Secretion and deposition of non-collagen ECM components is an elusive topic. Albacete-Albacete et al. identify exosome biogenesis and exosomal cargo sorting as novel mechanisms by which the stromal remodeling regulator caveolin-1 modulates ECM deposition, through the control of multivesicular body cholesterol content and plasticity.
Abstract
The composition and physical properties of the extracellular matrix (ECM) critically influence tumor progression, but the molecular mechanisms underlying ECM layering are poorly understood. Tumor–stroma interaction critically depends on cell communication mediated by exosomes, small vesicles generated within multivesicular bodies (MVBs). We show that caveolin-1 (Cav1) centrally regulates exosome biogenesis and exosomal protein cargo sorting through the control of cholesterol content at the endosomal compartment/MVBs. Quantitative proteomics profiling revealed that Cav1 is required for exosomal sorting of ECM protein cargo subsets, including Tenascin-C (TnC), and for fibroblast-derived exosomes to efficiently deposit ECM and promote tumor invasion. Cav1-driven exosomal ECM deposition not only promotes local stromal remodeling but also the generation of distant ECM-enriched stromal niches in vivo. Cav1 acts as a cholesterol rheostat in MVBs, determining sorting of ECM components into specific exosome pools and thus ECM deposition. This supports a model by which Cav1 is a central regulatory hub for tumor–stroma interactions through a novel exosome-dependent ECM deposition mechanism.
Introduction
The tumor microenvironment is a key factor in multiple stages of disease progression, including local invasion and distant metastasis (Chen et al., 2015; Quail and Joyce, 2013; Quail et al., 2013). Cancer-associated fibroblasts (CAFs) are an important cell subpopulation and constitute a major source of paracrine growth factors and ECM components (Augsten et al., 2014; Kalluri, 2016). However, the molecular mechanisms driving ECM deposition are incompletely understood. Recent studies confirm that ECM deposition ultimately depends on a functional core secretion machinery (Unlu et al., 2014), but further mechanistic characterization is warranted.
Exosomes are vesicles secreted by most cell types, generated from intraluminal vesicles (ILVs) of multivesicular bodies (MVBs), membrane-bound compartments linked to the endosomal system (Colombo et al., 2014; Gruenberg and Maxfield, 1995; van Niel et al., 2006). Exosomes enable a given cell to modulate the behavior of other cell populations through the transfer of effector or regulatory molecules, often over long (interorgan) distances (Braicu et al., 2015; Raposo and Stoorvogel, 2013), and can mediate tumor–stroma communication and premetastatic niche formation (Costa-Silva et al., 2015; Hoshino et al., 2015; Peinado et al., 2012).
Exosomal cargo composition is complex, containing proteins, small ligand peptides, and regulatory nucleic acids (Colombo et al., 2014), but its regulation is poorly understood. Endosomal sorting complexes required for transport (ESCRT) machinery–dependent and ESCRT-independent mechanisms coexist (Babst, 2011; Villarroya-Beltri et al., 2014), reflecting the heterogeneity of MVBs and exosomes and their potential differential impact on health and disease. Exosome biogenesis is also modulated by the organization of membrane subdomains through lipid composition of MVB membranes (Record et al., 2018). Cholesterol is likely relevant, given its higher abundance at MVBs compared with other compartments (Möbius et al., 2003) and its involvement in the formation of high-curvature membrane structures such as caveolae and synaptic vesicles. Cholesterol, long-chain sphingolipids, and phosphatidylserine (PS) 18:0/18:1 are also significantly enriched in exosome membranes (Skotland et al., 2019; Wang et al., 2007; Wubbolts et al., 2003). ER-endosome/MVB contacts allow for the regulated transfer of proteins and lipids, especially cholesterol, and are modulated through specific protein tethers, especially members of the oxysterol-binding protein (ORP) family (ORP1L and ORP5). Cholesterol accumulation decreases ER-MVB contacts (Raiborg et al., 2015; Rocha et al., 2009). Still, we lack a complete picture of the mechanisms modulating cholesterol levels at MVBs and their impact on exosome biology.
Endomembrane trafficking is regulated by caveolin-1 (Cav1), an essential scaffolding component of invaginated plasmalemmal nanodomains (caveolae). Cav1 participates in several essential cell functions, including mechanoadaption, endocytosis, spatiotemporal control of signal transduction, cholesterol homeostasis (Parton and del Pozo, 2013; Pol et al., 2020), and interorganelle communication, being an integral component of mitochondria-associated ER membranes, a membrane contact site (MCS) of specialized ER regions and mitochondria (Sala-Vila et al., 2016). Cav1 plays multiple, yet context-dependent, roles in tumor biology and is required for ECM biomechanical remodeling by CAFs promoting tumor invasion and metastasis (Goetz et al., 2011). Exosomal secretion of Cav1 occurs (Logozzi et al., 2009), but its relevance for Cav1-dependent stromal remodeling is unknown.
Here, we demonstrate that Cav1 is a constitutive component of exosomes produced by several cell types, including primary CAFs. N-terminal ubiquitin acceptor lysines are essential for Cav1 sorting into the MVB compartment. Comparative proteomics and lipidomics revealed that Cav1 modulates exosome cargo sorting and segregation of exosome subpopulations through a cholesterol-dependent mechanism to enable incorporation of ECM components, including Tenascin-C (TnC), an important driver of tumor progression, and exosome secretion is strictly required for their deposition. Exosomes derived from Cav1 knockout (KO) fibroblasts failed to deposit ECM and promote tumor cell invasiveness. Cav1-dependent remote exosomal ECM deposition occurs in vivo, suggesting the relevance of stromal Cav1 for distant ECM niche nucleation. We propose that Cav1 is a key regulatory hub also fostering stromal environments through the modulation of MVB membrane plasticity and exosome cargo specification, which depend on cholesterol content and organization.
Results
Cav1 redistributes to late endosome (LE)/MVB to its sorting in exosomes
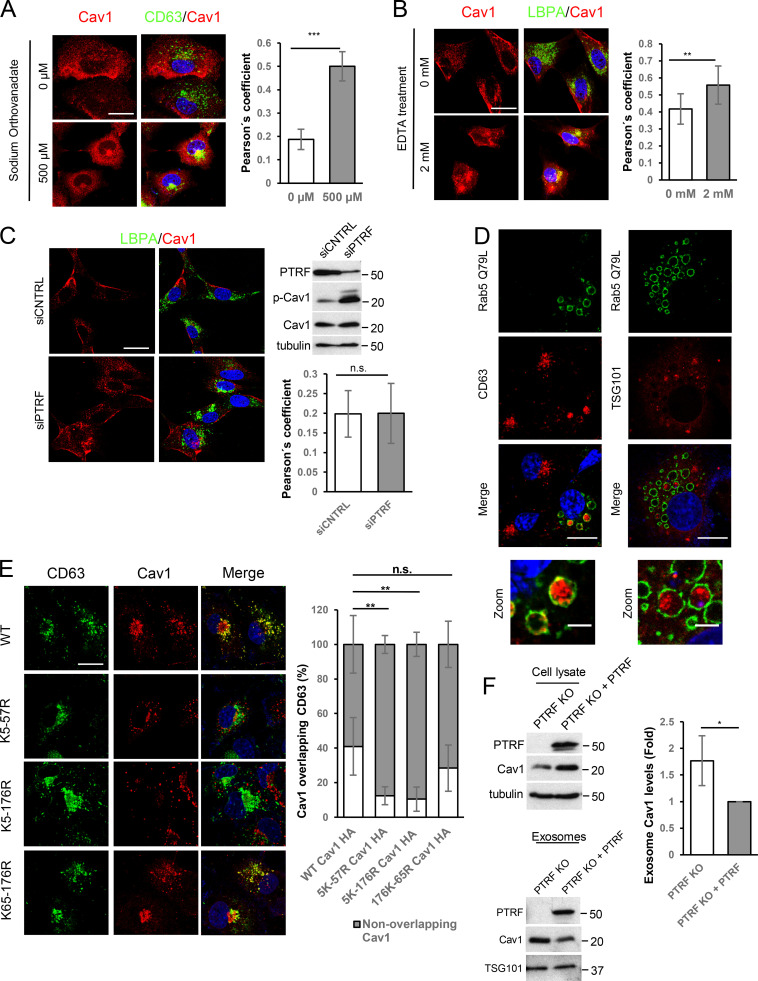
Cav1 internalization is tightly regulated (del Pozo et al., 2005; Nabi and Le, 2003; Zimnicka et al., 2016). Although endocytosed Cav1 has commonly been regarded as committed to degradation (Botos et al., 2008; Hayer et al., 2010), alternative fates cannot be excluded (Mundy et al., 2012). We analyzed the trafficking dynamics of endogenous Cav1 upon treatment with sodium orthovanadate, which induces Cav1 internalization by protecting Cav1 phosphorylation at tyrosine 14 (del Pozo et al., 2005). Orthovanadate exposure increased Cav1 phosphorylation and colocalization with LEs and MVBs (anti–lyso-bis-phosphatidic acid [LBPA] and anti-CD63 immunostainings, respectively; Fig. 1 A and Fig. S1 A). Similarly, a significant fraction of Cav1 relocalized to the LE/MVB compartment upon exposure of cells to EDTA, which induces Cav1 endocytosis by promoting cell detachment (Fig. S1 B; Grande-García and del Pozo, 2008). RNAi-mediated depletion of polymerase I and transcript release factor (PTRF/Cavin-1), an essential caveolar coat component required for caveolae stabilization (Hill et al., 2008), also induced partial Cav1 relocalization—albeit below significance threshold—and increased Cav1 phosphorylation (Fig. S1 C). High-resolution microscopy confirmed the constitutive presence of Cav1 in the MVB lumen, colocalizing with the exosomal marker CD63 (Fig. 1 B). Discrete patches of Cav1 localizing to ILVs within MVBs were clearly observable by artificially enlarging endosomes (Fig. 1 C and Fig. S1 D; Trajkovic et al., 2008). These observations demonstrate that MVBs are one of the destinies of endocytosed Cav1.
Figure 1.
Internalized Cav1 is sorted to exosomes. (A) Confocal images of Cav1 (red) and LBPA (green) in WT MEFs after treatment with sodium orthovanadate (NaV; 2 h). White arrows: plasma membrane–localized Cav1. Gray arrows: Cav1 relocalized to perinuclear area (scale bar, 25 µm). Charts show Pearson´s correlation coefficient for Cav1-LBPA colocalization. Error bars: mean ± SD; n = 80 cells. Western blot: phospho-Cav1 (pCav1) and total Cav1 in sodium orthovanadate–treated cells. (B) Cav1-CD63 colocalization. Right panels show high-resolution images of an MVB compartment (scale bar, 1 µm). (C) Cav1 distribution in Rab5(Q79L)-expressing COS7 cells (scale bar, 10 µm). Right: zoomed view of an endosome (scale bar, 2.5 µm). (D) Top: cartoon of assayed Cav1 constructs. Ubiquitination-target lysine residues (Ub) and residues mutated to arginine (X) are indicated. Bottom: confocal analysis of COS7 cells transfected with Rab5(Q79L) (green) and indicated Cav1 constructs (scale bar, 10 µm). Chart: endosomal lumen localization of Cav1 variants (internal, INT; white) versus the endosomal membrane (peripheral, MEMB; gray), expressed as percentage of total endosomal localization. Error bars are mean ± SD; n > 20 endosomes. (E) Western blot analysis of sodium orthovanadate–induced phosphorylation in WT Cav1 (pCav1), ubiquitination-target lysine mutants, and nonphosphorylatable Y14F Cav1. (F) Cav1 inclusion in exosomes derived from fibroblasts (MEFs). Representative Western blots of exosomal proteins floated on a continuous sucrose gradient (0.25–2 M). Individual 1-ml fractions were collected and after protein precipitation were loaded on electrophoresis gels and analyzed for Cav1 and the exosome marker Tsg101. Red rectangles highlight fractions with detectable amounts of the exosome marker Tsg101 (3–7). (G) Western blot analysis of cell lysates and exosomes of COS7 cells expressing HA-tagged Cav1 ubiquitination mutants. CNTRL, control; n.s., not significant. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S1.
Internalization of Cav1 favors its entry into exosomes. (A) Colocalization analysis of Cav1 (red) and CD63 (green) in WT MEFs after 2-h treatment with sodium orthovanadate (scale bar, 25 µm). Chart shows colocalization between CD63 and Cav1 as measured by Pearson’s correlation coefficient (mean ± SD; n = 2, total 80 cells). (B) Colocalization analysis of Cav1 (red) and LBPA (green) after 10-min EDTA treatment (scale bar, 25 µm). Chart shows Pearson’s correlation coefficient of the colocalization between the two labels (mean ± SD; n = 2, total 80 cells). (C) Confocal microscopy of Cav1 (red) and LBPA (green) in PTRF-KD MEFs (scale bar, 25 µm). A representative Western blot of phosphorylated Cav1 is shown for PTRF-KD cells. Chart shows colocalization analysis as measured by Pearson’s correlation coefficient (mean ± SD; n = 2, total 40 cells). (D) Confocal analysis of COS7 cells transfected with Rab5(Q79L) (green) and the exosomal markers CD63 and Tsg101 (red; scale bar, 15 µm). Zoomed views show the intraluminal accumulation of both markers (scale bar, 5 µm). (E) Colocalization analysis of lysine-arginine Cav1 mutants (red) and CD63 (green; scale bar, 25 µm). Chart shows the percentage of labeled colocalization (mean ± SD; n = 3). (F) Western blot analysis of Cav1 in cell lysates and released exosomes of control and PTRF-reconstituted PTRFKO cells. Tubulin and Tsg101 were used as loading controls. Chart shows Cav1 levels in exosomes produced by either control or PTRF-reconstituted PTRFKO cells (mean ± SD; n = 4). CNTRL, control; n.s., not significant. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Phosphorylation often triggers subsequent ubiquitination of adjacent acceptor residues (Hunter, 2007). We analyzed the subcellular distribution of Cav1 mutants in which ubiquitin acceptor lysine residues are replaced by arginine (Fig. 1 D, upper panel). Mutation of Cav1 N-terminal lysines impairs Cav1 ubiquitination (Kirchner et al., 2013). Cav1 constructs nicely decorated the outer enlarged-Rab5 endosome membrane/MVB; however, mutation of the N-terminal lysine cluster (K5-57R and K5-176R mutants) abolished Cav1 sorting into ILVs (Fig. 1 D), correlating with lower overall colocalization of these mutants with CD63 (Fig. S1 E). In contrast, mutation of C-terminal lysine residues (K65-176R) did not affect the ability of Cav1 to localize in MVBs (Fig. 1 D). Phosphorylation was not significantly altered in Cav1 mutants (Fig. 1 E), reinforcing the notion of a two-step mechanism for Cav1 sorting into MVBs: phosphorylation first elicits Cav1 internalization toward the outer MVB membrane, and ubiquitination acts as a checkpoint for Cav1 sorting into ILVs.
To assess the exosomal sorting of the ILV-localized Cav1 pool, we analyzed density gradient–purified exosomes from different cell lines. Endogenous Cav1 mainly partitioned with Tsg101, confirming that exosomes can carry Cav1 (Fig. 1 F). Cav1 sorting to exosomes was enhanced in PTRF-depleted mouse embryonic fibroblasts (MEFs; Fig. S1 F). In contrast, N-terminal ubiquitination mutants exhibited impaired exosomal sorting (Fig. 1 G). These data suggest that a significant proportion of internalized Cav1 is sorted to the MVB lumen in a ubiquitin-dependent manner, favoring its incorporation into exosomes. This process is constitutive in all Cav1-expressing cell lines tested.
Cav1 regulates exosomal biogenesis through modulation of MVB cholesterol content
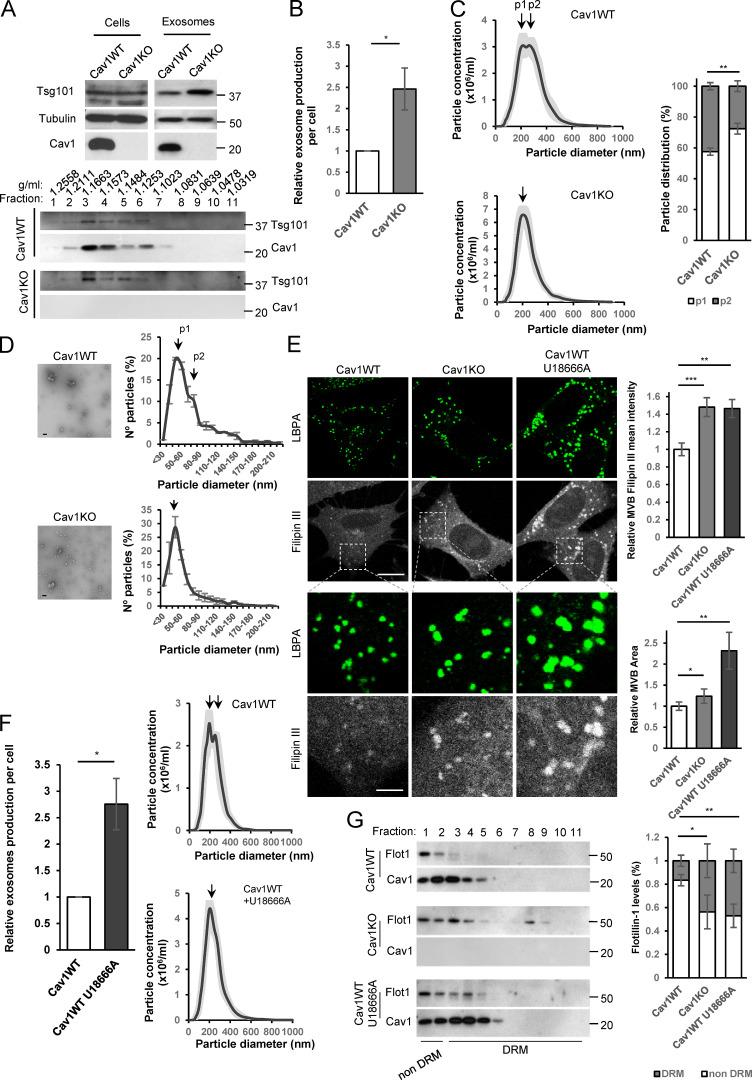
The regulated composition and organization of MVB membranes yield different ILV and exosome populations (Colombo et al., 2014). We explored whether Cav1, a regulator of endomembrane compartments (Pol et al., 2020), plays an active role in exosome biogenesis. Cav1KO fibroblasts were not impaired for exosome biogenesis and displayed higher rates of exosome secretion, as determined by Western blot of total purified exosomes from equal cell numbers (Fig. 2, A and B, upper panel in A), sucrose density gradient sedimentation (Fig. 2 A, lower panel; and Fig. 3 D), and physical characterization (Nanosight technology, transmission EM [TEM]; Fig. 2, C and D; Dragovic et al., 2011). We identified two exosome subpopulations of different sizes in WT fibroblasts, whereas Cav1KO fibroblasts produced a homogeneous population of small exosomes (Fig. 2, C and D).
Figure 2.
Cav1 regulates exosome biogenesis by modulating MVB cholesterol content. (A–G) Exosomes were isolated from supernatants of Cav1WT and Cav1KO fibroblast cultures. (A) Top: Western blots for indicated proteins in exosomes derived from equal cell numbers. Bottom: Western blots showing the distribution of Cav1 and Tsg101 in exosomes floated on a continuous sucrose gradient. (B) Quantification of exosome particles relative to cell number. Error bar are means ± SD; n = 10. (C) Left: Nanosight distribution profiles of exosome preparations from WT and Cav1KO fibroblasts, showing greater morphological and size heterogeneity of WT-derived exosomes. Right: Quantification of the two exosome populations identified in WT and Cav1KO fibroblasts. Error bars are mean ± SEM; n = 10. (D) Representative TEMs of WT and Cav1KO exosomes (scale bar, 200 nm). Distribution profiles measured from TEMs of WT and Cav1KO fibroblast-derived exosomes. (E) Filipin staining (gray) and LBPA (green) of WT, Cav1KO, and U18666A-treated Cav1WT fibroblasts (scale bar, 20 µm). Lower panel rows show zoomed views (scale bar, 6 µm). Charts on the right show quantitative analysis of filipin mean fluorescence intensity in MVBs (upper) and total MVB area (lower). Error bar are means ± SD; n = 3. (F) Exosome production per cell. Error bars are means ± SD; n = 4 (left) and Nanosight distribution profiles of exosome preparations (right) from WT fibroblasts without treatment (control) and treated with U18666A. (G) Western blots of sucrose density gradient fractions in the presence of TritonX-100 of exosomes produced by untreated WT and Cav1KO fibroblasts and U18666A-treated WT fibroblasts. The chart shows the proportion of flotillin (Flot1) in detergent-resistant and nonresistant membranes (DRM). Error bars are mean ± SD; n = 3. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 3.
Cav1 specifies protein cargo into exosomes. (A) Representation of hierarchical analysis of proteins equally expressed in the two parental cell populations (cell lysates, shaded in yellow) and differentially expressed in exosomes. Blue: proteins up-regulated in WT exosomes; red: proteins up-regulated in Cav1KO exosomes. Graphics on the right show STRING network analysis. Top: interactions among identified proteins up-regulated in Cav1WT MEF-derived exosomes. Highlighted groups are related to ECM components (blue) and extracellular vesicle/exosome biogenesis and lysosome components (green). Bottom: interactions among identified proteins up-regulated in Cav1KO MEF-derived exosomes. Highlighted groups are related to histones (red) and DNA/RNA binding (orange). (B) Clustered heatmap of extracellular proteins with significant changes in ECM protein abundance between exosomes produced by WT and Cav1KO fibroblasts (top) or in histone-related proteins (bottom). (C) Heatmap comparing annexin isoform enrichment in exosomes produced by Cav1KO and WT fibroblasts. (D) Western blot analysis showing the distribution of Cav1 and some of the proteins identified by quantitative proteomics in WT and Cav1KO-derived exosomes floated on an Optiprep (iodixanol) density gradient. Tsg101, Alix, and flotillin-1 were used as exosomal markers, and GM130 was included as a negative control.
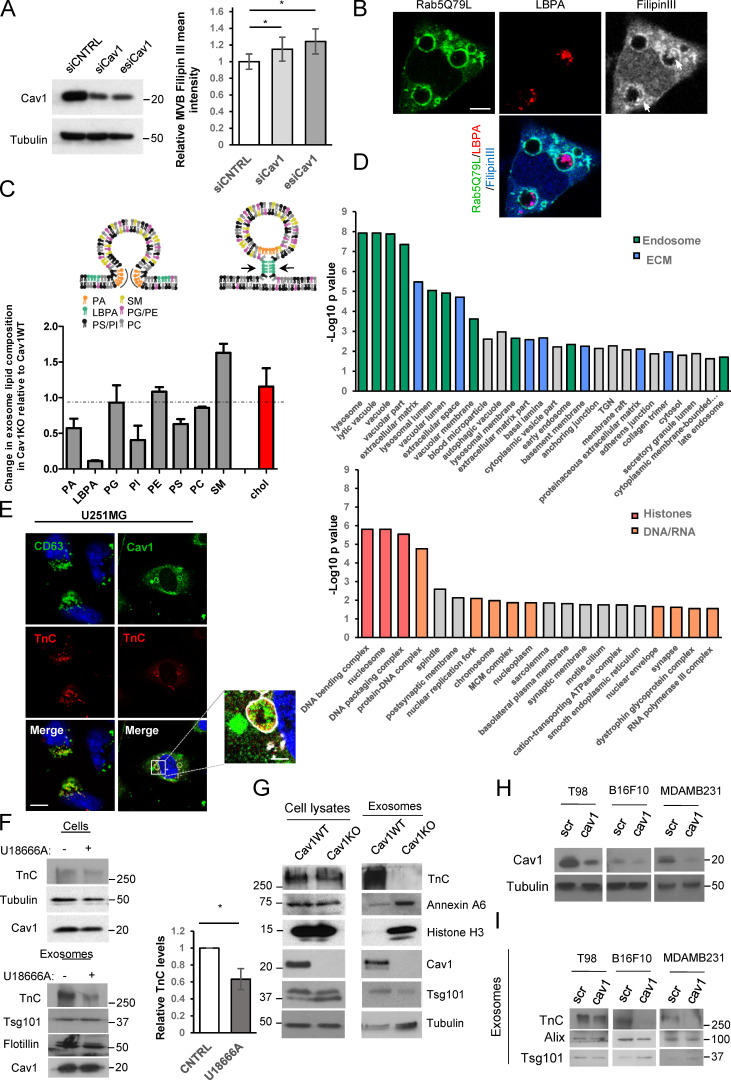
Cav1 binds cholesterol and organizes specialized membrane organization domains (Murata et al., 1995; Pol et al., 2020). We studied whether Cav1 promotes exosome heterogeneity by modulating cholesterol levels at MVBs. While the number of LBPA-positive structures did not differ between WT and Cav1KO fibroblasts, MVB area and filipin intensity (a proxy for mean cholesterol content) were significantly higher in Cav1KO cells (Fig. 2 E). This phenotype was recapitulated by siRNA depletion of Cav1 in WT cells (Fig. S2 A). Treatment with U18666A, which promotes cholesterol accumulation in the LE/MVB compartment (Cenedella, 2009), also increased exosome secretion while reducing exosome heterogeneity in WT cells (Fig. 2 F).
Figure S2.
Cav1-positive exosomes display a specific lipid/protein composition. (A) Western blot confirming Cav1KD in WT MEFs. Chart shows filipin III mean fluorescence intensity at MVBs of WT MEFs transfected with either control (siCNTRL, silencing RNA duplex control) or Cav1-targeting siRNAs (siCav1, siRNA duplex targeting Cav1; esiCav1, enzymatically prepared siRNAs targeting Cav1; mean ± SD; n = 4). (B) Distribution of LBPA (red) and filipin III (grey and blue) in Rab5(Q79L)-expressing MEFs (green). Arrows indicate LBPA and filipin III colocalization at ILVs (scale bar, 5 µm). (C) Upper panel: lipids involved in exosome biogenesis are summarized in the scheme (adapted from Subra et al., 2007). Lower panel: MS analysis of lipid composition profiles of Cav1KO MEF-derived exosomes compared with WT-derived exosomes. Values denote relative content in Cav1KO-derived exosomes as normalized to the relative content measured in WT exosomes for each lipid class (mean ± SD; n = 2 for determination of all lipid species except for cholesterol; for cholesterol determination, n = 3). PA, phosphatidic acid; SM, sphingomyelin; PG, phosphatidylglycerol; PI, phosphatidylinositol; PC, phosphatidylcholine; PE, phosphatidylethanolamine. (D) Enrichr diagram of “cellular component” category for proteins upregulated in either WT MEF–derived exosomes (top) or Cav1KO MEF–derived exosomes (bottom). MCM complex, minichromosome maintenance protein complex. (E) Colocalization analysis of TnC (red) with CD63 or Cav1 (green) in U251 glioblastoma cells expressing Rab5(Q79L) (white; scale bar, 25 µm; zoomed view scale bar, 2.5 µm). (F) Western blot analysis of TnC in cell lysates and released exosomes of untreated or U18666A-treated WT MEFs. Tsg101 was used as exosome loading control. Chart shows the relative amount of TnC in exosomes produced by WT control and U18666A-treated cells (mean ± SD; n = 4). (G) Western blot analysis confirming protein expression changes identified by quantitative proteomics in WT fibroblast lysates and purified exosomes compared with Cav1KO counterparts. (H) Validation of lentivirus-mediated KD (scr, scrambled control sequence; Cav1, Cav1-targeting shRNA) of Cav1 mRNA across tumor cell lines (T98: glioblastoma; B16F10: melanoma; MDAMB231: breast cancer). (I) Western blot analysis of TnC secretion in exosomes produced by the different tumor cell lines described in H. Tsg101 and Alix were used as exosomal markers. CNTRL, control. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
A major share of MVB cholesterol is contained in ILVs (Möbius et al., 2003). We confirmed this cholesterol accumulation in enlarged endosomes by overexpressing constitutively active Rab5(Q79L) (Fig. S2 B). To confirm whether altered cholesterol content in Cav1KO MVBs induces exosome membrane reorganization, we isolated cold-detergent–resistant membranes (exosome DRMs) from exosomes produced by either WT cells (control or U18666A treated) or Cav1KO cells. Flotillin-1, a marker for exosomes and liquid ordered domains, showed a wider distribution in Cav1KO cells (fractions 1 to 8) or U18666A-treated WT cells (fractions 1 to 6); in contrast, flotillin accumulation in untreated WT cell–derived exosomes was restricted to fractions 1 to 3 (Fig. 2 G), reflecting the increased highly ordered membrane content of Cav1KO-derived exosomes. Lipidomic analyses of exosome preparations confirmed a significant enrichment in PS—in particular PS (18:0/18:1) species—in agreement with previous data (Skotland et al., 2019). Of note, exosomes derived from WT cells invariably exhibited higher relative amounts of phosphatidic acid, phosphatidylinositol, and LBPA (which is known to play a relevant role in ILV biogenesis and dynamics; Cenedella, 2009; Kowal et al., 2014; Subra et al., 2007) compared with Cav1KO exosomes, which conversely displayed an increased amount of cholesterol (Fig. S2 C), in agreement with the observed accumulation of cholesterol at LE/MVB compartments of Cav1KO cells. These results suggest that Cav1 regulates exosome biogenesis by acting as a “cholesterol rheostat” at LE/MVBs, conferring plasticity to this compartment and enabling the segregation of different exosome populations.
Cav1 promotes the sorting of specific ECM cargoes into exosomes
We quantitated the proteomes of WT and Cav1KO fibroblasts and their corresponding exosome preparations. This analysis identified a subproteome that is equally abundant in WT and Cav1KO whole-cell lysates, but whose components are differentially sorted into exosomes depending on the Cav1 genotype (Fig. 3 A, left). With a threshold of |Zq| > 1.5, in Cav1KO exosomes 152 proteins were more abundant and 163 less abundant than in WT exosomes. Interaction network and functional annotation enrichment analysis revealed that WT-derived exosomes were significantly enriched in ECM components (including TnC, fibronectin [FN], nidogen, emilin, EDIL3, and heparan sulfate proteoglycans) and endosomal/endomembrane system components (Fig. 3 A, right upper panel; Fig. 3 B; Fig. S2 D; and Data S1). Conversely, a marked enrichment for DNA/RNA binding and chaperone proteins such as histones in Cav1KO exosomes was observed (Fig. 3 A, right lower panel; Fig. 3 B; Fig. S2 D; and Data S1). Consistent with our data supporting increased cholesterol content in Cav1KO MVBs, various annexin family members were substantially enriched in Cav1KO exosomes, despite a decreased abundance of these proteins in Cav1KO whole-cell lysates (Fig. 3 C). Annexins are Ca2+-dependent proteins that bind cholesterol-rich membranes, providing lipid-clustering activity, and may constitute a mechanism to accommodate cholesterol excess in Cav1KO exosomes (Lizarbe et al., 2013).
Differential exosome sorting of selected proteins was validated by Western blot analysis of exosome fractions (Fig. S2 G and Fig. S3 I) and iodixanol density gradients (Fig. 3 D). As shown in Fig. 3 D, the distributions of exosome markers and cargoes markedly correlated. A particular difference was observed for TnC (Zq = 3.02), an ECM protein involved in development, stem cell niche formation, and aggressive tumor metastasis (Lowy and Oskarsson, 2015; Midwood et al., 2016; Midwood and Orend, 2009). TnC was prominent in WT-derived exosomes but virtually absent in Cav1KO exosomes (Data S1). TnC colocalized with Cav1 within MVBs in fibroblasts and U251 cells (a TnC-secreting glioblastoma cell line; Fig. S2 E; Brösicke et al., 2013). Knockdown (KD) of Cav1 across different tumor cell lines (breast, glioblastoma, and melanoma) recapitulated the phenotype of blunted TnC secretion observed in Cav1KO MEFs (Fig. S2, H and I).
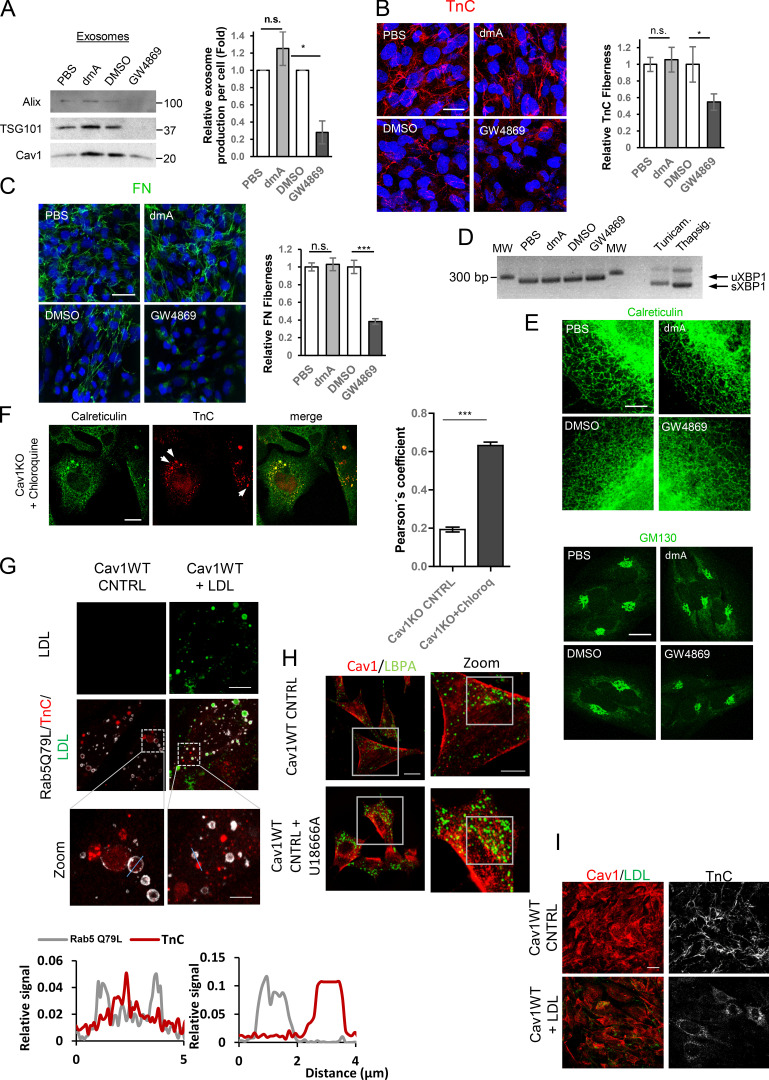
Figure S3.
Exosome inhibition favors intracellular TnC accumulation. (A) Western blot analysis (left) and quantification of exosome particle production per cell (right graph) showing reduced exosome secretion by WT fibroblasts after treatment with dmA or GW4869 (mean ± SEM; n = 3). (B) Pipeline of the fiberness macro (see Materials and methods). The original image, a, is first preprocessed (b), and nearly unidimensional tubular structures are enhanced to obtain an image for fiberness scoring (c). M0 measures the mean fiberness score in the whole image. a′ and c′ show magnifications of indicated region (red box) in panels a and c. (C) Representative Western blot and quantification (mean ± SEM; n = 9) showing the effect of indicated small-compound inhibitors on intracellular TnC accumulation. (D) Assessment of ER stress in WT MEFs treated with exosome secretion inhibitors dmA and GW4689. Upper panel:agarose gel for RT-PCR XBP1 mRNA splicing analysis. sXBP1, spliced mRNA species; uXBP1, unspliced mRNA species; Thapsig., Thapsigargin; Tunicam., Tunicamycin. Lower panel: quantitative (q) RT-PCR assessment of CHOP mRNA expression across indicated treatments. (E) Confocal microscopy analysis of Golgi (in green) labeled with GM130 in WT MEFs treated with exosome secretion inhibitors dmA and GW4689 (scale bar, 15 µm). (F and G) qRT-PCR assessment of KD efficiency of indicated siRNA treatments in WT fibroblasts (F) and Western blot (G). (H) Representative Western blot and quantification (mean ± SEM; n = 9) showing the effect of siRNA targeting Tsg101 and nSMase1 and 2 on intracellular TnC accumulation. (I) Representative Western blot of FN and pentraxin across both lysates and purified exosomes from WT and Cav1KO fibroblasts. (J) Representative Z-stack projection confocal microscopy images showing the effect of siRNAs targeting Tsg101 and nSMase1 and 2 on MEF FN matrix deposition (green; scale bar, 50 µm; mean ± SEM; n = 5). (K) FN deposition of WT MEFs treated with U18666A, as determined by Z-stack projection confocal microscopy (scale bar, 40 µm). Chart shows extracellular FN deposition in fibers produced by either untreated or U18666A-treated WT MEFs, as measured using custom software (mean ± SD; n = 8). (L) Depiction of an MCS between the endosomal compartment and the ER. Pairs of proteins described as key players for tethering and cholesterol transfer are indicated by dashed lines. Yellow arrows indicate the direction of cholesterol transfer as regulated by each tethering complex. From the ER toward the endosomes (box I) or from the endosomes to the ER (box II). n.s., not significant. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
This supports that Cav1 modulates exosome biogenesis and ECM cargo sorting across different cell types and is a prominent regulator of exosome-mediated intercellular communication.
Exosome biogenesis/secretion modulates ECM deposition
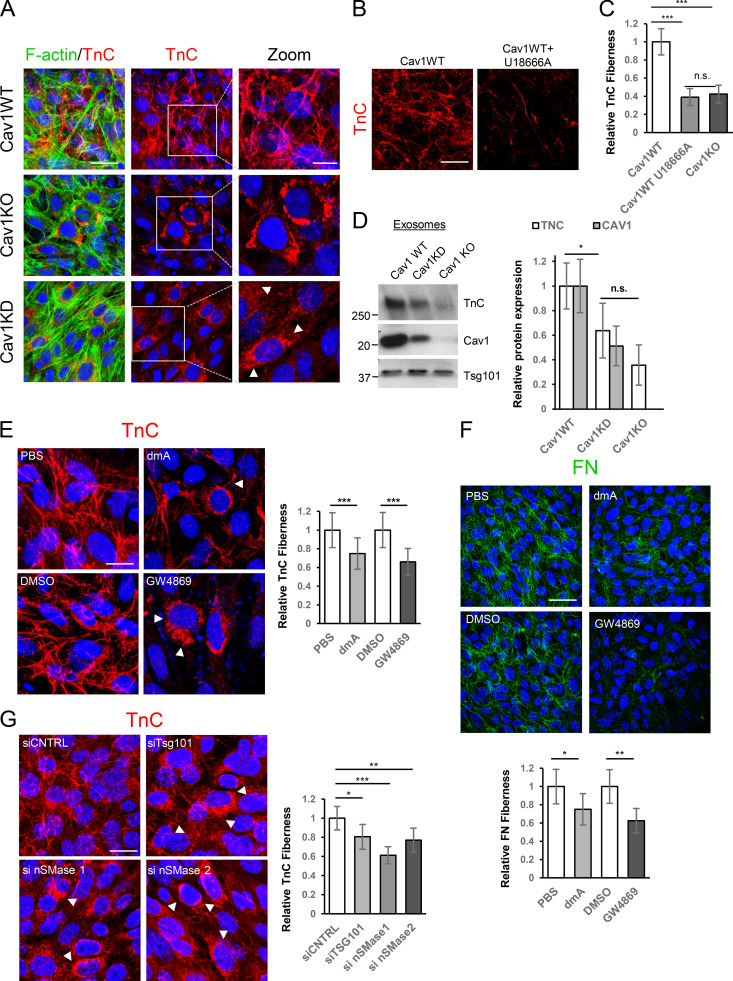
We examined whether exosome-delivered ECM components can initiate de novo ECM deposition. Cav1-deficient fibroblasts deposited less TnC fiber matrix than WT cells (Fig. 4 A). TnC fiber deposition was also reduced in U18666A-treated cells (Fig. 4, B and C; and Fig. S3 B). Reduced TnC deposition correlated with intracellular TnC accumulation in Cav1-deficient cells; this effect was less evident in U18666A-treated cells, probably reflecting a higher TnC degradation rate (Fig. 4 B). TnC was less abundant in exosomes derived from Cav1KO and Cav1KD MEFs or U18666A-treated WT MEFs (Fig. 4 D and Fig. S2 F). We used two different exosome secretion inhibitors to assess the contribution of exosomes to extracellular TnC fiber deposition: dimethylamiloride (dmA; a H+/Na+ and Na+/Ca2+ channel inhibitor) and GW4869 (a neutral sphingomyelinase [nSMase]-2 inhibitor; Savina et al., 2003; Trajkovic et al., 2008). Net exosome release by WT fibroblasts was reduced by GW4869 and to a lesser extent by dmA (Fig. S3 A), correlating with impaired TnC fiber deposition (Fig. 4 E). Both treatments led to intracellular accumulation of TnC aggregates (Fig. 4 E and Fig. S3 C). Deposition of FN, another key ECM component underrepresented in Cav1KO exosomes (Fig. 3 B, Fig. S3 I, and Data S1), was also reduced by exposure to exosome secretion inhibitors, with GW4869 having a much stronger effect (Fig. 4 F), although no significant differences could be observed upon U18666A treatment (Fig. S3 K). Relevance of exosome secretion for ECM deposition was also confirmed in primary CAFs, in which exosome release was largely inhibited by GW4869, whereas dmA had virtually no effect (Fig. S4 A). Accordingly, TnC and FN deposition was specifically affected by nSMase2 blockade, whereas dmA had no effect (Fig. S4 B and Fig. 4 C). Neither GW4869 nor dmA affected ER-Golgi homeostasis in MEFs or CAFs (Fig. S3 D, Fig. 3 E, Fig. S4 D, and Fig. 4 E), ruling out compromised ER-Golgi secretory function as a potential cause of defective deposition of ECM components.
Figure 4.
Exosome-mediated ECM deposition is a Cav1-dependent process. (A) Z-stack projection of confocal images showing TnC (red) in matrix deposited by WT, Cav1KD, and Cav1KO MEFs. F-actin is shown in green (scale bar, 40 µm). Zoomed views reveal an absence of TnC matrix deposition and a corresponding increased intracellular accumulation (white arrowheads) in Cav1KD and Cav1KO MEFs (scale bar, 10 µm). (B) TnC deposition of WT MEFs treated with U18666A determined by Z-stack projection confocal microscopy (scale bar, 40 µm). (C) Extracellular TnC deposition in fibers produced by Cav1WT, Cav1KO, and U18666A-treated Cav1WT MEFs (mean ± SD; n = 8). (D) Western blot analysis of TnC expression in exosomes secreted by indicated genotypes. Quantification: mean ± SEM; n = 6. (E) Effect of 5-d exposure to the exosome inhibitors dmA (75 nM) and GW4869 (10 µM) on TnC matrix deposition (red) by WT MEFs (scale bar, 20 µm). Right: chart shows extracellular TnC fiber deposition (mean ± SEM; n = 12). (F) Z-stack of confocal images showing the effect of 5-d exposure to dmA (75 nM) and GW4869 (10 µM) on FN matrix deposition (green) by WT MEFs (scale bar, 50 µm). Bottom: chart shows extracellular FN fiber deposition (mean ± SEM; n = 5). (G) Representative Z-stack projection of confocal images showing the effect of siRNAs targeting Tsg101 and neutral nSMase1 and 2 on TnC matrix deposition by WT MEFs (red; scale bar, 20 µm). The chart to the right shows extracellular TnC deposition in fibers measured as in C (mean ± SEM; n = 6). n.s., not significant. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S4.
Exosomes derived from CAFs modulate ECM deposition. (A) Western blot analysis (left) and exosome particle production per cell (right) show reduced exosome secretion by CAFs after treatment with dmA or GW4869 (mean ± SD; n = 3). (B and C) Representative Z-stack projection confocal microscopy images showing the effect of exosome inhibitor drugs (dmA and GW4689) on CAF TnC matrix deposition (B; scale bar, 20 µm; mean ± SEM; n = 5) and CAF FN matrix deposition (C; mean ± SEM; n = 5; scale bar, 50 µm). (D) ER stress was assessed by analysis of XBP1 mRNA splicing in CAFs treated with inhibitors of exosome biogenesis (dmA or GW4869). sXBP1, spliced mRNA species; uXBP1, unspliced mRNA species; Thapsig., Thapsigargin; Tunicam., Tunicamycin. (E) Confocal microscopy assessment of ER (immunolabeled for calreticulin; left panels; scale bar, 5 µm) and Golgi subcellular distribution (immunolabeled for GM130; right panels; scale bar, 20 µm) in CAFs treated with either dmA or GW4869. (F) Confocal microscopy showing accumulation of TnC (red; white arrows) within the ER (green) in chloroquine-treated Cav1KO MEFs (scale bar, 20 µm). Chart shows Pearson’s correlation coefficient for colocalization between TnC and calreticulin in Cav1KO cells across indicated conditions (mean ± SD; n = 3). (G) Impact of exogenous supplementation of Alexa Fluor 647–LDLs (in green) on TnC distribution (in red) in WT MEFs overexpressing Rab5(Q79L) mutant (in grey). Scale bar, 20 µm. Zoomed views show intraluminal accumulation of TnC upon treatment (scale bar, 5 µm). Histogram shows pixel intensities for Rab5(Q79L) (grey) and TnC (red) along the lines indicated on the images. (H) Confocal microscopy analysis of Cav1 distribution in U18666A-treated WT fibroblasts (scale bar, 20 µm). Zoomed images (scale bar, 10 µm). (I) Z-stack projection of confocal microscopy images show TnC (red) in matrices deposited by WT fibroblasts treated with LDLs (scale bar, 20 µm). CNTRL, control; n.s., not significant. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ESCRT machinery-regulated trafficking and ceramide biogenesis pathways generate exosomes with different cargoes (Colombo et al., 2014). We analyzed the effect of transient KD of either Tsg101 (a core regulator of the ESCRT-dependent pathway) or nSMase1 and 2 (key regulators of ceramide metabolism) on TnC matrix deposition (Fig. S3, F and G). Consistent with small inhibitor experiments (Fig. 4 E), KD of either pathway impaired TnC deposition, with ceramide synthesis disruption yielding stronger effects (Fig. 4 G and Fig. S3 H). These treatments also reduced FN deposition (Fig. S3 J). Thus, exosomes are a general ECM deposition mechanism, and ceramide-dependent exosome biogenesis is a major limiting step in this process.
TnC incorporation into exosomes has an intracellular origin and depends on cholesterol modulation
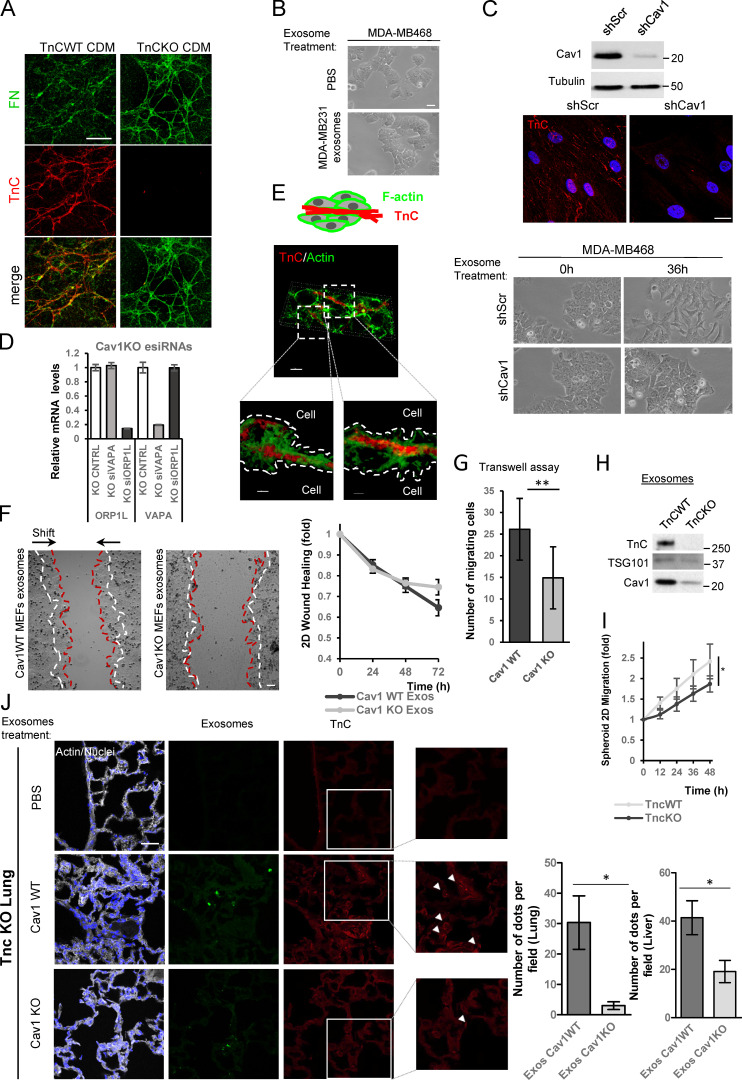
To rule out an external origin of exosome-incorporated TnC through endocytic trafficking, we developed an assay based on cell-derived matrices (CDMs). CDMs were generated by decellularization of ECMs synthesized in vitro by WT or TnCKO MEFs. We confirmed that TnCKO cells were specifically impaired for TnC deposition; other components, such as FN, did not exhibit such dramatic dependence and showed a slight up-regulation, likely because of the existence of alternative compensatory routes (Fig. 5 A and Fig. S5 A; see also Fig. S3 I). TnCKO or WT cells were then seeded on each of these CDMs (Fig. 5 B), and TnC internalization was determined. No intracellular TnC was detected inside TnCKO fibroblasts grown on TnC-rich CDMs, thus excluding endocytosis as an active TnC pool for exosome cargo sorting.
Figure 5.
Exosomal incorporation of TnC occurs intracellularly via the ER-MVB route. (A) Left: Western blot analysis of TnC and FN expression in lysates from TnCWT and TnCKO fibroblasts. Right: CDMs were generated from TnCWT and TnCKO cells as described in Materials and methods, and TnCWT and TnCKO cells were plated on these CDMs for further 24 h as depicted. CDMs were labeled with TnC (in red), and the plated cells were stained with F-actin (in gray; scale bar, 50 µm). (B) Western blot analysis of TnC and FN in total lysates (CDM + plated cells) and cells plated on CDM and isolated by trypsinization. Tubulin was used as a loading control. Cells were cultured on decellularized CDMs for 24 h prior to analysis. (C) Interaction assay of purified exosomes with TnC-rich or TnCKO CDMs. The scheme shows the assay protocol. Western blot shows analysis of TnC and FN binding to exosomes. (D) Subcellular fractionation analysis of intracellular TnC distribution in WT (treated with either vehicle or GW4869) and Cav1KO cells, using indicated markers. The boxed area denotes MVB-enriched fractions. Plots show the relative amount of TnC in each fraction with respect to the total TnC (normalized to 1) present in MVB-enriched fractions across conditions (mean ± SD; n = 3). (E) Colocalization between TnC (red) and calreticulin (green) was analyzed in Cav1WT MEFs treated with GW4869 alone or in combination with the lysosomal inhibitor chloroquine (scale bar, 50 µm; zoomed views scale bar, 20 µm). Chart shows Pearson´s correlation coefficient for colocalization TnC and calreticulin (mean ± SD; n = 3). CNTRL, control. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S5.
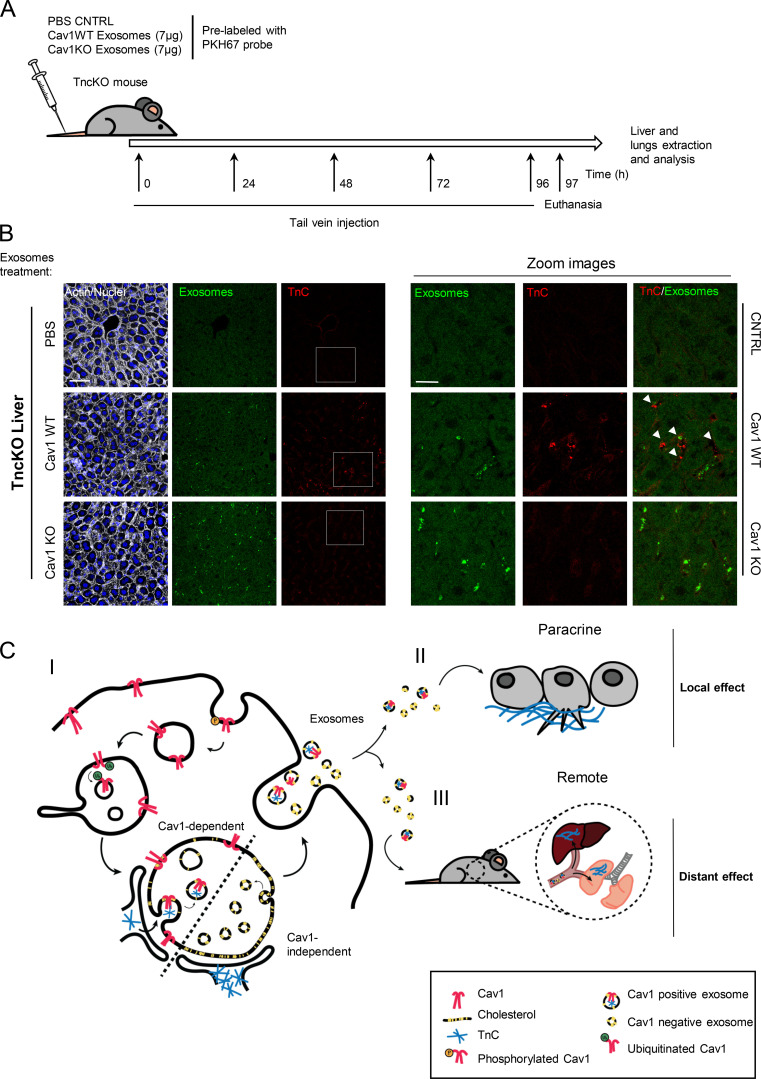
Disruption of ER-MVB contact sites modulates TnC matrix deposition. (A) Representative confocal microscopy of either WT or TnCKO CDMs showing FN (green) and TnC (red; scale bar, 20 µm). (B) Phase-contrast microscopy showing morphological changes in MDA-MB468 cells 36 h after exposure to MDA-MB231–derived exosomes (scale bar, 10 µm). (C) Top:Western blot analysis of Cav1KD efficiency in CCD-1069S CAFs. Middle: Z-stack projection of confocal microscopy images showing TnC (red) in matrix deposited by CAFs stably expressing either scrambled shRNA (shScr) or Cav1-targeting shRNA (shCav1; scale bar, 40 µm). Bottom:Phase-contrast microscopy showing morphological changes in MDA-MB468 cells 36 h after exposure to exosomes derived from either shScr or shCav1 CAFs (scale bar, 10 µm). (D) Quantitative RT-PCR assessment of VAPA and ORP1L mRNA KD efficiency in Cav1KO fibroblasts. (E) Top: Graphical scheme. Bottom: 3D reconstruction of TnC fibers deposited by exosomes produced by WT MEFs. (F) Wound closure assays performed with MDA-MB468 cells treated with fibroblast-derived WT and Cav1KO exosomes. The graph plots relative wound area (n = 6). (G) Number of invasive MDA-MB468 cells in transwell assays after exposure to exosomes derived from WT or Cav1KO fibroblasts (mean ± SD; n = 3). (H) Representative Western blot analysis of WT and TnCKO MEF-derived exosomes. (I) Phase-contrast microscopy of MDA-MB468 migration in spheroids generated in the presence of either WT or TnCKO MEF-derived exosomes over 72 h. Graph shows quantitative analysis of spheroid migration (mean ± SEM; n = 11). (J) Immunofluorescence microscopy of lung sections from TnCKO mice injected in the tail vein with WT or Cav1KO fibroblast-derived exosomes. Control mice were injected with PBS. Exosome foci are green. White arrowheads indicate regions of TnC deposition (red). Immunofluorescence images are representative of seven random fields from two independent experiments. The charts show exosome foci accumulation in liver and in lungs after WT or Cav1KO exosome injection. CNTRL, control. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To exclude the possibility that TnC detected in purified exosome fractions came from passive, nonspecific binding of extant extracellular TnC fibers to secreted exosomes, purified exosomes from TnCKO cells were incubated with TnC-rich matrix produced by WT cells (Fig. 5 C and Materials and methods section). We failed to detect TnC protein in TnCKO exosomes after their incubation with TnC-rich matrix (Fig. 5 C).
We then separated MVBs from the ER and Golgi compartments (which fractionate together). In untreated WT cells, TnC cosedimented both with MVB and ER markers (Fig. 5 D); however, upon treatment with GW4869, TnC was largely excluded from MVB fractions, a distribution shift recapitulated in Cav1KO cells (Fig. 5 D). Accordingly, a major fraction of cellular TnC colocalized with the ER in Cav1KO cells or GW4869-treated WT cells (Fig. 5 E and Fig. S4 F). This colocalization was enhanced upon chloroquine supplementation (Fig. S4 F), suggesting that defective transfer of TnC from the ER to MVBs results in ER retention of TnC and subsequent targeting for lysosomal degradation, supporting that lysosomal degradation enables TnC removal when its exosome sorting fails. Degradation rate differences could explain the variability in TnC accumulation in Cav1KO cells and WT cells treated with exosome inhibitor drugs and the lack of ER stress upon severe disruption of exosome biogenesis and secretion.
Thus, the presence of TnC in exosomes reflects true regulated intracellular sorting of TnC into MVBs, likely through a mechanism involving ER-MVB contact sites. This exosomal TnC pool is sourced in the cell from de novo translation and not from endocytosed material.
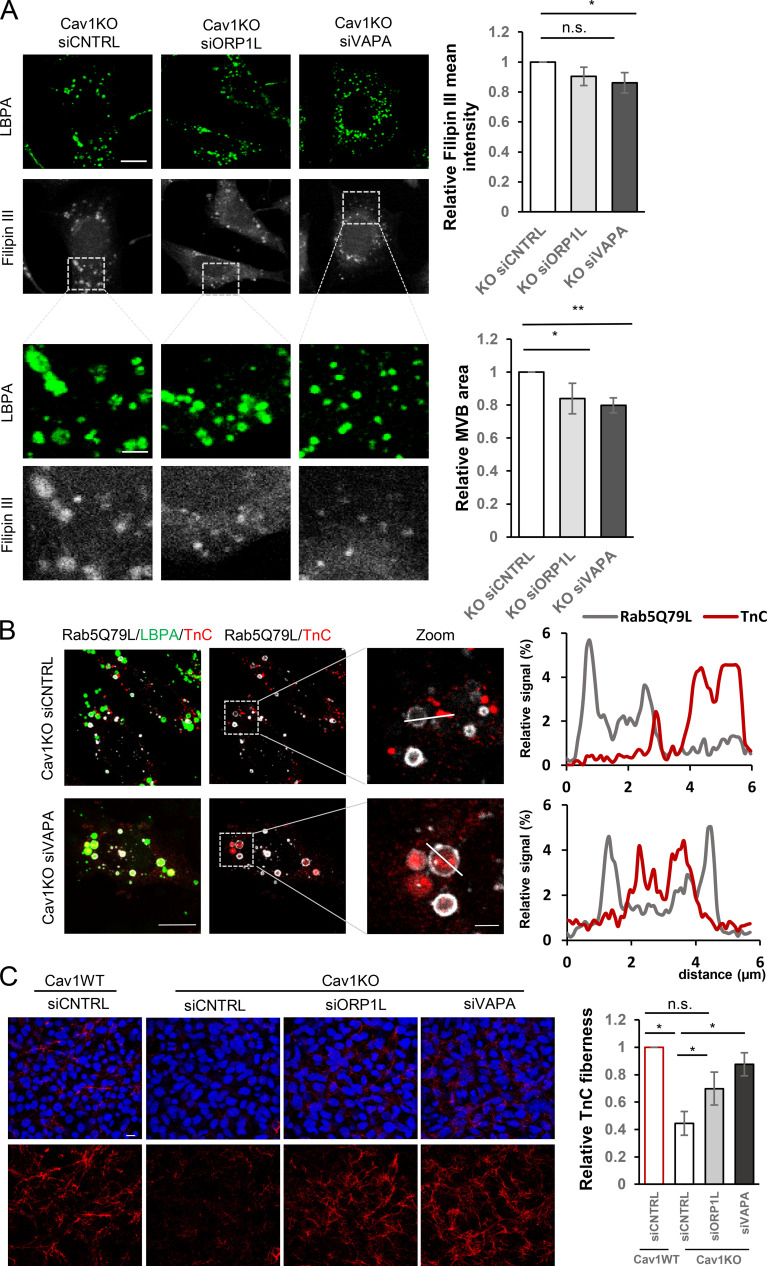
MCSs are small, specialized domains through which different membrane compartments communicate, allowing the active transfer of ions and lipids, including cholesterol in the case of ER-endosomal MCSs (summarized in a graphical scheme in Fig. S3 L; Rocha et al., 2009). We hypothesized that ER-MVB MCS disruption alleviates cholesterol accumulation in Cav1KO cells and restores the impaired sorting of ECM proteins onto exosomes. We knocked down ORP1L and vesicle-associated protein A (VAPA), as these components of the ER-endosome tether complex VAPA:ORP1L are relevant regulators of ER-to-endosome cholesterol transfer. Disruption of these contact sites in Cav1KO cells significantly reduced LE/MVB size and filipin staining intensity (Fig. 6 A and Fig. S5 D). Cholesterol content reduction upon disruption of these contact sites correlated with a rescue of TnC incorporation into MVBs (Fig. 6 B) and extracellular TnC fiber deposition to levels comparable to those observed in WT cells (Fig. 6 C).
Figure 6.
Cholesterol modulation at endosomal compartment/MVBs controls exosome-mediated TnC deposition. (A) Disruption of cholesterol transfer from the ER to the endosomes upon knocking down VAPA or ORP1L tethers in Cav1KO MEFs attenuates cholesterol accumulation as assessed by filipin (grayscale) and LBPA (green) staining (scale bar, 20 µm). Lower panel rows show zoomed thumbnails of the indicated regions of interest (scale bar, 6 µm). Charts on the right show quantitative analysis of filipin mean fluorescence intensity in MVBs (upper) and total MVB area (lower). Error bar are means ± SD; n = 3. (B) Confocal analysis of TnC distribution (in red) and LBPA (in green) in VAPA-depleted Cav1KO MEFs electroporated with Rab5(Q79L) (gray; scale bar, 10 µm). Rightmost panels show zoomed views of endosomes/MVBs (scale bar, 5 µm). Plots show pixel intensities for Rab5(Q79L) (gray) and TnC (red) along the indicated lines. (C) Z-stack projection of confocal images showing the effect on TnC fiber deposition (red) upon RNAi-mediated depletion for VAPA and ORP1L (scale bar, 10 µm). The chart shows extracellular TnC fiber deposition by either WT or Cav1KO MEFs upon depletion of indicated tethers (mean ± SEM; n = 3). n.s., not significant. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further characterize the role of endosomal cholesterol accumulation in the sorting of ECM proteins onto exosomes, we assessed the impact of loading WT cells with exogenous low-density lipoprotein (LDL)-cholesterol on TnC trafficking into LE/MVBs. Importantly, cholesterol-loaded cells showed a drastic reduction in the accumulation at LE/MVBs and extracellular deposition of TnC (Fig. S4, G and I).
Thus, the control of cholesterol content in the endosomal compartment is a key mechanism regulating exosome biogenesis and exosomal cargo specification, of which Cav1 is a relevant player.
Cav1-dependent exosome biogenesis directly drives ECM deposition and subsequent increase in tumor cell invasiveness.
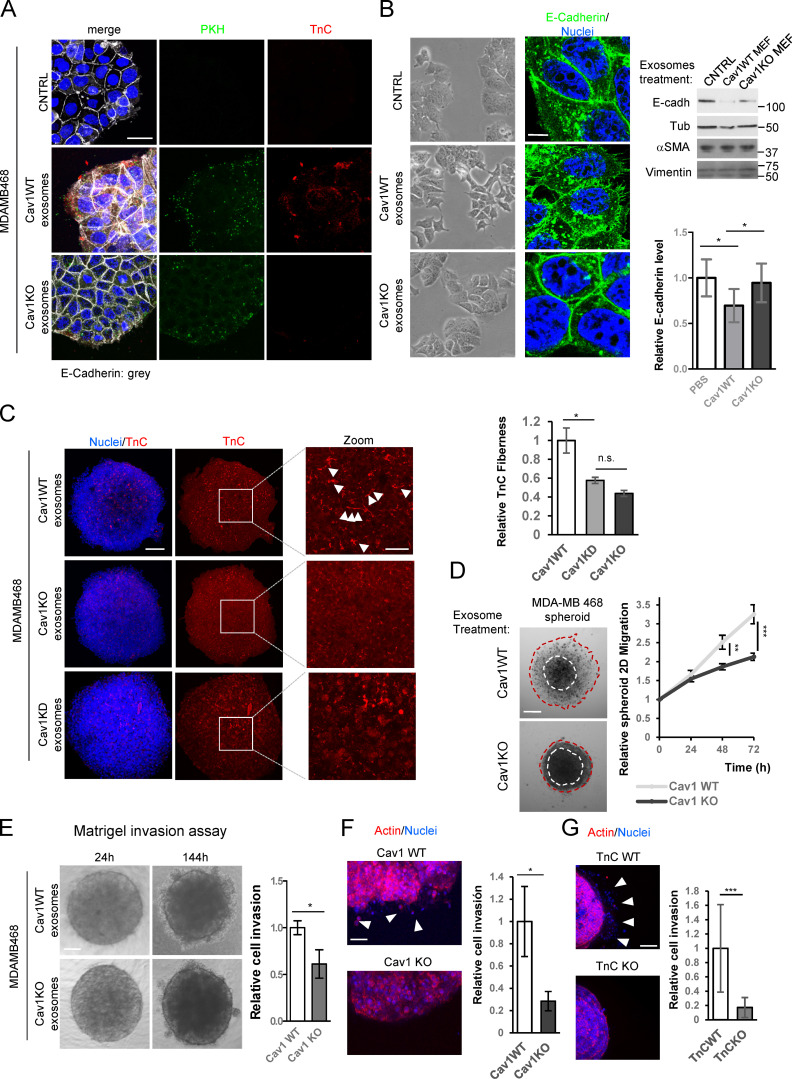
We compared the ability of exosomes purified from either WT or Cav1KO MEFs to deposit TnC on 2D cultures of the noninvasive breast cancer tumor cell line MDA-MB468, which naturally expresses negligible levels of Cav1 and TnC (Kang et al., 2016). After 24 h, tumor cells internalized prelabeled WT and Cav1KO exosomes with similar efficiency, as shown by similar accumulation of the general membrane dye PKH67 (Fig. 7 A, green). However, only WT exosomes led to detectable deposition of TnC in areas close to the periphery (Fig. 7 A, in red). In contrast, MDA-MB468 cultures exposed to Cav1KO exosomes showed TnC deposition levels comparable to those displayed by control cultures (exposed to vehicle). Exosomes thus have an intrinsic ability to deposit specific ECM material in vitro that requires Cav1-dependent regulation of exosome biogenesis.
Figure 7.
TnC deposition mediated by exosomes promotes tumor cell invasiveness. (A–G) MDA-MB468 breast tumor cells were incubated with PKH67-labeled exosomes derived from WT or Cav1KO MEFs or with PBS (control). (A) Microscopy images of exosome (green) uptake by MDA-MB468 cells and deposition of exosome-delivered TnC (red) in areas surrounding tumor cells. Scale bar, 30 µm. (B) Phenotypic changes in MDA-MB468 cells 36 h after exposure to fibroblast-derived WT and Cav1KO exosomes. Cells were stained for the epithelial-mesenchymal transition marker E-cadherin; nuclei were stained with Hoechst. Zoomed images show E-cadherin redistribution from cell-cell contact sites to intracellular compartments (scale bar, 10 µm). E-cadherin and other epithelial-mesenchymal transition markers were determined by Western blot. Chart shows the changes in E-cadherin upon treatment with PBS (CNTRL), WT, or Cav1KO exosomes (mean ± SD; n = 3). (C) Z-stack confocal images of TnC deposition (red) within 3D MDA-MB468 spheroids generated in the presence of WT, Cav1KO, or Cav1KD exosomes. Nuclei are stained with Hoechst (blue; scale bar, 150 µm). Zoomed images show a clear distribution of TnC deposits (scale bar, 50 µm). White arrowheads: TnC fibers. Right: chart shows extracellular TnC fiber deposition (mean ± SEM; n = 3). (D) Phase-contrast image of MDA-MB468 spheroid migration (scale bar, 175 µm). Spheroids were generated in the presence of the indicated exosomes and cultured for 72 h. Graph shows quantitative analysis of spheroid migration (mean ± SEM; n = 11). (E and F) Invasiveness of MDA-MB468 spheroids generated in the presence of fibroblast-derived WT or Cav1KO exosomes. Invasiveness was assessed in Matrigel (E; phase-contrast microscopy and quantified; mean ± SEM; n = 26 spheroids per condition; scale bar, 150 µm) or collagen type I gel (F; immunofluorescence; arrowheads indicate areas of cell invasion; mean ± SEM; n = 24 spheroids per condition; scale bar, 100 µm). (G) Invasiveness of MDA-MB468 spheroids generated in the presence of fibroblast-derived TnCWT and TnCKO exosomes. Arrowheads indicate areas of cell invasion. The chart shows quantification of MDA-MB468 spheroid invasiveness into collagen (mean ± SD; n = 15 spheroids per condition; scale bar, 150 µm). CNTRL, control; n.s., not significant. For all graphs, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Cav1-dependent exosomal deposition of TnC induced important morphological changes in MDA-MB468 tumor cells, eliciting cell-cell adhesion loss and internalization of the epithelial marker E-cadherin, associated with increased protrusiveness and emission of filopodia (Fig. 7 B). A similar phenotype was observed when using CAF lines transduced with lentiviral shRNA-encoding vectors (Fig. S5 C). Accordingly, MDA-MB468 cells exhibited increased migration, as assessed by wound healing and transwell migration assays (Fig. S5, F and G). Interestingly, exosomes purified from MDA-MB231 tumor cells, which contain low TnC compared with exosomes derived from WT fibroblasts despite having comparable amounts of Cav1 protein, failed to up-regulate protrusiveness and motility of MDA-MB468 cells (Fig. S5 B; Fig. S2, G and H; and Data S2). Thus, exosomal TnC/ECM cargo derived from stromal fibroblasts, and not exosomal Cav1 per se, drives the induction of invasive phenotypes in tumor cells.
We studied the ability of exosomes to deposit ECM on MDA-MB468 spheroids, scaffold-free 3D assemblies in which cells self-organize ECM and engage in intercellular communication and which have been validated as superior in vitro experimental proxies of live tissues for many applications (Fennema et al., 2013; Zanoni et al., 2016). WT-derived TnC-containing exosomes, but not exosomes derived from Cav1KO or Cav1KD fibroblasts (i.e., TnC depleted), efficiently deposited fibrillar TnC structures within cell spheroids (Fig. 7 C). 3D reconstruction demonstrated that TnC fibrils were preferentially located in the interstitial space (Fig. S5 E).
To characterize Cav1-dependent, exosome-induced phenotypic changes in tumor cells in this setting, spheroids generated in the presence of exosomes were plated and cells were allowed to migrate for 72 h. WT exosomes promoted significantly more migration than exosomes derived from Cav1KO MEFs (Fig. 7 D). To confirm the role of TnC in this process, we tested the effect of exosomes derived from TnCKO MEFs (Fig. 7 G and Fig. S5, H and I). TnCKO exosomes were poor stimulators of cell migration, supporting that the observed phenotypic impact derives largely from the sorting of specific ECM cargoes to exosomes.
We next examined whether exosomes increased tumor cell invasiveness. Cav1KO exosomes consistently exhibited a blunted ability to induce invasiveness in Matrigel invasion assays (Fig. 7 E) or 3D collagen matrix invasion assays (Fig. 7 F), similar to the reduced invasion promoted by exosomes derived from TnCKO cells (Fig. 7 G). These results further support the relevance of Cav1- and TnC-dependent exosome-induced ECM deposition and phenotypic alteration for tumor progression.
Cav1WT MEF-derived exosomes create TnC-rich niches in vivo
To evaluate whether exosomes deposit TnC in vivo over large (interorgan) distances, exosomes purified from WT or Cav1KO fibroblasts were fluorescently labeled and injected into the caudal vein of TnCKO mice every day for 1 wk (Fig. 8 A). Histopathological analysis revealed specific exosome accumulation in liver and, to a lesser extent, in lung (Fig. 8 B and Fig. S5 J). However, associated TnC deposition was only consistently detectable in liver and lung of mice injected with WT exosomes, contrasting with the residual TnC staining in the organs of animals injected with Cav1KO exosomes or vehicle (PBS), despite Cav1KO exosomes being able to reach this organ with comparable efficiency. Thus, Cav1 has a key role in exosome-dependent ECM deposition in vivo. Because TnC is a positive regulator of metastasis through the assembly of favorable distant microenvironments, Cav1 might specifically modulate cancer outcome through exosome-dependent stroma–tumor communication.
Figure 8.
Cav1-loaded fibroblast-derived exosomes deposit TnC in vivo in a tissue-specific manner. (A) Treatment protocol. (B) Fluorescence microscopy of liver sections from mice injected in the tail vein with Cav1WT or Cav1KO fibroblast-derived exosomes or with PBS (scale bar, 60 µm; zoomed views, scale bar, 30 µm). Exosome foci are green. White arrowheads indicate regions of TnC deposition. All images are representative of nine random fields from two independent experiments. (C) Proposed role of Cav1 in exosome-mediated ECM deposition. (C I) Endocytosed Cav1 enters the MVB compartment, where it promotes exosome heterogeneity in size and composition, favoring the entry of specific ECM components. (C II) Cav1WT fibroblast-derived exosomes stimulate protrusive activity and motility of breast cancer cells by nucleating local ECM (TnC) deposition in areas surrounding the tumor cells. (C III) Cav1WT fibroblast-derived exosomes also generate ECM-rich deposits at long distances from their source in vivo, generating sites of possible future metastasis.
Discussion
Exosomes constitute a means by which tumor cells and associated stroma not only communicate locally but also influence and transform distant locations or “niches” and even modulate organismal responses systemically (Costa-Silva et al., 2015; Hoshino et al., 2015; Peinado et al., 2012). Despite their ubiquity, the mechanisms underlying exosome biogenesis, cargo sorting, and function remain elusive. Here, we identify Cav1 as a modulator of exosome biogenesis through the regulation of cholesterol content in MVBs, which allows the generation of different exosome subpopulations and exosome cargo specification (mainly specific ECM proteins) with subsequent functional impact. Our data demonstrate that the presence of Cav1 within exosomes is favored by its endocytosis through Tyr14 phosphorylation. This promotes Cav1 targeting to the MVB outer membrane, where a second mechanism is activated that requires the presence of ubiquitination-targeting N-terminal domain lysine residues; this second mechanism sorts Cav1 into ILVs for exosome generation. Cav1 would act as a “cholesterol rheostat” in MVBs, thus increasing the plasticity of this compartment and enabling the segregation of different exosome populations. Cholesterol titration in this compartment could also determine exosome cargo specification by modulating ER-MVB contacts. Cav1-dependent exosomal ECM deposition elicits both local ECM deposition and the generation of distant ECM-enriched regions, supporting a novel role of Cav1 in stromal remodeling. This exosomal ECM deposition promotes protrusiveness and active migration of targeted tumor cells by increasing cell scattering, suggesting that these mechanisms contribute to tumor cell invasiveness and favor the nucleation of new ECM stromal niches (Fig. 8 C).
TnC is a key component involved in promoting exosome-dependent tumor invasion. TnC matrix deposition by Cav1-expressing fibroblasts strictly required intact exosome biogenesis and secretion. Our data support a model whereby the ER/Golgi compartment is a major direct supplier of TnC to exosomes and rule out incorporation from routes bypassing de novo synthesis, such as internalization from the extracellular space. Supported by studies performed in breast cancer cell lines (Campos et al., 2018), our results demonstrate that Cav1 regulates a general mechanism for exosome cargo specification, especially relevant for TnC sorting and secretion. Our results suggest that Cav1-dependent exosomal TnC secretion has a dual effect on tumor progression (Fig. 7): (1) local deposition of TnC at the tumor-invasive front presumably induces tumor cell migration away from the tumor mass (see Fig. 7), as observed upon direct addition of recombinant soluble TnC (Nagaharu et al., 2011); additionally, (2) Cav1-dependent exosome–mediated TnC deposition over large distances might contribute to the “priming” of stromal niches before the arrival of cancer cells, forming a “landing dock” for subsequent metastatic growth (Costa-Silva et al., 2015; Sceneay et al., 2013). We show that IV injection of exosomes derived from WT fibroblasts, but not from Cav1KO cells, can efficiently promote direct TnC–matrix deposition in distant organs (Fig. 8). Further investigation is required to confirm whether these ECM stromal niches have an impact on metastasis in vivo.
A pathway for caveolae internalization (Chaudhary et al., 2014) has been described in which caveolae are disassembled and Cav1 is internalized and ubiquitinated, followed by accumulation at the internal membrane of LEs/MVBs (Hayer et al., 2010; Hill et al., 2008). Interestingly, Cav1 is also targeted to LEs by perturbations of lysosomal pH and changes in cholesterol content, with no changes in Cav1 degradation rate, raising the possibility of alternative fates for this Cav1 pool (Mundy et al., 2012; Pol et al., 2020). The changes in Cav1-MVB colocalization we observed (Fig. 1 and Fig. S1) are compatible with a scenario whereby a significant proportion of internalized Cav1 is sorted into MVBs, in part to be incorporated into exosomes. However, at present we cannot rule out that other intracellular compartments, such as the ER, may also be a source of exosomal Cav1.
Cav1 binds cholesterol with high affinity (Murata et al., 1995), and its ability to move between different compartments might contribute to the regulation of cholesterol fluxes in cells (Pfrieger and Vitale, 2018; Pol et al., 2005). Our data support that Cav1 dictates the levels of free cholesterol in LE/MVB membranes, conferring the required plasticity for the generation of ILVs of varying size and favoring the sorting of ECM components onto exosomes for their secretion (Fig. 2, Fig. S2, and Fig. S3). Furthermore, artificial manipulation of cholesterol levels at LE/MVB, either by disrupting ER-endosome MCSs (Fig. 6 and Fig. S5 D), by pharmacologically blocking cholesterol trafficking at the endosomal compartment (Fig. 2; Fig. 4, B and C; Fig. S2 F; and Fig. S4 H), or by exogenous loading with LDL-cholesterol (Fig. S4, G and I), had a profound effect on the intracellular trafficking and exosomal secretion of ECM exosome cargo. This suggests a model whereby the control of cholesterol levels at endosomes/MVBs is integral to the regulation of exosome-mediated ECM deposition, as recently described for other signaling events and functions at these intracellular compartments (Lim et al., 2019; Eden et al., 2016). Of note, Cav1 at ER-mitochondria MCS regulates the recruitment of sterol metabolism complexes and cholesterol accumulation (Sala-Vila et al., 2016). Further investigation will be required to better define the mechanistic details of the interplay between cholesterol trafficking and exosome biogenesis and downstream functions, of which Cav1 appears to be a key player.
Interestingly, among identified Cav1-dependent exosomal cargoes, collagens are less abundant than other ECM components. Collagen secretion requires specific assembly mechanisms operating through the COPII biosynthetic/secretory machinery (Malhotra and Erlmann, 2015) and is thus unavailable to the exosome pathway (Fig. 3, Fig. 4, and Fig. S3). These pathways might in fact be oppositely regulated by Cav1, since a negative correlation has been described between Cav1 expression and collagen deposition in chronologically aged skin and during fibrosis (Gvaramia et al., 2013; Lee et al., 2015). Supporting our observation that exosomes can act as carriers of some ECM proteins, a recent study demonstrated exosomal secretion of FN through its binding to integrin α5β1; however, the function and underlying mechanism of this sorting were not determined (Lobert et al., 2010; Sung et al., 2015). Our data point to incorporation of TnC into ILVs through its sorting from the biosynthetic route to the MVB compartment, excluding the possibility of an extracellular origin through internalization or by direct binding to exosomes once they are released (Fig. 5).
Previous studies demonstrated high levels of Cav1-expressing exosomes in the plasma of melanoma patients (Logozzi et al., 2009). We propose that relative levels of Cav1-expressing exosomes could be a valuable prognostic marker of metastasis and tumor malignancy, as well as other cholesterol-related diseases. Our findings suggest new potential opportunities for diagnosis, prevention, and therapeutic intervention through exosomal tumor–stroma communication.
Materials and methods
Antibodies and reagents
Monoclonal anti-Cav1 (#3267) and anti-Alix (#2171) antibodies were from Cell Signaling Technology. Anti-LBPA was from Echelon (Z-SLBPA). Anti-CD63 (ab1318), antiflotillin (ab13349), antiperiostin (ab14041), anticalreticulin (ab2907), anti-PTRF (ab48824), and anti-Tsg101 (ab83) were from Abcam. Anti-GM130 (610823), anti–py14-Cav1 (611339), anticadherin (610181), and antivimentin (550513) were from BD Transduction. Antitubulin (T-9026), anti-FN (F3648), anti-Tenascin (Clone Mtn-12, T3413), and anti–smooth muscle actin (A5228) were from Sigma. Anti-Tenascin was also from Millipore (AB19011). Anti-nSMase1 (sc-377135) and anti-nSMase2 (sc-166637) were from Santa Cruz. Alexa Fluor 488-, 546-, and 647-conjugated phalloidin were from Life Technologies. GW4869 (567715) was from Calbiochem, and dmA (A4562) and U18666A (U3633) were from Sigma. Matrigel (354230) was from Becton Dickinson, and Collagen I from rat tail (354249) was from Corning. Filipin was from Sigma (F4767).
Cell culture
MEFs derived from Cav1WT and Cav1KO littermate mice used throughout this study have been previously described (del Pozo et al., 2005). TnCWT and TnCKO MEFs were provided by our collaborator Dr. Gertraud Orend. PTRFKO cells were provided by Dr. Parton (Institute for Molecular Bioscience, University of Queensland, St. Lucia, Australia) and were reconstituted with the retroviral expression vector pMIGR1. COS7 cells were from ATCC. MEFs and COS7 cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified 5% CO2–95% air incubator. The breast cancer–associated fibroblast line CCD-1069S and the breast tumor cell line MDA-MB468 were from ATCC and were cultured in DMEM/F-12 supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. CCD-1069S fibroblasts stably expressing shRNA targeting Cav1 or a scramble siRNA were generated by lentiviral infection as reported in Goetz et al. (2011). All the cell lines were routinely checked for mycoplasma infection.
siRNA silencing
siRNA silencing was performed using 20 nM siRNA and Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol, and cells were cultured for 2–3 d. siRNAs targeting mouse Tsg101, nSMase1, and nSMase2 were SMARTpool ON-TARGET plus reagents from Dharmacon (Tsg101 Cat. L-049922-01-0005; nSMase1 or Smpd2 Cat. L-044206-01-0005; and nSMase2 or Smpd3 Cat. L-059400-01-0005). The control siRNA was ON-TARGET plus nontargeting pool (Dharmacon Cat. D-001810-10-05). Cav1 was silenced with an siRNA from Dharmacon (seq. 5′-GAGCUUCCUGAUUGAGAUU-3′) and an enzymatically prepared siRNA from Sigma (EHU031251). esiRNAs targeting the VAPA:ORP1L tether complex were purchased from Sigma (EMU053971 and EMU012501, respectively).
Isolation and characterization of exosomes released by fibroblasts
Exosomes were isolated from cultured fibroblasts grown in exosome-free culture medium. To remove detached cells, conditioned medium was collected and centrifuged at 300 g for 10 min at 4°C. The supernatant was collected and centrifuged at 2,000 g for 20 min at 4°C. The supernatant was then centrifuged at 10,000 g at 4°C for 30 min to completely remove contaminating apoptotic bodies, microvesicles, and cell debris. The clarified medium was then ultracentrifuged at 110,000 g at 4°C for 70 min to pellet exosomes. The supernatant was carefully removed, and crude exosome-containing pellets were washed in ice-cold PBS. After a second round of ultracentrifugation, the resulting exosome pellets were resuspended in the desired volume of PBS.
For further purification, exosomes were ultracentrifuged on a discontinuous sucrose gradient including sucrose concentrations of 0.25 M, 0.5 M, 0.8 M, 1.16 M, 1.3 M, and 2 M in 20 mM Hepes, pH 7.4. Exosome samples were laid on the bottom of the gradient in the 2-M sucrose fraction, followed by centrifugation at 200,000 g for 18 h. 11 individual 1-ml gradient fractions were manually collected. Fractions were diluted in PBS and centrifuged at 110,000 g for 1 h at 4°C, and the resulting pellets were resuspended and analyzed. Alternatively, an equal volume of cold acetone was added to each fraction, and the proteins were precipitated for 2 h at −20°C. Protein pellets were collected by centrifugation at 16,000 g for 10 min and air-dried to eliminate acetone traces. The protein precipitates were monitored by Western blot for the expression of the exosomal markers Alix and Tsg101.
Alternatively, a discontinuous iodixanol gradient was used. To prepare the discontinuous iodixanol gradient, 40% (wt/vol), 20% (wt/vol), 10% (wt/vol), and 5% (wt/vol) solutions of iodixanol were made by diluting stock solutions of Optiprep (60% [wt/vol] aqueous iodixanol) with 0.25 M sucrose/10 mM Tris, pH 7.5. The gradient was formed by adding 3 ml of 40% iodixanol solution to a 14 × 89–mm polyallomer tube (Beckman Coulter), followed by careful layering of 3 ml each of 20% and 10% solutions and 2.5 ml of 5% solution. Exosome preparation was overlaid onto the top of the gradient and centrifugation performed at 100,000 g for 18 h at 4°C. 12 individual 1-ml gradient fractions were collected manually from the top to the bottom. Fractions were diluted with 1 ml PBS and precipitated with 2 ml cold acetone for 3 h at −20°C. Protein pellets were collected by centrifugation at 16,000 g for 10 min and air-dried to eliminate acetone traces. The protein precipitates were monitored by Western blot for the expression of exosomal markers Alix, Tsg101, flotillin-1, and the negative control GM130 (Golgi).
In all experiments, the exosomes used corresponded to the total exosome pellets resulting from the serial centrifugation steps; usually, 106 exosome particles were added per cell. Exosome uptake was performed in exosome-free medium.
Exosome concentrations and size distributions were determined by Nanoparticle Tracking Analysis (Nanosight).
Western blot analysis
Western blot samples were prepared with conventional 5× Laemmli sample buffer brought to 1× when mixed with the sample. The 5× Laemmli sample buffer is composed of 60 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.01% bromophenol blue.
Cell or exosome proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Blots were incubated for 2 h in PBS containing 5% skimmed milk and overnight with primary antibodies (typically diluted 1:1,000). After incubation (2 h) with horseradish peroxidase–conjugated goat anti-rabbit or goat anti-mouse antibody, signal on washed blots was detected by enhanced chemiluminescence (GE Healthcare). Band intensities were quantified with Image J software.
Intracellular lysate analysis
To prepare lysates containing only proteins from the cell interior, plated cells were treated with trypsin to eliminate proteins in the extracellular space. Detached cells were washed with PBS and resuspended in loading buffer for Western blot analysis. For total lysates comprising cell and ECM proteins, cells were scraped directly from the plate in loading buffer.
PKH67 labeling of exosomes
Purified exosomes were labeled with the green fluorescent dye PKH67 (Sigma; MIDI67) according to the manufacturer’s protocol. After staining, unbound PKH67 was removed with columns from Thermo Fisher Scientific (4484449).
EM
Exosomes were visualized by TEM according to the method of Théry et al. (2006). The exosome suspension was fixed in 2% PFA and transferred to formvar/carbon–coated EM grids. After 20 min, grids were placed sample-face down for 2 min in a 100-μl drop of PBS on a sheet of parafilm. Grids were then transferred to 1% glutaraldehyde for 5 min and then washed with distilled water. Samples were contrasted in 2% uranyl acetate and examined by TEM.
Spheroid formation assay
After trypsinization, MDA-MB468 cells were resuspended in exosome-free DMEM, counted, and adjusted to the desired concentration (usually 5 × 103 cells/ml). A 20-µl final volume was prepared with 5× methyl cellulose prepared in exosome-free medium, the MDA-MB468 cell suspension, and the desired number of exosomes (usually 1 × 106 particles). The mixture was carefully deposited in a drop on the inner surface of a 100-mm dish lid. The plate lid was placed on a plate containing 10 ml PBS to humidify the culture chamber. Under gravity, cells aggregated at the bottom of the hanging drop. After 24 h, the resulting cell aggregates were lifted with a pipette. For 2D spheroid migration assays, the aggregates were seeded on a 96-well plate to monitor cell migration from the spheroid by phase-contrast time-lapse video microscopy. Alternatively, aggregates were seeded on collagen or Matrigel gels as described below. In all assays, no additional exosomes were added to the medium apart from the exosomes used during spheroid generation.
Collagen embedding
High-density rat-tail type I collagen (Corning) was diluted to 2 mg/ml. This solution self-polymerizes to form a gel after 20–30 min at 37°C. Tumor cell spheroids were transferred to 25 µl unpolymerized collagen in the wells of a μ-slide angiogenesis plate (ibidi). After 30 min, 40 µl medium was added over the top of the gel. Medium was replaced every 48 h. Spheroids were imaged every 24 h for the duration of the experiment. Spheroid diameter was measured from captured images with ImageJ.
For Matrigel embedding assays, spheroids were transferred to a 25-µl 1:1 mix of Matrigel and medium.
Wound closure assay
For the wound healing assay, confluent monolayers were scraped with a 0.1–2-µl pipette tip in the presence of mitomycin C, and wound closure was monitored by capturing images at 10-min intervals.
CDM generation
3D matrices reminiscent of in vivo ECM were prepared as published (Beacham et al., 2007; Castelló-Cros and Cukierman, 2009; Goetz et al., 2011). Briefly, 2.5 × 105 cells/ml were plated on chemically cross-linked gelatin on tissue culture dishes or coverslips and maintained in confluence for 6 d. Cells were supplemented every 48 h with 50 mg/ml fresh L-ascorbic acid to stabilize ECM components, thus facilitating collagen production and polymerization. The resulting 3D cultures were checked for quality by immunofluorescence, or cells were removed from the matrix by alkaline detergent extraction, yielding cell-free 3D matrices for further analysis.
For analysis of exosome binding to preexisting CDMs, CDMs from TnCWT and TnCKO MEFs were obtained as previously indicated. Cell-free 3D matrices were scrapped in 1 ml PBS and homogenized passing several times through a syringe (0.5 × 16 mm). Cell-free 3D matrix corresponding to a p150 plate was incubated with 1 × 108 exosomes per condition for 1 h at 37°C. After that, samples were ultracentrifuged at 10,000 g for 30 min to remove unbound cell-free 3D matrix components followed by an ultracentrifugation at 110,000 g for 70 min to recover exosomes. Obtained exosomes were loaded with equal amount of Laemmli buffer and analyzed by SDS-PAGE.
For assessing potential TnC internalization through endocytic trafficking, cells from the indicated genotypes were plated onto previously isolated decellularized CDMs (obtained as described above) and cultured for 24 h in the incubator at 37°C. Cells were then trypsinized and rinsed with PBS in order to remove any extracellular nonendocytosed ECM proteins. Finally, cells were resuspended into denaturing, reducing SDS-PAGE loading buffer to obtain “cell lysates” ready for Western blot analysis.
Proteomic analysis
Lysates and exosomes derived from WT and Cav1−/− MEFs were digested using the filter-aided sample preparation protocol (Wiśniewski et al., 2009). Briefly, samples were dissolved in 50 mM Tris-HCl, pH 8.5, 4% SDS, and 50 mM DTT, boiled for 10 min, and centrifuged. Protein concentration in the supernatant was measured with the Direct Detect Spectrometer (Millipore). About 100 µg protein was diluted in 8 M urea in 0.1 M Tris-HCl, pH 8.5 (urea buffer; UA), and loaded onto 30-kD centrifugal filter devices (filter-aided sample preparation protein digestion kit). The denaturation buffer was removed by washing three times with UA. Proteins were then alkylated by incubation in 50 mM iodoacetamide in UA for 20 min in the dark, and excess alkylation reagents were eliminated by washing three times with UA and three additional times with 50 mM ammonium bicarbonate. Proteins were digested overnight at 37°C with modified trypsin (Promega) in 50 mM ammonium bicarbonate at a 50:1 protein:trypsin (wt/wt) ratio. The resulting peptides were eluted by centrifugation with 50 mM ammonium bicarbonate (twice) and 0.5 M sodium chloride. Trifluoroacetic acid was added to a final concentration of 1%, and the peptides were finally desalted onto C18 Oasis-HLB cartridges and dried down for further analysis.
Multiplexed isobaric labeling
For the quantitative analysis, tryptic peptides were dissolved in triethylammonium bicarbonate buffer, and the peptide concentration was determined by measuring amide bonds with the Direct Detect system. Equal amounts of each peptide sample were labeled using the 4-plex iTRAQ Reagents Multiplex Kit (Applied Biosystems). Briefly, each peptide solution was independently labeled at room temperature for 1 h with one iTRAQ reagent vial (mass tag 114, 115, 116, or 117) previously reconstituted with ethanol. After incubation at room temperature for 1 h, the reaction was stopped with diluted trifluoroacetic acid and peptides were combined. Samples were concentrated in a Speed Vac, desalted onto C18 Oasis-HLB cartridges, and dried down for mass spectrometry (MS) analysis.
MS
Digested peptides were loaded into the liquid chromatography–tandem MS (LC-MS/MS) system for on-line desalting onto C18 cartridges and analyzed by LC-MS/MS using a C-18 reversed-phase nano-column (75 µm I.D. × 25 cm, 2 µm particle size, Acclaim PepMap RSLC, 100 C18; Thermo Fisher Scientific) in a continuous acetonitrile gradient consisting of 0%–30% B for 240 min and 50%–90% B for 3 min (A = 0.5% formic acid; B = 90% acetonitrile, 0.5% formic acid). A flow rate of 200 nl/min was used to elute peptides from the reverse phase nano-column to an emitter nanospray needle for real-time ionization and peptide fragmentation in an Orbitrap QExactive mass spectrometer (Thermo Fisher Scientific). During the chromatography run, we examined an enhanced Fourier transform-resolution spectrum (resolution = 70,000) followed by the higher-energy collisional dissociation MS/MS spectra from the 20 most intense parent ions. Dynamic exclusion was set at 40 s. For increased proteome coverage, labeled samples were also fractioned by cation exchange chromatography (Oasis HLB-MCX columns); fractions were desalted and analyzed using the same system and conditions described before.
Protein identification and quantification
All spectra were analyzed with Proteome Discoverer (version 1.4.0.29; Thermo Fisher Scientific) using SEQUEST-HT (Thermo Fisher Scientific). The Uniprot database, containing all mouse sequences (March 3, 2013), was searched with the following parameters: trypsin digestion with two maximum missed cleavage sites; precursor and fragment mass tolerances of 2 D and 0.03 D, respectively; methionine oxidation as a dynamic modification; and carbamidomethyl cysteine and N-terminal and Lys iTRAQ modifications as fixed modifications. Peptides were identified using the probability ratio method (Martínez-Bartolomé et al., 2008), and false discovery rate was calculated using inverted databases and the refined method (Navarro et al., 2014) with an additional filtering for a precursor mass tolerance of 15 ppm (Bonzon-Kulichenko et al., 2015). Identified peptides had a false discovery rate ≤1%.
Proteins were quantified from reporter ion intensities and quantitative data analyzed with QuiXoT, based on the weighted spectrum, peptide, and protein statistical model (García-Marqués et al., 2016). In this model, protein log2-ratios are expressed as standardized variables (i.e., in units of SD according to their estimated variances [Zq values]). Functional class enrichment was assessed using the Enrichr package (Chen et al., 2013), and network depiction of annotated relationships among hits was retrieved from the STRING v.11 resource).
MS raw files and msf search files have been deposited in the PeptideAtlas repository (accession no. PASS01614; http://www.peptideatlas.org/PASS/PASS01614).
LC-MS/MS analysis of exosome glycerophospholipids
Exosome samples from WT and KO mice corresponding to 2.5 µg protein were used. Before lipid extraction, the following internal standards were added: 200 pmol each of 1,2-dipentadecanoyl-sn-glycero-3-phosphocholine, 1,2-dilauroyl-sn-glycero-3-phosphoethanolamine, 1,2-dipalmitoyl-sn-glycero-3-phosphoinositol, 1,2-dimiristoyl-sn-glycero-3-phosphoserine, 1,2-dimiristoyl-sn-glycero-3-phosphate, and 1-heptadecanoyl-2-arachidonoyl-3-phosphoglycerol, according to the method of Bligh and Dyer (1959). After evaporation of organic solvent under vacuum, the lipids were redissolved in 50 µl solvent mixture (75% A, 25% B), and 40 µl was injected into an Agilent 1260 Infinity high-performance liquid chromatograph equipped with an Agilent G1311C quaternary pump and an Agilent G1329B Autosampler. The column was a FORTIS HILIC (150 × 3 mm, 3-µm particle size; Fortis Technologies) protected with a Supelguard LC-Si (20 mm × 2.1 mm) guard cartridge (Sigma). Glycerophospholipids and sphingomyelin were separated according to the procedure described by Axelsen and Murphy (2010), with minor modifications. Briefly, the mobile phase consisted of a gradient of solvent A (hexanes:isopropanol, 30:40, by vol) and solvent B (hexanes:isopropanol:20 mM AcONH4 in water, 30:40:7 by vol). The gradient started at 75% A from 0 to 5 min, then decreased from 75% A to 40% A at 15 min, from 40% A to 5% A at 20 min, holding at 5% until 40 min, and increasing to 75% at 41 min. The column was then reequilibrated by holding at 75% A for an additional 14 min before the next sample injection. The flow rate through the column was fixed at 0.4 ml/min, and this flow entered into the electrospray interface of an API2000 triple quadrupole mass spectrometer (Applied Biosystems). The source parameters were set as follows: ion spray voltage, −4,500 V; curtain gas, 20 psi; nebulizer gas, 35 psi; desolvation gas, 65 psi; and temperature, 400°C. Phospholipid species were analyzed in Q1 with negative ionization. Compound parameters were fixed for all analyzed species: declustering potential, −65 V; entrance potential, −10 V; focusing potential, −300 V; and collision cell exit potential, −10 V. Phospholipid species were detected as [M-H]− ions except for phosphatidylcholine and sphingomyelin species, which were detected as [M+OAc]− adducts. For quantification, chromatographic peaks of each species were integrated and compared with the peak area of the internal standard corresponding to each phospholipid class. Since bis(monoacylglycerol)phosphate and sphingomyelin internal standards were unavailable at the time of analysis, these species were quantified using 1-heptadecanoyl-2-arachidonoyl-3-phosphoglycerol and 1,2-dipentadecanoyl-sn-glycero-3-phosphocholine standards, respectively. MS source raw data have been deposited in the MetaboLights public repository under accession no. MTBLS1969.
Cholesterol quantification by LC-MS/MS
To quantify the amount of cholesterol in exosomes, 25 µl of 19.68 µg/ml cholesterol (d7) solution in methanol (SPLASH LipidoMIX Internal Standard; Avanti) was added to each sample. Cholesterol was extracted, and proteins were precipitated by adding 475 µl cold methanol–ethanol (1:1 vol/vol). Samples were vortex-mixed, sonicated during 3 min, and incubated on ice for 20 min. Finally, supernatants were collected by centrifugation at 18,400 g for 20 min at 8°C and dried in SpeedVac (Savant SPD131DDA concentrator; Thermo Fisher Scientific) for 2 h at room temperature.
Metabolomics analysis was performed using an Ultimate 3000 HPLC system coupled to an Orbitrap ELITE Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). For chromatographic separation, an Agilent RP, Hi Recovery column (100 × 0.5 mm, 5 µ) was used. Cholesterol was identified by LC-MS/MS mode and quantified according to the response factor of the respective internal standard.
Subcellular fractionation
Intracellular compartments were separated by sucrose density gradient centrifugation. Cells were washed three times in 10 mM Tris-HCl, pH 7.5, and 0.5 mM MgCl2 and then scraped and resuspended in 50 mM Tris-HCl, pH 7.5, and 0.25 M sucrose. Cells were homogenized with 30 strokes using a Dounce homogenizer. The homogenates were then centrifuged at 3,000 g for 15 min at 4°C. The supernatant was placed over a continuous sucrose gradient (2 M at the bottom; 0.25 M at the top), and samples were separated at 100,000 g for 18 h in an SW40 rotor (Beckman Coulter) at 4°C. After centrifugation, 1-ml fractions were collected from the tube, yielding a total of 12 fractions. Cold acetone (1 ml) was added to each tube, and the homogeneous mixture was allowed to precipitate at −20°C for 2 h. The samples were centrifuged at 16,000 g at 4°C, and the protein pellets were allowed to dry for 2 h. The protein precipitates were then analyzed by SDS-PAGE for proteins, including the ER marker calreticulin and the MVB marker CD63.
Preparation of DRM fractions
DRMs from exosomes derived from Cav1WT and Cav1KO MEFs were purified as described in Navarro-Lérida et al. (2002), with the Triton X-100 concentration reduced from 1% to 0.5%.
Transwell migration assay
The transwell migration assay was performed with Boyden chambers containing an 8-µm pore-size polycarbonate filter (Corning Costar). Serum-free medium (300 µl) with or without exosomes was placed in the wells of a 24-well plate. The transwell inserts were placed in the wells, and 5 × 104 MDA-MB468 cells in 200 µl serum-free medium were added to the upper chamber. After incubation for 24 h, cells on the lower membrane surface were fixed and stained with crystal violet and counted under a microscope.
Animal model
All animal procedures conformed to European Union Directive 86/609/EEC and Recommendation 2007/526/EC regarding the protection of animals used for experimental and other scientific purposes, enforced in Spanish law under Real Decreto 1201/2005. Cav1WT and Cav1−/− exosomes were labeled with PKH67 and intravenously injected every day for 1 wk into the tail vein of TnC−/− mice (Talts et al., 1999) bred into the FVB background. After 4 d, lungs and livers were collected and embedded sections were analyzed by immunohistochemistry with anti-TnC antibody (Sigma).
Immunofluorescence microscopy
Cells grown on glass coverslips were washed twice with PBS and fixed for 15 min at room temperature in 4% PFA in PBS. Fixed cells were washed extensively with PBS and permeabilized for 5 min in PBS containing 0.1% TritonX-100 and 2% BSA to reduce nonspecific binding. Cells were incubated for 1 h at room temperature in PBS containing 0.2% BSA with primary antibodies (typically diluted 1:200). Subsequent washes and incubations with Alexa Fluor 647 or 546 phalloidin or fluorescent secondary antibodies were performed in PBS for 1 h at room temperature. Coverslips were mounted in PermaFluor aqueous mounting medium and examined with a Leica SPE, SP5, or SP8 confocal microscope, typically through 40× (ACS APO 40.0× 1.15 oil) or 63× (ACS APO 63.0× 1.30 oil) objectives. A 100× objective (HC PL Apo CS2 100×/NA 1.4 oil) was used for high-resolution imaging in an SP8 confocal microscope.
Filipin staining and quantification
Cells were grown on glass coverslips. After fixing for 10 min at 37°C with 4% PFA in PBS, cells were permeabilized with 0.2% BSA and 0.1% saponin in PBS (blocking solution) for 30 min. Samples were then incubated with primary antibody (LBPA) and fluorescently labeled–conjugated secondary antibodies for 1 h each, interspersed with three washes in blocking solution. After an additional wash in PBS, cells were incubated for 1 h with filipin (50 µg/ml in PBS). After three washes in PBS, slides were mounted in Fluoromount. Fiji software was used to quantify filipin fluorescence intensity (http://fiji.sc/). Cells were segmented based on LBPA staining. Filipin staining intensity in LBPA-positive vesicles was measured after intensity thresholding.
Quantitative real-time PCR
Frozen tissues or cell lines were analyzed for specific gene expression using Sybr Green PCR Reagents (Applied Biosystems) and specific primers: mouse Tsg101 sense 5′-TCTAACCGTCCGTCAAACTGT-3′, antisense 5′-TTGTACCAGTGAGGTTCACCA-3′; mouse nuclear sphingomyelinase (nSMase) 2 sense 5′-ACACGACCCCCTTTCCTAATA-3′, antisense 5′-GGCGCTTCTCATAGGTGGTG-3′; and mouse nSMase 1 sense 5′-TGGGACATCCCCTACCTGAG-3′, antisense 5′-TAGGTGAGCGATAGCCTTTGC-3′. Total RNA was extracted from tissues or cells using the RNeasy kit (Qiagen) and reverse-transcribed. Quantitative real-time PCR was performed with a 7500Fast Real-Time PCR System. Relative expression was normalized to GAPDH and hypoxanthine guanine phosphoribosyl transferase.
Immunohistochemical staining
Harvested tissues were fixed in 4% PFA, processed through graded sucrose, embedded in O.C.T. medium (Tissue-Tek), and stored at −80°C. Frozen sections (20 µm) were dried for 10 min at room temperature and blocked for 1 h in PBS containing 4% chicken serum and 0.3% TritonX-100. Sections were stained with anti-TnC antibody diluted 1:200 in blocking solution overnight at 4°C. Samples were washed extensively in PBS, 0.15% TritonX-100 for at least for 3 h. Samples were then incubated overnight with anti-rabbit Alexa Fluor 647 secondary antibody (1:200), Alexa Fluor 546 phalloidin (1:500), and Hoechst 33342 in blocking solution. After extensive washing, sections were mounted in PermaFluor mounting medium. Confocal Z-stack images were captured, and TnC deposits were quantified.
Extracellular TnC and FN fiber quantification
The procedure for image processing and quantification of “fiberness” was developed as a macro in Fiji (ImageJ 1.50e x64). Fiberness measures the amount of fiber-like structures in an image, providing a readout that takes into account both the density of fibers and their length independently of orientation. Noise reduction first stabilizes the image background. The structural information from the eigen values of the Hessian matrix is then used to apply a Frangi vesselness filter76 to enhance very thin, almost unidimensional tubular structures (filter scale: σ = 0.179 µm, close to pixel size). The output is a fiber-enhanced image in which each pixel contains a fiberness score. M0 readout was computed as the mean fiberness score in the whole image. The processing pipeline and extracted M0 measures are illustrated in Fig. S3 B.
LDL incorporation in vitro
Blood for LDL isolation was obtained from healthy volunteers after informed consent in accordance with local ethics committee guidelines (Comité de Ética de la Investigación del Instituto de Salud Carlos III: CEI PI 12_2016-v2).
Fluorescently labeled human LDL was prepared as described (Steffensen et al., 2015). Briefly, human blood from healthy donors was collected in K3EDTA-containing tubes. After centrifugation, plasma was collected, mixed with KBr, and layered on a KBr density gradient column. The column was centrifuged at 256,000 g for 18 h in an Optima L-100 ultracentrifuge (Beckman Coulter). The 1.063-g/ml density layer containing LDL was collected, purified on a PD10 column (17085101; GE Healthcare), and conjugated to the fluorochrome Alexa Fluor 647. Alexa Fluor 647–LDL was purified on a PD10 column, and purity was tested at the Centro Nacional de Investigaciones Cardiovasculares Carlos III (CNIC) Proteomics Unit by LC-MS.
Cav1WT MEFs were cultured in DMEM supplemented with 2% FBS and loaded with Alexa Fluor 647–LDL to a final concentration of 200 µg/ml for 24 h (for TnC analysis at MVBs) or for 48 h (for TnC deposition studies).
Statistical analysis
Error bars depict SEM. Statistical significance was determined with GraphPad Prism by unpaired Student’s t test or Mann-Whitney test, as indicated; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material
Fig. S1 shows that internalization of Cav1 favors its entry into exosomes. Fig. S2 shows that Cav1-positive exosomes display a specific lipid/protein composition. Fig. S3 shows that exosome inhibition favors intracellular TnC accumulation. Fig. S4 shows that exosomes derived from CAFs modulate ECM deposition. Fig. S5 shows that disruption of ER-MVB contact sites modulates TnC matrix deposition. Data S1 presents high-throughput comparative proteomics of cell lysates and exosomes derived from either WT or Cav1KO fibroblasts. Data S2 presents high-throughput comparative proteomics of exosomes derived from MDA-MB231 or MDA-MB468 cells.
Supplementary Material
presents high-throughput comparative proteomics of cell lysates and exosomes derived from either WT or Cav1KO fibroblasts.
presents high-throughput comparative proteomics of exosomes derived from MDAMB231 or MDAMB468 cells.
Acknowledgments
We thank the CNIC Microscopy and Histology Units for technical assistance. We thank Daniel Jiménez-Carretero (CNIC Cellomics Unit) for developing the software for TnC fiber quantification, Dr. Kirchner of Centre for Medical Biotechnology (University of Duisburg-Essen, Duisburg, Germany) for plasmids encoding ubiquitinated Cav1 mutants, Fátima Sánchez-Cabo (CNIC Bioinformatics Unit) for statistical analysis of exosome populations, Dr. Germán Andrés (Centro de Biologia Molecular Severo Ochoa) for help with electron microscopy, Dr. Fidel Lolo (CNIC, Madrid, Spain) for PTRF-reconstituted PTRFKO cells, Mauro Catalá for technical assistance, and Giulio Fulgoni (CNIC, Madrid, Spain) for fluorescently labeled LDLs. Miguel Sánchez-Álvarez provided help with manuscript editing.
This study was supported by the Ministerio de Ciencia, Innovación y Universidades (CSD2009-0016, SAF2014-51876-R, SAF2017-83130-R, BFU2016-81912-REDC, and IGP-SO-MINSEV1512-07-2016); the Fundació La Marató de TV3 (385/C/2019); the Worldwide Cancer Research Foundation (AICR 15-0404); and Fondo Europeo de Desarrollo Regional “Una manera de hacer Europa” (to M.Á. del Pozo). M.Á. del Pozo’s group received funding from the European Union Horizon 2020 research and innovation program under Marie Sklodowska-Curie grant agreement no. 641639. M.Á. del Pozo is a member of the Tec4Bio consortium (ref. S2018/NMT4443; Actividades de I+D entre Grupos de Investigación en Tecnologías, Comunidad Autónoma de Madrid/FEDER, Spain). J. Balsinde was supported by Ministerio de Ciencia, Innovación y Universidades grants SAF2013-48201-R and SAF2016-80883-R, and G. Orend was supported by Institut National de la Santé et de la Recherche Médicale, the University of Strasbourg, the Ligue Contre le Cancer, and the Institut National du Cancer (ref. TENPLAMET). L. Albacete-Albacete was supported by a Ministerio de Ciencia, Innovación y Universidades predoctoral fellowship associated with the Severo Ochoa Excellence program (ref. SVP-2013-06789). I. Navarro-Lérida was supported by a postdoctoral fellowship from the Asociación Española Contra el Cáncer (ref. INVES191NAVA). The Centro Nacional de Investigaciones Cardiovasculares Carlos III is supported by the Ministerio de Ciencia e Innovación and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
The authors declare no competing financial interests.
Author contributions: I. Navarro-Lérida and M.Á. del Pozo Barriuso conceived and designed the project and wrote the manuscript. L. Albacete-Albacete and I. Navarro-Lérida performed most of the experiments and analyzed and interpreted the data. L. Albacete-Albacete prepared the manuscript figures. J.A. López and J. Vázquez performed and analyzed proteomic studies. A.M. Astudillo, J. Balsinde, and A. Ferrarini performed and analyzed lipidomic experiments. I. Martín-Padura helped with in vivo experiments. G. Orend and M. Van-Der-Heyden provided reagents, TnCKO animals essential for the study, and scientific feedback.
References
- Augsten M., Sjöberg E., Frings O., Vorrink S.U., Frijhoff J., Olsson E., Borg Å., and Östman A.. 2014. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Res. 74:2999–3010. 10.1158/0008-5472.CAN-13-2740 [DOI] [PubMed] [Google Scholar]
- Axelsen P.H., and Murphy R.C.. 2010. Quantitative analysis of phospholipids containing arachidonate and docosahexaenoate chains in microdissected regions of mouse brain. J. Lipid Res. 51:660–671. 10.1194/jlr.D001750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. 2011. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 23:452–457. 10.1016/j.ceb.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beacham D.A., Amatangelo M.D., and Cukierman E.. 2007. Preparation of extracellular matrices produced by cultured and primary fibroblasts. Curr. Protoc. Cell Biol. Chapter 10:Unit 10.9 [DOI] [PubMed] [Google Scholar]
- Bligh E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Bonzon-Kulichenko E., Garcia-Marques F., Trevisan-Herraz M., and Vázquez J.. 2015. Revisiting peptide identification by high-accuracy mass spectrometry: problems associated with the use of narrow mass precursor windows. J. Proteome Res. 14:700–710. 10.1021/pr5007284 [DOI] [PubMed] [Google Scholar]
- Botos E., Klumperman J., Oorschot V., Igyártó B., Magyar A., Oláh M., and Kiss A.L.. 2008. Caveolin-1 is transported to multi-vesicular bodies after albumin-induced endocytosis of caveolae in HepG2 cells. J. Cell. Mol. Med. 12(5a, 5A):1632–1639. 10.1111/j.1582-4934.2007.00167.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braicu C., Tomuleasa C., Monroig P., Cucuianu A., Berindan-Neagoe I., and Calin G.A.. 2015. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 22:34–45. 10.1038/cdd.2014.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brösicke N., van Landeghem F.K., Scheffler B., and Faissner A.. 2013. Tenascin-C is expressed by human glioma in vivo and shows a strong association with tumor blood vessels. Cell Tissue Res. 354:409–430. 10.1007/s00441-013-1704-9 [DOI] [PubMed] [Google Scholar]
- Campos A., Salomon C., Bustos R., Díaz J., Martínez S., Silva V., Reyes C., Díaz-Valdivia N., Varas-Godoy M., Lobos-González L., et al. . 2018. Caveolin-1-containing extracellular vesicles transport adhesion proteins and promote malignancy in breast cancer cell lines. Nanomedicine (Lond.). 13:2597–2609. 10.2217/nnm-2018-0094 [DOI] [PubMed] [Google Scholar]
- Castelló-Cros R., and Cukierman E.. 2009. Stromagenesis during tumorigenesis: characterization of tumor-associated fibroblasts and stroma-derived 3D matrices. Methods Mol. Biol. 522:275–305. 10.1007/978-1-59745-413-1_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenedella R.J. 2009. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids. 44:477–487. 10.1007/s11745-009-3305-7 [DOI] [PubMed] [Google Scholar]
- Chaudhary N., Gomez G.A., Howes M.T., Lo H.P., McMahon K.A., Rae J.A., Schieber N.L., Hill M.M., Gaus K., Yap A.S., et al. . 2014. Endocytic crosstalk: cavins, caveolins, and caveolae regulate clathrin-independent endocytosis. PLoS Biol. 12 e1001832 10.1371/journal.pbio.1001832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhuang X., Lin L., Yu P., Wang Y., Shi Y., Hu G., and Sun Y.. 2015. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med. 13:45 10.1186/s12916-015-0278-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G., and Théry C.. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30:255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. . 2015. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17:816–826. 10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo M.A., Balasubramanian N., Alderson N.B., Kiosses W.B., Grande-García A., Anderson R.G., and Schwartz M.A.. 2005. Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat. Cell Biol. 7:901–908. 10.1038/ncb1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic R.A., Gardiner C., Brooks A.S., Tannetta D.S., Ferguson D.J., Hole P., Carr B., Redman C.W., Harris A.L., Dobson P.J., et al. . 2011. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine (Lond.). 7:780–788. 10.1016/j.nano.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E.R., Sanchez-Heras E., Tsapara A., Sobota A., Levine T.P., and Futter C.E.. 2016. Annexin A1 Tethers Membrane Contact Sites that Mediate ER to Endosome Cholesterol Transport. Dev. Cell. 37:473–483. 10.1016/j.devcel.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennema E., Rivron N., Rouwkema J., van Blitterswijk C., and de Boer J.. 2013. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 31:108–115. 10.1016/j.tibtech.2012.12.003 [DOI] [PubMed] [Google Scholar]
- García-Marqués F., Trevisan-Herraz M., Martínez-Martínez S., Camafeita E., Jorge I., Lopez J.A., Méndez-Barbero N., Méndez-Ferrer S., Del Pozo M.A., Ibáñez B., et al. . 2016. A Novel Systems-Biology Algorithm for the Analysis of Coordinated Protein Responses Using Quantitative Proteomics. Mol. Cell. Proteomics. 15:1740–1760. 10.1074/mcp.M115.055905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz J.G., Minguet S., Navarro-Lérida I., Lazcano J.J., Samaniego R., Calvo E., Tello M., Osteso-Ibáñez T., Pellinen T., Echarri A., et al. . 2011. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 146:148–163. 10.1016/j.cell.2011.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande-García A., and del Pozo M.A.. 2008. Caveolin-1 in cell polarization and directional migration. Eur. J. Cell Biol. 87:641–647. 10.1016/j.ejcb.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Gruenberg J., and Maxfield F.R.. 1995. Membrane transport in the endocytic pathway. Curr. Opin. Cell Biol. 7:552–563. 10.1016/0955-0674(95)80013-1 [DOI] [PubMed] [Google Scholar]
- Gvaramia D., Blaauboer M.E., Hanemaaijer R., and Everts V.. 2013. Role of caveolin-1 in fibrotic diseases. Matrix Biol. 32:307–315. 10.1016/j.matbio.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Hayer A., Stoeber M., Ritz D., Engel S., Meyer H.H., and Helenius A.. 2010. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J. Cell Biol. 191:615–629. 10.1083/jcb.201003086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M.M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S.J., Walser P., Abankwa D., Oorschot V.M., Martin S., et al. . 2008. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 132:113–124. 10.1016/j.cell.2007.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. . 2015. Tumour exosome integrins determine organotropic metastasis. Nature. 527:329–335. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 28:730–738. 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Kalluri R. 2016. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer. 16:582–598. 10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- Kang J., Park J.H., Lee H.J., Jo U., Park J.K., Seo J.H., Kim Y.H., Kim I., and Park K.H.. 2016. Caveolin-1 Modulates Docetaxel-Induced Cell Death in Breast Cancer Cell Subtypes through Different Mechanisms. Cancer Res. Treat. 48:715–726. 10.4143/crt.2015.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner P., Bug M., and Meyer H.. 2013. Ubiquitination of the N-terminal region of caveolin-1 regulates endosomal sorting by the VCP/p97 AAA-ATPase. J. Biol. Chem. 288:7363–7372. 10.1074/jbc.M112.429076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J., Tkach M., and Théry C.. 2014. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29:116–125. 10.1016/j.ceb.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Lee J.A., Choi D.I., Choi J.Y., Kim S.O., Cho K.A., Lee J.B., Yun S.J., and Lee S.C.. 2015. Methyl-β-cyclodextrin up-regulates collagen I expression in chronologically-aged skin via its anti-caveolin-1 activity. Oncotarget. 6:1942–1953. 10.18632/oncotarget.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.Y., Davis O.B., Shin H.R., Zhang J., Berdan C.A., Jiang X., Counihan J.L., Ory D.S., Nomura D.K., and Zoncu R.. 2019. ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat. Cell Biol. 21:1206–1218. 10.1038/s41556-019-0391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarbe M.A., Barrasa J.I., Olmo N., Gavilanes F., and Turnay J.. 2013. Annexin-phospholipid interactions. Functional implications. Int. J. Mol. Sci. 14:2652–2683. 10.3390/ijms14022652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert V.H., Brech A., Pedersen N.M., Wesche J., Oppelt A., Malerød L., and Stenmark H.. 2010. Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev. Cell. 19:148–159. 10.1016/j.devcel.2010.06.010 [DOI] [PubMed] [Google Scholar]
- Logozzi M., De Milito A., Lugini L., Borghi M., Calabrò L., Spada M., Perdicchio M., Marino M.L., Federici C., Iessi E., et al. . 2009. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 4 e5219 10.1371/journal.pone.0005219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy C.M., and Oskarsson T.. 2015. Tenascin C in metastasis: A view from the invasive front. Cell Adhes. Migr. 9:112–124. 10.1080/19336918.2015.1008331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V., and Erlmann P.. 2015. The pathway of collagen secretion. Annu. Rev. Cell Dev. Biol. 31:109–124. 10.1146/annurev-cellbio-100913-013002 [DOI] [PubMed] [Google Scholar]
- Martínez-Bartolomé S., Navarro P., Martín-Maroto F., López-Ferrer D., Ramos-Fernández A., Villar M., García-Ruiz J.P., and Vázquez J.. 2008. Properties of average score distributions of SEQUEST: the probability ratio method. Mol. Cell. Proteomics. 7:1135–1145. 10.1074/mcp.M700239-MCP200 [DOI] [PubMed] [Google Scholar]
- Midwood K.S., and Orend G.. 2009. The role of tenascin-C in tissue injury and tumorigenesis. J. Cell Commun. Signal. 3:287–310. 10.1007/s12079-009-0075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midwood K.S., Chiquet M., Tucker R.P., and Orend G.. 2016. Tenascin-C at a glance. J. Cell Sci. 129:4321–4327. 10.1242/jcs.190546 [DOI] [PubMed] [Google Scholar]
- Möbius W., van Donselaar E., Ohno-Iwashita Y., Shimada Y., Heijnen H.F., Slot J.W., and Geuze H.J.. 2003. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic. 4:222–231. 10.1034/j.1600-0854.2003.00072.x [DOI] [PubMed] [Google Scholar]
- Mundy D.I., Li W.P., Luby-Phelps K., and Anderson R.G.. 2012. Caveolin targeting to late endosome/lysosomal membranes is induced by perturbations of lysosomal pH and cholesterol content. Mol. Biol. Cell. 23:864–880. 10.1091/mbc.e11-07-0598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M., Peränen J., Schreiner R., Wieland F., Kurzchalia T.V., and Simons K.. 1995. VIP21/caveolin is a cholesterol-binding protein. Proc. Natl. Acad. Sci. USA. 92:10339–10343. 10.1073/pnas.92.22.10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabi I.R., and Le P.U.. 2003. Caveolae/raft-dependent endocytosis. J. Cell Biol. 161:673–677. 10.1083/jcb.200302028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaharu K., Zhang X., Yoshida T., Katoh D., Hanamura N., Kozuka Y., Ogawa T., Shiraishi T., and Imanaka-Yoshida K.. 2011. Tenascin C induces epithelial-mesenchymal transition-like change accompanied by SRC activation and focal adhesion kinase phosphorylation in human breast cancer cells. Am. J. Pathol. 178:754–763. 10.1016/j.ajpath.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P., Trevisan-Herraz M., Bonzon-Kulichenko E., Núñez E., Martínez-Acedo P., Pérez-Hernández D., Jorge I., Mesa R., Calvo E., Carrascal M., et al. . 2014. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 13:1234–1247. 10.1021/pr4006958 [DOI] [PubMed] [Google Scholar]
- Navarro-Lérida I., Alvarez-Barrientos A., Gavilanes F., and Rodriguez-Crespo I.. 2002. Distance-dependent cellular palmitoylation of de-novo-designed sequences and their translocation to plasma membrane subdomains. J. Cell Sci. 115:3119–3130. [DOI] [PubMed] [Google Scholar]
- Parton R.G., and del Pozo M.A.. 2013. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 14:98–112. 10.1038/nrm3512 [DOI] [PubMed] [Google Scholar]
- Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., et al. . 2012. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18:883–891. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger F.W., and Vitale N.. 2018. Cholesterol and the journey of extracellular vesicles. J. Lipid Res. 59:2255–2261. 10.1194/jlr.R084210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A., Martin S., Fernández M.A., Ingelmo-Torres M., Ferguson C., Enrich C., and Parton R.G.. 2005. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol. Biol. Cell. 16:2091–2105. 10.1091/mbc.e04-08-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A., Morales-Paytuví F., Bosch M., and Parton R.G.. 2020. Non-caveolar caveolins - duties outside the caves. J. Cell Sci. 133 jcs241562 10.1242/jcs.241562 [DOI] [PubMed] [Google Scholar]
- Quail D.F., and Joyce J.A.. 2013. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19:1423–1437. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., Siegers G.M., Jewer M., and Postovit L.M.. 2013. Nodal signalling in embryogenesis and tumourigenesis. Int. J. Biochem. Cell Biol. 45:885–898. 10.1016/j.biocel.2012.12.021 [DOI] [PubMed] [Google Scholar]
- Raiborg C., Wenzel E.M., and Stenmark H.. 2015. ER-endosome contact sites: molecular compositions and functions. EMBO J. 34:1848–1858. 10.15252/embj.201591481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., and Stoorvogel W.. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200:373–383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M., Silvente-Poirot S., Poirot M., and Wakelam M.J.O.. 2018. Extracellular vesicles: lipids as key components of their biogenesis and functions. J. Lipid Res. 59:1316–1324. 10.1194/jlr.E086173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J.. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185:1209–1225. 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Vila A., Navarro-Lérida I., Sánchez-Alvarez M., Bosch M., Calvo C., López J.A., Calvo E., Ferguson C., Giacomello M., Serafini A., et al. . 2016. Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 6:27351 10.1038/srep27351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A., Furlán M., Vidal M., and Colombo M.I.. 2003. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 278:20083–20090. 10.1074/jbc.M301642200 [DOI] [PubMed] [Google Scholar]
- Sceneay J., Smyth M.J., and Möller A.. 2013. The pre-metastatic niche: finding common ground. Cancer Metastasis Rev. 32:449–464. 10.1007/s10555-013-9420-1 [DOI] [PubMed] [Google Scholar]
- Skotland T., Hessvik N.P., Sandvig K., and Llorente A.. 2019. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 60:9–18. 10.1194/jlr.R084343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen L.B., Mortensen M.B., Kjolby M., Hagensen M.K., Oxvig C., and Bentzon J.F.. 2015. Disturbed Laminar Blood Flow Vastly Augments Lipoprotein Retention in the Artery Wall: A Key Mechanism Distinguishing Susceptible From Resistant Sites. Arterioscler. Thromb. Vasc. Biol. 35:1928–1935. 10.1161/ATVBAHA.115.305874 [DOI] [PubMed] [Google Scholar]
- Subra C., Laulagnier K., Perret B., and Record M.. 2007. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 89:205–212. 10.1016/j.biochi.2006.10.014 [DOI] [PubMed] [Google Scholar]
- Sung B.H., Ketova T., Hoshino D., Zijlstra A., and Weaver A.M.. 2015. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 6:7164 10.1038/ncomms8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talts J.F., Wirl G., Dictor M., Muller W.J., and Fässler R.. 1999. Tenascin-C modulates tumor stroma and monocyte/macrophage recruitment but not tumor growth or metastasis in a mouse strain with spontaneous mammary cancer. J. Cell Sci. 112:1855–1864. [DOI] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., and Clayton A.. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., and Simons M.. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 319:1244–1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Unlu G., Levic D.S., Melville D.B., and Knapik E.W.. 2014. Trafficking mechanisms of extracellular matrix macromolecules: insights from vertebrate development and human diseases. Int. J. Biochem. Cell Biol. 47:57–67. 10.1016/j.biocel.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G., Porto-Carreiro I., Simoes S., and Raposo G.. 2006. Exosomes: a common pathway for a specialized function. J. Biochem. 140:13–21. 10.1093/jb/mvj128 [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Baixauli F., Gutiérrez-Vázquez C., Sánchez-Madrid F., and Mittelbrunn M.. 2014. Sorting it out: regulation of exosome loading. Semin. Cancer Biol. 28:3–13. 10.1016/j.semcancer.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang L., and Huang H.W.. 2007. Evidence of cholesterol accumulated in high curvature regions: implication to the curvature elastic energy for lipid mixtures. Biophys. J. 92:2819–2830. 10.1529/biophysj.106.097923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., and Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Wubbolts R., Leckie R.S., Veenhuizen P.T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J.W., Geuze H.J., and Stoorvogel W.. 2003. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 278:10963–10972. 10.1074/jbc.M207550200 [DOI] [PubMed] [Google Scholar]
- Zanoni M., Piccinini F., Arienti C., Zamagni A., Santi S., Polico R., Bevilacqua A., and Tesei A.. 2016. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 6:19103 10.1038/srep19103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimnicka A.M., Husain Y.S., Shajahan A.N., Sverdlov M., Chaga O., Chen Z., Toth P.T., Klomp J., Karginov A.V., Tiruppathi C., et al. . 2016. Src-dependent phosphorylation of caveolin-1 Tyr-14 promotes swelling and release of caveolae. Mol. Biol. Cell. 27:2090–2106. 10.1091/mbc.E15-11-0756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
presents high-throughput comparative proteomics of cell lysates and exosomes derived from either WT or Cav1KO fibroblasts.
presents high-throughput comparative proteomics of exosomes derived from MDAMB231 or MDAMB468 cells.