Abstract
The enormous magnitude of scientific research carried out in the field of NSAIDs and cyclooxygenases (COXs) is known. They are crucial in pain management. COX-2 inhibitors have evolved over the years; from traditional NSAIDs to isoform-specific. The present study is aimed to identify a cluster of amino acids in the catalytic site whose energy contribution can better explain COX-2 inhibitory activity accurately than the binding energy of the whole protein. Initially, MD simulations (25 ns) and MM-PBSA calculations were performed for 8 diarylheterocyclic inhibitors. Per-residue energy decomposition studies were carried out to elucidate the energy contribution of each amino acid, and their correlation with COX-2 inhibitory activity was enumerated. A cluster of catalytic amino acids whose free energy sum has a high correlation with biological data was identified. The cluster of Gln178, Ser339, Tyr341, Arg499, Phe504, Val509 and Ala513 showed the correlation of -0.60. Further, the study was extended to a total of 26 COX-2 inhibitors belonging to different classes to validate the applicability of the cluster of amino acids identified. Results clearly suggest that the cluster of amino acids identified provide accurate screening method, and can be applied to predict COX-2 inhibitory activity of small molecules.
Keywords: Theoretical chemistry, Cyclooxygenase-2, Diarylheterocyclic compounds, Molecular dynamics simulations, Structure-activity relationship, Per-residue energy decomposition
Theoretical chemistry, Cyclooxygenase-2; Diarylheterocyclic compounds; Molecular dynamics simulations; Structure-activity relationship; Per-residue energy decomposition
1. Introduction
Non-steroidal anti-inflammatory drugs (NSAIDs) constitute a vital class of drugs that inhibit cyclooxygenases (COXs) [1]. COXs are important enzymes in the arachidonic acid metabolism involved in prostaglandin (PG) biosynthesis [2, 3]. The inducible isoform of COX was reported by Needleman, Simmons and Herschman's group, and described as COX-2 [4, 5, 6]. COX-2 has been recognized as a well-known drug target because of its well-characterized role in inflammatory disorders and various cancers [7, 8, 9].
Traditional NSAIDs suppress the activities of both isoforms; the constitutive cytoprotective COX-1 and the inducible COX-2. This led to adverse GI toxicities [10, 11, 12]. In order to develop better anti-inflammatory inhibitors with minimum adverse effects, efforts were made by various research communities for the development of selective COX-2 inhibitors [13, 14, 15]. The structural differences between COX-1 and COX-2 at the active site were exploited [16] for the same. Mutagenesis experiments illustrated that single amino acid substitution i.e. Ile to Val509 (Val523 in PGHS-1 numbering) in the COX-2 enzyme is crucial for its selectivity [17]. The COX-2 binding site is extensively studied by various research groups and it has been found that His90, Arg120, Tyr355, and Glu524 form a hydrogen bond network at the entrance of the binding site, known as gate residues [18]. Other amino acids such as Arg513, Gln192, Phe518, Trp387, Tyr385, Tyr348, Leu359, Tyr355, Leu531, Ser530 and Leu534 (in PGHS-1 numbering) are present towards the interior of the active site [19].
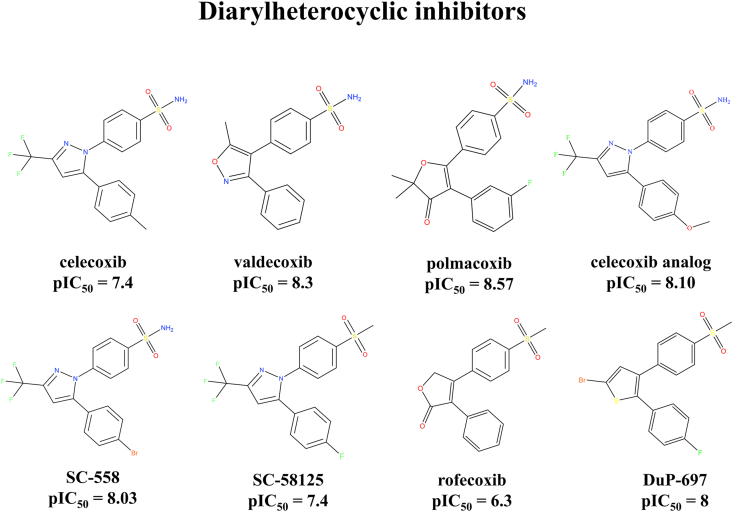
Numerous COX-2 inhibitors have been developed over the years. Structural details of the COX-2 enzyme and the binding mode of its inhibitors were explained previously employing various in silico methods like molecular docking and molecular dynamics simulation studies [14, 20, 21, 22, 23, 24]. Generally, COX-2 inhibitors are classified into two major classes on the basis of number of ring structures: (1) Tricyclic or Diarylheterocyclic compounds which possess two proximal diaryl moieties linked to a central heterocyclic or carbocyclic ring; the compounds in this class mainly differ from one another in the central aromatic ring which can either be 4-membered (cyclobutene), 5-membered (pyrazole, isoxazole, furanone) or 6-membered (pyridine, pyranone) [25]; (2) Non-tricyclic compounds: This class of compounds lacks the central cyclic ring. The acyclic core may contain 2 or 3-membered chain e.g. 1, 2-diarylethenes, acetylenes, and chalcone derivatives [25, 26, 27]. The diarylheterocyclic compounds can be further divided into (a) sulfonamide and (b) non-sulfonamides. It has been reported that –SO2NH2/SO2Me moiety at the para position of one of the aryl rings is crucial for selective and potent inhibition of COX-2 [19,25,28]. Celecoxib, polmacoxib, and valdecoxib are sulfonamide containing COX-2 inhibitors, whereas rofecoxib, etoricoxib, and SC-58125 have sulfomethyl group.
Apart from the structural classification of COX-2 inhibitors, they may be segregated into four groups on the basis of their selectivity index (SI) (ratio of COX-1 IC50/ratio of COX-2 IC50). Group 1 NSAIDs suppress COX activity with little selectivity e.g., aspirin, ibuprofen, diclofenac, indomethacin, naproxen, and piroxicam. Inhibitors in Group 2 show 5–50 fold COX-2 selectivity such as celecoxib, etodolac, meloxicam, and nimesulide. Group 3 consists of inhibitor with >50 fold selectivity like rofecoxib, NS-398, and valdecoxib, whereas Group 4 includes those NSAIDs which are weak inhibitors of both isoforms (5-aminosalicylic acid, sodium salicylate, nabumetone, and sulfasalazine) [19].
MM-PBSA Molecular Mechanics-Poisson Boltzmann Surface Area/MM-GBSA (Molecular Mechanics-Generalized Born Surface Area) approaches have become an integral part of structure-based drug design strategies and are being widely employed by various researchers [29, 30, 31, 32].
Earlier we have applied molecular dynamics simulations, and various in silico methods for COX-2 and other enzymes [14, 33, 34, 35, 36, 37]. MD simulations and per-residue decomposition studies were performed in order to identify crucial amino acid residues for COXIB binding at the COX-2 active site [38]. In continuation of our previous work, in the present study, we performed MD simulations on an extended set of COX-2 inhibitors in order to identify a group of amino acids whose cumulative free energy can be used to predict COX-2 inhibitory activity. Molecular docking, detailed Structure-activity relationship (SAR) studies and molecular dynamics simulations methods were employed. Per-residue free energy decomposition analysis was also included to elucidate the individual contribution of amino acids involved.
2. Materials and methods
2.1. Inhibitor data set
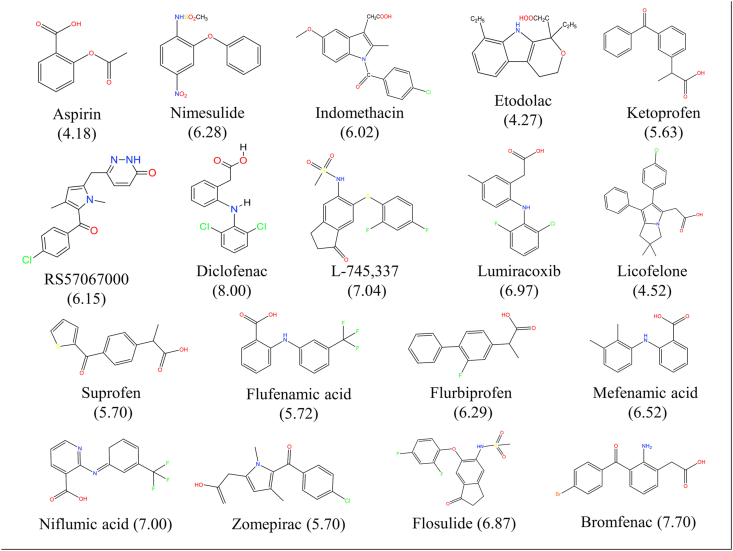
A dataset of 26 COX-2 inhibitors belonging to different classes was considered in the study. Among these 26 compounds, 8 belong to the diarylheterocyclic class of compounds, 18 compounds belonged to different classes and were randomly selected from the literature. The diarylheterocyclic inhibitors considered were celecoxib, polmacoxib, valdecoxib, SC-558, celecoxib-analog, rofecoxib, DuP-697, and SC-58125. The chemical structures of the considered inhibitors were given in Figure 1 and Figure 2 with their corresponding pIC50 values. The structures of the inhibitors were obtained from the PubChem database [39]. The RED server was used to optimize the structures (RED server uses the Hartree-Fock method for optimization) [40, 41, 42, 43, 44]. Out of the 26 inhibitors discussed in the current manuscript, 3 were studied and reported earlier [38]. They are included and discussed in the current study for better understanding and representation.
Figure 1.
Structures of diarylheterocyclic compounds considered in the study.
Figure 2.
Structures of other inhibitors considered in the study.
2.2. Preparation of protein-ligand complexes
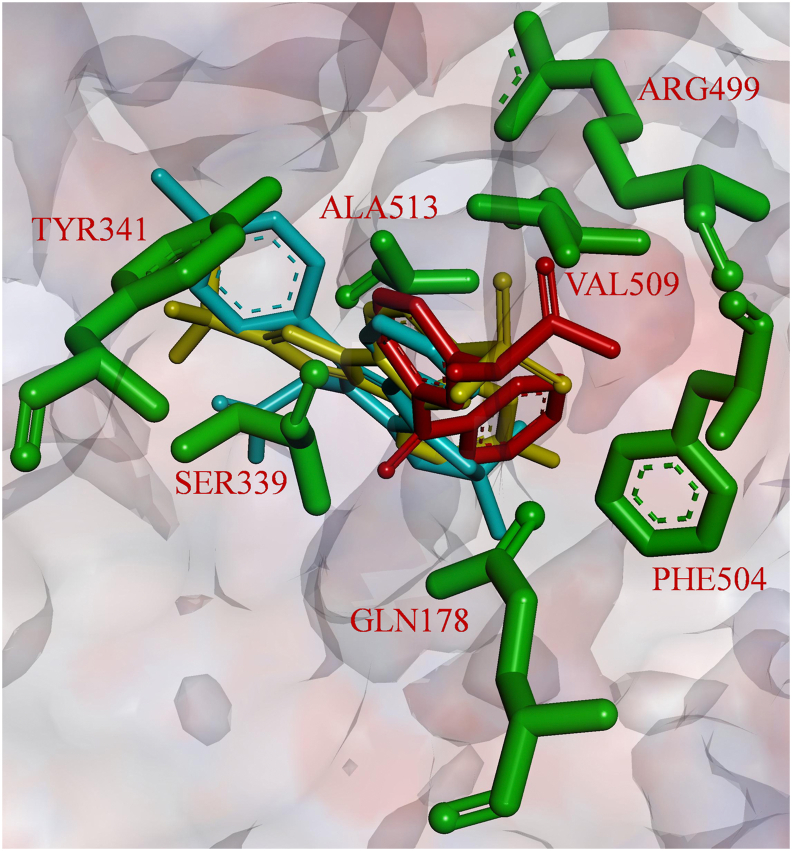
The structure of COX-2 was obtained from the protein data bank (PDB ID: 3LN1). It is a co-crystallized structure of COX-2 and celecoxib. Protein-inhibitor complexes for the other compounds were prepared by implementing molecular docking. The structure of docked celecoxib with two other inhibitors belonging to different scaffolds at the active site of COX-2 is given in Figure 3. The Molecular docking and simulation procedures followed are the same as discussed in our previous work [38], but for a better understanding of the readers, the same is presented in detail here.
Figure 3.
Structure of docked inhibitors at the active site of COX-2; celecoxib (yellow), licofelone (cyan) and ketoprofen (red).
2.3. Molecular docking
The protein-ligand complexes were prepared using AutoDock [45]. Active site residues were obtained from the bound celecoxib. The accuracy of AutoDock in the prediction of ligand conformation was assured using the re-docking procedure (explained in [38]).
100 docking poses for each inhibitor (with a population size 1000) were calculated. Kollman united atom partial charges, AutoDock atom types and polar only hydrogen atoms were taken into account while preparing protein. Empirical scoring function and Lamarckian genetic algorithm were used for ligand conformational search. All other parameters for docking were set to their default values. The interactions of the best energy ranked conformations were analyzed using Accelrys DS visualizer [46]. The prepared complexes were used as starting structures for further energy minimization and MD calculations.
2.4. Energy refinement and molecular dynamics (MD) simulations of the complexes
GROMACS [47, 48] was used in the study. GROMOS96 53a6 [49] force field was employed. Ligand parameters and topology were generated using SwissParam [50]. spc216 [51] water model was used to solvate each system using a cubic box having an edge length of 10 Ǻ. Each system was then neutralized by adding counter-ions. Long-range interactions were treated using Particle mesh Ewald (PME) [52]. Leap-frog integrator [53] was used for MD simulations.
Periodic boundary conditions (PBC) were considered while minimization. Two successive steps of energy minimization were performed using GROMACS. In the first step, 3000 minimization steps were carried out using a steepest descent method. In the second step, 5000 steps of minimization were performed using a conjugate gradient method. The energy step size was set to 0.001 nm in both cases. Each system was then heated from 0 to 300 K for 100 ps in NVT (constant Number of atoms, Volume, and Temperature) ensemble. In NPT (constant Number of atoms, Volume, and Pressure) ensemble, each system was equilibrated using a constant pressure of 1 bar for 100 ps with a time step of 2 fs per step. The coordinates obtained after equilibrating the system were used for MD simulations (25 ns) for each system.
2.5. Binding free energy and per-residue decomposition studies
The binding energies for all the systems were calculated using the MM-PBSA method (developed by Srinivasan et al. [54]). MM-PBSA method combines the molecular mechanics and continuum solvent models and is explained in our previous report [38]. A total of 250 snapshots from each simulated trajectory of 25 ns were extracted evenly after every 100 ps. The electrostatic energy, van der Waals energy, and polar solvation energy contributions were calculated using Adaptive Poisson-Boltzmann Solver (APBS) [55, 56]. Solvent-accessible surface area (SASA) was used to approximate the non-polar energy contributions. A value of 0.5 Ǻ and 1.4 Ǻ was set for grid spacing and probe radius (for SASA estimation). The solvent dielectric constant was set to 80, whereas the solute dielectric constant was set to 2.
Further, per-residue decomposition analysis was performed to obtain the energetic contribution of the amino acids involved in inhibitor binding. Binding free energy decomposition was performed using the g_mmpbsa tool. This tool decomposes the overall binding energy of the protein-ligand complex [57]. Python scripts “MmPbSaStat.py” and “MmPbSaDecomp.py” were employed for MM-PBSA calculations and individual contribution of different amino acids. In our previous study, all the amino acids i.e., His75, Arg106, Gln178, Leu335, Leu338, Ser339, Tyr341, Leu345, Leu370, Tyr371, Trp373, Arg499, Ala502, Ile503, Phe504, Met508, Val509, Glu510, Gly512, Ala513, Ser516 and Leu517 (present within 8 Ǻ of the active site) were considered. In the present study, Gln178, Val335, Leu338, Ser339, Tyr341, Tyr371, Arg499, Ile503, Phe504, Val509, Ala513 and Ser516 were considered. These amino acids formed interactions consistently with most of the inhibitors. Correlation studies between pIC50 values of these inhibitors and per-residue decomposition energies were also carried out. Amino acids showing negative correlation were selected for further study.
2.6. Identification of a cluster of amino acids to estimate the ligand-binding affinity
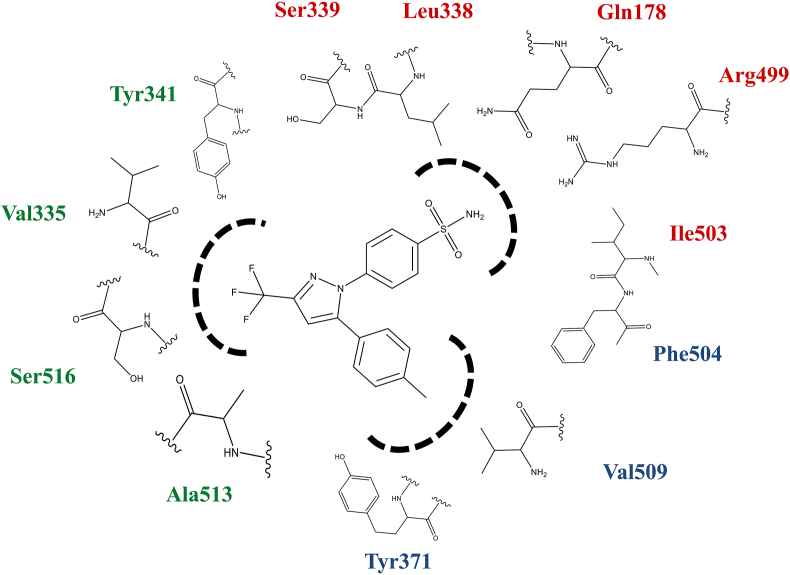
Site points are points in the active site of a drug target (adjacent to various interacting groups of amino acids) which can be occupied by an inhibitor for favorable binding [58, 59]. The potential of diarylheterocyclic compounds against COX-2 is well known. There are many inhibitors of COX-2 derived from this scaffold. The substituents on these aryl ring hugely contribute to their varied activity. Thorough SAR analysis was performed and three sites (Site-1, Site-2, and Site-3) were identified as shown in Figure 4. Amino acids contributing to the site points corresponding to the substituents at the para position of the phenyl ring (at 3-position) constituted Site-1. For example, hydrophobic –CH3 group present in celecoxib and –OCH3 in celecoxib analog. Site-2 included the amino acids interacting with sulfonamide/sulfomethyl moiety at the para position of another phenyl ring. Amino acids forming interactions with the central 5/6 membered ring and their functional groups were taken as Site-3. Among all the amino acids present at Site-1, 2 and 3 the ones which have a negative correlation with pIC50 were taken. The cumulative binding energy of these amino acids was computed and correlated with biological activity. All the calculations were performed initially for 8 diarylheterocyclic compounds and further extended in 18 other inhibitors. The correlation obtained was compared with that of MM-PBSA energies.
Figure 4.
Amino acid sites identified on the basis of interacting atom(s) of the inhibitor, the blue color represents Site-1, red color represents Site-2 and green color represents Site-3.
3. Results and discussion
In the present work, a variety of COX-2 inhibitors belonging to different structural classes were considered. Molecular docking, molecular dynamics simulations, and MM-PBSA based binding energy calculations were performed.
3.1. Docking studies and interaction analysis
After molecular docking, detailed interaction analysis was performed to get insights into the amino acids involved in various interactions.
3.2. Molecular dynamics simulations and binding free energy calculations
MD simulations allow a system to interact flexibly, so the dynamics of protein-ligand complexes can be monitored throughout the simulation in order to check protein flexibility and any other conformational changes. MD also facilitates to develop atomistic insights while explaining binding mechanisms. In order to observe the behavior of various inhibitors at the active site of COX-2 during simulation, different properties from simulated trajectories like energy profiles, stability, residue fluctuations, and energy contributions were analyzed.
3.3. Interaction analysis after MD simulations
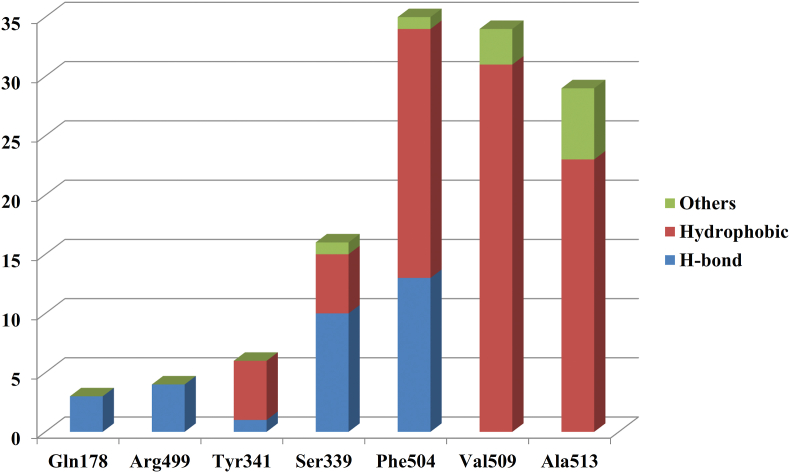
In order to understand the interactions at the active site of COX-2 all the amino acids present in the 8 Ǻ radius (discussed in the material and method section) were considered in the present study. Detailed interactions were studied and analyzed (Table 1). The interaction analysis provided in-depth understanding regarding the binding mechanisms of diverse inhibitors at the COX-2 active site along with further highlighting the importance of amino acids which are consistently involved in the formation of almost every class of COX-2 inhibitor considered. For the present study, amino acids making significant hydrogen bonds and a large number of hydrophobic interactions (pi-alkyl or pi-pi) were taken. We included a total of 12 amino acids as mentioned earlier. To further quantify the contribution of these amino acids towards COX-2 inhibition, MM-PBSA based binding energy calculations and per-residue decomposition energy analysis were performed. The binding free energy calculation experiments were aimed at elucidating the individual energy contributions of thee amino acids towards the overall binding energy and also to estimate the relationship between these energies and experimental biological activities (pIC50) of the investigational compounds. The amino acids included Gln178, Val335, Leu338, Ser339, Tyr341, Tyr371, Arg499, Ile503, Phe504, Val509, Ala513 and Ser516. Among these amino acids Gln178, Ser339, Tyr341, Arg499, Phe504, Val509, and Ala513 were showing negative correlation while Val335, Leu338, Tyr371, Ile503, and Ser516 were exhibiting positive correlation with pIC50 (Table 2). We selected these 7 amino acids with a negative correlation for further analysis. To understand the reason for significant correlation detailed interactions of these amino acids across the dataset were thoroughly studied and discussed. Gln178 was forming 3 conventional hydrogen bonds with 3 compounds; Ser339 was forming 10 conventional hydrogen bonds, 5 hydrophobic interactions and 1 other interaction in 12 compounds. Tyr341 formed 1 conventional hydrogen bond and 5 hydrophobic bonds with 5 compounds, Arg499 made 4 hydrogen bonds with 4 inhibitors, and Phe504 formed 13 hydrogen bonds, 21 hydrophobic interactions and 1 other interaction with 25 compounds. Val509 showed 31 hydrophobic interactions and 3 others with 25 inhibitors whereas Ala513 formed 23 hydrophobic and 6 other interactions with 19 compounds (Figure 5).
Table 1.
Various Interactions formed by inhibitors after MD simulations.
| S. No. | Ligand | Hydrogen | Hydrophobic |
Others (pi-sulfur, amide-pi stacked/halogen) | |||
|---|---|---|---|---|---|---|---|
| Pi-Sigma | Alkyl | Pi-Alkyl | Pi-Pi | ||||
| 1 | Aspirin | Trp373 | - | - | Leu338 | Phe504 | Met508 |
| Ser516 | Val509 | Val509 | |||||
| 2 | Diclofenac | Arg499 | Ser339 | Val335 | Leu338 | - | His75 |
| Phe504 | Val509 | Leu345 | |||||
| Ser339 | Leu517 | ||||||
| 3 | Etodolac | Tyr371 | Leu338 | Ala513 | Trp373 | - | - |
| Ser516 | Phe504 | Val335 | Leu338 | ||||
| Met508 | Val509 | ||||||
| 4 | Indomethacin | Ser339 | Val509 | Ala513 | Trp373 | Tyr371 | - |
| Ser516 | Met508 | Phe504 | Phe504 | ||||
| Ala502 | |||||||
| Val509 | |||||||
| Leu338 | |||||||
| 5 | RS57067000 | Ser339 | Val509 | Val102 | Tyr371 | His75 | - |
| Tyr371 | Leu517 | Trp373 | Trp373 | ||||
| Ser516 | Val335 | Phe504 | |||||
| Ala513 | |||||||
| Leu517 | |||||||
| Val509 | |||||||
| Ala502 | |||||||
| 6 | Nimesulide | Phe504 | - | - | Val509 | Phe504 | Phe504 |
| Tyr341 | Ala513 | Tyr341 | Gly512 | ||||
| Leu338 | Ala513 | ||||||
| 7 | Ketoprofen | Tyr371 | - | Val509 | Tyr341 | - | Met508 |
| Ser516 | Phe504 | Val509 | |||||
| Val335 | |||||||
| Ala513 | |||||||
| Leu517 | |||||||
| 8 | L-745, 337 | Ile503 | Val509 | - | Leu338 | Tyr371 | - |
| Phe504 | Phe504 | ||||||
| Ser339 | |||||||
| 9 | Lumiracoxib | Ser339 | - | Ala513 | Val509 | Phe504 | Gly512 |
| Gln336 | Val335 | Ala513 | Met508 | ||||
| Leu338 | |||||||
| Val509 | |||||||
| Val102 | |||||||
| 10 | Licofelone | Val330 | Val335 | Val335 | Tyr334 | - | - |
| Ala513 | Tyr371 | ||||||
| Leu338 | Arg106 | ||||||
| Met99 | Ala513 | ||||||
| Val102 | Ile331 | ||||||
| Leu103 | Val335 | ||||||
| Leu345 | Leu345 | ||||||
| Leu517 | |||||||
| 11 | Flufenamic Acid | - | Ser339 | Leu338 | Trp373 | - | Met508 |
| Val509 | Phe504 | ||||||
| Ala502 | |||||||
| Val335 | |||||||
| Leu338 | |||||||
| 12 | Flurbiprofen | Gly505 | - | Ala502 | Phe504 | - | Leu338 |
| Val335 | Ser339 | ||||||
| Ala513 | Arg106 | ||||||
| Val509 | |||||||
| 13 | Suprofen | His337 | Val509 | Val509 | His75 | Phe504 | - |
| Leu338 | Tyr341 | ||||||
| Phe504 | |||||||
| Leu338 | |||||||
| Ala513 | |||||||
| Met508 | |||||||
| 14 | Mefenamic Acid | Leu338 | - | Leu345 | Tyr341 | Phe504 | Met521 |
| Val509 | Val335 | ||||||
| Ala513 | |||||||
| Val509 | |||||||
| 15 | Niflumic Acid | Ser516 | - | Leu370 | Phe367 | Phe504 | Leu370 |
| Val335 | Trp373 | Gly512 | |||||
| Leu338 | Met508 | ||||||
| Val509 | Val509 | ||||||
| Ala513 | |||||||
| 16 | Zomepirac | Phe504 | Ser339 | Ala513 | Tyr341 | - | Met508 |
| Gly505 | Val509 | Trp373 | |||||
| Met508 | Val509 | ||||||
| 17 | Flosulide | Tyr371 | - | - | Val509 | Tyr371 | Leu370 |
| Ser516 | Ala513 | Phe504 | Met508 | ||||
| Leu338 | |||||||
| 18 | Bromfenac | Ser339 | Val509 | - | Leu338 | Phe504 | Gly512 |
| Ile503 | Ala513 | Ala513 | |||||
| Phe504 | Met508 | ||||||
| Leu338 | |||||||
| 19 | Gln178 | Ala513 | Tyr371 | ||||
| celecoxib | Leu338 | Ser339 | Leu370 | Phe504 | Tyr371 | Gly512 | |
| Ser339 | Val509 | Leu338 | Ala513 | ||||
| Phe504 | Ala513 | His75 | |||||
| 20 | celecoxib-analog | Arg499 | Ser339 | Val335 | - | His75 | |
| Phe504 | Val509 | Leu345 | Leu338 | ||||
| Ser339 | Leu517 | ||||||
| 21 | Phe504 | - | Met508 | Tyr371 | Phe504 | Arg106 | |
| SC-558 | Gln178 | Phe504 | |||||
| Arg499 | Val509 | ||||||
| Ile503 | |||||||
| 22 | Arg499 | - | Ala513 | - | Met508 | ||
| polmacoxib | Phe504 | Val335 | Val509 | ||||
| Ser516 | Leu517 | ||||||
| Ser339 | |||||||
| Ile503 | |||||||
| 23 | Gln178 | Ala513 | Ala513 | Val335 | Phe504 | His75 | |
| valdecoxib | Phe504 | Val335 | Val509 | Tyr371 | Gly512 | ||
| Ser516 | Ala513 | ||||||
| Ser339 | |||||||
| 24 | Ile503 | - | - | Leu338 | - | - | |
| rofecoxib | Phe504 | Val509 | |||||
| Ser516 | |||||||
| 25 | Ile503 | Val509 | - | - | Ala513 | ||
| SC-58125 | Phe504 | Leu338 | |||||
| Leu517 | |||||||
| 26 | Ile503 | Ala513 | Ala513 | Leu338 | |||
| DuP-697 | Phe504 | Val335 | Val509 | - | - | ||
| Leu517 | Val335 | ||||||
| Ala513 | |||||||
Table 2.
List of amino acids showing positive and negative correlation with the biological activities of the inhibitors.
| Amino Acid | Celecoxib |
Polmacoxib |
Valdecoxib |
Celecoxib-analog |
SC-558 |
SC-58125 |
Rofecoxib |
DUP-697 |
Correlation |
|---|---|---|---|---|---|---|---|---|---|
| 7.4 | 8.57 | 8.3 | 8.1 | 8.03 | 7.4 | 6.3 | 8 | ||
| GLN178 | -2.78 | -4.06 | -4.34 | -1.25 | -7.09 | 0.69 | 1.15 | -0.02 | -0.62 |
| SER339 | -4.41 | -5.66 | -8.08 | -8.51 | -6.03 | -4.81 | -3.91 | -4.29 | -0.61 |
| TYR341 | -3.47 | -4.47 | -4.03 | -5.72 | -4.41 | -2.10 | -1.93 | -4.67 | -0.79 |
| PHE504 | -10.22 | -12.88 | -13.90 | -15.15 | -13.76 | -13.31 | -13.23 | -13.46 | -0.28 |
| ARG499 | -7.01 | -6.97 | -2.82 | -13.00 | -7.58 | -1.95 | -3.32 | -1.21 | -0.29 |
| VAL509 | -11.56 | -10.76 | -10.93 | -13.66 | -12.10 | -13.54 | -11.63 | -10.50 | 0.20 |
| ALA513 | -5.78 | -4.29 | -4.28 | -6.77 | -5.41 | -6.08 | -3.76 | -6.35 | -0.22 |
| VAL335 | -4.70 | -5.49 | -4.89 | -6.07 | -6.01 | -4.15 | -6.18 | -5.93 | 0.03 |
| LEU338 | -9.22 | -8.10 | -9.80 | -10.33 | -9.11 | -10.90 | -11.86 | -9.46 | 0.80 |
| TYR371 | -2.18 | -1.21 | -2.62 | -4.38 | -4.00 | -3.10 | -5.43 | -3.19 | 0.63 |
| ILE503 | -5.38 | -5.84 | -6.38 | -4.39 | -6.62 | -5.36 | -8.13 | -7.36 | 0.44 |
| SER516 | 1.86 | -8.44 | -5.76 | -0.90 | 1.22 | 2.53 | -9.18 | 0.87 | 0.09 |
Figure 5.
Contribution of selected amino acids in establishing different interactions with various inhibitors.
Previous pharmacophore studies have shown the importance of hydrogen bonding and hydrophobic or aromatic interactions in the development of selective COX-2 inhibitors. Pharmacophore model generation for 2-(4-methylsulfonylphenyl)pyrimidine derivatives by Shah et al. [60] highlighted the importance of hydrogen bond acceptors and donors in the development of selective COX-2 inhibitors. Michaux et al. have generated a structure-based pharmacophore model for 16 COX-2 inhibitors and concluded the importance of a H-bond acceptor, an aromatic ring and two hydrophobic groups for the identification of novel candidates [61]. Palomer et al. have investigated pharmacophore features which can account for the activity of selective COX-2 inhibitors for diarylheterocyclic compounds. They have clearly shown the importance of aromatic features for the development of potent and specific COX-2 inhibitors for this class [62]. Another study highlights the significance of a hydrogen bond donor/acceptor, a hydrophobic and one ring aromatic feature in the development of a predictive pharmacophore model for COX-2 inhibitors [63].
3.4. Correlation studies
MD simulations and MM-PBSA calculations were performed for the 8 diarylheterocyclic group containing inhibitors. The correlation of ΔUele, ΔUvdW, ΔSASA/nm2 and ΔGbind with pIC50 was deduced. They showed correlation of - -0.04, -0.07, -0.27 and -0.27 respectively (Table 3). The units used to describe binding energy values are kJ/mol.
Table 3.
Correlation of various energy terms with pIC50 values.
| pIC50 | Van-der Waal energy | Electrostatic energy | Polar solvation energy | SASA energy | Binding energy (kJ/mol) | |
|---|---|---|---|---|---|---|
| Celecoxib | 7.40 | -224.66 | -134.98 | 182.56 | -20.95 | -198.03 |
| Celecoxib-analog | 8.10 | -267.01 | -165.37 | 183.58 | -20.43 | -269.24 |
| Valdecoxib | 8.30 | -223.09 | -141.71 | 158.65 | -18.30 | -224.45 |
| Polmacoxib | 8.57 | -227.50 | -135.45 | 166.50 | -20.16 | -216.61 |
| SC-558 | 8.03 | -264.45 | -150.19 | 167.53 | -20.66 | -267.77 |
| SC58125 | 7.40 | -257.63 | -97.15 | 159.46 | -20.57 | -215.89 |
| Rofecoxib | 6.30 | -236.89 | -136.90 | 167.51 | -18.19 | -224.48 |
| DuP-697 | 8.04 | -265.36 | -54.28 | 109.36 | -19.36 | -229.64 |
| -0.07 | -0.04 | -0.16 | -0.27 | -0.27 |
The bold values represent the correlation between the pIC50 values and the calculated energy terms.
Per-residue binding energy decomposition analysis revealed the contribution of various amino acids towards total binding energy. From the decomposition analysis, the contributions of the consistently interacting amino acids were extracted, and their correlation with experimental activity was deduced. Gln178, Ser339, Tyr341, Arg499, Phe504, Val509 and Ala513 showed correlation of -0.62, -0.61, -0.79, -0.29, -0.28, 0.20 and -0.22 respectively (Table 2). The energies of Val335, Leu338, Tyr371, Ile503, and Ser516 showed a correlation of almost 0, 0.80, 0.63, 0.44 and 0.09 respectively suggesting an inverse relation with biological activity. Among the amino acids Tyr371, Phe504 and Val509 interacting with the phenyl ring and its substituents (Site-1), Phe504 and Val509 were considered further. Among the amino acids present at Site-2, Gln178, Ser339, and Arg499 were used further. For the amino acids corresponding to the –CF3 binding region (Site-3), Tyr341 and Ala513 were used further. The cumulative binding free energy of the group of amino acids was computed. It showed a correlation of -0.60 (Table 4).
Table 4.
Correlation between pIC50 values of the inhibitors and sum of per-residue decomposition energies of the final cluster of amino acids.
| Sr. No. | Compounds | Correlation |
|---|---|---|
| 1. | Diarylheterocyclic Comps | -0.60 |
| 2. | Other Compounds | -0.47 |
| 3. | All Compounds | -0.70 |
To check the applicability of the method, 18 inhibitors of other classes were selected. The combined energy contributions of these amino acids showed a significant correlation of -0.47 with the pIC50 values of the structurally diverse COX-2 inhibitors (Table 4). The negative contribution of these amino acids towards the overall binding energy and significant correlation (between the combined energy terms and the pIC50) suggests their role in the effective binding of structurally diverse COX-2 inhibitors.
In the next phase, the energy contribution of the identified amino acids was summed together for the total dataset of 26 compounds considered in the study and their correlation with the pIC50 values was computed and a high correlation of -0.70 was observed (Table 4). These results are in accordance with our previous study wherein we performed MD based studies and reported the importance of Gln178, Val335, Ser339, Arg499 and Phe504 in the effective binding of COXIBs at the active site of COX-2 [64].
These results clearly suggest that the energy contributions of these amino acids can be a clear indication of COX-2 inhibitory activity.
4. Conclusion
In the present work, MD simulations, MM-PBSA and per-residue decomposition energy calculations were employed to investigate the binding mechanism of diarylheterocyclic and structurally diverse COX-2 inhibitors. The results obtained in each of the studies were thoroughly analyzed and cross-related. Detailed interaction analysis revealed the importance of Gln178, Val335, Leu338, Ser339, Tyr341, Tyr371, Arg499, Ile503, Phe504, Val509, Ala513 and Ser516 in forming a variety of interactions with the inhibitors considered in the present study. Further, their individual energy contributions were deduced using MM-PBSA and per-residue decomposition energy analysis. The individual energy terms for Gln178, Ser339, Tyr341, Arg499, Phe504, Val509 and Ala513 showed a good correlation with inhibitory activity. The cumulative energy contributions for these amino acids showed a good correlation of -0.60, -0.47 and -0.70 for the diarylheterocyclic, structurally diverse and the total dataset of 26 inhibitors (diarylheterocyclic and structurally diverse COX-2 inhibitors). These amino acids are reported as part of the active site in a number of previous reports but their role in effective inhibitor binding is not discussed. The results of the present study highlight the importance of Gln178, Ser339, Tyr341, Arg499, Phe504, Val509 and Ala513 in inhibitor recognition and binding at the COX-2 active site. These amino acids can be targeted for rational drug design targeting COX-2 and can be of significant importance for lead identification and optimization studies.
Declarations
Author contribution statement
Neha Chaudhary: Performed the experiments; Analyzed and interpreted the data.
P Aparoy: Conceived and designed the experiments.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank the Central University of Himachal Pradesh for the computational facilities.
References
- 1.Ong C. An evidence-based update on nonsteroidal anti-inflammatory drugs. Clin. Med. Res. 2007;5(1):19–34. doi: 10.3121/cmr.2007.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams C.S., Mann M., DuBois R.N. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene. 1999;18(55):7908–7916. doi: 10.1038/sj.onc.1203286. [DOI] [PubMed] [Google Scholar]
- 3.Simmons D.L., Botting R.M., Hla T. Cyclooxygenase isozymes: the Biology of prostaglandin Synthesis and inhibition. Pharmacol. Rev. 2004;56(3):387–437. doi: 10.1124/pr.56.3.3. [DOI] [PubMed] [Google Scholar]
- 4.Masferrer J.L. Selective regulation of cellular cyclooxygenase by dexamethasone and endotoxin in mice. J. Clin. Invest. 1990;86(4):1375–1379. doi: 10.1172/JCI114850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie W.L. Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc. Natl. Acad. Sci. U. S. A. 1991;88(7):2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kujubu D.A., Herschman H.R. Dexamethasone inhibits mitogen induction of the TIS10 prostaglandin synthase/cyclooxygenase gene. J. Biol. Chem. 1992;267(12):7991–7994. [PubMed] [Google Scholar]
- 7.Vane J.R., Bakhle Y.S., Botting R.M. Cyclooxygenases 1 and 2. Annu. Rev. Pharmacol. Toxicol. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 8.Gierse J.K. Valdecoxib: assessment of cyclooxygenase-2 potency and selectivity. J. Pharmacol. Exp. Therapeut. 2005;312(3):1206–1212. doi: 10.1124/jpet.104.076877. [DOI] [PubMed] [Google Scholar]
- 9.Shukla S. The bitter barricading of prostaglandin biosynthesis pathway: understanding the molecular mechanism of selective cyclooxygenase-2 inhibition by amarogentin, a secoiridoid glycoside from Swertia chirayita. PloS One. 2014;9(6) doi: 10.1371/journal.pone.0090637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell J.A., Warner T.D. COX isoforms in the cardiovascular system: understanding the activities of non-steroidal anti-inflammatory drugs. Nat. Rev. Drug Discov. 2006;5(1):75–86. doi: 10.1038/nrd1929. [DOI] [PubMed] [Google Scholar]
- 11.Urban M.K. COX-2 specific inhibitors offer improved advantages over traditional NSAIDs. Orthopedics. 2000;23(7 Suppl):S761–S764. doi: 10.3928/0147-7447-20000702-05. [DOI] [PubMed] [Google Scholar]
- 12.Mendes R.T. Selective inhibition of cyclooxygenase-2: risks and benefits. Rev. Bras. Reumatol. 2012;52(5):767–782. [PubMed] [Google Scholar]
- 13.Black W.C. From indomethacin to a selective COX-2 inhibitor: development of indolalkanoic acids as potent and selective cyclooxygenase-2 inhibitors. Bioorg. Med. Chem. Lett. 1996;6(6):725–730. [PubMed] [Google Scholar]
- 14.Reddy R.N. Computer aided drug design approaches to develop cyclooxygenase based novel anti-inflammatory and anti-cancer drugs. Curr. Pharmaceut. Des. 2007;13(34):3505–3517. doi: 10.2174/138161207782794275. [DOI] [PubMed] [Google Scholar]
- 15.Casturi S.R., Hegde P., Ramanujam R. Development of COX-2 selective inhibitors - therapeutic perspectives. Curr. Med. Chem. - Immunol. Endocr. Metabol. Agents. 2005;5(3):241–248. [Google Scholar]
- 16.Kalgutkar A.S. Biochemically based design of cyclooxygenase-2 (COX-2) inhibitors: facile conversion of nonsteroidal antiinflammatory drugs to potent and highly selective COX-2 inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2000;97(2):925–930. doi: 10.1073/pnas.97.2.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gierse J.K. A single amino acid difference between cyclooxygenase-1 (COX-1) and -2 (COX-2) reverses the selectivity of COX-2 specific inhibitors. J. Biol. Chem. 1996;271(26):15810–15814. doi: 10.1074/jbc.271.26.15810. [DOI] [PubMed] [Google Scholar]
- 18.Limongelli V. Molecular basis of cyclooxygenase enzymes (COXs) selective inhibition. Proc. Natl. Acad. Sci. U.S.A. 2010;107(12):5411–5416. doi: 10.1073/pnas.0913377107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao P., Knaus E.E. Evolution of nonsteroidal anti-inflammatory drugs (NSAIDs): cyclooxygenase (COX) inhibition and beyond. J. Pharm. Pharmaceut. Sci. 2008;11(2):81s–110s. doi: 10.18433/j3t886. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Nieto R., Perez C., Gago F. Automated docking and molecular dynamics simulations of nimesulide in the cyclooxygenase active site of human prostaglandin-endoperoxide synthase-2 (COX-2) J. Comput. Aided Mol. Des. 2000;14(2):147–160. doi: 10.1023/a:1008110924479. [DOI] [PubMed] [Google Scholar]
- 21.Neophytou N. Docking and molecular dynamics calculations of pyrrolidinone analog MMK16 bound to COX and LOX enzymes. EuroQSAR 2010. 2011;30:473–486. doi: 10.1002/minf.201000131. [DOI] [PubMed] [Google Scholar]
- 22.Habeeb A.G., Praveen Rao P.N., Knaus E.E. Design and synthesis of celecoxib and rofecoxib analogues as selective cyclooxygenase-2 (COX-2) Inhibitors: replacement of sulfonamide and methylsulfonyl pharmacophores by an azido bioisostere. J. Med. Chem. 2001;44(18):3039–3042. doi: 10.1021/jm010153c. [DOI] [PubMed] [Google Scholar]
- 23.Desiraju G.R. Computer-aided design of selective COX-2 Inhibitors: comparative molecular field analysis, comparative molecular similarity indices analysis, and docking studies of some 1,2-diarylimidazole derivatives. J. Med. Chem. 2002;45(22):4847–4857. doi: 10.1021/jm020198t. [DOI] [PubMed] [Google Scholar]
- 24.Reddy T.C. Kinetics and docking studies of a COX-2 inhibitor isolated from Terminalia bellerica fruits. Protein Pept. Lett. 2010;17(10):1251–1257. doi: 10.2174/092986610792231537. [DOI] [PubMed] [Google Scholar]
- 25.Zarghi A., Arfaei S. Selective COX-2 inhibitors: a review of their structure-activity relationships. Iran. J. Pharm. Res. 2011;10(4):655–683. [PMC free article] [PubMed] [Google Scholar]
- 26.Subbaramaiah K., Dannenberg A.J. Cyclooxygenase 2: a molecular target for cancer prevention and treatment. Trends Pharmacol. Sci. 2003;24(2):96–102. doi: 10.1016/S0165-6147(02)00043-3. [DOI] [PubMed] [Google Scholar]
- 27.Fiorucci S. Dual inhibitors of cyclooxygenase and 5-lipoxygenase. A new avenue in anti-inflammatory therapy? Biochem. Pharmacol. 2001;62(11):1433–1438. doi: 10.1016/s0006-2952(01)00747-x. [DOI] [PubMed] [Google Scholar]
- 28.Zarghi A., Kakhki S. Design, synthesis, and biological evaluation of new 2-phenyl-4H-chromen-4-one derivatives as selective cyclooxygenase-2 inhibitors. Sci. Pharm. 2015;83(1):15–26. doi: 10.3797/scipharm.1407-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin. Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massova I., Kollman P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000;18(1):113–135. [Google Scholar]
- 31.Hou T. Assessing the performance of the MM/PBSA and MM/GBSA methods: I. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou T. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2011;32(5):866–877. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh O. Understanding the dual inhibition of COX-2 and carbonic anhydrase-II by celecoxib and CG100649 using density functional theory calculations and other molecular modelling approaches. Protein Pept. Lett. 2015;22(10):903–912. doi: 10.2174/0929866522666150622102131. [DOI] [PubMed] [Google Scholar]
- 34.Reddy M.R. Free energy calculations to estimate ligand-binding affinities in structure-based drug design. Curr. Pharmaceut. Des. 2014;20(20):3323–3337. doi: 10.2174/13816128113199990604. [DOI] [PubMed] [Google Scholar]
- 35.Rathore R.S. Minimum MD simulation length required to achieve reliable results in free energy perturbation calculations: case study of relative binding free energies of fructose-1,6-bisphosphatase inhibitors. J. Comput. Chem. 2011;32(10):2097–2103. doi: 10.1002/jcc.21791. [DOI] [PubMed] [Google Scholar]
- 36.Aparoy P., Reddy K.K., Reddanna P. Structure and ligand based drug design strategies in the development of novel 5- LOX inhibitors. Curr. Med. Chem. 2012;19(22):3763–3778. doi: 10.2174/092986712801661112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy R.N. An analysis of hydrophobic interactions of thymidylate synthase with methotrexate: free energy calculations involving mutant and native structures bound to methotrexate. J. Mol. Model. 2010;16(2):203–209. doi: 10.1007/s00894-009-0535-9. [DOI] [PubMed] [Google Scholar]
- 38.Chaudhary N., Aparoy P. Deciphering the mechanism behind the varied binding activities of COXIBs through Molecular Dynamic Simulations, MM-PBSA binding energy calculations and per-residue energy decomposition studies. J. Biomol. Struct. Dyn. 2016:1–15. doi: 10.1080/07391102.2016.1165736. [DOI] [PubMed] [Google Scholar]
- 39.Kim S. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanquelef E. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011;39:W511–W517. doi: 10.1093/nar/gkr288. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dupradeau F.Y. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010;12(28):7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F., B J.-P., Cieplak P., Dupradeau F.-Y., Python R.E.D. 2013. Object Oriented Programming for Amber Force fields. [Google Scholar]
- 43.Bayly C.I. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97(40):10269–10280. [Google Scholar]
- 44.Frisch M.J. Gaussian, Inc.; Wallingford, CT, USA: 2009. Gaussian 09. [Google Scholar]
- 45.Morris G.M. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dassault Systèmes BIOVIA Discovery studio modeling environment. Release. 2015;4:5. [Google Scholar]
- 47.Van Der Spoel D. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 48.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91(1–3):43–56. [Google Scholar]
- 49.Oostenbrink C. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25(13):1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 50.Zoete V. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen W.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79(2):926–935. [Google Scholar]
- 52.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- 53.Van Gunsteren W.F., Berendsen H.J.C. A leap-frog algorithm for stochastic dynamics. Mol. Simulat. 1988;1(3):173–185. [Google Scholar]
- 54.Srinivasan J. Continuum solvent studies of the stability of DNA, RNA, and Phosphoramidate−DNA helices. J. Am. Chem. Soc. 1998;120(37):9401–9409. [Google Scholar]
- 55.Gohlke H., Kiel C., Case D.A. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 2003;330(4):891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 56.Hou T. Characterization of domain-peptide interaction interface: prediction of SH3 domain-mediated protein-protein interaction network in yeast by generic structure-based models. J. Proteome Res. 2012;11(5):2982–2995. doi: 10.1021/pr3000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumari R., Kumar R., Lynn A. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 58.Murcko M.A. John Wiley & Sons, Inc.; 2007. Recent advances in ligand design methods; pp. 1–66. (Reviews in Computational Chemistry). [Google Scholar]
- 59.Reddy N.P. Structure based drug design, synthesis and evaluation of 4-(benzyloxy)-1-phenylbut-2-yn-1-ol derivatives as 5-lipoxygenase inhibitors. Eur. J. Med. Chem. 2012;47(1):351–359. doi: 10.1016/j.ejmech.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Shah U.A. Pharmacophore generation and atom-based 3D-QSAR of novel 2-(4-methylsulfonylphenyl)pyrimidines as COX-2 inhibitors. Mol. Divers. 2010;14(3):559–568. doi: 10.1007/s11030-009-9183-3. [DOI] [PubMed] [Google Scholar]
- 61.Michaux C. Structure-based pharmacophore of COX-2 selective inhibitors and identification of original lead compounds from 3D database searching method. Eur. J. Med. Chem. 2006;41(12):1446–1455. doi: 10.1016/j.ejmech.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 62.Palomer A. Identification of novel cyclooxygenase-2 selective inhibitors using pharmacophore models. J. Med. Chem. 2002;45(7):1402–1411. doi: 10.1021/jm010458r. [DOI] [PubMed] [Google Scholar]
- 63.Chopra M. Molecular modeling study on chemically diverse series of cyclooxygenase-2 selective inhibitors: generation of predictive pharmacophore model using Catalyst. J. Mol. Model. 2008;14(11):1087–1099. doi: 10.1007/s00894-008-0350-8. [DOI] [PubMed] [Google Scholar]
- 64.Chaudhary N., Aparoy P. Deciphering the mechanism behind the varied binding activities of COXIBs through Molecular Dynamic Simulations, MM-PBSA binding energy calculations and per-residue energy decomposition studies. J. Biomol. Struct. Dyn. 2017;35(4):868–882. doi: 10.1080/07391102.2016.1165736. [DOI] [PubMed] [Google Scholar]