Summary
Background
The coronavirus disease 2019 (COVID-19) continues to spread around the world. In addition to community-acquired infections, nosocomial infections are also a major social concern. The likelihood of environmental contamination and transmission of the virus based on disease severity is unknown.
Methods
We collected nasopharyngeal, environmental and air samples from patients with COVID-19 admitted to the National Centre for Global Health and Medicine between January 29th and February 29th, 2020. The patients were classified by severity of disease. The collected samples were tested using severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) real-time reverse transcription polymerase chain reaction (real-time RT-PCR).
Results
SARS-CoV-2 was not detected in a subset of 11 air samples. Of the 141 environmental samples collected from three patient bays and two single rooms, four samples tested positive for SARS-CoV-2 by real-time RT-PCR. Detections were made on the surface of a stethoscope used in the care of a patient with severe disease, on the intubation tube of a patient classified as critical (and on ventilator management), and on the surface of a gown worn by the nurse providing care.
Conclusions
Regardless of the patients' disease severity, SARS-CoV-2 was detected on very few environmental surfaces. However, detection of SARS-CoV-2 on stethoscopes used in the care of multiple patients and on the surface of gowns worn by clinical staff indicates that medical devices may be linked to the spread of infection.
Keywords: Severe acute respiratory syndrome coronavirus 2, Environment, Contamination, Hospital
Introduction
The initial reports of coronavirus disease 2019 (COVID-19) came from Wuhan City, China in December 2019. COVID-19 continues to spread throughout the world as of early April 2020. [1] An outbreak of COVID-19 occurred in Japan in February 2020. Human-to-human transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has already been reported both in hospitals and in the community. [[2], [3], [4]] It was suggested that environmental contamination with Middle East Respiratory Syndrome coronavirus (MERS-CoV) was associated with nosocomial infection. [5] Environmental and air contamination with SARS-CoV-2 has been reported, [6,7] but there are few reports of environmental contamination by severity of disease.
Investigating environmental contamination according to disease severity may reveal the mode of transmission of infection in each clinical scenario. If links between likelihood of transmission and disease severity are identified, and given situations of limited medical resources (e.g., personal protective equipment [PPE]), disease severity may be a guide to allocation of those resources.
Methods
Patients with SARS-CoV-2 PCR-positive infections who were admitted to the National Centre for Global Health and Medicine from January 29th to February 29th, 2020 were classified by disease severity to determine whether room contamination varied with severity. Previous studies defined COVID-19 as moderate if the lung was involved and severe if it required adjunctive oxygen therapy. [6] However, in this study, the severity of each patient was defined as follows; mild disease was defined as upper respiratory infection (URI) and fever. Patients with moderate disease were defined as those with URI, fever and pneumonia, while patients with severe disease had URI, fever, pneumonia and also required supplemental oxygen therapy. Critical patients were defined as those with URI, fever, pneumonia and requiring supplemental oxygen on a ventilator. Patients with COVID-19 ranging from mild to severe were admitted to a four-bed negative pressure bay in a general ward, while critical patients were admitted to a single negative pressure room in a general ward and then transferred to a single negative pressure room in an isolation ward. We investigated contamination by collecting air samples and swabbing environmental surfaces in three bays, a single negative pressure room in a general ward and a single negative pressure room in an isolation ward associated with seven patients. The samples were tested by real-time reverse transcription polymerase chain reaction (real-time RT-PCR) and viral RNA load was quantified.
This study was reviewed and approved by the Ethics Committee of the Centre Hospital of the National Centre for Global Health and Medicine (Approval No. NCGM-G-003491-00) on the condition that a document that declares an opt-out policy by which any possible patient and/or relatives could refuse to be included in this study was uploaded on the Web page of the Centre Hospital of the National Centre for Global Health and Medicine.
Sample collection
To examine the possibility of airborne transmission of SARS-CoV-2, we collected 11 air samples in three negative pressure bays (Bay 1 to Bay 3), a single negative pressure room in a general ward (Room 1) and a single negative pressure room in an isolation ward (Room 2) using an MD8 airscan sampling device (Sartorius, Goettingen, Germany) and sterile gelatin filters (80 mm diameter and 3 μm pores; Sartorius). We placed the device on the floor about 1.5 meters–2 meters away from the patient's head. Air was sampled twice, at a speed of 50 L/minute for 20 minutes, in the negative pressure rooms and its associated restrooms. [5] The filters were dissolved aseptically and stored at -80 °C until they were analysed. Cotton swabs premoistened with viral transport medium (VTM) were used to swab surfaces aseptically. The following types of surfaces were swabbed in each bay and room, where applicable: (1) fomites (e.g., smart phones, tablets, masks, stethoscopes, blood pressure cuffs, intubation tubes, infusion pump, pillows, TV remote controls, bed remote controls, syringes, patient clothes, personal data assistants, personal computers, computer mouse, personal protective equipment [PPE; e.g., gown, face shield with mask], consent form paper, patient palm, towel under the intubation tube, front and back of patient G's wife's mask, pulse oximeter probe); (2) fixed structures in the bays and rooms and associated restrooms (e.g., door knobs, bed guardrails, over tables, touch screen of ventilator, monitor, nurse call buttons, TV, curtains, toilet seats, hand soap dispensers, window sill, exhaust port, door sensor); and (3) the ventilation exits on the ceilings in the negative pressure bays and rooms. Cotton swabs with polystyrene shafts (FB57835, Fisher Scientific, Hampton, NH, USA) were moistened with VTM and then rubbed across a maximum area of 4 × 5 cm2 in three different directions, applying even pressure. Immediately after sampling, swabs in VTM were put in the refrigerator before being stored at -80 °C. This sampling method was validated in a prior study. [8] Environmental samples from all rooms (except Room 2) were collected after 6–8 hours of daily room cleaning and disinfection. Room 2 was cleaned and items were disinfected at least once a day. While it was unclear how many hours had passed since cleaning, sampling in Room 2 was done in the late evening. The disinfectant solution contained potassium peroxymonosulfate and sodium dodecylbenzene sulfonate.
Laboratory procedures
Respiratory specimen samples and air and environmental swab samples were sent to the National Institute of Infectious Diseases, Tokyo, Japan, for real-time RT-PCR of SARS-CoV-2. Real-time RT-PCR for SARS-CoV-2 was performed using the QuantiTect Probe RT-PCR Kit (Qiagen, Hilden, Germany) with the following probe and primer sets; WuhanCoV-N1f 5′-GGCCGCAAATTGCACAAT-3′, WuhanCoV-N1r 5′-CCAATGCGCGACATTCC-3′, and WuhanCoV-N1pr-fam 5′-FAM-CCCCCAGCGCTTCAGCGTTCT-TAMRA-3′ targeting nucleoprotein gene (29175–29235 in MN908947.3) for analysis of respiratory samples; NIID_2019-nCOV_N_F2 5′-AAATTTTGGGGACCAGGAAC-3′, NIID_2019-nCOV_N_R2 5′-TGGCAGCTGTGTAGGTCAAC-3′, and NIID_2019-nCOV_N_P2 5′-FAM-ATGTCGCGCATTGGCATGGA-TAMRA-3′ targeting nucleoprotein gene (29125–29282 in GenBank accession MN908947.3) for environmental samples.
Results
One patient (A) had a mild case of COVID-19, two patients (B and C) had moderate disease, three patients (D, E and F) had severe disease, and one patient (G) was in critical condition. We conducted an environmental survey in patient A's room (Bay 1), which he entered with his wife (who had no detectable SARS-CoV-2 infection). Patients B and C were hospitalised in Bay 2 and had been in the same bay for 6 days prior to the environmental study. Patients D and E were admitted to Bay 3 and had been in the same room for 7 days prior to the environmental study. Patient F, who had severe disease, entered Bay 2 nine days after patients B and C were admitted; all three had been in the same room for 1 day before the environmental study. Patient G entered Room 1 three days before the environmental study; he was moved to Room 2 eight days after he entered Room 1 and we conducted an environmental survey two days after the move. In addition, there were 1–2 nurses stationed in Room 2 at all times.
Respiratory samples
SARS-CoV-2 real-time RT-PCR results at the time of sampling of all seven patients were positive. The viral RNA loads of each patient are shown in Table I.
Table I.
Sampling time points in patient illness and viral RNA load
| Disease severity | Patient | Bay or room | Day of illness when samples were collected | Presence of symptoms during sampling | Symptoms | Viral RNA load of patient (Copies/μL) | Viral RNA load of environmental samples (copies/swab) |
|---|---|---|---|---|---|---|---|
| MILD | A | Bay 1 | 5 | Asymptomatic | Fever | 172284.72 (day 2 sputum), 3759.82 (day 5 sputum) | undetectable |
| MODERATE | B | Bay 2 | 9 | Yes | Cough, fever, sputum production, fatigue | 762912.44 (day 6 sputum), 8719.48 (day 10 sputum) | undetectable |
| C | 7 | Asymptomatic | Cough, fever | 1297.50 (day 4 sputum), 151.63 (day 8 sputum) | undetectable | ||
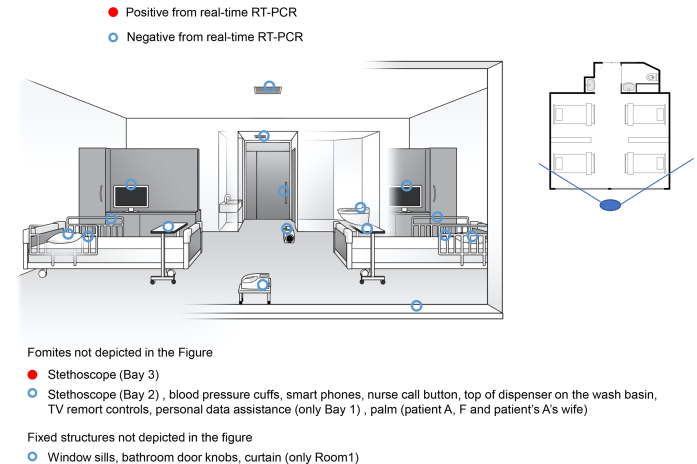
| SEVERE | D | Bay 3 | 8 | Yes | Cough, fever, shortness of breath | 6989.78 (day 6 sputum), 15.02 (day 10 nasopharyngeal) | 2.96 x 103 (Shared Stethoscope Membrane) |
| E | 16 | Yes | Cough, fever, shortness of breath | 49.10 (day 16 nasopharyngeal) | |||
| F | Bay 2; admission 9 days after patient B and C were admitted | 9 | Yes (Breathing 8L of oxygen through a mask) | Cough, fever, shortness of breath | 22780 (day 8 sputum), 4488 (day 8 nasopharyngeal) | undetectable | |
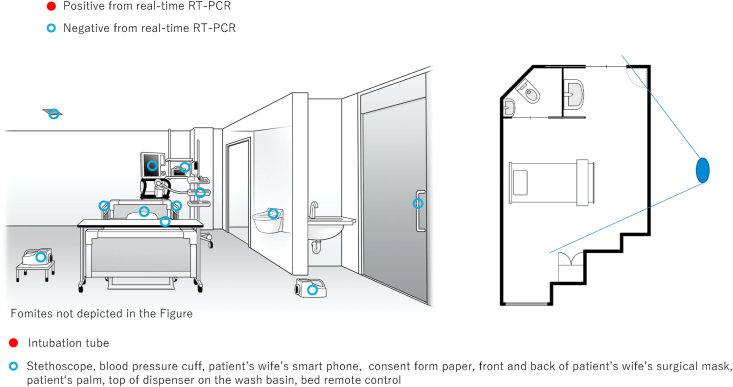
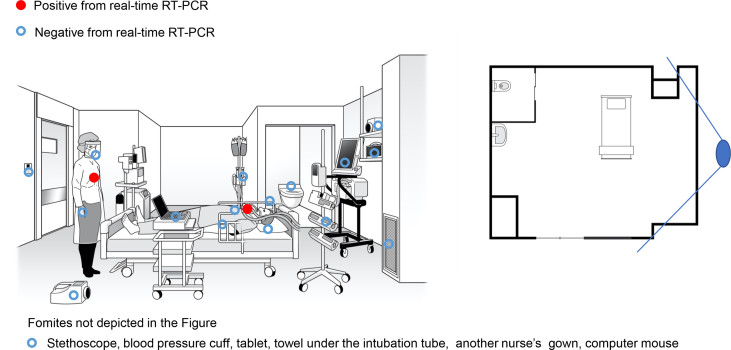
| CRITICAL | G | Room 1; moved to Room 2 on the 9th day after admission | 10, 17 | Yes (mechanical ventilation) | Cough, fever, shortness of breath, fatigue | 15986 (day 10 sputum), 24.85 (day 18 sputum) | 4.62 x 105 (day 10: intubation tube), 4.41 x 103 (day 17: intubation tube), 4.78 x 103 (day17: nurse's upper front part of gown) |
Air samples
All air samples were negative for SARS-CoV-2.
Environmental samples
Of the 141 swab samples collected from the three bays and two single rooms, four samples tested positive for SARS-CoV-2 by real-time RT-PCR (Figure 1, Figure 2, Figure 3). The first positive sample was collected from the Bay 3 (occupied by patients with severe disease), from the membrane surface of a stethoscope used in the care of patients D and E (viral RNA load: 2.96 x 103 copies/swab). The second positive sample was collected from the room of patient G, from the intubation tube on day 10 of admission (viral RNA load: 4.62 x 105 copies/swab). The remaining two positive samples were also collected from the room of the critical patient (G). One positive sample for SARS-CoV-2 (viral RNA load: 4.41 x 103 copies/swab) was from the intubation tube of patient G (on day 17 of admission), and the other (viral RNA load: 4.78 x 103 copies/swab) was from a nurse's PPE (upper front part of gown; Figure 3).
Figure 1.
Bays 1, 2, and 3, each with 4 beds. The floor plan of a four-person negative pressure room into which Patient A and A's wife, Patients B, C, and F, and Patients D and E were admitted. The solid line radiating from the large blue oval indicates the angle of observation used to illustrate the room.
Figure 2.
Single patient Room 1. The floor plan of the single negative pressure room where patient G was hospitalized. The solid line radiating from the large blue oval indicates the angle of observation used to illustrate the room.
Figure 3.
Single patient Room 2. The floor plan of the single negative pressure isolation room and in which patient G was admitted after transfer. The solid line radiating from the large blue oval indicates the angle of observation used to illustrate the room.
Discussion
The disease severity among patients with COVID-19 was classified into four categories (mild, moderate, severe, and critical) and airway, environmental surface, and air samples from rooms occupied by patients in each of these categories were examined. Previous studies have shown that SARS-CoV-2 environmental contamination is low. [9,10] Even in this study, the environmental contamination of SARS-CoV-2 was low. Out of 141 environmental samples, SARS-CoV-2 was detected on four items, including intubation tubes, a stethoscope, and a gown worn as PPE.
One of the most important findings demonstrated by this study was that SARS-CoV-2 was detected on the surface of a stethoscope and a gown. A prior study from Singapore showed that SARS-CoV-2 was detected on a stethoscope. [6] The stethoscope surface was used in direct contact with patients and may have been easily contaminated, as well as a potential route of transmission. Gown contamination was not reported in the prior study, [6] however, SARS-CoV-2 was detected on one of two nurses' gowns sampled in our study. These results suggest that medical devices that come into direct contact with patients can be easily contaminated and that PPE should be properly removed, and medical devices should be kept as clean as possible to prevent contact infections.
The second important finding was that SARS-CoV-2 was not detected in the environment of patients who had higher viral RNA load in the lower respiratory tract and more significant respiratory symptoms. For example, SARS-CoV-2 was not detected inside the oxygen mask of patient F (viral RNA load was 22780 Copy/μl in lower airway specimens), from whom environmental contamination was anticipated.
Thus, there was no association between the patients' viral RNA load or symptoms and environmental contamination. This finding is not consistent with those of previous studies [6,11] in which there was a correlation between environmental contamination and patients' symptoms/viral load in lower respiratory specimens. There were two implications. Firstly, critical care patients are handled by the staff frequently and in very close proximity (increasing risk for contamination). Additionally, critically ill patients have many IV catheters, endotracheal tubes, etc, again frequently handled by staff and the endotracheal tubes can become dislodged providing opportunity for contamination. Also, it is very difficult to clean the environment around these patients well in view of all the equipment. Secondly, patients who are less ill can be self-sufficient and require less equipment, IV, etc and therefore easier to maintain a clean environment. Therefore, it is important to cohort patients/staff, use disposable equipment where possible, and not to change PPE (other than hand hygiene) when cohorting COVID-19 patients in order not to breech and increase risk for contamination.
The third important finding was that SARS-CoV-2 was undetectable in all air samples (taken with an air sampler) and in all rooms ventilation exit surface environments. This was also shown in the study from Singapore. [6] However, the results of other SARS-CoV-2 studies have suggested airborne infection, [7] and this is supported by prior research that has shown aerosol and airborne transmission of MERS-CoV and the severe acute respiratory syndrome (SARS) coronavirus. [5,12] Further, an in vitro study showed that SARS-CoV-2 survived for 3 days in an experimental environment where artificial aerosols were generated. [13] There are three major implications of these data. As noted in the previously published study, the air taken from the hospital room was only a sample, allowing for the possibility that the virus was present but not captured. Another important consideration is that the patients who generated the aerosols may not have been doing so at the time of sampling. In other words, SARS-CoV-2 may not have been detected because the air survey was conducted 3 days after endotracheal intubation, which may have been an aerosol-generating event. It is also possible that some COVID-19 patients produce contaminated aerosols and others do not; additional studies are needed to explore the possibility of airborne transmission of SARS-CoV-2.
There are three limitations in this study. Firstly, this study includes a lack of testing of viral viability through culture after sampling. Without viral culture, it was unclear whether the detected virus was viable. Secondly, the air sampling was done in a negative pressure room, potentially reducing the concentration of the virus in the air to below the level of detection. Thirdly, the SARS-CoV-2 may have gone undetected due to frequent cleaning.
Conclusions
We investigated environmental surfaces and air contamination in SARS-CoV-2 patient bays and rooms by disease severity, and our findings show that environmental contamination by SARS-CoV-2 did not increase with increasing patient severity. SARS-CoV-2 was detected only on the surface of the stethoscope shared by two severe disease patients, the intubation tube of a critical patient who was on a ventilator and the gown of a nurse who cared for the patient. These results suggest that SARS-CoV-2 infection can occur in the environment and medical equipment around the SARS-CoV-2 patient, and therefore, it is necessary to properly clean the environment and medical equipment, and to appropriately don and doff PPE. Air contamination by SARS-CoV-2 was not confirmed in this study but the negative pressure environment may have affected the results, and further studies on air contamination are needed.
Credit author statement
Keiji Nakamura: Investigation, Methodology, Formal analysis, Data Curation, Writing - Original Draft, Project administration. Shinichiro Morioka: Investigation, Methodology, Writing - Review & Editing. Satoshi Kutsuna: Conceptualization. Shun Iida: Investigation, Resources, Writing - Review & Editing. Tadaki Suzuki: Investigation, Resources, Funding acquisition. Noriko Kinoshita: Tetsuya Suzuki: Investigation Yuko Sugiki: Ayako Okuhama: Kohei Kanda: Yuji Wakimoto: Mugen Ujiie: Kei Yamamoto: Resources. Masahiro Ishikane: Yuki Moriyama: Masayuki Ota: Takato Nakamoto: Satoshi Ide: Hidetoshi Nomoto: Yutaro Akiyama: Yusuke Miyazato: Kayoko Hayakawa: Sho Saito: Writing - Review & Editing, Funding acquisition. Norio Ohmagari: Supervision.
Acknowledgements
We would like to thank Takafumi Kurogi for illustrator; Editage (www.editage.com) for English language editing; and National Center for Global Health and Medicine Hospital COVD-19 nursing team.
Conflicts of interest
All authors report no potential conflicts.
Funding
This work was partly supported by funding research grants from the Emerging/Re-emerging Infectious Diseases Project of Japan from the Japan Agency for Medical Research and Development, AMED (20fk0108116h0001).
This study was supported in part by a Grant-in Aid from the Japan Agency for Medical Research and Development (AMED) under Grant Numbers JP19fk0108104 and JP19fk0108110.
References
- 1.World Health Organization Coronavirus disease 2019 (COVID-19) situation report-71. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reportsExternal Link [cited 2020 Mar 30]Available from:
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. NEJM. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q. Importation and human-to-human transmission of a novel coronavirus in Vietnam. NEJM. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S.H., Chang S.Y., Sung M., Park J.H., Bin Kim H., Lee H. Extensive viable Middle East Respiratory Syndrome (MERS) coronavirus contamination in air and surrounding environment in MERS isolation wards. Clin Infect Dis. 2016;63:363–369. doi: 10.1093/cid/ciw239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y. Air, surface environmental, and personal protective equipment contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323(16):1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Z.-D., Wang Z.-Y., Zhang S.-F., Li X., Li L., Li C. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China. Emerg Infect Dis. 2020 doi: 10.3201/eid2607.200885. 2020; Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Killingley B., Greatorex J., Cauchemez S., Enstone J.E., Curran M., Read R.C. Virus shedding and environmental deposition of novel A (H1N1) pandemic influenza virus: interim findings. Health Technol Assess. 2010;14:237–354. doi: 10.3310/hta14460-04. [DOI] [PubMed] [Google Scholar]
- 9.Colaneri M., Seminari E., Piralla A., Zuccaro V., Di Filippo A., Baldanti F. Lack of SARS-CoV-2 RNA environmental contamination in a tertiary referral hospital for infectious diseases in Northern Italy. J Hosp Infect. 2020;105:474–476. doi: 10.1016/j.jhin.2020.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colaneri M., Seminari E., Novati S., Asperges E., Biscarini S., Piralla A. Severe acute respiratory syndrome coronavirus 2 RNA contamination of inanimate surfaces and virus viability in a health care emergency unit. Clin Microbiol Infect. 2020 Aug;26(8):1094.e1–1094.e5. doi: 10.1016/j.cmi.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arons Melissa M., Hatfield Kelly M., Reddy Sujan C., Kimball Anne, James Allison, Jesica R. Presymptomatic SARS-CoV-2 Infection and Transmission in a Skilled Nursing Facility. NEJM. 2020 May 28;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu I.T., Li Y., Wong T.W., Tam W., Chan A.T., Lee J.H. Evidence of airborne transmission of the severe acute respiratory syndrome virus. NEJM. 2004;350:1731–1739. doi: 10.1056/NEJMoa032867. [DOI] [PubMed] [Google Scholar]
- 13.van Doremalen N., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. NEJM. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]