Abstract
The incorporation of non-canonical amino acids into proteins has emerged as a promising strategy to manipulate and study protein structure-function relationships with superior precision in vitro and in vivo. To date, fluorescent non-canonical amino acids (f-ncAA) have been successfully incorporated in proteins expressed in bacterial systems, Xenopus oocytes, and HEK-293T cells. Here, we describe the rational generation of a novel orthogonal aminoacyl-tRNA synthetase based on the E. coli tyrosine synthetase that is capable of encoding the f-ncAA tyr-coumarin in HEK-293T cells.
Keywords: Bioorganic chemistry, Bioinformatics, Proteins, Biochemistry, Molecular biology, Unnatural amino acids, Aminoacyl-tRNA synthetase, Coumarin, Fluorescence
Bioorganic Chemistry; Bioinformatics; Proteins; Biochemistry; Molecular Biology; Unnatural amino acids, aminoacyl-tRNA synthetase, coumarin, fluorescence.
1. Introduction
The emergence of chemical conjugation and genetic encoding techniques to label proteins with fluorescent probes has enabled significant advances in the mechanistic understanding of proteins in biochemical and cellular environments [1]. However, encoding large fluorescent proteins (e.g. GFP) as fusion protein products is experimentally straightforward but the relative size of the probes can alter the function and biology of the protein being studied. Alternatively, chemical conjugation of an expressed protein requires the labeling sites are solvent accessible and labeling of cytoplasmic sites often comes with significant background reactivity [2]. A possible solution to these issues is the use of genetic code expansion to introduce a relatively compact fluorescent side chain as a non-canonical amino acids directly into the target protein in a site-specific fashion [3]. Indeed, the rapid development of genetically encoded fluorophores as a non-canonical amino acids (ncAA) is emerging as a promising strategy to describe protein function under minimal perturbations in eukaryotic cells [3, 4]. The experimental strategy employs an orthogonal suppressor tRNA and an evolved aminoacyl tRNA synthetase (RS), in the current example based upon the tyrosine pair, which can be used to encode the ncAA at virtually any site in the reading frame of the target gene. This pair is orthogonal to the translation system employed, meaning that the evolved TyrRs cannot acylate endogenous Tyr-tRNA molecules and the suppressor tRNAtyr is minimally acylated by host cell synthetases. The orthogonal tRNA has the appropriate anticodon to suppress the nonsense codon, thus allowing for an introduced amber codon of target genes in both prokaryote and eukaryotic systems [3, 5, 6]. This approach has been successfully used for site-specific incorporation of f-ncAA into a soluble proteins in prokaryote cells (Dansyl and Hydroxycoumarin) [4, 7], membrane proteins expressed in Xenopus leavis oocytes (Bodipy and Anap) [8, 9], and proteins expressed in mammalian cells (Dansyl and Anap) [10, 11]. Hydroxycoumarin is notable because of its small size and high environmental sensitivity [4, 12, 13], however, no system yet exists for its incorporation via encoding in mammalian cells.
2. Results
The incorporation of a 7-hydroxycoumarin amino acid (TyrCoum) has been previously accomplished in E. coli using a Methanococcus jannaschii tyrosyl tRNA (MjtRNATyrCUA) and a tyrosyl-tRNA synthetase (MjTyrRS) [4]. To expand the possible applications for TyrCoum we sought to develop an tRNA/RS pair that is amenable to mammalian expression systems and thus started with the tyrosyl-tRNA synthetase from E. coli (EcTyrRS; PDBID 1X8X; Figure 1a) [14], in combination with a suitable tRNA from B. stearothermophilus (BstRNACUA) [6]. Evolved versions of this particular orthogonal pair allow for the efficient incorporation of a variety or aromatic ncAA, including p-benzoyl-L-phenylalanine (Bzp) and acetylphenylalanine (Azi) moieties in HEK-293T cells [15, 16].
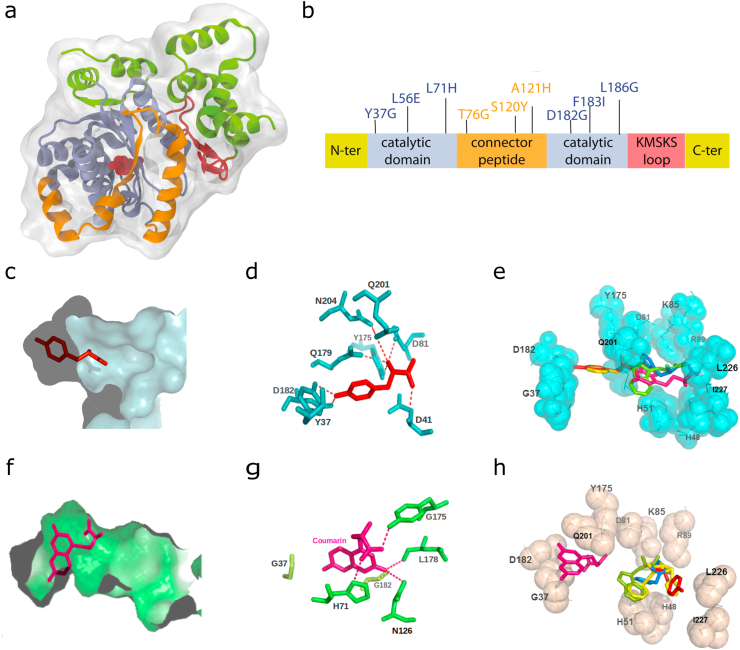
Figure 1.
Homology modeling of the CoumRS enzyme and molecular docking studies. (a) Structure of the EcTyrRS enzyme (PDB: 1X8X) used to compute the CoumRS homology model. The Tyr ligand is shown in red. Color code depicts the different regions of the enzyme. C- and N- terminal regions in green, catalytic domains in blue, the linker between them in brown, and the KMSKS region in red. (b) Linear scheme of TyrRS highlighting the nine engineered mutations present in the CoumRS. Color code is the same as in panel a. (c) Tyr ligand (red) in the binding pocket of TyrRS. (d) Network of amino acids stabilizing Try within the binding region. (e) Molecular docking showing that TyrRS binds Tyr leaving Phe, Trp, and Tyr-coumarin (yellow, green, and pink respectively) out of the principal binding region. (f) Tyr-coumarin ligand (pink) docked in the molecular model for CoumRS. The new enzyme presents a wider ligand binding region when compared to the wildtype enzyme in c. (g) The network of amino acids stabilizing Tyr-coumarin is not well conserved when compared to the wildtype enzyme. Nevertheless, the lowest energy configuration suggests that the orientation of both ligands is similar. (h) View of the lowest energy configuration of Tyr/TyrCoum molecular docking shows that CoumRS binds Tyr-coumarin leaving the natural aromatic amino acids (Phe, yellow; Tyr, red; Trp, green) out of the principal binding region.
Structural and structure-function information of the TyrRS was used as starting point for our de-novo design of a TyrCoum RS [14, 16, 17, 18, 19]. First, comparable residues were identified within the amino acid binding region of the EcTyrRS (PDB 1X8X), MjTyrRS (PDB 1ZH6) [16] and structural models of the modified tyrosyl-tRNA synthetases MjCoumRS, EcAziRS, and EcBpaRS (Figure 1). These comparisons were useful as they informed the subsequent selection of RS mutations predicted to increase the size and flexibility of the catalytic domain. The analysis identified four mutations that are predicted to participate in substrate coordination (Y37G, L71H, D182G, and L186G) while five sites (L56E, T76G, S120Y, A121H, and F183I) are located near the catalytic domain and connector peptide region (Figure 1b). The Y37G mutation was introduced to destabilize tyrosine binding [14, 17] and the additional D182G and L186G mutations served to further expand the binding pocket. The L71H and T76G are were reported in MjCoumRS (i.e. L65H and H70G) as crucial on the recognition of coumarin's phenolic ring [4]. S120Y and A212H, whose side chains are pointing out of the binding pocket, were introduced to promote flexibility within the binding region. The final engineered RS containing these nine mutations was termed EcCoumRS (Figure 1b).
As a first pass, f-ncAA docking in silico to the EcCoumRS was examined by generating a molecular model using the structure of EcTyrRS as template. As expected according to the design strategy, the incorporated mutations produce a wider binding pocket with the potential to accommodate larger aromatic sidechains (Figure 1 c and f). This structural analysis suggests that the f-ncAA substrate, TyrCoum, relies on a new set of hydrogen bonds to bind the catalytic site (Figure1 d and g) and that may preserve the orientation needed to form the end tRNA-amino acid complex (Figure 1 e and h). In silico derived estimates of binding energy (ΔGbinding = ΔGapo-ΔGligand-bound), a value grossly related to the binding affinity [20], suggests that the novel EcCoumRS could also be selective for TyrCoum (ΔGbinding = -10 kCal/mol) over natural aromatic side chains (ΔGbinding = -8.6, -8.3, -8.0 kCal/mol for Trp, Phe, and Tyr respectively).
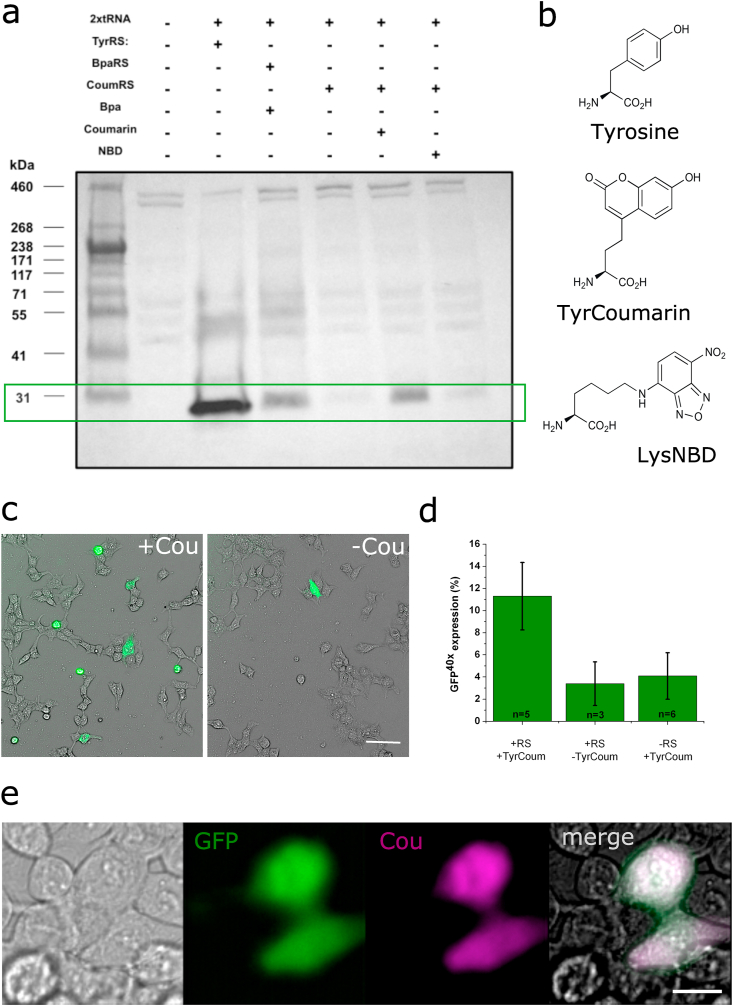
To test whether this new EcCoumRS/BstRNAtag pair was able to selectively incorporate TyrCoum, western blot experiments were performed on protein extracts from HEK-293T cells expressing a fluorescent protein (GFP40TAG) containing a mutation (Y40TAG) that precedes the protein chromophore [16]. This strategy was used to test for rescue of the full-length fluorescent protein in the presence of the f-ncAA (500 nM; Figure 2a). Because of its usefulness for biophysical studies [21], we tested the possibility of encoding LysNBD (7-nitrobenz-2-oxa-1,3-diazol-4-yl) a somewhat larger fluorescent aromatic side chain (Figure 2b). However, we did not observe evident encoding when compared to coumarin (Figure 2a). To confirm that TyrCoum can be encoded, we expressed the EcCoumRS/BstRNAtag pair together with GFP40TAG in HEK293T cells (Figure 2c). We observed that the suppressor pair rescues GFP40TAG only in the presence of TyrCoum (Figure 2d), where a clear double staining is observed in cells (Figure 2e).
Figure 2.
Amber codon suppression and incorporation of a coumarinyl aminoacid in HEK-293T cells. (a) Western blots for the detection of amber suppression in HEK-293T cells expressing the GFP40TAG gene in the absence or presence of Tyr-coumarin and Lys-NBD (lower band at 30 kDa, green box). 2nd lane is transfected with GFP40TAG alone and the 3rd lane represent the effect of expressing a TyrRS and the correspondant tRNA. The rescue of GFP40TAG of the BpaRS (lane 4) is comparable to the rescue in the presence of CoumRS (lane 6). (b) Structures corresponding to the f-ncAAs used in this study and the natural amino acid tyrosine. (c) The suppression of the amber codon was estimated by analyzing the expression of GFP40TAG on cultured HEK-293T cells. Images correspond to a representative filed of view in which transmitted light and fluorescent signals are merged for both the condition with (left) or without (right) Tyr-coumarin in the culture media. Bar = 40um (d) Quantification of GFP40TAG expression in the absence or presence of Tyr-coumarin in the culture media or absence of CoumRS in the transfection mix. Bars correspond to SD. (e) Colocalization of coumarin and GFP signals. Images taken from transiently transfected HEK293T cells expressing GFP rescued with a coumarin side chain at position 40. The signal of coumarin fluorescence is absent in the cells not expressing GFP. Bar = 10 um.
3. Discussion
The present study describes the engineering of an amber suppressor tRNA/aminoacyl-tRNA synthetase pair for encoding Tyr-coumarin, EcCoumRS. The approach takes advantage of the available structural and functional data on RS proteins from bacterial and eukaryotic systems that have been used previously to generate amber suppression-based genetic code expansion approaches.
It has been reported that the substrate binding process in EcTyrRS is supported by polar interactions where D182 binds the natural substrate (i.e. Tyrosine) and when the substrate is absent D182 is stabilized by polar interactions with neighbor residues [17]. For the case of TyrCoum, the mutations on residues D182G, F183I and L186G not only widen the cavity hosting the bulkier substrate but also provide internal contacts in which G182 is limited to interact with L178 alone; I183 is at short distance from Q179, G180, and N187; and G186 can be stabilized by contacting Y190, G191 and V192. Our docking simulations suggest that G182 changes the orientation of the alpha helix containing it, favoring ligand binding. Deeper within the binding pocket, I183 and G186 are facing away from the substrate binding site, where the modeling suggests that they are coordinated by neighboring residues. Overall, the data support the feasibility of our de-novo enzyme design where EcCoumRS yields hydroxycoumarin-containing proteins for single cell imaging and biochemical scale expression.
Further approaches including mutations on residues that participate in shaping binding sites [[22]23] and/or in vitro guided evolution selection of the EcCoumRS should be considered [24, 25]. Such approximation might produce more efficient enzymes for this and other similar synthetic side chains.
In this context, numerous analogs of the canonical amino acids tyrosine and tryptophan have been used as ncAAs because of their spectroscopic properties [26, 27]. However, their utility is restricted to auxotroph-based systems and are not yet available for orthogonal translation methods implemented in vitro or in living mammalian cells. The performance of the system detailed here is certainly a suitable starting point for selecting orthogonal translation systems for other very attractive fluorescent side chains with diverse spectroscopic properties [28, 29, 30].
Among the f-ncAAs that have been genetically encoded in eukaryotes, the most notable so far are dansylalanine and L-Anap [27]. A useful feature of these ncAAs is that their spectroscopic properties varies depending on the polarity of the medium. This has been exploited to investigate the dynamics of different protein structures that are resistant to labeling otherwise because of the poor accessibility of the probes to narrow spots. For example, L-Anap was successfully used to map dynamic rearrangements in membrane proteins such as Shaker potassium channel and the voltage-gated phospahatse Ci-VSP [31, 32]. Moreover, a good example of the usability of the toolkit described here are the single molecule recordings -obtained in living mammalian cells-describing structural rearrangements occurring during the activation of the capsaicin receptor TRPV1 [33].
In summary, we are reporting a general strategy to produce a tool to incorporate hydroxycoumarin in eukaryotic proteins of living cells. The toolkit is useful for live cell imaging and spectroscopic studies in native systems, without the need for additional purification procedures.
4. Methods
4.1. Molecular Biology
The gene encoding CoumRS was constructed with overlapping PCRs, using BpaRS as primary template. Oligonucleotides used for the aminoacyl-tRNA synthetase mutations were (mutated nucleotides are underlined):
-
a)
5′- CTTGTTCCATTGGAATGCCTGAAACGC - 3’; (L56E).
-
b)
5′- AAGCCGGTTGCGCACGTAGGCGGCGCGGGGGGTCTGATTGGC - 3’; (H71L, T76G).
-
c)
5′- TGTGGAGAAAACTATCATATCGCGGCGAAC - 3’; (S120Y, T121H).
-
d)
5′- TTGCAGGGTTATGGCATCGCCTGTGGGAACAAACAGTACG - 3’; (D182G, M183I, A186G).
Oligonucleotides used for the incorporation of restriction sites into the cassette encoding the synthetase were (initiation and termination codons are underlined):
C-terminal e) 5′- GTTTAAACTTAAGCTTGGTACCCCACCATG -3’; (HindIII).
N-terminal f) 5′- GACGACAAGTAATCTAGAGGGCCCGTTTAA -3’; (XbaI).
The plasmid encoding the tRNAUAG (pSVBpUC) was obtained from Thomas P. Sakmar Laboratory. PCRs were done with PfuUltra II Fusion HSD DNA polymerase from Agilent Technologies, according to the manufacturer's instructions. The gene encoding CoumRS was cloned into pCDNA3.1 and deposited in Addgene (plasmid # 64882).
4.2. Synthesis of f-ncAAs
4.2.1. Synthesis of TyrCoum
L-(7-hydroxycoumarin-4-yl) ethylglycine was obtained as described before [4]. Synthesis of NBD-lysine. Na-(t-Butoxyccarbonyl)-L-lysine (308 mg, 1.25 mmol) was dissolved in a 3% solution of sodium bicarbonate in water (13 mL) in a 100 mL roundbottom flask equipped with a stir bar, and heated to 37 °C. A solution of NBD-Cl (7- chloro-4-nitrobenz-2-oxa-1,3-diazole) (500 mg, 2.5 mmol) in 95% ethanol (35 mL) was slowly introduced via Pasteur pipet over 20 min. Following ~8 h of stirring the reaction mixture was acidified to pH ~1 by the addition of 6M HCl and extracted 3 times with dichloromethane. The organic phase was washed with saturated sodium bicarbonate and the aqueous layer retained. 6M HCl was added with rapid stirring in the presence of dichloromethane to lower the pH until the desired product came out as a yellow cloud. The acid mixture was extracted with more dichloromethane, the organic fractions combined and dried over anhydrous magnesium sulfate and placed on a rotary evaporator until a brownish residue was obtained. The purity of the product was estimated at ~90% based on TLC analysis. Without further purification the product was dissolved in dichloromethane (5 mL) and trifluoroacetic acid was slowly added (20 mL) and the solution was stirred at room temperature for 2 h until TLC showed no t-BOC remained on the compound. The reaction was then concentrated under vacuum to yield a dark, oily solid.
4.3. Cell culture
HEK293T cells, were cultured in DMEM (Gibco Inc.) supplied with 10% FBS (Gibco Inc.). Cells were prepared at (60–70)% confluence and transfected with lipofectamine 2000 following the instructions from the provider (Life technologies). The target gene was transfected 2–3 h after the initial transfection of the pair tRNAtag/EcCoumRS. The f-ncAA was added to cell cultures to a final concentration of 0.5–5 μM together with the second transfection procedure.
4.3.1. Imaging
24 h after transfection of the target gene, cells are disaggregated and plated on poly-L-lysine treated glass covers. The f-ncAA-containing media was removed at least 12h prior the experiment allowing cells to clear the soluble f-ncAA. Generally, the cells are recorded 36–48 h after the second transfection. Percentage of expression was calculated by counting the number of green cells on random fields of the sample, three fields per sample were counted and averaged.
4.4. Bioinformatics
Structure models for AziRS, BpaRS and CoumRS were made with Modeller 9.13 (Andrej Sali Lab, UCSF) [34], using as template the X-ray structure of E. coli TyrRS in complex with Tyr-AMS (PDBID 1X8X). The sequence identity obtained from the pairwise alignment of E. coli TyrRS and AziRS, BpaRS and CoumRS were 98.6% 98.8% and 95.8%.
Docking simulations were performed in parallel using two approaches: i) Autodock Vina docking software (Molecular Graphics Lab, The Scripps Research Institute) and ii) the online server SwissDock (Swiss Institute of Bioinformatics, Molecular modeling group). Docking was guided within a grid calculated with Autodock Vina as: volume coordinates dimensions X = 46, Y = 24, Z = 34, centered on X = 12.825, Y = 29.431, Z = 19.000, with 0.436Å spacing.
To first evaluate our docking strategy, we performed docking studies using Tyr ligand on the TyrRS crystal structure (1X8X, ligand removed) and calculate the RMSD for the position of the docked Tyr when compared to the Tyr at the crystal structure. To find a list of potential mutations implied in the ligand binding, an alanine screening was performed into Tyr binding pocket. The structures mutated were used as new receptors of Tyr and TyrCoum in docking calculations. From the obtained set of ΔGbinding, we chose the best scores for docked ligands taking in consideration both the RMSD from Autodock Vina (RMSDd/l and RMSDu/b) together with the full-fitness value from Swiss-Dock. Molecular graphics and analyses of ligand structures were performed with the UCSF Chimera package (University of California, San Francisco).
4.5. Western blots
The existing media was aspirated, and the cells washed with ice-cold PBS containing a cocktail of protease inhibitors (Roche), dispatched from the dish using a cell lifter and transferred to pre-chilled 1.5 mL tubes. The cells were then centrifuged (21000 x g; 4 °C) for 5 min and the supernatant discarded. The cell pellets were vortexed in 100 μL of cell lysis buffer, and protease inhibitors for 10 min in a cold room. The samples were then centrifuged (21,000 x g; 4 °C) for 20 min. Protein quantification was done using Bradford Protein Assay. 80 μg total protein of each sample (20 μg of TyrRS positive control) run on a 4–20% PAGE gel (pre-cast, Biorad). Proteins were then transferred onto a nitrocellulose membrane with cold pack (room temp); 5% milk blocking for 2h at room temp was followed by overnight incubation with the primary antibody (1:10,000; anti-GFP rabbit polyclonal; Synaptic Systems). Secondary incubation was performed for 2 h at RT (1:10,000; Goat Anti-Rabbit IgG HRP Conjugate; Millipore). Membranes were incubated with house ECL for 2 min and exposed every 1 min for 20 min.
Declarations
Author contribution statement
X. Steinberg: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data J. Galpin and E. de Guevara: Performed the experiments; Contributed reagents, materials, analysis tools or data.
G. Nasir and R. Sepulveda: Performed the experiments.
F. Gonzalez-Nilo: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
L. Islas: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
C. Ahern: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
S. Brauchi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by DRI-CONICYT/CONACYT grant to S. Brauchi and L.D.Islas (Grant number PCCI12023), Iniciativa Cientifica Milenio (ANID, Chilean Government), and the National Institutes of Health (U.S. Government). F. Gonzalez-Nilo was supported by FONDECYT (grant number 1170733) and Grand Laureate. L. Islas was supported by DGAPA-PAPIIT-UNAM (grant number IN209515 and IN203318). C. Ahern was supported by National Institutes of Health (NIGMS grant numbers GM106569and GM087519). S. Brauchi was supported by FONDECYT (grant number 1191868) and Anillo Cientifico (grant number ACT-1401).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
X. Steinberg was a MECESUP and CONICYT fellow; R.V. Sepulveda was a CONICYT fellow. C.A.Ahern is a member of the Membrane Protein Structural Dynamics Consortium, which is funded by NIH. S.E.Brauchi is a member of the Millennium Nucleus of Ion-Channel Associated Diseases (MiNICAD), which is funded by ANID.
Footnotes
The study demonstrates the feasibility of the de-novo design of tRNA synthetases. The approach provides a non-invasive tool for protein imaging and structure-function studies of proteins in live cells.
References
- 1.Tsien R. The green fluorescent protein. Biochemistry. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Giepmans B., Adams S., Ellisman M., Tsien R. The fluorescent toolbox for assessing protein location and function. Science. 2006;312(5771):217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Drabkin H.J., Park H.J., RajBhandary U.L. Amber suppression in mammalian cells dependent upon expression of an Escherichia coli aminoacyl-tRNA synthetase gene. Mol. Cell Biol. 1996;16:907–913. doi: 10.1128/mcb.16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J., Xie J., Schultz P.G. A genetically encoded fluorescent amino acid. J. Am. Chem. Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 5.Kiga D., Sakamoto K., Kodama K., Kigawa T., Matsuda T., Yabuki T., Shirouzu M., Harada Y., Nakayama H., Takio K., Hasegawa Y., Endo Y., Hirao I., Yokoyama S. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99:9715–9720. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakamoto K., Hayashi A., Sakamoto A., Kiga D., Nakayama H., Soma A., Kobayashi T., Kitabatake M., Takio K., Saito K., Shirouzu M., Hirao I., Yokoyama S. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summerer D., Chen S., Wu N., Deiters A., Chin J.W., Schultz P.G. A genetically encoded fluorescent amino acid. Proc. Natl. Acad. Sci. Unit. States Am. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantoja R., Rodriguez E.A., Dibas M.I., Dougherty D.A., Lester H.A. Single-molecule imaging of a fluorescent unnatural amino acid incorporated into nicotinic receptors. Biophys. J. 2009;96:226–237. doi: 10.1016/j.bpj.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalstrup T., Blunck R. Dynamics of internal pore opening in K(V) channels probed by a fluorescent unnatural amino acid. Proc. Natl. Acad. Sci. U.S.A. 2013;110:8272–8277. doi: 10.1073/pnas.1220398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen B., Xiang Z., Miller B., Louie G., Wang W., Noel J.P., Gage F.H., Wang L. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cell. 2011;29:1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee A., Guo J., Lee H.S., Schultz P.G. A genetically encoded fluorescent probe in mammalian cells. J. Am. Chem. Soc. 2013;135:12540–12543. doi: 10.1021/ja4059553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zinsli P.E. Investigation of rate parameters in chemical reactions of excited hydroxycoumarins in different solvents. J. Photochem. 1974;3:55–69. [Google Scholar]
- 13.Wagner B.D. The use of coumarins as environmentally-sensitive fluorescent probes of heterogeneous inclusion systems. Molecules. 2009;14:210–237. doi: 10.3390/molecules14010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T., Takimura T., Sekine R., Vincent K., Kamata K., Sakamoto K., Nishimura S., Yokoyama S. Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase. J. Mol. Biol. 2005;346:105–117. doi: 10.1016/j.jmb.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 15.Hino N., Hayashi A., Sakamoto K., Yokoyama S. Site-specific incorporation of non-natural amino acids into proteins in mammalian cells with an expanded genetic code. Nat. Protoc. 2006;1:2957–2962. doi: 10.1038/nprot.2006.424. [DOI] [PubMed] [Google Scholar]
- 16.Ye S., Köhrer C., Huber T., Kazmi M., Sachdev P., Yan E.C.Y., Bhagat A., RajBhandary U.L., Sakmar T.P. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 2008;283:1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi T., Sakamoto K., Takimura T., Sekine R., Kelly V.P., Kamata K., Nishimura S., Yokoyama S. Structural basis of non-natural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1366–1371. doi: 10.1073/pnas.0407039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin J.W., Cropp T.A., Anderson J.C., Mukherji M., Zhang Z., Schultz P.G. An expanded eukaryotic genetic code. Science. 2003;301(5635):964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Wang L., Schultz P.G., Wilson I.A. Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O -methyl-L-tyrosine. Protein Sci. 2005;14:1340–1349. doi: 10.1110/ps.041239305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldar S., Chattopadhyay A. Springer Series on Fluorescence. 2012. Application of NBD-Labeled Lipids in Membrane and Cell Biology; pp. 37–50. [Google Scholar]
- 22.Stokes A.L., Miyake-Stoner S.J., Peeler J.C., Nguyen D.P., Hammer R.P., Mehl R. Enhancing the utility of unnatural amino acid synthetases by manipulating broad substrate specificity. Mol. Biosyst. 2009;5:1032–1038. doi: 10.1039/b904032c. [DOI] [PubMed] [Google Scholar]
- 23.Baumann T., Hauf M., Richter F., Albers S., Möglich A., Ignatova Z., Budisa N. Computational aminoacyl-tRNA synthetase library design for photocaged tyrosine. Int. J. Mol. Sci. 2019;20:2343. doi: 10.3390/ijms20092343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rauch B.J., Porter J.J., Mehl R.A., Perona J.J. Improved incorporation of noncanonical amino acids by an engineered tRNATyr suppressor. Biochemistry. 2016;55:618–628. doi: 10.1021/acs.biochem.5b01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnefond L., Giegé R., Rudinger-Thirion J. Evolution of the tRNATyr/TyrRS aminoacylation systems. Biochimie. 2005;87:873–883. doi: 10.1016/j.biochi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Merkel L., Hoesl M.G., Albrecht M., Schmidt A., Budisa N. Blue fluorescent amino acids as in vivo building blocks for proteins. Chembiochem. 2010;11:305–314. doi: 10.1002/cbic.200900651. [DOI] [PubMed] [Google Scholar]
- 27.Cheng Z., Kuru E., Sachdeva A., Vendrell M. Fluorescent amino acids as versatile building blocks for chemical biology. Nat. Rev. Chem. 2020;4:275–290. doi: 10.1038/s41570-020-0186-z. [DOI] [PubMed] [Google Scholar]
- 28.Bae J.H., Rubini M., Jung G., Wiegand G., Seifert M., Azim M.K., Kim J., Zumbusch A., Holak T.A., Moroder L., Huber R., Budisa N. Expansion of the genetic code enables design of a novel “gold” class of green fluorescent proteins. J. Mol. Biol. 2003;328:1071–1081. doi: 10.1016/s0022-2836(03)00364-4. [DOI] [PubMed] [Google Scholar]
- 29.Lepthien S., Hoesl M.G., Merkel L., Budisa N. Azatryptophans endow proteins with intrinsic blue fluorescence. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105:16095–16100. doi: 10.1073/pnas.0802804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumann T., Hauf M., Schildhauer F., Eberl K.B., Durkin P.M., Deniz E., Löffler J.G., Acevedo-Rocha C.G., Jaric J., Martins B.M., Dobbek H., Bredenbeck J., Budisa N. Site-resolved observation of vibrational energy transfer using a genetically encoded ultrafast heater. Angew. Chem. Int. Ed. 2019;58:2899–2903. doi: 10.1002/anie.201812995. [DOI] [PubMed] [Google Scholar]
- 31.Kalstrup T., Blunck R. Dynamics of internal pore opening in KV channels probed by a fluorescent unnatural amino acid. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110:8272–8277. doi: 10.1073/pnas.1220398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakata S., Jinno Y., Kawanabe A., Okamura Y. Voltage-dependent motion of the catalytic region of voltage-sensing phosphatase monitored by a fluorescent amino acid. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:7521–7526. doi: 10.1073/pnas.1604218113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinberg X., Kasimova M.A., Cabezas-Bratesco D., Galpin J.D., Ladron-de-Guevara E., Villa F., Carnevale V., Islas L., Ahern C.A., Brauchi S.E. Conformational dynamics in TRPV1 channels reported by an encoded coumarin amino acid. eLife. 2017;6:2017. doi: 10.7554/eLife.28626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb B., Sali A. Current Protocols in Bioinformatics. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2014. Comparative protein structure modeling using MODELLER; pp. 5.6.1–5.6.32. 47, 5.6.1–5.6.32. [DOI] [PubMed] [Google Scholar]