largest node, located at the centre of a major component, denotes a patient from Mysuru district who infected 47 target nodes. Transmission between districts was limited, occurring chiefly from Mysuru to Mandya, a geographically adjacent district. The network figure also has two large-sized grey nodes that represent two patients with outdegree 29 and 25, from districts Vijayapura and Uttara Kannada, respectively. Bangalore had the highest number of cases, followed by Belagavi, Kalaburagi and Mysuru districts (Supplementary Table S2).
Fig. 2.
Aggregate network graphic created in Gephi. Arrowheads indicate direction of transmission from source node to target node. Node size determined by outdegree. Edges inherit colour from parent nodes.
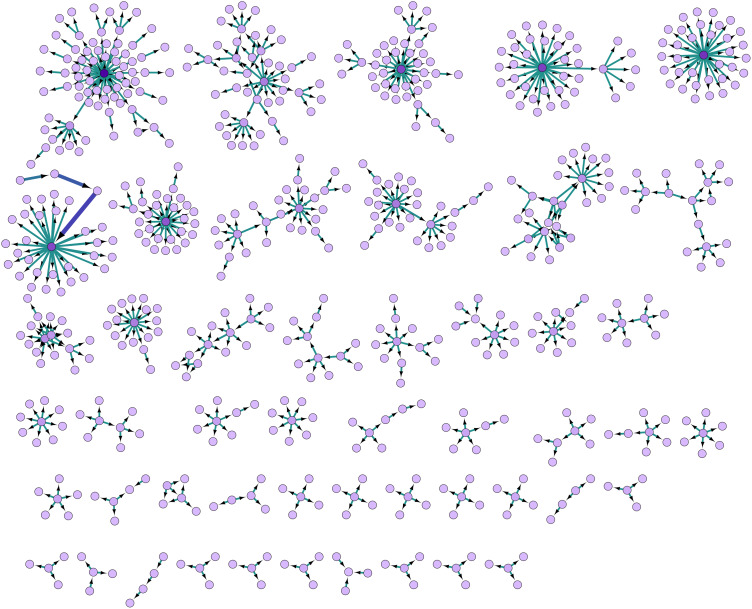
The aggregate network contained 93 clusters of connected nodes (components), of which 37 components, made up of five or more nodes each, had more than half of all the nodes (613, 53.44%) and four-fifths of the edges (611, 86.54%) concentrated within them (Fig. 3). The distribution of these clusters by district and type of origin is shown in Supplementary Table S3.
Fig. 3.
Major network components organised by size, created in Cytoscape. Arrowheads indicate direction of transmission from source node to target node. Edge betweenness determines the thickness and colour intensity of the edges.
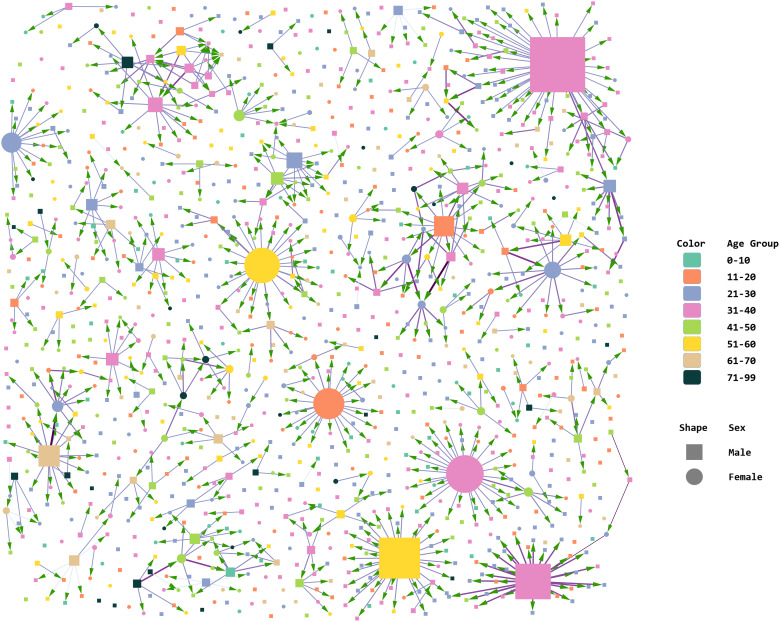
Figure 4 shows nodes coloured by age group and sized by outdegree. Figure 5 shows nodes coloured by source of infection and sized by betweenness centrality. We have considered patients with a history of travel from Delhi in a separate category as their count was comparable to the combined number of travellers from all other states of India. It is noteworthy that travellers from abroad did not contribute to the formation of any major cluster.
Fig. 4.
Age–sex attributes of nodes and clusters, created in Cytoscape. Node size determined by outdegree. Arrowheads indicate direction of transmission from source node to target node. Edge betweenness determines the thickness and colour intensity of the edges.
Fig. 5.
Network analysis by sources of infection (Cytoscape). Node size determined by betweenness. Arrowheads indicate direction of transmission from source node to target node. Edge betweenness determines the thickness and colour intensity of the edges.
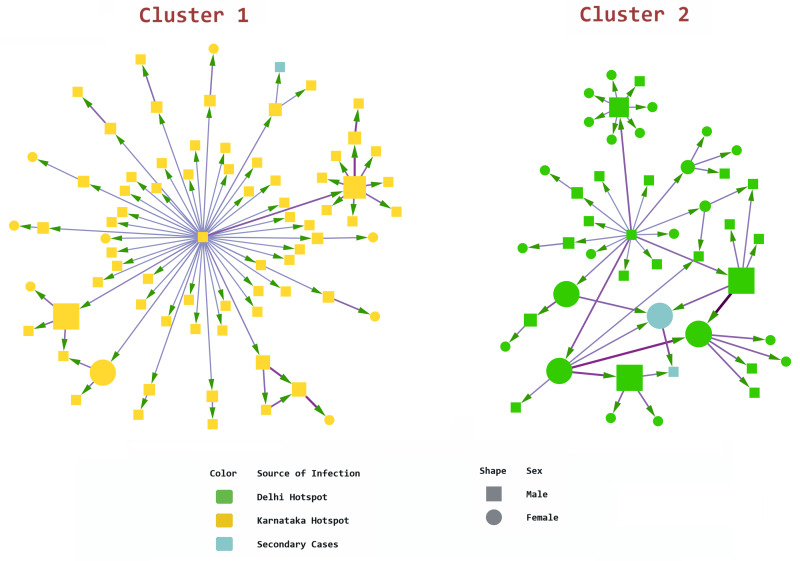
Comparing Figures 4 (nodes sized by outdegree) and 5 (nodes sized by betweenness), we find that in clusters with nodes that had multiple interconnections, relatively low outdegree and high betweenness, the key nodes were females. This indicates that women played a significant bridging role in these clusters. This differs from clusters with edges radiating from a central node with high outdegree and low betweenness, where typically, a young male was the nidus. The largest and second-largest components illustrate this difference in transmission (Fig. 6). The largest component had 75 nodes and 76 edges, and the second-largest component had 45 nodes and 50 edges. The largest cluster originated in the district of Mysuru; its source node was a male with high outdegree who spread the infection to many contacts. However, secondary transmission from those contacts was limited. This cluster is star shaped. The second-largest component resembles a spiderweb with multiple interconnected nodes and many female actors. This cluster was in Belagavi, and its network density was nearly twice that of the largest cluster (0.025 vs. 0.014), with a shorter average path length (1.314 vs. 1.321).
Fig. 6.
Comparing the two largest components (Cytoscape). Node size determined by betweenness. Arrowheads indicate direction of transmission from source node to target node. Edge betweenness determines the thickness and colour intensity of the edges.
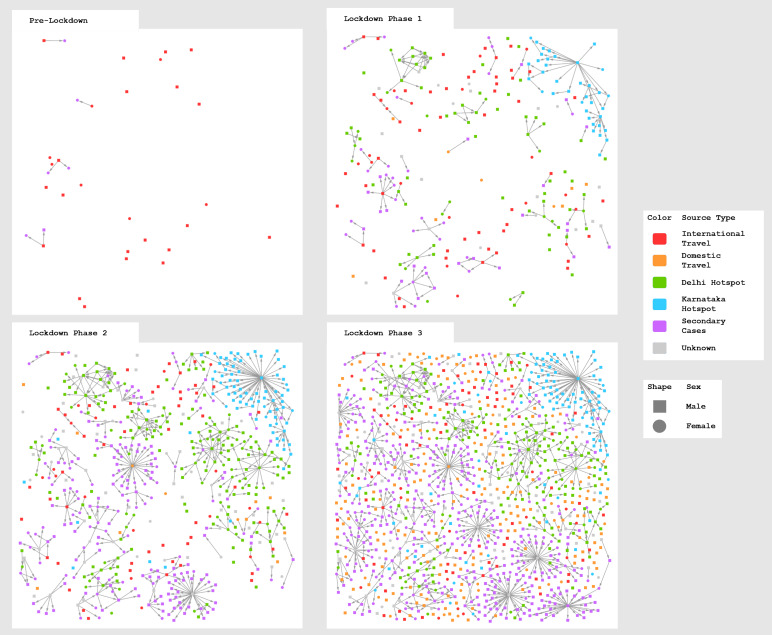
Dynamic evolution of the network
Figure 7 shows how the network began with the first detected cases and how it expanded in each phase of the lockdown. We see that in the initial, pre-lockdown phase, the cases were mostly isolated nodes with minimal occurrence of secondary cases. They are all travellers returning from abroad (red nodes, source type ‘International Travel’). By the time the first and strictest phase of the lockdown (24 March to 14 April) was declared, however, cluster formation had already begun and most of the new cases had a history of travel to Delhi or contact with returnees from Delhi (green nodes, source type ‘Delhi Hotspot’). The origin of the largest cluster, which was labelled as a Karnataka (in-state) hotspot (blue nodes), could not be traced either to travel or to contact with any known positive case. Clusters continued to form and grow during the second phase of the lockdown (15 April to 3 May). However, due to continuing curbs on travel and transportation, the fresh cases were mostly found among contacts of existing cases. In the beginning of May 2020, the government arranged special transportation facilities by road and rail, so that migrant labourers in distress could return to their home states. This resulted in many new cases with source traced to travel from out of state (orange nodes) in phase 3 of the lockdown (4 May to 17 May). We have shown the weekly increase in cases in the form of a line graph in Supplementary Figure S1.
Fig. 7.
Evolution of the network at each phase of lockdown. Node colour denotes infection source type. Arrowheads indicate direction of transmission from source node to target node.
Discussion
While we have performed a basic conventional analysis of the data, our chief objectives were to create social network graphics from the empirical contact tracing data, and derive insights into disease transmission therefrom.
Our study reveals that most cases of COVID-19 in Karnataka were young and middle-aged men. Deaths, however, occurred overwhelmingly among elderly patients. The age and sex profile of our study set matches nationwide surveillance data from India, with median age and age-distribution close to our sample, and a similar high attack rate in males [17].
Bangalore, the capital of Karnataka, is a densely populated metropolis, housing one-sixth of Karnataka's population in 1% of its area [15, 28]. The city airport is a major transit point for domestic and international travellers. These factors may explain Bangalore's relatively heavy burden of COVID-19 cases (229/1147). Despite accounting for nearly a fifth of the state's caseload, however, Bangalore did not have notably large or numerous clusters compared to other districts (Fig. 2 and Supplementary Tables S2 and S3). Most of the cases detected here were isolated nodes. Bangalore's low transmission may have been due to the disciplined observance of lockdown measures, and rigorous contact tracing and quarantine activities by its healthcare workforce [29, 30].
The presence of two large nodes (where size denotes outdegree) in districts that had a minor contribution to the total caseload (Fig. 2) points to the risk of cluster formation even in relatively unaffected areas if, for example, physical distancing measures are not followed scrupulously.
Shortly after the World Health Organization confirmed the novel coronavirus as the cause of the outbreak in China [31], public health authorities started precautionary screening and quarantine of passengers arriving from areas of concern at Bangalore International Airport [32]. These early steps may explain why we found no major clusters originating from international travellers. Conversely, we noted several clusters formed by people with a history of travel to the national capital, Delhi (Fig. 5 and Supplementary Tables S2 and S3). By 19 April 2020, the entire city of Delhi had been declared a COVID-19 hotspot [33] in the wake of a mass religious gathering that was found to be linked to nearly a third of the country's caseload earlier in the month [34]. Clusters of cases that originated from Delhi tended to be closely interconnected, with women playing an active transmission role. This could reflect close community ties between these individuals, or residence in underprivileged areas where strict physical distancing may not have been observed.
Most of the clusters in our network had a man with high outdegree as the nidus. Women, however, played an important role in transmission by bridging multiple nodes within clusters even though men outnumbered women in the 95th percentile region of betweenness. Further study is warranted into the social and behavioural characteristics of men and women that drive these differences.
The low density of our network, the presence of 948 nodes with zero outdegree, and the fact that only 34 source cases had infected close to two-thirds of all target cases, indicate that community transmission was negligible. A similar transmission pattern was reported from Shenzhen, China, where 8.9% of the cases had caused 80% of all infections [35]. Another recent analysis of detailed contact tracing data from Hunan, China, traced 80% of secondary cases back to 14% of infections [36]. Network analysis of COVID-19 patients in Henan, China, [14] revealed a non-uniform pattern of clustering (208/1105 patients in clusters) with a skewed distribution of patients in different cities. The Henan study also indicated a strong correlation of confirmed cases with travel to Wuhan (the epicentre of the pandemic), which corresponds to our observation that a fair proportion (17.44%) of the Karnataka patients had travelled to Delhi. These similarities indicate that our findings may be generalisable across populations.
Researchers have analysed network properties from different perspectives, depending on the type and complexity of networks. Entropy-based analysis has been used to identify influential nodes using local information dimensionality [37]. Fractal dimensions are being explored to determine the vulnerability of complex networks [38]. Mathematical modelling has been used to simulate and predict transmission dynamics in various types of networks [39]. These models are informed, and their predictions are influenced, by the types of data processing decisions that are made prior to collecting and analysing the contact data [40]. A dynamic simulation of this nature, such as was done by the Hunan researchers [36], would require data at a granular level, including educational, occupational and socioeconomic status of patients, their mobility patterns, severity of infection, and the duration and intensity of contact events. This information was unavailable in the anonymised secondary data that we used. The data available to us allowed only a limited dynamic analysis to be done.
Limitations
Our SNA findings may not universally reflect field realities. Some findings such as eccentricity and mean path length are theoretical constructs computed by software algorithms, but in practice, these metrics remain indeterminate as our network had very few inter-district connections and many isolated nodes and components. Our dataset included many patients with contact history still under investigation at the time of analysis. We were not able to analyse the role of type and duration of contact, as these data were unavailable for many patients. Although we have attempted to faithfully reproduce all the information that we could extract from the daily bulletins, the quality of our data is necessarily limited by the constraints of secondary data sources.
Conclusion
Our conventional analysis indicates that mortality due to COVID-19 is highest among senior citizens. We recommend that the elderly should be advised strict physical distancing, and older patients from rural or underserved areas should be pre-emptively transferred to tertiary centres with intensive care facilities. This may help in early detection and treatment of complications, mitigating their mortality risk.
The findings from our network analysis suggest that geographical, demographical and community characteristics could influence the spread of COVID-19. Gender influences the morphology of clusters, with men seeding the clusters and women propagating them.
Our results also highlight the need for recording, on an ongoing basis, high granularity contact tracing data in a uniform format. We believe that outbreak control task forces should be provided with requisite software and training in SNA techniques, and should directly receive contact tracing information from workers in the field. This would enable SNA in real time with the ability to visualise and flag evolving networks with alacrity. It would also help pinpoint nodes with high outdegree, betweenness and closeness scores, which imply an active role in the transmission and bridging of infection. Real-time SNA could thus help identify the super-spreaders responsible for a large proportion of transmission. In particular, close tracking of betweenness scores would allow detection of individuals who might be missed by conventional tracing methods. These actors may not themselves spread the infection to many contacts, but their bridging characteristic accelerates transmission in the community. Public health authorities could prioritise these individuals and clusters for immediate and rigorous containment, and formulate control measures tailored to the network characteristics of each locality. These measures could help minimise resource outlay, and potentially facilitate a significant reduction in the spread of COVID-19.
Acknowledgements
We thank the Government of Karnataka for their punctual and detailed bulletins summarising data of COVID-19 cases in the state. We are deeply grateful to the grassroots health workers who selflessly carried out the contact tracing surveys that made our analysis possible. We extend our sincere thanks to the reviewers and the editor for their valuable suggestions and support.
Author contributions
S. Saraswathi: conceptualisation, study design, data retrieval from government bulletins; A. Mukhopadhyay and H. Shah: data analysis; all authors: writing, editing, review and final approval of manuscript.
Financial support
We received no financial support for this study.
Conflict of interest
We have no conflict of interest to declare.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S095026882000223X.
click here to view supplementary material
Data availability statement
Our datasets were constructed using contact tracing details available in the daily bulletins released online by the Government of Karnataka. The complete archives of these bulletins can be accessed at the COVID-19 portal at the address https://covid19.karnataka.gov.in/govt_bulletin/en under the heading ‘Health Department Bulletins’.
References
- 1.Cheong KH and Jones MC (2020) Introducing the 21st century's New four horsemen of the coronapocalypse. BioEssays 42, 2000063. doi: 10.1002/bies.202000063. [DOI] [PubMed] [Google Scholar]
- 2.India Today. Coronavirus in India: Tracking country's first 50 COVID-19 cases; what numbers tell. https://www.indiatoday.in/india/story/coronavirus-in-india-tracking-country-s-first-50-covid-19-cases-what-numbers-tell-1654468-2020-03-12 (Accessed 26 August 2020).
- 3.The New Indian Express. Karnataka fares well in war against COVID-19. https://www.newindianexpress.com/states/karnataka/2020/apr/14/karnataka-fares-well-in-war-against-covid-19-2129751.html (Accessed 17 May 2020).
- 4.The Times of India. New tech solutions aiding Karnataka's battle against COVID-19. https://timesofindia.indiatimes.com/city/bengaluru/new-tech-solutions-aiding-karnatakas-battle-against-covid-19/articleshow/75782619.cms (Accessed 17 May 2020).
- 5.Health department bulletin – COVID-19 information portal. https://covid19.karnataka.gov.in/new-page/Health%20Department%20Bulletin/en (Accessed 18 May 2020).
- 6.Ndaïrou F et al. (2020) Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos, Solitons & Fractals 135, 109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang HM et al. (2020) Mathematical model describing CoViD-19 in São Paulo, Brazil – evaluating isolation as control mechanism and forecasting epidemiological scenarios of release. Epidemiology and Infection 148, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh A, Dey J and Bhardwaj S (2020) Is this the beginning or the end of COVID-19 outbreak in India? A data driven mathematical model-based analysis. medRxiv. 10.1101/2020.04.27.20081422. [DOI] [Google Scholar]
- 9.Ma X et al. (2020) Development and validation of prognosis model of mortality risk in patients with COVID-19. Epidemiology and Infection 148, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa GJB et al. (2020) Mortality and survival of COVID-19. Epidemiology and Infection 148, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman MEJ (2003) The structure and function of complex networks. SIAM Review 45, 167–256. [Google Scholar]
- 12.O'Malley AJ and Marsden PV (2008) The analysis of social networks. Health Services & Outcomes Research Methodology 8, 222–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yum S (2020) Social network analysis for coronavirus (COVID-19) in the United States. Social Science Quarterly 101, 1642–1647. doi: 10.1111/ssqu.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P et al. (2020) Statistical and network analysis of 1212 COVID-19 patients in Henan, China. International Journal of Infectious Diseases 95, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnataka Government. https://karnataka.gov.in/new-page/Karnataka%20at%20a%20Glance/en (Accessed 26 August 2020).
- 16.Government circulars – COVID-19 information portal. https://covid19.karnataka.gov.in/new-page/Government%20Circulars/en (Accessed 26 August 2020).
- 17.ICMR COVID Study Group et al. (2020) Laboratory surveillance for SARS-CoV-2 in India: performance of testing & descriptive epidemiology of detected COVID-19, January 22–April 30, 2020. Indian Journal of Medical Research 151, 424–437. doi: 10.4103/ijmr.IJMR_1896_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastian M, Heymann S and Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Web and Social Media; Third International AAAI Conference on Weblogs and Social Media Published online: 2009. https://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154 (Accessed 31 May 2020).
- 19.Bhatia R (2018) Top 7 Network Analysis Tools For Data Visualisation. Analytics India Magazine. https://analyticsindiamag.com/top-7-network-analysis-tools-for-data-visualisation/ (Accessed 18 May 2020).
- 20.Shannon P et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Degree Centrality – an overview, ScienceDirect Topics. https://www.sciencedirect.com/topics/computer-science/degree-centrality (Accessed 14 May 2020).
- 22.Betweenness Centrality – an overview, ScienceDirect Topics. https://www.sciencedirect.com/topics/computer-science/betweenness-centrality (Accessed 14 May 2020).
- 23.Rochat Y. Closeness centrality extended to unconnected graphs: the harmonic centrality index. https://core.ac.uk/download/pdf/148005918.pdf (Accessed 30 August 2020).
- 24.Girvan M and Newman MEJ (2002) Community structure in social and biological networks. Proceedings of the National Academy of Sciences of the United States of America 99, 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang F and Reilly C. Terminology – social network analysis. https://sites.google.com/a/umn.edu/social-network-analysis/terminology (Accessed 3 June 2020).
- 26.Hanneman RA and Riddle M. Introduction to social network methods: chapter 8: more properties of networks and actors. https://faculty.ucr.edu/~hanneman/nettext/C8_Embedding.html (Accessed 3 June 2020).
- 27.Karnataka COVID+ analysis: 17 May 2020. https://covid19.karnataka.gov.in/storage/pdf-files/GOK_Scorecard_V3%20(1).pdf (Accessed 28 August 2020).
- 28.Karnataka government. https://karnataka.gov.in/district/en (Accessed 26 August 2020).
- 29.The Times of India. Welcome indicator: Only 2% of COVID-19 beds occupied in Karnataka. https://timesofindia.indiatimes.com/city/bengaluru/welcome-indicator-only-2-of-covid-19-beds-occupied-in-karnataka/articleshow/75653647.cms (Accessed 2 June 2020).
- 30.News18 (2020) Trace, test and treat: armed with 3ts, how Bengaluru is on course to flattening the curve of COVID-19. https://www.news18.com/news/india/trace-test-and-treat-armed-with-3ts-how-bengaluru-is-on-course-to-flattening-the-curve-of-covid-19-2619117.html (Accessed 2 June 2020).
- 31.World Health Organization. WHO statement regarding cluster of pneumonia cases in Wuhan, China. https://www.who.int/china/news/detail/09-01-2020-who-statement-regarding-cluster-of-pneumonia-cases-in-wuhan-china (Accessed 3 June 2020).
- 32.The News Minute. Screening begins at Bengaluru airport over Coronavirus scare. https://www.thenewsminute.com/article/screening-begins-bengaluru-airport-over-coronavirus-scare-116502 (Accessed 3 June 2020).
- 33.Business Insider. No lockdown relaxation in Delhi as all 11 districts are coronavirus hotspots. https://www.businessinsider.in/india/news/no-lockdown-relaxation-in-delhi-as-all-11-districts-are-coronavirus-hotspots/articleshow/75232679.cms (Accessed 2 June 2020).
- 34.The Times of India. 1,445 cases linked to Tablighi Jamaat event; total cases rise to 4,281, death toll 111. https://timesofindia.indiatimes.com/india/1445-cases-linked-to-tablighi-jamaat-event-total-cases-rise-to-4067-death-toll-109/articleshow/75010939.cms (Accessed 2 June 2020).
- 35.Bi Q et al. (2020) Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. The Lancet Infectious Diseases 20, 911–919. doi: 10.1016/S1473-3099(20)30287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun K et al. (2020) Transmission heterogeneities, kinetics, and controllability of SARS-CoV-2. medRxiv. 10.1101/2020.08.09.20171132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen T and Deng Y (2020) Identification of influencers in complex networks by local information dimensionality. Information Sciences 512, 549–562. [Google Scholar]
- 38.Wen T, Song M and Jiang W (2018) Evaluating topological vulnerability based on fuzzy fractal dimension. International Journal of Fuzzy Systems 20, 1956–1967. [Google Scholar]
- 39.Wang C et al. (2017) A rumor spreading model based on information entropy. Scientific Reports 7, 9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson DE, et al. (2019) Transmission on empirical dynamic contact networks is influenced by data processing decisions. Epidemics 26, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S095026882000223X.
click here to view supplementary material
Data Availability Statement
Our datasets were constructed using contact tracing details available in the daily bulletins released online by the Government of Karnataka. The complete archives of these bulletins can be accessed at the COVID-19 portal at the address https://covid19.karnataka.gov.in/govt_bulletin/en under the heading ‘Health Department Bulletins’.