Abstract
Breast cancer (BC) has a poor prognosis and a high number of visceral metastases. Serine protease inhibitor, clade E member 1 (SERPINE1) is a molecule involved in several human malignancies. However, it remains unknown if SERPINE1 plays a role in the development of taxane resistance in TNBC cells. In the present study, the role and mechanism of SERPINE1 in the development of paclitaxel (PTX) resistance in TNBC cells were investigated. A bioinformatics analysis of gene expression profiles in PTX-resistant cells indicated that SERPINE1 was significantly associated with PTX resistance. Furthermore, the levels of SERPINE1 mRNA and protein were higher in PTX-resistant cells with respect to those in PTX-sensitive parent cells. Knockdown of SERPINE1 significantly inhibited cell survival and induced cell apoptosis in vitro. In addition, SERPINE1 silencing led to downregulation of the key angiogenetic vascular endothelial growth factor A (VEGFA). Furthermore, suppression of SERPINE1 markedly attenuated tumor growth in vivo. Collectively, these findings indicated that SERPINE1 significantly contributed to the proliferation and apoptosis of TNBC cells by regulating VEGFA expression. The present study demonstrated SERPINE1 as an oncogene in PTX drug resistance of breast cancer, and revealed that it may serve as a possible target for treating BC.
Keywords: triple-negative breast cancer, paclitaxel, drug resistance, cell apoptosis, vascular endothelial growth factor A
Introduction
Triple-negative breast cancer (TNBC) is a subtype of breast cancer in which estrogen receptor (ER) and the progesterone receptor (PgR), as well as the epidermal growth factor receptor 2 (HER2) are all negative (1). As one of the most aggressive breast cancer subtypes, TNBC accounts for 10–20% of all malignant breast tumors, and frequently has a worse prognosis and greater risks for recurrence and metastasis than other types of breast cancer (2).
While the number of potential therapeutic agents being tested in clinical trials with metastatic breast cancer patients continues to increase, currently, chemotherapy is the standard therapeutic strategy for TNBC (3,4). Paclitaxel (PTX), a natural taxane diterpene, was initially isolated from the bark of the Pacific yew (Taxus brevifolia), and is currently widely used in chemotherapy for TNBC (5,6). PTX specifically binds to and stabilizes the β-subunit of tubulin, thereby inhibiting the disassembly of microtubules in dividing cells, resulting in mitotic arrest and subsequent cell death (7,8). However, while PTX is effective for treating several types of cancer, >50% of patients with TNBC become resistant to chemotherapy, typically within 6 to 10 months (9,10). The frequent development of PTX drug resistance in TNBC patients underscores the importance of exploring the underlying mechanisms of PTX resistance, and identifying the critical molecules involved in this process. A better understanding of the mechanism for PTX resistance may improve our ability to treat BC.
In the present study, the related signaling pathways and genes in the process of PTX resistance in BC cell lines were investigated. Notably, a serine protease inhibitor, clade E member 1 (SERPINE1) was identified as a critical factor that mediated PTX drug resistance in BC cells. SERPINE1, also known as plasminogen activator inhibitor, type 1 (PAI-1), acts as a vital inhibitor of serine proteases that play important roles in signal transduction, cell adhesion, and cell migration (11–13). SERPINE1 also regulates urokinase and plasminogen activators that transform the pro-enzyme plasminogen to plasmin, which subsequently promotes cellular invasion via activation of matrix metalloproteinases and degradation of the extracellular matrix (12). A high level of SERPINE1 has been revealed to be associated with a poor prognosis of breast cancer (11). However, whether SERPINE1 plays a role in the development of drug resistance in BC is unknown.
Materials and methods
Bioinformatics analysis
Gene expression datasets used for bioinformatics analyses were downloaded from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (14) by importing the accession numbers GSE28784 and GSE90564. GSE28784 contains the gene expression data of sensitive, docetaxel-resistant, and PTX-resistant MDA-MB-231 cells. GSE90564 is a gene expression profiling dataset consisting of 5 BC cell lines (BT20, SUM149, MDA-MB-231, MDA-MB-436, and MDA-MB-468), which are resistant to PTX after being exposed to increased concentrations of PTX, and their gene expression patterns were compared to those of parental PTX-sensitive cells. We used the 151 genes that were shared by both datasets to perform Gene Ontology (GO) (15) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses (16) for the purpose of identifying gene functions and pathways related to PTX resistance. Gene interaction analysis and Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) were used to analyze the functional associations between genes that may be responsible for PTX resistance, and then to identify the core regulatory genes on the list.
K-means clustering method
Based on the previous studies (17,18), K-means clustering method was applied to calculate the different clusters of the gene interaction network.
Cell line generation and cell culture
MDA-MB-231 and MCF-7 cells were obtained from the ATCC. Paclitaxel-resistant cells were created as previously described (19,20). The parent PTX-sensitive cells were continuously maintained in a paclitaxel containing medium in which the paclitaxel concentration gradient was between 2 and 30 nM. Both cell lines were grown in RPMI-1640 medium (Thermo Fisher Scientific, Inc.) containing 1% penicillin/streptomycin and 10% fetal bovine serum (Thermo Fisher, Scientific, Inc.). Cells were incubated at 37°C and 5% CO2. For PTX cells, the culture medium contained 30 nM paclitaxel.
MTT assay
Cells (1×104 cells/well) were plated into 96-well plates. After culture for 24 h at 37°C, the original medium was replaced with fresh medium containing 0, 0.1, 0.5, 1, 5, and 10 µM paclitaxel. After 48 h, the cells in each well were treated with 20 µl of MTT (0.5 mg/ml), and incubation was continued for an additional 4 h at 37°C, and then 100 µl of DMSO was applied to dissolve the formazan in each well for 15 min at 37°C. Subsequently, the optical density (OD) was examined at a wavelength of 492 nm using a plate reader (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
The total RNAs were isolated from the treated cells or clinical tissues using the TRIzol reagent (cat no. 15596-026; Invitrogen; Thermo Fisher Scentific, Inc.). The purified RNA was used as a template to carry out reverse transcription using the Prime Script RT Master Mix Perfect Real Time kit (Takara Biotechnology Co., Ltd.). Quantitative analysis of target genes was performed using SYBR® GreenER™ qPCR SuperMix (Invitrogen; Thermo Fisher Scientific, Inc.) on an ABI 7700 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The thermocycling procedures were as follows: Incubation at 95°C for pre-denaturation for 2 min, followed by 40 cycles with denaturation at 95°C for 22 sec, and annealing at 59°C for 20 sec. The primer sequences used were as follows: SERPINE1 forward, 5′-GCAAGGCACCTCTGAGAACT-3′ and reverse, 5′-GGGTGAGAAAACCACGTTGC-3′. β-actin was utilized for the standardization, and its primer sequences were: Forward, 5′-AGAGCTACGAGCTGCCTGAC-3′ and reverse, 5′-AGCACTGTGTTGGCGTACAG-3′.
Western blot analysis
The antibodies in this experiment were obtained from Proteintech. Treated cells were lysed in cell lysis buffer containing 1 mM PMSF (Beyotime Institute of Biotechnology) on ice for 15 min and centrifuged at 16,100 × g for 5 min at 4°C. Next, 50-µg samples of total protein were dispersed on 10% SDS-PAGE gels by electrophoresis and transferred onto cut-out nitrocellulose membranes (Beyotime Institute of Biotechnology). The membranes were blocked for 1 h with 5% skimmed milk at room temperature and incubated with anti-SERPINE1 antibody (cat. no. A00637-1; 1:1,000 dilution; Boster Biological Technology), VEGFA (cat. no. BA0407; 1:500 dilution; Wuhan Boster Biological Technology Co., Ltd.), cleaved caspase-3 (product code ab32042; 1:500 dilution; Abcam), Bax (cat. no. A00183; 1:1,000 dilution; Wuhan Boster Biological Technology Co., Ltd.) and β-actin (product code ab8224; 1:1,000 dilution; Abcam) overnight at 4°C. Subsequently, they were incubated with horseradish peroxidase (HRP)-labeled secondary antibodies (1:1,000 dilution; product code ab7090; Abcam) for 1 h at room temperature. After incubation, the membranes were developed using chemiluminescent substrates (Beyotime Institute of Biotechnology). The densitometric quantification of the bands were calculated using the ImageJ software (version 1.50b; National Institutes of Health; http://imagej.nih.gov/ij/).
Gene knockdown with short hairpin RNAs (shRNAs)
shRNA (5′-CCGGCCTGAAGGTGAAGAACATCATCTCGAGATGATGTTCTTCACCTTCAGGTTTTTG-3′) that specifically targeted SERPINE1 and control shRNA plasmids (5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG-3′) were acquired from Sigma-Aldrich; Merck KGaA. For transient transfection, the treated MDA-MB-231/PTX and MCF-7/PTX cells (6×105 cells/well) were plated into 6-well plates and cultured overnight at 37°C. The adherent cells were then transfected with the shRNAs using Lipofectamine 2000™ reagent (Thermo Fisher Scientific, Inc.) according to the instructions supplied by the manufacturer. Briefly, MDA-MB-231/PTX and MCF-7/PTX cells were transfected with control and SERPINE1 shRNA at concentration of 100 ng/µl for 5 min at room temperature. After being cultured for 5 h at 37°C, the cells were washed with PBS and cultured with fresh culture medium. Finally, the transfected cells were cultured for an additional 48 h at 37°C prior to being treated or harvested for further evaluation.
Flow cytometric assay
The treated MDA-MB-231/PTX and MCF-7/PTX cells (1×106 cells/well) were double-stained with an Annexin V-FITC/PI apoptosis detection kit (Hangzhou Multi Sciences (Lianke) Biotech Co., Ltd.) according to the manufacturer's instructions. The results were monitored by flow cytometry (FACScan; BD Biosciences). The data was analyzed using the CellQuest™ Pro software (version 5.1; BD Biosciences). Early-stage apoptotic cells contained Annexin V-positive and PI-negative cells, while late-stage apoptotic cells included both Annexin V-positive and PI-positive cells. All experiments were performed independently in triplicate.
In vivo tumorigenic assay
A total of 30 male nude mice (6 weeks old, 20.6±2.3 g) were provided from Department of Laboratory Animals of Central South University, and the housing conditions of the nude mice were as follows: 22±1°C temperature, 50–60% humidity, 12-h light/dark cycle, and ad libitum access to food and water. To evaluate the tumorigenic capacity of BC cells and MDA-MB-231 cells, they were first transfected with either the control shRNA construct or shSERPINE1 and then cultured to achieve a sufficient population of SERPINE1-knockdown cells. Next, 5×106 cells were harvested, re-suspended, and subcutaneously injected into the 30 nude mice (5 mice in each group). The tumor growth was periodically monitored by examining the tumor size. At the end of the study, the mice were sacrificed after being anesthetized with 1% pentobarbital sodium (i.p.) at a dose of 50 mg/kg and then sacrificed by decapitation. Successful anesthesia was considered mice breathing and with a heartbeat, and mice that did not have a heartbeat or breath were presumed euthanized. Then tumors were removed for further evaluation. Animal experiments were approved by the Animal Ethics Committee of Xiangnan University Affiliated Hospital (approval no. 2019sydw0821).
Statistical analysis
All results were expressed as a mean value ± standard deviation (SD). Statistical analyses were conducted using GraphPad Prism (version 6.0; GraphPad Software, Inc.) or SPSS 14.0 (SPSS, Inc.). Unpaired Student's t-test was used to analyze the differences between two groups. One-way analysis of variance (ANOVA) with Tukey's post hoc test was used for comparisons among three or more groups. A P-value <0.05 was considered to indicate a statistically significant difference. All experiments were repeated at least three times.
Results
Genes and signaling pathways involved in PTX resistance in BC cell lines
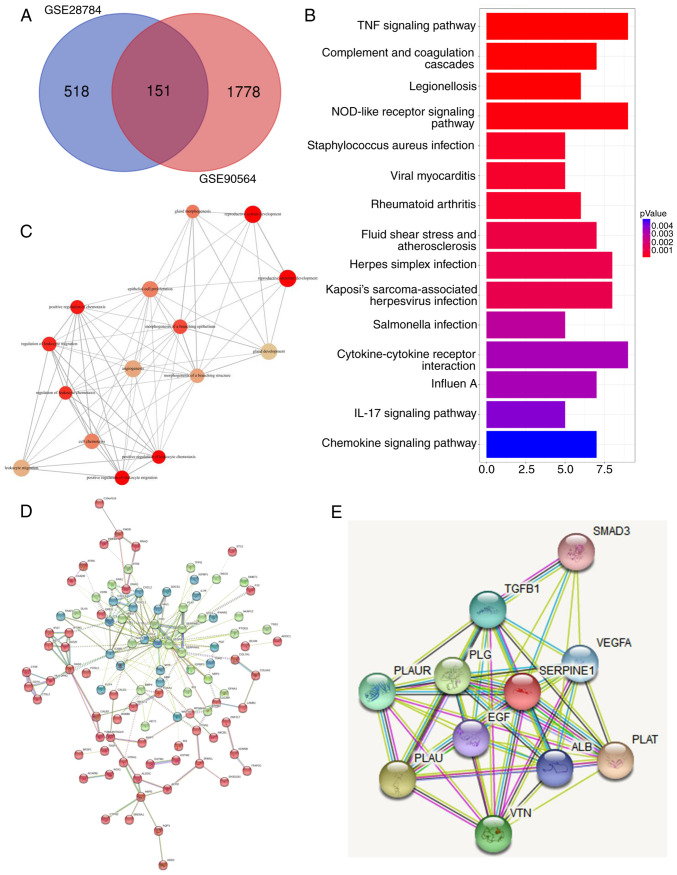
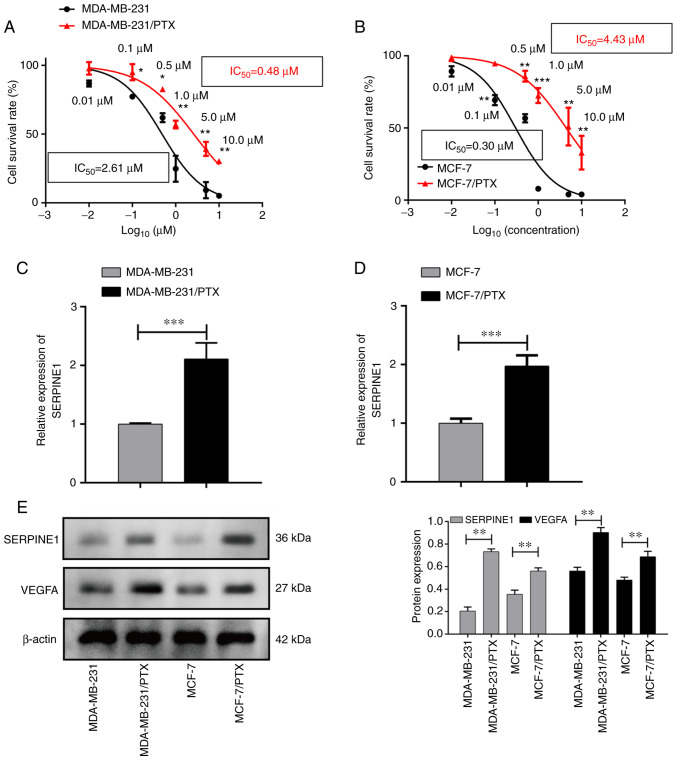
In order to identify common mechanisms for PTX resistance among BC cells, two datasets that were generated by comparing the gene expression patterns of PTX-resistant cells with those of PTX-sensitive cells were examined. A list of differentially expressed genes that may be related to PTX resistance was generated for each dataset. A Venn diagram revealed that 151 overlapping genes were differentially regulated in both datasets (Fig. 1A). Pathway analysis indicated that the differentially expressed molecules were highly involved in TNF and NOD signaling (Fig. 1B), which are pathways known to regulate cancer metastasis (21,22). In addition, a subsequent annotation analysis also revealed that these differentially expressed genes mainly participated in multiple biological processes including cell migration and chemotaxis (Fig. 1C). Next, based on the differentially expressed genes, various genes with high core connectivity were identified through the gene interaction network (Fig. 1D). In addition, STRING was applied to reveal the interaction of SERPINE1-generated proteins, among which, it was observed that SERPINE1 exhibited close associations with several genes (SMAD3, TGF-β1, VEGFA, PLAUR, PLG, PLAU, VTN, ALB, PLAT and EGF) (Fig. 1E). To further examine the role of SERPINE1 in the development of PTX resistance in TNBC cells, SERPINE1 expression in PTX-resistant and -sensitive cells was evaluated. As revealed in Fig. 2A and B, two breast cancer cell lines were revealed to develop PTX resistance, characterized by increased IC50 to PTX, as demonstrated by their higher survival rates when compared to their PTX-sensitive parental cells after treatment with specific doses of PTX. qPCR results revealed that SERPINE1 was highly expressed in PTX-resistant cells (Fig. 2C and D), and was overexpressed in the PTX-resistant cells when compared to the parental cells. Notably, a concomitant increase in VEGFA expression in the PTX-resistant cells was also detected (Fig. 2E).
Figure 1.
Identification of signaling pathways and genes related to PTX resistance in breast cancer cell lines. (A) Venn diagram distribution of differentially expressed genes from 2 arrays. As revealed, 518 genes were specifically differentially expressed in GSE28784 cells and 1,778 genes were specifically differentially expressed in GSE90564 cells, while 151 genes were expressed in both datasets. (B) GO annotations and a functional pathway analysis of 151 genes performed using GO-terms and KEGG. (C) Gene-set enrichment analysis of common differentially regulated genes from A. (D) Gene interaction network constructed based on the shared gene list from A. Different colors represent different clusters as calculated using the K-means clustering method. Core genes in the network were identified based on their edge connectivity number. (E) Gene interaction map related to SERPINE1 generated using STRING. PTX, paclitaxel; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and genomes; SERPINE1, serine protease inhibitor, clade E member 1; STRING, Search Tool for the Retrieval of Interacting Genes.
Figure 2.
SERPINE1 is upregulated in PTX-resistant BC cell lines. (A) Survival curves for MDA-MB-231 PTX-resistant and -sensitive cells treated with various concentrations of PTX. *P<0.05, **P<0.01. (B) Survival curves for MCF-7 PTX-resistant and -sensitive cells treated with various concentrations of PTX. **P<0.01, ***P<0.001. Some points have no error bars because the standard deviation is too small in A and B. (C) SERPINE1 mRNA levels in MDA-MB-231 PTX-resistant vs. -sensitive cells as assessed by RT-qPCR. ***P<0.001. (D) SERPINE1 mRNA levels in MCF-7 PTX-resistant vs. -sensitive cells as assessed by RT-qPCR. ***P<0.001. (E) SERPINE1 protein expression in MDA-MB-231 and MCF-7 PTX-resistant vs. -sensitive cells as measured by western blotting. **P<0.01. SERPINE1, serine protease inhibitor, clade E member 1; PTX, paclitaxel; BC, breast cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction. *P<0.05, **P<0.01, ***P<0.001.
Suppression of SERPINE1 abolishes PTX resistance and promotes BC cell apoptosis
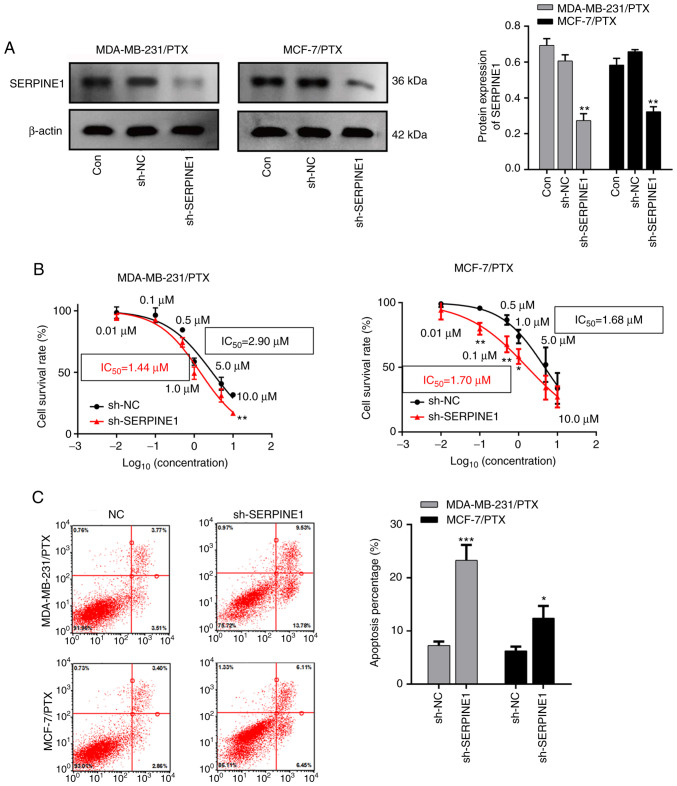
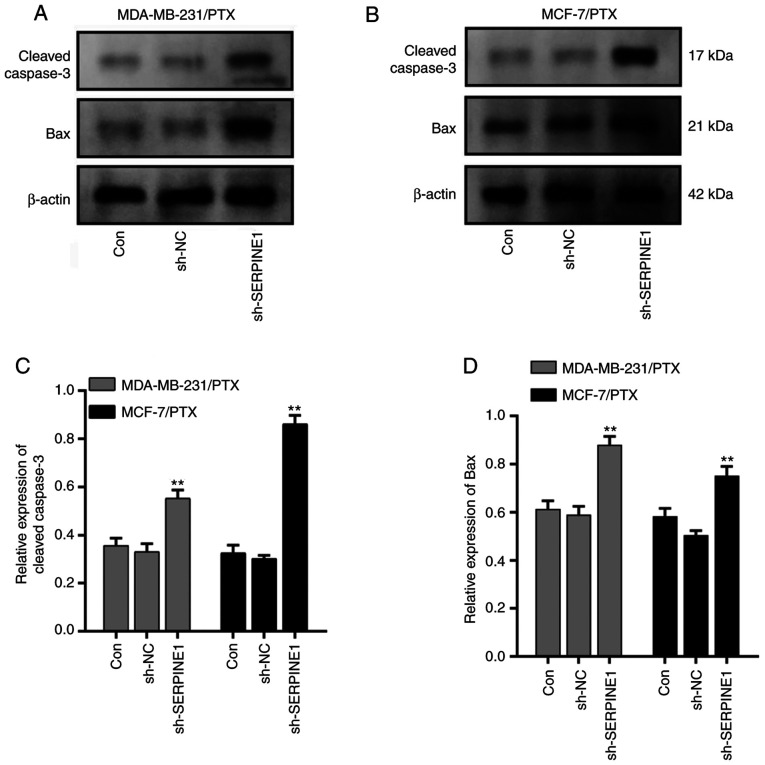
Given that SERPINE1 levels were significantly increased in PTX resistant cells, it was investigated whether knockdown of SERPINE1 impaired resistance of BC to PTX. A shRNA that specifically targeted SERPINE1 RNA was used to efficiently suppress SERPINE1 expression in MDA-MB-231 and MCF-7 PTX-resistant cells (Fig. 3A), and it was determined that knockdown of SERPINE1 significantly inhibited the survival of PTX-resistant BC cells (Fig. 3B) with attenuated the IC50 to PTX. Furthermore, a flow cytometric analysis indicated that SERPINE1 suppression significantly induced apoptosis in large populations of both PTX-resistant cell lines (Fig. 3C). Moreover, cleaved caspase-3 and Bax, which are important markers of cell apoptosis activation (23), were both significantly upregulated following SERPINE1 suppression (Fig. 4) in MDA-MB-231/PTX (Fig. 4A and C) and MCF-7/PTX (Fig. 4B and D) cells.
Figure 3.
Knockdown of SERPINE1 suppresses PTX-resistant BC cell survival and promotes apoptosis. (A) Western blots revealing the levels of SERPINE1 protein in knockdown cells. **P<0.01. (B) Survival curves for MDA-MB-231 and MCF-7 PTX-resistant cells with/without SERPINE1 suppression and treated with various concentrations of PTX. *P<0.05, **P<0.01. Some points have no error bars because the standard deviation is too small. (C) Flow cytometric detection of cell apoptosis among BC cells with/without SERPINE1 suppression. *P<0.05, ***P<0.001. SERPINE1, serine protease inhibitor, clade E member 1; PTX, paclitaxel; BC, breast cancer.
Figure 4.
SERPINE1 suppression induces apoptosis in PTX-resistant TNBC cells. (A) Levels of cleaved caspase-3 and Bax proteins in the control and SERPINE1-knockdown MDA-MB-231 PTX-resistant cells. (B) Levels of cleaved caspase-3 and Bax proteins in the control and SERPINE1-knockdown MCF-7 PTX-resistant cells. (C) Quantification of cleaved caspase-3 protein levels in the control and SERPINE1-knockdown PTX-resistant cells. **P<0.01. (D) Quantification of Bax protein levels in the control and SERPINE-knockdown PTX-resistant cells. **P<0.01. SERPINE1, serine protease inhibitor, clade E member 1; PTX, paclitaxel; TNBC, triple-negative breast cancer.
Knockdown of SERPINE1 suppresses the tumorigenic capacity of PTX-resistant TNBC cells in vivo
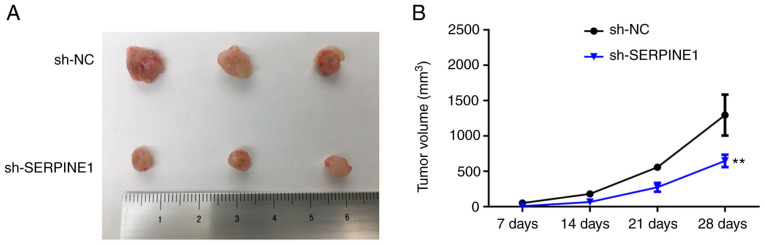
To explore how SERPINE1 may regulate the survival of PTX-resistant cells in vivo, PTX-resistant BC cells with or without SERPINE1 knockdown were inoculated into mice. Consistent with the in vitro observations, the tumors generated by SERPINE1-knockdown cells were significantly smaller than those generated by the control cells (Fig. 5), indicating that suppression of SERPINE1 may be an effective strategy for counteracting PTX-resistant BC cell proliferation in vivo.
Figure 5.
Downregulation of SERPINE1 decreases the tumorigenic capacity of PTX-resistant BC cells in vivo. (A) Image revealing the appearance of tumors from mice that were inoculated with MDA-MB-231/PTX cells with or without SERPINE1 knockdown. (B) Quantification of tumor volumes at the end of the in vivo tumorigenic assays. **P<0.01. SERPINE1, serine protease inhibitor, clade E member 1; PTX, paclitaxel; BC, breast cancer.
VEGFA may mediate SERPINE1 in promoting PTX resistance
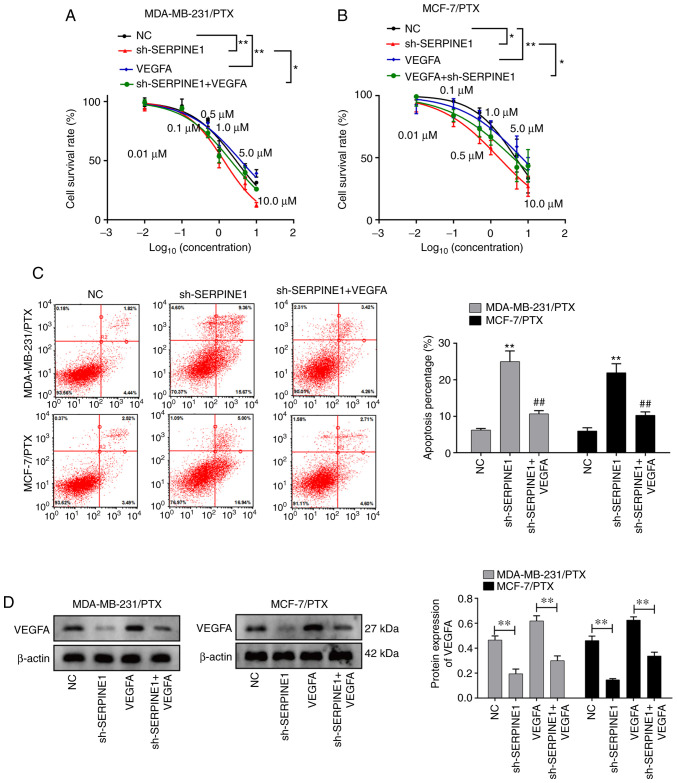
Since concomitant increases in VEGFA and SERPINE1 were detected in PTX-resistant BC cell lines, and given the central role of VEGFA in promoting tumor angiogenesis and growth (23–25), it was hypothesized that SERPINE1 may promote PTX resistance through induction of VEGFA. To test this hypothesis, PTX-resistant cells with or without SERPINE1 knockdown were further treated with VEGFA. Notably, it was revealed that VEGFA treatment abolished the effect of SERPINE1 suppression in regulating cell survival (Fig. 6A and B), suggesting VEGFA induction as a potential mechanism for SERPINE1-regulated PTX resistance. Consistent with this observation, the addition of VEGFA also significantly prevented cells from entering apoptosis (Fig. 6C). The ability of SERPINE1 to regulate VEGFA in TNBC cell lines was also confirmed. As revealed in Fig. 6D, knockdown of SERPINE1 significantly decreased VEGFA expression in both BC cell lines, indicating that SERPINE1 acts to upregulate VEGFA in BC cells.
Figure 6.
VEGFA is regulated by SERPINE1 and mediates PTX resistance in BC cells. (A) Survival curves for MDA-MB-231 PTX-resistant cells with/without SERPINE1 suppression and supplemented with VEGFA. *P<0.05, **P<0.01. (B) Survival curves for MCF-7/PTX cells with/without SERPINE1 suppression and supplemented with VEGFA. Some points have no error bars because the standard deviation was too small in A and B. *P<0.05, **P<0.01. (C) Flow cytometric evaluation of apoptosis among BC cells with/without SERPINE1 suppression and treated with VEGFA. **P<0.01 vs. NC group; ##P<0.01 vs. sh-SERPINE1 group. (D) Expression of VEGFA protein in BC cells with/without SERPINE1 suppression and treated with VEGFA. **P<0.01. VEGFA, vascular endothelial growth factor A; SERPINE1, serine protease inhibitor, clade E member 1; PTX, paclitaxel; BC, breast cancer.
Discussion
Breast cancer, as a heterogeneous disease, possesses multifarious molecular subtypes. The most well recognized BC markers are hormonal receptors (ER, PGR, and HER2), and breast cancers that express specific receptors can be treated with drugs that specifically target those receptor molecules (26–29). Conversely, TNBC, which lacks specific receptors, has emerged as the most aggressive BC subtype and is challenging to treat (30–35). To date, no single treatment has proven to be effective in all BC subtypes. Chemotherapeutic drugs are commonly used to treat receptor-negative subtypes, and taxanes are considered to be the first-line treatment for TNBC (36–38). However, treatment with taxanes (such as PTX) usually leads to a short-lasting benefit due to the development of PTX resistance (36,39). Therefore, gaining a better understanding of the underlying mechanisms for BC PTX resistance is critical for improving the efficacy of chemotherapy and developing new strategies for the treatment of BC.
In the present study, SERPINE1 was identified as an important factor that mediates development of PTX resistance in TNBC cells. SERPINE1 expression was significantly increased in PTX-resistant cells when compared with PTX-sensitive cells. Overexpression of SERPINE1 has been reported to enhance the migration of cancer cells (40). Although the genetic and environmental determinants of SERPINE1 expression are not fully understood, several studies have suggested that SERPINE1 levels can be regulated by cytokines, growth factors, and hormones (41,42). Higgins (43) reported that SERPINE1 was localized at the tumor invasive front, and its expression could be induced by TGF-β1 during the early progression stage of squamous cell carcinoma. Other findings suggested involvement of the EGFR/MEK/Rho-ROCK signaling pathway in SERPINE1 induction (44).
In PTX-resistant TNBC cells, it was revealed that knockdown of SERPINE1 by shRNA significantly decreased cell survival and promoted cell apoptosis. Previous study indicated that an increase of SERPINE1 expression protects against programmed cell death (45). Conversely, studies revealed that SERPINE1 deficiency promotes apoptosis in multiple cancer types, which is consistent with our observation (45–47). The underlying mechanism for SERPINE1-mediated cell survival may involve activation of the Akt and ERK signaling pathways and suppression of Fas/FasL-dependent apoptosis (48). The present study demonstrated that knockdown of SERPINE1 disrupted tumorigenesis, which suggests the value of targeting SERPINE1 to treat BC and prevent PTX drug resistance in vivo.
Several mechanisms may contribute to SERPINE1-mediated cancer progression and PTX resistance. According to previous studies, SERPINE1 functions to prevent excessive degradation of the extracellular matrix, modulates cell adhesion, and stimulates cell proliferation and angiogenesis (49,50). Tumor growth and metastasis are heavily dependent on angiogenesis (51). Notably, PTX displays antiangiogenic activity via its antiproliferative effect on activated endothelial cells, and that effect is achieved by downregulating the levels of VEGF and Ang-1 in tumor cells (52). It was hypothesized that PTX resistance mediated by SERPINE1 may involve a VEGF factor. When SERPINE1 expression was knocked down in BC cells, VEGFA expression was significantly decreased, suggesting that SERPINE1 may function as an inducer of VEGFA in order to promote PTX resistance in cancer cells. VEGFs are known to play important roles in angiogenic processes that are critical for tumor cell survival, proliferation, and migration (24). Loss of VEGFA expression was reported to increase the sensitivity of colorectal cancer cells to 5-fluorouracil by inducing apoptosis (53,54). Humanized monoclonal antibodies directed against VEGFA were used for antiangiogenic therapy in the clinical treatment of cancer (55). Moreover, accumulating evidence indicates that VEGFA plays an important role in enhancing cancer cell resistance to chemotherapeutic drugs by protecting cancer cells from the cytotoxic effects of those agents (56). For example, the efficacy of nanoparticle albumin-bound PTX (nab-PTX) in treating human breast cancer was significantly enhanced by the concurrent administration of anti-VEGFA (57). Although potential mechanisms must be further explored, studies have suggested the involvement of apoptosis regulatory molecules and the PI3K/AKT signaling pathway (58), which are associated with SERPINE1 function.
In conclusion, the present data demonstrated that increased levels of SERPINE1 expression contributed to the resistance of BC to treatment with PTX. In addition, SERPINE1-mediated drug resistance is mediated by an upregulation of VEGFA and subsequent suppression of cell apoptosis. The present findings also suggest SERPINE1 as a potential target for eliminating PTX resistance during cancer treatment. However, certain limitations still remain in the present study, such as the specific mechanism of SERPINE1 in BC with PTX resistance, the influences of SERPINE1 on the migration and invasion abilities of BC with PTX resistance.
Acknowledgements
We would like to thank Dr Edward C. Mignot, Shandong University, for linguistic advice.
Funding
The present study was supported by Hunan Provincial Natural Science Foundation of China (grant no. 2018JJ6123), the Planned Science and Technology Project of Hunan Province (grant no. 2017SK4010), the Scientific Research Fund of Hunan Provincial Health and Family Planning Commission (grant no. C2017014), Chenzhou Science and Technology Project, and the Natural Science Foundation of Xiangnan University (grant no. 2015XB12).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QZ and DJ designed the experiments. QZ performed most of the experiments with the assistance of LL. QZ and LL collected and analyzed the data. DJ validated the data analysis. QZ drafted the manuscript and LL and DJ revised the draft. All authors approved the final manuscript before submission.
Ethics approval and consent to participate
Animal experiments were approved by the Animal Ethics Committee of Xiangnan University Affiliated Hospital (approval no. 2019sydw0821).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gong Z, Wang J, Wang D, Buas MF, Ren X, Freudenheim JL, Belinsky SA, Liu S, Ambrosone CB, Higgins MJ. Differences in microRNA expression in breast cancer between women of African and European ancestry. Carcinogenesis. 2019;40:61–69. doi: 10.1093/carcin/bgy134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomao F, Papa A, Zaccarelli E, Rossi L, Caruso D, Minozzi M, Vici P, Frati L, Tomao S. Triple-negative breast cancer: New perspectives for targeted therapies. Onco Targets Ther. 2015;8:177–193. doi: 10.2147/OTT.S67673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoso F, Costa A, Norton L, Senkus E, Aapro M, André F, Barrios CH, Bergh J, Biganzoli L, Blackwell KL, et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2) Breast. 2014;23:489–502. doi: 10.1016/j.breast.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, Zackrisson S, Cardoso F, ESMO Guidelines Committee Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v8–v30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Wang F, Sun D, Wang R. A review of the ligands and related targeting strategies for active targeting of paclitaxel to tumours. J Drug Target. 2016;24:590–602. doi: 10.3109/1061186X.2016.1154561. [DOI] [PubMed] [Google Scholar]
- 6.Paridaens R, Biganzoli L, Bruning P, Klijn JG, Gamucci T, Houston S, Coleman R, Schachter J, Van Vreckem A, Sylvester R, et al. Paclitaxel versus doxorubicin as first-line single-agent chemotherapy for metastatic breast cancer: A European organization for research and treatment of cancer randomized study with cross-over. J Clin Oncol. 2000;18:724–733. doi: 10.1200/JCO.2000.18.4.724. [DOI] [PubMed] [Google Scholar]
- 7.Gascoigne KE, Taylor SS. How do anti-mitotic drugs kill cancer cells? J Cell Sci. 2009;122:2579–2585. doi: 10.1242/jcs.039719. [DOI] [PubMed] [Google Scholar]
- 8.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10:194–204. doi: 10.1038/nrc2830. [DOI] [PubMed] [Google Scholar]
- 9.Koudelka S, Turánek J. Liposomal paclitaxel formulations. J Control Release. 2012;163:322–334. doi: 10.1016/j.jconrel.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Yuan P, Shentu J, Xu J, Burke W, Hsu K, Learoyd M, Zhu M, Xu B. Pharmacokinetics and safety of olaparib tablets as monotherapy and in combination with paclitaxel: Results of a Phase I study in Chinese patients with advanced solid tumours. Cancer Chemother Pharmacol. 2019;83:963–974. doi: 10.1007/s00280-019-03799-1. [DOI] [PubMed] [Google Scholar]
- 11.Azimi I, Petersen RM, Thompson EW, Roberts-Thomson SJ, Monteith GR. Hypoxia-induced reactive oxygen species mediate N-cadherin and SERPINE1 expression, EGFR signalling and motility in MDA-MB-468 breast cancer cells. Sci Rep. 2017;7:15140. doi: 10.1038/s41598-017-15474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeberg MAT, Farhat YM, Easa A, Kallenbach JG, Malcolm DW, Buckley MR, Benoit DSW, Awad HA. Serpine1 knockdown enhances MMP activity after flexor tendon injury in mice: Implications for adhesions therapy. Sci Rep. 2018;8:5810. doi: 10.1038/s41598-018-24144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K, Zhang S, Zhang D, Tao Q, Zhang T, Liu G, Liu X, Zhao T. Identification of SERPINE1, PLAU and ACTA1 as biomarkers of head and neck squamous cell carcinoma based on integrated bioinformatics analysis. Int J Clin Oncol. 2019;24:1030–1041. doi: 10.1007/s10147-019-01435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41((Database Issue)):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Gene Ontology Consortium, corp-author. The gene ontology resource: 20 Years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei Y, Yu D, Bin Z, Yang Y. Interactive K-means clustering method based on user behavior for different analysis target in medicine. Comput Math Methods Med. 2017;2017:4915828. doi: 10.1155/2017/4915828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang TN, Li TJ, Shao GF, Wu SX. An improved K-means clustering method for cDNA microarray image segmentation. Genet Mol Res. 2015;14:7771–7781. doi: 10.4238/2015.July.14.3. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Dong Q, Hu S, Cai J, Zhang W, Sun J, Wang T, Xie J, He H, Xing J, et al. Proteomic analysis of the proteins that are associated with the resistance to paclitaxel in human breast cancer cells. Mol Biosyst. 2014;10:294–303. doi: 10.1039/C3MB70428A. [DOI] [PubMed] [Google Scholar]
- 20.Panayotopoulou EG, Müller AK, Börries M, Busch H, Hu G, Lev S. Targeting of apoptotic pathways by SMAC or BH3 mimetics distinctly sensitizes paclitaxel-resistant triple negative breast cancer cells. Oncotarget. 2017;8:45088–45104. doi: 10.18632/oncotarget.15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edilova MI, Abdul-Sater AA, Watts TH. TRAF1 signaling in human health and disease. Front Immunol. 2018;9:2969. doi: 10.3389/fimmu.2018.02969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu S, Jin J, Gokhale S, Lu AM, Shan H, Feng J, Xie P. Genetic alterations of TRAF proteins in human cancers. Front Immunol. 2018;9:2111. doi: 10.3389/fimmu.2018.02111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewangan J, Srivastava S, Mishra S, Divakar A, Kumar S, Rath SK. Salinomycin inhibits breast cancer progression via targeting HIF-1α/VEGF mediated tumor angiogenesis in vitro and in vivo. Biochem Pharmacol. 2019;164:326–335. doi: 10.1016/j.bcp.2019.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273:114–127. doi: 10.1111/joim.12019. [DOI] [PubMed] [Google Scholar]
- 25.Jia J, Zhang H, Zhang H, Du H, Liu W, Shu M. Activated androgen receptor accelerates angiogenesis in cutaneous neurofibroma by regulating VEGFA transcription. Int J Oncol. 2019;55:157–166. doi: 10.3892/ijo.2019.4797. [DOI] [PubMed] [Google Scholar]
- 26.Alluri P, Newman LA. Basal-like and triple-negative breast cancers: Searching for positives among many negatives. Surg Oncol Clin N Am. 2014;23:567–577. doi: 10.1016/j.soc.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carotenuto P, Roma C, Rachiglio AM, Botti G, D'Alessio A, Normanno N. Triple negative breast cancer: From molecular portrait to therapeutic intervention. Crit Rev Eukaryot Gene Expr. 2010;20:17–34. doi: 10.1615/CritRevEukarGeneExpr.v20.i1.20. [DOI] [PubMed] [Google Scholar]
- 28.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 29.Kalimutho M, Parsons K, Mittal D, López JA, Srihari S, Khanna KK. Targeted therapies for triple-negative breast cancer: Combating a stubborn disease. Trends Pharmacol Sci. 2015;36:822–846. doi: 10.1016/j.tips.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jhan JR, Andrechek ER. Triple-negative breast cancer and the potential for targeted therapy. Pharmacogenomics. 2017;18:1595–1609. doi: 10.2217/pgs-2017-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng CK, Schultheis AM, Bidard FC, Weigelt B, Reis-Filho JS. Breast cancer genomics from microarrays to massively parallel sequencing: Paradigms and new insights. J Natl Cancer Inst. 2015;107:djv015. doi: 10.1093/jnci/djv015. [DOI] [PubMed] [Google Scholar]
- 34.Podo F, Buydens LM, Degani H, Hilhorst R, Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J, Monleon D, et al. Triple-negative breast cancer: Present challenges and new perspectives. Mol Oncol. 2010;4:209–229. doi: 10.1016/j.molonc.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardner ER, Dahut WL, Scripture CD, Jones J, Aragon-Ching JB, Desai N, Hawkins MJ, Sparreboom A, Figg WD. Randomized crossover pharmacokinetic study of solvent-based paclitaxel and nab-paclitaxel. Clin Cancer Res. 2008;14:4200–4205. doi: 10.1158/1078-0432.CCR-07-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci USA. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ten Tije AJ, Verweij J, Loos WJ, Sparreboom A. Pharmacological effects of formulation vehicles: Implications for cancer chemotherapy. Clin Pharmacokinet. 2003;42:665–685. doi: 10.2165/00003088-200342070-00005. [DOI] [PubMed] [Google Scholar]
- 39.Menderes G, Lopez S, Han C, Altwerger G, Gysler S, Varughese J, Schwartz PE, Santin AD. Mechanisms of resistance to HER2-targeted therapies in HER2-amplified uterine serous carcinoma, and strategies to overcome it. Discov Med. 2018;26:39–50. [PubMed] [Google Scholar]
- 40.Leik CE, Su EJ, Nambi P, Crandall DL, Lawrence DA. Effect of pharmacologic plasminogen activator inhibitor-1 inhibition on cell motility and tumor angiogenesis. J Thromb Haemost. 2006;4:2710–2715. doi: 10.1111/j.1538-7836.2006.02244.x. [DOI] [PubMed] [Google Scholar]
- 41.Erickson LA, Ginsberg MH, Loskutoff DJ. Detection and partial characterization of an inhibitor of plasminogen activator in human platelets. J Clin Invest. 1984;74:1465–1472. doi: 10.1172/JCI111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh AK, Murphy SB, Kishore R, Vaughan DE. Global gene expression profiling in PAI-1 knockout murine heart and kidney: Molecular basis of cardiac-selective fibrosis. PLoS One. 2013;8:e63825. doi: 10.1371/journal.pone.0063825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgins PJ. Balancing AhR-dependent pro-oxidant and Nrf2-responsive anti-oxidant pathways in age-related retinopathy: Is SERPINE1 expression a therapeutic target in disease onset and progression? J Mol Genet Med. 2014;8:101. doi: 10.4172/1747-0862.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freytag J, Wilkins-Port CE, Higgins CE, Higgins SP, Samarakoon R, Higgins PJ. PAI-1 mediates the TGF-beta1+EGF-induced ‘scatter’ response in transformed human keratinocytes. J Invest Dermatol. 2010;130:2179–2190. doi: 10.1038/jid.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavón MA, Arroyo-Solera I, Téllez-Gabriel M, León X, Virós D, López M, Gallardo A, Céspedes MV, Casanova I, López-Pousa A, et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget. 2015;6:29016–29033. doi: 10.18632/oncotarget.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pavón MA, Arroyo-Solera I, Céspedes MV, Casanova I, León X, Mangues R. uPA/uPAR and SERPINE1 in head and neck cancer: Role in tumor resistance, metastasis, prognosis and therapy. Oncotarget. 2016;7:57351–57366. doi: 10.18632/oncotarget.10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu DM, Wang S, Wen X, Han XR, Wang YJ, Fan SH, Zhang ZF, Shan Q, Lu J, Zheng YL. MircoRNA-1275 promotes proliferation, invasion and migration of glioma cells via SERPINE1. J Cell Mol Med. 2018;22:4963–4974. doi: 10.1111/jcmm.13760. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Yao H, He G, Chen C, Yan S, Lu L, Song L, Vijayan KV, Li Q, Xiong L, Miao X, Deng X. PAI1: A novel PP1-interacting protein that mediates human plasma's anti-apoptotic effect in endothelial cells. J Cell Mol Med. 2017;21:2068–2076. doi: 10.1111/jcmm.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCann JV, Xiao L, Kim DJ, Khan OF, Kowalski PS, Anderson DG, Pecot CV, Azam SH, Parker JS, Tsai YS, et al. Endothelial miR-30c suppresses tumor growth via inhibition of TGF-β-induced Serpine1. J Clin Invest. 2019;130:1654–1670. doi: 10.1172/JCI123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takayama Y, Hattori N, Hamada H, Masuda T, Omori K, Akita S, Iwamoto H, Fujitaka K, Kohno N. Inhibition of PAI-1 limits tumor angiogenesis regardless of angiogenic stimuli in malignant pleural mesothelioma. Cancer Res. 2016;76:3285–3294. doi: 10.1158/0008-5472.CAN-15-1796. [DOI] [PubMed] [Google Scholar]
- 51.Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis. 2017;20:409–426. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 52.Luan X, Guan YY, Lovell JF, Zhao M, Lu Q, Liu YR, Liu HJ, Gao YG, Dong X, Yang SC, et al. Tumor priming using metronomic chemotherapy with neovasculature-targeted, nanoparticulate paclitaxel. Biomaterials. 2016;95:60–73. doi: 10.1016/j.biomaterials.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Samuel S, Fan F, Dang LH, Xia L, Gaur P, Ellis LM. Intracrine vascular endothelial growth factor signaling in survival and chemoresistance of human colorectal cancer cells. Oncogene. 2011;30:1205–1212. doi: 10.1038/onc.2010.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iqbal S, Lenz HJ. Integration of novel agents in the treatment of colorectal cancer. Cancer Chemother Pharmacol. 2004;54(Suppl 1):S32–S39. doi: 10.1007/s00280-004-0884-0. [DOI] [PubMed] [Google Scholar]
- 55.Fallah A, Sadeghinia A, Kahroba H, Samadi A, Heidari HR, Bradaran B, Zeinali S, Molavi O. Therapeutic targeting of angiogenesis molecular pathways in angiogenesis-dependent diseases. Biomed Pharmacother. 2019;110:775–785. doi: 10.1016/j.biopha.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 56.Tsai JL, Lee YM, Pan CY, Lee AY. The novel VEGF121- VEGF165 fusion attenuates angiogenesis and drug resistance via targeting VEGFR2-HIF-1α-VEGF165/Lon signaling through PI3K-AKT-mTOR pathway. Curr Cancer Drug Targets. 2016;16:275–286. doi: 10.2174/156800961603160206125352. [DOI] [PubMed] [Google Scholar]
- 57.Tonissi F, Lattanzio L, Merlano MC, Infante L, Lo Nigro C, Garrone O. The effect of paclitaxel and nab-paclitaxel in combination with anti-angiogenic therapy in breast cancer cell lines. Invest New Drugs. 2015;33:801–809. doi: 10.1007/s10637-015-0249-z. [DOI] [PubMed] [Google Scholar]
- 58.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.